Abstract
Processing bodies (P-bodies) are cytoplasmic foci implicated in the regulation of mRNA translation, storage, and degradation. Key effectors of microRNA (miRNA)-mediated RNA interference (RNAi), such as Argonaute-2 (Ago2), miRNAs, and their cognate mRNAs, are localized to these structures; however, the precise role that P-bodies and their component proteins play in small interfering RNA (siRNA)-mediated RNAi remains unclear. Here, we investigate the relationship between siRNA-mediated RNAi, RNAi machinery proteins, and P-bodies. We show that upon transfection into cells, siRNAs rapidly localize to P-bodies in their native double-stranded conformation, as indicated by fluorescence resonance energy transfer imaging and that Ago2 is at least in part responsible for this siRNA localization pattern, indicating RISC involvement. Furthermore, siRNA transfection induces up-regulated expression of both GW182, a key P-body component, and Ago2, indicating that P-body localization and interaction with GW182 and Ago2 are important in siRNA-mediated RNAi. By virtue of being centers where these proteins and siRNAs aggregate, we propose that the P-body microenvironment, whether as microscopically visible foci or submicroscopic protein complexes, facilitates siRNA processing and siRNA-mediated silencing through the action of its component proteins.
INTRODUCTION
RNA interference (RNAi) is a powerful homology-based gene silencing mechanism directed by small RNAs, including small interfering RNAs (siRNAs) and microRNAs (miRNAs). RNAi is a posttranscriptional process, leading to either translational repression of the target mRNA, typical of miRNA-mediated silencing, or degradation of the target via siRNA-mediated silencing (Hammond, 2005). Recently, several components of the RNAi pathway, including Argonaute proteins (Sen and Blau, 2005), miRNAs, and their targets (Liu et al., 2005b; Pillai et al., 2005) have been localized to processing bodies (P-bodies). P-bodies are recognized as important cytoplasmic mRNA processing centers where nontranslating mRNA is sorted and either stored, repressed, or degraded (for review, see Eulalio et al., 2007a). Enzymes associated with mRNA degradation, such the CCR4-CAF-1-Not complex involved in deadenlyation (Cougot et al., 2004); DCP1 and -2 (van Dijk et al., 2002) involved in decapping; and XRN-1, a 5′-3′ exonuclease (Ingelfinger et al., 2002), have all been localized to P-bodies. These foci also contain proteins involved in nonsense-mediated decay (Sheth and Parker, 2006) and AU-rich element-mediated mRNA decay pathways (Vasudevan and Steitz, 2007).
Although it is clear that RNAi and P-bodies are associated, the precise role of P-bodies in RNAi remains unclear. There is evidence that P-body components are essential for miRNA-based gene silencing. Depletion of certain P-body components, including RCK/P54 (Chu and Rana, 2006) and GW182 in human (Liu et al., 2005a) and Drosophila (Rehwinkel et al., 2005) cells leads to a loss of silencing. Furthermore, the decay of miRNA targets seems to require the deadenylation complex, decapping enzyme, and 5′-3′ exonuclease, all of which are P-body components (Rehwinkel et al., 2005). Knockdown of Drosha and DGCR8, proteins involved in miRNA production, leads to the loss of P-bodies in human cells, indicating that miRNAs are crucial components of P-bodies (Pauley et al., 2006). For silencing to occur, it has been suggested that miRNAs and the RNAi proteins direct the target mRNAs to P-bodies, where the general degradation/repression machinery is localized (Eulalio et al., 2007a,b) and that P-bodies are formed as a consequence of silencing.
