Abstract
Fluorescent tagging is a powerful tool for imaging proteins in living cells. However, the steric effects imposed by fluorescent tags impair the behavior of many proteins. Here, we report a novel technique, Instant with DTT, EDT, And Low temperature (IDEAL)-labeling, for rapid and specific FlAsH-labeling of tetracysteine-tagged cell surface proteins by using prion protein (PrP) and amyloid precursor protein (APP) as models. In prion-infected cells, FlAsH-labeled tetracysteine-tagged PrP converted from the normal isoform (PrPsen) to the disease-associated isoform (PrPres), suggesting minimal steric effects of the tag. Pulse-chase analysis of PrP and APP by fluorescent gel imaging demonstrated the utility of IDEAL labeling in investigating protein metabolism by identifying an as-yet-unrecognized C-terminal fragment (C3) of PrPsen and by characterizing the kinetics of PrPres and APP metabolism. C3 generation and N-terminal truncation of PrPres were inhibited by the anti-prion compound E64, a cysteine protease inhibitor. Surprisingly, E64 did not inhibit the synthesis of new PrPres, providing insight into the mechanism by which E64 reduces steady-state PrPres levels in prion-infected cells. To expand the versatility of tetracysteine tagging, we created new Alexa Fluor- and biotin-conjugated tetracysteine-binding molecules that were applied to imaging PrP endocytosis and ultrastructural localization. IDEAL-labeling extends the use of biarsenical derivatives to extracellular proteins and beyond microscopic imaging.
INTRODUCTION
Surface proteins serve important functions in all cells in interactions that control cell survival. They also participate in pathological processes through improper cell signaling, acting as receptors for pathogens, or, as in Alzheimer's and prion diseases, converting into misfolded forms that may be directly toxic to cells. To understand how cell surface proteins participate in the above-mentioned processes, scientists often want to specifically label surface proteins and monitor changes in their distribution, structure, or metabolism over time by using biochemical and microscopy techniques. Achieving rapid and specific labeling of this crucial class of proteins at desired locations within the target proteins without altering their function has been a significant challenge of interest to many fields, including those studying prions.
Prions are unconventional pathogens lacking a nucleic acid genome and are the infectious agents of transmissible spongiform encephalopathies (TSEs) such as scrapie in sheep and bovine spongiform encephalopathy in cattle. Prions consist primarily of oligomers of a β-sheet–rich isoform (PrPres) of the prion protein (PrP), a glycosylphosphatidylinositol (GPI)-anchored glycoprotein. PrP is normally expressed in an α-helix–rich isoform (PrPsen) in many cell types. Prion propagation is thought to involve the PrPres-directed conformational refolding of PrPsen into new PrPres by using a templating mechanism requiring highly specific interactions between PrPsen and PrPres. Strain-associated pathogenic properties of prions are postulated to be enciphered in the strain-specific conformations of PrPres (Kocisko et al., 1994; Bessen et al., 1995; Telling et al., 1996; Safar et al., 1998). PrPsen and PrPres have different biochemical properties; for example, PrPsen is sensitive to protease digestion, but a “protease-resistant core” of PrPres, encompassing residues 89–231, resists digestion. PrPsen binds a variety of cellular components in interactions that are likely involved in executing its diverse physiological functions (Caughey and Baron, 2006). Some of these ligands have been shown to affect PrPres formation and can influence the pathogenesis of prion disease. Therefore, investigation of the behavior of PrPsen and PrPres is important to advance our understanding of prion pathogenesis.
To facilitate studies of PrP cell biology, we sought to develop methods to label both PrPsen and PrPres isoforms with fluorescent tags in live cells. Such methods would aid investigation into critical unresolved questions in the field, including mechanisms of uptake, propagation, and intercellular spread of prions. Unfortunately, attempts thus far to generate PrP fusions to fluorescent proteins (e.g., green fluorescent protein [GFP]) have failed to create fusion proteins capable of converting to GFP-tagged PrPres likely due in part to the large size and/or location of the fluorescent tag (Barmada and Harris, 2005; Bian et al., 2006). To fluorescently label PrPres, a fluorescent tag must be protease resistant and inserted into the region that ultimately forms the protease-resistant core because the N-terminal region (residues 23–88) is removed by endogenous proteases after PrPsen converts to PrPres (Caughey et al., 1991). However, this region of PrP is so intolerant to modification that even a single mutation in a critical position can severely impair conversion of PrPsen (Priola and Chesebro, 1995; Kaneko et al., 1997). This places additional constraints on tags that can be used to label PrP molecules, some of which apply to many other proteins.
To circumvent these problems, we have used one of the smallest tags available, the tetracysteine (TC) motif (CCPGCC), which preferentially binds to biarsenical compounds such as the fluorescein derivative called FlAsH (Griffin et al., 1998; Adams et al., 2002). FlAsH/TC-motif labeling has its own inherent problems, including very high background staining due to FlAsH binding to off-target cysteine-rich proteins (Stroffekova et al., 2001; Berens et al., 2005; Langhorst et al., 2006; Hearps et al., 2007). In addition, extracellular proteins have been poor candidates for FlAsH-labeling because the cysteine residues of the TC-motif are oxidized in extracellular environments and/or during export through the endoplasmic reticulum and consequently lose their affinity for FlAsH. The only report of FlAsH labeling of extracellular proteins (Adams et al., 2002) required a custom-made, membrane-impermeant variant of FlAsH and extended preincubation of the cells with reductants to recover the affinity of the TC-motif for FlAsH, making the procedure complex and impractical.
We have developed a novel technique, termed Instant with DTT, EDT, And Low temperature (IDEAL)-labeling, for very rapid and specific labeling of cell surface PrPsen that converts into fluorescent PrPres in live cells. Using this approach, we show that the TC-motif can be used to label specific internal sites in proteins with minimal effects on PrP behavior. With pulse-chase analyses of FlAsH-labeled PrP, we expand the characterization of PrPsen metabolism by demonstrating a new proteolytic fragment derived from the C-terminus and provide new insight into the mechanism of action of the antiprion compound E64. Furthermore, we have synthesized two new FlAsH derivatives, consisting of conjugates to Alexa Fluor dye or biotin, each of which displays unique advantages over FlAsH, such as improved fluorescence properties, versatility of detection, and ease of affinity purification. We provide evidence that IDEAL-labeling is generally applicable to studies of cell surface proteins by the successful labeling and analysis of another protein of broad interest, amyloid precursor protein (APP). The novel IDEAL-labeling and biarsenical conjugate technologies will encourage more widespread use of TC-motif tagging, especially in the context of cell surface proteins.
MATERIALS AND METHODS
Reagents
FlAsH in-cell labeling kit was purchased from Invitrogen (Carlsbad, CA). Triton X-100, Triton X-114 (TX114), deoxycholate, dithiothreitol (DTT), ethanedithiol (EDT), 2-mercaptoethanesulfonate (MES), tris-(2-carboxyethyl)phosphine (TCEP), ammonium chloride, and chymotrypsin were purchased from Sigma-Aldrich (St. Louis, MO). E64, Pefabloc SC, and Complete were from Roche Diagnostics (Indianapolis, IN). All the media, buffers, reagents, and fetal bovine serum (FBS) for cell culture were purchased from Invitrogen (Carlsbad, CA). All the restriction enzymes and PNGase F were from New England Biolabs (Ipswich, MA).
TC-PrP Constructs
The TC-motif was inserted in PrP with the QuikChange site-directed mutagenesis kit (Stratagene, Cedar Creek, TX) according to the manufacturer's instructions. Mouse PrP open-reading frame (gift from Dr. Ryuichiro Atarashi, Nagasaki University, Nagasaki, Japan) flanked by a HindIII site and an XbaI site on the 5′ and 3′ sides, respectively, and cloned in pCR cloning vector (Invitrogen) was used as the template. Primers used to construct PrP mutants were as follows: for 90TC sense (5′-TGCCCCGGGTGTTGTCAAGGAGGGGGTACCCACAATC-3′) and antisense (5′-ACAACACCCGGGGCAGCAGCCCCATCCACCGCCATG-3′) primers were used; and for 230TC sense (5′-TGCCCCGGGTGTTGTGATTCCAGCAGCACCGTGCTTTTC-3′) and antisense (5′-ACACCCGGGGCAGCAGCCTCTTCTCCCGTCGTAATAGGC-3′) primers were used. The 230TC construct encodes a TC motif flanked by glycine and aspartate residues (GCCPGCCD). Addition of these residues was based on theoretical considerations. The glycine was inserted to add conformational flexibility, and aspartate was inserted to act as a spacer to ensure proper addition of the GPI anchor. A variant of the 230TC construct (230TC*) containing the minimal TC motif (CCPGCC) also was created (5′-TGCTGCCCCGGGTGTTGTTCCAGCAGCACCGTGCTTTTC-3′, sense primer; and 5′-TGCTGCCCCGGGTGTTGTTCCAGCAGCACCGTGCTTTTC-3′, antisense primer), and this was used in the experiments described in Supplemental Figure 1. The mutated PrP genes were cleaved with HindIII and XbaI and subsequently cloned in an expression vector, pcDNA3.1(+) (Invitrogen).
TC-APP Construct
Human APP695 construct (EX-F0776-M02) was purchased from Genecopoeia (Germantown, MD). Based on this construct, we made a construct of human APP695 cloned in pcDNA3.1, flanked by HindIII and XbaI sites, and used this construct as the template in site-directed mutagenesis. Sense and antisense primers to insert the TC-motif between residues 597 and 598 were 5′-GCTGCCCCGGGTGTTGTGCAGAATTCCGACATGACTCAGG-3′ and 5′-AACACCCGGGGCAGCAATCCATCTTCACTTCAGAGATCTCC-3′, respectively.
