Abstract
COMATOSE (CTS), the Arabidopsis homologue of human Adrenoleukodystrophy protein (ALDP), is required for import of substrates for peroxisomal β-oxidation. A new allelic series and a homology model based on the bacterial ABC transporter, Sav1866, provide novel insights into structure-function relations of ABC subfamily D proteins. In contrast to ALDP, where the majority of mutations result in protein absence from the peroxisomal membrane, all CTS mutants produced stable protein. Mutation of conserved residues in the Walker A and B motifs in CTS nucleotide-binding domain (NBD) 1 resulted in a null phenotype but had little effect in NBD2, indicating that the NBDs are functionally distinct in vivo. Two alleles containing mutations in NBD1 outside the Walker motifs (E617K and C631Y) exhibited resistance to auxin precursors 2,4-dichlorophenoxybutyric acid (2,4-DB) and indole butyric acid (IBA) but were wild type in all other tests. The homology model predicted that the transmission interfaces are domain-swapped in CTS, and the differential effects of mutations in the conserved “EAA motif” of coupling helix 2 supported this prediction, consistent with distinct roles for each NBD. Our findings demonstrate that CTS functions can be separated by mutagenesis and the structural model provides a framework for interpretation of phenotypic data.
INTRODUCTION
ATP-binding cassette (ABC) transporters are ubiquitous, integral membrane proteins that mediate vectorial transport of a diverse range of molecules across membranes and consequently are involved in a wide variety of biological processes (Higgins, 1992; Rea, 2007). All ABC transporters share the same basic architecture, comprising two transmembrane domains (TMDs), each composed of several α-helices that are involved in substrate recognition and translocation across the lipid bilayer, and two nucleotide-binding domains (NBDs) that bind and hydrolyze ATP, providing the driving force for transport. In prokaryotes, these domains are usually present on separate polypeptides, whereas in eukaryotes, fusion of a TMD and NBD module to form a “half transporter,” or indeed fusion of all modules to form a “full transporter” is more common. The past decade has seen the determination of high-resolution crystal structures for several isolated NBDs and, more recently, complete ABC transporters (reviewed in Dawson et al., 2007).
The NBDs characteristically contain several highly conserved features common to nucleotide hydrolases, including the Walker A motif (GxxGxGKS/T, where x is any amino acid), forming the P-loop, which binds to the α- and β-phosphates of nucleotides, and the Walker B motif (ΦΦΦΦDE, where Φ is hydrophobic), thought to coordinate the Mg2+ ion or polarize the attacking water molecule (Walker et al., 1982; Hung et al., 1998; Schneider and Hunke, 1998; Hopfner et al., 2000). ABC transporters additionally contain the signature motif (LSGGQ), which contacts the γ-phosphate of ATP; the Q-loop (or “lid”) containing a glutamine residue, which interacts with γ-phosphate through a water molecule; and the switch region (or H-loop), originally suggested to polarize the attacking water molecule for nucleotide hydrolysis but more recently proposed to act as a linchpin during catalysis (Walker et al., 1982; Hung et al., 1998; Schneider and Hunke, 1998; Hopfner et al., 2000; Zaitseva et al., 2005).
In the ATP-bound state, the NBDs form a dimer with two composite ATP-binding sites each comprising the Walker A, Walker B, and switch motifs of one NBD and the signature motif from the second NBD (Hopfner et al., 2000; Moody et al., 2002; Smith et al., 2002). In the ATP switch model, formation and dissociation of the NBD dimer associated with ATP binding and hydrolysis are proposed to provide a regulatable switch that induces conformational changes in the TMDs to drive transport (Higgins and Linton, 2004; Linton and Higgins, 2007). Structural and biochemical evidence indicates that conformational changes in the NBDs are transmitted to the TMDs via the coupling helices, short α-helices located in a cytoplasmic loop between two transmembrane segments of each TMD. The coupling helices are embedded in a surface groove on the NBDs, making contacts with side chains around the Q-loop (Kerppola and Ames, 1992; Dawson and Locher, 2006; Daus et al., 2007; Zolnerciks et al., 2007). Thus, the structural arrangement of the TMD-NBD “transmission interface” appears to be a common feature of ABC transporters (Dawson et al., 2007).
Eukaryotic ABC proteins have been classified into eight subfamilies (A–H; Dean and Annilo, 2005; Dean et al., 2001; Verrier et al., 2008). ABC subfamily D is represented in all eukaryotic taxa and contains a group of integral peroxisomal membrane proteins considered to play roles in the import of substrates for the β-oxidation pathway (Theodoulou et al., 2006). Yeast contains two ABCD genes, PXA1 and PXA2, which encode “half-size” ABC proteins with the domain organization TMD-NBD and which form a heterodimeric transporter required for the peroxisomal transport of long-chain fatty acyl-coenzyme A esters (Hettema et al., 1996; Shani and Valle, 1996; Verleur et al., 1997). The human ABC transporter subfamily D comprises four “half-size” members, including Adrenoleukodystrophy protein (ALDP), which is defective in X-linked Adrenoleukodystrophy (X-ALD), the most common peroxisomal neurodegenerative disease (reviewed in Berger and Gärtner, 2006; Theodoulou et al., 2006; Kemp and Wanders, 2007). More than 480 mutant alleles of the gene encoding ALDP are associated with X-ALD, including more than 250 mis-sense mutations, many of which result in protein instability (http://www.x-ald.nl; Berger and Gärtner, 2006). X-ALD is characterized biochemically by the accumulation of saturated, straight-chain very-long-chain fatty acids (VLCFA; C ≥22:0) in plasma and tissue, and ALDP has recently been shown function as a homodimeric transporter that accepts fatty acyl-CoAs (Van Roermund et al., 2008).
Arabidopsis contains a single peroxisomal ABC transporter with the systematic name AtABCD1 (Verrier et al., 2008), which has been identified in no less than five independent forward genetic screens (Russell et al., 2000; Zolman et al., 2001; Hayashi et al., 2002; Eastmond, 2006; Hooks et al., 2007) and is known as CTS/AtPXA1/PED3/ACN2, referred to hereafter as COMATOSE (CTS). CTS encodes a full-size ABC protein with the topology TMD-NBD-TMD-NBD. The two halves of the protein are each around 35% identical to ALDP but are distinct from each other; this protein structure, which can be considered as a “fused heterodimer” appears to be conserved in bryophytes and angiosperms (Theodoulou et al., 2006; Rensing et al., 2008; Verrier et al., 2008).
CTS plays key roles in breaking of seed dormancy, germination, and mobilization of triacylglycerol (TAG) seed storage reserves (Footitt et al., 2002; Hayashi et al., 2002; Russell et al., 2000). Dormancy is a physiological state whereby seeds will not germinate even under optimal conditions until they have undergone a period of after-ripening. This serves to disperse seed germination in space and time, increasing seedling survival from a single seed set (Bewley, 1997). After-ripened cts mutants cannot germinate even after classical dormancy-breaking treatments. However, artificial removal of surrounding endosperm/testa tissues allows germination, but subsequently mutants are unable to establish in the absence of exogenous sucrose, due to a failure to break down stored TAG, which normally provides both energy and carbon skeletons for early seedling development. Similar phenotypes observed in some other β-oxidation mutants point to CTS (and β-oxidation) having distinct roles in the completion of germination and in subsequent establishment (Pinfield-Wells et al., 2005; Pracharoenwattana et al., 2005; Footitt et al., 2006).
Auxin is a hormone that controls many aspects of plant development (Teale et al., 2006). One effect of exogenously applied auxins such as 2,4-dichlorophenoxyacetic acid (2,4-D) and indole acetic acid (IAA) is the inhibition of primary root growth. The auxin precursors, 2,4-dichlorophenoxybutyric acid (2,4-DB) and indole butyric acid (IBA), which are converted by peroxisomal β-oxidation to 2,4-D and IAA, respectively, have been used to isolate mutants in this pathway by virtue of their long root phenotype when grown on 2,4-DB and IBA (Hayashi et al., 1998; Zolman et al., 2000). The isolation of the cts alleles pxa1 and ped3 in these screens indicates a role for CTS in mediating transport of these compounds into the peroxisome for conversion to the active auxin (Zolman et al., 2001; Hayashi et al., 2002).
Jasmonic acid (JA) is a lipid-derived hormone. JA biosynthesis begins in the chloroplast and is completed in the peroxisome via β-oxidation of 12-oxophytodieonoic acid (OPDA; Schaller et al., 2005). cts mutants have reduced levels of both basal and wound-induced JA, consistent with a partial block in transport of OPDA into the peroxisome (Theodoulou et al., 2005). Taken together, these findings strongly suggest that CTS is a broad specificity transporter that mediates uptake of diverse substrates for β-oxidation into the peroxisome, although it is possible that CTS acts indirectly, for example, as a transport regulator.
In the current study, we present the characterization of a new allelic series of mis-sense mutants, which has allowed the separation of different physiological roles of CTS in planta and has afforded insights into the structure-function relationship of the protein. The potential structural and catalytic consequences of single amino acid substitutions are examined using a homology-based structural model and implications of these results are discussed in terms of kinetics and substrate specificity.