In siRNA-based gene silencing, the role of P-bodies is less well understood. Because siRNAs direct Argonaute-2 (Ago2) to cleave the homologous target RNA (Hammond et al., 2000; Martinez et al., 2002), the close presence of mRNA degrading machinery might not be essential to achieve silencing. Indeed, depletion of P-body components such as LSm1 and RCK/p54 (Chu and Rana, 2006) in human cells has no effect on siRNA silencing, leading to a view that P-bodies are dispensable for siRNA-mediated RNAi. However, depletion of the P-body structural component GW182 has shown mixed results; three reports show that silencing GW182 does inhibit siRNA silencing to a certain extent (Jakymiw et al., 2005; Liu et al., 2005a; Lian et al., 2007), whereas others show no requirement of GW182 for siRNA function (Rehwinkel et al., 2005; Chu and Rana, 2006; Eulalio et al., 2007b). However, siRNAs have been found to localize to P-bodies (Jakymiw et al., 2005), and the number and size of P-bodies were found to increase upon siRNA transfection, in a target-dependant manner (Lian et al., 2007). Moreover, after knockdown of Lsm1 or RCK/p54, functional siRNAs are capable of inducing P-body reassembly (Lian et al., 2007). This suggests that although microscopic P-bodies are not necessarily required for RNAi, silencing could occur in submicroscopic complexes that might then trigger the assembly of larger, microscopic structures. A recent study noted that P-body disassembly was induced by a range of siRNAs, whose targets were not related to mRNA metabolism and P-body components (Serman et al., 2007), indicating that the structure and function of P-bodies are more complex than currently believed. These conflicting results led us to further examine the relationship between siRNA and P-bodies. It has been suggested that it is more important to understand the individual contributions of P-body components to RNAi, rather than the ability to aggregate/functions of the aggregate (Wu and Belasco, 2008). Here, we show that in the presence of microscopically visible P-bodies, double-stranded siRNAs rapidly localize to these aggregates with a requirement for Ago2 and that GW182 and Ago2, key RNAi machinery components that localize to P-bodies, are up-regulated upon siRNA transfection into cells.
MATERIALS AND METHODS
Cell Culture and Transfection
HeLa cells were cultured in DMEM (10% fetal bovine serum) in a 37°C incubator with 5% CO2. Lipofectamine 2000 (Invitrogen, Paisley, United Kingdom) was used for all transfections.
siRNAs.
siAgo2 was obtained from Bioneer (Hørsholm, Denmark), siLuc was from Dharmacon RNA Technologies (Lafayette, CO), and siMAPK was from QIAGEN (Dorking, Surrey, United Kingdom). All other siRNAs were obtained from Eurogentec (Seraing, Belgium). All sequences 5′-3′, sense strand only, are as follows: siPPIB, GGA-AAG-ACU-GUU-CCA-AAA-AUU (Dharmacon RNA Technologies); siLA, GGU-GGU-GAC-GAU-CUG-GGC-UUU (Dharmacon RNA Technologies); siAgo2, GCA-CGG-AAG-UCC-AUC-UGA-AUU (Chu and Rana, 2006); siLuc, UAA-GGC-UAU-GAA-GAG-AUA-CUU (Dharmacon RNA Technologies); siGW, GAA AUG CUC UGG UCC GCU AUU (Lian et al., 2007); siRCK, GCA GAA ACC CUA UGA GAU UUU (Chu and Rana, 2006); siSMN, GCA UGC UCU AAA GAA UGG UUU; and siMAPK (AllStars Positive control Mm/Hs MAPK1 control siRNA; QIAGEN).
DNA Constructs.
pNEGFP-hDCP1a (van Dijk et al., 2002; Pillai et al., 2005) was kind gift from Dr. Jens Lykke-Andersen (University of Colorado, Boulder, CO), transfected as 0.4 μg/well (24-well plate).
Immunocytochemistry
Cells were fixed using 4% paraformaldehyde and permeabilized in 0.3% Triton X-100. All blocking and incubation performed in 2% bovine serum albumin, 2.5% goat serum, 0.3% Triton-X 100. Antibody concentrations were 1:5000 anti-GWB serum and 1:1000 Alexa 488 goat anti-human immunoglobulin G (Invitrogen). Human anti-GW182 serum 18033 was a kind gift from Dr. Marvin Fritzler (University of Calgary, Calgary, AB, Canada).