Generation of Cell Lines Stably Producing TC-PrPres
Neuro2a (N2a) cells, a mouse neuroblastoma cell line, and 22L-scrapie-infected N2a cells (22L/N2a) were reported previously (Kocisko et al., 2003). The 22L/N2a cells were transfected with the pcDNA3.1–90TC with Effectene (QIAGEN, Valencia, CA) according to the manufacturer's instructions, and then stably transfected cells were selected in medium containing G418. The cells were single-cell cloned and screened for high levels of FlAsH-PrPres(90TC), resulting in the 22L(90TC) line. For cells persistently producing TC-PrPres(230TC), at first noninfected N2a cells stably expressing TC-PrP(230TC) were made, and then the cells were cocultured with 22L/N2a for 1 wk. After elimination of 22L/N2a cells in G418-containing medium, the remaining cells were single-cell cloned and screened for high levels of FlAsH-PrPres(230TC), resulting in the 22L(230TC) line. The two scrapie-infected cell lines were cured of infection by PPS-treatment (Birkett et al., 2001), and the cured cell lines, N2a(90TC) and N2a(230TC), were used as noninfected counterparts.
IDEAL-Labeling
Culture media for cell growth and labeling and all solutions should be 0.2-μm filtered to remove particles that may nonspecifically bind FlAsH. First, the necessary amount of biarsenical compound was calculated based on the required volumes of labeling medium and desired final concentration of the fluorophore. The calculated amount of biarsenical compound was mixed with an equal volume of 50 mM EDT and one-half volume of 2 M DTT and incubated in the dark for ∼10 min at room temperature (RT). Meanwhile, Opti-MEM-I with 1% FBS and 20 mM NH4Cl [OM(1% FBS)] was added to rinse the cells. Then, OM(1% FBS) was added to the fluorophore mixture to make the final biarsenical compound concentration to be 1.3–1.4 μM and supplemented with 2 M DTT to make the final DTT concentration 15–20 mM. The labeling medium was kept on ice until use. After removal of the rinse medium, the cells were treated for 3 min with a minimal volume of labeling medium sufficient to cover the cells. After removing the labeling medium, OM(1% FBS) was added to the cells and after 5–10 min in the dark, the OM(1% FBS) was replaced with Opti-MEM-I with 10% FBS, and the cells were placed in a CO2 incubator until 30 min elapsed since the beginning of labeling. Finally, the labeled cells were harvested or used for FlAsH-pulse chase.
Fluorescent Gel Analysis/FlAsH Pulse-Chase Experiments
To enhance TC-PrP expression, the cells were treated with 1.5 mM sodium butyrate (in media) for 10–12 h followed by overnight incubation without sodium butyrate. After IDEAL-labeling, cells were rinsed with Hanks' balanced salt solution and harvested with 210 μl of phosphate-buffered saline 0.5% Triton X-100/0.5% deoxycholate (TX100/DOC) lysis buffer with protease inhibitor cocktail Complete. After centrifugation, 10 μl of supernatant was taken for “lysate” samples, and the rest was subjected to phosphotungstate (PTA) precipitation. Lysate samples were deglycosylated with PNGase F according to the manufacturer's instructions, except that 0.25× sample buffer without reductant (0.5% SDS, 12.5 mM Tris-HCl, 2% glycerol, and 0.00025% bromphenol blue, pH 7.1) was used for denaturation. For PTA-precipitated samples, N-lauroylsarcosine (2% final concentration) was added to the lysate followed by shaking at 37°C for 5–10 min. If protease digestion was required, lysates were made without Complete and chymotrypsin or proteinase K (PK) digestion (15 μg/ml; 30 min) was performed before PTA precipitation. Digestion was stopped with 2 mM Pefabloc SC. Next, 1/12.5-volume of PTA/Mg stock solution (4% sodium phosphotungstate and 170 mM magnesium chloride, pH 7.4) was added followed by shaking at 37°C for 30 min. Thereafter, the sample was centrifuged at 22,500 × g, 26°C for 40 min. In the lysates from 22L(230TC) cells, a “wash” was necessary to reduce FlAsH-PrPsen in PTA precipitates (Supplemental Figure 2). Pellets were deglycosylated with PNGase F. The samples were run on 10% Bis-Tris NuPAGE precast gels with 2-(N-morpholino)ethanesulfonic acid running buffer (Invitrogen). Gels were scanned on a Typhoon scanner (GE Healthcare, Piscataway, NJ) with excitation at 488 nm/520BP40 emission (FlAsH) or excitation at 532 nm/610BP30 emission (AF568-FlAsH). Images were analyzed with ImageQuant software (GE Healthcare).
C3-like Fragment in Wild-Type N2a Cells
N2a cells on a 10 cm dish were harvested with TX114 lysis buffer and then endogenous PrP was partially purified by phase-separation. Samples prepared from the detergent phase were precipitated with methanol/chloroform. The pelleted protein was dissolved in 50 μl of 6M guanidine hydrochloride (GdnHCl). The GdnHCl solution was diluted with 950 μl of 2% TX114 lysis buffer, subjected to phase separation at 37°C, and then centrifuged at 22,500g at 26°C for 10 min. After removal of the aqueous phase, 1 ml of 60 mM of sodium acetate (pH 5.2) was added to the detergent phase, vortexed rigorously, and incubated on ice until it became clear. The cleared lysate was centrifuged at 22,500g at 4°C for 15 min. The supernatant was transferred to a new tube, subjected to phase separation, centrifuged as above, and the aqueous phase was discarded. Then 800 μl of wash buffer (0.1% TX114 in Tris-buffered saline, pH 7.5) was added to the detergent phase, vortexed rigorously, and incubated on ice for 5 min. The clear lysate was centrifuged again at 22,500g at 4°C for 15 min. The supernatant was transferred to a new tube, phase-separated at 37°C for 10 min, centrifuged as above, and the aqueous phase was removed. Then 200 μl of wash buffer was added and the lysate was subjected to methanol/chloroform precipitation. The pellet was resuspended in 16 μl of 0.25X sample buffer, deglycosylated, and 5 μl of 5X sample buffer was added and boiled. Out of 25 μl, 10 μl was subjected to SDS-PAGE and transferred to PVDF membrane by a semi-dry method. In immunoblotting, R1 anti-PrP monoclonal antibody (mAb) (Williamson et al., 1998) (kind gift from Dennis Burton and Anthony Williamson, Scripps Research Institute, La Jolla, CA) was used for detection of the C-terminal epitope (220–231) of PrP, and the secondary antibody was alkaline phosphatase-conjugated anti-human Fab (Sigma- Aldrich). Blots were developed using Attophos substrate (Promega, Madison, WI) and scanned on a Storm 860 PhosphorImager (GE Healthcare). Images were analyzed with ImageQuant software (GE Healthcare).
E64 Treatment
To assess the effects of prolonged E64 treatment on steady-state PrPres levels, 22L(230TC) cells were plated on 24-well plates and kept in medium containing E64 at the indicated concentration for 3 d, with daily media changes to maintain the reagent concentration. The cells were harvested with TX100/DOC lysis buffer, digested with PK (20 μg/ml) for 30 min at 37°C, and the digestion was stopped with 2 mM Pefabloc SC. After addition of 5× sample buffer to 1× concentration, the lysates were boiled. The samples were subjected to SDS-PAGE and immunoblotting as described above using the anti-PrP mAb D13 (InPro, South San Francisco, CA). In FlAsH-pulse-chase experiments to measure the effects of E64 specifically on PrPres biosynthesis, E64 (80 μg/ml) treatment was initiated after completion of IDEAL-labeling.
Confocal Microscopy Studies of Fluorescent Biarsenical-labeled Cells
Cells were plated on precleaned (30-min bath sonication in ethanol followed by rinses with sterile water) coverslip-bottomed dishes (Willco Wells B.V., Amsterdam, The Netherlands) and labeled with the designated biarsenical compound as described above. Cells expressing TC-PrP (Figures 1B and 5C) were fixed (4% paraformaldehyde/5% sucrose in phosphate-buffered saline [PBS] at RT for 30 min) before imaging in PBS for purely technical reasons to minimize PrP endocytosis before and during imaging and to allow neutralization of the pH of intracellular compartments to ensure our images of FlAsH-labeled cells (Figure 1B) would account for all populations of the pH-sensitive FlAsH dye. Imaging of live or fixed cells was performed using a Nikon LiveScan Swept-field confocal scan head mounted on a Nikon TE2000E microscope with a 60× 1.49 numerical aperture (NA) TIRF objective and an ASI XY Piezo Z scanning stage. Emission filters ET525BP50 and ET620BP60 were from Chroma Technology (Brattleboro, VT). The microscope was enclosed in an environmentally controlled chamber (In Vivo Scientific, St. Louis, MO) maintained at 37°C and (for live cells) supplemented with humidified 5% CO2. Images were captured with a Cascade II:512 camera. Some images (Supplemental Figure 1A) were captured using an MRC1024 laser-scanning confocal system (Bio-Rad, Hercules, CA) coupled to a Zeiss microscope using a 40× 1.2 NA water immersion objective. Experimental and control images were always acquired and analyzed with identical settings. Images were analyzed using Nikon NIS-Elements (Nikon Instruments, Melville, NY), ImageJ (W. S. Rasband, National Institutes of Health, Bethesda, MD), Imaris (Bitplane, St. Paul, MN), and AutoQuant (Media Cybernetics, Bethesda, MD) software. Images were deconvolved using three-dimensional-blind iterative deconvolution in the AutoDeblur module of AutoQuant. Figures were assembled in Adobe Photoshop (Adobe Systems Mountain View, CA).
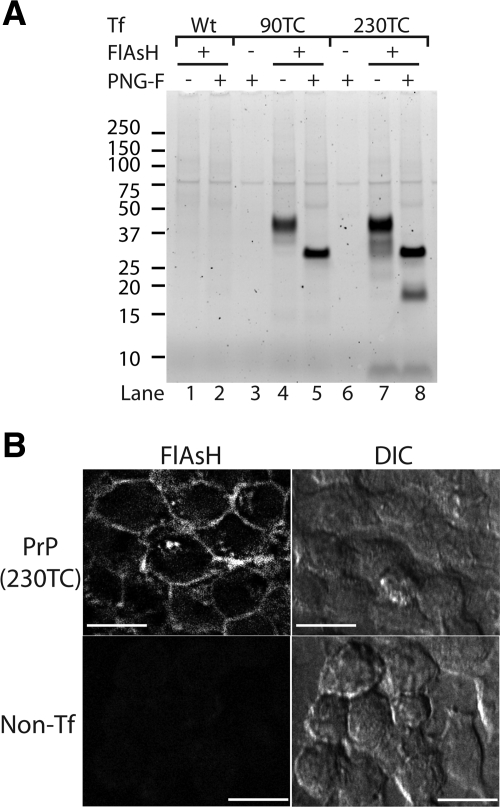
Figure 1.
