Abstract
Mps1 is a protein kinase that plays essential roles in spindle checkpoint signaling. Unattached kinetochores or lack of tension triggers recruitment of several key spindle checkpoint proteins to the kinetochore, which delays anaphase onset until proper attachment or tension is reestablished. Mps1 acts upstream in the spindle checkpoint signaling cascade, and kinetochore targeting of Mps1 is required for subsequent recruitment of Mad1 and Mad2 to the kinetochore. The mechanisms that govern recruitment of Mps1 or other checkpoint proteins to the kinetochore upon spindle checkpoint activation are incompletely understood. Here, we demonstrate that phosphorylation of Mps1 at T12 and S15 is required for Mps1 recruitment to the kinetochore. Mps1 kinetochore recruitment requires its kinase activity and autophosphorylation at T12 and S15. Mutation of T12 and S15 severely impairs its kinetochore association and markedly reduces recruitment of Mad2 to the kinetochore. Our studies underscore the importance of Mps1 autophosphorylation in kinetochore targeting and spindle checkpoint signaling.
INTRODUCTION
Faithful segregation of chromosomes is essential for genome stability and organism development (Lengauer et al., 1997; Nicklas, 1997; Nasmyth, 2002). Aberrant chromosome segregation generates aneuploid cells, a hallmark frequently associated with cancer cells (Lengauer et al., 1997). It has been speculated that aneuploidy may be a driving force for cellular transformation. Aneuploidy is primarily caused by errors during mitosis. In normal cells, correct segregation of chromosomes is ensured by an evolutionarily conserved surveillance signal transduction pathway called the mitotic spindle checkpoint (McIntosh, 1991). Defects in chromosome separation elicit checkpoint signal(s) to delay the onset of anaphase until every chromosome has successfully attached to the spindles (Amon, 1999; Yu, 2002; Draviam et al., 2004; Weaver and Cleveland, 2005). The spindle checkpoint response is highly robust because a single unoccupied kinetochore is sufficient to cause mitotic arrest until proper attachment of microtubules is reestablished (Rieder et al., 1994).
The molecular components of the spindle checkpoint pathway were first identified in yeast through various genetic screens (Hoyt et al., 1991; Li and Murray, 1991; Weiss and Winey, 1996; Amon, 1999). Subsequent studies revealed that most of the key checkpoint proteins are conserved from yeast to vertebrate systems. Homologues of these components in mammalian cells include Bub1, BubR1, Bub3, Mad1, Mad2, and Mps1 (Wassmann and Benezra, 2001; Kops et al., 2005). Delay of mitotic progression upon triggering of the spindle checkpoint is apparently achieved by inhibition of the anaphase promoting complex/cyclosome (APC/C), an E3 ubiquitin ligase that is responsible for ubiquitination and degradation of securing and cyclin B (Nasmyth, 2001). Degradation of securin activates the separase protease, which removes the cohesion protein Scc1 from the held sister chromatids, allowing their subsequent separation in anaphase (Uhlmann et al., 1999; Nasmyth, 2001). The current paradigm for turning on spindle checkpoint signaling invokes production of diffusible inhibitors of CDC20, an activator and substrate specificity selector for APC/C (Yu, 2002). Inhibitors of the APC may include activated Mad2, BubR1, or Bub1 or a complex of Cdc20, Mad2, BubR1, and Bub3 (Fang et al., 1998; Sudakin et al., 2001; Tang et al., 2004). The inhibition is released upon proper attachment of kinetochores to the spindle, although the molecular mechanism(s) underlying extinguishment of the checkpoint signal remains to be elucidated.
Mps1 is among the several protein kinases implicated in transducing the checkpoint signal. Originally identified as a dual-specificity kinase whose levels are elevated in a variety of tumor cell lines (Mills et al., 1992; Lindberg et al., 1993), Mps1 seems to be an essential mitotic kinase that regulates normal mitotic progression, chromosome congression, and cytokinesis from yeast to vertebrate cells (Fisk et al., 2004; Jelluma et al., 2008b). In yeast, Mps1 is essential for spindle pole body duplication and has been implicated in centrosome duplication in mammalian cells (Winey et al., 1991; Fisk and Winey, 2001). Mps1 is distributed diffusely throughout the cell and relocates to kinetochores in early mitosis and upon activation of the spindle checkpoint (Stucke et al., 2002, 2004; Liu et al., 2003). The protein kinase activity of Mps1 is strongly elevated in mitosis and correlates with increased autophosphorylation of Mps1 (Stucke et al., 2002; Liu et al., 2003; Kang et al., 2007; Mattison et al., 2007). Indeed, autophosphorylation of Mps1 at T676 of the activation loop has been shown to contribute to the elevated kinase activity (Kang et al., 2007; Mattison et al., 2007). Whether autophosphorylation also regulates other aspects of Mps1 biology remains unknown.
A common feature shared by the checkpoint proteins is that they all localize to kinetochores upon activation of the spindle checkpoint. Recruitment of checkpoint proteins to kinetochores seems to be a hierarchical process (Martin-Lluesma et al., 2002; Vigneron et al., 2004). For example, kinetochore localization of Mad1 and Mad2 requires Mps1, and Mad2 kinetochore localization depends on Mad1 but not vice versa (Martin-Lluesma et al., 2002). This result is consistent with the notion that Mps1 functions upstream in the spindle checkpoint pathway (Abrieu et al., 2001). Kinetochore localization of Mps1 requires Hec1/Ndc80, a core component of the kinetochore outer plate essential for organizing microtubule attachment sites (DeLuca et al., 2005). Recently, PRP4, a serine-threonine kinase, also has been linked to kinetochore localization of Mps1, Mad1, and Mad2 (Montembault et al., 2007). In addition, other protein kinases also have been implicated in Mps1 kinetochore recruitment (Zhao and Chen, 2006; Montembault et al., 2007).
In this report, we investigate the functional relevance of Mps1 kinase activity and autophosphorylation in kinetochore localization of spindle checkpoint proteins. We found that the kinase activity of Mps1 is essential for its kinetochore recruitment and autophosphorylation of Mps1 at T12 and that S15 is necessary for kinetochore targeting and subsequent recruitment of other spindle checkpoint proteins.
MATERIALS AND METHODS
Cell Culture, Transfections, and Antibodies
SW480 and 293T cells were purchased from American Type Culture Collection (Manassas, VA) and were maintained at 37°C in a 5% CO2 atmosphere in DMEM supplemented with 10% fetal calf serum (Invitrogen, Carlsbad, CA), penicillin, streptomycin (100 IU/ml and 100 mg/ml, respectively), and l-glutamine. Mirus transfection reagent (Mirus, Madison, WI) was used for transfection of 293T cells and retroviral packaging. Small interfering RNA (siRNA) transfection was performed using SiLentFect (Bio-Rad, Hercules, CA). The 21-nucleotide RNA duplexes targeting Mps1 were purchased from Dharmacon RNA Technologies (Lafayette, CO). Antibodies against Mps1 (C19) or Mps1-NT were from Santa Cruz Biotechnology (Santa Cruz, CA) or Millipore (Billerica, MA). Anti-phospho-Smad2 was a gift of Drs. P. Ten Djike, C. Heldin, and A. Moustakas. Anti-Mad2 and CREST antibodies were purchased from Covance Research Products (Princeton, NJ) and Antibodies (Davis, CA), respectively.
DNA Manipulation and Stable Cell Line Generation
Mammalian Mps1 and Smad2 expression vectors have been described previously (Zhu et al., 2007). Point mutations in Mps1 were constructed using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA). Deletion mutants of Mps1 were constructed by polymerase chain reaction (PCR), and the relevant fragments of Mps1 were subcloned into pREX-IRES-Hygromycin (Hygro), a derivative of the bicistronic retroviral vector pREX-IRES-GFP described previously (Liu et al., 2000). The Mps1 siRNA-insensitive allele (Mps1R) was created by scrambling the coding sequence using QuikChange mutagenesis with a pair of oligonucleotides (oligos) (5′-CAAGAGCCAGATGATGCCAGAGATTATTTTCAAATGGCCAGAGC-3′ and 5′-GCTCTGGCCATTTGAAAATAATCTCTGGCATCATCTGGCTCTTG-3′). All constructs and mutations were confirmed by DNA sequencing. Retroviral expression constructs were transfected into 293T cells with the pCL amphotrophic helper plasmid (Naviaux et al., 1996). Forty-eight hours after transfection, virus-containing supernatant was collected and used to infect SW480 cells as described previously (Liu et al., 1997). Cells expressing a defined level of green fluorescent protein (GFP) were isolated using a MoFlo cell sorter (Dako Colorado, Fort Collins, CO) as described previously (Liu et al., 1997).