Confocal Microscopy
An LSM 510 laser scanning confocal microscope (Carl Zeiss, Jena, Germany) was used. Images were taken with the 40× and 63× oil immersion objective lenses. The following filter sets were used: MBS NT 80/20 or HFT UV 488/543/633 with DBS NFT 490 or NFT 545 or mirror or none. An argon laser at 488 nm was used to excite enhanced green fluorescent protein (eGFP), with emission collected using a 505- to 550-nm band pass filter. A helium-neon laser was used to excite Cy3 at 543 nm, with emission collected using band pass 560–615 nm; and to excite Cy5 at 633 nm, with emission collected using long pass filter 650 nm. Fluorescence resonance energy transfer (FRET) signal was collected by excitation at 543 nm and emission collection by using long pass filter 650 nm. Wavelength spectra were collected by excitation at 543 nm using the lambda scan setting, collecting images at approximately 10-nm intervals between 560 and 732 nm. For all images, pixel time of 1.6 ms was used. All lasers were used at 100% intensity. Constant levels of gain and offset were maintained for slides imaged on the same day. Pix FRET software was used to present images corrected for spectral bleed-through and background elimination (Feige et al., 2005). For quantification of FRET, data were obtained using the lambda spectrum setting and LSM 510 software. Fluorescence emission intensity from the lambda scans for Cy3 (Fd, read at 560–570 nm) and Cy5 (Fd, read at 660–670 nm) were recorded from approximately 20 P-bodies from a total of three images for each time point. Fa/Fd ratios were calculated from these readings, as described in Raemdonck et al. (2006).
Quantification of Colocalization
Images were analyzed for colocalization using the Just Another Colocalization Program (Jacop) plugin on ImageJ (National Institutes of Health, Bethesda, MD), and statistical data are reported from the Costes' randomization based colocalization module (Bolte and Cordelieres, 2006).
Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
RNA was harvested using the RNeasy kit (QIAGEN) and DNAse treated with the RNase-free DNAse kit (Promega, Madison, WI). cDNA was prepared from the RNA by using the High Capacity Reverse transcription kit (Applied Biosystems, Foster City, CA). qPCRs were performed using the Power SYBR Green master mix on the ABI7000 thermal cycler (Applied Biosystems). A DDCt method of relative quantification was used for data analysis. Primer sequences were as follows: human Ago2, forward 5′-CGCGTCCGAAGGCTGCTCTA-3′ and reverse 5′-TGGCTGTGCCTTGTAAAACGCT-3′; human cyclophilin-B, forward 5′-AAAGTCACCGTCAAGGTGTATTT-3′ and reverse 5′-TCACCGTAGATGCTCTTTCCTC-3′; human glyceraldehyde-3-phosphate dehydrogenase (GAPDH), forward 5′-AAG-GTG-AAG-GTC-GGA-GTC-AA-3′ and reverse, 5′-GAA-GAT-GGT-GAT-GGG-ATT-TC-3′; human RCK/p54, forward AGAGGCCCTGTGAAACCCA and reverse: CTTGCTGCTGAGTGCCATTA; and human GW182, forward CTCTGTGGATGCTCCTGAAAG and reverse TGCTTGGATTTAACCCTCCATTT.
Western Blotting
Total cell protein was extracted with radioimmunoprecipitation assay buffer (Sigma-Aldrich) and 10% Protease Inhibitor complex (Sigma Chemical, Poole, Dorset, United Kingdom) and quantified using the Bradford method. Five, 15, and 30 μg of total protein was loaded onto an 8% polyacrylamide gel and run at 100V. The protein was transferred to a nitrocellulose membrane overnight at 50 V. Blocking and incubation with antibodies were in 5% milk in 0.1% phosphate-buffered saline (PBS)-Tween.