MATERIALS AND METHODS
Isolation and Generation of New CTS Alleles
A TILLING screen of the first NBD of CTS was carried out via the Arabidopsis TILLING Project (ATP; http://tilling.fhcrc.org:9366/), using primers 5′ tgctggcaacacatcatcttctgg-3′ and 5′-ctagctgcagcaaacgctcgttga-3′. The screen identified 12 mutations, including seven unique mis-sense alleles designated cts-3-cts-9; see Figure 1; Table 1. The remaining mutations were either located in introns or were silent. E617K (cts-7) was recovered twice independently. EMS-mutagenized seeds from the lines identified by the screen (Till et al., 2003) were obtained from the Nottingham Arabidopsis Stock Centre (NASC). Four backcrosses to their wild type (Col-Big Mama) were made. Mutations were followed with molecular markers (dCAPS and selective primers) during crossing (Supplementary Table S2). For site-directed mutagenesis (SDM), a plant transformation vector containing the CTS open reading frame (ORF) downstream of a 2.6-kb CTS promoter sequence was used (Footitt et al., 2007). Mutations were introduced using the QuikChange II XL SDM kit (Stratagene, La Jolla, CA) following the manufacturer's instructions (primers are listed in Supplementary Table S3). The plasmids pCRBlunt ORF5′ and pSK ORF3′ (Footitt et al., 2007) were used as template, and mutated sequences were verified by sequencing. Mutagenized fragments were excised from pCRBlunt ORF5′ using SacII and PstI and ligated into the SacII/PstI sites of pSK ProORF (Footitt et al., 2007). The promoter-ORF cassette was then excised with KpnI and SmaI from pSK ProORF and ligated into KpnI/SmaI sites of pG0229-T (Footitt et al., 2007). Mutagenized fragments of the 3′ part of the ORF were excised with PstI and SmaI from pSK ORF3′ and ligated directly into PstI/SmaI-cut pG0229-T/CTS promORF (Footitt et al., 2007). Agrobacterium tumefaciens GV3101 pMP90 transformed with these constructs were used for floral-dip transformation of Arabidopsis thaliana accession Ler (Landsberg erecta). Transformants were selected by repeated spraying with a 150 mg/l solution of glufosinate ammonium (Bayer CropScience, Cambridge, United Kingdom) and used as pollen donors for crossing to cts-1 plants. F2 plants were screened for cts-1 homozygosity by PCR, as described in Footitt et al. (2007). Homozygosity of the transgene was confirmed by observation of complete resistance of F3 seeds to 10 mg/l glufosinate ammonium.
Figure 1.
CTS model and protein expression in mutant alleles. (A) Schematic model of CTS showing position of mutations. Transmembrane domains (TMD) are shown in dark gray and nucleotide-binding domains (NBD) in light gray. Conserved motifs within the NBDs are represented by boxes and are in the following order: Walker A (A), ABC signature motif (ABC), Walker B (B). (B) Immunoblot analysis of total protein isolated from 4-d-old seedlings. Equal amounts of protein (60 μg TILLING mutants and controls, 40 μg SDM mutants and controls; 25 μg other alleles) were loaded and transfer confirmed by staining of membranes with Ponceau S. Protein expression was detected with an antibody raised to the C-terminal 225 amino acids of CTS. No signal was obtained with preimmune serum (data not shown). Wild-type accessions for the site-directed and TILLING alleles are Landsberg erecta (Ler) and Columbia-Big Mama (Col-BM), respectively.
Table 1.
Mutant alleles used in this study
| Allele | Mutation type | Effect | Amino acid conservation | Germinates? | Establishment minus sucrose? | Oil mobilization defect?e | IBA and 2,4-DB response of roots |
|---|---|---|---|---|---|---|---|
| cts-1a | Fast neutron | No protein | No | No | Yes | Resistant | |
| cts-2a | T-DNA | No protein | No | No | Yes | Resistant | |
| pxa1-1b | EMS | Mis-spliced | No | No | Yes | Resistant | |
| ped3-1c | EMS | W1242stop | No | No | Yes | Resistant | |
| ped3-2c | EMS | R1035W | P | No | No | Yes | Resistant |
| ped3-3c | EMS | W1007stop | No | No | Yes | Resistant | |
| ped3-4c | EMS | S810N | P | Yes (slow) | No | Yes | Resistant |
| cts-3 | EMS | G503E | P, Y, H | No | No | Yes | Resistant |
| cts-4 | EMS | L514F | N | Yes | Yes | No | Sensitive |
| cts-5 | EMS | P539L | P, Y, H | No | No | Yes | Resistant |
| cts-6 | EMS | D568N | P (Asp or Glu) | Yes | Yes | No | Sensitive |
| cts-7 | EMS | E617K | P, Y, H | Yes | Yes | No | Intermediate |
| cts-8 | EMS | C631Y | Φ in all | Yes (slow) | Yes | No | Intermediate |
| cts-9 | EMS | S657N | N | Yes | Yes | No | Sensitive |
| SDM | E300D | P, Y, H | Yes | Yes | No | Sensitive | |
| SDM | K487A | P, Y, H | No | No | Yes | Resistant | |
| SDM | D606N | P, Y, H | No | No | Yes | Resistant | |
| SDM | E954D | P, Y, H | Yes | Yes | No | Intermediate | |
| SDM | K1136A | P, Y, H | Yes | Yes | No | Sensitive | |
| SDM | D1276N | P, Y, H | Yes | Yes | No | Intermediate |
Abbreviations: EMS, ethane methyl sulfonate-induced; SDM, site-directed mutagenesis; P, plants; Y, yeast; H, humans; N, not conserved; Φ, hydrophobic amino acid residue.
a. Footitt et al., 2002; b. Zolman et al., 2001; c. Hayashi et al., 2002; d. 19 aa encoded by intron replace terminal 32 aa; e. assayed by retention of hypocotyl oil bodies (Nile Red staining) and hypocotyl length in presence and absence of sucrose.
Protein expression in all cts alleles was analyzed as described in Footitt et al. (2002), and equal protein loading on membranes was confirmed by staining with Ponceau S.
Subcellular Localization
The CTS ORF was amplified in two overlapping halves using primer pairs FT59/FT35 and FT36/FT60 (Supplementary Table S3), with pG0229-T/CTS promORF (Footitt et al., 2007) as template. The fragments were cloned into pCR4Blunt-TOPO (Invitrogen, Paisler, United Kingdom), excised with KpnI-PstI and PstI-BamHI, respectively, and cloned into the KpnI-BamHI sites of pBIN1935S-GFP in a three-way ligation. Mutations were introduced by fragment exchange, as described above. Agro-infiltration of tobacco leaves was preformed as described in Sparkes et al. (2005, 2006), and peroxisomal localization was assessed by cotransformation with mRFP-SRL (Pracharoenwattana et al., 2005). Confocal imaging was performed using a Zeiss inverted LSM510 laser scanning microscope with a 63× oil immersion objective (Thornwood, NY). Fluorescence was detected using a 488-nm/543-nm dichroic beam splitter, a 505–530-nm band-pass filter for green fluorescent protein (GFP) and a 560–615-nm bandpass filter for red fluorescent protein (RFP). The pinhole was usually set to give confocal sections between 1 and 2.5 μm. After acquisition image processing was done using the LSM 5 browser software (Zeiss) and Adobe Photoshop (San Jose, CA) elements.
Phenotypic Characterization
Germination and Establishment Assays.
All seeds used for phenotypic characterization were from plants grown synchronously in controlled environment chambers (23°C, 16 h light, 100 μE light). Seeds of mutants and their corresponding wild types were stored at 24°C in the dark until completely after-ripened. For germination and establishment assays, seeds were plated on 0.5× MS medium, as previously described (Footitt et al., 2006), and germination and establishment were recorded over a period of 7 d. Germination sensu stricto is defined as emergence of the radicle (embryonic root) from the seed coat (Bewley, 1997) and establishment as the development of photosynthetic competence (i.e., greening), after depletion of seed reserves. In all cases, experiments were carried out in triplicate, using 50–60 seeds per replicate. For the assay of hypocotyl growth, sterilized seeds were plated on 0.5× MS medium with or without 0.5% (wt/vol) sucrose, and seed coats were ruptured with a needle. Seeds were chilled at 4°C for 2 d and then transferred to a growth chamber. After 24 h in the light to induce germination, plates were wrapped in aluminum foil, and the length of hypocotyls was measured after a total of 7 d in the growth chamber. Experiments were carried out in triplicate, using 10–15 seeds per replicate. To determine the presence of lipid bodies in seedlings, seedlings were grown for 5 d on 0.5× MS medium with 0.5% (wt/vol) sucrose and then stained for 5 min in a 1 μg/ml solution of Nile Red (Molecular Probes, Invitrogen, Paisley, United Kingdom) and washed twice with water. Hypocotyl images were recorded with a Leica TCS SP2 (Leica Microsystems, Milton Keynes, United Kingdom), with the following settings: excitation 488 nm and emission 556–580 nm (Nile Red) and 718–749 nm (chlorophyll autofluorescence).
Root Growth Assays.
Seeds were plated on 0.5× MS medium with 0.5% (wt/vol) sucrose containing the indicated concentrations of 2,4-DB or IBA or no supplements. For nongerminating alleles, seed coats were ruptured with a needle to allow germination. Seeds were chilled for 2 d at 4°C and then placed vertically in a growth chamber for 5–8 d. Experiments were carried out in triplicate with 10–15 seeds per replicate. Mean root length of seedlings grown on medium with supplements is expressed as percentage of mean growth of the same line on medium without supplements.