Mass Spectrometry Protein Sequencing Analysis
Eight micrograms of GST-TEV-6xHis-Mps1 purified from insect cell were autophosphorylated under the conditions described previously (Zhu et al., 2007). The reaction mixture was processed with standard dithiothreitol reduction, iodoacetimide alkylation, and in-solution tryptic digestion. The digestion mixture was fractionated by a microcapillary reverse phase high-performance liquid chromatography column directly coupled to an electron ion-spray mass spectrometer (QSTAR Pulsar; Applied Biosystems/MDS Sciex, Foster City, CA). Phosphopeptides were analyzed using a polarity switching method to alternate between detection and sequencing of the phosphopeptides in the same run (Williamson et al., 2006). First a negative mode precursor ion scan is acquired, monitoring for the marker ion PO3− at −79 m/z, over a mass range of 500-1800 m/z, with Q1 set to low resolution and Q3 set to unit resolution. When the signal intensity of the precursor ion scan is above a threshold of 1000 cps, polarity is switched to positive mode, and a high-resolution scan is acquired for charge determination and accurate mass measurement of the three most intense ions, followed by positive mode MS/MS sequencing. Tandem mass spectrometry (MS/MS) was searched with MASCOT version 2.0 (MatrixScience, London, United Kingdom) by using a small database of 50 standard proteins, including the sequence of the MPS1 fusion protein. Parent mass tolerance was 1.2 Da, MS/MS tolerance was 0.6 Da, with fixed modifications set to carbamidomethyl on cysteine, and variable modifications set for methionine oxidation and phosphorylation on Ser, Thr, and Tyr. MS/MS identifications with mascot scores above 20 were the manually validated for quality and phosphorylation site determination.
Orthophosphate Labeling of Cells and Two-dimensional (2D) Phosphopeptide Mapping
For 2D phosphopeptide analysis of Mps1, pEXL-FLAG-Mps1 and pEXL-FLAG-Mps1T12S15→AA were transient transfected into 293T cells. Forty-eight hours after transfection, wild-type and mutant kinases were immunoprecipitated with the FLAG antibody from cell lysates and labeled with [γ-32P]ATP under the autophosphorylation conditions before SDS-polyacrylamide gel electrophoresis (PAGE). Radiolabeled Mps1 or Mps1T12S15→AA was digested with trypsin and subjected to 2D phosphopeptide mapping as described previously (Boyle et al., 1991). SW480 cells stably expressing yellow fluorescent protein (YFP)-Mps1 and YFP-Mps1T12S15→AA grown in 10-cm plates were synchronized by double thymidine block. After washing twice with phosphate-free media (DME; Invitrogen), cells were released in 4 ml of phosphate-free media with 10% dialyzed fetal bovine serum and 0.1 μg/ml nocodazole for 12 h plus 1 mCi/ml [32P]orthophosphate (PerkinElmer Life and Analytical Sciences, Boston, MA) for 4 h. Cells were harvested and YFP-Mps1 or YFP-Mps1T12S15→AA were immunoprecipitated with an Mps1 CT antibody (Millipore) in radioimmunoprecipitation assay buffer and resolved by 12% SDS-PAGE gel before transferring to a nitrocellulose membrane. Radiolabeled YFP-Mps1 and YFP-Mps1T12S15→AA were analyzed as described above.
siRNA Knockdown and Immunofluorescence Microscopy
Mps1 knockdown was achieved using the siRNA (GCACGUGACUACUUU CAAAUU) synthesized by Dharmacon RNA Technologies. To achieve high knockdown efficiency, SW480 cells were transfected twice with 20 nM Mps1 siRNA with SiLentFect (Bio-Rad) every 24 h according to the manufacturer's protocol. For immunostaining, cells were grown on coverslips and washed three times with Dulbecco's phosphate-buffered saline (D-PBS) before fixing for 10 min in D-PBS plus 1% paraformaldehyde at room temperature. After fixation, the cells were blocked with 5% nonfat dry milk in PBS/Tween 20 for 45 min and probed with primary antibody diluted in 5% nonfat dry milk for 90 min. Anti-Mps1, NT (Millipore), and anti-Mad2 (Covance Research Products) were used at 1:100 dilution. Kinetochores were identified by staining cells with the human autoimmune serum CREST (Antibodies) at 1:300 dilution. After extensive wash, the secondary antibodies conjugated with Alexa Fluor 596 (Invitrogen) were applied with 1:600 dilution. The slides were prepared according to standard procedure.
Image Acquisition and Analysis
Pictures of immunofluorescence-stained cells were taken on a TE2000-S microscope (Nikon, Tokyo, Japan) equipped with MetaMorph image analysis software (Molecular Devices, Sunnyvale, CA). Acquired images were sized, scaled, pseudocolored, and overlaid by using MetaMorph software. For quantitation of the relative amount of YFP-Mps1 and related mutants on the kinetochores, a method described by Hoffman et al. (2000) was adopted, with minor modifications. The primary 16-bit images were analyzed using ImageJ software (http://rsbweb.nih.gov/ij/). Briefly, the kinetochores were centered by a circle with 3-pixel radius (Rin) (0.86 μm in diameter, which is large enough to cover a majority of kinetochore fluorescence in SW480 cell), and the total integrated fluorescence counts within this region (Fin) were measured. To subtract the background within this area, an outer circle with 4 pixel radius (Rin) was centered on the same kinetochore and the integrated fluorescence counts (Fout) was obtained (a detailed illustration of the method is described in figure 3 of Hoffman et al., 2001). The background of fluorescence (Fbackground) can be calculated as Fbackground = (Fout − Fin)(π Rin2/π(Rout − Rin)2). The integrated intensity of YFP-Mps1 or its related mutants on a given kinetochore was obtained using the equation Fkinetochore = Fin − Fbackground. Because the expression levels of the fusion protein in each given cell could affect the fluorescence intensity of the kinetochore, cells with similar overall fluorescence intensity were chosen for quantitation. In addition, the average values of kinetochore fluorescence was normalized to the relative expression levels of YFP-Mps1 proteins in the cytosol in each cell, which was calculated as a ratio of average cytoplasmic fluorescence intensity of a given cell (Bi) (calculated by an average of fluorescence intensity of 3-pixel radius circles randomly chosen in the cytoplasmic region with the number of circles picked equaling to the number of kinetochores quantified in a given cell) versus average cytoplasmic fluorescence from at least ten cells (B̄). Finally, kinetochore fluorescence intensity of YFP-Mps1 or related mutants in a given cell is normalized to the average fluorescence staining intensity of CREST in the same cell (Ci). Thus relative fluorescence intensity of YFP-Mps1 or related mutants is defined as F'kinetochore = (Fin − Fbackground)/Ki, where Ki = (Bi/B̄) × Ci. For the endogenous Mps1 kinetochore localization, average values of kinetochore fluorescence of Mps1 were normalized to the CREST fluorescence intensity. The statistical analysis was performed using GraphPad software (GraphPad Software, San Diego, CA).
RESULTS
Identification of Mps1 Autophosphorylation Sites by Mass Spectrometry
Mps1 is hyperphosphorylated during mitosis with a concomitant increase in kinase activity (Stucke et al., 2002; Liu et al., 2003; Kang et al., 2007). Phosphorylation could be a compelling mechanism to regulate Mps1 function during mitosis. To determine the role of Mps1 phosphorylation, we expressed and purified recombinant Mps1 from insect cells by using the baculoviral expression system. To identify Mps1 phosphorylation sites in vitro, purified Mps1 kinase was autophosphorylated by incubation with cold ATP and subjected to tryptic digestion. Multiple electrospray ionization-liquid chromatography/tandem mass spectrometry runs were carried out to identify the phosphopeptides. With ∼90% coverage of Mps1 from three independent runs, we were able to identify eight distinct phosphopeptides, with a total of 14 serines or threonines that were phosphorylated (Table 1 and Supplemental Figure 1A). The phosphorylation sites occupied in vitro are predominantly in the N- and C-terminal regions of Mps1 in the activation loop of the kinase domain. Two types of Mps1 autophosphorylation sites were detected. The first type is phosphoserine or phosphothreonine (pS/T) followed by a hydrophobic residue (e.g., Ile, Leu, or Val). We have previously found that Mps1 can transphosphorylate two distal serine residues of Smad2/3 at the carboxyl terminal SSXS motif (Zhu et al., 2007). Thus, this type of site can be targeted for either trans- or autophosphorylation by Mps1. The second type of phosphorylation site for Mps1 is phosphoserine or phosphothreonine (pS/T) followed by a negatively charged residue (e.g., D or E).
Table 1.