Antibodies used were as follows: polyclonal rabbit anti-cyclophilin-B (Abcam, Cambridge, United Kingdom) used at 1:100,000 overnight 4°C; monoclonal mouse anti-GAPDH (Abcam) used at 1:1,000,000 dilution overnight 4°C; and horseradish peroxidase-conjugated goat anti-rabbit or mouse secondary antibodies (Millipore Bioscience Research Reagents, Temecula, CA) were used at 1:5000 dilution at room temperature for 1 h.
Microarray Analysis
The data sets for the microarrays listed in Supplemental Table 1 were downloaded from the National Center for Biotechnology Information (NCBI) GEO website, consolidated on Excel (Microsoft, Redmond, WA) and then analyzed on Theiler's murine encephalomyelitis virus (TMEV) to detect genes that were significantly changed in expression levels across all arrays. Expression levels corresponding to each gene of interest were recorded for all the arrays, and we used p ≤ 0.05 for significance.
Statistical Tests
Student's t test (two-tailed) in Excel (Microsoft) or GraphPad Prism (GraphPad Software, San Diego, CA) used for all statistical analyses. Statistical analyses from microarrays were generated using TMEV and also from the original data set analysis available on NCBI GEO. For siRNA knockdown and real-time PCR experiments, three biological replicates were tested in triplicate. For quantification of FRET, 20 P-bodies from a total of three images were analyzed.
RESULTS
siRNAs Localize Rapidly to P-Bodies
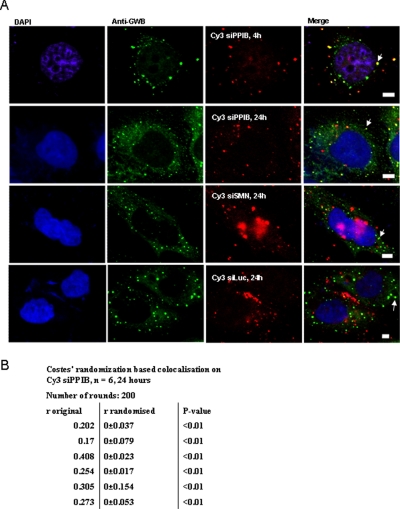
To study the pattern of intracellular siRNA localization, we visualized siRNAs targeted against cyclophilin-B (siPPIB, with Cy3 label on 5′ antisense strand), SMN (siSMN with Cy3 label on 5′ sense strand), or firefly luciferase (siLuc with Cy3 label on 5′ sense strand) transfected into HeLa cells. The cells were fixed after 4 or 24 h and immunostained with human anti-GW182 serum 18033, and colocalization of the siRNAs with P-bodies was studied. Significant colocalization of the antisense-labeled siRNA was observed (Fig. 1A). Colocalization of siPPIB to P-bodies was statistically quantified using Costes' randomization-based colocalization, with a highly significant (p < 0.01) colocalization coefficient of 0.268 ± 0.038 (SE) (Figure 1B) (Costes et al., 2004; Bolte and Cordelieres, 2006). Quantification of siLuc and siSMN colocalization to P-bodies showed a lower but again highly significant colocalization coefficient (siLuc, 0.159 ± 0.09; siSMN, 0.15 ± 0.03, p < 0.01). On monitoring siRNA localization in live cells expressing the P-body decapping enzyme component hDcp1a tagged with eGFP (eGFP-hDcp1a) to visualize P-bodies (van Dijk et al., 2002; Pillai et al., 2005), we saw that siRNAs could be found in P-bodies within 30 min of transfection (Supplemental Figure 1). Some large bodies containing intense siRNA signals were also present; these have been described previously to be vesicles/endosomes that remained from transfection (Jakymiw et al., 2005).
Figure 1.
