Abstract
Functional proteins of complex eukaryotes within the same species are rather invariant. A single catalytic component of telomerase TERT is essential for an active telomerase complex that maintains telomeres. Surprisingly, we have identified two paralogous SpTERT-L and SpTERT-S genes with novel domains in Strongylocentrotus purpuratus (purple sea urchin). The SpTERT-S and SpTERT-L genes were differentially expressed throughout embryogenesis. An unusual germline nucleotide substitution and amino acid variation was evident in these TERTs. The hypervariability of SpTERT-S haplotypes among different individuals reached unprecedented levels of π > 0.2 in exon 11 region. The majority of nucleotide changes observed led to nonsynonymous substitutions creating novel amino acids and motifs, suggesting unusual positive selection and rapid evolution. The majority of these variations were in domains involved in binding of SpTERT to its RNA component. Despite hypervariability at protein level, SpTERT-S conferred telomerase activity, and its suppression during early embryogenesis led to arrest at late mesenchymal blastula. Domain exchange and embryo rescue experiments suggested that SpTERT may have evolved functions unrelated to classic telomerase activity. We suggest that telomerase has a specific and direct function that is essential for integration of early polarity signals that lead to gastrulation. Identification of these unique hypervariable telomerases also suggests presence of a diversity generation mechanism that inculcates hypervariable telomerases and telomere lengths in germline.
INTRODUCTION
Early studies of Theodore Boveri and others in sea urchin embryos led to the formation of a theoretical and practical framework that established the requirement of normal chromosome ploidy for correct development (Boveri, 1889). Further work by Muller and McClintock established importance of chromosome ends in maintenance of correct ploidy (McClintock, 1938; Muller, 1938). These end-units in eukaryotes are characterized by long tracts of repeating, noncoding DNA sequences called telomeres (Blackburn and Gall, 1978; Szostak and Blackburn, 1982).
As a result of the end replication problem and telomere erosion (Olovnikov, 1971; Watson, 1972), telomeres must be resynthesized by an adjunct to the usual replication machinery: the ribonucleoprotein reverse transcriptase complex called telomerase. Telomerase was first identified in Tetrahymena (Greider and Blackburn, 1985), and it has since been characterized in many eukaryotes (Harrington et al., 1997; Kilian et al., 1997; Lingner et al., 1997; Meyerson et al., 1997; Nakamura et al., 1997). The telomerase holoenzyme is composed of an RNA subunit and several protein subunits, including the major catalytic subunit Telomerase Reverse Transcriptase (TERT). Telomeres are protected by a specialized protein complex (de Lange, 2005). Loss of this end protection may lead to telomere shortening, and this event is associated with entry into senescence (Harley et al., 1990). Therefore, telomere length needs to be reset in germ cells. Indeed, germ cells have long telomeres that do not shorten with age (Allsopp et al., 1992). Furthermore, immortalized cell lines (Counter et al., 1992; Kim et al., 1994) express telomerase to maintain functional telomeres. Therefore, to prevent telomere dysfunction between generations, telomere shortening needs to also be circumvented during development. Consistent with this notion, telomerase activity has been found in eggs and embryos of many species (Mantell and Greider, 1994; Wright et al., 1996; Betts and King, 1999; Schaetzlein et al., 2004), and telomerase knockout mice show telomere shortening but remain viable (Blasco et al., 1997).
Like vertebrates, purple sea urchin telomeres terminate in TTAGGG repeats (Lejnine et al., 1995). In addition, sea urchins are deuterostomes and hence share close ancestry with vertebrates (Gaskell, 1890; Berrill, 1955). The purple sea urchin embryo in particular has been used as a versatile developmental model system to study fundamental gene regulation (Levine and Davidson, 2005; Byrum et al., 2006).
The invariant identity of crucial catalytic enzymes within the same species is thought to be a requirement to maintain fundamental cell survival functions. Contrary to this notion, here we describe unanticipated results during cloning of the catalytic subunit of TERT, which we termed SpTERT from a complex basal deuterostome Strongylocentrotus purpuratus. Our results suggest that evolutionary pressure at the SpTERT locus has resulted in positive selection for unique hypervariable telomerases. Here, we describe in detail the unusual results of their expression patterns, intraspecific genetic variation, and early essential embryonic functions.
MATERIALS AND METHODS
Contig Identification and Primer Design
Initial polymerase chain reaction (PCR) primers were chosen based on genomic sequences from early drafts of the Sea Urchin Genome Project (Baylor College of Medicine, Houston, TX). Specifically, the contig AAGJ01196796 showed homology to hTERT (NP_003210) on the basis of a TBLASTN alignment. The homologous region spanned 283 amino acids (575–794 on hTERT). This contig was analyzed using the European Molecular Biology Open Software Suite (EMBOSS; http://emboss.sourceforge.net/) software package to identify coding sequences.
Potential exons of the contig were found by translating all reading frames and performing pairwise alignments with known TERT peptide sequences (Xenopus laevis [AAG43537.1], Canis familiaris [AAQ02791.1], Mus musculus [NP_033380.1], Gallus gallus [AAV35463.1], Takifugu rubripes [AAX59693.1], Rattus norvegicus [AAT09125.1], Mesocricetus auratus [AAF17334.1], Aspergillus fumigatus Af293 [EAL87013.1], Cryptococcus neoformans var. neoformans JEC21 [AAW43582.1], Aspergillus nidulans FGSC A4 [EAA59961.1], Arabidopsis thaliana [CAC01849.1], Schizosaccharomyces pombe [AAC49803.1], Moneuplotes crassus [AAM95622.1], Oryza sativa [AAK35007.1], and Theileria annulata [CAI75739.1]). In this manner, the orientation of SpTERT within the contig was deduced, and regions of highest homology were used as a basis for primer design. All primers were optimized using the Primer3 software (Rozen and Skaletsky, 2000) and synthesized by Operon Biotechnologies (Huntsville, AL). The prediction of intron and exon boundaries for SpTERT was done with the EMBOSS software, est2genome (Mott, 1997).
Rapid Amplification of cDNA Ends (RACE)
In 3′ RACE, first-strand cDNA was synthesized using an oligo(dT)anchor primer on 2 μg of total RNA. A PCR performed with a gene-specific primer SPT-AL and the anchor primer was resolved on a 1% TAE agarose gel and strained with ethidium bromide. A 0.7-kb fragment was extracted with QIAquik gel extraction kit (QIAGEN, Valencia, CA), cloned into pCR 2.1 vector with the TOPO-TA cloning kit (Invitrogen, Carlsbad, CA), and sequenced by single pass primer extension (ACGT DNA Technologies, Toronto, ON, Canada). The 5′ cDNA end was obtained by RNA-ligase mediated RACE (Ambion, Austin, TX), which involves ligating a known anchor sequence to the decapped 5′ end of the RNA. A primer specific to this anchor sequence and a gene-specific primer close to the 5′ end (SPT-K26) were used. PCR products were cloned and sequenced as described for 3′ RACE (above).
Full-Length Reverse Transcriptase (RT)-PCR and Cloning of SpTERT
The sequence results from 5′/3′ RACE were used to synthesize primers for obtaining the full-length sequence. The forward primer SPT-X2L (just upstream of the start codon) and the reverse primer (just downstream of the stop codon) was SPT-32R. First-strand synthesis was done on 4 μg of total RNA with Superscript III (Invitrogen), primed with oligo(dt)18, at 55°C for 1 h. The PCR was carried out on a PTC-200 thermal cycler (MJ Research, Watertown, MA) with Platinum High Fidelity Taq polymerase (Invitrogen) according to the manufacturer's specifications (0.2 mM dNTPs, 1.5 mM MgCl2, 0.4 μM of each a forward and reverse primer, 2 U of Taq, and 200 ng of cDNA). The template was denatured at 94°C for 3 min, followed by 35 cycles of 94°C for 30 s, 30 s annealing at the 5°C above the melting temperature calculated for the primer pair, and extension at 72°C for 5 min. The PCR product was cloned and sequenced in the same method described for the RACE products.
Primer Design for Targeting SpTERT-S and SpTERT-L
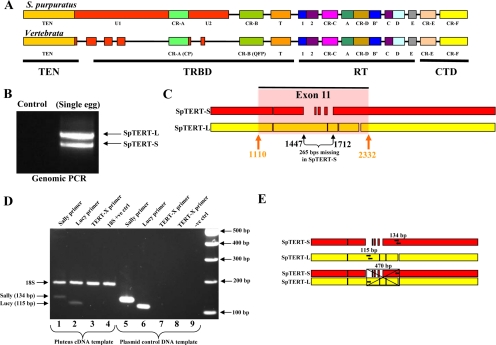
Full-length cloned short and long SpTERT nucleotide sequences were aligned using multiple sequence alignment software with ClustalW (Chenna et al., 2003) and then analyzed in MEGA3 version 3.1 (Kumar et al., 2004) for regions of homology. Using Primer3 (Rozen and Skaletsky, 2000) to assist in the optimization of design, primers were targeted for SpTERT-S and SpTERT-L, aptly named “Sally” and “Lucy,” respectively. The multiple sequence alignment revealed a conserved 13-base region found exclusively in SpTERT-S and occurred as a missing gap in SpTERT-L, of which the reverse primer (Sally_R1) annealed to, whereas the forward primer (Sally_F1 or Sally_F2) targeted a sequence common to both short and long versions. When combined for use in PCR, Sally_F1 with Sally_R1 and Sally_F2 with Sally_R1 primers amplified a 392- and 134-base pair region of SpTERT-S, respectively. SpTERT-L primers (Lucy_F2 and Lucy_R2) were designed to target the highly conserved sequence found within the 265-base region exclusive to SpTERT-L and together amplified a 115-base pair fragment. Furthermore, to confirm the presence of two discrete transcripts a combination of Lucy_F2 primer (specific to a unique region of SpTERT-L) with Sally_R1 primer (specific to a unique region of SpTERT-S) together formed what we named “TERT-X.” If SpTERT existed as a single transcript or alternatively spliced gene it would then amplify a 470-base pair fragment of SpTERT by PCR according to the alignment of SpTERT-S and SpTERT-L (Figure 1E). The TERT-X primer set was applied to both cDNA synthesized from Pluteus stage RNA and individual plasmid DNA containing full-length SpTERT-S or SpTERT-L. All primers were synthesized by Operon Biotechnologies.
Figure 1.
