Abstract
The translational regulator CPEB1 plays a major role in the control of maternal mRNA in oocytes, as well as of subsynaptic mRNAs in neurons. Although mainly cytoplasmic, we found that CPEB1 protein is continuously shuttling between nucleus and cytoplasm. Its export is controlled by two redundant NES motifs dependent on the nuclear export receptor Crm1. In the nucleus, CPEB1 accumulates in a few foci most often associated with nucleoli. These foci are different from previously identified nuclear bodies. They contain Crm1 and were called Crm1 nucleolar bodies (CNoBs). CNoBs depend on RNA polymerase I activity, indicating a role in ribosome biogenesis. However, although they form in the nucleolus, they never migrate to the nuclear envelope, precluding a role as a mediator for ribosome export. They could rather constitute a platform providing factors for ribosome assembly or export. The behavior of CPEB1 in CNoBs raises the possibility that it is involved in ribosome biogenesis.
INTRODUCTION
Cytoplasmic polyadenylation element–binding protein (CPEB) is one of the best known translational regulators. It has been extensively studied in Xenopus oocytes, as this regulation is essential for oocyte maturation and first embryonic divisions (Richter, 2007). Regulated mRNAs identified in this context encode proteins directly involved in meiosis progression and mitosis, such as cyclins, cdk, histones, Aurora A kinase, and Eg5 kinesin. Maternal mRNAs are synthesized with a long polyA tail (250 Å), then deadenylated in the cytoplasm, and stored in a translationally dormant state. Readenylation in the cytoplasm follows progesterone stimulation and increases translation, allowing meiotic progression. CPEB is specifically bound to the CPE motif (cytoplasmic polyadenylation element) in the 3′ untranslated region of target mRNAs. The domains responsible for RNA binding are contained in its C-terminal moiety, which contains two RNA recognition motifs (RRMs) and a zinc-finger domain. CPEB represses translation by binding to eIF4E-T (Minshall et al., 2007) and maskin (Stebbins-Boaz et al., 1999) in the early and late oocyte, respectively. In turn, these proteins bind and block the translation initiation factor eIF4E on the capped extremity of the mRNA. Stimulation of the oocyte by progesterone triggers CPEB and maskin phosphorylation, which leads to release of eIF4E on the 5′ end and to polyA extension on the 3′ end, both contributing to resumption of translation.
Mammalian genomes contain four CPEB genes, CPEB1 being the closest homolog to Xenopus CPEB. In addition, four protein isoforms are produced from CPEB1 gene, which differ by the length of their N-terminal region (Welk et al., 2001) and by the insertion of five amino acids, GNMPK, in the first RRM, which may affect the RNA-binding properties of the protein (Wilczynska et al., 2005). Beyond its involvement in oocytes, the CPEB1 regulation pathway is also important in the brain. It controls translation of CPE-containing mRNAs in the dendrites, at the level of synapses, in response to neurotransmitters (Richter, 2007). In addition, it has recently been shown to contribute to cell migration in astrocytes (Jones et al., 2008). Interestingly, the isoforms with or without the GNMPK motif are differentially expressed in ovary and brain, raising the possibility that CPEB1 mRNA targets are different in both tissues (Wilczynska et al., 2005). At the cellular level, CPEB1 is enriched in GW bodies (called P-bodies in yeast), which are cytoplasmic ribonucleoproteic granules containing 5′ to 3′ mRNA degradation machinery, as well as translational repressors, such as Rck/p54 and eIF4E-T. This localization may indicate that mRNAs repressed by CPEB1 are then targeted for degradation (Wilczynska et al., 2005). Overall, all studies published so far concerning CPEB1 document its activity on polyadenylation and translation in the cytoplasm.
Although in eukaryotes translation is physically compartmentalized in the cytoplasm, translational regulators are often present in both cytoplasmic and nuclear compartments. For instance, Rck/p54 which is essential for the repression of maternal mRNA in the cytoplasm in combination with CPEB1 (Minshall et al., 2001), is both nuclear and cytoplasmic in early Xenopus oocytes (Smillie and Sommerville, 2002). Such a dual localization is observed for various factors, including CUG-BP/EDEN-BP (Fujimura et al., 2008), TIA1 (Lopez de Silanes et al., 2005; Zhang et al., 2005), TIAR (Mazan-Mamczarz et al., 2006), FMRP, FXR1, and FXR2 (Eberhart et al., 1996; Tamanini et al., 1999). In some cases, for instance CUG-BP/EDEN-BP, TIA1, and TIAR, it reflects a dual function of the protein in both translation and splicing (Philips et al., 1998; Le Guiner et al., 2001). When the nuclear function is unknown, as is the case for Rck/p54, it is postulated that the protein binds the nascent mRNA at the site of transcription, ensuring it exerts its control from the moment they enter the cytoplasm (Smillie and Sommerville, 2002). Here we found that CPEB1 can shuttle between nucleus and cytoplasm and identified the two nuclear export signals (NESs) responsible for its export to the cytoplasm. In the nucleus, CPEB1 accumulates along with the nuclear export receptor Crm1 in new nucleolus-associated bodies. The dynamics of these bodies indicates a role in ribosomal biogenesis and suggests a new function for CPEB1 in ribosome assembly.
MATERIALS AND METHODS
Cell Culture and Transfection
Epithelioid carcinoma HeLa cells were routinely maintained in DMEM, supplemented with 10% fetal calf serum. For Crm1 inhibition, cells were treated with 5 ng/ml LMB (Calbiochem, La Jolla, CA) for 5 h, unless otherwise indicated. Inhibition of both RNA polymerases I and II, and selective inhibition of RNA polymerases I, were achieved by treating the cells with 5 μg/ml and 0.1 μg/ml actinomycin D (Roche Applied Science, Indianapolis, IN) for 2 h, respectively. Control experiments attesting the differential activity of these doses have been published elsewhere (Chen et al., 2005).
Transient transfections were performed with 1.5 μg plasmid DNA per 35-mm-diameter dish by a standard calcium phosphate procedure.
Plasmids and Mutagenesis
Green fluorescent protein (GFP)-tagged and untagged CPEB1, and red fluorescent protein (RFP)-tagged p54 contain the full open reading frames of human CPEB1-lg and Rck/p54, respectively, as described previously (Wilczynska et al., 2005). In CPEB1-C, the CPEB1 sequence starts at an internal BamHI site, so that the first AUG encodes M227. In CPEB1-N, the CPEB1 sequence stops at an internal HpaI site, so that the last CPEB1 amino acid is V328. NES1* (L99L102L104AAA) and NES2* (I202L205I207AAA) point mutations were created in the CPEB1-short expression vector, whereas RRM1* (F314A) and RRM2* (F433A) point mutations were created in the CPEB1-long expression vector, using the QuickChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). GFP-tagged polypyrimidine tract binding protein (PTB) and TIA1 (Zhang et al., 2005) were a kind gift of David Spector (CSH Laboratories, Cold Spring Harbor, NY) and Veronique Kruys (Institute of Biology and Molecular Medicine, Charleroi-Gosselies, Belgium), respectively.
In Situ Fractionation and Nuclear Matrix Isolation
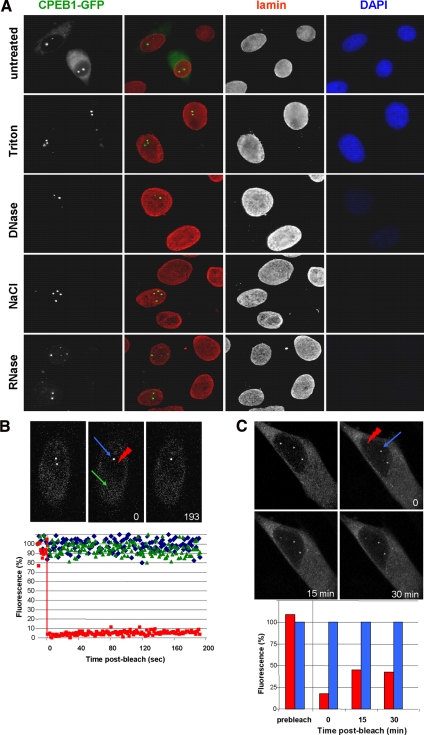
Cells grown on polylysine-coated coverslips were washed three times with ice-cold PBS. Cells were then sequentially extracted using the method described by Buckler-White et al. (1980) slightly modified. In brief, cells were sequentially extracted with 1) 1% Triton X-100 in TMS buffer (50 mM Tris-HCl, pH 7.4, 5 mM MgSO4, and 250 mM sucrose) for 5 min at room temperature; 2) 50 U/ml RNase-free DNase RQ1 (Promega, Madison, WI) in TMS at 37°C for 45 min, followed by addition of ammonium sulfate to a final concentration of 0.25 M; 3) 2 M NaCl in TM buffer (10 mM Tris-HCl, pH 7.4, and 0.02 mM MgSO4) twice for 15 min at room temperature; and 4) 50 μg/ml RNase A in TM for 15 min at room temperature. Monolayers of extracted cells at different steps of the nuclear matrix preparation were fixed with −20°C methanol for 3 min before staining.