IDEAL-labeling of cell surface TC-PrP with FlAsH. (A) Fluorescent gel analysis of cell lysates from IDEAL-labeled cells labeled with FlAsH. Cells were transfected with either wild-type PrP (lanes 1 and 2) or the indicated TC-PrP (lanes 3–8) expression construct. Samples where FlAsH was omitted show autofluorescent proteins (lanes 3 and 6). PNG-F indicates samples deglycosylated with PNGase F. (B) Confocal microscopy images of IDEAL-labeled cells labeled with FlAsH. PrP(230TC), cells expressing PrP with TC motif at position 230. Non-Tf, nontransfected control cells. DIC, differential interference contrast. Bars = 20 μm.
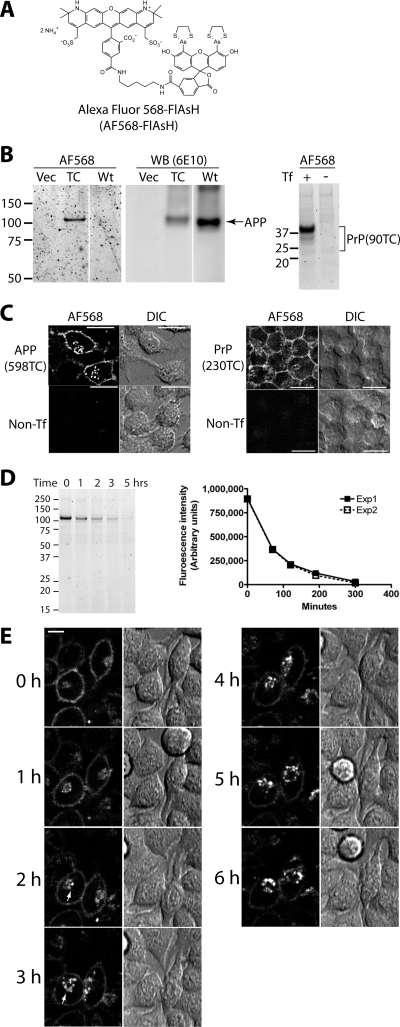
Figure 5.
Specific labeling of TC-APP and TC-PrP with AF568-FlAsH. (A) Structure of Alexa Fluor 568-carboxy-FlAsH (AF568-FlAsH). (B) APP(598TC) and PrP(90TC) are specifically labeled with AF568-FlAsH. Samples from transfected and IDEAL-labeled cells were subjected to fluorescent gel analysis (AF568). The APP gel was then analyzed by immunoblotting (WB) with 6E10 anti-APP antibody. Vec, cells transfected with pcDNA3.1. TC, sample from cells expressing APP(598TC). Wt, cells expressing wild-type APP. Tf, transfection. (C) Confocal microscopy images of AF568-FlAsH-labeled APP(598TC) and TC-PrP(230TC). APP(598TC), cells expressing APP(598TC) labeled with AF568-FlAsH. Non-Tf, nontransfected cells labeled with AF568-FlAsH. PrP(230TC), cells expressing PrP(230TC) labeled with AF568-FlAsH. Bars, 20 μm. (D) Kinetics of AF568-FlAsH-APP(598TC) metabolism. AF568-FlAsH-pulse chase on N2a cells transiently expressing APP(598TC). The fluorescent gel shows representative results of an experiment performed in duplicate. The graph shows kinetics of catabolism of full-length AF568-FlAsH-labeled APP(598TC). Two independent experiments (each performed in duplicate) gave similar results. Average band intensities of the duplicate samples within each experiment are plotted and the results from both experiments are shown. (E) Live cell imaging of endocytic trafficking of PrP(230TC). Stably-transfected cells expressing PrP(230TC) were IDEAL-labeled with AF568-FlAsH and selected fields of view were subsequently monitored hourly by live cell confocal microscopy. Arrows indicate newly-endocytosed PrP(230TC) trafficked to a perinuclear compartment. A representative example of the events observed is shown. Fluorescent images correspond to a single confocal Z-slice located 7 μm above the adherent surface of the cells. Bar, 10 μm.
Detection of Bio-FlAsH-labeled TC-PrPs
Transiently (Figure 6B) or stably (Figure 6, C and D)-transfected N2a cells were labeled with Bio-FlAsH. For gel analysis, the labeled cells were harvested immediately, and cell lysate samples were made as described above. The samples were fractionated by SDS-PAGE, and a fluorescence image of the gel was captured on a Typhoon scanner (GE Healthcare) before semidry transfer to a PVDF membrane. The membrane was blocked with Superblock (Pierce Chemical, Rockford, IL) supplemented with 0.05% Tween 20 for 4 h, incubated with alkaline phosphatase-conjugated NeutrAvidin (Pierce Chemical) diluted 1:15,000 in the blocking solution for 1 h, and then was washed in 0.05 Tween 20 in Tris-buffered saline. Detection of blots and analysis were as above.
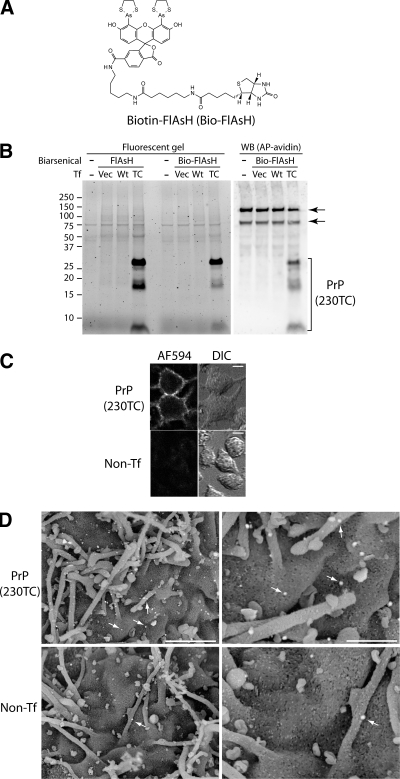
Figure 6.
Specific labeling of TC-PrP with Bio-FlAsH. (A) Structure of biotin-carboxy-FlAsH (Bio-FlAsH). (B) Comparable labeling specificity and efficiency of Bio-FlAsH (lanes 6–8) to FlAsH (lanes 2–4) and specific detection of Bio-FlAsH-PrP by immunoblotting with NeutrAvidin (lanes 9–12). (−), unlabeled, nontransfected cells. Vec, cells transfected with pcDNA3.1. Wt, cells transfected with wild-type PrP. TC, cells transfected with PrP(230TC). WB(AP-avidin), immunoblot with alkaline phosphatase-conjugated NeutrAvidin. Bracket indicates PrP(230TC) bands. Arrows indicate endogenous biotinylated proteins. (C) Visualization of Bio-FlAsH-labeled PrP(230TC) with Alexa Fluor 594 streptavidin-FluoroNanogold. Cells were labeled sequentially with Bio-FlAsH and then with streptavidin-FluoroNanogold before confocal microscopy. PrP(230TC), cells stably expressing PrP(230TC). Non-Tf, nontransfected cells. Bars, 10 μm. (D) Visualization of PrP(230TC) by scanning electron microscopy. Cells were labeled sequentially with Bio-FlAsH and then with streptavidin-FluoroNanogold before imaging by scanning electron microscopy. Arrows indicate gold particles. Only rare gold particles were observed on nontransfected control cells (bottom). PrP(230TC), cells stably expressing PrP(230TC). Non-Tf, nontransfected cells. Bars, 1.5 μm (left) and 600 nm (right).
For analysis by confocal microscopy, the cells were grown on precleaned coverslip-bottom dishes. Immediately after Bio-FlAsH labeling, the cells were rinsed once with Hanks' balanced Salt solution and chilled to 4°C in PBS. The cells were labeled for 20 min at 4°C with a 1:20 dilution of Alexa Fluor 594-streptavidin-FluoroNanogold (Nanoprobes, Yaphank, NY) in PBS + 2% bovine serum albumin (BSA) + 0.1% BSA-c (Electron Microscopy Sciences, Hatfield, PA). After multiple washes with ice-cold PBS + 0.1% BSA-c, one wash of which included 10 mM d-biotin, the cells were fixed with 4% paraformaldehyde/5% sucrose in PBS for 20 min. Finally, the cells were rinsed with PBS before imaging.
For analysis by scanning electron microscopy, cells were grown on clean silicon chips and labeled with Bio-FlAsH as described above. After the final wash, the cells were fixed in Karnovsky's fixative (Electron Microscopy Sciences) at RT. Samples were then processed for gold enhancement (GOLDENHANCE-EM formulation; Nanoprobes) as per the manufacturer's instructions with a reaction time of 8 min. Using microwave-assisted processing, samples were then postfixed with an OTO method with 1% osmium tetroxide in cacodylate buffer followed by saturated thiocarbohydrazide, washed twice with water, dehydrated with ethanol, and critical-point dried through carbon dioxide. The silicon chips were then mounted on aluminum stubs, lightly coated with chromium using an ion beam sputterer (South Bay Technology, Arlington, VA), and imaged at 5 kV on a Hitachi S4500 field emission scanning electron microscope (Hitachi High Technologies America, Schaumburg, IL) in mixed mode to simultaneously detect backscattered and secondary electrons.
Additional Methods
Descriptions of the synthesis of the Alexa Fluor 568-FlAsH and Bio-FlAsH biarsenical compounds are available in Supplemental Material.
RESULTS
IDEAL-Labeling of Cell Surface TC-PrP with FlAsH
To develop methods for labeling cell surface-expressed TC-tagged proteins, we first created PrP constructs containing TC insertions (TC-PrPs) at either residue 90 [PrP(90TC)] or 230 [PrP(230TC)]. Initially, we tried labeling cells expressing TC-PrPs with FlAsH by using protocols based on one published for labeling extracellular proteins (Adams et al., 2002). We failed to detect any TC-PrP-specific staining and observed very high background labeling as reported by others (Stroffekova et al., 2001; Berens et al., 2005; Langhorst et al., 2006; Hearps et al., 2007) (see Supplemental Figure 1).