Creation of a Homology-based Structural Model
A homology model of CTS was made using the crystal structure of the Staphylococcus aureus multidrug transporter Sav1866 (PDB accession 2HYD;(Dawson and Locher, 2006) as a template. Alignment of the N- and C-terminal halves of the CTS sequence with the structural template, which is much less similar to the plant protein in the TMD than in the NBD regions, was optimized using a combination of techniques. An initial alignment was created using the profile-to-profile based Multiple Mapping Method (Rai and Fiser, 2006). This was then adjusted in the light of analysis of the aligned sequences of 85 ABC transporter subfamily D members and, separately, of 148 Sav1866 homologues for patterns of residue conservation (using the ConSeq method; Berezin et al., 2004), and of hydrophobicity. Final adjustments to the alignment were informed by the results of threading analysis performed using the LOOPP program (Teodorescu et al., 2004). The resultant alignment (Supplementary Figure S2) excluded regions 1–93 and 677–747, i.e., the N-terminus and the linker between the two halves of CTS, which are not found in the template. Modeler version 8.2 (Accelrys, San Diego, CA) (Fiser and Sali, 2003) was used to create 100 models, and the five of lowest energy were analyzed using ProCheck (http://www.biochem.ucl.ac.uk/∼roman/procheck/procheck.html; Laskowski et al., 1993). That selected for further analysis had only two residues in the disallowed region of the Ramachandran plot.
RESULTS
Creation of a CTS Allelic Series
A collection of new alleles containing single amino acid exchanges was created via SDM and TILLING, in order to analyze the structure-function relationships of CTS (Figure 1A; Table 1). Six amino acids known to be important for ABC transporter function were selected for SDM: lysine residues in the Walker A motifs (K487 and K1136), aspartate residues in the Walker B motifs (D606 and D1276; Hung et al., 1998; Schneider and Hunke, 1998; Gaudet and Wiley, 2001; Frelet and Klein, 2006), and residues E300 and E954, which reside in the coupling helix “EAA” motifs (Kerppola and Ames, 1992; Mourez et al., 1997; Daus et al., 2007). Mutated versions of the CTS ORF were expressed under control of its own promoter in the cts-1 background, which is phenotypically null (Footitt et al., 2007). In a second, complementary approach, seven new alleles containing EMS-induced mutations within NBD1 were obtained after screening of a TILLING population (Till et al., 2003). None of these mutations was present in previously defined functionally significant motifs of NBD1 (Table 1; Supplementary Figure S1). However, two of the mutations falling in the region between the Walker A and ABC signature motifs, G503E and P539L, and one of the mutations in the region following Walker B, E617K, altered amino acid residues which are conserved in CTS homologues found in plants, Saccharomyces cerevisiae and Homo sapiens (Supplementary Figure S1). Several of the mutations generated in the CTS allelic series have equivalents in X-linked adrenoleukodystrophy patients (Supplementary Table S1; http:/www.x-ald.nl; Kemp et al., 2001).
As mutations in ALDP have been shown to affect protein stability in many cases (Watkins et al., 1995; Feigenbaum et al., 1996; Kemp et al., 2001), we probed Western blots with proteins from 4-d-old seedlings with an antibody raised against the C-terminus of CTS (residues 1112–1337; Figure 1B). For previously published alleles, this analysis detected ped3-2 and ped3-4 mutant proteins only, which contain the single amino exchanges R1035W and S810N, respectively (Hayashi et al., 2002). The cts-1, cts-2, ped3-1, ped3-3, and pxa1-1 alleles produce truncated proteins (Zolman et al., 2001; Footitt et al., 2002; Hayashi et al., 2002; Table 1), which either lack the relevant epitopes or are presumably unstable and below detectable levels. Protein expression was evident for all the novel alleles described here, with the TILLING alleles expressing CTS protein amounts that were comparable to the wild type. The SDM alleles exhibited variations in protein amount that are probably due to copy number or possibly integration site effects. This novel allelic series was used, together with selected preexisting alleles, to analyze the functions of CTS during germination and seedling establishment.
Nucleotide-binding Domains 1 and 2 of COMATOSE Are Functionally Distinct
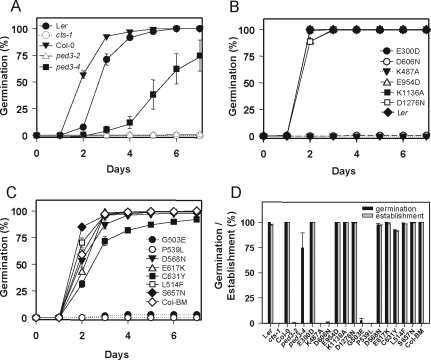
Germination and seedling establishment of all after-ripened cts alleles were assayed over 7 d of imbibition. Under these conditions, seeds of ped3-4 showed an increased germination potential compared with other previously published alleles that did not germinate (Figure 2A). The two mutations in NBD1, K487A, and D606N, prevented germination of seeds (Figure 2B) whereas mutation of the equivalent amino acids in NBD2, K1136A, and D1276N, did not, indicating that the two NBDs of CTS are functionally distinct. Mutations introduced into the EAA motifs of CTS (E300D and E954D) did not affect germination potential (Figure 2B).
Figure 2.
Germination and establishment assays. (A–C) Germination of seeds on 0.5× MS. (A) Previously described alleles, (B) SDM alleles, and (C) TILLING alleles. (D) Germination (■) and establishment (▩) after 7 d on 0.5× MS. All germination and establishment data are expressed as mean with SEM. If error bars are not visible, they are smaller than the symbols.
Two mutations derived by TILLING, G503E, and P539L prevented seeds from germinating, whereas all other TILLING mutations did not influence germination potential, except C631Y, which had a minor effect on the speed of germination (Figure 2C). Analysis of the establishment of germinated seeds after 7 d showed that, with the exception of ped3-4, seeds that had germinated also established (Figure 2D), establishment being defined as the production of expanded green cotyledons when germinated in the light. ped3-4 was the only allele able to germinate but unable to establish, demonstrating that these two functions of CTS can be separated (Hayashi et al., 2002).
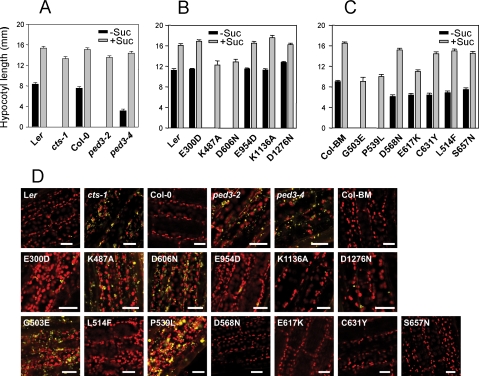
The ability of seeds to utilize stored energy reserves was examined indirectly by measuring hypocotyl expansion in the dark (Figure 3, A–C). Mutants K487A, D606N, G503E, and P539L, in common with the cts-1 null allele and ped 3–2, were unable to expand hypocotyls in the absence of an external source of energy, and, even in the presence of sucrose, hypocotyls of mutant alleles were consistently shorter than their corresponding wild type. In contrast, ped3-4 and the remaining site-directed and TILLING alleles did not require exogenous sucrose for hypocotyl expansion in the dark, although some of these mutants had shorter hypocotyls in the absence of sucrose than the corresponding controls. This was particularly striking for ped3-4, which germinates but fails to establish (Figure 2A and Hayashi et al., 2002). Mobilization of energy reserves was examined directly by assay for the disappearance of lipid bodies following germination (Figure 3D). Lipid bodies were visualized by staining seedlings with the lipophilic dye Nile Red (Kimura et al., 2004). This analysis showed that seedlings of mutant alleles in which germination completion is prevented maintained considerable numbers of hypocotyl lipid bodies (Figure 3D). In addition, ped3-4, which is able to germinate but not establish in the absence of sucrose, also retains a similar number of lipid bodies as those alleles unable to germinate, further demonstrating the separation of the germination and establishment phenotypes in this allele and indicating that storage lipid breakdown is not required for germination.
Figure 3.
Mobilization of energy reserves. (A–C) Hypocotyl length in presence (▩) or absence (■) of external energy source (sucrose). (A) Previously described alleles, (B) SDM alleles, and (C) TILLING alleles. All germination and establishment data are expressed as mean with SEM. (D) Confocal laser scanning microscope pictures of hypocotyls from 5-d-old seedlings stained with Nile Red. Lipid bodies are false-colored in yellow, and chlorophyll autofluorescence is shown in red. Scale bar, 30 μm.
To exclude the possibility that the K487A and D606N mutants resulted in a null phenotype due to a failure of peroxisomal targeting, both mutations were introduced into a CTS-GFP fusion protein and coexpressed transiently in tobacco epidermal cells with the peroxisomal marker mRFP-SRL and examined by confocal laser scanning microscopy (Figure 4). The left-hand column of panels shows peroxisomes marked by monomeric RFP (mRFP) fluorescence. Wild-type CTS with GFP fused to the C terminus (top row) locates to the same structures as mRFP (GFP fluorescence center panel; merged image on the right). This is expected as this construct can complement the cts-1 null mutant (D. Dietrich, unpublished data). By the same criteria, both mutants also correctly targeted to peroxisomes (middle and bottom rows).
Figure 4.
