Abstract
The completion of chromosome segregation during anaphase requires the hypercondensation of the ∼1-Mb rDNA array, a reaction dependent on condensin and Cdc14 phosphatase. Using systematic genetic screens, we identified 29 novel genetic interactions with budding yeast condensin. Of these, FOB1, CSM1, LRS4, and TOF2 were required for the mitotic condensation of the tandem rDNA array localized on chromosome XII. Interestingly, whereas Fob1 and the monopolin subunits Csm1 and Lrs4 function in rDNA condensation throughout M phase, Tof2 was only required during anaphase. We show that Tof2, which shares homology with the Cdc14 inhibitor Net1/Cfi1, interacts with Cdc14 phosphatase and its deletion suppresses defects in mitotic exit network (MEN) components. Consistent with these genetic data, the onset of Cdc14 release from the nucleolus was similar in TOF2 and tof2Δ cells; however, the magnitude of the release was dramatically increased in the absence of Tof2, even when the MEN pathway was compromised. These data support a model whereby Tof2 coordinates the biphasic release of Cdc14 during anaphase by restraining a population of Cdc14 in the nucleolus after activation of the Cdc14 early anaphase release (FEAR) network, for subsequent release by the MEN.
INTRODUCTION
Faithful chromosome transmission requires the precise coordination between chromosome segregation and cell division. In Saccharomyces cerevisiae, mitotic exit is tightly coupled with the late segregation of the repetitive ribosomal DNA (rDNA) locus through the timely release of the Cdc14 phosphatase, a key regulator of mitotic exit (for recent reviews, see D'Amours and Amon, 2004; Pereira and Schiebel, 2004). Cdc14 performs diverse functions at the closing of mitosis (Stegmeier and Amon, 2004). First, Cdc14 promotes the late segregation of the ∼1 Mb rDNA array through the anaphase recruitment of condensin, which serves to both resolve cohesin-independent linkages and compact the array (D'Amours et al., 2004; Sullivan et al., 2004; Torres-Rosell et al., 2004; Wang et al., 2004; Machin et al., 2005). Second, Cdc14 is required for anaphase spindle integrity through the localization of Sli15/Ipl1 kinase (Pereira and Schiebel, 2003). Finally, Cdc14 phosphatase functions to reset the cell cycle to the G1 state through the inactivation of mitotic cyclin-dependent kinases (CDK) and the reversal of CDK phosphorylation events (Visintin et al., 1998).
Given these diverse functions, the spatiotemporal regulation of Cdc14 function is tightly controlled throughout the cell cycle. From G1 through anaphase onset, Cdc14 is sequestered in the nucleolus in a complex with its inhibitor Net1/Cfi1 (Shou et al., 1999; Straight et al., 1999; Visintin et al., 1999; Traverso et al., 2001; Huang and Moazed, 2003). In budding yeast, the nucleolus is assembled around the ∼1–1.5 Mb RDN locus, a highly specialized region owing to its repetitive nature, large size, and compartmentalization within the cell. The RDN locus comprises 100–150 copies of the 9.1-kb ribosomal DNA repeat on chromosome XII (Petes, 1979). Each unit encodes the 35S and 5S ribosomal RNAs and contains two nontranscribed spacer elements, NTS1 and NTS2. These regions contain multiple specialized cis-acting sequences that control replication initiation (ARS in NTS2) and pausing (replication fork block [RFB] in the NTS1) as well as transcription from PolI (35S rRNA; NTS2), PolII (E pro, NTS1), and PolIII (5S rRNA) promoters. The replication fork block binding protein, Fob1, binds directly to the RFB site in NTS1 (Mohanty and Bastia, 2004) and tethers a complex termed RENT (regulator of nucleolar silencing and telophase exit) consisting of Cdc14, its inhibitor Net1/Cfi1 and the silencing factor Sir2 (Shou et al., 1999; Straight et al., 1999; Visintin et al., 1999; Huang and Moazed, 2003). The RENT complex regulates Cdc14 activity and cellular localization, promotes DNA silencing within the rDNA, and inhibits hyperrecombination between repeats, thus promoting the overall stability of the array (Shou et al., 1999; Straight et al., 1999; Visintin et al., 1999; Huang and Moazed, 2003; Huang et al., 2006). In addition, a recent analysis of the protein interaction network inhibiting rDNA recombination also identified Tof2, Csm1, and Lrs4 as physically interacting with RENT complex components (Huang et al., 2006). Whether these factors also function in rDNA segregation and/or the regulation of Cdc14 activity during mitosis was not investigated.
As cells progress from metaphase into anaphase, chromosome segregation ensues. In budding yeast, active Cdc14 is released from the nucleolus during anaphase in two sequential waves. Two cell cycle–regulated pathways termed the FEAR (Cdc14 early anaphase release network) and the MEN (mitotic exit network) coordinate this release (see D'Amours and Amon, 2004). FEAR activation causes an early and partial release of Cdc14 into the nucleus, which is required to complete rDNA segregation (D'Amours et al., 2004; Sullivan et al., 2004; Torres-Rosell et al., 2004; Wang et al., 2004). The subsequent full release of Cdc14 occurs under the control of the MEN and promotes Cdc14 function on both nuclear and cytoplasmic substrates (Visintin et al., 1998). Although Cdc14 release from Net1/Cfi1 is modulated through the phosphorylation of both the phosphatase and its inhibitor (Traverso et al., 2001; Shou et al., 2002; Visintin et al., 2003; Azzam et al., 2004), the mechanism through which the FEAR network effects only a partial release of nucleolar Cdc14 in response to early anaphase remains unknown.
Recent studies have begun to address both why the segregation of the rDNA array is delayed until anaphase and how Cdc14 functions to promote rDNA segregation before mitotic exit. First, the high rate of transcription by PolI throughout the budding yeast cell cycle appears to impede rDNA segregation by promoting cohesin-independent linkages (Elliott and McLaughlin, 1979; D'Amours et al., 2004; Sullivan et al., 2004; Torres-Rosell et al., 2004; Wang et al., 2004; Machin et al., 2006; Tomson et al., 2006). Second, the large size of the rDNA array itself (approximately half the size of the largest yeast chromosome) necessitates additional compaction relative to the other yeast chromosomes in order to avoid breakage during cytokinesis, and this hypercondensation takes place during anaphase (Lavoie et al., 2004; Machin et al., 2005). The phosphatase Cdc14, under the control of the FEAR network, promotes both of these functions through the timely enrichment of the condensin complex to the nucleolus (Freeman et al., 2000; Bhalla et al., 2002; D'Amours et al., 2004; Sullivan et al., 2004; Wang et al., 2004).
Condensin is an evolutionarily conserved structural maintenance of chromosome (SMC) complex comprised of five subunits, which in budding yeast are named Smc2, Smc4, Brn1, Ycg1/Ycs5, and Ycs4 (for a recent review see Hirano, 2005). In mitosis, condensin is required for chromosome transmission fidelity and functions to both organize sister chromatids into discrete segregating units and shorten them to avoid chromosomal breakage during cell division. To date, budding yeast condensin has been implicated in numerous aspects of chromosome metabolism during both interphase and mitosis, including sister chromatid cohesion, mitotic chromosome condensation, rDNA stability, and silencing and the faithful segregation of both unique and repetitive loci (Strunnikov et al., 1995; Freeman et al., 2000; Lavoie et al., 2000, 2002, 2004; Ouspenski et al., 2000; Bhalla et al., 2002; Machin et al., 2004, 2005; Johzuka et al., 2006; Lam et al., 2006; Wang et al., 2006; Tsang et al., 2007; Yong-Gonzalez et al., 2007). Within the rDNA, condensin localizes to the nontranscribed spacer 1 and both Fob1-dependent and independent binding sites has been reported (Freeman et al., 2000; Wang et al., 2005; Johzuka et al., 2006; Johzuka and Horiuchi, 2007). By chromatin immunoprecipitation (ChIP), the association of condensin with the rDNA varies little from S phase through mitosis (Freeman et al., 2000; Johzuka et al., 2006), yet cytological observations demonstrate a dramatic enrichment of condensin in the nucleolus of anaphase cells (Freeman et al., 2000; Bhalla et al., 2002; Machin et al., 2004). This redistribution of condensin correlates with the existence of two cell cycle pathways for mitotic rDNA condensation, distinguishable by their cofactor requirements and rDNA structure (Lavoie et al., 2004; Machin et al., 2005). From early M phase until metaphase, the repeated rDNA array sequentially adopts a series of distinct morphologies, culminating in a characteristic rDNA loop, which extends away from the bulk chromosomes (Guacci et al., 1994; Lavoie et al., 2004; Machin et al., 2005). In addition to condensin, the metaphase condensation pathway requires cohesin, a second SMC-based complex, which is essential for the pairing of newly replicated sister chromatids from S phase until anaphase onset (Guacci et al., 1997; Lavoie et al., 2002). In anaphase however, when cohesin is removed from chromosomes, the successful segregation of the rDNA requires two additional cofactors: the Ipl1/Aurora B kinase as well as Cdc14 phosphatase, thereby interlinking chromosome segregation and the exit from mitosis (Machin et al., 2005).