To improve the labeling procedure, we used DTT as a reducing agent instead of MES (Adams et al., 2002), expecting that DTT might reduce cystine residues of the TC-motif more efficiently. After testing many conditions varying all aspects of the labeling protocol (e.g., labeling temperature, time, reagents, reagent concentrations, and pre- and postincubation treatments to reduce nonspecific binding; see Supplemental Figure 1 for a few selected examples), we found incubation of the cells with DTT plus FlAsH for only 2–3 min achieved a high labeling efficiency and specificity (Figure 1A, lane 1 vs. lanes 4 and 7). Incubation for >3 min did not result in additional labeling (data not shown). This was not due to exhaustion of the labeling reagents as a single aliquot of labeling medium could be used to sequentially label duplicate cell cultures and achieve equivalent labeling efficiency for each culture (data not shown). Together, these observations suggested that maximal labeling had been achieved within the context of these rapid labeling conditions. The banding pattern for FlAsH-PrP(90TC) represented the three glycoforms of PrPsen as shown by deglycosylation (Figure 1A, lane 4 vs. lane 5). FlAsH-labeled PrP(230TC) showed additional lower molecular weight (MW) bands that were more clearly resolved after deglycosylation (Figure 1A, lane 7 vs. lane 8), which corresponded to metabolic fragments of PrP (see below). We also observed characteristic cell surface PrP staining by confocal microscopy (Lee et al., 2001) (Figure 1B). Some FlAsH-PrP seemed localized to intracellular vesicles, perhaps due to internalization of cell surface FlAsH-PrP after labeling (see below). These data represent the first demonstration that specific FlAsH labeling of cell surface TC-tagged proteins can be achieved without using membrane-impermeable derivatives of FlAsH (Adams et al., 2002). We termed this new rapid technique IDEAL-labeling.
FlAsH-PrPsen Is Converted to FlAsH-PrPres
To evaluate the influence of biarsenical tagging on protein folding behavior, FlAsH-labeled PrPsen(90TC) (Figure 2A) was tested for conversion to FlAsH-PrPres. Conversion of PrPsen to PrPres is associated with acquisition of partial resistance to protease digestion. N-Terminal truncation of new PrPres is mediated either by endogenous cellular proteases or by exogenous protease digestion of cell lysates (Figure 2B, lane 3 vs. lane 4). In FlAsH-pulse-chase experiments of cells expressing PrP(90TC), an ∼21-kDa fragment accumulated over time specifically in cells infected with mouse scrapie prions (Figure 2C, arrowhead), consistent with the expected apparent MW of deglycosylated FlAsH-PrPres that had been truncated by cellular proteases. This 21-kDa fragment was efficiently precipitated by PTA (Figure 2D, arrowhead), a procedure to enrich for PrPres (Safar et al., 1998). The scrapie infection-specific 21-kDa fragment resisted digestion with PK, showing that it is FlAsH-PrPres (Figure 2E). The absence of the 21-kDa fragment immediately after labeling (e.g., Figure 2D, lane 1) demonstrates that pre-existing PrPres was not labeled; hence, the FlAsH-PrPres must have been newly synthesized from a FlAsH-PrPsen precursor.
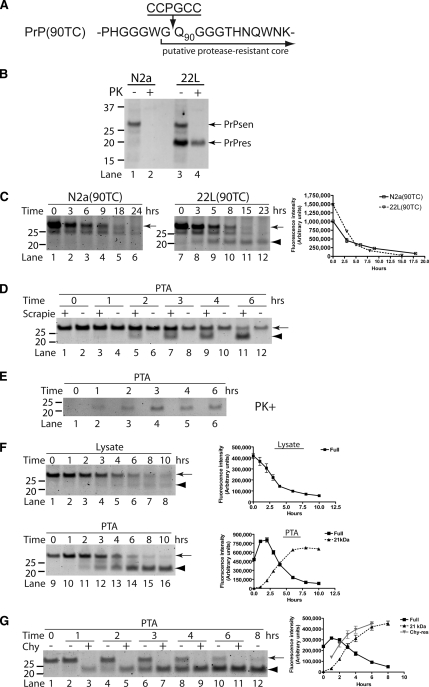
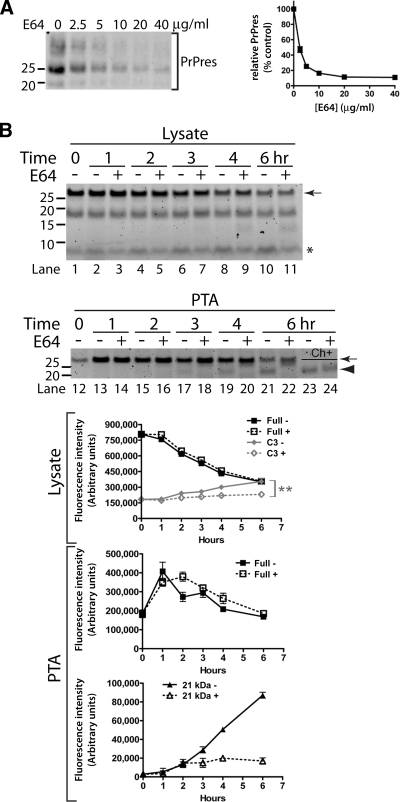
Figure 2.
FlAsH-PrPsen conversion to FlAsH-PrPres and kinetic analysis of FlAsH-PrP. In the experiments presented, scrapie-infected [22L(90TC)] and uninfected [N2a(90TC)] N2a cells expressing PrP(90TC) were used unless otherwise noted. All the samples were deglycosylated with PNGase F. The arrows indicate full-length FlAsH-PrP(90TC), and the arrowheads indicate the scrapie infection-specific 21-kDa fragment corresponding to proteolytically truncated PrPres. Representative gels are shown. Error bars are included on all graphs but in many cases are small enough to be obscured by the plot symbols. (A) Structure of PrP(90TC). The putative proteolytic cleavage site as occurs after conversion to PrPres is indicated. (B) Proteolytic truncation of wild-type PrPres. Immunoblot (D13 antibody) of uninfected (N2a) and scrapie-infected (22L) N2a cells expressing only wild-type PrP. The ∼21-kDa PrPres band is specific to infected cells and is smaller than full-length PrPsen due to proteolytic truncation. (C) Scrapie infection-specific fragment, 21-kDa band. FlAsH-pulse chase of FlAsH-PrP(90TC) in detergent-phase of TX114 lysates from 22L(90TC) and N2a(90TC) cells. Replicate experiments were performed (n = 4). The graph shows the quantitative analysis of the band intensities of full-length (arrow) FlAsH-PrP (average ± SEM). (D) Comparison of FlAsH-PrP in PTA precipitates from 22L(90TC) (scrapie “+”) and N2a(90TC) (scrapie “−”) cells. The fluorescent gel shows FlAsH-PrPs in PTA-precipitated samples from FlAsH-pulse-chase analysis of each cell line. Experiments were done in quadruplicate. (E) TC-PrPres is not directly labeled with FlAsH but FlAsH-PrPsen converts to FlAsH-PrPres. Scrapie-infected cells transiently transfected with pcDNA3.1–90TC were labeled with FlAsH and harvested after various incubation times. Samples correspond to PTA-precipitated pellets of PK-digested lysates. (F) Kinetics of FlAsH-PrP(90TC) in cell lysate and PTA-precipitated samples from 22L(90TC) cells. The fluorescent gels and graphs show results of FlAsH-pulse-chase analysis of 22L(90TC). Lysate fractions correspond to 1/20 the cell equivalents loaded in the PTA fractions. Lysate, kinetics of FlAsH-PrP(90TC) in cell lysate fraction. PTA, kinetics of FlAsH-PrP(90TC) in PTA-precipitated fraction. ■, Full, full-length FlAsH-PrP(90TC). ▴, 21 kDa, 21-kDa band. The experiments were performed in triplicate and the curves show the average band intensities (± SEM). (G) Kinetics of 21-kDa band and Chy-res metabolism. The fluorescent gel shows FlAsH-PrP in PTA precipitates from FlAsH-pulse chase of 22L(90TC) cells. ■, Full, full-length FlAsH-PrP(90TC) in samples without digestion. ▴, 21 kDa, 21-kDa band in samples without digestion. ( ) Chy-res, 21-kDa band in samples from lysates digested with chymotrypsin before PTA precipitation. The experiments were performed in triplicate and the curve shows the average band intensity ± SEM.
) Chy-res, 21-kDa band in samples from lysates digested with chymotrypsin before PTA precipitation. The experiments were performed in triplicate and the curve shows the average band intensity ± SEM.
To further characterize the FlAsH-labeled PrP molecules, we assessed the kinetics of FlAsH-PrPsen metabolism and conversion to FlAsH-PrPres. In uninfected cells (Figure 2C, arrow, lanes 1–6), FlAsH-PrPsen had a half-life of ∼3 h, similar to that determined previously by radioactive pulse-labeling experiments (Caughey and Raymond, 1991). A similar half-life also was observed for FlAsH-PrPsen in infected cells (Figure 2C, arrow, lanes 7–12 and Figure 2F, lysate), which was likely a reflection of the fact that FlAsH-PrPres constituted only a small fraction of the total FlAsH-PrP in these cells (see below).