Subcellular localization of K487A and D606N mutants. Transient coexpression of CTS-GFP and the peroxisomal marker mRFP-SRL in tobacco epidermal cells, visualized by confocal laser scanning microscopy. Top row, wild-type CTS sequence fused to GFP; middle row, CTS-GFP with the K487A mutation; bottom row, CTS-GFP with the D606N mutation. Left-hand column, peroxisomes visualized by fluorescence emitted by the mRFP-SRL reporter (false-colored magenta). Center panel, GFP fluorescence. Right panel, merged image. Scale bar, 10 μm.
Response to Auxin Precursors Is Differentially Affected in the Allelic Series
Resistance to the auxin precursors, 2,4-DB and IBA, was used as an indirect test of β-oxidation of these compounds. Both 2,4-D and IAA treatment led to a drastic decrease in root growth in all plant lines tested, regardless of the state of the CTS locus (Figure 5, A and B), demonstrating that they retain their sensitivity to the end product of the pathway. Consistent with a block in β-oxidation, cts-1 and both ped3 alleles were resistant to 2,4-DB and IBA (Figure 5, C and D, and Hayashi et al., 2002). In contrast, the new alleles reported here showed varying degrees of resistance to both IBA and 2,4-DB (Figure 5, E–H). The site-directed mutants, D606N and K487A, showed resistance across a range of IBA and 2,4-DB concentrations that was similar to the null allele cts-1 (Figure 5, E and F) as did the TILLING alleles, P539L and G503E (Figures 5, G and H). The site-directed mutants, D1276N and E954D, showed intermediate resistance to 2,4-DB at all concentrations tested and to IBA at the intermediate concentration of 10 μM. Likewise from the TILLING series, C631Y and E617K, exhibited intermediate resistance to 2,4-DB and IBA, which varied depending on the concentration (Figure 5, G and H). Because E617K, C631Y, D1276N, and E954D were able to germinate and establish, these amino acid changes apparently affect the response to auxin precursors but not the ability to utilize fatty acids. Finally, the site-directed alleles, E300D and K1136A and the TILLING alleles D568N, L514F, and S657N behaved comparably to wild type (Figure 5, E–H).
Figure 5.
Response of roots to auxin precursors. (A and B) Site-directed cts mutants and the respective wild-type accession (Ler) are presented as an exemplar of the sensitivity of Arabidopsis roots to natural (IAA) and synthetic (2,4-D) auxins. Graphs show root response to 0.2 μM 2,4-D (A) and 100 nM IAA (B). (C–H) graphs show root response to 2,4-DB (C, E, and G) and IBA (D, F, and H). Results were standardized against growth on unsupplemented medium [0.5 x MS plus 0.5% (wt/vol) sucrose]. (C and D) previously described alleles, (E and F) SDM alleles, (G and H) TILLING alleles. Data are expressed as mean with SE of the mean (n = 3 plates; 15–20 seedlings per plate).
Arabidopsis root growth is extremely sensitive to the inhibitory effects of IAA (Zolman et al., 2000; Figure 5) and roots of seedlings heterozygous for CTS exhibit IBA resistance (Zolman et al., 2001). Therefore, it is possible that a relatively minor loss of CTS function might result in IBA resistance in the root assay. To attempt to distinguish further between the cts alleles with intermediate phenotypes, we measured the elongation of dark-grown hypocotyls, which is considerably less sensitive to IAA than roots (Rashotte et al., 2003). Hypocotyls of wild-type accessions exhibited sensitivity to both IAA and IBA (Figure 6, A–D). A representative null allele, cts-1 was resistant to 100 μM IBA (Figure 6C); however, elongation was inhibited at higher concentrations, suggesting that toxicity did not result from CTS-dependent conversion of IBA to IAA beyond 100 μM. As in Figure 3A, control hypocotyls were longer in the presence of sucrose, but interestingly, sucrose potentiated the inhibitory effect of IBA on hypocotyl elongation (Figure 6, C and D), an effect that was not observed with IAA (Figure 6, A and B).
Figure 6.
Response of dark-grown hypocotyls to IBA and IAA. Seeds were plated on 0.5× MS medium ± 0.5% (wt/vol) sucrose and stratified for 2 d. After 24-h light to permit germination, plates were covered and incubated for a further 5 d in the dark. (A and B) Response of wild-type hypocotyls to IAA. (C and D) Response of wild-type (Ler and Col-BM) and cts-1 hypocotyls to IBA. (E–H) Response of intermediate cts alleles to IBA in the absence (E and F) or presence (G and H) of sucrose. (E and G) SDM alleles; (F and H) TILLING alleles. All data are expressed as mean with SEM (20–30 hypocotyls).
In the absence of sucrose, hypocotyls of E954D and D1276N exhibited modest resistance to 30 and 100 μM IBA (Figure 6E) but, as seen for the wild type, the inclusion of sucrose increased the sensitivity of these alleles to IBA (i.e., decreased hypocotyl length compared with the same IBA concentration in the absence of sucrose; Figure 6G). D1276N had the lowest level of IBA resistance, consistent with the root data and both D1276N and E954D showed similar levels of hypocotyl inhibition to wild type (Ler) at 100 μM IBA in the presence of sucrose (Figure 6G). In contrast, E617K and C631Y were much less sensitive to IBA in the presence of sucrose, retaining appreciable resistance to 100 μM IBA (Figure 6, F and H).
Effect of Mutations on Protein Structure: Insights from a Homology Model
The CTS structure was modeled on that of the homodimeric bacterial multidrug ABC transporter Sav1866 (Dawson and Locher 2006), to which it displays low, but significant, sequence similarity (Supplementary Figure S2). Each monomer of the latter possesses six transmembrane (TM) helices followed by an NBD, such that the dimer structure is akin to that predicted for the six TM-NBD-6 TM-NBD, full-length CTS protein. Moreover, predicted TM helices 2 and 3 and 4 and 5 in each of the two TMDs of CTS are linked by long hydrophilic loops, similar to the intracellular loops found in the corresponding region of Sav1866. The latter contain coupling helices 1 and 2 (CH1 and 2), respectively, which interact primarily with the NBD of the opposite subunit and appear to play a role in transmitting conformational changes arising from ATP binding and hydrolysis to the TMDs (Dawson and Locher, 2006). Alignment of the distantly related CTS and template sequences in the TM regions was assisted by comparison of the patterns of residue conservation and polarity in multiple sequence alignments of 85 CTS and 148 Sav1866 homologues, as detailed in Materials and Methods. We have previously demonstrated the utility of this approach for successful homology modeling of membrane proteins to distantly related templates (Holyoake et al., 2006). Although the ADP-bound form of Sav1866 (PDB accession 2HYD) was used as a template for modeling, a subsequent, lower-resolution structure of the transporter complexed to AMP-PNP indicates that the template structure represents the ATP-bound (outward-facing) conformation (Dawson and Locher, 2007). Because the NBDs of subfamily D ABC transporters face the cytoplasm (Contreras et al., 1996), in the CTS model the central cavity faces the peroxisomal lumen (Figure 7A).
Figure 7.
Homology model of CTS. CTS was modeled on the structure of the bacterial multidrug transporter Sav1866 complexed with ADP (PDB accession 2HYD). AMP-PNP has been shown within the NBDs at positions equivalent to those found in the AMPPNP-bound form of Sav1866 (PDB accession 2ONJ), to illustrate the putative location of bound nucleotide. (A) Cartoon representation of the complete model, showing the N- and C-terminal halves of the CTS sequence in green and blue, respectively, and space-filling representations of AMP-PNP. (B) NBD1 (green), showing AMP-PNP and the locations of residues subjected to SDM. Part of the cytoplasmic loop connecting TM10 and TM11, including the CH2 region, is shown in blue. (C) NBD2 (blue), showing AMP-PNP and the locations of residues subjected to SDM. Part of the cytoplasmic loop connecting TM4 and TM5, including the CH2 region, is shown in green. (D) NBD1 (green), showing AMP-PNP and the locations of TILLING mutants and additional residues that may contribute to the phenotype of the E617K mutation. The α-carbon of G503 is shown in space-filling representation. Part of the cytoplasmic loop connecting TM10 and TM11, including the CH2 region, is shown in blue.
Active Site Mutants
The locations of residues subjected to site-directed mutagenesis in NBD1 are shown in Figure 7B. The severe phenotype resulting from mutation of K487 to alanine is consistent with a considerable body of literature supporting roles for this highly conserved residue, located in the P-loop/Walker A motif, in binding the β- and γ-phosphates of ATP and thus in ATP hydrolysis (Schneider and Hunke, 1998; Frelet and Klein, 2006). Similarly, even the relatively conservative mutation of D606 to asparagine abolished function. This highly conserved residue, located in the β-strand that contains the Walker B motif, has a predicted role in Mg2+ binding, and its mutation in other ABC transporters has typically been found to impair function severely (Frelet and Klein, 2006). The residues mutated in CTS NBD2, K1136, and D1276 occupy positions in the model analogous to their counterparts in NBD1 (Figure 7C) and would be expected to play similar roles in the catalytic cycle. The relative lack of phenotype seen when these residues are mutated is consistent with a functional difference between the two NBDs.