MATERIALS AND METHODS
Reagents
Yeast media, sugars, α-factor, and G418 were purchased from Bioshop Canada (Burlington, ON, Canada). clonNAT was from Werner BioAgents (Jena, Germany) and nocodazole from Sigma (Oakville, ON, Canada). Protease inhibitors and antibodies were from Boehringer Mannheim (Laval, QC, Canada; EDTA-free Complete protease inhibitors, mouse anti-DIG, pig anti-goat-FITC, goat anti-mouse HRP). Additional antibodies were from Jackson ImmunoResearch (West Grove, PA; goat anti-mouse FITC, goat anti-rat FITC, and goat anti-mouse Cy3), Serotec (Raleigh, NC; Yol1/34 rat mAB-tubulin) and BabCO (Richmond, CA; 16B12 mouse mAb anti-HA), and Sigma (Raleigh, NC; peroxidase anti-peroxidase). IgG Sepharose was from Amersham-Pharmacia Canada (Oakville, ON, Canada). Restriction enzymes and Taq were from New England Biolabs (Beverly, MA).
Strains
Yeast strains are listed in Table 1 and were generated using standard genetic techniques (Guthrie and Fink, 1991). All gene deletions were generated by the Saccharomyces Genome Deletion Project (Open Biosystems, Huntsville, AL). Primers 200–300 base pairs upstream and downstream of each open reading frame (ORF) were used to PCR amplify individual deletions and integrate into additional genetic backgrounds: YPH499, YBL304-1, W303, MN70, and YBL194-1. Integrations were confirmed using specific primers 5′ of the upstream amplification primer and an internal KAN primer. YBL304-1 (cdc15-1:NAT) was created by integrating a 7.1-kb SpeI-PvuI fragment from pBL276 into YPH499. YBL265-1 (tof2Δ::KAN) was mated to S288C backcrosses of mob1-77 and dbf2-2 (originally from Mike Tyers, University of Edinburgh) for MEN suppression analysis. 3XHA (hemagglutinin)-tagged Cdc14 from DD48 (gift from Damien D'Amours, Université de Montréal) was PCR amplified and used to complement cdc14-1 temperature-sensitive growth by genomic integration. Query strains for systematic genetic analysis (SGA) analysis were generated by crossing or integration of previously characterized condensin mutant alleles marked with NAT into the Y2454 MATx parent (Tong et al., 2001), and the correct integration was verified by temperature sensitivity, colony PCR, and linkage analysis before performing screens. Yeast two-hybrid analysis was performed the AH109 reporter strain using pGBKT7 and pGADT7 bait/prey plasmids (Clontech's Matchmaker system, Palo Alto, CA).
Table 1.
S. cerevisiae strains used in this study
| Strain | Mating type | Genotype | Background | Reference |
|---|---|---|---|---|
| BY4741 | MATa | his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | S288C | Brachmann et al. (1998) |
| Y2454 | MATx | mfa1Δ::MFA1pr-HIS3 can1Δ his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | S288C | Tong et al. (2001) |
| YBL82#4 | MATx | Y2454 isogenic brn1-9:natR | S288C | This study |
| YBL83#1 | MATx | Y2454 isogenic YCG1:natR | S288C | This study |
| YBL84#3 | MATx | Y2454 isogenic ycg1-2:natR | S288C | This study |
| YBL100 | MATx | Y2454 isogenic smc2-8:natR | S288C | This study |
| YBL186-16B | MATx | mfa1Δ::MFA1pr-HIS3 can1Δ his3Δ leu2Δ lys2 ura3 ycs4-2:natR | S288C | This study |
| YBL247 | MATx | Y2454 isogenic mcd1-1:natR | S288C | This study |
| YPH499 | MATa | ade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ63 ura3-52 | S288C | Sikorski and Heiter (1989) |
| YBL257-1 | MATa | YPH499 isogenic pat1Δ::KAN | S288C | This study |
| YBL258-1 | MATa | YPH499 isogenic csm1Δ::KAN | S288C | This study |
| YBL259-1 | MATa | YPH499 isogenic fob1Δ::KAN | S288C | This study |
| YBL260-1 | MATa | YPH499 isogenic rtt103Δ::KAN | S288C | This study |
| YBL262-1 | MATa | YPH499 isogenic sac7Δ::KAN | S288C | This study |
| YBL263-1 | MATa | YPH499 isogenic smi1Δ::KAN | S288C | This study |
| YBL264-1 | MATa | YPH499 isogenic rmd11Δ::KAN | S288C | This study |
| YBL265-1 | MATa | YPH499 isogenic tof2Δ::KAN | S288C | This study |
| YBL267-1 | MATa | YPH499 isogenic rad52Δ::KAN | S288C | This study |
| YBL268-1 | MATa | YPH499 isogenic ynl254cΔ::KAN | S288C | This study |
| YBL269-1 | MATa | YPH499 isogenic ase1Δ::KAN | S288C | This study |
| YBL270-1 | MATa | YPH499 isogenic ctf4Δ::KAN | S288C | This study |
| YBL271-1 | MATa | YPH499 isogenic kar3Δ::KAN | S288C | This study |
| YBL272-1 | MATa | YPH499 isogenic rad50Δ::KAN | S288C | This study |
| YBL315-1 | MATa | YPH499 isogenic yml013c-aΔ::KAN | S288C | This study |
| YBL363-1 | MATa | YPH499 isogenic lrs4Δ::KAN | S288C | This study |
| YBL364-1 | MATa | YPH499 isogenic top1Δ::KAN | S288C | This study |
| YBL304-1 | MATa | YPH499 isogenic cdc15-1:natR | S288C | This study |
| YBL290-1 | MATa | YPH499 isogenic cdc15-1:natR pat1Δ::KAN | S288C | This study |
| YBL291-1 | MATa | YPH499 isogenic cdc15-1:natR csm1Δ::KAN | S288C | This study |
| YBL292-1 | MATa | YPH499 isogenic cdc15-1:natR fob1Δ::KAN | S288C | This study |
| YBL294-1 | MATa | YPH499 isogenic cdc15-1:natR sac7Δ::KAN | S288C | This study |
| YBL295-1 | MATa | YPH499 isogenic cdc15-1:natR smi1Δ::KAN | S288C | This study |
| YBL296-1 | MATa | YPH499 isogenic cdc15-1:natR rmd11Δ::KAN | S288C | This study |
| YBL297-1 | MATa | YPH499 isogenic cdc15-1:natR tof2Δ::KAN | S288C | This study |
| YBL298-1 | MATa | YPH499 isogenic cdc15-1:natR rad52Δ::KAN | S288C | This study |
| YBL299-1 | MATa | YPH499 isogenic cdc15-1:natR rad50Δ::KAN | S288C | This study |
| YBL300-1 | MATa | YPH499 isogenic cdc15-1:natR ynl254cΔ::KAN | S288C | This study |
| YBL301-1 | MATa | YPH499 isogenic cdc15-1:natR ase1Δ::KAN | S288C | This study |
| YBL302-1 | MATa | YPH499 isogenic cdc15-1:natR ctf4Δ::KAN | S288C | This study |
| YBL303-2 | MATa | YPH499 isogenic cdc15-1:natR kar3Δ::KAN | S288C | This study |
| YBL368-1 | MATa | YPH499 isogenic cdc15-1:natR lrs4Δ::KAN | S288C | This study |
| YBL314-1 | MATa | YPH499 isogenic cdc15-1:natR yml013c-aΔ::KAN | ||
| YBL418-19D | MATa | YPH499 isogenic 3xHA-CDC14 | S288C | This study |
| YBL418-30A | MATa | YPH499 isogenic 3xHA-CDC14 tof2Δ::KAN | S288C | This study |
| YBL418-33C | MATa | YPH499 isogenic 3xHA-CDC14 cdc15-1:natR | S288C | This study |
| YBL418-22A | MATa | YPH499 isogenic 3xHA-CDC14 cdc15-1:natR tof2Δ::KAN | S288C | This study |
| W303 | MATa | ade2-1 can1-100 his3-11,15 leu1-3,112 trp1-1 ura3-1 | W303 | Thomas and Rothstein (1989) |
| MN70 | MATa | W303 isogenic rdn1Δ/pRDN-wt-U RDN1 URA3 | W303 | Nierras et al. (1997) |
| YBL553-1 | MATa | W303 isogenic csm1Δ::KAN | W303 | This study |
| YBL554-1 | MATa | W303 isogenic fob1Δ::KAN | W303 | This study |
| YBL559-1 | MATa | W303 isogenic tof2Δ::KAN | W303 | This study |
| YBL624 | MATx | W303 isogenic ycs4-2:natR | W303 | This study |
| YBL562-1 | MATa | MN70 isogenic csm1Δ::KAN | W303 | This study |
| YBL563-1 | MATa | MN70 isogenic fob1Δ::KAN | W303 | This study |
| YBL564-1 | MATa | MN70 isogenic lrs4Δ::KAN | W303 | This study |
| YBL568-1 | MATa | MN70 isogenic tof2Δ::KAN | W303 | This study |
| YBL625 | MATx | MN70 isogenic ycs4-2:natR | W303 | This study |