TERT schematics. (A) Schematic representation of SpTERT and its comparison to vertebrate TERT. The U1 and U2 motifs are defined based on their uniqueness to sea urchins, whereas vertebrate TERT also displays smaller unique domains as well. The CR-A, CR-B, CR-C, CR-D, CR-E, motifs 1, A–E, and T are conserved domains. (B) Genomic PCR carried out on a single egg by using primers targeted toward the full-length exon 11 region by using SPT-EX11–10F and SPT-BR primers. The two products represent SpTERT-S and SpTERT-L. (C) Schematic showing an alignment of full-length SpTERT-S (red) and SpTERT-L (yellow) reveals gaps and the highlighted exon 11 region attributing to most of the size difference. The black arrows denote unique primer target sites for specific gene PCR amplification. (D) Confirmation of two independent TERT transcripts. Using primers specific to SpTERT-S (Sally_F2 and Sally_R1), SpTERT-L (Lucy_F2 and Lucy_R2), and the TERT-X primer set (combination of SpTERT-S reverse and SpTERT-L forward primers) set, a PCR was carried out using both pluteus source cDNA (lanes 1–4) and plasmid DNA (lanes 5–8) as templates. The Sp18S was run as an internal and positive control. The PCR products in lanes 5 and 7 used SpTERT-S plasmid DNA template, whereas lanes 6 and 8 used SpTERT-L plasmid DNA template. (E) Representative schematics of all three primer combinations shown in D. The black arrows indicate where the specific primer sets were targeted and their sizes are shown directly above. For the third TERT-X primer set, the reverse SpTERT-S primer was combined with the forward SpTERT-L primer to yield a predicted target size of 470 base pairs. The crossed box represents the nonamplified 470 base pairs region using the TERT-X primer set.
Calculations of Nucleotide and Amino Acid Variability
Multiple aligned nucleotide sequences were set into defined groupswithin DnaSP 4.0 (Rozas et al., 2003) software. The determination of nucleotide diversity, π, for our sequences measured the average number of nucleotide differences per site between aligned sequence pairs (Nei, 1987) and was defined by
 |
whereby n, xi, and πij are the number of DNA sequences examined, the population frequency of the ith type of DNA sequence, and the proportion of different nucleotides between the ith and the jth types of DNA sequences, respectively. The SD of nucleotide diversity was obtained by applying a square root to the sampling variance, V(π̂), defined by
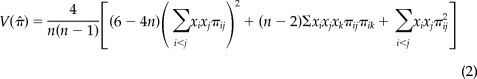 |
assuming that πij values are constant (Nei and Tajima, 1981). DnaSP allows for the grouping of several nucleotide sites into a user-defined scan window (nucleotide range) such that all π values determined are represented by a midpoint nucleotide position on a plotted graph (Supplemental Figure 2A).
Similarly, multiple aligned amino acid sequences were grouped together using BioEdit (Hall, 1999) software. The determination of entropy, Ω, for our sequences measured the lack of predictability for individual amino acid alignment sites. A modified version of the original mathematical theory devised by Claude Shannon (Shannon, 1948; Pierce, 1980; Schneider and Stephens, 1990) by using
gives Ω as a measure of uncertainty, where b is an amino acid residue, l is a position, and f(b,l) is the frequency at which residue b is found at position l. Therefore, the information obtained at position l is the entropy (uncertainty) at that site; and as an alignment improves in quality, the entropy at each position decreases (Supplemental Figure 2B). ln(f(b,l)) is a correction factor that converts the values to nits rather than bits and the data points remain relative to each other. For the analysis and comparison of the π and Ω, independent graphs were generated and then superimposed upon one another.
A quick comparison of the nucleotide variability profiles for SpTERT-S and SpTERT-L from the exon 11 region of Figure 2 to the profiles of Figure 4 may appear as a discrepancy in π values, especially with SpTERT-L. However, it must be noted that the calculation of π inherently takes an average over a defined scan window of several nucleotides that is not calculated on a base-per-base basis. Therefore, any given set of aligned sequences, with increased amount of sequences analyzed and the increased amount of mutations found within a defined range that are not located at the same site will contribute to a higher overall value of π.
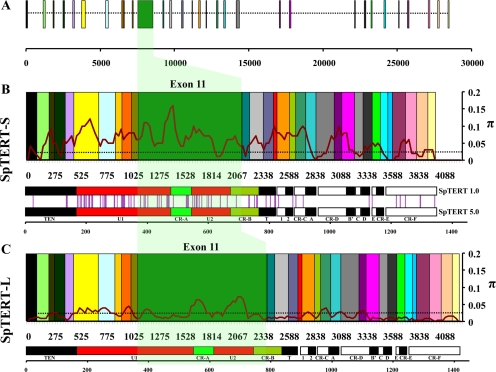
Figure 2.
Exon boundaries assembled from genomic contig. (A) The full-length cloned sequence of SpTERT-S was subjected to an alignment against a genomic contig using the EMBOSS software, est2genome. The introns (dotted line) and individual exons (colored boxes) boundaries were predicted. The unique exon 11 region is shown in green. (B) The predicted exons from A are assembled without introns according to a nucleotide scale. A multiple sequence alignment of all cloned full-length SpTERT-S was used to measure nucleotide diversity (π, pairwise differences). The data output shown for π is the midpoint value of a 15 nucleotide sliding window (steps of 5 bases) scanning the length of the entire sequence. An arbitrary threshold manually set at 0.025 allows for comparison of areas with high nucleotide variability to low nucleotide variability. The corresponding protein domains and motifs of SpTERT are shown immediately below the assembled exons on an amino acid scale for comparison. The vertical purple lines seen between genes represent sites of amino acid substitution relative to their domain. (C) Same as in B but for SpTERT-L.
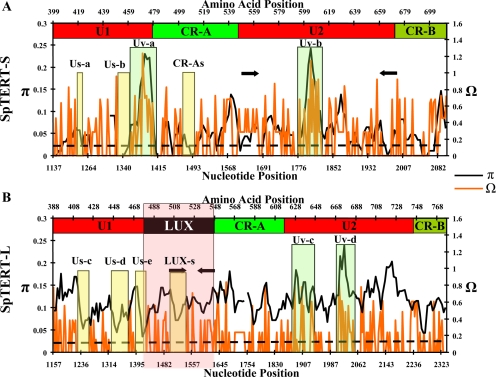
Figure 4.
Detailed nucleotide and amino acid variability analysis of exon 11 sequences of SpTERT-S and SpTERT-L. (A) Exon 11 sequences (n = 15) of SpTERT-S were analyzed for nucleotide diversity (π, black). The amino acid variation was measured as entropy by using aligned amino acid sequences (Ω, orange). Ω represents lack of predictability for an alignment position at each residue. The amino acid domains and motifs coded for by exon 11 are shown at the top of each graph along the amino acid scale. The LUX subdomain (black box) of U1 region is the major contributor to the size difference observed between SpTERT-L and SpTERT-S. The black arrows shown on each plot display the primer targets used for quantitative RT-PCR in subsequent experiments. US-a and CRA-s represent two silent domains with invariable amino acid composition. The dotted line represents the approximate level of currently known nucleotide variation. (B) Same as in A except for SpTERT-L (n = 16) sequences were used for variability analysis. Broken line out of parameter range. The US-b, US-c, US-d, and US-e represent four silent regions in the U1 region of SpTERT-L.
The Ka/Ks value is a ratio of nonsynonymous rate (codon mutation) to synonymous rate (silent codon mutation) at a given site within a multiple amino acid sequence alignment that is used to estimate positive or purifying selection. These rates are normalized, so that in the case of no selection, Ka/Ks = 1, in purifying selection Ka decreases so that Ka/Ks <1, and in positive selection Ks decreases so that Ka/Ks >1. Positively selected sites may suggest molecular adaptation or rapid evolutionary replacement of an amino acid. Negatively selected sites suggest elimination of mutations that create novel amino acids. All Ka/Ks ratios were calculated using DnaSP software by using the aligned translated amino acid sequences of exon 11.
Single Egg Genomic PCR of SpTERT
Individual eggs were collected and washed twice in phosphate-buffered saline (PBS)−/− and put in a 50-μl volume of PCR master mix. The PCR amplification included 5 μl of 10× PCR buffer, 2 mM MgCl2, 0.5 μM each primers, 0.2 mM deoxynucleotide triphosphate (MBI Fermentas, Hanover, MD) with primers SPT-EX11–10F and SPT-BR. PCR was performed on an MJ-PTC200 thermal cycler (MJ Research) at 100°C for 10 min for denaturation and 80°C for 1 min (adding Taq 1.25 U) followed by 35 cycles of 94°C for 30 s, 56°C for 45 s, and 72°C for 1 min. The PCR products were run on agarose gels and visualized by staining with ethidium bromide.
Single Embryo Telomere Repeat Amplification Protocol (SE-TRAP)
Embryo TRAP was carried out in three stages for optimal sensitivity and to allow temporal analysis of telomerase activity during development: 1) lysis, 2) extension, and 3) amplification. For stage 1, a single egg or embryo was selected in 2 μl under a stereoscope, rinsed twice in 5 ml of ice-cold PBS−/−, and immediately picked up in 2 μl and dropped into 38 μl of ice-cold lysis solution, effectively a 1× TRAP buffer (final conc. 20 mM Tris, pH 8.0, 6.3 mM KCl, 1.5 mM MgCl2, 0.05% Tween, and 1 mM EGTA) with 1× protease inhibitor (Complete Mini EDTA-Free; Roche Diagnostics, Indianapolis, IN) and 8 U of Protector RNase inhibitor (Roche Diagnostics). The 45-μl extension solution was identical to the lysis solution but with the addition of [γ-32P]ATP-labeled TS primer and 50 μM each of dATP, dTTP, dGTP, and dCTP. TS primer was radiolabeled with T4 polykinase (New England Biolabs, Ipswich, MA) in the provided buffer for 20 min at 37°C and then 5 min at 85°C. For stage 2, extension was carried out in a MJ-PTC200 thermal cycler (MJ Research) for 30 min at 30°C followed by 10 min at 85°C to inactivate the reaction. The reaction was stored at −20°C until all samples were ready for amplification. For stage 3, The 45-μl extension reaction was thawed on ice and brought up to 50 μl by the addition of 2 U of recombinant Taq polymerase (MBI Fermentas) and Primer Mix (containing the reverse primer and internal control primers; Millipore Bioscience Research Reagents, Temecula, CA) in a 1× TRAP buffer. A two-step PCR was performed at 94°C for 30 s and 59°C for 30 s for 21–28 cycles in a thermal cycler. The reaction product was resolved on a 15% polyacrylamide gel. The gel was exposed to a phosphorous screen and visualized on a PhosphorImager (Typhoon; GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom).
Expression Levels as Determined by Standard RT-PCR
For all RT-PCR experiments, three independent batches of eggs from three animals were fertilized with the same sperm in separate 2-liter screw-cap, plastic rotating flasks (Corning Life Sciences, Acton, MA) with rotation at 60 rpm and at 12°C. Embryos at each stage were then removed for analysis. Total RNA was isolated from different stages of development by using the TRIzol reagent (Invitrogen) and quantified by spectrophotometry. Residual DNA was removed with DNase I (MBI Fermentas) treatment as per manufacturer's recommendations. Equal RNA amounts (1.0 μg) were used for first-strand synthesis using SuperScript II reverse transcriptase (Invitrogen), primed with random hexamer, at 42°C for 1 h. A multiplex PCR reaction was performed using gene-specific primers Sally_F1 and Sally_R1 for SpTERT-S and Lucy_F2 and Lucy_R2 for SpTERT-L.