Coupling Helix 2 Mutants
The “EAA motif” residues E300, and E954, which are absolutely conserved in all 85 ABCD sequences investigated in the present study, reside in the putative CH2 regions (Dawson and Locher, 2006) linking TMs 4 and 5, and 10 and 11, in the N- and C-terminal TMDs of CTS, respectively. In the homology model of CTS, the side chains of these residues lie within ∼3 Å of R1264 in NBD2 and R594 in NBD1, respectively, and are thus predicted to be involved in salt-bridge interactions (Figures 7, B and C). The likely importance of these putative interactions is supported by the fact that these arginine residues, which are located a few residues downstream of the signature motif, are completely conserved in the 85 ABC subfamily D sequences examined. The lack of phenotypic effect of E300D and the relatively mild phenotype of E954D (intermediate resistance to 2,4-DB and IBA; Figure 5, E and F) may reflect preservation of the interaction in the mutants by virtue of the conservative nature of the mutation. It is noteworthy that a phenotype was only observed for the TMD2 EAA motif, predicted to contact NBD1, again pointing to the functional significance of this NBD.
TILLING Mutants with Strong Phenotypes
CTS residue G503 is very highly conserved in ABC transporter subfamily D, and the corresponding position is also almost completely conserved in the 148 homologues examined of Sav1866, where it is G396. In the model of CTS, this residue lies within a beta strand (see Figure 7D) and its mutation to glutamate is predicted to result in clashes with the adjacent β-strands for all possible side-chain rotamers. Consequent disruption of this β-sheet network is probably responsible for the strong phenotype of the G503E mutant. CTS residue P539 is absolutely conserved in ABC transporter subfamily D, and the corresponding position is also predominantly occupied by a small residue in homologues of Sav1866, where it is glycine (G438). In the model of CTS it lies at the C-terminal end of the helix that follows the Q-loop and its main function may be to allow the subsequent turn and thus maintain structural integrity of the NBD (Figure 7D). Interestingly this residue is adjacent to L540, which corresponds to F508 of the cystic fibrosis transmembrane conductance regulator (CFTR). Deletion of F508 (a mutation responsible for two thirds of cystic fibrosis disease cases) causes the protein to be incompletely folded and retained in the endoplasmic reticulum (ER; Du et al., 2005; Gadsby et al., 2006).
Tilling Mutants with Intermediate Phenotypes
Mutation of E617 to lysine yielded a mild phenotype (intermediate resistance to auxin precursors; Figures 5, G and H, and 6H). This residue, located between the D and H-loops of NBD1, is absolutely conserved in ABC transporter subfamily D, and the corresponding position in the Sav1866 homologues examined in the present study is similarly almost always occupied by glutamate. In the model of CTS, the E617 side chain lies within 3.5 Å of the almost absolutely conserved H-loop residue R637, with which it is predicted to form a salt bridge (Figure 7D). The corresponding conserved glutamate and arginine residues in Sav1866 also interact in this manner. Loss of the salt bridge in the CTS E617K mutant might affect activity via an effect on the conserved H-loop residue H636, which is adjacent to the arginine. The C631Y mutant also showed intermediate resistance to 2,4-DB and IBA (Figures 5, G and H, and 6H) and additionally exhibited a slower rate of germination (Figure 2C). The origin of such a phenotype is not obvious from the CTS model: although the corresponding position, in the β-strand that precedes the H-loop (Figure 7D), is occupied by leucine in the majority of ABC transporter subfamily D members, phenylalanine or tyrosine are found at this location in a number of the CTS homologues.
TILLING Mutants with No Detectable Mutant Phenotype
The TILLING mutants L514F, D568N, and S657N had no detectable mutant phenotype under the conditions tested in this study. In agreement with this, none of these residues is highly conserved in ABC transporter subfamily D, and their locations in the model suggest that mutation is unlikely to interfere with function. L514F is located in a short loop at the periphery of the NBD, linking the Walker A/P-loop and the Q-loop (see Figure 7D). D568 is located at the end of a helix at the periphery of NBD1 and does not appear to be in a position to form any significant interactions (see Figure 7D); similarly, S657 is located in a strand on the periphery of the NBD (see Figure 7D).
DISCUSSION
The aim of this study was to probe the structure-function relations of CTS and to determine whether the different physiological functions of this ABC transporter could be separated by mutagenesis. Analysis of preexisting and novel mutant alleles allowed us to reach several major conclusions.
CTS Mis-sense Mutants Produce Stable Protein
All TILLING and site-directed mutants expressed full-length immunoreactive CTS at close to wild-type levels, in marked contrast to ALDP, in which about 70% of mutants, including several that correspond to the cts alleles generated in this study, do not produce immunoreactive protein at the peroxisomal membrane (Supplementary Table S1; Watkins et al., 1995; Feigenbaum et al., 1996; Kemp et al., 2001). Intriguingly, this implies that there are different mechanisms for sensing mutant peroxisomal membrane proteins in humans and plants and/or that such proteins with altered functionality have different fates in the cell. This is surprising, because many features of peroxisomal protein targeting appear to be conserved between eukaryotes (Halbach et al., 2005; Landgraf et al., 2003; Van Ael and Fransen, 2006). The apparent stability and correct targeting of mutant proteins potentially makes CTS an attractive model for the structure-function analysis of subfamily D transporters, which may provide useful insights into ALDP. For example, if X-ALD–related mutations can be shown to have benign effects on function in CTS or in ALDP expressed in planta, this may point to the utility of therapeutic chaperones for ALDP, a strategy used for CFTR mutations that result in protein mistargeting (Loo et al., 2005).
The Two Nucleotide-binding Sites of CTS Are Not Functionally Equivalent
A striking result from this study is the nonequivalence of the two NBDs as revealed by mutagenesis. Mutation of the lysine of the Walker A motif of NBD1 and the conserved aspartate of the Walker B motif of NBD1 led to a complete loss-of-function phenotype. Surprisingly, the equivalent mutations in NBD2 led to a wild-type phenotype (the modest auxin precursor resistance of D1276N may reflect the low level of cts protein in this allele; Figure 1). This is the first report of functional asymmetry in a plant ABC transporter protein and is the first example in the ABCD subfamily.
A further indication of the different functional roles of CTS-NBD1 and NBD2 is supplied by the EAA motif mutants. Dawson et al. (2007) propose that the structural arrangement of the TMD-NBD “transmission interface” is a common feature of ABC transporters. In the prokaryotic MDR transporter, Sav1866, the coupling helices are domain-swapped, i.e., the coupling helix of one subunit contacts the NBD of the other (Dawson and Locher, 2006), and cross-linking studies have indicated that a similar arrangement exists in the homologous human ABC transporter, P-glycoprotein (Zolnerciks et al., 2007). Our mutagenesis data and structural model are also consistent with domain-swapping for CTS: the effect of mutating the EAA motif in TMD1 (E300D, which contacts NBD2) was indistinguishable from wild type, whereas the equivalent TMD2 mutation (E954D) resulted in an auxin precursor resistance phenotype, in agreement with an interaction between NBD1 and TMD2 and the functional importance of NBD1 versus NBD2.
The ATP-binding site of ABC transporters is composed of the Walker A, Walker B, and H-loops of one NBD and the signature motif of the other. Functional asymmetry has been reported for several eukaryotic ABC transporters (TAP; Karttunen et al., 2001; CFTR, Aleksandrov et al., 2002; Pdr5, Ernst et al., 2008; MRP, Gao et al., 2000; Hou et al., 2000) and has been suggested to result from substitution of conserved residues within these motifs, resulting in one composite consensus site and one composite degenerate site (Ernst et al., 2006; Procko et al., 2006). Although both NBDs of CTS have a consensus signature motif (LSLGE), the histidine residue of the H-loop, which has been suggested to act as the linchpin for catalysis (Zaitseva et al., 2005) is replaced by glutamine in NBD2. Thus mutations in Walker A or B of NBD1 will result in both hybrid ATP-binding sites being degenerate, whereas mutations in the Walker A or B of NBD2 would still allow one consensus site (Figure 8). A corresponding H-to-Q substitution is present in the NBD of human TAP1, the N-terminal NBD of CFTR contains a serine in this position and the N-terminal NBD1 of Pdr5 has tyrosine (Figure 8B; Ernst et al., 2006, 2008; Procko et al., 2006). Reintroduction of a His residue (in the presence of the Walker B catalytic carboxylate residue, also present in CTS NBD2) increased ATPase activity dramatically in the isolated TAP1 NBD homodimer (Ernst et al., 2006; Procko et al., 2006), but when the linchpin His was mutated to Q in the consensus site of the full-length TAP, only a small defect of class I loading was seen in vivo (Procko et al., 2006). Similarly, when the corresponding His of Pdr5 NBD2 was mutated to A, only rhodamine transport was affected, and the effect on ATPase activity of the full-length protein was minor (Ernst et al., 2008). In summary, although the presence of substitutions within conserved residues of the ATP-binding site is an attractive explanation of functional asymmetry, the available data suggest this may be too simplistic a view. In both CFTR and MRP, nucleotide is bound but turned over very slowly at the degenerate site whereas nucleotide hydrolysis is rapid at the consensus site. However, both NBDs are required for function in these asymmetric ABCs (Gadsby et al., 2006; Gao et al., 2000; Hou et al., 2000; Vergani et al., 2005), in contrast to CTS, in which canonical Walker motif residues in NBD2 appear to be dispensable for function. A number of roles have been proposed for the degenerate site of asymmetric ABC transporters (Alberts et al., 2001; Gadsby et al., 2006), but the fact that the NBD2 composite nucleotide binding site is not crucial in planta indicates that the CTS degenerate site does not have additional accessory roles in the native setting.