Immunofluorescence
For all antibodies except anti-fibrillarin, cells grown on glass coverslips were fixed in methanol at −20°C for 3 min. Cells were rehydrated in phosphate-buffered saline (PBS) and incubated with the primary antibody for 1 h, rinsed with PBS, incubated with the secondary antibody for 30 min, rinsed with PBS, and stained with 0.12 μg/ml DAPI for 1 min, all steps being performed at room temperature. Slides were mounted in Citifluor (Citifluor Labs, Birmingham, United Kingdom). For fibrillarin antibodies, cells were fixed by incubation in PBS with 4% paraformaldehyde for 10 min and permeabilized in PBS with 0.5% Triton X-100 for 10 min.
Mouse monoclonal anti-CPEB1 were produced, as previously described (Wilczynska et al., 2005). Rabbit polyclonal anti-p54 antibody was purchased from Bethyl Laboratories (Montgomery, TX); mouse monoclonal anti-exportin-1/Crm1 from BD Biosciences (San Jose, CA); rabbit polyclonal anti-Crm1, mouse monoclonal anti-lamin A/C(636), and mouse monoclonal anti-Sam68 from Santa Cruz Biotechnology (Santa Cruz, CA); and rabbit polyclonal anti-fibrillarin from Aviva Systems Biology (San Diego, CA). Rabbit polyclonal anti-hDcp1 antibody was a kind gift from Bertrand Séraphin (Centre de Génétique Moléculaire, Gif-sur-Yvette, France), rabbit polyclonal anti-PML from Mounira Chelbi-Alix (Institut André Lwoff, Villejuif, France), and mouse monoclonal anti-coilin and anti-fibrillarin antibodies from Gérard Pierron (Institut André Lwoff, Villejuif, France). Secondary antibodies conjugated to rhodamine and FITC were purchased from Jackson ImmunoResearch Laboratories (Immunotech, Marseille, France).
Microscopy
Standard microscopy was performed on a Leica DMR microscope (Leica, Heidelberg, Germany) using a 63× 1.32 NA oil immersion objective. Photographs were taken using a Micromax CCD camera (Princeton Instrument, Trenton, NJ) driven by Metamorph software (Universal Imaging, West Chester, PA). Confocal images were obtained on a Leica TCS-NT/SP1 inverted confocal laser-scanning microscope (Leica) using an Apochromat 63× 1.32 NA oil immersion objective. Fluorescence signals were acquired in 0.16-μm optical sections using Leica software. A single section is shown in all figures. All images were processed using Adobe Photoshop software (San Jose, CA).
For fluorescence recovery after photobleaching (FRAP) experiments, cells were grown on glass coverslips, mounted in a POC chamber system (Helmut Saur, Reutlingen, Germany) with 2 ml culture medium, and analyzed with the confocal equipment described above. Confocal sections were acquired using an excitation wavelength of 488 nm at 4% of its power, at a rate of one frame per second. Selected nuclear foci were photobleached using excitation wavelength of 488 nm at maximal power. Prebleach, bleach, and postbleach steps were linked and analyzed using Leica software. For fluorescence analysis 15 and 30 min after bleach, a pile of 0.16-μm optical sections was acquired through the whole nucleus. The maximal projection is shown on the figure.
For videomicroscopy, cells were grown on glass coverslips and mounted in a POC chamber system (Helmut Saur) with 2 ml culture medium maintained at 37°C and 5% CO2. Cells were observed on a Zeiss inverted microscope Axiovert (Carl Zeiss SAS, Le Pecq, France) equipped with a DG4 Lambda switcher (Sutter Instrument, Novato, CA) and driven by the Metamorph software (Universal Imaging). Timed series were acquired using a 63× 1.32 NA oil immersion objective.
Western Blotting
Cells were scraped in PBS, resuspended in lysis buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 1 mM EDTA, 2.5 mM EGTA, and 1% Nonidet P-40) supplemented with Complete Protease Inhibitor cocktail (Roche Diagnostics, Meylan, France) and incubated on ice for 30 min. Total soluble proteins were recovered after centrifugation at 15,000 × g and 4°C for 10 min, whereas nuclear and cytoplasmic proteins were separated by centrifugation at 500 × g and 4°C for 10 min. Total and cytoplasmic proteins were quantified by the Coomassie blue protein assay (Pierce, Rockford, IL). Seventy-five micrograms of total and cytoplasmic proteins, and nuclear proteins corresponding to the same number of cells as cytoplasmic ones, were separated on a 8.5% polyacrylamide SDS-PAGE gel and transferred to a PVDF membrane (Perkin Elmer, Villebon-sur-Yvette, France). Nonspecific protein-binding sites were blocked by incubation in PBS-T (PBS, 0.1% Tween-20) containing 5% (wt/vol) nonfat dry milk for 1 h at room temperature. The membrane was then incubated with the primary antibody for 1 h at 37°C. After washing in PBS-T, the blot was incubated with horseradish peroxidase–conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories; Immunotech) for 1 h at room temperature. After washing in PBS-T, immune complexes were detected using the Supersignal West Pico Chemiluminescent Signal kit (Pierce) and visualized by exposure to CL-XPosure film (Pierce).
RESULTS
Presence of CPEB1 Foci in the Nucleus
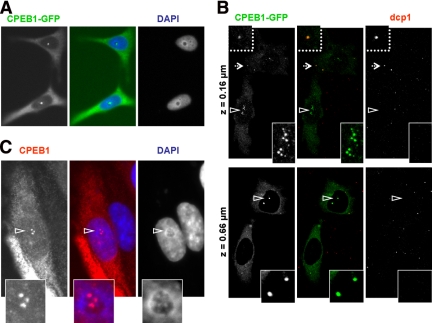
We have previously reported that the CPEB1 protein is diffusely localized in the cytoplasm of mammalian cells and is enriched in GW bodies and stress-induced stress granules. Moreover, expression of green fluorescent protein (GFP)-tagged CPEB1 can induce the assembly of stress granules (Wilczynska et al., 2005). In the course of our studies, we also noted the presence of CPEB1 in the nucleus of transfected cells, as illustrated in Figure 1. HeLa cells were transfected with an expression vector for CPEB1-GFP and observed 24 h later after DAPI staining of the nuclei. In addition to cytoplasmic fluorescence, half of the cells harbored fluorescent foci in the nucleus (Figure 1A). To confirm that these foci are not GW bodies located under the nucleus, GW bodies were immunostained with anti-dcp1 antibody, and cells were observed by confocal microscopy (Figure 1B). Cytoplasmic foci contained Dcp1, as expected for GW bodies, whereas foci clearly within the nucleus did not. Importantly, this nuclear localization was not restricted to cells expressing high levels of CPEB1. Some cells harbored CPEB1 nuclear foci, whereas the protein was hardly visible in the cytoplasm, suggesting that it is a primary site of accumulation for CPEB1. Because of the low level of endogeneous CPEB1, we could not detect unambiguously such foci in untransfected cells. However, they were present in cells transfected with untagged CPEB1, as detected using anti-CPEB1 antibody (Figure 1C), indicating that the GFP moiety is not involved in this localization. Generally, the number of nuclear foci was less than six. Strikingly, most of them appeared closely associated with nucleoli, as identified upon DAPI staining of the DNA (Figures 1, A and C).
Figure 1.
Presence of CPEB1 foci in the nucleus. (A) CPEB1-GFP accumulation in nuclear foci. HeLa cells were transfected with an expression vector for GFP-tagged CPEB1. After 24 h, cells were fixed, stained with DAPI, and observed by fluorescence microscopy. (B) Intranuclear localization of CPEB1 foci. Cells transfected as in A were stained with anti-dcp1 antibody and observed by confocal microscopy. Two focal planes are shown, at the base (top panel) and the middle (bottom panel) of the cell. The dotted arrow points to a GW body that has been enlarged above. Arrowheads point to CPEB1 nuclear foci that have been enlarged (below). (C) Untagged CPEB1 accumulation in nuclear foci. HeLa cells were transfected with an expression vector for CPEB1, stained with anti-CPEB1 antibody and DAPI, and observed by fluorescence microscopy. The CPEB1 nuclear foci indicated by the arrowhead are enlarged below.