The higher abundance of ribosomal mRNA often confounds the linearity of PCR reaction. To overcome this obstacle, we used an excess proportion of chain terminated (lacking 3′OH) 18S primers to normal 18S primers (18SF and 18SR). These modified primers, which can bind the template but do not allow polymerase extension, are blended with normal primers at an empirically determined ratio of 4:1 (modified:normal). The PCR was carried out on an MJPTC-200 thermal cycler with Taq polymerase (MBI Fermentas) according to the manufacturer's specifications (0.2 mM dNTPs, 1.5 mM MgCl2, 0.8 μM each of a forward and reverse primer, and 2 U of Taq). The template was denatured at 94°C for 3 min, followed by 40 cycles of 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s, and followed by a final step of 72°C for 5 min. Ten microliters of the PCR product was separated by agarose gel (Amresco, Solon, OH) electrophoresis and stained with ethidium bromide (Invitrogen) for visualization.
Relative RNA Expression Levels as Determined by Real-Time Quantitative (Q)-PCR
For Q-PCR experiments, three independent in vitro fertilizations (IVFs) were performed, and each stage was collected and mRNA was prepared. This mRNA was then used to generate at each stage two cDNA products that were then used independently in triplicates (total of six per stage of embryogenesis). SpTERT amplification levels were determined by quantitative-PCR on an ABI 7900HT sequence detection system (Applied Biosystems, Foster City, CA) with SYBR Green chemistry (QIAGEN). The cDNA preparation was similar to that of RT-PCR use; however, the template was used at a final concentration of 500 ng/reaction in a 20-μl total reaction volume. Each sample had been run through the Q-PCR analysis in triplicate on two different sets of freshly synthesized cDNA, by using a no-template negative control for each sample set of cDNA and primers. Each 20-μl reaction contained 10 μl of SYBR Green master mix, 2 μl of template cDNA or water, 1 μl of forward and reverse primer mix at 0.6 μM each/reaction, and 7 μl of nuclease-free water. Thermal cycling parameters were 95°C for 15 min for the enzyme activation, followed by 40 cycles of 94°C for 15 s, 60°C for 30 s, and 72°C for 30 s, after which a denaturation step was applied to verify a single product.
By applying the egg-stage samples as our calibrator for all other stages to be compared with, the SDS 2.1 software (Applied Biosystems) determined the number of cycles required to attain the threshold concentration (Ct). With the threshold set to 0.2, this ensured that it crossed each of the amplification curves during the exponential phase of growth, thus eliminating the condition of limiting reagents affecting the efficiency of the reaction. Using the Ct determined for the calibrator and the 18S endogenous control, the relative quantity for any particular target was determined by applying the formula relative quantitation (RQ) = 2−ΔΔCt. The logs of the relative quantities are plotted such that values with varying maxima and minima are easily visualized on an appropriate scale.
IVF
All S. purpuratus obtained from Santa Barbara Marine Bio (Santa Barbara, CA) were all collected near Santa Barbara within a 200-ft radius of coal oil point (15-ft average depth). Animals were maintained at 12°C and fed with frozen Egregia kelp regularly. The animals were induced to spawn by 0.1- to 0.5-ml injection with sterile 0.5 M KCl. Eggs were collected into an artificial antibiotic/seawater preparation that we termed IOPS (Instant Ocean; specific gravity, 1.024; 0.2 μM filtered, with 20 U/ml penicillin and 50 μg/ml streptomycin). Sperm was pipetted dry into 1.5-ml Eppendorf tubes and diluted 1:500 in IOPS before use. Eggs were washed and filtered immediately after collection to remove debris and contaminants. Fertilization of embryos occurred at a density of 1500 embryos per ml. Embryos were then cultured at 12°C in a 2000-ml screw-cap, plastic rotating flasks (Corning Life Sciences) at 60 rpm.
Microinjection Method
Gametes are collected in the same manner for IVF use. In preparation for microinjection, 1% (wt/vol) protamine sulfate was poured into 60- × 15-mm dishes (Corning Life Sciences) for 5–10 s. After rinsing well and drying the dishes, they retain a charged coating of protamine sulfate, which is suitable for immobilizing eggs. The dish is filled with 5 ml of IOPS containing 2 mM p-aminobenzoic acid. A hand-pulled rowing pipette with a 100-μm end diameter was used to draw up eggs and deposit them onto the dish. The eggs were fertilized with the minimum amount of diluted (1:500 typical) sperm required to achieve 95–100% fertilization.
The microinjection needle was pulled from a borosilicate capillary (with filament) (1.0 mm o.d., 0.78 i.d. mm, 10-cm length; Sutter Instrument, Novato, CA). A P-97 micropipette puller (Sutter Instrument) was used for all capillaries. We empirically generated a needle with a sealed tip and an outer diameter of <2 μm. Micropipette pulling parameters were set at p = 300, heat = 500, pull = 155, vel = 80, and time = 200). The needle was back-loaded, and fertilized eggs were injected using a Picospritzer III pressure injector (Parker Hannifin, Mayfield Heights, OH). All micromanipulations were performed on an Eclipse TE2000-S inverted microscope (Nikon, Tokyo, Japan).
Generation of Chimeric Protein Constructs and Dominant Negatives
SpTERT Dominant-Negative Constructs.
SpTERT1.0-DN Construct.
The SpTERT1.0-DN construct was designed by site-directed mutagenesis (Stratagene, La Jolla, CA). The dominant-negative point mutation at D1090A (orthologous to D868A of hTERT) was first created in pBluesript and subsequently subcloned into pTNT vector. The SpTERT4.0-DN1 construct contained a point mutation at D996A (orthologous to D712A of hTERT). The SpTERT4.0-DN2 construct contained a point mutation D1154A (orthologous to D868A of hTERT). All constructs were subsequently subcloned back into pTNT. All constructs were sequenced in the targeted region for verification.
SpTERT4.0-L-ΔLUX.
The pTNT-SpTERT4.0-L-ΔLUX construct was constructed as follows. pCR-2.1-TOPO-SpTERT-4.0 was used as a template in a PCR reaction; the N-terminal portion of SpTERT-4.0 was first amplified using platinum high-fidelity DNA polymerase (Invitrogen). This fragment was void of Lucy Unknown eXclusive (LUX) domain. The amplification included 5 μl of 10× PCR buffer, 1.5 mM MgSO4, 0.5 μM each primer, 0.2 mmol of dNTPs (MBI Fermentas) High-Fi DNA polymerase (1.0 U), and 1 ng of template DNA (PCR2.1-spTERT4.0). PCR was performed at 94°C for 2 min followed by 25 cycles of 94°C for 30 s, 56°C for 30 s, and 68°C for 2 min. This gave rise to an ∼1.4-kb N-terminal fragment. The remaining portion of SpTERT-L was also amplified in the same manner. First, the N-terminal fragment was cloned in pTNT vector using XhoI and XbaI sites. Then, the second remaining was excised with XbaI and NotI and cloned back in the pTNT+N-terminus vector lacking LUX.
SpTERT1.0-S+LUX.
The N-terminal portion of SpTERT-1.0-S was amplified by PCR (∼1.4 kb), restricted with XhoI and XbaI, and subsequently cloned in the pTNT vector. In the second step, the remaining portion of SpTERT-1.0-S was amplified by PCR and restricted with XbaI and Not and also cloned in the N-terminal containing pTNT vector. In the third step, we used PCR to amplify the LUX domain and cut the fragment with XbaI. This fragment was sequentially cloned in the third step into pTNT vector containing the remaining SpTERT-1.0-S. The resulting SpTERT-1.0-S+LUX was sequenced to find a clone with the correct orientation of the LUX domain.
SpTERT1.0-SΔU1.
The N-terminal portion of pCR-TOPO-SpTERT1.0-S was amplified by PCR and subsequently cloned in pTNT vector. Cut with XhoI and XbaI (∼500-base pair fragment). A second PCR performed (∼2.5 kb) using the same SpTERT1.0-S as template. This fragment was cut with NotI and XbaI and also sequentially cloned in the same pTNT vector containing the 500-base pair N-terminal fragment. In this manner, U1 was excluded from the final resulting plasmid.
In Vitro Transcription of mRNA and Antisense Morpholino Design
All mRNA transcripts were generated using mMessage mMachine T7 kit (Ambion). The mRNA product is capped at the 5′ end with 7-methyl guanosine and has a poly-A tail as defined in the vector; hence, it has the two key traits of eukaryotic mRNA. The mRNA generated in this way was diluted in nuclease-free water. GFP-SpTERT-S+L (containing green fluorescent protein [GFP] fused C-terminally in frame to the first 100 nucleotides of SpTERT), and wild-type (wt)-SpTERT1.0 were first subcloned into pTNT vector (Promega. Madison, WI), which is suitable for in vitro expression with T7 RNA polymerase with an upstream kozak sequence. RNA concentrations assessed by spectrophotometry were aliquoted and injected at several concentrations ranging from 500 to 750 ng/μl for wt-SpTERT1.0 and 5′ untranslated region (UTR)-SpTERT-S-enhanced (e)GFP, and 750-1000 ng/μl for GFP-SpTERT-S+L RNA. For SE-TRAP experiments, diluted RNA was added to a final injection solution containing 10 mM Tris, pH 8.0, and 100 mM Lissamine dye. The constructs used in the SE-TRAP assays were wt-SpTERT4.0, wt-SpTERT5.0, SpTERT4.0-DN1, SpTERT4.0-DN2, SpTERT1.0-DN, SpTERT1.0+LUX, SpTERT4.ΔLUX, and SpTERT1.0ΔU1, and they were all at a final concentration in the range of 500–750 ng/μl in solution.
The antisense morpholino to SpTERT (S-morpholino) was obtained from Gene Tools (Philomath, OR), and its sequence was 5′-TGAATCCTCCTCACGTTCACAGTGC. The standard control morpholino from Gene Tools was used as the negative control. All morpholinos were dissolved in 10 mM Tris, pH 8.0, for injection. The S-morpholino was injected at 150–200 μM, for a final concentration of ∼1.0–2.0 μM per zygote.
Injection Volume Calculation
To estimate the final concentration of RNA or DNA in the injected egg, we used our microinjection apparatus to produce a drop of injection solution suspended at the end of the injection needle. To produce a visible drop, we increased the time of the injection pulse by a factor of 100, which has the effect of multiplying the drop volume by the same factor. Using an eyepiece micrometer under 200× magnification we were able to estimate the diameter of the drop within 2 μm. We calculated the volume of the roughly spherical drop and divided it by 100, giving an injection volume of 2–4 pl, depending on the size of the needle tip. We then estimated that the ratio of the injection volume to the egg volume multiplied by DNA/RNA concentration to calculate the final concentration in the egg (Supplemental Figure 1B).