Figure 8.
Cartoon showing amino acid composition of CTS ATP-binding sites. (A) Schematic diagrams of CTS-NBD dimers for wild type and the D606N and D1276N mutants. NBD1 (white) and NBD2 (gray) form two ATP-binding sites (black). The upper ATP-binding site has the consensus sequence, whereas the lower site is degenerate (Q instead of H in H-loop). After Procko et al. (2006). (B) Alignment of conserved NBD motifs of AtCTS, human TAP1/2, CFTR, and Pdr5. Amino acids shown in A are shaded in color. Asterisks mark the amino acids changed by SDM in CTS.
Sucrose Potentiates the Inhibitory Effect of IBA on Hypocotyl Elongation
An interesting and novel observation is that the inhibitory effect of IBA (but not IAA) on hypocotyl elongation is potentiated by sucrose (Figure 6, A–D). Because inclusion of sucrose in the media reduces the rate of TAG breakdown during seedling establishment (Fulda et al., 2004), a plausible explanation is that fatty acids and IBA compete with one another for transport via CTS into the peroxisome for β-oxidation. Thus, in the presence of sucrose, the flux of fatty acids through CTS would be reduced, allowing an increase in the rate of IBA transport and subsequent conversion to IAA.
Mutants Separate Different Functions of CTS
A detailed side-by-side characterization revealed three classes of mutants: 1) effectively null (i.e., nonfunctional in all tests), 2) indistinguishable from wild type, and 3) differentially affected in several of the known functions of CTS. Two novel NBD1 mutants generated by TILLING (G503E and P539L) abolished function in all assays tested, underscoring the importance of NBD1 for all functions. Of the third category, the NBD1 mutants E617K and C631Y exhibited auxin precursor resistance, but were essentially wild type for all other functions tested. One interpretation of this finding is that substrate binding and/or translocation is affected in these mutants, i.e., they are competent in the β-oxidation of fatty acids but not 2,4-DB or IBA. The observation that both these mutants retain an appreciable resistance to 100 μM IBA, even in the presence of sucrose (Figure 6H), yet are still able to degrade their oil bodies (Figure 3D) supports this interpretation. Transport experiments with purified and reconstituted protein would be necessary to determine unequivocally whether this is the case. The current consensus is that the substrate binding sites of ABC transporters are located in the TMDs (e.g., Martin et al., 2000; Karwatsky and Georges, 2004; Ambudkar et al., 2006; Herget et al., 2007). However, mis-sense mutations in NBDs of P-glycoprotein, Pdr5, and the prokaryotic hemolysin transporter, HlyB, have been shown to affect substrate specificity, probably by affecting transduction to the TMDs (Beaudet and Gros, 1995; Sheps et al., 1995; Egner et al., 1998). An alternative possibility is that these mutants have partial activity, or in the case of D1276N, low expression, therefore reducing the rate of transport of auxin precursors into peroxisomes. Dependent on the threshold level of 2,4-DB or IBA conversion required to generate an effect on root (or hypocotyl) elongation, even partial loss of activity may be sufficient to generate a resistance phenotype. The observation that heterozygous CTS/cts plants show resistance to 2,4-DB and IBA but can elongate hypocotyls in the dark in the absence of sucrose supports this hypothesis (Hayashi et al., 1998; Zolman et al., 2001).
Another residue potentially affecting substrate specificity is S810N, located in helix 2 of TMD2 (helix 8 in the whole molecule). This mutant (ped3-4) germinates but fails to establish (Hayashi et al., 2002), which is intriguing because it is consistent with the notion that the function of CTS in germination is β-oxidation of an as-yet unknown substrate and that mobilization of lipids is not required for germination (Pinfield-Wells et al., 2005; Baker et al., 2006; Footitt et al., 2006). However it is possible that β-oxidation of TAG-derived fatty acids is required for both germination and establishment, but that the energetic threshold for establishment is much higher than for germination. As argued above for auxin precursor resistance, the S810N mutation may simply reduce β-oxidation of fatty acids by a subtle alteration in transport kinetics such that the two physiological processes are differentially affected. Indeed the two explanations, altered substrate specificity and altered kinetics may not be mutually exclusive, because Ernst et al. (2008) have proposed that substrate selection by the yeast multidrug transporter Pdr5 may be determined by both structural features and kinetics. In vivo experiments are unable to distinguish clearly between these possibilities and understanding the kinetic effects of the mutations ultimately awaits the purification and reconstitution of CTS for transport studies.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Arabidopsis TILLING project for carrying out the original TILLING screen, Dr. Makoto Hayashi (NIBB, Okazaki, Japan) for seeds of the ped3 allelic series, and Professor Bonnie Bartel (Rice University, Texas) for pxa1-1. The authors are grateful to Julietta Marquez, Barbara Johnson, and Fiona Moulton for expert technical assistance; Dr. Gareth Howell and Dr. Laura-Anne Brown for advice on the confocal imaging; Dr. Salvador Gezan for statistical advice; and Dr. Stephan Kemp for permission to cite unpublished data from http://www.x-ald.nl. This work was supported by the Biotechnology and Biological Sciences Research Council (Grants P19769 and P19770). H.S. was funded by a postdoctoral fellowship from the University of Nottingham. R. Res. receives grant-aided support from the Biotechnology and Biological Sciences Research Council of the United Kingdom.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-07-0745) on November 19, 2008.
REFERENCES
- Alberts P., Daumke O., Deverson E. V., Howard J. C., Knittler M. R. Distinct functional properties of the TAP subunits coordinate the nucleotide-dependent transport cycle. Curr. Biol. 2001;11:242–251. doi: 10.1016/s0960-9822(01)00073-2. [DOI] [PubMed] [Google Scholar]
- Aleksandrov L., Aleksandrov A. A., Chang X. B., Riordan J. R. The first nucleotide binding domain of cystic fibrosis transmembrane conductance regulator is a site of stable nucleotide interaction, whereas the second is a site of rapid turnover. J. Biol. Chem. 2002;277:15419–15425. doi: 10.1074/jbc.M111713200. [DOI] [PubMed] [Google Scholar]
- Ambudkar S. V., Kim I. W., Sauna Z. E. The power of the pump: mechanisms of action of P-glycoprotein (ABCB1) Eur. J. Pharm. Sci. 2006;27:392–400. doi: 10.1016/j.ejps.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Baker A., Graham I. A., Holdsworth M., Smith S. M., Theodoulou F. L. Chewing the fat: β-oxidation in signalling and development. Trends Plant Sci. 2006;11:124–132. doi: 10.1016/j.tplants.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Beaudet L., Gros P. Functional dissection of P-glycoprotein nucleotide-binding domains in chimeric and mutant proteins—modulation of drug-resistance profiles. J. Biol. Chem. 1995;270:17159–17170. doi: 10.1074/jbc.270.29.17159. [DOI] [PubMed] [Google Scholar]
- Berezin C., Glaser F., Rosenberg J., Paz I., Pupko T., Fariselli P., Casadio R., Ben-Tal N. ConSeq: the identification of functionally and structurally important residues in protein sequences. Bioinformatics. 2004;20:1322–1324. doi: 10.1093/bioinformatics/bth070. [DOI] [PubMed] [Google Scholar]
- Berger J., Gärtner J. X-linked adrenoleukodystrophy: clinical, biochemical and pathogenetic aspects. Biochim. Biophys. Acta Mol. Cell Res. 2006;1763:1721–1732. doi: 10.1016/j.bbamcr.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Bewley J. D. Seed germination and dormancy. Plant Cell. 1997;9:1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras M., Sengupta T. K., Sheikh F., Aubourg P., Singh I. Topology of ATP-binding domain of adrenoleukodystrophy gene product in peroxisomes. Arch. Biochem. Biophys. 1996;334:369–379. doi: 10.1006/abbi.1996.0467. [DOI] [PubMed] [Google Scholar]
- Daus M. L., Grote M., Muller P., Doebber M., Herrmann A., Steinhoff H. J., Dassa E., Schneider E. ATP-driven Malk dimer closure and reopening and conformational changes of the “EAA” motifs are crucial for function of the maltose ATP-binding cassette transporter (MalFGK(2)) J. Biol. Chem. 2007;282:22387–22396. doi: 10.1074/jbc.M701979200. [DOI] [PubMed] [Google Scholar]
- Dawson R.J.P., Hollenstein K., Locher K. P. Uptake or extrusion: crystal structures of full ABC transporters suggest a common mechanism. Mol. Microbiol. 2007;65:250–257. doi: 10.1111/j.1365-2958.2007.05792.x. [DOI] [PubMed] [Google Scholar]