Nucleocytoplasmic Transport Signals in CPEB1 Protein
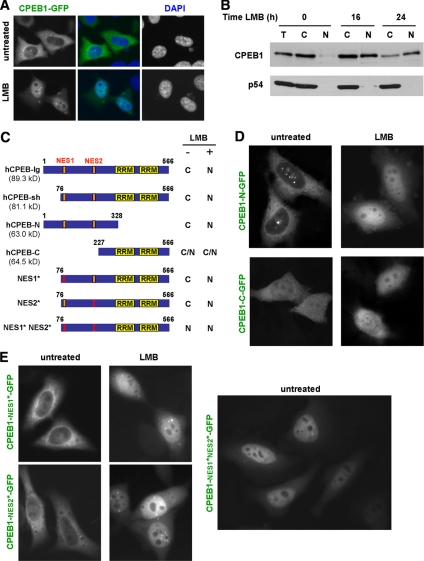
The observation of CPEB1 in the nucleus raised the question of its transport between nucleus and cytoplasm. Although we could not identify any obvious consensus nuclear localization signal (NLS) in the sequence, two candidate CRM1-dependent NESs were present at positions 95–104 (LCLGLQSLSL) and 198–207 (LSDLISSLRI; with respect to protein NP_085097). We therefore investigated the effect of leptomycin B (LMB), which is a specific inhibitor of this export pathway. HeLa cells were transfected with CPEB1-GFP and, 24 h later, treated with 5 ng/ml LMB for 5 h. In most cells, CPEB1 relocated to the nucleus (Figure 2A), demonstrating that CPEB1 is continuously exported to the cytoplasm via the CRM1-dependent pathway. The protein spread diffusely over the nucleoplasm, as well as accumulated in a few nuclear granules, and was excluded from nucleoli. As pointed out above, endogeneous CPEB1 expression is too low for immunofluorescence studies. We therefore investigated its nucleocytoplasmic partition by cell fractionation and Western blot analysis. Untransfected HeLa cells were treated with LMB for 16 and 24 h. Cytoplasmic and nuclear fractions were then separated and proteins were analyzed by Western blot with anti-CPEB1 antibody. Although CPEB1 was nearly undetectable in the nuclear fraction of untreated cells, it accumulated in the nucleus after LMB treatment (Figure 2B), confirming that the endogeneous protein enters the nucleus, and is exported via the CRM1 machinery. As a control, we analyzed the Rck/p54 DEAD box helicase, which belongs to the same cytoplasmic ribonucleoprotein complex as CPEB1 in oocytes (Minshall et al., 2007). In concordance with previous reports (Smillie and Sommerville, 2002), Rck/p54 was cytoplasmic and not sensitive to LMB treatment.
Figure 2.
Crm1-dependent nuclear export of CPEB1. (A) LMB-sensitive export of CPEB1-GFP. HeLa cells were transfected with GFP-tagged CPEB1 and treated 24 h later with LMB for 5 h. Cells were fixed, stained with DAPI, and observed by fluorescence microscopy. (B) LMB-sensitive export of endogenous CPEB1. Untransfected HeLa cells were treated with LMB for the indicated times. Total (T), cytoplasmic (C), and nuclear (N) protein extracts were analyzed by Western blot using anti-CPEB1 and anti-p54 antibodies. (C) LMB-sensitivity of CPEB1 mutants. The fragment of CPEB1 cloned upstream of GFP is schematically represented, with RRMs boxed in yellow. For each construct, the numbers refer to the first and last amino acid with respect to CPEB1 long isoform, and the molecular weight of the GFP fusion is given in parentheses. Analysis was performed as in A. C and N, cytoplasmic and nuclear localization, respectively. (D) LMB sensitivity of CPEB1-N and -C. Analysis was performed as in A. (E) Point mutations of CPEB1 NES. Point mutations were introduced into NES1 (NES1*) or NES2 (NES2*) of the GFP-fused CPEB1-sh construct. Sensitivity to LMB was tested as in A (top panel). The double mutant (NES1*-NES2*) was analyzed in the absence of LMB (bottom panel).
To delineate import and export sequences, we then analyzed the localization and LMB sensitivity of truncated CPEB1 proteins fused to GFP (Figure 2C). The short isoform of CPEB1 protein behaved like the long isoform, being mostly cytoplasmic in untreated cells and accumulating in the nucleus after LMB treatment (data not shown). CPEB1-N, where the C-terminal region is truncated including the two RRMs and the zinc finger, gave similar results (Figure 2D). However, CPEB1-C, where the N-terminal region is deleted, was both cytoplasmic and nuclear in untreated cells and was insensitive to LMB treatment (Figure 2D). Altogether, the four proteins were competent for nuclear import, whether in the presence (CPEB1-lg, CPEB1-sh, and CPEB1-N) or in the absence (CPEB1-C) of LMB treatment. This requires an active import process, as the size of all GFP-fused proteins is >63 kDa, which precludes passive entry into the nucleus. If the region involved in import is unique, then it must be located between aa 227 and 328, because this is the sequence common to all tested CPEB1 constructs. As to the export, all constructs except CPEB1-C were sensitive to LMB, indicating that the region involved in export is located between aa 76 and 227. As it contains the two candidate NES, we assayed their activity by introducing mutations in each of them separately. Three leucine or isoleucine residues were replaced by alanine in either NES95–104 (LCLGLQSLSL) or NES198–207 (LSDLISSLRI). Both mutants were mostly cytoplasmic in untreated cells and were sensitive to LMB treatment (Figure 2E). However, when both mutations were combined, the double mutant protein accumulated in the nucleus in the absence of LMB. Therefore, both NES95–104 and NES198–207 are functional separately, and CPEB1 is exported by the CRM1-dependent machinery under the exclusive control of these two NESs.
CPEB1 Nuclear Foci Do Not Correspond to Known Nuclear Bodies
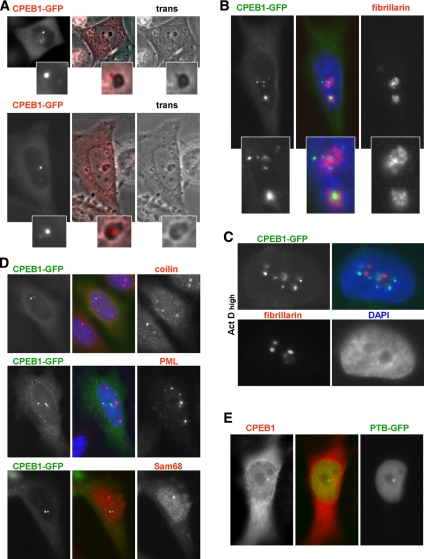
To obtain insight into the nature of CPEB1 nuclear foci, we analyzed their localization with respect to known nuclear structures. As mentioned above, CPEB1 nuclear foci were most often associated with nucleoli. HeLa cells were transfected with CPEB1-GFP and analyzed 24 h later by microscopy. In live cells, nucleoli appear as darker area of the nucleus, and CPEB1 foci were located either at their periphery (Figure 3A, top; note that in the figure CPEB1-GFP is colored in red for better visualization) or in their center (Figure 3A, bottom). Transfected cells were then fixed and immunostained with monoclonal anti-fibrillarin antibody, which identify the fibrillar component of the nucleolus. CPEB1 foci were close to, but did not overlap with fibrillarin staining (Figure 3B). Nucleoli also contain a granular component and fibrillar centers that are intermingled with the fibrillar component. These three components segregate into distinct compartments after treatment with the transcriptional inhibitor actinomycin D (Puvion-Dutilleul et al., 1992). HeLa cells transfected with CPEB1-GFP were therefore treated with 5 μg/ml actinomycin D for 2 h. As expected, the fibrillarin became more compact (Figure 3C). In the same time, CPEB1 foci dispersed into more numerous and smaller foci, which became distant from the nucleolus. This indicated that they correspond neither to the granular component, which is adjacent to the fibrillar compartment in these conditions, nor to fibrillar centers, which coalesce into a single compartment (Puvion-Dutilleul et al., 1992).
Figure 3.