Summary of Mps1 phosphopeptides
| Peptide | Sites identified | Auto vs. Trans |
|---|---|---|
| 10ELTIDSIMNK19 | T12 | A |
| 10ELTIDSIMNK19 | T12 and S15 | A |
| 27FKNEDLTDELSLNK40 | T33 and S37 | A |
| 75NSVPLSDALLNK86 | S80 | A |
| 274VPVNLLNSPDCDVKTDDSVVPCFMK298 | T288 | A |
| 358NKTESSLLAK367 | T360, S362, S363 | A |
| 358NKTESSLLAK367 | T360, S363 | A |
| 681DSQVGTVNYMPPEAIK696 | T686 | A |
| 662LIDFGIANQMQPDTTSVVK680 | T675 | A |
| 662LIDFGIANQMQPDTTSVVK680 | T677 | A |
| 811YVLGQLVGLNSPNSILK827 | S821 | T |
One phosphopeptide containing S821 near the carboxy terminus of Mps1 does not fall into these two categories. Because insect cells are eukaryotes, there is a possibility that some of the sites identified are independent of Mps1 kinase and phosphorylated by other kinases in insect cells. To address this issue, we also purified catalytically inactive Mps1 kinase (Mps1KD) from insect cells. As shown in our previous study (Zhu et al., 2007), purified Mps1KD can neither autophosphorylate itself nor phosphorylate other substrates, such as Smad2, in vitro. Because no label can be incorporated in the purified Mps1KD in the presence of labeled ATP, it is unlikely there are other kinases capable of phosphorylating Mps1 copurified with Mps1KD in vitro. Mps1KD was incubated with or without ATP and subjected to mass spectrometry analysis by using identical conditions as wild-type Mps1. Four phosphopeptides were identified (Supplemental Figure 1B) in both samples, suggesting no additional phosphorylation in vitro with purified Mps1KD. All of them except one feature phosphoserine or phosphothreonine (pS/T) followed by a proline residue. Interestingly, all of these sites except S821 were not detected in wild-type Mps1 in vitro, suggesting that kinase-dead Mps1 is preferentially targeted by other kinases in insect cells. The only phosphopeptide shared by the wild-type and the kinase dead Mps1 is the one containing pSer821. This type of site is known to be targeted by mitogen-activated protein (MAP) kinase (Zhao and Chen, 2006; Cui and Guadagno, 2008). Thus, by comparing the phosphorylation profile of wild-type and kinase-dead Mps1, we can definitively assign Mps1 autophosphorylation sites in vitro. Identification of autophosphorylation sites of Mps1 suggests that Mps1 exhibits a rather broad spectrum of specificity of phosphorylation site choice. This begs the question of whether any of these autophosphorylation sites are physiologically relevant.
Cell Cycle-dependent Mps1 Subcellular Localization
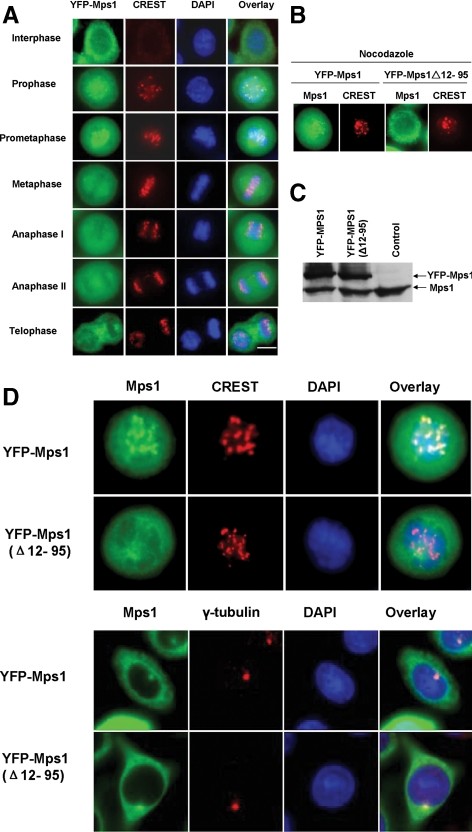
Subcellular localization of Mps1 is tightly regulated during the cell cycle (Fisk and Winey, 2001; Stucke et al., 2002, 2004; Liu et al., 2003). In the G1 phase of the cell cycle, endogenous Mps1 is distributed diffusely throughout cells and relocates to the nucleus and centrosomes during the G2/M transition. During prophase and prometaphase, Mps1 is targeted to the kinetochores; it comes off the kinetochores in metaphase. On completion of mitosis, Mps1 returns to the cytoplasm. To determine whether Mps1 phosphorylation plays a regulatory role for cell cycle-dependent dynamic localization, we stably expressed a YFP-Mps1 fusion protein in SW480 colon cancer cells. Localization of YFP-Mps1 during cell cycle progression was tracked by fluorescence microscopy of live cells. As shown in Figure 1A, YFP-Mps1 is targeted to the kinetochore, identified by staining with CREST antisera, during prophase and prometaphase. The staining pattern of YFP-Mps1 is in excellent agreement with previously described endogenous Mps1 localization (Fisk and Winey, 2001; Stucke et al., 2002, 2004; Liu et al., 2003), suggesting that YFP-Mps1 is a valid system to study Mps1 localization properties during mitosis.
Figure 1.
Subcellular localization of YFP-Mps1 during the cell cycle. (A) YFP-Mps1 is stably expressed in SW480 cells using the bicistronic retroviral vector pREX-IRES-Hygro. YFP positive cells were selected by fluorescence-activated cell sorting sorting. Asynchronous cells were fixed with 1% paraformaldehyde and stained with a CREST antiserum (Antibodies) to identify kinetochores (red) and 4,6-diamidino-2-phenylindole (DAPI) to identify DNA. (B) Deletion of aa 12-95 abrogates Mps1 kinetochore targeting in nocodazole-arrested cells. YFP-Mps1Δ12-95 stably expressed in SW480 cells was compared with YFP-Mps1 cells. Wild-type and mutant YFP-Mps1 cells were treated with 0.1 μg/ml nocodazole for 8 h after release from synchronization with double thymidine treatment. Cells were fixed with 1% paraformaldehyde and stained with a CREST antiserum and DAPI. (C) Immunoblot of the endogenous and YFP-Mps1 in SW480 cells. (D) Deletion of aa 12-95 of Mps1 does not affect Mps1 centrosome localization in interphase cells. Asynchronous wild-type and mutant Mps1 cells were fixed with 1% paraformaldehyde and centrosomes were stained with an anti-γ-tubulin antibody and DAPI. Bar, 8 μm.
Previous studies suggested that the N-terminal 301 amino acids of Mps1 are sufficient for targeting Mps1 to the kinetochore (Liu et al., 2003; Stucke et al., 2004). Subsequent studies further confirmed the importance of the N-terminal region of Mps1 in mediating kinetochore localization of Mps1. To further confirm the essential role of the N-terminal region of Mps1 in targeting full length Mps1 to the kinetochore, we expressed a small deletion mutant of Mps1 (Δ amino acids [aa] 12-95) in SW480 cells. Kinetochore localization of YFP-Mps1 was determined by treating cells expressing wild-type or mutant YFP-Mps1 with nocodazole, which causes prometaphase arrest by activating spindle checkpoint signaling. Although YFP-Mps1 shows robust kinetochore localization, removal of aa 12-95 does not affect the expression levels of the fusion protein but almost abrogates Mps1 targeting to kinetochores (Figure 1, B–D). Even though this deletion represents a relatively small perturbation in Mps1, there is a possibility that such a truncation may disrupt Mps1 structure and result in a misfolded protein. To rule out this possibility, we examined centrosome localization of wild-type and mutant Mps1. As shown in Figure 1D, mutant YFP-Mps1 is indistinguishable from wild-type Mps1 in centrosome localization, suggesting that it is unlikely that such a deletion causes the protein to be misfolded. This result also suggests that the kinetochore targeting signal of Mps1 is separable from the centrosome targeting signal.
N-Terminal Autophosphorylation Sites of Mps1 Are Required for Mps1 Kinetochore Targeting
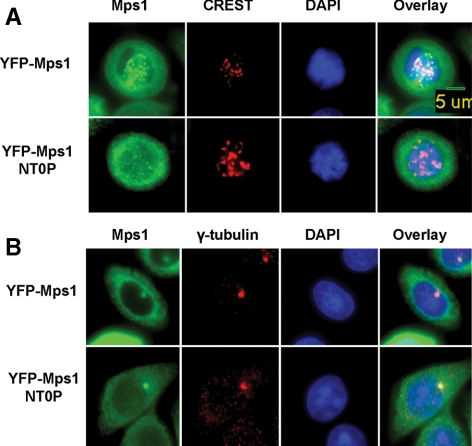
Five phosphopeptides encompassing nine different Ser/Thr residues identified from our mass spectrometry analysis of Mps1 autophosphorylation sites in vitro are located in the N-terminal region of Mps1 outside the kinase domain. Given the important role of the N-terminal region of Mps1 in kinetochore targeting, we hypothesized that phosphorylation of these sites may regulate Mps1 kinetochore recruitment upon activation of spindle checkpoint signaling. To test this hypothesis, we created YFP-Mps1NT0P by changing all nine Ser/Thr residues to alanines and stably expressed this mutant in SW480 cells. As shown in Figure 2A, whereas wild-type YFP-Mps1 shows robust kinetochore targeting upon treatment with nocodazole, mutation of all nine N-terminal autophosphorylation sites markedly reduces YFP-Mps1 kinetochore localization. To determine whether removal of these phosphorylation sites also perturbs centrosome targeting of Mps1, we analyzed localization of wild-type and mutant YFP-Mps1 on centrosomes in interphase cells. No difference was observed between wild-type and mutant Mps1 (Figure 2B), suggesting that these N-terminal autophosphorylation sites are very important in regulating Mps1 recruitment to the kinetochore but dispensable for centrosome targeting.