- Dawson R.J.P., Locher K. P. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443:180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- Dawson R.J.P., Locher K. P. Structure of the multidrug ABC transporter Sav1866 from Staphylococcus aureus in complex with AMP-PNP. FEBS Lett. 2007;581:935–938. doi: 10.1016/j.febslet.2007.01.073. [DOI] [PubMed] [Google Scholar]
- Dean M., Annilo T. Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu. Rev. Genomics Hum. Genet. 2005;6:123–142. doi: 10.1146/annurev.genom.6.080604.162122. [DOI] [PubMed] [Google Scholar]
- Dean M., Rzhetsky A., Allikmets R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 2001;11:1156–1166. doi: 10.1101/gr.184901. [DOI] [PubMed] [Google Scholar]
- Du K., Sharma M., Lukacs G. L. The ΔF508 cystic fibrosis mutation impairs domain–domain interactions and arrests post-translational folding of CFTR. Nat. Struct. Mol. Biol. 2005;12:17–25. doi: 10.1038/nsmb882. [DOI] [PubMed] [Google Scholar]
- Eastmond P. J. SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell. 2006;18:665–675. doi: 10.1105/tpc.105.040543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egner R., Rosenthal F. E., Kralli A., Sanglard D., Kuchler K. Genetic separation of FK506 susceptibility and drug transport in the yeast Pdr5 ATP-binding cassette multidrug resistance transporter. Mol. Biol. Cell. 1998;9:523–543. doi: 10.1091/mbc.9.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst R., Keuppers P., Klein C. M., Schwarzmueller T., Kuchler K., Schmitt L. A mutation of the H-loop selectively affects rhodamine transport by the yeast multidrug ABC transporter Pdr5. Proc. Natl. Acad. Sci. USA. 2008;105:5069–5074. doi: 10.1073/pnas.0800191105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst R., Koch J., Horn C., Tampe R., Schmitt L. Engineering ATPase activity in the isolated ABC cassette of human TAP1. J. Biol. Chem. 2006;281:27471–27480. doi: 10.1074/jbc.M601131200. [DOI] [PubMed] [Google Scholar]
- Feigenbaum V., LombardPlatet G., Guidoux S., Sarde C. O., Mandel J. L., Aubourg P. Mutational and protein analysis of patients and heterozygous women with X-linked adrenoleukodystrophy. Am J. Hum. Genet. 1996;58:1135–1144. [PMC free article] [PubMed] [Google Scholar]
- Fiser A. S., Sali A. MODELLER: generation and refinement of homology-based protein structure models. Methods Enzymol. 2003;374:461–491. doi: 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed] [Google Scholar]
- Footitt S., Dietrich D., Fait A., Fernie A. R., Holdsworth M. J., Baker A., Theodoulou F. L. The COMATOSE ATP-binding cassette transporter is required for full fertility in Arabidopsis. Plant Physiol. 2007;144:1467–1480. doi: 10.1104/pp.107.099903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Footitt S., Marquez J., Schmuths H., Baker A., Theodoulou F. L., Holdsworth M. Analysis of the role of COMATOSE and peroxisomal beta-oxidation in the determination of germination potential in Arabidopsis. J. Exp. Bot. 2006;57:2805–2814. doi: 10.1093/jxb/erl045. [DOI] [PubMed] [Google Scholar]
- Footitt S., Slocombe S. P., Larner V., Kurup S., Wu Y. S., Larson T., Graham I., Baker A., Holdsworth M. Control of germination and lipid mobilization by COMATOSE, the Arabidopsis homologue of human ALDP. EMBO J. 2002;21:2912–2922. doi: 10.1093/emboj/cdf300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frelet A., Klein M. Insight in eukaryotic ABC transporter function by mutation analysis. FEBS Lett. 2006;580:1064–1084. doi: 10.1016/j.febslet.2006.01.024. [DOI] [PubMed] [Google Scholar]
- Fulda M., Schnurr J., Abbadi A., Heinz E., Browse J. Peroxisomal Acyl-CoA synthetase activity is essential for seedling development in Arabidopsis thaliana. Plant Cell. 2004;16:394–405. doi: 10.1105/tpc.019646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadsby D. C., Vergani P., Csanady L. The ABC protein turned chloride channel whose failure causes cystic fibrosis. Nature. 2006;440:477–483. doi: 10.1038/nature04712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Cui H. R., Loe D. W., Grant C. E., Almquist K. C., Cole S.P.C., Deeley R. G. Comparison of the functional characteristics of the nucleotide binding domains of multidrug resistance protein 1. J. Biol. Chem. 2000;275:13098–13108. doi: 10.1074/jbc.275.17.13098. [DOI] [PubMed] [Google Scholar]
- Gaudet R., Wiley D. C. Structure of the ABC ATPase domain of human TAP1, the transporter associated with antigen processing. EMBO J. 2001;20:4964–4972. doi: 10.1093/emboj/20.17.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbach A., Lorenzen S., Landgraf C., Volkmer-Engert R., Erdmann R., Rottensteiner H. Function of the PEX19-binding site of human adrenoleukodystrophy protein as targeting motif in man and yeast—PMP targeting is evolutionarily conserved. J. Biol. Chem., 2005;280:21176–21182. doi: 10.1074/jbc.M501750200. [DOI] [PubMed] [Google Scholar]
- Hayashi M., Nito K., Takei-Hoshi R., Yagi M., Kondo M., Suenaga A., Yamaya T., Nishimura M. Ped3p is a peroxisomal ATP-binding cassette transporter that might supply substrates for fatty acid beta-oxidation. Plant Cell Physiol, 2002;43:1–11. doi: 10.1093/pcp/pcf023. [DOI] [PubMed] [Google Scholar]
- Hayashi M., Toriyama K., Kondo M., Nishimura M. 2,4-dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid beta-oxidation. Plant Cell. 1998;10:183–195. doi: 10.1105/tpc.10.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herget M., Oancea G., Schrodt S., Karas M., Tampé R., Abele R. Mechanism of substrate sensing and signal transmission within an ABC transporter—use of a trojan horse strategy. J. Biol. Chem. 2007;282:3871–3880. doi: 10.1074/jbc.M608480200. [DOI] [PubMed] [Google Scholar]
- Hettema E. H., vanRoermund C.W.T., Distel B., vandenBerg M., Vilela C., RodriguesPousada C., Wanders R.J.A., Tabak H. F. The ABC transporter proteins Pat1 and Pat2 are required for import of long-chain fatty acids into peroxisomes of. Saccharomyces cerevisiae. EMBO J. 1996;15:3813–3822. [PMC free article] [PubMed] [Google Scholar]
- Higgins C. F. ABC transporters—from microorganisms to man. Annu. Rev. Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- Higgins C. F., Linton K. J. The ATP switch model for ABC transporters. Nat. Struct. Mol. Biol. 2004;11:918–926. doi: 10.1038/nsmb836. [DOI] [PubMed] [Google Scholar]
- Holyoake J., Caulfeild V., Baldwin S. A., Sansom M.S.P. Modeling, docking, and simulation of the major facilitator superfamily. Biophys. J. 2006;91:L84–L86. doi: 10.1529/biophysj.106.093971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooks M. A., Turner J. E., Murphy E. C., Johnston K. A., Burr S., Jaroslawski S. The Arabidopsis ALDP protein homologue COMATOSE is instrumental in peroxisomal acetate metabolism. Biochem. J. 2007;406:399–406. doi: 10.1042/BJ20070258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner K. P., Karcher A., Shin D. S., Craig L., Arthur L. M., Carney J. P., Tainer J. A. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- Hou Y. X., Cui L. Y., Riordan J. R., Chang X. B. Allosteric interactions between the two non-equivalent nucleotide binding domains of multidrug resistance protein MRP1. J. Biol. Chem. 2000;275:20280–20287. doi: 10.1074/jbc.M001109200. [DOI] [PubMed] [Google Scholar]
- Hung L. W., Wang I.X.Y., Nikaido K., Liu P. Q., Ames G.F.L., Kim S. H. Crystal structure of the ATP-binding subunit of an ABC transporter. Nature. 1998;396:703–707. doi: 10.1038/25393. [DOI] [PubMed] [Google Scholar]
- Karttunen J. T., Lehner P. J., Gupta S. S., Hewitt E. W., Cresswell P. Distinct functions and cooperative interaction of the subunits of the transporter associated with antigen processing (TAP) Proc. Natl. Acad. Sci. USA. 2001;98:7431–7436. doi: 10.1073/pnas.121180198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karwatsky J. M., Georges E. Drug binding domains of MRP1 (ABCC1) as revealed by photoaffinity labeling. Curr. Medic. Chem. Anticancer Agents. 2004;4:19–30. doi: 10.2174/1568011043482133. [DOI] [PubMed] [Google Scholar]
- Kemp S., Pujol A., Waterham H. R., van Geel B. M., Boehm C. D., Raymond G. V., Cutting G. R., Wanders R.J.A., Hugo H. W. ABCD1 mutations and the X-linked adrenoleukodystrophy mutation database: role in diagnosis and clinical correlations. Hum. Mutat. 2001;18:499–515. doi: 10.1002/humu.1227. [DOI] [PubMed] [Google Scholar]
- Kemp S., Wanders R.J.A. X-linked adrenoleukodystrophy: very long-chain fatty acid metabolism, ABC half-transporters and the complicated route to treatment. Mol. Genet. Metab. 2007;90:268–276. doi: 10.1016/j.ymgme.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Kerppola R. E., Ames G. F. Topology of the hydrophobic membrane-bound components of the histidine periplasmic permease—comparison with other members of the family. J. Biol. Chem. 1992;267:2329–2336. [PubMed] [Google Scholar]
- Kimura K., Yamaoka M., Kamisaka Y. Rapid estimation of lipids in oleaginous fungi and yeasts using Nile red fluorescence. J. Microbiol. Methods. 2004;56:331–338. doi: 10.1016/j.mimet.2003.10.018. [DOI] [PubMed] [Google Scholar]