Relationship of CPEB1 nuclear foci with other nuclear granules. (A) Relationship with nucleoli in live cells. HeLa cells were transfected with GFP-tagged CPEB1 and observed 24 h later by fluorescence and phase-contrast (trans) microscopy. Note that CPEB1-GFP is colored in red for better visualization. Top and bottom panels show CPEB1 foci at the periphery and inside the nucleolus, respectively. (B) Relationship with nucleoli in fixed cells. HeLa cells transfected as in A were fixed, stained with anti-fibrillarin antibody and DAPI, and observed by fluorescence microscopy. (C) Relationship with nucleoli in actinomycin D–treated cells. Transfected cells were treated with 5 μg/ml actinomycin D for 2 h, fixed, and stained as in B. (D) Relationship with Cajal, PML, and Sam68 bodies. Transfected cells were immunostained with the indicated antibodies. (E) Relationship with PNC. Cells were cotransfected with GFP-tagged PTB and untagged CPEB1, fixed, and immunostained with anti-CPEB1 antibody.
We next investigated whether CPEB1 foci were related to other known nuclear bodies, such as Cajal bodies, PML bodies, Sam68 bodies, or the perinucleolar compartment (PNC). HeLa cells transfected with CPEB1-GFP were analyzed for the three first structures by immunofluorescence using anti-coilin, anti-PML, and anti-Sam68 antibodies, respectively (Figure 3D). In parallel, cells were cotransfected with untagged CPEB1 and PTB-GFP, as a marker of the PNC (Huang et al., 1997), and CPEB1 foci were localized using anti-CPEB1 antibody (Figure 3E). No colocalization with any of these markers was observed. In conclusion, CPEB1 foci do not overlap with known nuclear compartments. In addition, their localization and appearance is sensitive to the transcriptional activity of the cell.
The Nuclear and Cytoplasmic CPEB1 Complexes Differ in Composition
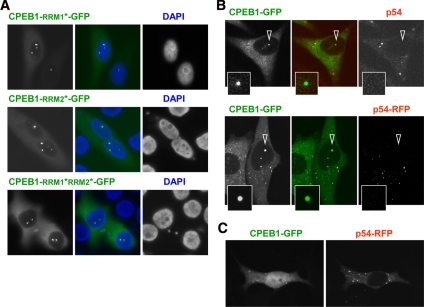
Translational regulation by CPEB1 in the cytoplasm involves its binding to CPE motifs in the 3′UTR of target mRNA. We therefore investigated whether the accumulation of CPEB1 in nuclear foci depends on its mRNA-binding capacity. We have previously shown the existence of an alternative CPEB1 isoform that contains a GNMPK insert in the first RNA recognition motif (RRM1), which is likely to affect its RNA-binding specificity or affinity (Wilczynska et al., 2005). Both CPEB1 isoforms, with and without GNMPK, were similarly present in nuclear foci, indicating no impact of this motif on this localization (data not shown). Next, we analyzed the localization of an RNA-binding deficient mutant of CPEB1. A Phe-to-Ala mutation was introduced in the rnp2 motif of RRM1 and RRM2, separately and in combination (Jessen et al., 1991; Wilczynska et al., 2005). Nuclear CPEB1 foci were observed both with the single and double mutants (Figure 4A). Accordingly, the complete truncation of the two RRMs and zinc finger did not prevent accumulation of CPEB1 in nuclear foci (Figure 2D). Therefore, this localization does not depend on the RNA-binding capacity of CPEB1.
Figure 4.
Difference between CPEB1 nuclear foci and CPEB1 cytoplasmic complex. (A) Nuclear localization of CPEB1 is independent of mRNA binding. Point mutations were introduced into RRM1 (RRM1*), RRM2 (RRM2*), or both RRMs (RRM1*-RRM2*) of the GFP-fused CPEB1-lg construct. Cells were transfected and observed 24 h later by fluorescence microscopy. (B) Absence of Rck/p54 in nuclear CPEB1 foci. In the top panel, cells were transfected with GFP-tagged CPEB1 for 24 h and immunostained with anti-p54 antibody. In the bottom panel, cells were cotransfected with GFP-tagged CPEB1 and RFP-tagged Rck/p54. The CPEB1 nuclear foci indicated by arrowheads are enlarged below. (C) No coimport of Rck/p54 with CPEB1. Cells cotransfected with GFP-tagged CPEB1 and RFP-tagged Rck/p54 were treated with LMB for 5 h and observed by fluorescence microscopy.
Beside mRNA, the cytoplasmic CPEB1 complex contains a set of proteins, including the DEAD-box helicase Rck/p54, which is required for mRNA repression (Minshall et al., 2007). To assess its presence in nuclear CPEB1 foci, cells were transfected with CPEB1-GFP and 24 h later, were immunostained with anti-p54 antibody. Endogeneous Rck/p54 was readily detected in GW bodies, but not in CPEB1 nuclear foci (Figure 4B, top). Even overexpression of p54-RFP did not lead to any accumulation in these foci (Figure 4B, bottom). Furthermore, accumulation of CPEB1 in the nucleus upon LMB treatment did not lead to the concomitant displacement of Rck/p54, whether endogeneous (Figure 2B) or overexpressed (Figure 4C). These data indicate that CPEB1 traffics to the nucleus independently of Rck/p54. Similarly, the Sm-like protein Lsm14/RAP55, which also belongs to the cytoplasmic CPEB1 complex, and the translation initiation factor eIF4E, which is bound to CPEB1 via eIF4E-T, were absent from nuclear foci (data not shown). In conclusion, partners of CPEB1 in the nuclear foci appear different from the well-characterized CPEB1 cytoplasmic complex, in terms of both mRNA and protein components.
CPEB1 Foci Are Stably Associated with the Nuclear Matrix
The fact that CPEB1 accumulates in nuclear foci irrespectively of its mRNA-binding capacity raised the possibility that it is associated with non-RNA components of the nucleus. We addressed this issue by analyzing CPEB1 foci during nuclear matrix preparation. Monolayers of HeLa cells transfected with CPEB1-GFP were submitted to in situ sequential extraction with Triton X-100, DNase I, 2 M NaCl, and RNase A before fixation. The fractionation procedure was controlled by immunostaining with antibody directed against lamin A/C, two nuclear matrix–associated proteins, and DNA staining with DAPI (Figure 5A). The efficiency of chromatin digestion was attested by the disappearance of the DAPI staining, whereas the integrity of the nuclear matrix was verified by lamin A/C detection. Although Triton solubilized CPEB1 protein in the cytoplasm, CPEB1 nuclear foci, like lamin, resisted all extractions and remained as numerous and bright in the final nuclear matrix as they were in intact cells. This indicated that CPEB1 foci are associated with the nuclear matrix and not to chromatin or nascent mRNA.
Figure 5.
Stable association of CPEB1 nuclear foci with the nuclear matrix. (A) Association with the nuclear matrix. Cells transfected with GFP-tagged CPEB1 were treated successively as indicated. At each step, a slide was fixed and stained with anti-lamin antibody and DAPI. Images were acquired using the same exposure time at each step. (B) Photobleaching of CPEB1 in CPEB1 nuclear foci. Cells were transfected as in A. After 24 h, a single CPEB1 focus was photobleached, and the fluorescence recovery was recorded over 193 s using a confocal microscope. In the top panel, images at indicated times were selected for illustration, with the red arrowhead pointing to the photobleached focus, the blue arrow to an unbleached nuclear focus, and the green arrow to an area of the cytoplasm, within the same cell. In the bottom panel, the fluorescence associated to the bleached (red) and unbleached (blue) nuclear foci, and to the cytoplasm (green), were quantified and plotted as a function of time. (C) Fluorescence recovery over 30 min. Photobleaching was performed as in B, and an image stack through the whole nucleus was acquired at 15 and 30 min. A maximal projection of all focal planes is presented for each time point (top panel). The fluorescence associated to the bleached (red) and unbleached (blue) foci was quantified and plotted as a function of time (bottom panel).
We next investigated the traffic of CPEB1 between these foci and the nucleoplasm by FRAP. A single focus was photobleached and fluorescence was monitored over time (Figure 5B). Strikingly, no significant recovery in the bleached foci was observed within 193 s, despite high levels of fluorescence in neighboring nuclear foci and in cytoplasm. We repeated the photobleaching experiment and analyzed fluorescence 15 and 30 min after the bleach (Figure 5C). Only 30% of the fluorescence was recovered in the foci 15 min after bleach, and no further recovery was observed in the subsequent 15 min. By comparison, photobleaching of the same CPEB1-GFP protein in cytoplasmic stress granules, which are induced in some cells of the same culture, led to full fluorescence recovery with a half-life of ∼30 s (Mollet et al., 2008). Altogether, these data show that CPEB1 in nuclear foci is tightly, and stably, associated with the nuclear matrix.