Immunohistochemistry and Confocal Microscopy
We injected 150–200 embryos with either S-morpholino or control morpholino and fixed them with 1% paraformaldehyde in artificial seawater (+Mg, +Ca) at mesenchymal blastula for 1 h. After washing twice in PBST (1× PBS +Mg, +Ca, and 0.1% Tween), the embryos were treated with ice-cold methanol briefly and then directly transferred into a mix of 7% goat and 4% sheep serum PBST blocking solution for an additional hour. After washing the embryos twice more in PBST, they were then treated in 1:50 dilution of phospho-(Ser/Thr) (ATM/ATR) substrate (Cell Signaling Technology, Danvers, MA), or anti-phospho-histone H2A.X (Ser139) (Millipore, Billerica, MA), or 53BP1 (Cell Signaling Technology) antibody for 1 h. After primary antibody incubation, the embryos were washed twice again in PBST and then conjugated with a 1:200 dilution AlexaFluor 488 goat anti-mouse or anti-rabbit immunoglobulin G (H+L) (cross-absorbed) secondary antibody for 30 min. After an additional two more washes in PBST, the embryos are incubated in 1:50 4,6-diamidino-2-phenylindole (DAPI) counterstain (Sigma-Aldrich, St. Louis, MO) for 30 min. The embryos are then given a final wash in PBST and visualized using a laser scanning confocal microscope (Carl Zeiss, Thornwood, NY) with 63× plan-apo water immersion objective. Images were then manipulated using the LSM Image Browser software (Carl Zeiss). For DNA damage response experiments, blastula stage embryos in 5-cm Petri dishes placed on ice were subjected to ionizing radiation for a total dose of 12 Gy (13 min, 57 s) in a Gammacell 40 Exactor (Nordion International, Kanata, ON, Canada). After 15-min postirradiation, embryos were immediately fixed as described above.
RESULTS
Identification and Cloning of SpTERT-S
We cloned an echinoderm TERT by using RNA from blastula-stage embryos of S. purpuratus. This was accomplished by first searching the early contig assemblies of sea urchin genome for homology to hTERT. Using RACE, we then generated a complete SpTERT cDNA sequence. The resulting open reading frame consisted of 4047 bp, corresponding to a predicted protein of 1349 amino acids and a predicted molecular mass of 155 kDa. Therefore, this protein has a predicted size that is ∼28 kDa larger than a typical vertebrate hTERT. Across the region covering the CR-B domain to the C terminus (an alignment length of 697 residues), SpTERT showed 33% identity to that of hTERT. SpTERT was highly conserved over several critical domains known to be required for TERT catalytic activity. These include RNA template-binding (TRB) domain (Jacobs et al., 2006; Moriarty et al., 2002), RT domains required for catalytic activity (Nakamura et al., 1997), TRBD domain required for binding to telomerase RNA subunit (Lai et al., 2001) and a C-terminal domain (CTD) required for in vivo telomerase activity (Banik et al., 2002). In addition to several other conserved regions (Figure 1A) (including motifs TEN, Telomerase RNA binding domain [TRBD], CTD, CR-A(CP) to CR-F, T, 1, 2, A, B′, C, D, and E), the SpTERT protein contains at least two distinct unique domains that are not present in other species. These unknown domains, which we termed U1 and U2, seem to be located in the TRBD domain a region known also to be required for interaction with telomerase RNA component (TR) (Lai et al., 2001). The U1 and U2 (collectively named U domain) are located at critical sites between regions TEN and CR-A(CP) or CR-A and CR-B (QFP), respectively (Figure 1A). These U1/U2 regions showed no identity to other proteins based on a BLAST analysis. We designated this gene as SpTERT-S (SpTER1.0-S).
Identification and Cloning of SpTERT-L
To verify the sequence of SpTERT in an independent experiment, we amplified SpTERT from egg mRNA from a second animal. To our surprise, we amplified a second longer SpTERT of 4233 bp (designated SpTERT-L or SpTERT-L2.0) encoding a protein with a predicted size of 160 kDa. To verify that these two forms do not simply represent alternative spliced forms of SpTERT, we amplified SpTERT by genomic PCR (by using SPT-10F and SPT-BR primers) from individual eggs (Figure 1B) or genomic sperm DNA (Supplemental Figure 1A). These results consistently indicated the presence of the two genes in the same animal, SpTERT-S and SpTERT-L. The SpTERT-L was 265 bp longer than SpTERT-S (Figure 1, B and C). To confirm these results, we cloned full-length SpTERT from five additional wild-caught animals. These SpTERTs were further designated as SpTERT 4.0(L), 5.0(S), 6.0(L), 7.0(L), and 8.0(L). These results consistently revealed the presence of two distinct short (2) and long (5) SpTERTs by using seven total full-length cDNAs of SpTERT. Using primer combinations specific to SpTERT-S and SpTERT-L, we found that SpTERT-S and SpTERT-L represent distinct transcripts (Figure 1, D and E).
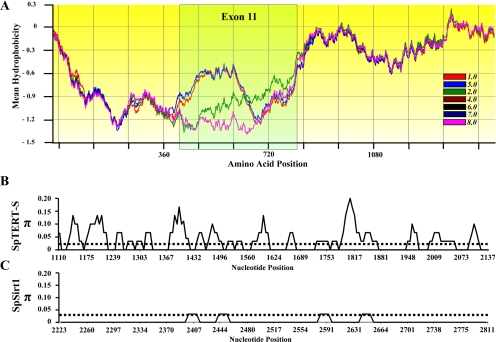
Interestingly, we observed an unusual intraindividual nucleotide variation (Figure 2) and changes in mean hydrophobicity profile (gapped Kyte and Doolittle scale; scan window size, 170) between these seven full-length SpTERTs (Figure 3A). The majority of this variation was present around exon 11 of the SpTERT-S (Figure 2, B–C). The overall nucleotide variation led to 90 nonsilent amino acid changes and a small deletion in the full-length protein (Figure 2C). These protein changes were mostly focused in the TRBD domain, the RNA binding domain of telomerase catalytic subunit. However, many changes were also observed in regions that are located in the conserved catalytic domain such as CR-B(QFP), motif 1, E, and CR-F (C-terminal) (Figure 2C). These unusual observations prompted us to investigate in detail the source of this variation.
Figure 3.
Comparison of SpTERTs. (A) A multiple sequence alignment of seven translated full-length short and long SpTERTs. The protein sequences of SpTERTs were used to generate a gapped, mean hydrophobicity plot (BioEdit). Areas of amino acid superimposition suggest similar hydrophobic properties of the amino acid at any given position (4.0, 6.0, 7.0, and 8.0 are superimposed). The highlighted exon 11 region reveals a divergence of protein similarity with distinct profiles. Note that regions of similar hydrophobicity were superimposed by the program and masked the presence of all seven versions in some areas. (B) A nucleotide diversity (π) scale for n = 4 SpTERT-S exon 11. (C) Nucleotide diversity (n = 4) of a nonconserved region (outside the conserved catalytic core domain) of SpSIRT1 region analyzed through DNAsp software.
Organization and Hypervariability of SpTERTs
Both SpTERT-S and SpTERT-L genes consist of 31 exons with a long exon 11 (Figure 2). Analysis of intraspecific nucleotide variation (π, the average number of nucleotide differences per site between two sequences) among all full-length SpTERT-S gene sequences revealed an unexpected nucleotide variation, particularly in the exon 11 region (Figures 2–4 and Table 1; for details of π calculations, see Materials and Methods and Supplemental Figures 2 and 3).
Table 1.
Statistical significance among variable gene regions
| Gene region | n | Avg. π/site | Avg. Ω/amino acid | p for π or Ω |
|---|---|---|---|---|
| Exon 11 SpTERT-S | 15 | 0.062 | 0.263 | <0.0001 |
| Exon 11 SpTERT-L | 16 | 0.114 | 0.028 | |
| Exon 11 SpTERT-S | 2 | 0.333 | 0.360 | <0.0001 |
| Exon 6 SpTERT-S | 2 | 0.088 | 0.101 | |
| Exon 11 SpTERT-L | 5 | 0.051 | 0.147 | =0.0001* |
| Exon 6 SpTERT-L | 5 | 0.030 | 0.055 | |
| SpTERT-S U-region | 4 | 0.037 | 0.107 | <0.0001 |
| SpSIRT1 U-region | 4 | 0.004 | 0.006 |
Summary of nucleotide diversity (π) for several groupings of SpTERT by using multiple sequence alignments analyzed through DNAsp software in the same manner for Fig. 2. SpSIRT1 was used as a control to refute the presence of hypervariability across all genes of S. purpuratus and shows an evident contrast with respect to any grouping of SpTERTs. Statistical analysis for comparison among means was carried out using a two-tailed t test, whereby a value of p < 0.0001 is considered to be significant.
* p value for π = 0.0001 and Ω < 0.0001.
Intriguingly, the exon 11 region codes for most of the U domain of the protein that consists of two subdomains U1 and U2 encompassing the CR-A(CP) motif (Figures 1A and 2A). The nucleotide polymorphism was unusually high in this region, with a peak in exon 11, π > 0.15 (Figure 2B). Although this nucleotide variation in SpTERT-S was highest in the exon 11 region, it was not restricted to the exon 11 (Figure 2B).
To gain a better understanding of the significance of this variation, we generated an exon 11 database of sequences as we performed genomic PCR (n = 24) and RT-PCR from oligo(dT)-primed RNA (n = 7) spanning the exon 11/U region of SpTERT from germline cells. For SpTERT-S, we obtained gametes (sperm DNA, n = 2; single egg, n = 11; and egg RNA, n = 2) from 15 different individuals. The primers used for all PCR amplifications were SPT-EX11–10F and SPT-BR. Compilation of these sequences and subsequent analysis showed high variation (Figure 4A). Similar results were obtained for exon 11 sequences of the SpTERT-L genes for the individuals (with addition of 1, n = 16) (sperm DNA, n = 2; single egg, n = 9; and egg RNA, n = 5) (Figure 4B). Overall, there was a highly significant variation of π = 0.124 among all (n = 31) sequences obtained. The nucleotide variation in both genes mostly created in-frame SpTERT proteins (Figure 4).
Distinct Hypervariable and Silent Regions of the U Domain
When we analyzed the exon 11 region of different animals, we observed that the high variation (π > 0.2) observed in SpTERT-S is found largely in the distinct U domain (Figures 2B and 3A). We measured nucleotide variation (π, shown in black) and amino acid entropy (Figure 4) (Ω, shown in orange; see Materials and Methods for details). The overall amino acid variability for the entire exon 11 region was deduced by considering all Ω > 0. The nucleotide variation in exon 11 of SpTERT-L was significantly higher than SpTERT-S (Table 1). Intriguingly, the opposite was true for amino acid variation. There were significantly more silent mutations in the exon 11 of SpTERT-L in comparison with SpTERT-S (Table 1). Therefore, SpTERT-S contained significantly more nonsynonymous substitutions. Multiple hypervariable peaks were present in SpTERT-S, with two major hypervariable domains with π > 0.1, which were named Uv-a (U region variable a) and UV-b. We also identified three silent region Us-a (U region silent a) and Us-b and CR-As in SpTERT-S. Analysis of SpTERT-L, in contrast, defined four silent regions (regions with low amino acid variation): Us-c, Us-d, Us-e, and LUX-s (Figure 4B).
As shown (Figure 4), the major difference between SpTERT-S and SpTERT-L is the LUX subdomain of U1 (defined in the box, Figure 4B). The LUX subdomain was only present in SpTERT-L and distinguishes it from SpTERT-S.
We then asked whether the hypervariability observed in SpTERT-S was unique to telomerase. As a control for our experiments, we amplified a region of SpSIRT1 gene. Comparison of sequences from four independent sequences (8 total) showed that nucleotide diversity of SpSIRT1 did not surpass π = 0.04 at the most variable positions (Figure 3C) (π = 0.003 overall average; Table 1), whereas same random number of SpTERT-S sequences peaked at π = 0.2, with an average π = 0.1 (Figure 3B). Therefore, the nucleotide variability in the SpTERT-S is highly significant (p < 0.0001, t test two-tailed; Table 1). This indicates that the nucleotide diversity observed in SpTERT is unique to telomerase and not a common feature of another S. purpuratus gene. We also measured Ka/Ks ratios as a measure of strength of positive versus purifying selection in all exon 11 sequences (n = 31). We found Ka/Ks = 5.3 (significant positive selection) for SpTERT-S and 3.6 for SpTERT-L.