- Landgraf P., Mayerhofer P. U., Polanetz R., Roscher A. A., Holzinger A. Targeting of the human adrenoleukodystrophy protein to the peroxisomal membrane by an internal region containing a highly conserved motif. Eur. J. Cell Biol. 2003;82:401–410. doi: 10.1078/0171-9335-00331. [DOI] [PubMed] [Google Scholar]
- Laskowski R. A., Macarthur M. W., Moss D. S., Thornton J. M. PROCHECK—a program to check the stereochemical quality of protein structures. J. Appl. Crystallog. 1993;26:283–291. [Google Scholar]
- Linton K. J., Higgins C. F. Structure and function of ABC transporters: the ATP switch provides flexible control. Pfluegers Arch. 2007;453:555–567. doi: 10.1007/s00424-006-0126-x. [DOI] [PubMed] [Google Scholar]
- Loo T. W., Bartlett M. C., Clarke D. M. Rescue of folding defects in ABC transporters using pharmacological chaperones. J. Bioenerg. Biomembr. 2005;37:501–507. doi: 10.1007/s10863-005-9499-3. [DOI] [PubMed] [Google Scholar]
- Martin C., Berridge G., Higgins C. F., Mistry P., Charlton P., Callaghan R. Communication between multiple drug binding sites on P-glycoprotein. Mol. Pharmacol. 2000;58:624–632. doi: 10.1124/mol.58.3.624. [DOI] [PubMed] [Google Scholar]
- Moody J. E., Millen L., Binns D., Hunt J. F., Thomas P. J. Cooperative, ATP-dependent association of the nucleotide binding cassettes during the catalytic cycle of ATP-binding cassette transporters. J. Biol. Chem. 2002;277:21111–21114. doi: 10.1074/jbc.C200228200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourez M., Hofnung N., Dassa E. Subunit interactions in ABC transporters: A conserved sequence in hydrophobic membrane proteins of periplasmic permeases defines an important site of interaction with the ATPase subunits. EMBO J. 1997;16:3066–3077. doi: 10.1093/emboj/16.11.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinfield-Wells H., Rylott E. L., Gilday A. D., Graham S., Job K., Larson T. R., Graham I. A. Sucrose rescues seedling establishment but not germination of Arabidopsis mutants disrupted in peroxisomal fatty acid catabolism. Plant J. 2005;43:861–872. doi: 10.1111/j.1365-313X.2005.02498.x. [DOI] [PubMed] [Google Scholar]
- Pracharoenwattana I., Cornah J. E., Smith S. M. Arabidopsis peroxisomal citrate synthase is required for fatty acid respiration and seed germination. Plant Cell. 2005;17:2037–2048. doi: 10.1105/tpc.105.031856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko E., Ferrin-O'Connell I., Ng S. L., Gaudet R. Distinct structural and functional properties of the ATPase sites in an asymmetric ABC transporter. Mol. Cell. 2006;24:51–62. doi: 10.1016/j.molcel.2006.07.034. [DOI] [PubMed] [Google Scholar]
- Rai B. K., Fiser A. Multiple mapping method: a novel approach to the sequence-to-structure alignment problem in comparative protein structure modeling. Proteins Struct. Funct. Bioinform. 2006;63:644–661. doi: 10.1002/prot.20835. [DOI] [PubMed] [Google Scholar]
- Rashotte A. M., Poupart J., Waddell C. S., Muday G. K. Transport of the two natural auxins, indole-3-butyric acid and indole-3-acetic acid, in Arabidopsis. Plant Physiol. 2003;133:761–772. doi: 10.1104/pp.103.022582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea P. A. Plant ATP-Binding cassette transporters. Annu. Rev. Plant Biol. 2007;58:347–375. doi: 10.1146/annurev.arplant.57.032905.105406. [DOI] [PubMed] [Google Scholar]
- Rensing S. A., et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- Russell L., Larner V., Kurup S., Bougourd S., Holdsworth M. The Arabidopsis COMATOSE locus regulates germination potential. Development. 2000;127:3759–3767. doi: 10.1242/dev.127.17.3759. [DOI] [PubMed] [Google Scholar]
- Schaller F., Schaller A., Stintzi A. Biosynthesis and metabolism of jasmonates. J. Plant Growth Regul. 2005;23:179–199. [Google Scholar]
- Schneider E., Hunke S. ATP-binding-cassette (ABC) transport systems: Functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol. Rev. 1998;22:1–20. doi: 10.1111/j.1574-6976.1998.tb00358.x. [DOI] [PubMed] [Google Scholar]
- Shani N., Valle D. A Saccharomyces cerevisiae homolog of the human adrenoleukodystrophy transporter is a heterodimer of two half ATP-binding cassette transporters. Proc. Natl. Acad. Sci. USA. 1996;93:11901–11906. doi: 10.1073/pnas.93.21.11901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheps J. A., Cheung I., Ling V. Hemolysin transport in Escherichia coli- point mutants in HlyB compensate for a deletion in the predicted amphiphilic helix region of the HlyA signal. J. Biol. Chem. 1995;270:14829–14834. doi: 10.1074/jbc.270.24.14829. [DOI] [PubMed] [Google Scholar]
- Smith P. C., Karpowich N., Millen L., Moody J. E., Rosen J., Thomas P. J., Hunt J. F. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol. Cell. 2002;10:139–149. doi: 10.1016/s1097-2765(02)00576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I. A., Hawes C., Baker A. AtPEX2 and AtPEX10 are targeted to peroxisomes independently of known endoplasmic reticulum trafficking routes. Plant Physiol. 2005;139:690–700. doi: 10.1104/pp.105.065094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I. A., Runions J., Kearns A., Hawes C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006;1:2019–2025. doi: 10.1038/nprot.2006.286. [DOI] [PubMed] [Google Scholar]
- Teale W. D., Paponov I. A., Palme K. Auxin in action: signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 2006;7:847–859. doi: 10.1038/nrm2020. [DOI] [PubMed] [Google Scholar]
- Teodorescu O., Galor T., Pillardy J., Elber R. Enriching the sequence substitution matrix by structural information. Proteins Struct. Funct. Bioinform. 2004;54:41–48. doi: 10.1002/prot.10474. [DOI] [PubMed] [Google Scholar]
- Theodoulou F. L., Holdsworth M., Baker A. Peroxisomal ABC transporters. FEBS Lett. 2006;580:1139–1155. doi: 10.1016/j.febslet.2005.12.095. [DOI] [PubMed] [Google Scholar]
- Theodoulou F. L., Job K., Slocombe S. P., Footitt S., Holdsworth M., Baker A., Larson T. R., Graham I. A. Jasmonoic acid levels are reduced in COMATOSE ATP-binding cassette transporter mutants. Implications for transport of jasmonate precursors into peroxisomes. Plant Physiol. 2005;137:835–840. doi: 10.1104/pp.105.059352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till B. J., et al. Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res. 2003;13:524–530. doi: 10.1101/gr.977903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ael E., Fransen M. Targeting signals in peroxisomal membrane proteins. Biochim. Biophys. Acta Mol. Cell Res. 2006;1763:1629–1638. doi: 10.1016/j.bbamcr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Van Roermund C.W.T., Visser W., Ijlst L., van Cruchten A., Boek M., Kulik W., Waterham H. R., Wanders R.J.A. The human peroxisomal ABC half transporter ALDP functions as a homodimer and accepts acyl-CoA esters. FASEB J. 2008;22:4201–4208. doi: 10.1096/fj.08-110866. [DOI] [PubMed] [Google Scholar]
- Vergani P., Lockless S. W., Nairn A. C., Gadsby D. C. CFTR channel opening by ATP-driven tight dimerization of its nucleotide-binding domains. Nature. 2005;433:876–880. doi: 10.1038/nature03313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verleur N., Hettema E. H., VanRoermund C.W.T., Tabak H. F., Wanders R.J.A. Transport of activated fatty acids by the peroxisomal ATP-binding-cassette transporter Pxa2 in a semi-intact yeast cell system. Eur. J. Biochem. 1997;249:657–661. doi: 10.1111/j.1432-1033.1997.00657.x. [DOI] [PubMed] [Google Scholar]
- Verrier P. J., et al. Plant ABC proteins—a unified nomenclature and updated inventory. Trends Plant Sci. 2008;13:151–159. doi: 10.1016/j.tplants.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. distantly related sequences in the alpha-subunits and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins P. A., Gould S. J., Smith M. A., Braiterman L. T., Wei H. M., Kok F., Moser A. B., Moser H. W., Smith K. D. Alterred expression of ALDP in X-linked adrenoleukodystrophy. Am. J. Hum. Genet. 1995;57:292–301. [PMC free article] [PubMed] [Google Scholar]
- Zaitseva J., Jenewein S., Jumpertz T., Holland I. B., Schmitt L. H662 is the linchpin of ATP hydrolysis in the nucleotide-binding domain of the ABC transporter HlyB. EMBO J. 2005;24:1901–1910. doi: 10.1038/sj.emboj.7600657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman B. K., Yoder A., Bartel B. Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics. 2000;156:1323–1337. doi: 10.1093/genetics/156.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman B. K., Silva I. D., Bartel B. The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid beta-oxidation. Plant Physiol. 2001;127:1266–1278. [PMC free article] [PubMed] [Google Scholar]
- Zolnerciks J. K., Wooding C., Linton K. J. Evidence for a Sav1866-like architecture for the human multidrug transporter P-glycoprotein. FASEB J. 2007;21:3937–3948. doi: 10.1096/fj.07-8610com. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.