VI CPEB1 Nuclear Foci Contain Crm1
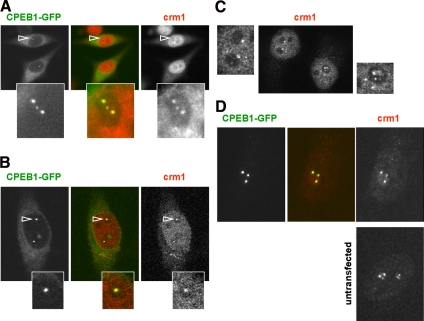
Intriguingly, mutation of either NES1 of NES2 abolished accumulation of CPEB1 in nuclear foci (Figure 2E). The mutant proteins could nevertheless enter the nucleus, as attested by their accumulation in the nucleoplasm after LMB treatment. Therefore, the accumulation in specific foci was possible only in the presence of two functional NESs. As Crm1 is the only protein known to bind the NES, we investigated whether it is present in CPEB1 foci. HeLa cells transfected with CPEB1-GFP were fixed and stained with anti-Crm1 antibodies. Although both a monoclonal and a polyclonal anti-Crm1 antibody gave similar results, only results obtained with the monoclonal are shown. Crm1 was detected in all CPEB1 nuclear foci (Figure 6A). This colocalization was confirmed by confocal microscopy (Figure 6B). However, Crm1 also accumulated in bodies distant from nucleoli, which did not contain CPEB1 (data not shown). Importantly, a similar number of nucleoli-associated Crm1 bodies were detected in untransfected HeLa cells (Figure 6C), indicating that these foci are not assembled due to CPEB-GFP overexpression. They were called CNoBs for Crm1 Nucleolar Bodies.
Figure 6.
Evidence for CNoBs. (A and B) Presence of Crm1 in nuclear CPEB1 foci. Cells transfected with GFP-tagged CPEB1 for 24 h were immunostained with anti-Crm1 antibody and observed by fluorescence microscopy (A) and confocal microscopy (B). The CPEB1 nuclear foci indicated by arrowheads are enlarged below. (C) Presence of CNoBs in untransfected cells. Untransfected HeLa cells were immunostained with anti-Crm1 antibody and observed by confocal microscopy. CNoBs present in each nucleus were enlarged on each side. Note the additional presence of nonnucleolar Crm1 bodies on the right image. (D) Association of CNoBs with the nuclear matrix. Cells transfected with GFP-tagged CPEB1 (top panel) or untransfected (bottom panel) were treated as in Figure 5A and stained with anti-Crm1 antibody. One nucleus at the final nuclear matrix stage is shown for each. Cells shown in B and C were taken from the same experiment, before treatment.
We next investigated if the Crm1 protein present in CNoBs was associated with the nuclear matrix, like CPEB1. Cells transfected with CPEB1-GFP were submitted to in situ sequential extraction as described above and immunostained with anti-Crm1 antibody (Figure 6D). Diffuse nucleoplasmic Crm1 was mostly extracted by these treatments, whereas Crm1 in CNoBs was not. This was true in cells expressing CPEB1-GFP (top panel) as well as in untransfected cells (bottom panel). Therefore Crm1 in these bodies is tightly associated with the nuclear matrix.
CNoBs Depend on RNA Polymerase I Transcription
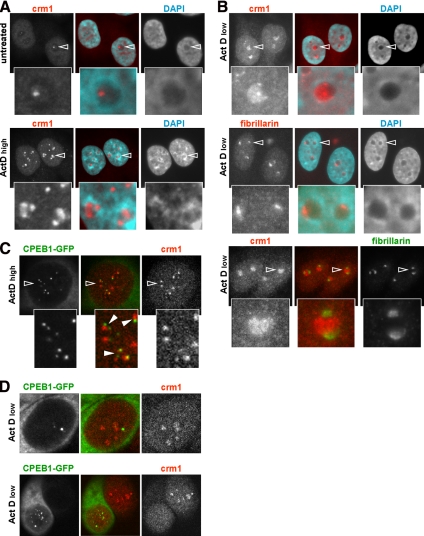
Because CPEB1 foci dispersed upon inhibition of RNA synthesis by actinomycin D (Figure 3C), we analyzed the behavior of Crm1 in these conditions. Untransfected HeLa cells were treated with 5 μg/ml actinomycin D for 2 h and analyzed with monoclonal anti-Crm1 antibody. In control culture, half of the cells harbored CNoBs (Figure 7A, top panel). After actinomycin D treatment, intense DAPI staining around nucleoli indicated that dense chromatin was excluded from nucleoli, as previously reported (Puvion-Dutilleul et al., 1992). Crm1 accumulated in the nucleoli of most cells, forming two to three patches at the periphery of each nucleolus (Figure 7A, bottom panel). In some cells, Crm1 also accumulated in additional small foci distant from nucleoli. At the dose of 5 μg/ml, actinomycin D inhibits both RNA polymerase I and II. To determine which polymerase is important for CNoBs, the experiment was repeated using 0.1 μg/ml actinomycin D, which specifically inhibits RNA polymerase I (Chen et al., 2005). Crm1 localization was strongly disrupted with respect to untreated cells. It filled nucleoli, as judged by DAPI staining, generally with the exception of a small crescent (Figure 7B, top panel). As a control, cells were also stained with monoclonal anti-fibrillarin antibody. At this low dose of actinomycin D, fibrillarin was concentrated in a crescent shape region, potentially corresponding to the region where Crm1 was absent (Figure 7B, middle panel). This was proved by immunostaining with anti-CPEB1 antibody used in combination with a polyclonal anti-fibrillarin antibody (Figure 7B, bottom panel). In conclusion, CNoBs are sensitive to actinomycin D, and Crm1 relocalizes to the nonfibrillar compartment of the nucleolus after selective inhibition of RNA polymerase I.
Figure 7.
Sensitivity of CNoBs to RNA polymerase I inhibition. (A) Sensitivity to high dose of actinomycin D. Untransfected cells were treated with 5 μg/ml actinomycin D for 2 h, fixed, and stained with anti-Crm1 antibody and DAPI. Cells were observed by fluorescence microscopy. (B) Sensitivity to a low dose of actinomycin D. Untransfected cells were treated with 0.1 μg/ml actinomycin D for 2 h, fixed, and stained with DAPI and either anti-Crm1 (top panel), anti-fibrillarin (middle panel), or a combination of both (bottom panel) antibodies. Cells were observed by fluorescence microscopy. The CNoBs indicated by arrowheads are enlarged below. (C) Dissociation of CPEB1 and Crm1 at high dose of actinomycin D. Cells transfected with CPEB1-GFP were treated with 5 μg/ml actinomycin D for 2 h, fixed, and stained with anti-Crm1 antibody and DAPI. Cells were observed by confocal microscopy. The area indicated by an open arrowhead is enlarged below. The filled arrowheads point to CPEB1 foci that remain in contact with Crm1 foci. (D) Dissociation of CPEB1 and Crm1 at low dose of actinomycin D. Cells were stained and observed as in C. Two cells were chosen for illustration, showing dissociation of CPEB1 foci and Crm1 (top panel), and absence of Crm1 in nucleoli of transfected cells, when compared with neighboring untransfected cells (bottom panel).
We next studied the respective localization of CPEB1 and Crm1 after actinomycin D treatment. HeLa cells transfected with CPEB1-GFP were treated 2 h with high and low dose of actinomycin D and stained with anti-Crm1 and anti-fibrillarin antibodies. CPEB1 clearly dissociated from Crm1 at 5 μg/ml actinomycin D (Figure 7C). Nevertheless, CPEB1 foci remained frequently in contact with Crm1 patches, as illustrated on the enlarged area. Similarly, CPEB1 dissociated from Crm1 at 0.1 μg/ml actinomycin D (Figure 7D, top panel). Therefore, CPEB1 colocalization with Crm1 is sensitive to RNA polymerase I activity. We also noted that Crm1 accumulated less in the nucleoli of cells expressing CPEB1-GFP than in untransfected cells (Figure 7D, bottom panel; compare two adjacent cells), even though fibrillarin condensed as in untransfected cells (data not shown), indicating that overexpression of CPEB1-GFP did not alter cellular sensitivity to actinomycin D. To investigate if this effect is secondary to general translational repression by overexpressed CPEB1-GFP in the cytoplasm, we repeated the experiment with another translational inhibitor fused to GFP, TIA1 (Lopez de Silanes et al., 2005). TIA1-GFP did not disturb Crm1 relocalization after actinomycin D treatment (data not shown), suggesting that CPEB1 expression specifically interferes with Crm1 nucleolar accumulation once RNA polymerase I transcription is inhibited.