| YBL419-11A | MATa | YPH499 isogenic cdc14-1 | S288C | This study |
| YBL419-2A | MATa | ade2-101 his3Δ200 lys2-801 trp1Δ63 ura3-52 tof2Δ::KAN cdc14-1 | S288C | This study |
| YBL620-9C | MATa | ade2-101 his3Δ200 leu2Δ1 trp1Δ63 ura3-52 mob1-77 | S288C | This study |
| YBL613-1B | MATa | ade2-101 his3Δ200 trp1Δ63 ura3-52 tof2Δ:KAN mob1-77 | S288C | This study |
| YBL621-1C | MATa | ade2-101 his3Δ200 leu2Δ1 trp1Δ63 ura3-52 dbf2-2 | S288C | This study |
| YBL617-1A | MATa | ade2-101 his3Δ200 leu2Δ1 lys2-801 ura3-52 tof2Δ:KAN dbf2-2 | S288C | This study |
| YBL194-1 | MATa | his3Δ200 trp1Δ63 SMC4-HAx3:his5+ cdc15-1 | S288C | This study |
| YBL593-1 | MATa | his3Δ200 trp1Δ63 SMC4-HAx3:his5+ cdc15-1 tof2Δ::KAN | S288C | This study |
| YBL594-1 | MATa | his3Δ200 trp1Δ63 SMC4-HAx3:his5+ cdc15-1 fob1Δ::KAN | S288C | This study |
SGA Screens
The systematic construction of double mutants used an ordered array of G418-resistant yeast knock out strains (MATa xxxΔ::KAN) according to Tong et al. (2001). SGA screens were performed as previously described (Tong et al., 2001) except that all steps were performed at 23°C. Briefly, NAT-marked query strains were mated to an array of yeast gene deletions (xxxΔ::KAN) and sporulated on plates, and haploid progeny were selected (two rounds) using an MFA1pr-HIS3 selection plus canavanine to select against the CAN1/can1Δ diploid (SD-HIS+CAN) and one round of double mutant haploid selection with drug (SD-HIS+CAN+NAT+KAN). Each double mutant combination was generated in quadruplicate, and visual comparison of colony sizes between haploid and double mutant selection plates was used as a primary screen for possible genetic interactions. Approximately 200–300 double mutant combinations exhibited slow or no growth phenotypes on the double mutant plates at least two of four times at 23°C. It is noteworthy that growth of double mutants was also assessed at higher temperatures (28 and 30°C) to look for suppressors of condensin mutants; none however were identified. Of the ∼300 double mutants identified visually in the primary screen at 23°C, 30 interactions were confirmed by standard tetrad dissection/linkage analysis and/or random spore analysis. For random spore, 400 μl of freshly saturated diploid was sporulated in 0.3% KOAc supplemented with 20 μg/ml adenine, 5 μg/ml histidine, 20 μg/ml leucine, 30 μg/ml lysine, and 8.75 μg/ml uracil for 5–6 d. A fivefold serial dilution series was plated onto selective media to compare haploid and double mutant haploid growth. All identified ycs4-2 interactors were individually tested against additional mutants in condensin (brn1-9, ycg1-2, and smc2-8) and cohesin (mcd1-1; Tong et al., 2004) to determine the specificity of interactions. Suppression or enhancement of temperature sensitivity was assessed by growing fivefold serial dilutions on YPD at indicated temperatures for 2–4 d.
Cell Biology and Biochemistry
All cell synchronization experiments were performed with log phase cells in YPD media at 23°C unless specified otherwise. G1 synchronization was performed by adding 3 μM α-factor to cells for 3–3.5 h until >85% of the cells were unbudded or shmooed. To release cells into an unperturbed cell cycle or a mitotic block, G1 cells were pelleted and washed twice in 1 vol YPD supplemented with 0.1 mg/ml pronase E, followed by resupension in YPD+pronase. To arrest cells in metaphase, 15 μg/ml nocodazole (from a 1.5 mg/ml stock in DMSO) was added, and the cells were grown for an additional 2.5–3.5 h at 23°C, until >85% were large budded. Anaphase arrests used the cdc15-1 mutation: cells were shifted to 37°C without nocodazole for 2–3 h, until >90% of cells were large budded. For G1 to G1 arrests, cells were released from α-factor without pronase E and 5 μM α-factor was readded 75 min after release. Fluorescence in situ hybridization was performed exactly as previously described (Guacci et al., 1994; Lavoie et al., 2004) using mouse anti-DIG, FITC pig anti-goat (Boehringer Mannheim), and FITC goat anti-mouse (Jackson ImmunoResearch). Chromosome spreads (Michaelis et al., 1997) and indirect immunofluorescence were performed using 1:200 Yol1/34 rat mAB-tubulin (Serotec) and 16B12 mouse mAb-HA (BabCO) at 1:250 for spreads and 1:500 for IIF as in (Lavoie et al., 2004). Secondary antibodies Cy3 goat anti-mouse (1:1000), FITC goat anti-mouse (1:500), and FITC goat anti-rat (1:200) were from Jackson ImmunoResearch. Fluorescence microscopy was performed using a Nikon Eclipse E1000 microscope (Melville, NY; 1000×) with all scoring directly at the microscope with no integration. Images were captured with a Hamamatsu ORCA-ER digital camera (Bridgewater, NJ) using G2-A for Cy3, UV-1A for DAPI, and V-2A for FITC; gain was set to 1. Images were taken with Nikon Simple PCI software (ver. 5.1.0.0110). Image attributes (contrast and color choice) were modified in Adobe Photoshop 8.0 (San Jose, CA) for publication purposes only.
Sequence Alignment
Tof2 sequences from S. cerevisiae, S. mikatae, S. bayanus, and S. paradoxus were aligned using ClustalX ver. 1.83 (www.clustal.org). The Tof2 alignment was then compared with a similar alignment of Net1/Cfi1 sequences to identify four regions of conservation in the N-terminal portion of the proteins.
RESULTS
Novel Condensin-interacting Genes
To identify new genes required for condensin function, we performed parallel systematic synthetic enhancement screens (Tong et al., 2001) using previously characterized hypomorphic, temperature-sensitive alleles of condensin (Freeman et al., 2000; Tong et al., 2001; Lavoie et al., 2002). Thirty genes reproducibly interacted with the ycs4-2 mutant at the permissive temperature, as judged by synthetic growth defects of double mutant strains (Figure 1). Genes interacting with condensin fell into a small number of functional categories including genomic integrity, cytoskeletal function, rDNA stability, uncharacterized and other. Of these, CSM1, LRS4, TOF2, CIN8, KAR3, YDJ1, and RTT103 interacted with all four condensin alleles tested and exhibited among the most dramatic growth defects. CIN8 and KAR3 are microtubule motors involved in spindle dynamics and are therefore unlikely to directly modulate mitotic chromosome structure (Kashina et al., 1997; Wu et al., 2006). YDJ1 and RTT103 function in protein and RNA stability, respectively, and their synthetic lethal interactions with condensin likely reflect our use of temperature-sensitive DAmP alleles, which decrease the amount of mRNA and thus reduce the function of hypomorophic alleles (Caplan and Douglas, 1991; Kim et al., 2004; Schuldiner et al., 2005). In contrast, CSM1 (Chromosome Segregation in Meiosis 1; Rabitsch et al., 2001, 2003) , LRS4 (Loss of Ribosomal Silencing 4; (Smith et al., 1999) and TOF2 (Topoisomerase-interacting Factor 2; Park and Sternglanz, 1999) encode nucleolar proteins with no known role in rDNA condensation and thus were chosen for further analysis.