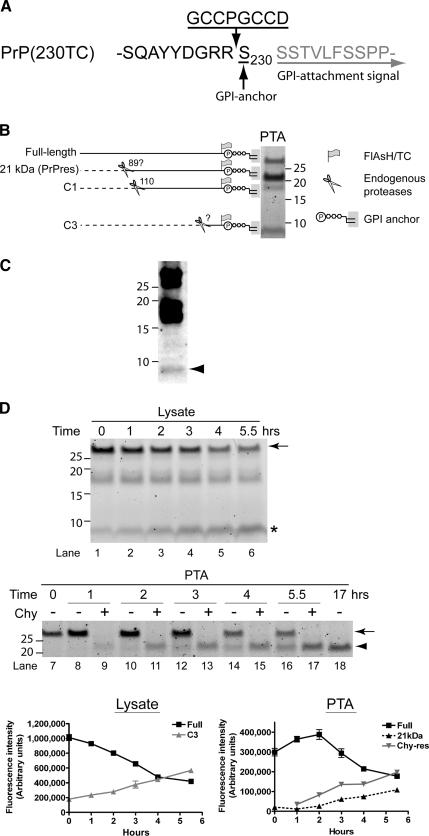
To assess the kinetics of FlAsH-PrPres metabolism, including the kinetics of processing by cellular proteases, we first needed a method to prepare PrPres from cells without using digestion by exogenous proteases as a means to remove PrPsen. PTA precipitation is a technique used to selectively concentrate PrPres (Safar et al., 1998), although a low level of PrPsen is known to be precipitated with PTA (Lee et al., 2005) (Supplemental Figure 2). The full-length FlAsH-PrP (arrow) detected in PTA fractions at 0 h likely represents this contaminating PrPsen (Figure 2F, lane 9). Over time, full-length FlAsH-PrP initially increased and then decreased quickly; reciprocally, the 21-kDa fragment appeared and increased (Figure 2F, PTA). The reciprocal nature of the kinetics of full-length FlAsH-PrP versus the 21-kDa fragment was consistent with the formation and subsequent truncation of FlAsH-PrPres by endogenous cysteine proteases (A). This construct enabled the detection of C-terminal products of PrP metabolism. Three FlAsH-labeled fragments were common to both uninfected (Figure 1A, lane 8) and scrapie-infected (Figure 3, B and D) PrP(230TC)-expressing cells. These consisted of full-length PrP; a fragment of ∼17–18 kDa, likely a proteolytic product called C1 (Chen et al., 1995; Taraboulos et al., 1995); and a previously unreported fragment of ∼9 kDa, which we called C3 (Figure 3B). A 21-kDa fragment was specific to scrapie-infected cells and corresponded to truncated FlAsH-PrPres (see below). A C3-like fragment was present in wild-type N2a cells, providing evidence that this newly identified fragment was not an artifact of FlAsH-PrP(230TC) (Figure 3C).
Figure 3.
Kinetic analysis of FlAsH-PrP(230TC) identifies a new metabolic fragment of PrP. (A) Structure of PrP(230TC). The GPI-attachment signal is removed from the nascent protein before GPI anchor addition to Ser230. (B) Summary of metabolic fragments of FlAsH-PrP(230TC). Proteolytic fragments observed were generated by endogenous proteases. The fluorescent gel shows FlAsH-PrP(230TC) fragments in PTA precipitates from 22L(230TC) cells chased for several hours after labeling. To create this specific image for reference purposes only, the precipitate was not washed with N-lauroylsarcosine buffer after PTA precipitation to retain contaminant fragments that are preferentially reduced by washing (e.g., full-length). (C) Nontransfected N2a cells have a C3-like fragment. Immunoblotting of wild-type PrP from detergent phase of TX114 lysates of nontransfected N2a cells with monoclonal R1 antibody, which recognizes C-terminus (residues 220–231) of PrP. Arrowhead indicates the ∼9-kDa C3-like fragment. (D) Kinetics of FlAsH-PrP(230TC) in 22L(230TC) cells. The fluorescent gels and the graphs show the results of FlAsH-pulse-chase analysis of 22L(230TC) cells. Lysate fractions correspond to 1/20 the cell equivalents loaded in the PTA fractions. The experiments were performed in quadruplicate and the curves show average band intensity ± SEM. The arrows indicate full-length FlAsH-PrP(230TC), the arrowhead indicates the 21-kDa truncated PrPres band, and the asterisk indicates the C3 fragment. Error bars are included on all graphs but in many cases are small enough to be obscured by the plot symbols. Lysate, kinetics of FlAsH-PrP(230TC) in cell lysate samples. PTA, kinetics of FlAsH-PrP(230TC) in PTA-precipitated samples. ■, Full, full-length FlAsH-PrP(230TC).  , C3, C3 fragment. ▴, 21 kDa, 21-kDa band.
, C3, C3 fragment. ▴, 21 kDa, 21-kDa band.  , Chy-res, samples digested with chymotrypsin.
, Chy-res, samples digested with chymotrypsin.
To better evaluate the kinetics of FlAsH-PrPres synthesis, truncation with an exogenous protease was necessary before gel analysis to include (in the 21-kDa band) amounts of FlAsH-PrPres not yet truncated by endogenous proteases. PK seemed too harsh for this purpose because the amount of FlAsH-PrPres was always lower in PK-treated samples (Supplemental Figure 4A). We discovered that chymotrypsin efficiently digested FlAsH-PrPsen while preserving FlAsH-PrPres (Supplemental Figure 4, B and C). Therefore, we used chymotrypsin treatment to assay for total FlAsH-PrPres in the following experiments. The kinetics of chymotrypsin-truncated FlAsH-PrPres (Chy-res) formation confirmed that the full-length FlAsH-PrP band in PTA precipitates contained some FlAsH-PrPres (Figure 2G). By comparison of levels of FlAsH-PrPsen in cell lysates and FlAsH-PrPres, we estimated that ∼7–11% of the initial FlAsH-PrPsen is converted into FlAsH-PrPres. This is similar to estimates made using radiolabeled PrPsen (Caughey and Raymond, 1991; Borchelt et al., 1992) (∼3–10%), suggesting that FlAsH-labeling does not impair the conversion efficiency of PrPsen. Collectively our analyses of the kinetics of FlAsH-PrPres metabolism are in accordance with observations based on metabolic labeling (Caughey et al., 1991; Caughey and Raymond, 1991; Borchelt et al., 1992; Taraboulos et al., 1992), providing strong evidence that FlAsH/TC motif tagging exerts a minimal impact on PrP metabolism and conversion.
C3 Is a New Metabolic Fragment of PrPsen
To explore the flexibility to insert the TC motif at desired locations within a target protein, we created a PrP construct [PrP(230TC)] with the TC-motif adjacent to the GPI attachment site (Figure 3 Supplemental Figure 3 and see below). These kinetics suggested N-terminal truncation of FlAsH-PrPres occurs shortly after conversion. This is similar to the truncation of wild-type PrPres observed by pulse-chase radiolabeling (Caughey et al., 1991).
Next, we determined the kinetics of FlAsH-PrP(230TC) metabolism. Full-length FlAsH-PrPsen(230TC) showed a half-life of ∼4 h (Figure 3D, lysate). The C3 fragment accumulated during the chase period (Figure 3D, asterisk, lysate). In PTA precipitates with and without chymotrypsin digestion to monitor the kinetics of FlAsH-PrP(230TC) conversion to FlAsH-PrPres, the kinetics of FlAsH-PrP(230TC) basically mirrored that of FlAsH-PrP(90TC) (Figure 3D). The estimated conversion efficiency for FlAsH-PrP(230TC) was somewhat lower than FlAsH-PrP(90TC) (∼1–2%). This might be attributed to differences in FlAsH-PrP expression levels, clonal differences between cell lines, or other as yet unknown factors. Together, the results showed that the TC motif is amenable to insertion at multiple sites within a protein (PrPsen) without dramatically affecting its metabolism or conformational refolding as induced by another protein (PrPres). Furthermore, by analyzing the proteolytic fragments generated in various TC-tagged constructs, a new fragment in the PrP metabolic pathway was identified.
E64 Inhibits C3 Generation but Not PrPres Biosynthesis
Having established that PrP(230TC) was a sensitive probe for proteolytic processing of PrP, the influence of various anti-prion compounds on the kinetics of FlAsH-PrP(230TC) was evaluated to determine whether the mechanisms of action of these compounds might involve alterations to PrP metabolism. One of these compounds was a cysteine protease inhibitor called E64 (Doh-Ura et al., 2000). When 22L(230TC) cells were treated for several days with E64 and assayed for total steady-state PrPres by immunoblotting, we observed a dose-dependent decrease in PrPres levels (Figure 4A). Because previous studies with E64 or other cysteine protease inhibitors made use of cells infected with the Chandler strain of mouse-adapted prions (Doh-Ura et al., 2000;Yadavalli et al., 2004), this shows that E64 exhibits anti-prion activity against multiple strains of mouse prions in the N2a cell model. In FlAsH-pulse-chase analysis of 22L(230TC) cells, E64 had no impact on the metabolism of full-length PrP, confirming that cysteine proteases are not the major protease mediating PrPsen degradation in N2a cells (Figure 4B, arrow, lysate) and consistent with previous reports (Doh-Ura et al., 2000;Luhr et al., 2004). Remarkably, C3 levels were significantly reduced in E64-treated cells, suggesting that this fragment is mainly produced by cysteine protease activity (Figure 4B, asterisk, lysate). In PTA precipitates, the initial increase in full-length FlAsH-PrP was similar between treated and untreated cells, but the 21-kDa truncated FlAsH-PrPres fragment did not occur in E64-treated samples even with a 6-h chase (Figure 4B, lanes 12–22). Surprisingly, the absence of the 21-kDa band was actually due to inhibition of the N-terminal truncation of FlAsH-PrPres by endogenous cysteine proteases rather than inhibition of FlAsH-PrPres biosynthesis as shown by analysis of chymotrypsin-digested samples (Figure 4B, arrowhead, lane 23 vs. lane 24). It is notable that treatment with E64 or leupeptin (Figure 4B, lane 22 and Supplemental Figure 3) gave rise to a new band of slightly lower apparent MW than full-length PrP, suggesting that proteases other than cysteine proteases may also participate in the processing of nascent FlAsH-PrPres. Because FlAsH-PrPres levels reach a plateau within 5.5–6 h after labeling (Figure 3D, arrowhead, lane 17 vs. lane 18), these observations demonstrated that the anti-prion activity of E64 is not achieved through inhibition of PrPres biosynthesis.
Figure 4.