Evolution of SpTERT
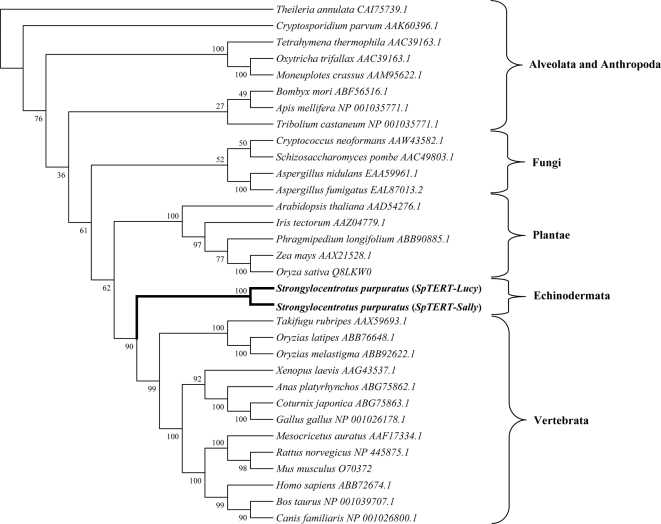
Multiple sequence alignment analysis with TERTs from other species indicated to us that SpTERT is highly conserved in regions that span the RNA template binding and the RT domain (Figure 1A). The RT motifs 1, 2, and A–E are present in all TERTs (Nakamura et al., 1997). The T motif is involved in binding to the RNA subunit (Bryan et al., 2000) and was also conserved in SpTERT (Figures 1A and 2). When the phylogenetic relationships were calculated among TERTs via a neighbor-joining matrix, we found that both SpTERTs shows a clear relationship with all other deuterostomes TERTs, consistent with the phylogenetic placement of sea urchins (outgrouping to vertebrates) (Figure 5). This analysis demonstrates that SpTERT-L and SpTERT-S are true orthologues of the vertebrate TERTs and represents the most divergent metazoan TERTs relative to vertebrates.
Figure 5.
Phylogeny tree of multiple TERTs. Cloned SpTERT-S and SpTERT-L along with full-length translated TERT sequences from 30 diverse species obtained from GenBank (accession numbers shown). Aligned using ClustalW and analyzed by the neighbor-joining method, the phylogeny tree was constructed using the Poisson correction model for amino acids and including complete deletion sites. Bootstrap values are shown. SpTERTs (in bold) are classified under Echinodermata.
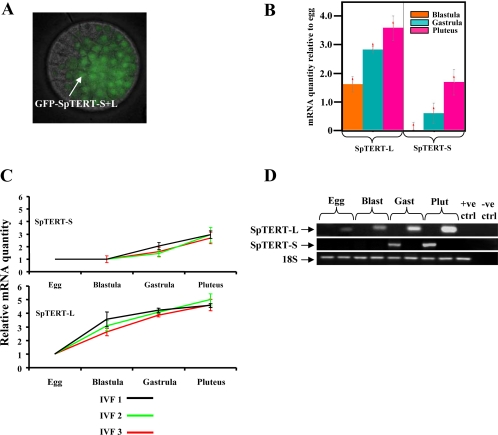
Developmental Regulation of SpTERT mRNA
To gain a better understanding of the function of SpTERT during embryogenesis, we next investigated the temporal regulation of SpTERT protein and mRNA expression. Expression of a GFP-SpTERT fusion mRNA (containing the 200-bp N-terminal portion of L+S forms) in eggs was traceable to the nuclei of early blastula embryo (Figure 6A), which is consistent with nuclear localization of TERT. To measure the endogenous levels of SpTERT mRNA during different stages of embryogenesis, we used a set of SpTERT-S–specific primers that amplified a 392-base pair fragment in the silent regions of exon 11 (Figures 4A and 6, B–D). We also designed a specific set of primers that exclusively amplified 115 bp of SpTERT-L found in the silent LUX subdomain of SpTERT-L (Figures 4B and 6, B–D).
Figure 6.
Temporal expression of SpTERT-S and SpTERT-L during embryogenesis. (A) Nuclear localization of eGFP-SpTERT S+L in a live S. purpuratus blastula. (B) RQ of three developmental stages of an IVF in triplicate measurements (RQ = 2−ΔΔCt). RQ was determined by Q-PCR methods using 18S as an endogenous control and set relative to egg-stage mRNA transcript of SpTERT-S and SpTERT-L; graphs are plotted as the log of relative quantity. (C) Relative mRNA levels shown across three independent IVFs, each developmental stage of three IVFs was measured in triplicate using Q-PCR to determine RQ for both SpTERT-S and SpTERT-L. All values are shown relative to egg-stage mRNA level. (D) Semiquantitative RT-PCR on an agarose gel stained with ethidium bromide. Total RNA extracted from each stage was treated with DNase I to prevent genomic contamination. The 18S was used as an internal control.
Differential and temporal expression of SpTERT-L and SpTERT-S were further investigated by performing three separate in vitro fertilizations (3 independent sources of eggs, same male sperm). We then quantified the mRNA in triplicate from each stage by Q-PCR at several developmental stages (Figure 6, B–D; see Supplemental Figure 4 for stages of embryogenesis in S. purpuratus).
When the SpTERT signal obtained via Q-PCR was normalized to that of an endogenous 18s control, we found that SpTERT-S transcript was expressed at low levels in egg and blastula and had increased expression at gastrula and pluteus (Figure 6B). In contrast, SpTERT-L mRNA was gradually increased from egg to pluteus (Figure 6, C and D). We found a similar differential expression pattern at blastula between SpTERT-S and SpTERT-L by using standard RT-PCR (Figure 6D).
Development of a SE-TRAP and Its Measurement in Ectoderm and Juvenile Rudiment
The differential temporal mRNA expression results suggested to us that SpTERT-S and SpTERT-L may have nonredundant functions. To gain a better understanding of their functional activities during embryogenesis, we developed a sensitive and quantitative three-stage SE-TRAP to analyze telomerase activity both quantitatively and temporally (Supplemental Figure 1, C and D). We found empirically that the SE-TRAP is linear up to at least four embryos/reaction (Supplemental Figure 1, C and D).
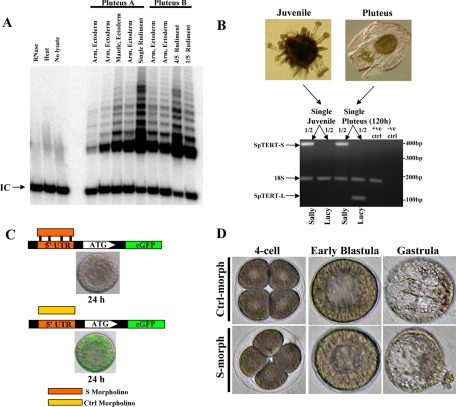
Next, we used 17-d-old plutei (post-IVF) with mature visible rudiments and microdissected finely the rudiment and ectodermal fragments. These fragments included the juvenile rudiment, arms and ciliary band fragments. Two genetically unrelated plutei were used throughout the experiment that follows. In pluteus, telomerase activity was lower in both ectodermal arms and differentiated ciliary band and highly elevated in the rudiment (Figure 7A). In Pluteus B, the rudiment was cut into four-fifth and one-fifth sections and subsequently subjected to SE-TRAP analysis. Telomerase activity was clearly present in both fragments and still higher in the rudiment and correlated with the amount dissected (Figure 7A). These results clearly indicated that telomerase activity is present in multiple ectodermal regions. However, the activity is particularly elevated in juvenile rudiment.
Figure 7.
Differential expression pattern of juvenile SpTERT and telomerase activity in mature plutei. (A) Two independent mature plutei (17 d old) were dissected to separate the arm, ciliary band, and rudiment. These fragments were then subjected to SE-TRAP. In pluteus B, the rudiment was cut into approximately four-fifth and one-fifth sections. (B) RT-PCR analysis of SpTERT-S and SpTERT-L by using Sally and Lucy primers on cDNA obtained from a single mature pluteus (120 h) and single juvenile. 18S was used as both the positive and internal control. (C) Top, S. purpuratus eggs were microinjected with a 5′ UTR-eGFP construct and the S-morpholino. The resulting blastula was then assayed for eGFP expression. Bottom, a control morpholino was coinjected with the 5′ UTR-eGFP construct. (D) Eggs were microinjected with 2 μM S-morpholino or control morpholino oligonucleotide, and embryos were followed until gastrula.
We next used a single juvenile pluteus with a grown rudiment and performed single-embryo RT-PCR by using either SpTERT-S– or SpTERT-L–specific primers. Results indicated that only SpTERT-S was expressed highly in the juvenile, whereas there was no detectable expression of SpTERT-L (Figure 7B). In contrast, 120-d plutei expressed both SpTERT-S and SpTERT-L. Therefore, presence of SpTERT-S mRNA strongly correlated with telomerase activity. To investigate these results further, we tested whether the hypervariable SpTERT-S was indeed an active protein and functional during embryogenesis.
Suppression of SpTERT-S during Development and SpTERT-S Activity
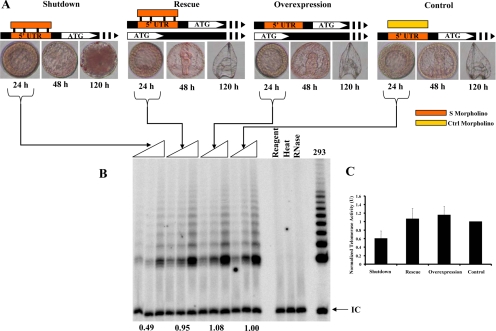
To investigate in detail telomerase function during embryogenesis, we identified the 5′ UTR region of SpTERT-S and designed an effective and specific UTR morpholino oligonucleotide (S-morpholino) (see Materials and Methods). When this morpholino was injected at low concentration of ∼1.5 μM into S. purpuratus embryos, it was able to suppress a 5′ UTR-SpTERT-S-eGFP reporter (Figure 7C). These SpTERT-S–suppressed embryos proceeded toward mesenchymal blastula; ingression of most primary mesenchymal cells occurred normally, but some cells at the vegetal pole lost polarity and underwent exogression (Figure 7D). At 1.5 μM concentrations of S-morpholino, ∼75% of embryos underwent exogression, and the remaining embryos failed to gastrulate. These phenotypes were successfully rescued with a wt-SpTERT-S mRNA indicating that it is a specific phenotype (Figure 8A). Overexpression of wt-SpTERT-S mRNA alone or a control morpholino did not have any effect on embryogenesis (Figure 8A). Suppression of SpTERT-S by the S-morpholino at 1.5 μM led to an ∼50% reduction in telomerase activity measured by quantitative SE-TRAP (Figure 8, B and C). This reduction in activity was fully restored by expression of SpTERT-S mRNA (Figure 8, B and C). Overexpression of SpTERT-S mRNA alone had no effect on telomerase activity, suggesting that SpTR, the RNA component of SpTERT-S, is limiting. These results suggested to us that SpTERT-S gives rise to telomerase activity in the embryo and is essential for embryogenesis. We next sought to understand the mechanism by which SpTERT-S regulates embryogenesis.