Figure 1.
Genetic interaction profiles of yeast condensin. (A) Synthetic genetic interactions between the hypomorphic condensin allele ycs4-2::NAT (YBL186-16b) and indicated yeast deletion strains (xxxΔ::KAN). The systematic generation of double mutant combinations was performed in parallel and in quadruplicate using systematic genetic analysis technology (Tong et al., 2001) using the yeast knock out collection. Synthetic growth defects were initially determined by comparing growth of single versus double mutants at 23°C, the permissive temperature for all the condensin mutants. Interactions were individually confirmed by tetrad analysis and/or random spore analysis plus PCR to confirm the identity of the deleted locus and its linkage with the KAN marker. Filled and open circles denote synthetic lethal and sick interactions, respectively. (B) Verification by random spore analysis. Sporulated cultures (genotypes as indicated) were digested to release spores, and fivefold serial dilutions were plated on media that selects for MATa haploid single or double mutants. Genetic interactions were observable as a decrease in viable double mutant progeny (haploid selection plus clonNAT and G418) versus single mutant progeny. (C) Comparison of genetic interaction profiles between condensin mutants.
CSM1, LRS4, and TOF2 Function in Mitotic rDNA Segregation
Because condensin localizes to the nucleolus during the mitotic cell cycle and is known to promote rDNA condensation and segregation, we reasoned that the observed synthetic lethality between condensin and csm1Δ, lrs4Δ, tof2Δ, as well as fob1Δ mutants could at least in part derive from defects in rDNA segregation. To test this, we generated double mutants in a background carrying a deletion of the rDNA array and which provides rRNA from a high copy plasmid (Figure 2A). Deletion of the rDNA array rescued the lethality of the double mutants as a high frequency of double mutant spores, which carried resistance markers for both G418 and clonNAT, were recovered and grew robustly on rich media. Thus, the lethality observed in the ycs4-2 csm1Δ, lrs4Δ, tof2Δ, and fob1Δ strains primarily resulted from defects in segregating the large rDNA array.
Figure 2.
Analysis of rDNA array stability and condensation. (A) Suppression of synthetic lethality by deletion of the rDNA array. ycs4-2::NAT RDN1 (YBL624) and rdn1Δ (YBL625) strains were crossed with indicated nucleolar gene deletions (xxxΔ::KAN in MN70; YBL562-1, YBL563-1, YBL564-1, and YBL568-1). In rdn1Δ strains, the rDNA was carried on a high copy plasmid. After dissection of meiotic progeny, double mutant combinations (circled) were identified or inferred using drug markers. (B) Deletion of the rDNA array did not suppress the temperature sensitivity of ycs4-2::NAT. Fivefold dilutions of indicated strains (W303, MN70, YBL624, and YBL625) were spotted on rich media and incubated at the indicated temperature for 2–3 d. (C and D) Fluorescence in situ hybridization of the yeast RDN locus. Deletion mutants in WT (YPH499) or cdc15-1 (YBL304-1) backgrounds as indicated were arrested in metaphase (nocodazole, 23°C, C) or anaphase (cdc15-1 mutation, 37°C, D) and processed for rDNA FISH (see Materials and Methods). Micrographs show the rDNA signal in green with the bulk chromosomes in red. In WT cells, condensed rDNA adopts a distinct loop (metaphase) or line (anaphase) morphology, whereas decondensed rDNA appears as a disorganized puff. Quantitation of the rDNA condensation in metaphase or anaphase arrested cells, respectively. Numbers reflect the percent nuclei with condensed rDNA loops (C) or lines (D). At least 100 nuclei were score per sample.
To next address if the condensin-interacting genes identified in our screen function in rDNA condensation, we performed fluorescence in situ hybridization (FISH) using the 9.1-kb ribosomal DNA as a probe. Deletion strains were synchronized in G1 and then released into a mitotic arrest either by treatment with the microtubule-depolymerizing drug nocodazole (Nz) or by temperature inactivation of the mitotic network exit kinase Cdc15 (cdc15-1) and then processed for rDNA FISH (Figure 2, C and D). In interphase, the rDNA exists as a disordered “puff” structure, which by metaphase forms a characteristic loop that extends away from the bulk chromosomes in a reaction requiring both cohesin and condensin. In csm1Δ, top1Δ, fob1Δ, and lrs4Δ strains, 18, 23, 31, and 34% of nuclei formed rDNA loops, respectively, (vs. 84% in wild type [WT]) with 4–8% of the total nuclei exhibiting morphologies consistent with partial condensation (clusters and lines; refer to Lavoie et al., 2004). This suggests that most if not all rDNA condensation is abrogated in the absence of these genes. In contrast, yml013c-aΔ and rtt103Δ strains accumulated a high level of condensation intermediates: of the 69 and 80% of nuclei with condensed rDNA, 28 and 14% of nuclei, respectively, were scored as having partially condensed rDNA.
In anaphase cells where the bulk of cohesion between sisters is removed and the chromosomes have segregated into each daughter cell, the rDNA array hypercondenses to yield a visibly shortened FISH signal, closely juxtaposed to the chromosomes. Under these conditions, CSM1, LRS4, and FOB1 were also required for condensation, with 8, 10, and 26% of nuclei, respectively, exhibiting the expected rDNA line morphology versus 88% in WT cells. Although the increased magnitude of the defects compared with that seen in metaphase-arrested cells could derive from the modified experimental regimen that included a 37°C temperature shift, the absence of temperature sensitivity in any of these strains (data not shown) rather suggests that these genes may function in both the metaphase and anaphase condensation pathways. In contrast, deletion of tof2Δ had a much more pronounced effect on rDNA condensation during anaphase (17% nuclei had condensed rDNA) versus metaphase (68% condensed). These data suggest that although Csm1, Lrs4, and Fob1 are likely to affect multiple aspects of rDNA structure and transmission during mitosis, Tof2 plays a specific role in anaphase rDNA compaction and partition.
Tof2 Genetically Interacts with Cdc14 Phosphatase
The observation that Tof2 specifically promotes anaphase hypercondensation of the rDNA suggested that Tof2 could modulate either condensin recruitment and/or function within the rDNA. To address this, we performed chromatin spreads and cytologically assessed condensin binding to the DAPI-poor nucleolar region in both metaphase and anaphase arrested cells (Figure 3). In the absence of Tof2, condensin was still enriched within the rDNA, indicating that bulk condensin recruitment is not itself sufficient to promote rDNA condensation. Consistent with this, condensin binding was also observed in fob1Δ cells, which sustain neither metaphase nor anaphase condensation. Because the N-terminal regions of Tof2 and the Cdc14 inhibitor Net1/Cfi1 share significant homology, we next asked if Tof2 could impinge on rDNA condensation by regulating Cdc14 (Figure 4). Both Cdc14 full-length and its N-terminal 375AA region, which contains both the phosphatase and Net1-binding activities (Park and Sternglanz, 1999; Traverso et al., 2001; Huang et al., 2006), promoted a robust two-hybrid interaction with the N-terminal regions of Tof2 (1–270) and Net1 (1–340). We next looked for genetic interactions between tof2Δ and cdc14, FEAR, and MEN mutants. No synthetic genetic interactions were observed between tof2Δ and spo12Δ, bns1Δ, and slk19Δ (FEAR components) or between cdc15-1, bub2Δ, amn1Δ, and lte1Δ (MEN components; data not shown). In contrast, tof2Δ exacerbated the temperature sensitivity of a cdc14-1 strain and partially rescued the lethality of two MEN mutants, dbf2-2 and mob1-77 (Figure 4, C and D). Because Dbf2 and Mob1 are two kinases that function immediately upstream of Cdc14 release during mitotic exit, these data suggest that like Net1, Tof2 may negatively regulate Cdc14 function and/or release.