E64 inhibits C3 generation and PrPres truncation but not PrPres biosynthesis. (A) E64 treatment lowers steady-state PrPres levels after a 3-d treatment of 22L(230TC) cells with the indicated concentration of inhibitor. PrPres was detected by immunoblot. The bands in each lane correspond to the three glycoforms of PrP. The graph shows the average intensity (± SEM) of the PrPres bands as a percentage of the untreated control (n = 3). (B) Influence of E64 on metabolism of FlAsH-PrP(230TC). The fluorescent gels and the graphs show the results of FlAsH-pulse-chase analysis of 22L(230TC) cells incubated in the absence or presence of E64 (80 μg/ml). The graphs show average band intensity ± SEM (n = 4). The double asterisk denotes a statistically significant difference in C3 levels at 6 h between E64-treated and untreated samples (two-tailed unpaired t test, p = 0.0089). The arrows indicate full-length FlAsH-PrP(230TC), the arrowhead indicates the 21-kDa truncated PrPres band, and the asterisk indicates the C3 fragment. Lysate, kinetics of FlAsH-PrP(230TC) in cell lysate samples (lanes 1–11). PTA, kinetics of FlAsH-PrP(230TC) in PTA-precipitated samples (lanes 12–24). Ch+, samples digested with chymotrypsin (lanes 23–24). Full, full-length FlAsH-PrP(230TC). C3, C3 fragment. 21 kDa, 21-kDa truncated PrPres band.
Labeling of APP and PrP with Alexa Fluor 568-FlAsH (AF568-FlAsH)
Some properties of FlAsH (e.g., pH sensitivity, poor photostability) limit its use in certain applications associated with the study of cell surface proteins, such as visualization of trafficking to endocytic compartments. To expand the tools available for the analysis of TC-tagged proteins, we developed two new FlAsH derivatives (Supplemental Material). The first compound, Alexa Fluor 568-FlAsH (AF568-FlAsH; Figure 5A), combines the preferable fluorescent properties of an Alexa Fluor dye with the TC-motif specificity of FlAsH. To establish the versatility of IDEAL-labeling, we made TC-tagged human amyloid precursor protein [APP(598TC)], which had the TC-motif between residues 597 and 598, immediately C-terminal to the beta-cleavage site. APP(598TC) and PrP(90TC) were labeled with AF568-FlAsH with similar specificity to FlAsH-labeled TC-tagged PrP (Figure 5B). Confocal microscopy studies of cells labeled with AF568-FlAsH showed specific cell-surface labeling characteristic of APP and PrP (Figure 5C). Some AF568-FlAsH-labeled PrP(230TC) and APP(598TC) seemed localized to intracellular vesicles. Because the highly charged AF568-FlAsH compound should be membrane impermeable, it is possible that this intracellular labeled protein was internalized from the cell surface after labeling. Indeed when IDEAL-labeled cells were kept cold to block endocytosis and fixed immediately after a series of quick rinses to remove unbound AF568-FlAsH, the intracellular population of labeled protein was not detected, suggesting that it originated from a cell surface population (Supplemental Figure 5). Pulse-chase of AF568-FlAsH-APP(598TC) showed a short half-life of ∼45 min (Figure 5D), similar to that of wild-type APP in cultured cells (Allinquant et al., 1995; Storey et al., 1999). These experiments demonstrate that modification of carboxy-FlAsH does not affect its binding properties to the TC-motif and that IDEAL-labeling can be applied to multiple cell surface proteins.
To provide an example of correlative biochemical and microscopy analysis of a TC-tagged protein, we visualized the endocytosis of AF568-FlAsH-labeled PrP(230TC) by live cell time lapse confocal microscopy at hourly time points comparable with those in Figure 3D (lysate). At the first time point (0 h), the majority of PrP(230TC) was present at the cell surface, although a small population localized to perinuclear vesicles also was observed (Figure 5E). After 2–3 h, the perinuclear population of protein seemed to increase accompanied by an apparent decrease in plasma membrane labeling. Through the remainder of the time lapse, there was a progressive increase in the intracellular population of PrP(230TC) with a concomitant decrease in cell surface fluorescence (Figure 5E and Supplemental Video 1). These observations correlated with the biochemical analysis of PrP(230TC) metabolism with the first demonstrable decrease in full-length PrP(230TC) occurring after ∼2–3 h of chase, suggesting that turnover of cell surface PrP(230TC) occurs after endocytic trafficking to a perinuclear compartment.
Labeling of PrP with Bio-FlAsH
The second novel compound is a FlAsH-biotin conjugate termed “Bio-FlAsH” (Figure 6A). We found that Bio-FlAsH-labeled TC-tagged PrP was detected with high specificity and sensitivity by fluorescent gel analysis (Figure 6B), immunoblotting (with a NeutrAvidin-alkaline phosphatase conjugate) (Figure 6B), and immunofluorescence using a streptavidin-FluoroNanogold conjugate detectable by both fluorescence (Figure 6C) and electron microscopy (Figure 6D). At the ultrastructural level, PrP(230TC) was localized to filopodia and intervening regions of the cell surface (Figure 6D). As reported previously (Howarth et al., 2005), bands representing endogenous intracellular biotinylated proteins were detected by immunoblot (Figure 6B, arrows). The reduced intensity of the Bio-FlAsH-labeled PrP(230TC) fragments (Figure 6B) was not unexpected because its carboxy-FlAsH moiety exhibits half the fluorescence intensity of FlAsH (Cao et al., 2006). Therefore, Bio-FlAsH allows efficient biochemical characterization of TC-tagged proteins and visualization by fluorescence and electron microscopy.
DISCUSSION
Despite its promise, biarsenical labeling of TC-tagged proteins has seen limited use thus far. One reason may be background presumably due to FlAsH binding to off-target cysteine-rich proteins (Stroffekova et al., 2001; Berens et al., 2005; Langhorst et al., 2006; Hearps et al., 2007), necessitating a postincubation step to reduce this background. Successful applications of this technology have used protocols similar to that originally reported for FlAsH (Perlman and Resh, 2006; Arhel et al., 2006; Roberti et al., 2007; Tour et al., 2007) but have not addressed the specificity issue to allow one-step labeling of any protein, especially those localized to cell surfaces. A previous attempt to label cell surface proteins required a custom-synthesized membrane-impermeable probe, which still showed some background labeling (Adams et al., 2002). Thus, the biarsenical labeling approach has not yet realized its full potential with the inability to conveniently target cell surface proteins.
Labeling PrPres with fluorescent tags in live cells presented an exceptional challenge because of the sensitivity of its formation to steric effects by foreign tags. Being one of the smallest fluorescent labels, FlAsH/TC-motif labeling seemed a promising option so we sought to develop strategies to overcome the above-mentioned limitations. Fortunately, DTT can rapidly reduce endoplasmic reticulum proteins in live cells without affecting cell viability (Braakman et al., 1992), suggesting it was a good candidate reducing agent to enhance biarsenical labeling efficiency while ensuring reduction of cystine residues within the TC motif of cell surface proteins. We discovered DTT worked and simplified the procedure by eliminating the preincubation and postincubation steps traditionally used to reduce background labeling. To our knowledge, the method reported here is among the most rapid to achieve specific fluorescent labeling of cell surface proteins, facilitating the time resolution of pulse-chase labeling for subsequent biochemical and/or live cell imaging experiments. Our success in fluorescent-tagging PrP and APP proves that extracellular proteins can be labeled specifically and efficiently with the commercially available FlAsH reagent.
Because of the potential power of TC-tagging, we and others have developed new biarsenical derivatives (Bhunia and Miller, 2007; Cao et al., 2007; Chen et al., 2007; Liu et al., 2007; Tour et al., 2007). We synthesized FlAsH conjugates containing either an Alexa Fluor dye or biotin that worked as efficiently and specifically as FlAsH, suggesting that conjugation to carboxy-FlAsH would not substantially affect the affinity of the biarsenical moiety for the TC-motif. This is important because such probes are useful only when they are specific. Biarsenical scaffolds conjugated to Alexa Fluor dye (Bhunia and Miller, 2007) or biotin (with an additional dopamine moiety) (Liu et al., 2007) have been described by other groups, but the affinity and specificity of these reagents for the minimal CCPGCC TC motif remain to be shown in the context of the highly challenging application of labeling TC-tagged proteins expressed in mammalian cells where the ratio of TC-tagged protein to total protein is usually very low and off-target biarsenical-binding proteins are common (Griffin et al., 2000; Stroffekova et al., 2001). Furthermore, without a method to label cell-surface TC-tagged proteins these membrane-impermeable biarsenical compounds would be restricted to cell-free or fixed/permeabilized cell applications. With the advent of IDEAL-labeling, biarsenical labeling becomes a technique to rapidly add a functional molecule onto a designated part of a cell surface target protein in situ on live cells. The molecule may have properties other than fluorescence. Bio-FlAsH is one example, which has tremendous potential for many applications via detection with the plethora of commercially available biotin-binding conjugates. Because carboxy-FlAsH is itself a fluorophore, it serves as a scaffold for the creation of multifunctional derivatives.
The successful conversion of FlAsH-PrPs to FlAsH-PrPres demonstrates that the steric effects of the FlAsH/TC-motif are small, encouraging its application to other proteins with steric sensitivities. It is difficult to modify the PK-resistant core region of PrP without impairing its convertibility, as demonstrated in part by barriers for prion transmission between species consisting of differences in only a few amino acid residues in this region between inoculated PrPres and host-encoded PrPsen. The precise structure of PrPres has not been determined. It seems to consist of PrP oligomers with tightly packed β-sheet strands, some of which are generated from refolding of either α-helical and/or unstructured regions of PrPsen. This ordered assembly of PrP molecules during the conversion process may contribute to its sensitivity to PrP modifications. Although PrP(230TC) could convert to PrPres, a mutant PrP containing a 15 amino acid tag at the same site as PrP(230TC) did not convert to PrPres (data not shown), suggesting a limitation to tag sizes even in relatively tolerant locations. We speculate that, besides the minimal size, the hairpin structure of the TC-motif might further limit its steric effects. Whatever the explanation, the minimal steric effect of the TC-motif would extend its application to many other proteins.