Figure 2.
Autophosphorylation of Mps1 at N-terminal residues is required for robust kinetochore targeting of Mps1. (A) Kinetochore targeting of wild-type and N-terminal autophosphorylation site mutant Mps1 (Mps1NT0P) in nocodazole-arrested mitotic cells. Cells were treated and analyzed as described in Figure 1B. (B) Asynchronous wild-type and mutant Mps1 cells were fixed with 1% paraformaldehyde and stained with an anti-γ-tubulin antibody and DAPI.
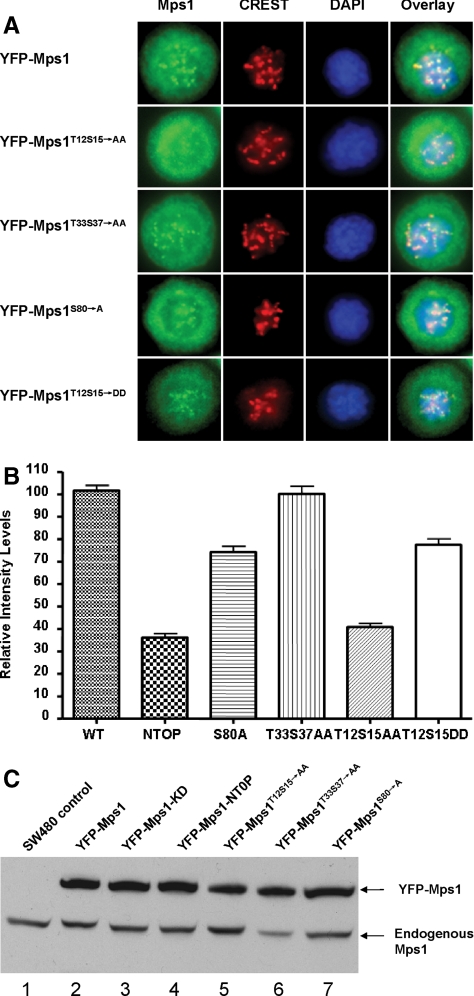
To further delineate which residues among the nine serine/threonine sites identified above are crucial in mediating kinetochore targeting of Mps1, we constructed three additional Mps1 point mutants and stably expressed them in SW480 cells. Because Mps11-301 has been shown to be sufficient for kinetochore targeting and deletion of Mps1 aa 12-95 abolishes Mps1 targeting (5 of 9 phosphorylation sites are located within this region), we focused our efforts on the five phosphorylation sites in the N-terminal region and did not test the effects of T288, T360, S362, and S363 on kinetochore localization of full-length Mps1. As shown in Figure 3A, whereas mutation of T33, S37 has no discernible effects on Mps1 kinetochore targeting, mutation of T12 and S15 to alanines severely impairs Mps1 kinetochore localization and displays a pattern almost identical to YFP-Mps1NT0P (Figure 3B). In contrast, mutation of S80 to alanine had only a minor effect (Figure 3, A and B). Again, none of these mutations affects Mps1 centrosome localization (data not shown). We independently confirmed expression of wild-type and mutant YFP-Mps1 fusions by immunoblotting analysis (Figure 3B). If phosphorylation of T12S15 is important, we would expect that substituting T12S15 with aspartic acids should partially mimic phosphorylation at these sites and the resulting mutant should exhibit little defects in kinetochore targeting. To test this hypothesis, YFP-Mps1T12S15→DD was created and stably expressed in SW480 cells. As expected, kinetochore targeting of this mutant is more resemble the wild-type (Figure 3, B and C), which support the hypothesis that phosphorylation at T12S15 is critical for Mps1 kinetochore localization.
Figure 3.
Identification of the N-terminal Mps1 autophosphorylation sites absolutely required for Mps1 kinetochore targeting. (A) YFP-Mps1T12S15→AA, YFP-Mps1T33S37→AA, and YFP-Mps1S80→A were stably expressed in SW480 cells. Kinetochore localization of these Mps1 mutants are analyzed as described in Figure 2. (B) Quantitation of fluorescent density of kinetochores labeled by YFP-Mps1 and YFP-Mps1 mutants in prometaphase cells. For YFP-Mps1, 115 kinetochores were determined in six cells randomly selected prometaphase cells; for YFP-Mps1NTOP, 103 kinetochores were counted in six randomly chosen prometaphase cells; for YFP-Mps1S80→A, 68 kinetochores were analyzed; for YFP-Mps1T33S37→AA, 83 kinetochores were counted; for YFP-Mps1T12S15→AA, 98 kinetochores were analyzed; for YFPMps1T12S15→DD, 55 kinetochore were counted. The difference is statistically significant between YFP-Mps1, YFP-Mps1NTOP (p < 0.001) and so is YFP-Mps1 and YFP-Mps1T12S15→AA. (C) Immunoblotting analysis of expression of wild-type and mutant Mps1 in SW480 cells. Cell lysates prepared from parental uninfected cells or cells stably expressing various mutants of Mps1 were blotted with the Mps1 antibody C19 (Santa Cruz Biotechnology).
There is a possibility that mutation of T12 and S15 could have affected Mps1 kinase activity. We addressed this issue by cotransfecting wild-type and mutant Mps1 with Smad2 in 293T cells followed by measuring Smad2 phosphorylation by using a phospho-specific antibody. In agreement with our previous result (Zhu et al., 2007), wild-type Mps1 phosphorylates Smad2 at SSMS motif, whereas little if any phosphorylation can be observed with kinase-dead Mps1 (Supplemental Figure 2). Phosphorylation of Smad2 by T12 and S15 mutants of Mps1 is as efficient as the wild-type Mps1, suggesting that these mutants do not affect Mps1 kinase activity. Taken together, our data suggests that phosphorylation of Mps1 at T12 and S15 is required for robust kinetochore targeting of full-length Mps1 but dispensable for its kinase activity.
In Vitro and in Vivo Phosphorylation of T12 and S15
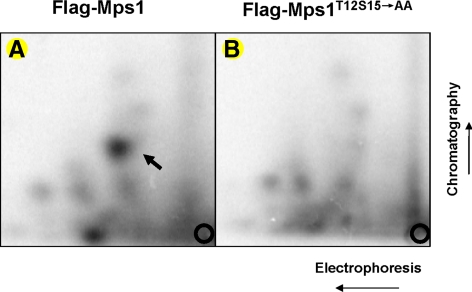
Even though Mps1 undergoes autophosphorylation at T12 and S15 in vitro and both residues prove to be critical for kinetochore targeting of Mps1, it is still important to demonstrate that these sites are also phosphorylated in cells. To further demonstrate phosphorylation of Mps1 at T12S15 expressed in mammalian cells, FLAG-tagged Mps1 and Mps1T12S15→AA were expressed in 293T cells. Wild-type and mutant Mps1 were immunoprecipitated with the anti-FLAG antibody, subsequently incubated with [γ-32P]ATP under kinase reaction conditions before SDS-PAGE and blotting to nitrocellulose membrane. 32P-labeled Mps1 or Mps1 mutant was excised and subjected to 2D tryptic mapping analysis (Boyle et al., 1991). Shown in Figure 4, FLAG-tagged Mps1 and Mps1T12S15→AA differs by only one major spot on the thin layer chromatography (TLC) plate. The missing spot in Mps1T12S15→AA but present in wild-type Mps1 is probably the phosphopeptide containing T12 and S15. This result further confirms the mass spectrometry data indicating that T12 and S15 are targeted for phosphorylation.
Figure 4.
2D tryptic mapping analysis of wild-type or T12 and S15 mutant Mps1 phosphorylation. FLAG-tagged Mps1 or Mps1T12S15→AA were expressed in 293T cells by transient transfection. Forty-eight hours after transfection, FLAG-Mps1 or FLAG-Mps1T12S15→AA were immunoprecipitated using an anti-FLAG antibody and incubated in the presence of [γ-32P]ATP under the in vitro kinase reaction conditions before SDS-PAGE. Bands corresponding to 32P-labeled Mps1 or Mps1T12S15→AA were excised and subjected to 2D phosphopeptide mapping analysis as described previously (Boyle et al., 1991).