Figure 8.
Suppression of SpTERT, its genetic rescue, and activity effects. (A) From top left to top right: shutdown-embryos in which SpTERT was shutdown by S-morpholino; Rescue, the S-morpholino–injected embryos were rescued by wt-SpTERT mRNA expression; Overexpression, the wt-SpTERT mRNA was injected by itself as a control; and Control, the control morpholino oligonucleotide-injected embryos. Times after injection are shown. The red lissamine tracking dye was used in these experiments. (B) After 24 h, one, two, and four blastula embryos were selected on the basis of dye tracking for three independent SE-TRAP reactions per experiment as in A and analyzed by polyacrylamide gel electrophoresis. (C) The experiment in A was repeated three times, and SE-TRAP was performed. The pooled quantified results are shown in the graph. Calculated densities of TRAP products were normalized to that of the control morpholino-injected embryos and set to 1 unit of activity.
SpTERT-S Suppression and an Atypical DNA Damage Response
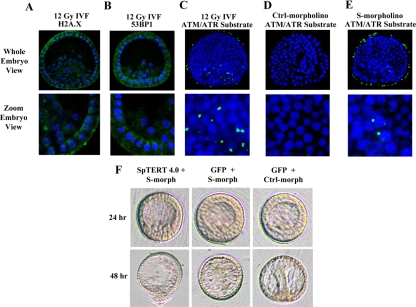
Telomere dysfunction as a result of telomerase suppression has been associated with activation of a DNA damage response in vertebrates. Therefore, it is reasonable to hypothesize that suppression of SpTERT-S in S. purpuratus embryos could result in activation of a DNA damage response in certain cells of mesenchymal blastula leading to its loss of polarity and exogression. To test this model, we used several antibodies against proteins known to be activated up on DNA damage, namely, 53BP1, gamma H2A.X and phospho-ATM/ATR substrates. The gamma H2A.X is present in S. purpuratus, however, it lacks the phosphor gamma Ser139 residue known to be phosphorylated up on DNA damage in vertebrates; hence, no signal can be detected by this antibody (Figure 9A). Furthermore, no region of an S. purpuratus 53BP1-like protein showed homology to the 53BP1 antibody; hence, no signal is obtained (Figure 9B). In contrast, when we used the phosphor-ATM/ATR substrate antibody, we detected a signal that was enhanced upon induction of DNA damage by ionizing radiation (Figure 9C). This signal was not detected in control nontreated embryos (Figure 9D). When SpTERT-S was suppressed we observed a signal for phosphor-ATM/ATR substrate suggesting that a DNA damage response is initiated upon telomerase inhibition (Figure 9E). On more detailed analysis and DAPI staining, it was clear, however, that the signal observed was often cytoplasmic and not nuclear as expected. Hence, it seems that the response observed up on SpTERT-S inhibition is rather nonclassical in S. purpuratus.
Figure 9.
Immunohistochemical analysis telomerase suppressed embryos and wt-SpTERT4.0-Lrescue phenotype. (A and B) Blastula-stage embryos were subjected to 12 Gy of radiation and subjected to gamma H2A.X or 53BP1 staining. No cross-reactivity with S. purpuratus was observed, and only nonspecific background was obtained. (C) Blastula-stage embryos were stained with an ATM/ATR phospho-substrate antibody (green) and DAPI counterstain (blue). Embryos were compressed at the time of mounting and visualization. Localization of the positive signal in the irradiated embryos was found to have a close, yet atypical association with the nucleus of the cell and possibly localized within the cytoplasm. (D) The nonirradiated control showed no positive staining. (E) The embryos injected with S-morpholino showed ATM/ATR substrate staining compared with controls. (F) Rescue attempt of telomerase suppressed embryos with wt-SpTERT4.0-L. Coinjection of wt-SpTERT4.0-L mRNA with the S-morpholino (that reduces telomerase activity) could not rescue the embryos. The control morpholino and GFP mRNA injected embryos showed no apparent aberration as it entered the gastrula stage of development.
Consequence of Variation on SpTERT-S Protein Activity
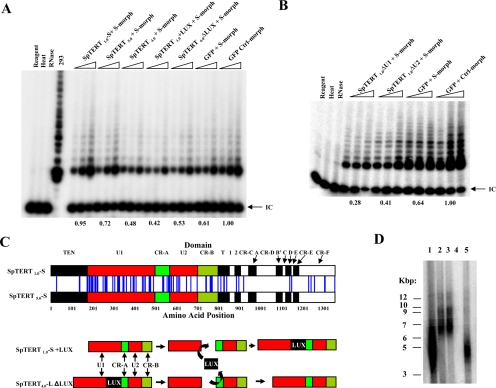
The hypervariability in SpTERT-S prompted us to ask the question whether these changes affect protein function. In particular, do these amino acid changes alter classic telomerase activity. To address this, we microinjected wt-SpTERT1.0-S and wt-SpTERT5.0-S, which represent two divergent SpTERTs, into S. purpuratus eggs, and we measured activity similar to that in Figure 8. The results indicated that wt-SpTERT1.0-S is ∼30% more active than wt-SpTERT5.0-S (Figure 10A). These two genetic variants of SpTERT differ by 90 amino acids and a small indel in the U1 region (Figure 10C, top schematic). Hence, at this point it is not clear which of these changes in isolation or in combination with one another reduce telomerase activity. However, what is clear from our results is that these variations result in significant changes in protein activity.
Figure 10.
SpTERT activities and interchanged LUX domain constructs. (A) Full length SpTERT1.0-S and SpTERT5.0-S mRNA were independently co-injected with S-morpholino and their relative efficiencies in rescuing the telomerase activity was measured. One, two, and four blastuals after microinjection were subjected to SE-TRAP and the product quantified. Values with respect to the control set are shown below each set of microinjections. The SpTERT4.0-L mRNA coninjected with S-morpholino showed no significant increase in activity over the S-morpholino control. In an attempt to construct a “pseudo-SpTERT-L like” SpTERT-S and a “pseudo-SpTERT-S” SpTERT-L, the LUX domain was inserted in frame into SpTERT-S and removed from SpTERT-L. Insertion of LUX into SpTERT-S effectively removes its function, while removing LUX from SpTERT-L does not render it an effective pseudo SpTERT-S. (B) Similar to (A), The mRNAs from SpTERT1.0-ΔU1-S, SpTERT1.0-ΔU2-S, and controls were co-injected with S-morpholiono. The resulting blastulas were subjected to SE-TRAP. Removal of the U1 or U2 domains from SpTERT1.0-S did not result in the rescue of activity back to endogenous levels of control. (C) Top, a schematic representation is shown for the aligned full length SpTERT-S variants, 1.0 and 5.0 that were used in (A). The vertical blue lines seen between both genes represent sites of amino acid substitution relative to the domain of which they are located in. Bottom, the strategy used in removing the differential LUX domain from SpTERT4.0-L and cloning it in-frame into SpTERT1.0-S. Only the domains contained within exon 11 are shown. (D) TRF length analysis of S. purpuratus sperm DNA. DNA samples from three individuals were restricted and subjected to southern blot analysis using a telomeric probe. Lanes 2 and 3 are from the same animal. Lane 4 is blank.
Differential Activities of SpTERT-S and SpTERT-L
Intriguingly, unlike SpTERT-S when we microinjected a wild-type wt-SpTERT4.0-L, it was unable to produce classic telomerase activity above background (Figure 10A). We reasoned that because one of the major differences between SpTERT-S and SpTERT-L genes is the LUX domain, perhaps the deletion of LUX domain can regulate telomerase activity. So, we proceeded with making in frame chimeric proteins. The chimeric protein SpTERT1.0-S+LUX was constructed in which the LUX domain was introduced after U1. We also made another chimeric protein (SpTERT4.0-L-ΔLUX) in which LUX domain was deleted (Figure 10C, bottom schematic). Addition of LUX domain to SpTERT-S eliminated its classical activity (Figure 10A). However, deletion of LUX domain in SpTERT4.0-L did not confer telomerase activity to SpTERT-L, and it still remained inactive (Figure 10A). Collectively these results suggest that natural absence of LUX domain in SpTERT-S protects it against loss of activity and furthermore suggest that SpTERT-L may have distinct activities beyond classical telomere synthesis functions. To gain better insight into function of U region, we also made a U1 deletion mutant of SpTERT1.0-S, and as before, we measured its activity and compared with a wt-SpTERT1.0-S. Results indicated that deletion of U1 region abolishes telomerase activity (Figure 10B). These results indicated to us that U1 is essential for SpTERT-S biological activity.
Variation in Germline Telomere Length
Maintenance of telomere length is achieved by cooperative effects of both telomere binding proteins and telomerase activities. Hence, we reasoned that perhaps these hypervariable changes in SpTERT and telomerase activity can affect telomere length. We obtained sperm DNA from several individuals and subjected them to restriction with HinfI and RsaI to release the telomere restriction fragments. These restricted DNA fragments (TRFs) were then run on a gel and subjected to Southern blot analysis by using a telomeric probe as described previously (Vaziri and Benchimol, 1998). When we measured telomere length in sperm DNA of several individuals, we found significant variation in germline mean telomere lengths (4–7 kbp).
DISCUSSION
To study telomerase function during embryogenesis, we cloned a novel telomerase reverse transcriptase gene that we termed SpTERT1.0-S and that upon overexpression in embryos was incapable of increasing telomerase activity beyond endogenous levels. To rule out the possibility of mutations during the cloning procedure, we recloned the SpTERT cDNA from a second animal. Surprisingly, we identified a second cDNA that was significantly different from SpTERT-S. This gene that we termed SpTERT-L contained an additional 265-base pair insertion and a significant number of amino acid substitutions. To our surprise, this enzyme was still incapable of restoring telomerase activity up on overexpression. We ruled out nonlinearity of telomerase activity assay by developing a modified two-stage SE-TRAP assay that is highly linear and can be used for temporal analysis. Yet, neither SpTERT-S or SpTERT-L seemed to give rise to telomerase activity up on overexpression in embryos. We hence recloned SpTERT from five additional unrelated animals. To our surprise, we found significant intraspecific germline variations among SpTERT-S and to a lesser extent in SpTERT-L. Furthermore, we found that lack of activity we originally observed was likely due to limiting levels of SpTR, the RNA component of telomerase RNP. As a result of these endeavors, we have uncovered the presence of several hypervariable telomerases with unusual properties and functions during embryogenesis.