Assembly and Mobility of CNoBs
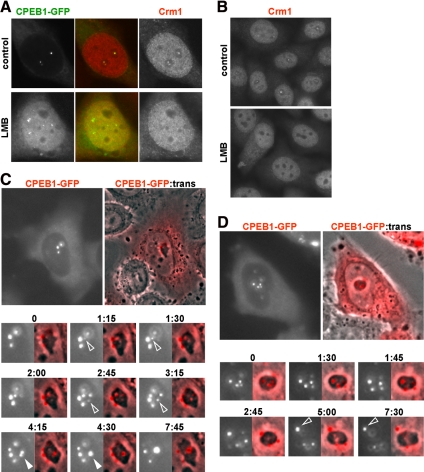
We have demonstrated that LMB treatment leads to CPEB1 accumulation in the nucleus. As mentioned above, CPEB1 was excluded from nucleoli, but still was enriched in small nucleoplasmic granules (Figure 2A). These granules did not derive from preexisting CNoBs. Indeed, after LMB addition, they appeared at a time when CPEB1 was still present in CNoBs, as observed by videomicroscopy (data not shown). Given its colocalization in untreated cells, we investigated if Crm1 was delocalized along with CPEB1. HeLa cells transfected with CPEB1-GFP were treated with LMB for 5 h and immunostained with anti-Crm1 antibody (Figure 8A). No Crm1 accumulation was observed in these CPEB1-containing aggregates, confirming the dissociation between CPEB1 and Crm1. Surprisingly, all Crm1 nuclear bodies, whether CNoBs or bodies distant from nucleoli, fully disappeared after LMB treatment. This was confirmed in untransfected cells (Figure 8B). As Crm1 was still present in the nucleoplasm, this suggests that CNoB maintenance requires the ability of Crm1 to bind NES motif.
Figure 8.
Dynamics of CNoBs. (A) Dissociation of CPEB1 foci by LMB. Cells transfected with CPEB1-GFP were treated or not with LMB for 5 h, fixed, and stained with anti-Crm1 antibody and DAPI. Imaging was performed by confocal microscopy. (B) Dissociation of CNoBs by LMB. Untransfected cells were treated or not with LMB for 5 h, fixed, and immunostained with anti-Crm1 antibody. Imaging was performed by fluorescence microscopy. (C and D) Formation of CNoBs in the nucleolus and migration to its periphery. HeLa cells were transfected with expression vectors for GFP-tagged CPEB1 (colored in red for better visualization). After 24 h, cells with nuclear foci were observed live by fluorescence and phase contrast (trans) videomicroscopy during a 8-h time lapse. Two cells have been chosen for illustration. Only the main nucleoli at indicated times are shown. In C, the successive assembly of two CNoBs and the fusion between two CNoBs are indicated by open and filled arrowheads, respectively. In D, Dissociation of a CNoB from the nucleolus is indicated by an open arrowhead.
To study the dynamics of CNoBs, we took advantage of the fact that CPEB1 is systematically colocalized with CNoBs and used it as a marker. HeLa cells were transfected with CPEB1-GFP and observed by videomicroscopy for up to 8 h (Figure 8, C and D; note that CPEB1-GFP was colored in red for better visualization). Overall, foci were rather stable over such a long period of time. Nevertheless, we could track their movements and observe changes within them. They formed in the center of nucleolus and slowly moved to the periphery. This is clearly visible in Figure 8C, where two CNoBs appear successively at time 1:15 (min:s) and 2:45 (open arrowheads) and increase in size in the next 45 min. The former then moves to the nucleolar periphery between time 2:45 and 3:15. This slow movement from interior to exterior is a general phenomenon. For instance, in the nucleus illustrated in Figure 8D, one CNoB is present at the periphery and three inside nucleolus at time 0. Within the after 45 min, one of the interior CNoBs migrated to the periphery, whereas the two others relocated between time 1:45 and 2:45. No movement from outside to inside was observed. Two CNoBs would occasionally fuse, as seen on Figure 8C, between time 4:15 and 4:30 (filled arrowhead). Finally, CNoBs sometimes detached from nucleoli, as visible on Figure 8D, between time 5:00 and 7:30 (open arrowhead), but they never reached the nuclear envelope. In conclusion, CNoBs assemble in the nucleolus, migrate to its periphery, and do not traffic further. Moreover, their maintenance is sensitive to LMB.
DISCUSSION
CPEB1 Nucleocytoplasmic Traffic
Although all reports hitherto have described CPEB1 as a cytoplasmic protein, here we report that human CPEB1 is in fact continuously shuttling between the nucleus and the cytoplasm. No consensus NLS could be detected in the sequence, but the localization of truncated versions of CPEB1 suggests that the region responsible for nuclear import is localized between aa 227 and 328. It may contain a nonconsensus import sequence or enable binding to a protein that contains an import sequence. In contrast, two consensus NES motifs are present at positions 95–104 and 198–207. We have shown that both are functional separately, as CPEB1 containing a point mutation in either one was competent for export, in a Crm1-dependent manner. Moreover, these NESs are the only sequences responsible for cytoplasmic export of the protein, as CPEB1 localization becomes nuclear when both NESs are mutated. This feature is likely to be conserved through evolution. Indeed, both NES95–104 (LCLGLQSLSL) and NES198–207 (LSDLISSLRI) are perfectly conserved in amphibians (Xenopus NP_001084072) and birds (Gallus XP_413713). In fish (zebrafish AAH45467), NES198–207 is identical, whereas essential leucines are conserved in NES95–104 (WGLGLQSLSL; la Cour et al., 2003). Though it is not an isolated case, one can wonder why a protein should contain two redundant NESs. A simple explanation would be that this enables a more efficient export of the protein or that this allows export when one of the NESs is masked upon interaction with proteins or RNA.
In the cytoplasm, CPEB1 is specifically associated with CPE-containing mRNA, within a complex of proteins that includes Rck/p54, which is required for translational repression. CPEB1 traffic to the nucleus does not involve a complex of the same composition. Indeed, during LMB treatment, Rck/p54 did not accumulate with overexpressed CPEB1 in the nucleoplasm, as would be expected if it were associated with it. Moreover, the C-terminal moiety of CPEB1, which is responsible for CPE binding, was dispensable for both import and export, indicating that traffic is not driven by CPE-containing mRNA.
CPEB1 Foci Colocalize with Crm1
Once in the nucleus, CPEB1 concentrates in a few foci. Several types of nuclear bodies of similar size have been described, including PNC, Sam68 bodies, Cajal bodies, and PML bodies, most of them being better defined in terms of components than function (reviewed in Zimber et al., 2004). However, CPEB1 foci did not colocalize with markers of any of these bodies. Strikingly, CPEB1 foci were most often associated to nucleoli, like PNC and Sam68 bodies. The fact that point mutations of CPEB1 NES resulted in disappearance of these foci led us to investigate their relationship with the only known NES ligand, the nuclear export receptor Crm1. Indeed, we found that CPEB1 foci contained Crm1. Importantly, similar Crm1-containing bodies associated with nucleoli (CNoBs) were present in untransfected cells. In some cells, beside this localization, Crm1 was also enriched in bodies distant from nucleoli, in agreement with a previous report that Crm1 is a component of Cajal bodies (Boulon et al., 2004).
The accumulation of CPEB1 in CNoBs cannot be simply explained by the redundant binding to Crm1 due to the presence of two NESs in CPEB1. Indeed, β-actin, which also contains two functional NESs (Wada et al., 1998), was observed in the nucleus, but not in CNoBs, when expressed in fusion with GFP in HeLa cells (data not shown). Nevertheless, both NESs are required for CPEB1 localization in CNoBs, suggesting the simultaneous binding of one CPEB1 protein to two Crm1 molecules. Although the C-terminal region of Crm1 has been reported to dimerize in solution, there is no evidence that the full-length protein does (Petosa et al., 2004). However, two Crm1 molecules could be brought closer together through interaction with other proteins. Interestingly, CPEB1 dissociated from Crm1 in the nucleoli of cells treated with actinomycin D. At the nuclear envelope, the only mechanism reported for Crm1 cargo release is the hydrolysis of Ran-GTP into Ran-GDP (Petosa et al., 2004). It remains to be assessed whether a similar mechanism can occur in nucleolus, and, in such a case, how it could be sensitive to ongoing RNA polymerase I transcription.