To determine whether Mps1 is phosphorylated at T12 and S15 in mitotic-arrested cells, SW480 cells expressing either YFP-Mps1 or YFP-Mps1T12S15→AA were left in nocodazole for 12 h before labeling with [32P]orthophosphate for 4 h. YFP-Mps1 or YFP-Mps1T12S15→AA was immunoprecipitated from mitotic-arrested cells and blotted to nitrocellulose membrane after resolution by SDS-PAGE. Both YFP-Mps1 and YFP-Mps1T12S15→AA can be labeled by [32P]orthophosphate in mitotic-arrested cells, although the intensity of YFP-Mps1T12S15→AA is lower than that of YFP-Mps1 (Supplemental Figure 3A). This could be a result of fewer occupied sites due to mutations in the potential phosphorylation sites. 32P-Labeled YFP-Mps1 or YFP-Mps1T12S15→AA bands from mitotic cells were excised and subjected to 2D tryptic phosphopeptide analysis. Five major labeled phosphopeptides are clearly visible in the YFP-Mps1 sample, and only four of the five are detectable in YFP-Mps1T12S15→AA. We interpret the missing spot on the TLC plate as the T12, S15 containing phosphopeptide (ELTIDSIMNK) because this spot is only present in wild-type but not in the mutant Mps1. Hence, phosphorylation of T12 and S15 is likely to be phosphorylated in mitotic-arrested cells.
The T12 and S15 Phosphorylation Sites Are Also Required for Kinetochore Targeting of the N-Terminal Domain of Mps1
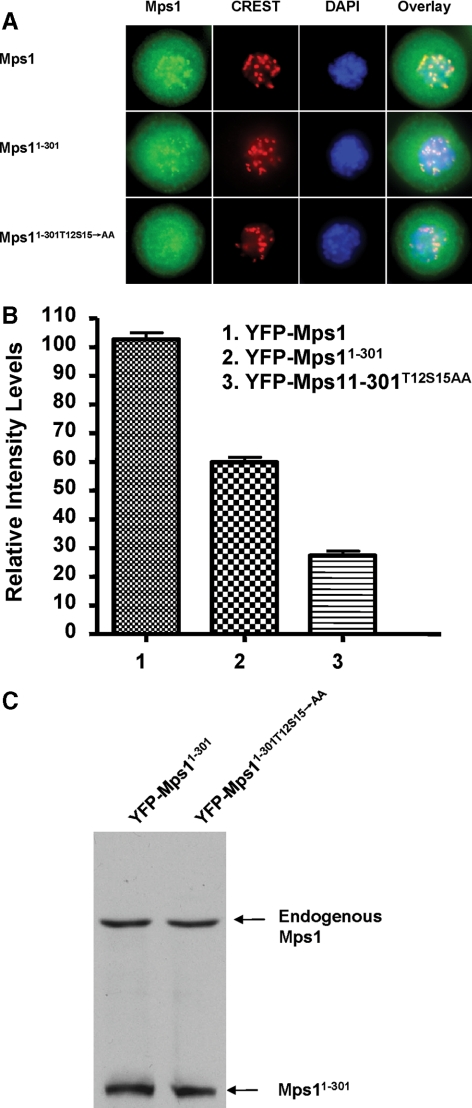
Previous studies suggest that the amino-terminal 301 residues of Mps1 are both necessary and sufficient for its kinetochore association (Liu et al., 2003; Stucke et al., 2004). Having established that T12 and S15 are important for full-length recruitment to the kinetochore, we also investigated whether these two sites are also critical for the N-terminal fragment of Mps1 to associate with the kinetochore. In agreement with previous observations (Liu et al., 2003; Stucke et al., 2004), YFP-Mps11-301 relocates to the kinetochore in response to nocodazole treatment when stably expressed in SW480 cells (Figure 5A). This result confirms that the N-terminal 301 amino acid residues of Mps1 indeed contain the kinetochore targeting signal(s).
Figure 5.
Kinetochore targeting of the N-terminal fragment of Mps1 requires phosphorylation at T12 and S15. (A) Kinetochore localization of YFP-Mps1, YFP-Mps11-301, and YFP-Mps11-301T12S15→AA in prometaphase-arrested cells induced by nocodazole treatment (0.1 μg/ml). The kinetochores and chromosomes are identified by staining with human CREST antisera and DAPI, respectively. (B) Quantitation of fluorescent density of YFP-Mps1 and related mutants on the kinetochores of prometaphase cells. More than 52 kinetochores in at least five randomly selected cells in each cell line were analyzed. YFP-Mps11-301T12S15→AA cannot relocalize to kinetochores in prometaphase cells. (C) Immunoblotting analysis of expression of YFP-Mps11-301, and YFP-Mps11-301T12S15→AA in SW480 cells.
Even though the N-terminal fragment of Mps1 does not contain the kinase domain, T12 and S15 can still be phosphorylated by Mps1 given that autophosphorylation of Mps1 occurs through an intermolecular mechanism (Kang et al., 2007; Mattison et al., 2007). In fact, the N-terminal domain is an excellent substrate for Mps1 in vitro (data not shown). To assess the role of T12 and S15 in the kinetochore targeting of the N-terminal fragment of Mps1, we introduced point mutations at these two positions into YFP-Mps11-301. The resulting mutant (YFP-Mps11-301/T12S15→AA) is defective in kinetochore association (Figure 5A). Mutation of these phosphorylation sites has no effect on their expression levels (Figure 5B). Together, these results suggest that Mps1 autophosphorylation sites T12 and S15 are required for both the full-length and the N-terminal region of Mps1 to associate with the kinetochore upon activation of spindle checkpoint signaling.
Phosphorylation of T12 and S15 Is Required for Robust Spindle Checkpoint Signaling and Recruitment of Mad2 to Kinetochore
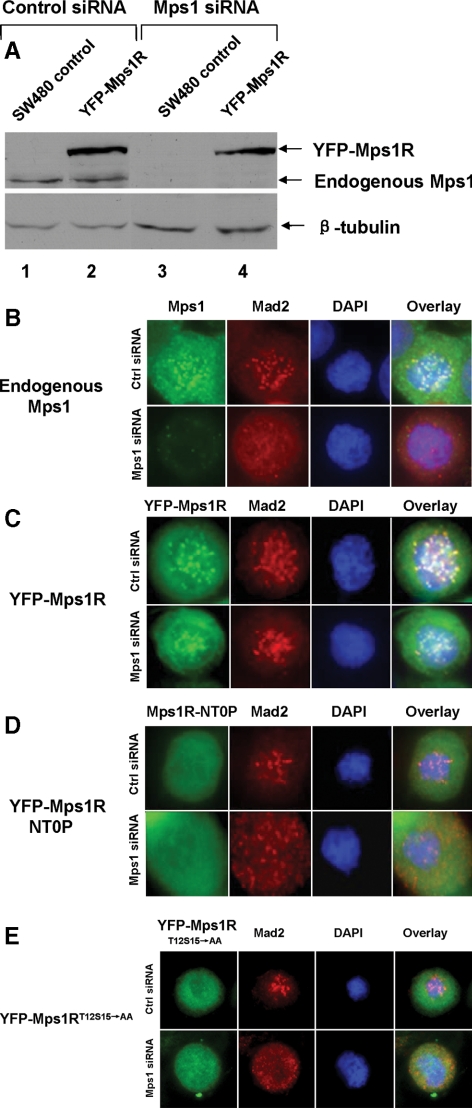
Activation of spindle checkpoint signaling leads to recruitment of checkpoint signaling components such as Mps1, Bub1, BubR1, Mad1, and Mad2 to the kinetochore to delay the onset of anaphase. Mps1 is required for kinetochore localization of Mad2 (Martin-Lluesma et al., 2002; Howell et al., 2004; Vigneron et al., 2004; Zhao and Chen, 2006). Whether phosphorylation of Mps1 at T12 and S15 is required for the checkpoint signaling response is unknown. Endogenous Mps1 can be depleted by treatment with a siRNA duplex (Figure 6A), and knockdown of Mps1 inhibits targeting of Mad2 to the kinetochore (Figure 6B). A siRNA-resistant allele of Mps1 was constructed by scrambling the targeted coding sequence (Mps1R). YFP-Mps1R can be efficiently expressed in the presence of the siRNA targeting endogenous Mps1 (Figure 6A). YFP-Mps1R can mediate kinetochore recruitment of Mad2 upon depletion of the endogenous Mps1 with the Mps1 siRNA (Figure 6C). To determine whether phosphorylation of T12 and S15 is required for efficient Mad2 localization to the kinetochore in response to spindle damage, we stably expressed YFP-Mps1RT12S15→AA and Mps1RNT0P in SW480 cells (Figure 6, D and E). The resulting cell lines were transfected with either control or Mps1 siRNA and treated with nocodazole for 12 h. In cells treated with control siRNA, Mad2 localized to kinetochores in prometaphase arrested cells due to the presence of the endogenous Mps1. In contrast, in cells with the Mps1 siRNA, Mad2 was not recruited to kinetochores, suggesting that neither YFP-Mps1RT12S15→AA nor Mps1RNT0P is able to mediate kinetochore localization of Mad2 in the absence of endogenous Mps1. These data suggest that phosphorylation of the N-terminal region of Mps1 or, more specifically, of T12 and S15 is required for Mps1 to activate spindle checkpoint signaling.