Multiple Hypervariable Telomerases
We observed significant intraspecific (among different individuals) nucleotide and amino acid variation in the telomerase reverse transcriptase of any complex species. Total interspecific nucleotide sequence variation among (n = 31) exon 11 sequences of SpTERT-S and SpTERT-L was estimated at an unprecedented average of π = 0.12. Furthermore, this variation was not restricted to exon 11 alone. Significant variation was evident in the evolutionary conserved RT domain and other regions of SpTERT-S as well. This nucleotide diversity led to significant amino acid variation. Although a few nucleotide variations in exon 11 resulted in silent mutations, the majority of these nucleotide changes led to significant nonsynonymous amino acid substitutions. Literature suggests that single copy DNA sequences of S. purpuratus differ from each other by ∼4% (Britten et al., 1978). Others have also estimated that this variation to be generally at π = 0.006–0.029 for multiple exons of the endo16 gene (Balhoff and Wray, 2005). Consistent with this, when we measured as a control nucleotide variation in another unrelated catalytic protein (SpSIRT1 deacetylase), it showed no significant variation of a region outside the catalytic core with π = 0.003. Hence, the unusually high levels of variability in SpTERT are not a common feature of another gene in S. purpuratus. We also measured Ka/Ks ratios as a measure of strength of positive versus purifying selection in all exon 11 sequences (n = 31). We found Ka/Ks = 5.3 for SpTERT-S and 3.6 for SpTERT-L. This indicates that SpTERT-S in particular is under intense positive selection that cannot be explained by purifying Darwinian selection, which predicts ratios to be less than 1. There is a possibility that SpTERT-S has undergone positive Darwinian selection which maybe the result of a large population gene pool. Alternatively, the mutations maybe actively directed through a novel diversity generation mechanism to exon 11 and other regions. Our studies also imply that the variation observed in certain subset of gene domains in S. purpuratus may far exceed what has been anticipated. It is also possible that an active diversity generation mechanism targets certain genomic hot spots, hence rendering the resident genes to hypervariation.
Effect of Hypervariability on Enzyme Activity
We were able to measure and compare telomerase activity of two variant SpTERT1.0-S and SpTERT5.0-S catalytic subunits by microinjection and expression at blastula stage. These two variants differ by ∼90 amino acids across the protein and a small deletion in the U1 domain. Despite this level of change in protein identity, SpTERT5.0-S is only 30% less active than SpTERT1.0-S. The majority of amino acid variation was focused in the exon 11 region spanning U1, CR-A (CP), U2, CR-B (QFP), motif T, and motif 2 and around motif D and CR-F. Small changes were also present in TEN domain. However, in contrast to these domains, others such as motif 1, motif A, B′, C, and CR-D remained either hypomutable or underwent silent mutations, therefore completely clear of any amino acid changes. These constant motifs, in particular motif A, are known to be involved in nucleotide selectivity. Mutations in certain residues in motif A can switch the enzyme nucleotide binding specificity. Other conserved domains such as A and C are involved in metal ion binding and are key structural motifs of RT palm domains. Hence, it seems that majority of domains such as T motif, CR-A (CP), and CR-B (QFP) and their proximal domains such as U1 and U2 that are involved in RNA component binding are highly hypermutable. Despite this hypermutablility and changes in amino acid, SpTERT-S remains active. Studies in the past have used Saccharomyces cerevisiae as a model to perform unigenic evolutionary mutational screens for identification of essential functional and structural domains of telomerase (Friedman and Cech, 1999). The hypervariable telomerases of purple sea urchin introduce a natural evolutionary contrivance by which one can study function of telomerase or other proteins subjected to hypervariability.
Evolutionary Race between SpTR and SpTERT
The hypervariability in exon 11 of SpTERT-S spans mostly crucial domains required for functional binding to telomerase RNA component. If these regions are hypermutable, then how does SpTR, the RNA component of S. purpuratus, remain in a functional complex with telomerase? Our data indirectly suggest that this interaction must occur in vivo because the hypervariable SpTERT-S genes are indeed active and RNase sensitive in embryos. One explanation could be that mutational changes in SpTERT-S are matched by SpTR. Therefore, SpTERT-S and SpTR may be in an evolutionary race. In the future, it would be interesting to investigate whether SpTR or SpTERT are the evolutionary driver for variation.
Distinct Telomerases and Their Evolution
What is the biological significance of having two SpTERTs? In S. cerevisiae and other eukaryotes, the primary function of telomerase is to maintain the integrity and synthesis of telomeric ends. Mutations in telomerase components lead to significant telomere shortening and eventual loss of viability. Our results suggest that a secondary noncanonical embryonic function for telomerase has occurred by gene duplication followed by positive selection and subsequent neofunctionalization. The two SpTERTs of S. purpuratus might have evolved by duplication; one SpTERT has retained canonical telomere synthesis function and the other SpTERT evolved noncanonical functions. A second explanation that may not be mutually exclusive for existence of two telomerase forms is that indirect developers such as S. purpuratus evolved two distinct telomerase reverse transcriptases with overlapping and yet distinct functions. One protein, such as SpTERT-S, for functions in early blastula/gastrula and rudiment and another protein, SpTERT-L, exclusively for larval/ectodermal functions. Consistent with this model, we find that microdissected rudiment has high telomerase activity compared with ectodermal regions and that this activity was strongly associated with presence of SpTERT-S mRNA in juveniles. Hence, these experiments suggested to us that SpTERT-S contributes to classic telomere synthesis activity. Further evidence for this came from experiment in which SpTERT-S was shut down by translational inhibition. SpTERT-S inhibition led to haploinsufficiency in telomerase activity, and this haploinsufficency was restored fully by microinjection of wild-type SpTERT-S. This indicates that the hypervariable SpTERT-S is indeed the active form and is the catalytic subunit giving rise to telomerase activity. Consistent with our observations, it has been shown that the single-cell ciliate Euplotes crassus also harbors multiple EcTERT genes that are expressed at different times after mating. These genes seem to have altered functions; one function for telomere maintenance and another function for de novo telomere formation (Karamysheva et al., 2003). Although our results currently provide evidence for telomere maintenance function of SpTERT-S, we have so far been unable to identify a telomere-related function for SpTERT-L. At this point we can't rule out the possibility that these SpTERTs represent mosaic maternal/paternal alleles.
As expected, phylogenetic analysis of SpTERT-S and SpTERT-L proteins showed the closest evolutionary relatedness of any invertebrate to the vertebrate TERTs. The two SpTERT-L and SpTERT-S genes that we have identified differ from one another in sequences spanning the largest exon 11 of the SpTERT. The SpTERT-L was 265 bp longer in the distinct U1 region compared with that of SpTERT-S. We have designated this region of unknown function as the LUX subdomain. Resistance to gene conversion between two paralogous genes is generally reached by evolution of unique nucleotide domains or variation. Both of these characteristics are fulfilled in SpTERT-L as an addition of LUX subdomain in the U1 region, along with other significant nucleotide variation that exists between other regions of the two genes. These variations may explain why in purple sea urchins, SpTERT-L and SpTERT-S have been maintained in the gene pool without conversion to a single TERT. Consistent with this notion, the two genes have distinct and overlapping expression patterns that correspond with the temporal protein activity observed. This model is further strengthened by the experiments in which we inserted the LUX domain of SpTERT4.0-L into wild-type SpTERT1.0-S, creating a chimeric protein. Insertion of LUX domain in the correct position led to inactivation of SpTERT1.0-S classic telomerase activity. This indicates that in the wild population presence of LUX and other changes in SpTERT-L ensure resistance to gene conversion that would eliminate SpTERT-S and are detrimental to embryos. Hence, differences in SpTERT-L and SpTERT-S may ensure that they both survive in the gene pool.
Distinct Expression Pattern and Activities of SpTERT-L
SpTERT-S and SpTERT-L are differentially expressed and also have overlapping expression patterns. When telomerase activity is suppressed during early blastula, it can be rescued by SpTERT-S; however, SpTERT-L is unable to rescue telomerase upon overexpression. Furthermore, the arrest at blastula caused by telomerase suppression can be further rescued by SpTERT-S but not SpTERT-L. In addition, SpTERT-L is incapable of conferring telomerase activity. One of the major differences between SpTERT-L and SpTERT-S is presence of LUX domain in SpTERT-L. We reasoned that perhaps deletion of LUX will convert SpTERT-L to an active telomerase. These experiments indicated that deletion of LUX from SpTERT4.0-L does not restore telomerase activity to SpTERT-L. Hence, other changes present in SpTERT-L must be responsible for rendering it inactive. Insertion of LUX domain in SpTERT-S, however, leads to inactivation of SpTERT-S. Hence, it seems that by evolving LUX domain and other mutational changes throughout evolution, SpTERT-L has undergone self-inactivation for classic telomerase activity. Yet, interestingly, SpTERT-L is overall much less variable across the whole protein compared with SpTERT-S. Second, generation of two potent dominant-negative mutations in SpTERT4.0-L and its higher than endogenous overexpression in embryos still does not affect telomerase activity. At this point, however, we cannot completely rule out the possibility that SpTERT-L also contains classic telomerase activity, and our results are not due to instability of SpTERT-L RNA or protein. These genetic studies in combination with its differential expression patterns and its inability to rescue telomerase suppressed embryos suggest that SpTERT-L may have undergone neofunctionalization throughout evolution.
Telomerase Is Essential for Embryogenesis and Correct Polarity
Our original goal throughout the course of this study was to evaluate the effect of telomerase inhibition on early development. Suppression of SpTERT-S by translational blockers was associated with loss of telomerase activity and developmental arrest at mesenchymal blastula. In most cases, the developmental arrest was the result of exogression of cells in the vegetal pole of the embryo. Instead of moving inward toward the blastocoel and forming the early archentron, the putative vegetal/endodermal cells reversed polarity and exogressed. In some cases, the mesenchyme blastula-like embryo simply was arrested and did not proceed further. Suppression of telomerase has been associated with telomere dysfunction and activation of a DNA damage response pathway. To test this possibility, we stained telomerase suppressed blastulae with several markers of DNA damage response. One such antibody was against the phosphor-substrates of ATM/ATR proteins that are known to be formed in forms of foci post-DNA damage response in the nucleus. We found that the arrested embryos activate an atypical DNA damage response by phosphorylating substrates of ATM/ATR proteins localized in foci. However, instead of nuclear localization, they seemed to have cytoplsmic/membrane foci localizations. These results do not conclusively support a model in which a classical telomere-dependent DNA damage response has been activated but point toward the possibility that ATM/ATR substrates are activated in these telomerase suppressed embryos in an atypical way. Whether these changes are causal remain to be determined.
Telomere Length Hypervariability
Inactivation of telomerase by knockout studies has been shown to produce viable offspring (Blasco et al., 1997). This implies that loss of telomerase in inbred mouse embryos does not interfere with embryogenesis. Our results in contrast show that telomerase is essential for embryogenesis in S. purpuratus. Even induction of haploinsufficiency in telomerase activity in the blastulae is not compatible with embryogenesis. S. purpuratus shares significant ancestry with vertebrates and most importantly as we show their telomere lengths are highly similar to that of H. sapiens. One explanation for the differences observed could be differences in telomere length. Mice have ultralong telomeres in the variable range of 50–150 kb (Kipling and Cook, 1990), yet purple sea urchin at 4–7 kbp. Therefore, perhaps in telomerase knockout mouse embryos with long telomeres lack of telomerase does not compromise integrity of telomeres. However, in species such as sea urchins and humans with short telomeres, even small changes in telomere length or dysfunction can result in growth arrest. A second and equally likely explanation is that lack of telomerase in inbred mouse embryos is compensated by a second unknown mechanism, such as alternative lengthening of telomeres activation or other changes that compensate for telomerase requirement.