Compatibility of FlAsH with fluorescent gel analysis allows observation of the same FlAsH-labeled molecules both microscopically and biochemically as effectively demonstrated in Figures 3D (lysate) and 5E. Whereas immunoblotting shows the steady-state of protein molecules collectively irrespective of their age, pulse-chase analysis can provide dynamic time-resolved analysis. Unlike conventional pulse-chase using radioisotopes, lack of immunoprecipitation in FlAsH-pulse chase minimizes loss of labeled proteins, and the fixed stoichiometry between FlAsH and fragments regardless of their sizes achieves more accurate quantification of labeled fragments. Altered behavior of the labeled proteins due to the tag and/or labeling procedures was a concern, but the results of FlAsH-pulse-chase experiments were consistent with reports for wild-type PrP and APP, confirming a minimal impact on the behavior of these proteins. Moreover, there were several novel results related to PrP biology.
First, C3 was identified as a new metabolic fragment of PrP. To date, PrP shedding possibly by metalloprotease-directed cleavage near the C terminus has been described (Parkin et al., 2004), although the short C-terminal by-product has never been noted. E64-sensitive proteolysis responsible for C3 formation (Figure 4B) might represent another shedding mechanism of PrP. Further experiments with E64 established that it is active against multiple strains of mouse-adapted prions but led to the surprising finding that the anti-prion activity of this compound in infected N2a cells is not mediated through inhibition of new PrPres biosynthesis. The anti-prion activity of cysteine protease inhibitors is somewhat controversial. Depending upon the host cell line, cysteine protease inhibitors can either reduce (Doh-Ura et al., 2000;Yadavalli et al., 2004) or increase (Luhr et al., 2004) steady-state PrPres levels. According to Yadavalli et al. (2004), N-terminal truncation of PrPres by calpain-mediated cleavage assists PrPres propagation in ScN2a and SMB models of prion infection. In the present study, PrPres biosynthesis was not inhibited by E64 even at concentrations that blocked complete N-terminal truncation of newly formed PrPres, suggesting that the anti-prion effects of E64 are independent of its effects on PrPres proteolysis. Interestingly, we did observe evidence of a very small N-terminal truncation of FlAsH-PrPres formed in the presence of cysteine protease inhibitors (Supplemental Figure 3), but the protease(s) involved remains to be identified. Our data are consistent with a time dependence to the inhibitory effect of E64, raising the possibility that cysteine proteases may indirectly contribute to PrPres propagation via the modification of other cellular factors analogous to the effects of a particular tyrosine kinase (Ertmer et al., 2004). This could explain the differential effects observed between different prion- infected cell lines. Alternatively, N-terminal proteolysis of PrPres may facilitate faithful transmission of PrPres aggregates between cells either during mitosis or by extracellular mechanisms (e.g., exosomes; Fevrier et al., 2004), which would be important for maintenance of PrPres formation in the context of rapidly dividing cell models of infection.
Next, based on quantification of the 21-kDa protease-resistant core of PrPres with and without exogenous protease digestion, we found chymotrypsin digestion allows a more accurate evaluation of total FlAsH-PrPres. Although PK resistance has been regarded as a hallmark of disease-associated PrP, PK is not necessarily the ideal protease to assess protease resistance of PrPres. Evidence for the existence of PK-sensitive forms of disease-associated PrP has come from experiments using mild PK digestion at low concentrations or low temperatures (Tremblay et al., 2004) or immunological assays relying on differential epitope exposure between PrPres and PrPsen (Safar et al., 1998). Therefore, defining PrPres based specifically on PK digestion as opposed to other proteases is arbitrary. The Chy-res might represent “PK-sensitive PrPres,” and this moderately protease-sensitive fragment might aid structural studies of PrPres. Although a previous report comparing PK versus chymotrypsin digestion of brain homogenates failed to detect a difference in protease sensitivity of PrPres, their work differs significantly from ours with respect to the method of analysis, the source material for digestion, and the use of very high enzyme concentrations (Pan et al., 2005).
Finally, the lack of direct labeling of TC-PrPres fortuitously enabled measurement of the “conversion rate” of FlAsH-PrPsen rather than the change in total TC-PrPres. This allowed rapid analysis of the effects of many compounds specifically on PrPres synthesis, permitting detection of early events not readily observed in analysis of steady-state PrPres levels (data not shown). This could be developed as a high-throughput screen for anti-prion therapeutics. Likewise, FlAsH-labeled TC-tagged APP constructs might assist the search for new Alzheimer's drugs by allowing rapid screening for modulators of APP processing.
In conclusion, we developed a specific method for fluorescent labeling of extracellular TC-tagged proteins, PrP and APP, and demonstrated its usefulness in imaging and protein metabolism studies. This sets the stage for important experiments in prion biology such as imaging of the formation and spread of PrPres. Such imaging experiments will themselves be challenging due to the fact only a small percentage of PrP-sen is converted to PrP-res, but our system provides valuable tools to begin these investigations. We also created powerful new biarsenical derivatives. The present report establishes the versatility of biarsenical compounds in imaging and pulse-chase studies and illustrates the potential of this technique to attach functional molecules to specific target proteins on live cells, encouraging the development of more biarsenical compounds.
Supplementary Material
ACKNOWLEDGMENTS
We thank Lynne and Greg Raymond, Danielle Offerdahl, and Lindsay Hohsfield for technical support. We thank Ted Hackstadt for access to the Typhoon instrument, Dennis Burton and Anthony Williamson for providing R1 antibody, and Gary Griffiths for overseeing the synthesis of the new biarsenical molecules. We are grateful to Evan Eisenberg, Byron Caughey, Bruce Chesebro, Suzette A. Priola, John Portis, and Valerie Sim for critical reading and helpful advice. This research was supported by the Intramural Research Program of the National Institutes of Health (National Institute of Allergy and Infectious Diseases and the Imaging Probe Development Center, National Heart Lung and Blood Institute).
Abbreviations used:
- IDEAL-labeling
instant with DTT, EDT, and low temperature-labeling
- PK
proteinase K
- PrP
prion protein
- PrPsen
normal protease-sensitive prion protein
- PrPres
prion disease-associated protease-resistant prion protein
- PrP(90TC)
prion protein with tetracysteine motif at residue 90
- PrP(230TC)
prion protein with tetracysteine motif at residue 230
- PTA
phosphotungstate
- TC
tetracysteine.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-06-0635) on November 5, 2008.
REFERENCES
- Adams S. R., Campbell R. E., Gross L. A., Martin B. R., Walkup G. K., Yao Y., Llopis J., Tsien R. Y. New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: synthesis and biological applications. J. Am. Chem. Soc. 2002;124:6063–6076. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- Allinquant B., Hantraye P., Mailleux P., Moya K., Bouillot C., Prochiantz A. Downregulation of amyloid precursor protein inhibits neurite outgrowth in vitro. J. Cell Biol. 1995;128:919–927. doi: 10.1083/jcb.128.5.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arhel N., Genovesio A., Kim K. A., Miko S., Perret E., Olivo-Marin J. C., Shorte S., Charneau P. Quantitative four-dimensional tracking of cytoplasmic and nuclear HIV-1 complexes. Nat. Methods. 2006;3:817–824. doi: 10.1038/nmeth928. [DOI] [PubMed] [Google Scholar]
- Barmada S. J., Harris D. A. Visualization of prion infection in transgenic mice expressing green fluorescent protein-tagged prion protein. J. Neurosci. 2005;25:5824–5832. doi: 10.1523/JNEUROSCI.1192-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berens W., Van Den B. K., Yoon T. J., Westbroek W., Valencia J. C., Out C. J., Marie N. J., Hearing V. J., Lambert J. Different approaches for assaying melanosome transfer. Pigment Cell Res. 2005;18:370–381. doi: 10.1111/j.1600-0749.2005.00263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessen R. A., Kocisko D. A., Raymond G. J., Nandan S., Lansbury P. T., Caughey B. Non-genetic propagation of strain-specific properties of scrapie prion protein. Nature. 1995;375:698–700. doi: 10.1038/375698a0. [DOI] [PubMed] [Google Scholar]
- Bhunia A. K., Miller S. C. Labeling tetracysteine-tagged proteins with a SplAsH of color: a modular approach to bis-arsenical fluorophores. Chembiochem. 2007;8:1642–1645. doi: 10.1002/cbic.200700192. [DOI] [PubMed] [Google Scholar]
- Bian J., Nazor K. E., Angers R., Jernigan M., Seward T., Centers A., Green M., Telling G. C. GFP-tagged PrP supports compromised prion replication in transgenic mice. Biochem. Biophys. Res. Commun. 2006;340:894–900. doi: 10.1016/j.bbrc.2005.12.085. [DOI] [PubMed] [Google Scholar]
- Birkett C. R., Hennion R. M., Bembridge D. A., Clarke M. C., Chree A., Bruce M. E., Bostock C. J. Scrapie strains maintain biological phenotypes on propagation in a cell line in culture. EMBO J. 2001;20:3351–3358. doi: 10.1093/emboj/20.13.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchelt D. R., Taraboulos A., Prusiner S. B. Evidence for synthesis of scrapie prion proteins in the endocytic pathway. J. Biol. Chem. 1992;267:16188–16199. [PubMed] [Google Scholar]