Figure 6.
Autophosphorylation of Mps1 is required for kinetochore recruitment of checkpoint protein Mad2. (A) Depletion of the endogenous but not YFP-tagged siRNA-resistant Mps1 (YFP-Mps1R) in SW480 cells. Uninfected or YFP-Mps1R-expressing SW480 cells were transfected with control or Mps1 siRNA. Expression of the endogenous Mps1 or YFP-Mps1 was determined by immunoblotting with an Mps1 antibody. (B) Depletion of the endogenous Mps1 abrogates Mad2 kinetochore localization. Wild-type SW480 cells were transfected with control or Mps1 siRNA. Two days after transfection, cells were treated nocodazole for 12 h and fixed in 1% paraformaldehyde. Fixed cells were stained with antibodies against Mps1 or Mad2 or DAPI. (C) YFP-Mps1R supports kinetochore recruitment of Mad2 upon depletion of the endogenous Mps1 by siRNA. (D) Mutation of autophosphorylation sites of Mps1 blocks kinetochore recruitment of Mps1 and Mad2. (E) Phosphorylation of T12 and S15 of Mps1 is required for kinetochore targeting of Mad2.
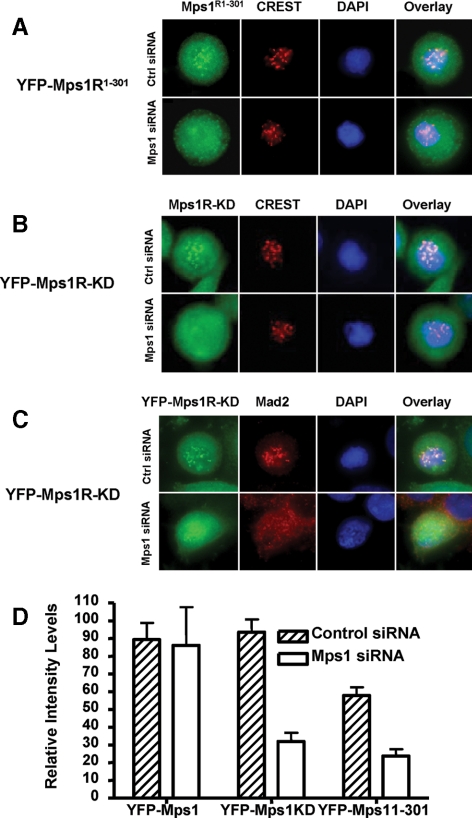
Previous studies suggest that Mps1 kinase activity is not required for kinetochore targeting of Mps1 as GFP-Mps1-KD (kinase dead) or GFP-Mps11-301 without the kinase domain can target to kinetochores (Liu et al., 2003; Stucke et al., 2004). Given that Mps1 can undergo transautophosphorylation, overexpression of GFP-Mps1-KD or GFP-Mps11-301 could still be phosphorylated by the endogenous Mps1. To further evaluate the requirement of Mps1 kinase activity in its kinetochore targeting, we stably expressed siRNA-resistant YFP-Mps1R-KD or YFP-Mps1R1-301 in SW480 cells. As expected, both of them can target to the kinetochore as described in cells transfected with control siRNA (Figure 7, A and B). However, when endogenous Mps1 was depleted by treatment with siRNA, neither YFP-Mps1R-KD nor YFP-Mps1R1-301 can accumulate at kinetochores. Consistent with the defects of Mps1 kinetochore targeting, Mad2 localization to the kinetochore was also abolished (Figure 7C). Thus, these results suggest that kinase activity-deficient Mps1 is incapable of relocating to the kinetochore in the absence of endogenous Mps1. Mps1 kinase activity is required for kinetochore localization of spindle checkpoint proteins including Mps1 itself.
Figure 7.
Mps1 kinase activity is required for its kinetochore targeting and recruitment of Mad2 upon activation of the mitotic spindle checkpoint in SW480 cells. (A) Kinetochore localization of the N-terminal fragment of Mps1 requires endogenous Mps1. SW480 cells stably expressing YFP-Mps1R1-301 were transfected with control or Mps1 siRNA. Localization of YFP-Mps1R1-301 was determined by immunofluorescence microscopy as described in text. B is the same as A except kinase-deficient Mps1 mutant (YFP-Mps1R-KD) was used. (C) Mps1 kinase activity is required for kinetochore recruitment of Mad2. SW480 cells stably expressing YFP-Mps1R-KD were transfected with control or Mps1 siRNA. Cells were fixed and stained with an antibody against Mad2 and with DAPI. (D) Quantitation of fluorescent density of kinetochores labeled by YFP-Mps1 and related mutants in control or Mps1 siRNA treated prometaphase cells. At least 42 kinetochores from prometaphase cells in each cell line were scored and differences between control and Mps1 siRNA treated samples from YFP-Mps1R-KD or YFP-Mps1R1-301 are statistically significant (p < 0.05).
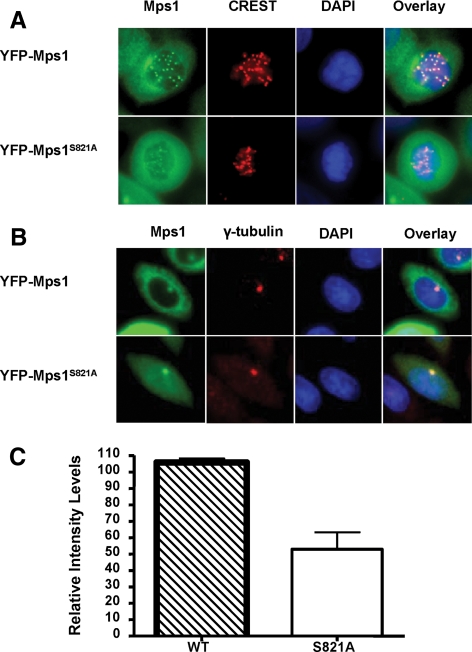
Phosphorylation of S821 Is Important for Kinetochore Targeting of Full-Length Mps1
Previous studies suggest that phosphorylation of Xenopus Mps1 at S844 by MAP kinase is essential for kinetochore targeting in Xenopus egg extracts. The equivalent site of S844 in human Mps1 is S821. Our mass spectrometry data suggests that S821 is phosphorylated in insect cells by unknown kinases. To address the significance of this phosphorylation in mammalian cells, we constructed a stable cell line expressing the S821A mutant of YFP-Mps1. In agreement with the Xenopus system results, kinetochore localization of YFP-Mps1S821→A is decreased by at least 50% compared with the wild-type control, suggesting that phosphorylation of this site play a significant role in kinetochore recruitment of Mps1 in mammalian cells (Figure 8, A and C). To determine whether S821 also affects centrosome localization of Mps1, we compared centrosome staining of YFP-Mps1 and YFP-Mps1S821→A in interphase cells. No significant difference is observed between control and the mutant. Thus, phosphorylation of S821 seems to play a role in regulating kinetochore but not centrosome localization of Mps1.
Figure 8.
Phosphorylation of S821 is important for kinetochore recruitment of Mps1 but not for centrosome localization. (A) Kinetochore targeting of YFP-Mps1 and YFP-Mps1S821→A in nocodazole-arrested mitotic cells. Cells were treated and analyzed as described in Figure 1B. (B) Centrosome localization of YFP-Mps1 and YFP-Mps1S821→A in interphase cells. (C) Quantitation of fluorescent density of YFP-Mps1 and YFP-Mps1S821→A on the kinetochores of prometaphase cells. The differences between YFP-Mps1 and YFP-Mps1S821→A are statistically significant (p < 0.001).
DISCUSSION
We report here that autophosphorylation of T12 and S15 at the N-terminal domain of Mps1 is a key regulatory event required for Mps1 kinetochore targeting and subsequent recruitment of Mad2 to the kinetochore upon activation of spindle checkpoint signaling. We showed that phosphorylation of T12 and S15 occurs in mitotic-arrested cells and that mutation of T12 and S15 abrogates Mps1 kinetochore association. We propose that phosphorylation of T12 and S15 may either create a recognition motif to interact with cellular machinery to transport Mps1 to the kinetochore or cause allosteric changes in Mps1 to expose the kinetochore targeting signal(s) embedded in the N-terminal region of Mps1.