Role of CNoBs
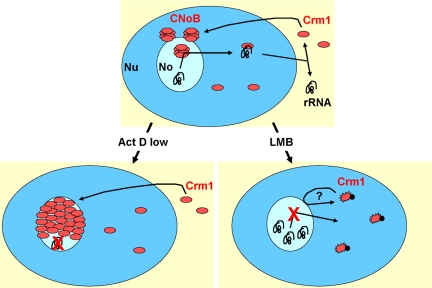
CNoBs are resistant to in situ sequential extraction with Triton X-100, DNase I, 2 M NaCl, and RNase A, indicating that they are part of the nuclear matrix, rather than being bound to chromatin or nascent RNA. Nevertheless, their presence is dependent on the continuous production of rRNA. We have shown that they disappear after treatment with a low dose of actinomycin D, which specifically inhibits RNA polymerase I. After this treatment, the fibrillar component, which contains nascent rRNA and fibrillarin, becomes condensed on the side of the nucleolus, whereas most of the nucleolus volume is filled with the granular component, which contains maturing ribosomes (Puvion-Dutilleul et al., 1992). In these conditions, CNoBs disappear, whereas Crm1 protein is redistributed over the granular part of the nucleolus, indicating that it is associated with maturing ribosomes and not with nascent rRNA. This localization is consistent with a role of CNoBs in processing or export of the ribosomes, as illustrated in the cartoon presented in Figure 9.
Figure 9.
Model recapitulating CNoB sensitivity to actinomycin D and LMB treatments. Nucleus (Nu) and nucleolus (No) are in dark and light blue, respectively. Crm1 protein (red) accumulates in CNoBs. There, it delivers factors required for ribosome biogenesis and/or uploads preribosomes to be exported to the cytoplasm. When RNA polymerase I is inhibited by actinomycin D, no new ribosome is to be processed and exported, and Crm1 accumulates in nucleoli. When LMB blocks Crm1, Crm1 disappears from nucleoli, and ribosome biogenesis is inhibited due to defective processing and/or export.
Crm1 plays a role in pre-60S export by triggering the transport of Nmd3, which binds to the ribosomal protein Rpl10p, through nuclear pores (Gadal et al., 2001; Thomas and Kutay, 2003). With respect to this function, the fact that Crm1 forms CNoBs and accumulates in the nucleolus when transcription is inhibited, would mean that Crm1 loads up pre-60S in the nucleolus, and not at the nuclear envelope. When RNA polymerase I transcription is on, CNoBs could be a platform where pre-60S associates to Crm1 for export (Figure 9). It can be noted that Nmd3 also contains two NESs (Gadal et al., 2001), raising the possibility that they play a role in its proper nucleolar localization, as observed for CPEB1. When RNA polymerase I transcription is off, Crm1 would further accumulate in the nucleolus and not be able to leave (Figure 9). The observation that blockage of Crm1 function by LMB leads to CNoB disappearance, but not to Crm1 spreading over the granular region of the nucleolus, as observed with actinomycin D, suggests that its anchoring to the granular component is mediated by binding to NES-containing proteins. Noteworthy, the nucleolar localization of Nmd3 in cells treated with actinomycin D is very similar to the one we observed for Crm1 (Trotta et al., 2003). Nmd3 could therefore anchor Crm1 or be anchored by Crm1.
Furthermore, Crm1 is also involved in pre-40S biogenesis (Rouquette et al., 2005). Crm1 inhibition by LMB leads to the accumulation of one of its precursors, of 26S, probably due to the blockage of cleavage at site A0. This default is associated with defective pre-40S export. Interestingly, A0 cleavage requires the snRNA U3, and LMB has been shown to prevent the routing of U3 from Cajal bodies, where it is processed, to the nucleolus (Boulon et al., 2004). In this context, CNoBs could also be a platform for processing factors. In cells treated with LMB, Crm1 would be unable to transport U3 to nucleoli and CNoBs would disappear (Figure 9). Conversely, when ribosomal transcription is off, Crm1 would continuously bring U3 and eventually fill the nucleolus.
Role of CPEB1 in CNoBs
Several RNA-binding proteins with a documented role in the control of mRNA in the cytoplasm have been shown to shuttle between nucleus and cytoplasm. As described in the Introduction, a common interpretation is that this enables binding to regulated mRNAs at the site of transcription, so that they are regulated as soon as they enter the cytoplasm (Smillie and Sommerville, 2002; Nielsen et al., 2003; Oleynikov and Singer, 2003). The best experimental evidence has been given for ZBP, which controls the transport of β-actin mRNA to the leading edge of chicken embryonic fibroblasts. ZBP is seen in the nucleus on actin alleles, provided that they are being transcribed (Oleynikov and Singer, 2003). In the case of CPEB1, our data are not consistent with such a function. First, inactivation of both RRMs by point mutation, or complete deletion of the C-terminal region responsible for CPE binding in the 3′ untranslated region of regulated mRNA, did not inhibit localization in CNoBs. Second, CPEB1 foci were resistant to RNase treatments. Third, FRAP demonstrated that CPEB1 association to CNoBs is stable, with 70% of the protein immobile over 30 min. For comparison, in similar experiments, half of ZBP present at the actin transcription site is replaced in 90 s (Oleynikov and Singer, 2003). All these data argue for a role of CPEB1 in CNoBs unrelated to CPE-containing mRNAs.
A new possibility is that CPEB1 plays a role in the genesis of ribosomes. Although its long residence time in CNoBs does not support a role in export to the cytoplasm, it could play a role in their assembly. Several observations argue for such a role. First of all, CPEB1 foci dissociate from nucleoli when synthesis of rRNA is specifically inhibited by a low dose of actinomycin D. Then, when Crm1 condenses at the periphery of nucleoli in cells treated with high dose of actinomycin D, CPEB1 foci are frequently in contact with Crm1 area, suggesting a functional link with Crm1 complex in this location. Finally, expression of CPEB1-GFP interferes with the localization of Crm1 in the granular region of nucleoli in cells treated with low dose of actinomycin D. Overall, these observations raise the interesting possibility that a translational regulator be also involved in the biogenesis of the translational apparatus.
CNoB Assembly and Maintenance
We next explored the dynamics of CNoBs in live cells. These bodies assemble in the center of nucleoli. They seem to disassemble after LMB treatment, suggesting that their mechanism of assembly requires interaction between Crm1 and an NES-containing protein. However, because this was assessed by the localization of only Crm1 and CPEB1, we cannot exclude that both proteins are removed from the foci, whereas other components are maintained. The number of CNoBs can decrease due to meeting and fusion. CNoBs are sometimes mobile, and, intriguingly, their movements are consistently from the interior of nucleolus to the outside. These movements are very slow, of the order of 25 nm per minute. Once at the nucleolar periphery, some of them were occasionally seen to detach from nucleoli. However, they never reached the nuclear envelope. This excludes a model where CNoBs would traffic between nucleoli and nuclear pores in order to transport mature ribosomes to the cytoplasm. If CNoBs play a role in transport, then they act as a platform enabling cargo uploading by Crm1, followed by release of the cargo-Crm1 complex (Figure 9). Similarly, because ribosome biogenesis is occurring throughout the whole volume of the nucleolus, a role in ribosomal maturation would be most likely as a platform providing or recycling ribosome processing factors.
ACKNOWLEDGMENTS
We thank Chantal Azerrad for her help in mutagenesis. Anna Wilczynska was supported by a Marie Curie fellowship (5th PCRD, contract QLGA-1999-50406) and a FEBS Collaborative Experimental Scholarship for Central and Eastern Europe. This work was supported by the Centre National de la Recherche Scientifique, the Ligue contre le Cancer, and the Agence Nationale pour la Recherche.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-09-0904) on October 15, 2008.