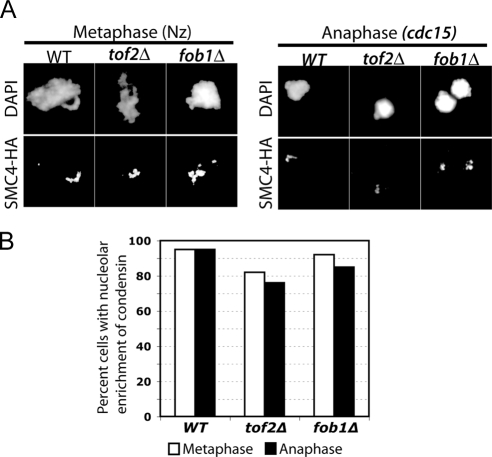
Figure 3.
Nucleolar enrichment of condensin in tof2Δ cells. (A) Chromatin spreads of the condensin subunit Smc4-HA in the presence (YBL194-1) or absence of Tof2 (YBL593-1) or Fob1 (YBL594-1) were performed in metaphase- versus anaphase-arrested cells. Micrographs show the localization of condensin predominantly in a subnuclear, DAPI-poor region characteristic of the rDNA. (B) Quantitation of the data shown in A. At least 100 nuclei is scored per sample.
Figure 4.
TOF2 interacts with CDC14. (A) Net1 and Tof2 sequence alignments. The N-terminal 341 AA of Net1 are sufficient to bind Cdc14 and share 25% sequence identity and 64% similarity with Tof2. Four regions of high homology were found to be conserved between Net1 and Tof2 in four closely related Saccharomyces yeast species (S. cerevisiae, S. mikatae, S. parodoxus, and S. bayanus). These correspond to region I-IV in Net1 (aa 1–14, 72–161, 209–220, and 494–505, respectively) versus Tof2 (aa 4–17, 82–172, 222–233, and 363–374, respectively). Dashed lines represent Net1-AA1–340 and Tof2-AA1–270 that were used for yeast two-hybrid analysis. (B) Yeast two-hybrid interactions between Tof2 (1–270) and Cdc14 (FL and 1–375). (C) tof2Δ exacerbates cdc14-1 temperature sensitivity. Fivefold serial dilutions of tof2Δ (YBL265-1), cdc14-1 (YBL419-11a) and tof2Δ cdc14-1 (YBL419-2a) were spotted on rich solid and incubated at the indicated temperatures for 2–3 d. (D) tof2Δ suppresses MEN mutants. Single and double mutants as indicated were spotted on rich media as fivefold serial dilutions and incubated 2–3 d at the indicated temperatures. The following strains were used: dbf2-2 (YBL621-1c), dbf2-2 tof2Δ (YBL617-1a), mob1-77 (YBL620-9c), and mob1-77 tof2Δ (YBL613-1b).
Tof2 Is Required for the Biphasic Release of Cdc14 during Anaphase
During mitotic exit Cdc14 is released from the nucleolus in two sequential waves; thus, Tof2 could function in one or both pathways. To address if the timing of Cdc14 release was affected in tof2Δ cells, we monitored Cdc14 localization in synchronized cycling cells (Figure 5). Cells were synchronized in G1 and released into an unperturbed cell cycle with additional α-factor added after 1 h to prevent reentry into the next cell cycle. At 15-min time points, cells were fixed and processed for indirect immunofluorescence to monitor HA-Cdc14 localization (nucleolar, nuclear, or cytoplasmic) versus spindle length (Figure 5). Although the cell cycle timing was nearly identical between WT and tof2Δ strains as judged by budding index as well as both cellular and nuclear morphology (data not shown), a dramatic difference in the kinetics of Cdc14 release was apparent as the cells entered anaphase. At 105 min after G1 release, which corresponds to anaphase entry, twice as many cells with fully released Cdc14 was observed in the tof2Δ versus WT cells (Figure 5, B and C). Thus, although the onset of Cdc14 release was similar in both TOF2 and tof2Δ strains, the partial release characteristic of the FEAR pathway was difficult to observe in cells lacking Tof2. To demonstrate this more clearly, we next quantified Cdc14 release versus spindle length in the 105-min sample (Figure 5C). The FEAR-dependent release of Cdc14 occurs in early anaphase cells with a short spindle typically ranging from 4 to 6 μm. Compared with WT cells, the full release of Cdc14 from the nucleolus occurred precociously in the absence of Tof2, in 23% of cells with very short spindles (2–4 μm) and 78% of cells with 4–6 μm spindles (vs. 8 and 57% in WT, respectively).
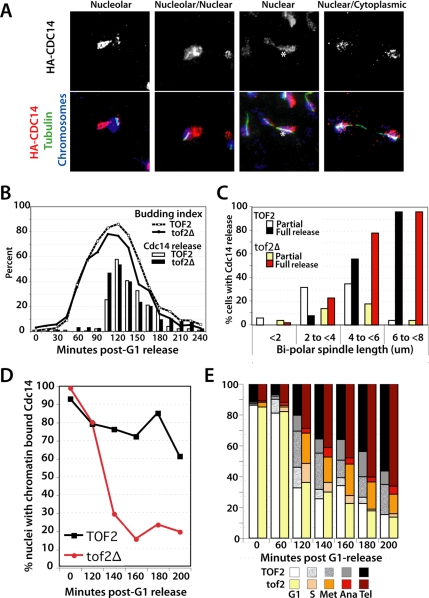
Figure 5.
Tof2 regulates Cdc14 release during anaphase. (A) HA-Cdc14 staining in mitotic cells. Indirect immunofluorescence (IIF) of HA-Cdc14 and tubulin versus chromosome DAPI staining in YBL418-9d. Micrographs in the top panels show the HA-Cdc14 signal in isolation. Bottom panels show a merged image of HA-Cdc14 (in red) versus mitotic spindles (tubulin, in green) and chromosomes (DAPI stain, in blue). Micrograph of HA-Cdc14 versus spindles and chromosomes during mitosis. (B) Total Cdc14 release (no nucleolar staining visible) as a function of time and cell cycle stage. G1-synchronized cells were released into an unperturbed cell cycle. Cells were taken every 15 min, monitored for budding index (top curves), and processed for IIF to detect HA-Cdc14 signal. Complete release of Cdc14 from nucleoli was scored as a function of time in TOF2 (YBL418-9d) versus tof2Δ (YBL418-30a) cells. Note that additional α-factor was added at 1 h after release to prevent reentry into the cell cycle. (C) Cdc14 release versus spindle length in TOF2 (YBL418-9d) versus tof2Δ (YBL418–30a) cells. Cdc14 signal was scored as a partial (nucleolar staining evident) or full (no nucleolar staining) release relative to spindle length. (D) Kinetics of HA-Cdc14 chromatin dissociation in TOF2 cdc15-1 (YBL418-33c) versus tof2Δ cdc15-1 (YBL418-22a) cells during early anaphase. Synchronized G1 cells were released at 37°C. At indicated time points, duplicate samples were taken for chromatin spreads of HA-Cdc14 and to monitor cell cycle progression (E). In all samples, greater than 100 cells or nuclei were scored per time point.
One possible explanation for the enhanced release of Cdc14 in the absence of Tof2 is that the MEN might be precociously activated in tof2Δ cells. To test this, we used a cdc15-1 mutant that is deficient in the MEN and therefore arrests the cell cycle in anaphase after FEAR activation but before mitotic exit. We again performed a synchrony-release time course experiment with cells progressing synchronously from G1 to anaphase (cdc15-1). To monitor the pool of Cdc14 remaining on chromatin after FEAR activation, we followed Cdc14 release by chromatin spreads (Figure 5D). In WT cells, the partial release of Cdc14 in the absence of the MEN function was readily observed: even after 200 min after release from G1, 60–70% of nuclei retained an HA-Cdc14 signal on chromatin. Furthermore, in cells where the chromatin remained relatively unspread, the Cdc14 signal was associated with the weakly stained DAPI mass characteristic of the nucleolus. In contrast, although the TOF2 and tof2Δ cultures initiated the release of Cdc14 at the same time, tof2Δ cells exhibited a more precipitous release of Cdc14 from chromatin (Figure 5D). By 140 min after release from G1, only 30% of nuclei stained positively for Cdc14, and this decreased to ∼20% by 200 min. The dramatic Cdc14 release occurred even in the absence of the MEN, arguing that anaphase and mitotic exit were not simply progressing more rapidly in tof2Δ cells. Indeed the cell cycle distribution of TOF2 and tof2Δ cultures was almost identical (Figure 5E). Together, these data indicate that Tof2 functions at the interface between the FEAR network and the MEN and serves to inhibit the full release of nucleolar Cdc14, thereby promoting the biphasic release of Cdc14 observed during a normal mitosis.