Hyperphosphorylation of Mps1 has been well documented in mitotic cells (Stucke et al., 2002, 2004; Liu et al., 2003; Fisk et al., 2004). The phosphorylation sites of Mps1 in mitotic cells have just begun to emerge (Zhao and Chen, 2006; Kang et al., 2007; Mattison et al., 2007; Cui and Guadagno, 2008). It seems that Mps1 is targeted by both autophosphorylation and transphosphorylation by other kinases. Consistent with previous observation, we found that the activation loop of Mps1 is targeted by autophosphorylation (T675, S678, and T686) (Kang et al., 2007; Mattison et al., 2007). Autophosphorylation of these sites has been shown to enhance Mps1 kinase activity by four- to sevenfold and enhances kinetochore targeting of Mps1 and Bub1 (Kang et al., 2007; Mattison et al., 2007). Mps1 purified from mitotic HeLa extracts showed phosphorylation at T33, S37, S80, S281, T436, T453, T468, T360, S363, T371, S382, and S821 (Kang et al., 2007; Kasbek et al., 2007; Cui and Guadagno, 2008; Jelluma et al., 2008a). Using Mps1 purified from insect cells, which possess robust kinase activity, we demonstrated that some of these sites are primarily Mps1 autophosphorylation sites (e.g., T33, S37, S80, T360, S363, T675, T676, T678, and T686). T12 and S15 phosphorylation has not been described in other studies. There is a possibility that these two sites are phosphorylated at low stoichiometry compared with other sites and only associate with active Mps1 localized to the kinetochores. Given that kinetochore targeting of Mps1 is a highly dynamic process and only a small fraction of Mps1 bound to kinetochores with the residence time of Mps1 being <10 s (Howell et al., 2004), dynamic or reversible phosphorylation of T12S15 would be consistent with its role in regulating Mps1 kinetochore recruitment.
Phosphorylation of the TP or SP sites in Mps1 can be attributed to MAP kinase (Zhao and Chen, 2006; Kang et al., 2007) or Cdk2 kinase activity (Kasbek et al., 2007; Cui and Guadagno, 2008), although a recent study using Mps1 purified from insect cells suggests that S821 may be an autophosphorylation site., It has been proposed that phosphorylation of these sites increases the stability of Mps1 during mitosis (Kasbek et al., 2007; Cui and Guadagno, 2008). However, S821 is also phosphorylated in Mps1 kinase-dead mutant purified from insect cells, suggesting that this site is more likely to be targeted by other kinases rather than an autophosphorylation site. The fact that TP or SP sites are readily detectable in mammalian cells with hyperactive MAP kinase pathway and Xenopus extracts with elevated MAP kinase activity suggests that the MAP kinase pathway may cross talk with the Mps1 pathway through hyperphosphorylation of Mps1 at the canonical MAP kinase phosphorylation sites.
Hyperphosphorylation of Mps1 at multiple sites occurs both in vitro and in vivo (Kang et al., 2007; Kasbek et al., 2007; Mattison et al., 2007; Cui and Guadagno, 2008; Jelluma et al., 2008a). There are considerable variations in the number of phosphorylation sites reported in the literature. For example, there are far more autophosphorylation sites with recombinant Mps1 purified from Escherichia coli than from insect cells. This observation may suggest the heterogeneity of Mps1 phosphorylation, which poses significant challenges to address the function of each individual site in vivo if functional redundancy exists among these phosphorylation sites. Throughout our studies we use the T12S15 double mutant to address the potential function of these sites in Mps1 kinetochore relocalization, it is very possible that only one of these sites is occupied in vivo for a given Mps1 molecule. Consistent with this notion, T12 singly phosphorylated peptide is more readily detectable than the T12S15 doubly phosphorylated peptide, suggesting T12 is a preferable phosphorylation site in wild-type Mps1. Because of the potential redundancy of S15 and presence of T12S15 double phosphorylated species in vivo, it is necessary to use double mutant to address the function of T12 S15 phosphorylation in Mps1 kinetochore recruitment.
The requirement for Mps1 kinase activity for its kinetochore recruitment has not been fully addressed. In agreement with previous observations, the kinase-deficient Mps1 mutant can be recruited to kinetochores as is the N-terminal region of Mps1 lacking the kinase domain (Stucke et al., 2002, 2004; Liu et al., 2003). However, kinase-deficient Mps1 is defective in kinetochore targeting in Xenopus egg extracts upon depletion of the endogenous Mps1 (Zhao and Chen, 2006). Given that Mps1 undergoes extensive intermolecular autophosphorylation (Kang et al., 2007; Mattison et al., 2007), it is quite possible that kinase-deficient Mps1 or the N-terminal domain of Mps1 can be phosphorylated by the endogenous Mps1 in mammalian cells. Our results showing that endogenous Mps1 is required for kinetochore localization of kinase-deficient Mps1 are consistent with this hypothesis although it cannot be ruled out at this point that another kinase whose activity depends on Mps1 is involved in this process. It has been previously noted that inhibition of Mps1 kinase activity abrogates spindle checkpoint responses in transform tumor cells but not normal cells (Schmidt and Medema, 2006), suggesting that normal cells may have multiple redundant pathways to ensure robust checkpoint responses. This scenario could explain the apparent differences in kinetochore targeting behavior of Mps1 kinase-deficient mutant between HeLa cells (Kang et al., 2007) and SW480 cells (this study) when the endogenous Mps1 is depleted. Whereas the spindle checkpoint is relatively weakened in SW480 cells (Tighe et al., 2001), HeLa cells possess a very robust spindle checkpoint control (Schmidt and Medema, 2006). We speculate that there could be an unidentified kinase(s) present in HeLa that is missing in SW480 cells and responsible for phosphorylating T12S15 when the autophosphorylation activity of Mps1 is disabled. Future studies are needed to determine whether this is case.
In this report, we found that Mps1 is autophosphorylated in vitro and in vivo at T12 and S15, and this phosphorylation is required for kinetochore targeting of Mps1. Disagreement exists in the literature as to the exact location of the Mps1 kinetochore targeting signal. Deletion mapping analyses suggest that the target signal may be located in the N-terminal region within the first 300 amino acids of Mps1 (Stucke et al., 2002, 2004; Liu et al., 2003). However, Chen and colleagues recently demonstrated that phosphorylation of S844 is crucial for kinetochore targeting of Mps1 and they proposed that phosphorylation of S844 (equivalent to S821 of human Mps1) by MAP kinase may create a phospho-epitope to serve as a kinetochore targeting signal (Zhao and Chen, 2006). This raises the possibility that the C terminus of Mps1 could generate a target signal upon phosphorylation. Our mutation analysis agrees with the importance of S821 phosphorylation in kinetochore targeting of Mps1. However, our results are also consistent with the notion that the N terminus of Mps1 contains a kinetochore localization signal and that this signal requires phosphorylation at T12 and S15. More importantly, the C-terminal region of Mps1, including the kinase domain, is insufficient for kinetochore targeting (data not shown). To reconcile all these data, we propose that kinetochore recruitment of Mps1 requires phosphorylation at both the N terminus and C terminus of the protein (Supplemental Figure 4). Whereas phosphorylation of the N terminus at T12 and S15 creates a kinetochore target signal to be recognized by kinetochore-associated proteins, phosphorylation of Mps1 at the C terminus at S821 causes allosteric changes in Mps1 conformation, which may expose the kinetochore targeting signal. Further experiments are needed to identify the cellular components that recognize the kinetochore targeting signal and whether a significant conformational change occurs in Mps1 upon phosphorylation and dephosphorylation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Mark Winey for advice and sharing unpublished results and Katheryn Resing for mass spectrometry advice. We thank Drs. David Clarke and Kristen Barthel for critical readings of the manuscript. This work was supported by National Institutes of Health grant CA-107098 and a Pilot grant from the Colorado Cancer Center (to X. L.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-03-0324) on October 15, 2008.