We also find variation in mean length of germline telomeres in S. purpuratus at ∼4–7 kpb. The hypervariability in telomere lengths maybe the result of germline variations in SpTERT protein we describe here. These variations in telomere length maybe the result of changes in telomerase processitivity or alterations in other unknown activities. However, it should be noted that regulation of telomere length in eukaryotes is complex and determined by interplay among telomere binding proteins, telomerase, and proteins regulating their activities.
In summary, our findings challenge commonly held notions that invariant enzymes are required for catalytic functions. Here, we provide evidence for the first known hypervariable gene of a multicellular complex animal that is functional and crucial for embryo survival. Generation of such highly diverse SpTERTs may reflect the evolutionary pressures that operate on the survival of adult or embryonic S. purpuratus in which case only embryos/juveniles with certain telomerases survive. To date, there has been no report of such germline variation in a crucial gene between individuals of the same species. Although counterintuitive, it suggests that depending on the enzyme, a large amount of flexibility in the structure maybe tolerated with minimal loss of function leading to even neofunctionalization. Most importantly, our findings point toward the presence of an active and unknown mechanism of diversity generation. Our results suggest that SpTERT of S. purpuratus may have undergone a gene duplication event followed by intense positive selection perhaps driven by SpTR for SpTERT-S and a more purifying selection for SpTERT-L. Throughout evolution, these forces may have caused neofunctionalization of SpTERT that may have contributed to generation of telomere length variation in germline and second led to a novel role for telomerase in early embryogenesis. The distinct and yet variable nature of these unique telomerases provides the telomerase field and others a natural contrivance by which novel telomerase functions and mechanisms of gene diversification can be studied.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Robert Weinberg and Dr. Christine Byrum for critical reading of the manuscript. We also thank Dr. Sam Benchimol, Dr. Yoshio Masui, and Dr. Ellen Larsen for discussions. We thank Dr. Daniel Durocher for antibodies and S.Narala for assistance with Figure 7A. We also are grateful to Dr. David McClay for reagents, advice, discussions, and critical reading of the manuscript. This work was supported by the Canadian Foundation for Innovation and Canada Research Chair program.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-07-0748) on October 22, 2008.
REFERENCES
- Allsopp R. C., Vaziri H., Patterson C., Goldstein S., Younglai E. V., Futcher A. B., Greider C. W., Harley C. B. Telomere length predicts replicative capacity of human fibroblasts. Proc. Natl. Acad. Sci. USA. 1992;89:10114–10118. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balhoff J., Wray G. Evolutionary analysis of the well characterized endo16 promoter reveals substantial variation within functional sites. Proc. Natl. Acad. Sci. USA. 2005;102:8591–8596. doi: 10.1073/pnas.0409638102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banik S. S., Guo C., Smith A. C., Margolis S. S., Richardson D. A., Tirado C. A., Counter C. M. C-Terminal regions of the human telomerase catalytic subunit essential for in vivo enzyme activity. Mol. Cell. Biol. 2002;22:6234–6246. doi: 10.1128/MCB.22.17.6234-6246.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrill N. J. The Origin of the Vertebrates. Oxford, United Kingdom: Oxford University Press; 1955. [Google Scholar]
- Betts D. H., King W. A. Telomerase activity and telomere detection during early bovine development. Dev. Genet. 1999;25:397–403. doi: 10.1002/(SICI)1520-6408(1999)25:4<397::AID-DVG13>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Blackburn E. H., Gall J. G. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J. Mol. Biol. 1978;120:33–53. doi: 10.1016/0022-2836(78)90294-2. [DOI] [PubMed] [Google Scholar]
- Blasco M. A., Lee H. W., Hande M. P., Samper E., Lansdorp P. M., DePinho R. A., Greider C. W. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- Boveri T. Ein geschlechtich erzeugter Organismus ohne mütterliche Eigenschaften. Am. Nat. 1889;27:222–232. [Google Scholar]
- Britten R., Cetta A., Davidson E. The single-copy DNA sequence polymorphism of the sea urchin Strongylocentrotus purpuratus. Cell. 1978;15:1175–1186. doi: 10.1016/0092-8674(78)90044-2. [DOI] [PubMed] [Google Scholar]
- Bryan T. M., Goodrich K. J., Cech T. R. Telomerase RNA bound by protein motifs specific to telomerase reverse transcriptase. Mol. Cell. 2000;6:493–499. doi: 10.1016/s1097-2765(00)00048-4. [DOI] [PubMed] [Google Scholar]
- Byrum C. A., Walton K. D., Robertson A. J., Carbonneau S., Thomason R. T., Coffman J. A., McClay D. R. Protein tyrosine and serine-threonine phosphatases in the sea urchin, Strongylocentrotus purpuratus: identification and potential functions. Dev. Biol. 2006;300:194–218. doi: 10.1016/j.ydbio.2006.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R., Sugawara H., Koike T., Lopez R., Gibson T. J., Higgins D. G., Thompson J. D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;32:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counter C. M., Avilion A. A., LeFeuvre C. E., Stewart N. G., Greider C. W., Harley C. B., Bacchetti S. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- Friedman K. L., Cech T. R. Essential functions of amino-terminal domains in the yeast telomerase catalytic subunit revealed by selection for viable mutants. Genes Dev. 1999;13:2863–2874. doi: 10.1101/gad.13.21.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskell W. H. On the origin of vertebrates from a crustacean-like ancestor. Q. J. Microsc. Soc. 1890;31:379–444. [Google Scholar]
- Greider C. W., Blackburn E. H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- Hall T. A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids. Symp. Ser. 1999;41:95–98. [Google Scholar]
- Harley C. B., Futcher A. B., Greider C. W. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- Harrington L., Zhou W., McPhail T., Oulton R., Yeung D. S., Mar V., Bass M. B., Robinson M. O. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev. 1997;11:3109–3115. doi: 10.1101/gad.11.23.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs S. A., Podell E. R., Cech T. R. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat. Struct. Mol. Biol. 2006;13:218–225. doi: 10.1038/nsmb1054. [DOI] [PubMed] [Google Scholar]
- Karamysheva Z., Wang L., Shrode T., Bednenko J., Hurley L. A., Shippen D. E. Developmentally programmed gene elimination in Euplotes crassus facilitates a switch in the telomerase catalytic subunit. Cell. 2003;113:565–576. doi: 10.1016/s0092-8674(03)00363-5. [DOI] [PubMed] [Google Scholar]
- Kilian A., Bowtell D. D., Abud H. E., Hime G. R., Venter D. J., Keese P. K., Duncan E. L., Reddel R. R., Jefferson R. A. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum. Mol. Genet. 1997;6:2011–2019. doi: 10.1093/hmg/6.12.2011. [DOI] [PubMed] [Google Scholar]
- Kim N. W., Piatyszek M. A., Prowse K. R., Harley C. B., West M. D., Ho P. L., Coviello G. M., Wright W. E., Weinrich S. L., Shay J. W. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- Kipling D., Cook H. Hypervariable ultra-long telomeres in mice. Nature. 1990;347:400–402. doi: 10.1038/347400a0. [DOI] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M. MEGA 3, Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lai C. K., Mitchell J. R., Collins K. RNA binding domain of telomerase reverse transcriptase. Mol. Cell. Biol. 2001;21:990–1000. doi: 10.1128/MCB.21.4.990-1000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejnine S., Makarov V. L., Langmore J. P. Conserved nucleoprotein structure at the ends of vertebrate and invertebrate chromosomes. Proc. Natl. Acad. Sci. USA. 1995;92:2393–2397. doi: 10.1073/pnas.92.6.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M., Davidson E. H. Gene regulatory networks for development. Proc. Natl. Acad. Sci. USA. 2005;102:4936–4942. doi: 10.1073/pnas.0408031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J., Hughes T. R., Shevchenko A., Mann M., Lundblad V., Cech T. R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- Mantell L. L., Greider C. W. Telomerase activity in germline and embryonic cells of. Xenopus. EMBO J. 1994;13:3211–3217. doi: 10.1002/j.1460-2075.1994.tb06620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The production of homozygous deficient tissues with mutant characteristics by means of aberrant mitotic behavior of ring-shaped chromosomes. Genetics. 1938;23:315–376. doi: 10.1093/genetics/23.4.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson M., et al. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- Moriarty T. J., Huard S., Dupuis S., Autexier C. Functional multimerization of human telomerase requires an RNA interaction domain in the N terminus of the catalytic subunit. Mol. Cell. Biol. 2002;22:1253–1265. doi: 10.1128/MCB.22.4.1253-1265.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott R. EST_GENOME: a program to align spliced DNA sequences to unspliced genomic DNA. Comput. Appl. Biosci. 1997;13:477–478. doi: 10.1093/bioinformatics/13.4.477. [DOI] [PubMed] [Google Scholar]
- Muller H. J. The remaking of chromosomes. Collecting Net. 1938;13:181–195. [Google Scholar]
- Nakamura T. M., Morin G. B., Chapman K. B., Weinrich S. L., Andrews W. H., Lingner J., Harley C. B., Cech T. R. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Nei M., Tajima F. DNA polymorphism detectable by restriction endonucleases. Genetics. 1981;97:145–163. doi: 10.1093/genetics/97.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olovnikov A. M. [Principle of marginotomy in template synthesis of polynucleotides] Dokl. Akad. Nauk SSSR. 1971;201:1496–1499. [PubMed] [Google Scholar]
- Pierce J. R. Signals and Noise. 2nd ed. New York: Dover Publications, Inc; 1980. Introduction to Information Theory: Symbols. [Google Scholar]
- Rozas J., Sanchez-DelBarrio J. C., Messeguer X., Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. J. Primer3 on the WWW for General Users and for Biologist Programmers. Totowa, NJ: Humana Press; 2000. [DOI] [PubMed] [Google Scholar]
- Schaetzlein S., Lucas-Hahn A., Lemme E., Kues W. A., Dorsch M., Manns M. P., Niemann H., Rudolph K. L. Telomere length is reset during early mammalian embryogenesis. Proc. Natl. Acad. Sci. USA. 2004;101:8034–8038. doi: 10.1073/pnas.0402400101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T. D., Stephens R. M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon C. E. A mathematical theory of communication. Bell Syst. Tech. J. 1948;27:379–423. 623–656. [Google Scholar]
- Szostak J. W., Blackburn E. H. Cloning yeast telomeres on linear plasmid vectors. Cell. 1982;29:245–255. doi: 10.1016/0092-8674(82)90109-x. [DOI] [PubMed] [Google Scholar]
- Vaziri H., Benchimol S. Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative life span. Curr. Biol. 1998;8:279–282. doi: 10.1016/s0960-9822(98)70109-5. [DOI] [PubMed] [Google Scholar]
- Watson J. D. Origin of concatemeric T7 DNA. Nat. New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- Wright W. E., Piatyszek M. A., Rainey W. E., Byrd W., Shay J. W. Telomerase activity in human germline and embryonic tissues and cells. Dev. Genet. 1996;18:173–179. doi: 10.1002/(SICI)1520-6408(1996)18:2<173::AID-DVG10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.