DISCUSSION
Nucleolar Integrity Genes Are Required for rDNA Condensation
The deletion of genes implicated in nucleolar and/or rDNA metabolism are especially detrimental for cell growth when condensin is compromised. Nucleolar factors such as FOB1, CSM1, LRS4, TOP1, and TOF2 genetically interact with multiple condensin mutants and are required for rDNA condensation (Figure 2). Of these, the replication Fork Blockingless 1 gene FOB1 is a key regulator of rDNA integrity (Kobayashi et al., 1998). Fob1 binds the RFB site in the rDNA and functions to prevent bidirectional replication, to regulate hyperrecombination and to promote rDNA silencing (Kobayashi and Horiuchi, 1996; Johzuka and Horiuchi, 2002; Huang and Moazed, 2003; Kobayashi, 2003; Mohanty and Bastia, 2004). FOB1 was previously shown to genetically interact with condensin, and consistent with this, it is required for condensin localization to the nontranscribed spacer region 1 (NTS1) in the rDNA (Johzuka et al., 2006; Machin et al., 2006). Fob1 also functions in the recruitment of the RENT complex to NTS1, which mediates both nucleolar silencing and regulates mitotic exit through its role in Cdc14 sequestration (Huang and Moazed, 2003; Stegmeier et al., 2004). In contrast to Fob1, much less is known about the functions of Tof2, Csm1, and Lrs4. Tof2 was initially identified as a topoisomerase I interacting factor and along with Csm1 and Lrs4, which form a complex, has recently been implicated in rDNA silencing (Park and Sternglanz, 1999; Huang et al., 2006). Consistent with this, all three proteins have been localized by ChIP to the NTS1/RFB region of the rDNA in a Fob1-dependent manner and physically associate with RENT complex components (Huang et al., 2006).
Why are Csm1, Lrs4, and Tof2 required for condensation? In the absence of Csm1 and Lrs4 (and also Fob1), the rDNA fails to condense in both metaphase and anaphase cells (Figure 2). The simplest explanation is suggested by the observation that in fob1Δ mutants, condensin fails to specifically localize to the RFB site (Johzuka et al., 2006). Because Fob1 is also required for the chromatin localization of the Csm1/Lrs4 complex (Huang et al., 2006), it seems likely that condensin recruitment to the RFB may also require these factors. Consistent with this, Johzuka and colleagues have demonstrated that Csm1, Lrs4, and Tof2 are required for the specific recruitment of condensin (K. Johzuka, National Institute for Basic Biology, Okazaki, Japan, personal communication). Recruitment to the RFB region can only be a part of the story, however, because condensin is still enriched within the rDNA in the absence of Fob1 or Tof2, and in the latter case rDNA condensation can still occur (this study). These data suggest that either condensin binding at the RFB site is not absolutely required for higher order folding of the rDNA array or that other factors could mitigate this requirement during the cell cycle. For instance, the cohesin complex is also bound within the nontranscribed spacer (Laloraya et al., 2000; Kobayashi and Ganley, 2005) and is required for rDNA condensation in metaphase, though not in anaphase cells (Guacci et al., 1997; Lavoie et al., 2002, 2004). We have previously proposed that the cohesin complex promotes condensation by delimiting domains of condensin function (Lavoie et al., 2004). During early anaphase, however, when cohesin is removed and the rDNA must be further compacted, condensin recruitment to the RFB through TOF2, CSM1, and LRS4 would ensure an even distribution of enzyme within the rDNA, and this is likely to contribute to an ordered compaction process.
Putting the Brake on Cdc14 Release
Genetically, TOF2 interacts with CDC14 and functions both as an inhibitor of condensation during metaphase, as well as an activator during anaphase (this work). This dual role mirrors that of Cdc14 phosphatase, which must be kept inactive during metaphase to prevent the precocious antagonization of CDK phosphorylation, but is activated in early anaphase to promote condensin recruitment and segregation of the rDNA (D'Amours and Amon, 2004; Stegmeier and Amon, 2004). In a recent study, Tof2 was proposed to function as a nucleolar activator of Cdc14 activity (Geil et al., 2008). Consistent with this, the temperature sensitivity of a cdc14-1 mutant is exacerbated in a tof2Δ background (Figure 4). In contrast, however, we show that the deletion of tof2Δ suppresses two MEN mutants, dbf2-2 and mob1-77, suggesting that Cdc14 activity during anaphase is enhanced in the absence of Tof2. These data are most readily be explained by postulating a role for Tof2 as a regulator of Cdc14 release (Figure 6). Cdc14 is released in two waves during a normal anaphase (D'Amours and Amon, 2004; Stegmeier and Amon, 2004). The first partial release from the nucleolus is under the control of the FEAR pathway and promotes rDNA resolution and segregation. The second, complete, release of Cdc14 is under the control of the MEN and promotes the closing of mitosis. Genetically TOF2 functions as an activator of FEAR-dependent Cdc14 release and an inhibitor of the MEN. In response to the activation of the FEAR pathway, an enhanced release of Cdc14 was observed in the absence of Tof2, even when the MEN was inactivated (this work). These data place Tof2 at the juncture between the FEAR and the MEN pathways and suggest that Tof2 promotes the biphasic release of Cdc14 by tempering the FEAR in early anaphase.
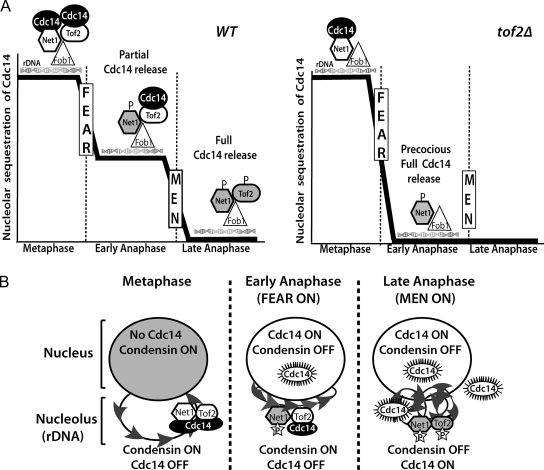
Figure 6.
Models for the biphasic release of Cdc14 and its role in rDNA condensation. (A) In WT cells (left), Cdc14 is restricted to the ribosomal DNA through interactions with its inhibitor, Net1, as well as the Cdc14-binding protein Tof2. Both proteins coimmunoprecipitate with Fob1, the RFB-binding protein. In response to FEAR network activation in early anaphase, Net1 phosphorylation promotes a partial release of Cdc14 into the nucleus. Cdc14 remaining in the nucleolus is dependent on Tof2, which may function by either directly restraining a population of Cdc14 within the nucleolus or alternatively, could inhibit Cdc14 release from Net1 in a subpopulation of RENT complexes. In late anaphase, the MEN promotes the full release of Cdc14, possibly through phosphorylation of Tof2. In the absence of Tof2 (right), FEAR network activation promotes the full release of Cdc14, independently of the MEN. Note that for simplicity, only a subset of Net1- and Tof2-interacting factors are included in the schematic. (B) Model for the spatiotemporal regulation of condensation during mitosis. In metaphase, Cdc14 activity is inhibited throughout the nucleus and nucleolus by sequestration and binding to its inhibitor Net1. In early anaphase, Net1 phosphorylation in the nucleolus causes the partial release of active Cdc14 into the nucleus, which promotes condensin enrichment to the nucleolus and antagonizes its activity in the nucleus. Tof2 bound to the rDNA serves as a second Cdc14 sequestration activity, thereby promoting condensin dependent hypercondensation of the rDNA. In late anaphase, full release of Cdc14 throughout the nucleolus and the cell favors rDNA decondensation and a resetting of the cell cycle to the G1 state.