REFERENCES
- Abrieu A., Magnaghi-Jaulin L., Kahana J. A., Peter M., Castro A., Vigneron S., Lorca T., Cleveland D. W., Labbe J. C. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell. 2001;106:83–93. doi: 10.1016/s0092-8674(01)00410-x. [DOI] [PubMed] [Google Scholar]
- Amon A. The spindle checkpoint. Curr. Opin. Genet. Dev. 1999;9:69–75. doi: 10.1016/s0959-437x(99)80010-0. [DOI] [PubMed] [Google Scholar]
- Boyle W. J., van der Geer P., Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- Cui Y., Guadagno T. M. B-Raf(V600E) signaling deregulates the mitotic spindle checkpoint through stabilizing Mps1 levels in melanoma cells. Oncogene. 2008;27:3122–3133. doi: 10.1038/sj.onc.1210972. [DOI] [PubMed] [Google Scholar]
- DeLuca J. G., Dong Y., Hergert P., Strauss J., Hickey J. M., Salmon E. D., McEwen B. F. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol. Biol. Cell. 2005;16:519–531. doi: 10.1091/mbc.E04-09-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draviam V. M., Xie S., Sorger P. K. Chromosome segregation and genomic stability. Curr. Opin. Genet. Dev. 2004;14:120–125. doi: 10.1016/j.gde.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Fang G., Yu H., Kirschner M. W. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk H. A., Mattison C. P., Winey M. A field guide to the Mps1 family of protein kinases. Cell Cycle. 2004;3:439–442. [PubMed] [Google Scholar]
- Fisk H. A., Winey M. The mouse Mps1p-like kinase regulates centrosome duplication. Cell. 2001;106:95–104. doi: 10.1016/s0092-8674(01)00411-1. [DOI] [PubMed] [Google Scholar]
- Hoffman D. B., Pearson C. G., Yen T. J., Howell B. J., Salmon E. D. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell. 2001;12:1995–2009. doi: 10.1091/mbc.12.7.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B. J., Moree B., Farrar E. M., Stewart S., Fang G., Salmon E. D. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr. Biol. 2004;14:953–964. doi: 10.1016/j.cub.2004.05.053. [DOI] [PubMed] [Google Scholar]
- Hoyt M. A., Totis L., Roberts B. T. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Jelluma N., Brenkman A. B., McLeod I., Yates J. R., 3rd, Cleveland D. W., Medema R. H., Kops G. J. Chromosomal instability by inefficient Mps1 auto-activation due to a weakened mitotic checkpoint and lagging chromosomes. PLoS ONE. 2008a;3:e2415. doi: 10.1371/journal.pone.0002415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelluma N., Brenkman A. B., van den Broek N. J., Cruijsen C. W., van Osch M. H., Lens S. M., Medema R. H., Kops G. J. Mps1 phosphorylates borealin to control aurora B activity and chromosome alignment. Cell. 2008b;132:233–246. doi: 10.1016/j.cell.2007.11.046. [DOI] [PubMed] [Google Scholar]
- Kang J., Chen Y., Zhao Y., Yu H. Autophosphorylation-dependent activation of human Mps1 is required for the spindle checkpoint. Proc. Natl. Acad. Sci. USA. 2007;104:20232–20237. doi: 10.1073/pnas.0710519105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasbek C., Yang C. H., Yusof A. M., Chapman H. M., Winey M., Fisk H. A. Preventing the degradation of mps1 at centrosomes is sufficient to cause centrosome reduplication in human cells. Mol. Biol. Cell. 2007;18:4457–4469. doi: 10.1091/mbc.E07-03-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops G. J., Weaver B. A., Cleveland D. W. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat. Rev. Cancer. 2005;5:773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- Lengauer C., Kinzler K. W., Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- Li R., Murray A. W. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Lindberg R. A., Fischer W. H., Hunter T. Characterization of a human protein threonine kinase isolated by screening an expression library with antibodies to phosphotyrosine. Oncogene. 1993;8:351–359. [PubMed] [Google Scholar]
- Liu S. T., Chan G. K., Hittle J. C., Fujii G., Lees E., Yen T. J. Human MPS1 kinase is required for mitotic arrest induced by the loss of CENP-E from kinetochores. Mol. Biol. Cell. 2003;14:1638–1651. doi: 10.1091/mbc.02-05-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Constantinescu S. N., Sun Y., Bogan J. S., Hirsch D., Weinberg R. A., Lodish H. F. Generation of mammalian cells stably expressing multiple genes at predetermined levels. Anal. Biochem. 2000;280:20–28. doi: 10.1006/abio.2000.4478. [DOI] [PubMed] [Google Scholar]
- Liu X., Sun Y., Constantinescu S. N., Karam E., Weinberg R. A., Lodish H. F. Transforming growth factor beta-induced phosphorylation of Smad3 is required for growth inhibition and transcriptional induction in epithelial cells. Proc. Natl. Acad. Sci. USA. 1997;94:10669–10674. doi: 10.1073/pnas.94.20.10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Lluesma S., Stucke V. M., Nigg E. A. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 2002;297:2267–2270. doi: 10.1126/science.1075596. [DOI] [PubMed] [Google Scholar]
- Mattison C. P., Old W. M., Steiner E., Huneycutt B. J., Resing K. A., Ahn N. G., Winey M. Mps1 activation loop autophosphorylation enhances kinase activity. J. Biol. Chem. 2007;282:30553–30561. doi: 10.1074/jbc.M707063200. [DOI] [PubMed] [Google Scholar]
- McIntosh J. R. Structural and mechanical control of mitotic progression. Cold Spring Harb. Symp. Quant. Biol. 1991;56:613–619. doi: 10.1101/sqb.1991.056.01.070. [DOI] [PubMed] [Google Scholar]
- Mills G. B., Schmandt R., McGill M., Amendola A., Hill M., Jacobs K., May C., Rodricks A. M., Campbell S., Hogg D. Expression of TTK, a novel human protein kinase, is associated with cell proliferation. J. Biol. Chem. 1992;267:16000–16006. [PubMed] [Google Scholar]
- Montembault E., Dutertre S., Prigent C., Giet R. PRP4 is a spindle assembly checkpoint protein required for MPS1, MAD1, and MAD2 localization to the kinetochores. J. Cell Biol. 2007;179:601–609. doi: 10.1083/jcb.200703133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 2001;35:673–745. doi: 10.1146/annurev.genet.35.102401.091334. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Segregating sister genomes: the molecular biology of chromosome separation. Science. 2002;297:559–565. doi: 10.1126/science.1074757. [DOI] [PubMed] [Google Scholar]
- Naviaux R. K., Costanzi E., Haas M., Verma I. M. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J. Virol. 1996;70:5701–5705. doi: 10.1128/jvi.70.8.5701-5705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas R. B. How cells get the right chromosomes. Science. 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- Rieder C. L., Schultz A., Cole R., Sluder G. Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J. Cell Biol. 1994;127:1301–1310. doi: 10.1083/jcb.127.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M., Medema R. H. Exploiting the compromised spindle assembly checkpoint function of tumor cells: dawn on the horizon? Cell Cycle. 2006;5:159–163. doi: 10.4161/cc.5.2.2309. [DOI] [PubMed] [Google Scholar]
- Stucke V. M., Baumann C., Nigg E. A. Kinetochore localization and microtubule interaction of the human spindle checkpoint kinase Mps1. Chromosoma. 2004;113:1–15. doi: 10.1007/s00412-004-0288-2. [DOI] [PubMed] [Google Scholar]
- Stucke V. M., Sillje H. H., Arnaud L., Nigg E. A. Human Mps1 kinase is required for the spindle assembly checkpoint but not for centrosome duplication. EMBO J. 2002;21:1723–1732. doi: 10.1093/emboj/21.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V., Chan G. K., Yen T. J. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Shu H., Oncel D., Chen S., Yu H. Phosphorylation of Cdc20 by Bub1 provides a catalytic mechanism for APC/C inhibition by the spindle checkpoint. Mol. Cell. 2004;16:387–397. doi: 10.1016/j.molcel.2004.09.031. [DOI] [PubMed] [Google Scholar]
- Tighe A., Johnson V. L., Albertella M., Taylor S. S. Aneuploid colon cancer cells have a robust spindle checkpoint. EMBO Rep. 2001;2:609–614. doi: 10.1093/embo-reports/kve127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F., Lottspeich F., Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- Vigneron S., Prieto S., Bernis C., Labbe J. C., Castro A., Lorca T. Kinetochore localization of spindle checkpoint proteins: who controls whom? Mol. Biol. Cell. 2004;15:4584–4596. doi: 10.1091/mbc.E04-01-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassmann K., Benezra R. Mitotic checkpoints: from yeast to cancer. Curr. Opin. Genet. Dev. 2001;11:83–90. doi: 10.1016/s0959-437x(00)00161-1. [DOI] [PubMed] [Google Scholar]
- Weaver B. A., Cleveland D. W. Decoding the links between mitosis, cancer, and chemotherapy: the mitotic checkpoint, adaptation, and cell death. Cancer Cell. 2005;8:7–12. doi: 10.1016/j.ccr.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Weiss E., Winey M. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J. Cell Biol. 1996;132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson B. L., Marchese J., Morrice N. A. Automated identification and quantification of protein phosphorylation sites by LC/MS on a hybrid triple quadrupole linear ion trap mass spectrometer. Mol. Cell Proteomics. 2006;5:337–346. doi: 10.1074/mcp.M500210-MCP200. [DOI] [PubMed] [Google Scholar]
- Winey M., Goetsch L., Baum P., Byers B. MPS1 and MPS2, novel yeast genes defining distinct steps of spindle pole body duplication. J. Cell Biol. 1991;114:745–754. doi: 10.1083/jcb.114.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. Regulation of APC-Cdc20 by the spindle checkpoint. Curr. Opin. Cell Biol. 2002;14:706–714. doi: 10.1016/s0955-0674(02)00382-4. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Chen R. H. Mps1 phosphorylation by MAP kinase is required for kinetochore localization of spindle-checkpoint proteins. Curr. Biol. 2006;16:1764–1769. doi: 10.1016/j.cub.2006.07.058. [DOI] [PubMed] [Google Scholar]
- Zhu S., Wang W., Clarke D. C., Liu X. Activation of Mps1 promotes transforming growth factor-beta-independent Smad signaling. J. Biol. Chem. 2007;282:18327–18338. doi: 10.1074/jbc.M700636200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.