REFERENCES
- Boulon S., et al. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol. Cell. 2004;16:777–787. doi: 10.1016/j.molcel.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Buckler-White A. J., Humphrey G. W., Pigiet V. Association of polyoma T antigen and DNA with the nuclear matrix from lytically infected 3T6 cells. Cell. 1980;22:37–46. doi: 10.1016/0092-8674(80)90152-x. [DOI] [PubMed] [Google Scholar]
- Chen C., Fossar N., Weil D., Guillaud-Bataille M., Danglot G., Raynal B., Dautry F., Bernheim A., Brison O. High frequency trans-splicing in a cell line producing spliced and polyadenylated RNA polymerase I transcripts from an rDNA-myc chimeric gene. Nucleic Acids Res. 2005;33:2332–2342. doi: 10.1093/nar/gki530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhart D. E., Malter H. E., Feng Y., Warren S. T. The fragile X mental retardation protein is a ribonucleoprotein containing both nuclear localization and nuclear export signals. Hum. Mol. Genet. 1996;5:1083–1091. doi: 10.1093/hmg/5.8.1083. [DOI] [PubMed] [Google Scholar]
- Fujimura K., Kano F., Murata M. Dual localization of the RNA binding protein CUGBP-1 to stress granule and perinucleolar compartment. Exp. Cell Res. 2008;314:543–553. doi: 10.1016/j.yexcr.2007.10.024. [DOI] [PubMed] [Google Scholar]
- Gadal O., Strauss D., Kessl J., Trumpower B., Tollervey D., Hurt E. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 2001;21:3405–3415. doi: 10.1128/MCB.21.10.3405-3415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Deerinck T. J., Ellisman M. H., Spector D. L. The dynamic organization of the perinucleolar compartment in the cell nucleus. J. Cell Biol. 1997;137:965–974. doi: 10.1083/jcb.137.5.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen T. H., Oubridge C., Teo C. H., Pritchard C., Nagai K. Identification of molecular contacts between the U1 A small nuclear ribonucleoprotein and U1 RNA. EMBO J. 1991;10:3447–3456. doi: 10.1002/j.1460-2075.1991.tb04909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K. J., Korb E., Kundel M. A., Kochanek A. R., Kabraji S., McEvoy M., Shin C. Y., Wells D. G. CPEB1 regulates beta-catenin mRNA translation and cell migration in astrocytes. Glia. 2008;56:1401–1413. doi: 10.1002/glia.20707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- la Cour T., Gupta R., Rapacki K., Skriver K., Poulsen F. M., Brunak S. NESbase version 1.0, a database of nuclear export signals. Nucleic Acids Res. 2003;31:393–396. doi: 10.1093/nar/gkg101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guiner C., Lejeune F., Galiana D., Kister L., Breathnach R., Stevenin J., Del Gatto-Konczak F. TIA-1 and TIAR activate splicing of alternative exons with weak 5′ splice sites followed by a U-rich stretch on their own pre-mRNAs. J. Biol. Chem. 2001;276:40638–40646. doi: 10.1074/jbc.M105642200. [DOI] [PubMed] [Google Scholar]
- Lopez de Silanes I., Galban S., Martindale J. L., Yang X., Mazan-Mamczarz K., Indig F. E., Falco G., Zhan M., Gorospe M. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol. Cell. Biol. 2005;25:9520–9531. doi: 10.1128/MCB.25.21.9520-9531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazan-Mamczarz K., Lal A., Martindale J. L., Kawai T., Gorospe M. Translational repression by RNA-binding protein TIAR. Mol. Cell. Biol. 2006;26:2716–2727. doi: 10.1128/MCB.26.7.2716-2727.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshall N., Reiter M. H., Weil D., Standart N. CPEB interacts with an ovary-specific eIF4E and 4E-T in early Xenopus oocytes. J. Biol. Chem. 2007;282:37389–37401. doi: 10.1074/jbc.M704629200. [DOI] [PubMed] [Google Scholar]
- Minshall N., Thom G., Standart N. A conserved role of a DEAD box helicase in mRNA masking. RNA. 2001;7:1728–1742. doi: 10.1017/s135583820101158x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollet S., Cougot N., Wilczynska A., Dautry F., Kress M., Bertrand E., Weil D. Translationally Repressed mRNA Transiently Cycles through Stress Granules during Stress. Mol. Biol. Cell. 2008;19:4469–4479. doi: 10.1091/mbc.E08-05-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J., Adolph S. K., Rajpert-De Meyts E., Lykke-Andersen J., Koch G., Christiansen J., Nielsen F. C. Nuclear transit of human zipcode-binding protein IMP1. Biochem. J. 2003;376:383–391. doi: 10.1042/BJ20030943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleynikov Y., Singer R. H. Real-time visualization of ZBP1 association with beta-actin mRNA during transcription and localization. Curr. Biol. 2003;13:199–207. doi: 10.1016/s0960-9822(03)00044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petosa C., Schoehn G., Askjaer P., Bauer U., Moulin M., Steuerwald U., Soler-Lopez M., Baudin F., Mattaj I. W., Muller C. W. Architecture of CRM1/Exportin1 suggests how cooperativity is achieved during formation of a nuclear export complex. Mol. Cell. 2004;16:761–775. doi: 10.1016/j.molcel.2004.11.018. [DOI] [PubMed] [Google Scholar]
- Philips A. V., Timchenko L. T., Cooper T. A. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- Puvion-Dutilleul F., Mazan S., Nicoloso M., Pichard E., Bachellerie J. P., Puvion E. Alterations of nucleolar ultrastructure and ribosome biogenesis by actinomycin D. Implications for U3 snRNP function. Eur. J. Cell Biol. 1992;58:149–162. [PubMed] [Google Scholar]
- Richter J. D. CPEB: a life in translation. Trends Biochem. Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Rouquette J., Choesmel V., Gleizes P. E. Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J. 2005;24:2862–2872. doi: 10.1038/sj.emboj.7600752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie D. A., Sommerville J. RNA helicase p54 (DDX6) is a shuttling protein involved in nuclear assembly of stored mRNP particles. J. Cell Sci. 2002;115:395–407. doi: 10.1242/jcs.115.2.395. [DOI] [PubMed] [Google Scholar]
- Stebbins-Boaz B., Cao Q., de Moor C. H., Mendez R., Richter J. D. Maskin is a CPEB-associated factor that transiently interacts with elF-4E. Mol. Cell. 1999;4:1017–1027. doi: 10.1016/s1097-2765(00)80230-0. [DOI] [PubMed] [Google Scholar]
- Tamanini F., Bontekoe C., Bakker C. E., van Unen L., Anar B., Willemsen R., Yoshida M., Galjaard H., Oostra B. A., Hoogeveen A. T. Different targets for the fragile X-related proteins revealed by their distinct nuclear localizations. Hum. Mol. Genet. 1999;8:863–869. doi: 10.1093/hmg/8.5.863. [DOI] [PubMed] [Google Scholar]
- Thomas F., Kutay U. Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J. Cell Sci. 2003;116:2409–2419. doi: 10.1242/jcs.00464. [DOI] [PubMed] [Google Scholar]
- Trotta C. R., Lund E., Kahan L., Johnson A. W., Dahlberg J. E. Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates. EMBO J. 2003;22:2841–2851. doi: 10.1093/emboj/cdg249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada A., Fukuda M., Mishima M., Nishida E. Nuclear export of actin: a novel mechanism regulating the subcellular localization of a major cytoskeletal protein. EMBO J. 1998;17:1635–1641. doi: 10.1093/emboj/17.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welk J. F., Charlesworth A., Smith G. D., MacNicol A. M. Identification and characterization of the gene encoding human cytoplasmic polyadenylation element binding protein. Gene. 2001;263:113–120. doi: 10.1016/s0378-1119(00)00588-6. [DOI] [PubMed] [Google Scholar]
- Wilczynska A., Aigueperse C., Kress M., Dautry F., Weil D. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J. Cell Sci. 2005;118:981–992. doi: 10.1242/jcs.01692. [DOI] [PubMed] [Google Scholar]
- Zhang T., Delestienne N., Huez G., Kruys V., Gueydan C. Identification of the sequence determinants mediating the nucleo-cytoplasmic shuttling of TIAR and TIA-1 RNA-binding proteins. J. Cell Sci. 2005;118:5453–5463. doi: 10.1242/jcs.02669. [DOI] [PubMed] [Google Scholar]
- Zimber A., Nguyen Q. D., Gespach C. Nuclear bodies and compartments: functional roles and cellular signalling in health and disease. Cell Signal. 2004;16:1085–1104. doi: 10.1016/j.cellsig.2004.03.020. [DOI] [PubMed] [Google Scholar]