What consequence might precocious Cdc14 release have for rDNA resolution and condensation? Given that chromosome condensation is tightly controlled during the cell cycle, the nuclear and nucleolar compartments must either be in a permissive or nonpermissive state for chromosome condensation (Figure 6B). In the permissive state, CDK activity is high while Cdc14 activity is low. This would allow the mitotic activation of condensin, a CDK substrate, and permit the establishment of mitotic chromosome structure throughout chromatin observed in metaphase cells. On anaphase onset, however, the FEAR network promotes the partial release of active Cdc14 into the nucleus, thereby distinguishing the nuclear and nucleolar compartments as two functionally distinct environments with respect to condensation potential. In early anaphase, the decondensation of euchromatin occurs in the nucleus (Guacci et al., 1994), presumably through Cdc14 activity and condensin removal from non-rDNA sites. In the nucleolus however, where Tof2 resides, the condensation potential of the rDNA is maintained in response to FEAR activation because Tof2 promotes the rDNA sequestration of Cdc14 to the rDNA. It is noteworthy that condensin is also posttranslationally modified at this time, both by the Ipl1 kinase and through sumoylation, and although the biochemical consequences of these modification have yet to be assessed, it is tempting to speculate that they may contribute to condensin's hypercondensation activity on the rDNA (D'Amours et al., 2004; Lavoie et al., 2004; Wang et al., 2004). By late anaphase, the full release of Cdc14 throughout the cell resets the cell cycle to an interphase state where the rDNA is decondensed.
How Tof2 tempers the early anaphase release of Cdc14 remains to be determined. Because Tof2 copurifies with Cdc14, Net1, Sir2, and Fob1 as well as the Csm1/Lrs4 complex and localizes to the RFB site in a Fob1-dependent manner (Huang et al., 2006), Tof2 could function either as an independent Cdc14 sequestering activity or regulate a subset of the Net1-based RENT complexes, perhaps by inhibiting the phosphorylation of Net1. An independent Cdc14-sequestering complex is consistent with the recent demonstration that purified Tof2 and Cdc14 proteins directly interact (Giel et al., 2008) and would explain the puzzling observation in the literature that the majority of Cdc14 localizes near the RFB site in the nontranscribed spacer 1 region, which does not correspond to the peak of Net1 binding (Huang and Moazed, 2003; Stegmeier et al., 2004). Alternatively, Tof2 could inhibit the release of Cdc14 from a subset of RENT complexes during early anaphase, which is consistent with the relatively low levels of Tof2 versus Net1 protein in cells (about five times less than Net1; Ghaemmaghami et al., 2003). In this model, Cdc14 sequestration to the nucleolus during metaphase would be independent of Tof2, thereby promoting the chromosome condensation observed in tof2Δ cells (this study). In either case, the existence of dual Cdc14 sequestering activities explains how the biphasic release of Cdc14 is effected during mitotic exit.
ACKNOWLEDGMENTS
We are indebted to Katsuki Johzuka and Takashi Horiuchi for communication of unpublished results and to Brenda Andrews (University of Toronto), Charlie Boone (University of Toronto) and members of their labs for strains for SGA, use of the pinning robots, and technical advice on SGA. We thank Mike Tyers for the dbf2 and mob1 mutants, Jack Greenblatt (University of Toronto) for the TOF2-TAP strain, Damien D'Amours for HA-tagged Cdc14 strain, and members of the Lavoie lab for critical reading of the manuscript. This work was funded by Canadian Institutes of Health Research operating (FRN 57913) and salary (MSH 63646) grants to B.D.L.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-08-0879) on October 15, 2008.
REFERENCES
- Azzam R., Chen S. L., Shou W., Mah A. S., Alexandru G., Nasmyth K., Annan R. S., Carr S. A., Deshaies R. J. Phosphorylation by cyclin B-Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus. Science. 2004;305:516–519. doi: 10.1126/science.1099402. [DOI] [PubMed] [Google Scholar]
- Bhalla N., Biggins S., Murray A. W. Mutation of YCS4, a budding yeast condensin subunit, affects mitotic and nonmitotic chromosome behavior. Mol. Biol. Cell. 2002;13:632–645. doi: 10.1091/mbc.01-05-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann C. B., Davies A, Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Caplan A. J., Douglas M. G. Characterization of YDJ1, a yeast homologue of the bacterial dnaJ protein. J. Cell Biol. 1991;114:609–621. doi: 10.1083/jcb.114.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amours D., Amon A. At the interface between signaling and executing anaphase—Cdc14 and the FEAR network. Genes Dev. 2004;18:2581–2595. doi: 10.1101/gad.1247304. [DOI] [PubMed] [Google Scholar]
- D'Amours D., Stegmeier F., Amon A. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell. 2004;117:455–469. doi: 10.1016/s0092-8674(04)00413-1. [DOI] [PubMed] [Google Scholar]
- Elliott S. G., McLaughlin C. S. Regulation of RNA synthesis in yeast. III. Synthesis during the cell cycle. Mol. Gen. Genet. 1979;169:237–243. doi: 10.1007/BF00382269. [DOI] [PubMed] [Google Scholar]
- Freeman L., Aragon-Alcaide L., Strunnikov A. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J. Cell Biol. 2000;149:811–824. doi: 10.1083/jcb.149.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geil C., Schwab M., Seufert W. A nucleolus-localized activator of Cdc14 phosphatase supports rDNA segregation in yeast mitosis. Curr. Biol. 2008;18:1001–1005. doi: 10.1016/j.cub.2008.06.025. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Guacci V., Hogan E., Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J. Cell Biol. 1994;125:517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V., Koshland D., Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Fink G. R. Guide to Yeast Genetics and Molecular Biology. New York: Academic Press; 1991. [Google Scholar]
- Hirano T. Condensins: organizing and segregating the genome. Curr. Biol. 2005;15:R265–R275. doi: 10.1016/j.cub.2005.03.037. [DOI] [PubMed] [Google Scholar]
- Huang J., Brito I. L., Villen J., Gygi S. P., Amon A., Moazed D. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev. 2006;20:2887–2901. doi: 10.1101/gad.1472706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Moazed D. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev. 2003;17:2162–2176. doi: 10.1101/gad.1108403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johzuka K., Horiuchi T. Replication fork block protein, Fob1, acts as an rDNA region specific recombinator in S. cerevisiae. Genes Cells. 2002;7:99–113. doi: 10.1046/j.1356-9597.2001.00508.x. [DOI] [PubMed] [Google Scholar]
- Johzuka K., Horiuchi T. RNA polymerase I transcription obstructs condensin association with 35S rRNA coding regions and can cause contraction of long repeat in Saccharomyces cerevisiae. Genes Cells. 2007;12:759–771. doi: 10.1111/j.1365-2443.2007.01085.x. [DOI] [PubMed] [Google Scholar]
- Johzuka K., Terasawa M., Ogawa H., Ogawa T., Horiuchi T. Condensin loaded onto the replication fork barrier site in the rRNA gene repeats during S phase in a FOB1-dependent fashion to prevent contraction of a long repetitive array in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006;26:2226–2236. doi: 10.1128/MCB.26.6.2226-2236.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashina A. S., Rogers G. C., Scholey J. M. The bimC family of kinesins: essential bipolar mitotic motors driving centrosome separation. Biochim. Biophys. Acta. 1997;1357:257–271. doi: 10.1016/s0167-4889(97)00037-2. [DOI] [PubMed] [Google Scholar]
- Kim M., Krogan N. J., Vasiljeva L., Rando O. J., Nedea E., Greenblatt J. F., Buratowski S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- Kobayashi T. The replication fork barrier site forms a unique structure with Fob1p and inhibits the replication fork. Mol. Cell. Biol. 2003;23:9178–9188. doi: 10.1128/MCB.23.24.9178-9188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Ganley A. R. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science. 2005;309:1581–1584. doi: 10.1126/science.1116102. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Heck D. J., Nomura M., Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 1998;12:3821–3830. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Horiuchi T. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells. 1996;1:465–474. doi: 10.1046/j.1365-2443.1996.d01-256.x. [DOI] [PubMed] [Google Scholar]