- Braakman I., Helenius J., Helenius A. Manipulating disulfide bond formation and protein folding in the endoplasmic reticulum. EMBO J. 1992;11:1717–1722. doi: 10.1002/j.1460-2075.1992.tb05223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Chen B., Squier T. C., Mayer M. U. CrAsH: a biarsenical multi-use affinity probe with low non-specific fluorescence. Chem. Commun. 2006:2601–2603. doi: 10.1039/b602699k. [DOI] [PubMed] [Google Scholar]
- Cao H., Xiong Y., Wang T., Chen B., Squier T. C., Mayer M. U. A red Cy3-based biarsenical fluorescent probe targeted to a complementary binding peptide. J. Am. Chem. Soc. 2007;129:8672–8673. doi: 10.1021/ja070003c. [DOI] [PubMed] [Google Scholar]
- Caughey B., Baron G. S. Prions and their partners in crime. Nature. 2006;443:803–810. doi: 10.1038/nature05294. [DOI] [PubMed] [Google Scholar]
- Caughey B., Raymond G. J. The scrapie-associated form of PrP is made from a cell surface precursor that is both protease- and phospholipase-sensitive. J. Biol. Chem. 1991;266:18217–18223. [PubMed] [Google Scholar]
- Caughey B., Raymond G. J., Ernst D., Race R. E. N-terminal truncation of the scrapie-associated form of PrP by lysosomal protease(s): implications regarding the site of conversion of PrP to the protease-resistant state. J. Virol. 1991;65:6597–6603. doi: 10.1128/jvi.65.12.6597-6603.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Cao H., Yan P., Mayer M. U., Squier T. C. Identification of an orthogonal peptide binding motif for biarsenical multiuse affinity probes. Bioconjug. Chem. 2007;18:1259–1265. doi: 10.1021/bc0603900. [DOI] [PubMed] [Google Scholar]
- Chen S. G., Teplow D. B., Parchi P., Teller J. K., Gambetti P., utilio-Gambetti L. Truncated forms of the human prion protein in normal brain and in prion diseases. J. Biol. Chem. 1995;270:19173–19180. doi: 10.1074/jbc.270.32.19173. [DOI] [PubMed] [Google Scholar]
- Doh-Ura K., Iwaki T., Caughey B. Lysosomotropic agents and cysteine protease inhibitors inhibit scrapie-associated prion protein accumulation. J. Virol. 2000;74:4894–4897. doi: 10.1128/jvi.74.10.4894-4897.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertmer A., Gilch S., Yun S. W., Flechsig E., Klebl B., Stein-Gerlach M., Klein M. A., Schatzl H. M. The tyrosine kinase inhibitor STI571 induces cellular clearance of PrPSc in prion-infected cells. J. Biol. Chem. 2004;279:41918–41927. doi: 10.1074/jbc.M405652200. [DOI] [PubMed] [Google Scholar]
- Fevrier B., Vilette D., Archer F., Loew D., Faigle W., Vidal M., Laude H., Raposo G. Cells release prions in association with exosomes. Proc. Natl. Acad. Sci. USA. 2004;101:9683–9688. doi: 10.1073/pnas.0308413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin B. A., Adams S. R., Tsien R. Y. Specific covalent labeling of recombitant protein molecules inside live cells. Science. 1998;281:269–272. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- Griffin B. A., Adams S. R., Jones J., Tsien R. Y. Fluorescent labeling of recombinant proteins in living cells with FlAsH. Methods Enzymol. 2000;327:565–578. doi: 10.1016/s0076-6879(00)27302-3. [DOI] [PubMed] [Google Scholar]
- Hearps A. C., Pryor M. J., Kuusisto H. V., Rawlinson S. M., Piller S. C., Jans D. A. The biarsenical dye lumiotrade mark exhibits a reduced ability to specifically detect tetracysteine-containing proteins within live cells. J. Fluoresc. 2007;17:593–597. doi: 10.1007/s10895-007-0225-x. [DOI] [PubMed] [Google Scholar]
- Howarth M., Takao K., Hayashi Y., Ting A. Y. Targeting quantum dots to surface proteins in living cells with biotin ligase. Proc. Natl. Acad. Sci. USA. 2005;102:7583–7588. doi: 10.1073/pnas.0503125102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko K., Zulianello L., Scott M., Cooper C. M., Wallace A. C., James T. L., Cohen F. E., Prusiner S. B. Evidence for protein X binding to a discontinuous epitope on the cellular prion protein during scrapie prion propagation. Proc. Natl. Acad. Sci. USA. 1997;94:10069–10074. doi: 10.1073/pnas.94.19.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocisko D. A., Baron G. S., Rubenstein R., Chen J., Kuizon S., Caughey B. New inhibitors of scrapie-associated prion protein formation in a library of 2000 drugs and natural products. J. Virol. 2003;77:10288–10294. doi: 10.1128/JVI.77.19.10288-10294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocisko D. A., Come J. H., Priola S. A., Chesebro B., Raymond G. J., Lansbury P. T., Caughey B. Cell-free formation of protease-resistant prion protein. Nature. 1994;370:471–474. doi: 10.1038/370471a0. [DOI] [PubMed] [Google Scholar]
- Langhorst M. F., Genisyuerek S., Stuermer C. A. Accumulation of FlAsH/Lumio Green in active mitochondria can be reversed by beta-mercaptoethanol for specific staining of tetracysteine-tagged proteins. Histochem. Cell Biol. 2006;125:743–747. doi: 10.1007/s00418-005-0136-3. [DOI] [PubMed] [Google Scholar]
- Lee I. S., Long J. R., Prusiner S. B., Safar J. G. Selective precipitation of prions by polyoxometalate complexes. J. Am. Chem. Soc. 2005;127:13802–13803. doi: 10.1021/ja055219y. [DOI] [PubMed] [Google Scholar]
- Lee K. S., Magalhaes A. C., Zanata S. M., Brentani R. R., Martins V. R., Prado M. A. Internalization of mammalian fluorescent cellular prion protein and N-terminal deletion mutants in living cells. J. Neurochem. 2001;79:79–87. doi: 10.1046/j.1471-4159.2001.00529.x. [DOI] [PubMed] [Google Scholar]
- Liu B., Archer C. T., Burdine L., Gillette T. G., Kodadek T. Label transfer chemistry for the characterization of protein-protein interactions. J. Am. Chem. Soc. 2007;129:12348–12349. doi: 10.1021/ja072904r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhr K. M., Nordstrom E. K., Low P., Ljunggren H. G., Taraboulos A., Kristensson K. Scrapie protein degradation by cysteine proteases in CD11c+ dendritic cells and GT1-1 neuronal cells. J. Virol. 2004;78:4776–4782. doi: 10.1128/JVI.78.9.4776-4782.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T., Wong P., Chang B., Li C., Li R., Kang S. C., Wisniewski T., Sy M. S. Biochemical fingerprints of prion infection: accumulations of aberrant full-length and N-terminally truncated PrP species are common features in mouse prion disease. J. Virol. 2005;79:934–943. doi: 10.1128/JVI.79.2.934-943.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin E. T., Watt N. T., Turner A. J., Hooper N. M. Dual mechanisms for shedding of the cellular prion protein. J. Biol. Chem. 2004;279:11170–11178. doi: 10.1074/jbc.M312105200. [DOI] [PubMed] [Google Scholar]
- Perlman M., Resh M. D. Identification of an intracellular trafficking and assembly pathway for HIV-1 gag. Traffic. 2006;7:731–745. doi: 10.1111/j.1398-9219.2006.00428.x. [DOI] [PubMed] [Google Scholar]
- Priola S. A., Chesebro B. A single hamster PrP amino acid blocks conversion to protease-resistant PrP in scrapie-infected mouse neuroblastoma cells. J. Virol. 1995;69:7754–7758. doi: 10.1128/jvi.69.12.7754-7758.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberti M. J., Bertoncini C. W., Klement R., Jares-Erijman E. A., Jovin T. M. Fluorescence imaging of amyloid formation in living cells by a functional, tetracysteine-tagged alpha-synuclein. Nat. Methods. 2007;4:345–351. doi: 10.1038/nmeth1026. [DOI] [PubMed] [Google Scholar]
- Safar J., Wille H., Itri V., Groth D., Serban H., Torchia M., Cohen F. E., Prusiner S. B. Eight prion strains have PrP(Sc) molecules with different conformations. Nat. Med. 1998;4:1157–1165. doi: 10.1038/2654. [DOI] [PubMed] [Google Scholar]
- Storey E., Katz M., Brickman Y., Beyreuther K., Masters C. L. Amyloid precursor protein of Alzheimer's disease: evidence for a stable, full-length, trans-membrane pool in primary neuronal cultures. Eur. J. Neurosci. 1999;11:1779–1788. doi: 10.1046/j.1460-9568.1999.00599.x. [DOI] [PubMed] [Google Scholar]
- Stroffekova K., Proenza C., Beam K. G. The protein-labeling reagent FLASH-EDT2 binds not only to CCXXCC motifs but also non-specifically to endogenous cysteine-rich proteins. Pflugers Arch. 2001;442:859–866. doi: 10.1007/s004240100619. [DOI] [PubMed] [Google Scholar]
- Taraboulos A., Raeber A. J., Borchelt D. R., Serban D., Prusiner S. B. Synthesis and trafficking of prion proteins in cultured cells. Mol. Biol. Cell. 1992;3:851–863. doi: 10.1091/mbc.3.8.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraboulos A., Scott M., Semenov A., Avrahami D., Laszlo L., Prusiner S. B. Cholesterol depletion and modification of COOH-terminal targeting sequence of the prion protein inhibit formation of the scrapie isoform. J. Cell Biol. 1995;129:121–132. doi: 10.1083/jcb.129.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telling G. C., Parchi P., DeArmond S. J., Cortelli P., Montagna P., Gabizon R., Mastrianni J., Lugaresi E., Gambetti P., Prusiner S. B. Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science. 1996;274:2079–2082. doi: 10.1126/science.274.5295.2079. [DOI] [PubMed] [Google Scholar]
- Tour O., Adams S. R., Kerr R. A., Meijer R. M., Sejnowski T. J., Tsien R. W., Tsien R. Y. Calcium Green FlAsH as a genetically targeted small-molecule calcium indicator. Nat. Chem. Biol. 2007;3:423–431. doi: 10.1038/nchembio.2007.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay P., Ball H. L., Kaneko K., Groth D., Hegde R. S., Cohen F. E., DeArmond S. J., Prusiner S. B., Safar J. G. Mutant PrPSc conformers induced by a synthetic peptide and several prion strains. J. Virol. 2004;78:2088–2099. doi: 10.1128/JVI.78.4.2088-2099.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson R. A., Peretz D., Pinilla C., Ball H., Bastidas R. B., Rozenshteyn R., Houghten R. A., Prusiner S. B., Burton D. R. Mapping the prion protein using recombinant antibodies. J. Virol. 1998;72:9413–9418. doi: 10.1128/jvi.72.11.9413-9418.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadavalli R., Guttmann R. P., Seward T., Centers A. P., Williamson R. A., Telling G. C. Calpain-dependent endoproteolytic cleavage of PrPSc modulates scrapie prion propagation. J. Biol. Chem. 2004;279:21948–21956. doi: 10.1074/jbc.M400793200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.