- Laloraya S., Guacci V., Koshland D. Chromosomal addresses of the cohesin component Mcd1p. J. Cell Biol. 2000;151:1047–1056. doi: 10.1083/jcb.151.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam W. W., Peterson E. A., Yeung M., Lavoie B. D. Condensin is required for chromosome arm cohesion during mitosis. Genes Dev. 2006;20:2973–2984. doi: 10.1101/gad.1468806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie B. D., Hogan E., Koshland D. In vivo dissection of the chromosome condensation machinery: reversibility of condensation distinguishes contributions of condensin and cohesin. J. Cell Biol. 2002;156:805–815. doi: 10.1083/jcb.200109056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie B. D., Hogan E., Koshland D. In vivo requirements for rDNA chromosome condensation reveal two cell-cycle-regulated pathways for mitotic chromosome folding. Genes Dev. 2004;18:76–87. doi: 10.1101/gad.1150404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie B. D., Tuffo K. M., Oh S., Koshland D., Holm C. Mitotic chromosome condensation requires Brn1p, the yeast homologue of Barren. Mol. Biol. Cell. 2000;11:1293–1304. doi: 10.1091/mbc.11.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machin F., Paschos K., Jarmuz A., Torres-Rosell J., Pade C., Aragon L. Condensin regulates rDNA silencing by modulating nucleolar Sir2p. Curr. Biol. 2004;14:125–130. [PubMed] [Google Scholar]
- Machin F., Torres-Rosell J., De Piccoli G., Carballo J. A., Cha R. S., Jarmuz A., Aragon L. Transcription of ribosomal genes can cause nondisjunction. J. Cell Biol. 2006;173:893–903. doi: 10.1083/jcb.200511129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machin F., Torres-Rosell J., Jarmuz A., Aragon L. Spindle-independent condensation-mediated segregation of yeast ribosomal DNA in late anaphase. J. Cell Biol. 2005;168:209–219. doi: 10.1083/jcb.200408087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C., Ciosk R., Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Mohanty B. K., Bastia D. Binding of the replication terminator protein Fob1p to the Ter sites of yeast causes polar fork arrest. J. Biol. Chem. 2004;279:1932–1941. doi: 10.1074/jbc.M309078200. [DOI] [PubMed] [Google Scholar]
- Nierras C. R., Liebman S. W., Warner J. R. Does Saccharomyces need an organized nucleolus? Chromosoma. 1997;105:444–451. [PubMed] [Google Scholar]
- Ouspenski I. I., Cabello O. A., Brinkley B. R. Chromosome condensation factor Brn1p is required for chromatid separation in mitosis. Mol. Biol. Cell. 2000;11:1305–1313. doi: 10.1091/mbc.11.4.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H., Sternglanz R. Identification and characterization of the genes for two topoisomerase I-interacting proteins from Saccharomyces cerevisiae. Yeast. 1999;15:35–41. doi: 10.1002/(SICI)1097-0061(19990115)15:1<35::AID-YEA340>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Pereira G., Schiebel E. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science. 2003;302:2120–2124. doi: 10.1126/science.1091936. [DOI] [PubMed] [Google Scholar]
- Pereira G., Schiebel E. Cdc14 phosphatase resolves the rDNA segregation delay. Nat. Cell Biol. 2004;6:473–475. doi: 10.1038/ncb0604-473. [DOI] [PubMed] [Google Scholar]
- Petes T. D. Yeast ribosomal DNA genes are located on chromosome XII. Proc. Natl. Acad. Sci. USA. 1979;76:410–414. doi: 10.1073/pnas.76.1.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabitsch K. P., Petronczki M., Javerzat J. P., Genier S., Chwalla B., Schleiffer A., Tanaka T. U., Nasmyth K. Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev. Cell. 2003;4:535–548. doi: 10.1016/s1534-5807(03)00086-8. [DOI] [PubMed] [Google Scholar]
- Rabitsch K. P., et al. A screen for genes required for meiosis and spore formation based on whole-genome expression. Curr. Biol. 2001;11:1001–1009. doi: 10.1016/s0960-9822(01)00274-3. [DOI] [PubMed] [Google Scholar]
- Schuldiner M., et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Shou W., Azzam R., Chen S. L., Huddleston M. J., Baskerville C., Charbonneau H., Annan R. S., Carr S. A., Deshaies R. J. Cdc5 influences phosphorylation of Net1 and disassembly of the RENT complex. BMC Mol. Biol. 2002;3:3. doi: 10.1186/1471-2199-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou W., Seol J. H., Shevchenko A., Baskerville C., Moazed D., Chen Z. W., Jang J., Charbonneau H., Deshaies R. J. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. S., Caputo E., Boeke J. D. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol. Cell. Biol. 1999;19:3184–3197. doi: 10.1128/mcb.19.4.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F., Amon A. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 2004;38:203–232. doi: 10.1146/annurev.genet.38.072902.093051. [DOI] [PubMed] [Google Scholar]
- Stegmeier F., Huang J., Rahal R., Zmolik J., Moazed D., Amon A. The replication fork block protein Fob1 functions as a negative regulator of the FEAR network. Curr. Biol. 2004;14:467–480. doi: 10.1016/j.cub.2004.03.009. [DOI] [PubMed] [Google Scholar]
- Straight A. F., Shou W., Dowd G. J., Turck C. W., Deshaies R. J., Johnson A. D., Moazed D. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell. 1999;97:245–256. doi: 10.1016/s0092-8674(00)80734-5. [DOI] [PubMed] [Google Scholar]
- Strunnikov A. V., Hogan E., Koshland D. SMC2, a 0 gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes Dev. 1995;9:587–599. doi: 10.1101/gad.9.5.587. [DOI] [PubMed] [Google Scholar]
- Sullivan M., Higuchi T., Katis V. L., Uhlmann F. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell. 2004;117:471–482. doi: 10.1016/s0092-8674(04)00415-5. [DOI] [PubMed] [Google Scholar]
- Thomas B. J., Rothstein R. The genetic control of direct-repeat recombination in Saccharomyces: the effect of rad52 and rad1 on mitotic recombination at GAL10, a transcriptionally regulated gene. Genetics. 1989;123:725–738. doi: 10.1093/genetics/123.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomson B. N., D'Amours D., Adamson B. S., Aragon L., Amon A. Ribosomal DNA transcription-dependent processes interfere with chromosome segregation. Mol. Cell. Biol. 2006;26:6239–6247. doi: 10.1128/MCB.00693-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A. H., et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Torres-Rosell J., Machin F., Jarmuz A., Aragon L. Nucleolar segregation lags behind the rest of the genome and requires Cdc14p activation by the FEAR network. Cell Cycle. 2004;3:496–502. [PubMed] [Google Scholar]
- Traverso E. E., Baskerville C., Liu Y., Shou W., James P., Deshaies R. J., Charbonneau H. Characterization of the Net1 cell cycle-dependent regulator of the Cdc14 phosphatase from budding yeast. J. Biol. Chem. 2001;276:21924–21931. doi: 10.1074/jbc.M011689200. [DOI] [PubMed] [Google Scholar]
- Tsang C. K., Li H., Zheng X. S. Nutrient starvation promotes condensin loading to maintain rDNA stability. EMBO J. 2007;26:448–458. doi: 10.1038/sj.emboj.7601488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R., Craig K., Hwang E. S., Prinz S., Tyers M., Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- Visintin R., Hwang E. S., Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus [see comments] Nature. 1999;398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- Visintin R., Stegmeier F., Amon A. The role of the polo kinase Cdc5 in controlling Cdc14 localization. Mol. Biol. Cell. 2003;14:4486–4498. doi: 10.1091/mbc.E03-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B. D., Butylin P., Strunnikov A. Condensin function in mitotic nucleolar segregation is regulated by rDNA transcription. Cell Cycle. 2006;5:2260–2267. doi: 10.4161/cc.5.19.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B. D., Eyre D., Basrai M., Lichten M., Strunnikov A. V. Condensin binding at distinct and specific chromosomal sites in the Saccharomyces cerevisiae genome. Mol. Cell. Biol. 2005;25:7216–7225. doi: 10.1128/MCB.25.16.7216-7225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B. D., Yong-Gonzalez V., Strunnikov A. V. Cdc14p/FEAR pathway controls segregation of nucleolus in S. cerevisiae by facilitating condensin targeting to rDNA chromatin in anaphase. Cell Cycle. 2004;3:960–967. doi: 10.4161/cc.3.7.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Xiang X., Hammer J. A., 3rd. Motor proteins at the microtubule plus-end. Trends Cell Biol. 2006;16:135–143. doi: 10.1016/j.tcb.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Yong-Gonzalez V., Wang B. D., Butylin P., Ouspenski I., Strunnikov A. Condensin function at centromere chromatin facilitates proper kinetochore tension and ensures correct mitotic segregation of sister chromatids. Genes Cells. 2007;12:1075–1090. doi: 10.1111/j.1365-2443.2007.01109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]