Rapid localization of siRNA to P-bodies. (A) HeLa cells grown on poly-l-lysine–coated coverslips were transfected with 50 nM Cy3 siPPIB, Cy3 siSMN, or Cy3 siLuc (Red). At the given time points after transfection, cells were fixed and immunostained with anti-GWB serum (green). Overlays of the red and green images show colocalization of siRNA and P-bodies, indicated by arrows. Bar, 10 μm. (B) Colocalization of Cy3 siPPIB to P-bodies was statistically quantified using the Jacop program from six images, three containing between three and five cells per image and three containing ∼10 cells per image. Significant colocalization was observed, with the average Pearson's colocalization coefficient, r = 0.268 (SE = 0.038), p value for statistically significant colocalization obtained with Costes randomization based colocalization for each individual image <0.01.
If siRNAs were recruited to P-bodies simply because they were already bound to the target in the RISC complex, only the antisense strand should have been present in P-bodies. However, not only the antisense siRNA but also sense strand-labeled siSMN, sense strand-labeled siPPIB, and siRNA with no complementary target, siLuc (Figure 1A), localized to P-bodies, confirming a similar previous report (Jakymiw et al., 2005). This led us to investigate whether the reason a complementary target was not required for siRNA localization to P-bodies is because the siRNA localized to P-bodies in double-stranded form, before binding the target in the RISC complex.
Imaging of siRNA Labeled with FRET Dyes Reveals That Double-stranded siRNA Localizes to P-Bodies
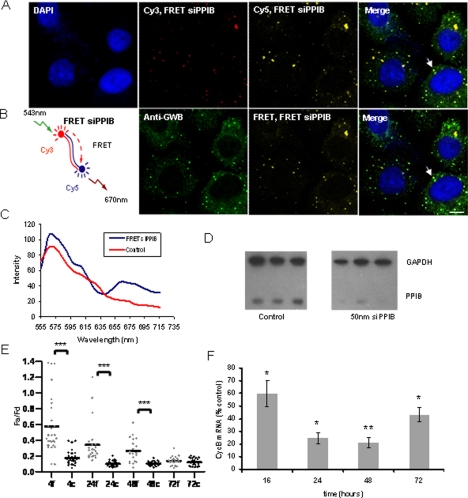
FRET technology has been shown to be a powerful real-time methodology to follow the intracellular fate and function of siRNAs where FRET has been successfully applied to discriminate between labeled siRNAs in single- or double-stranded conformation (Raemdonck et al., 2006; Jarve et al., 2007). Therefore, to test our hypothesis that double-stranded siRNA localizes to P-bodies, we decided to use FRET-labeled siRNA, which was siPPIB siRNA with 5′ sense strand Cy3 label and 5′ antisense strand Cy5 label (FRET siPPIB). When these fluorophores are <10 nm apart, upon excitation of the donor Cy3, due to energy transfer, emission from the acceptor, Cy5, is detected (Massey et al., 2006). In FRET siPPIB, the fluorophores are ∼6 nm apart in the native double-stranded siRNA and should lead to a FRET signal.
We first tested this siRNA in solution in vitro by using a fluorescent spectrophotometer, and we found upon excitation of the donor Cy3 that a reliable FRET signal could be detected from Cy5 at 670 nm. A much lower signal was obtained from siRNA tagged with Cy3 alone, showing that the bleed-through of Cy3 into the Cy5 emission region was much lower than any FRET signal collected (Supplemental Figure 2A). We transfected FRET siPPIB into HeLa cells, which were then fixed and immunostained with human anti-GWB serum after 24 h. It was possible to detect both sense and antisense strands using the respective Cy3 and Cy5 labels (Figure 2A) in P-bodies. In addition, we detected a strong FRET signal (Figure 2B), indicating that these dyes were in proximity, i.e., that the two siRNA strands were associated together. On examining the emission wavelength spectrum from a single P-body, we detected both the donor (Cy3 at ∼570 nm) and acceptor (Cy5 at ∼670 nm) spectra, arising only from donor excitation (543 nm) (Figure 2C). Several studies have shown that labeling the 5′ end of siRNAs with fluorescent dyes has a minimal effect on gene silencing activity; we therefore performed a Western blot assay on cells transfected with 50 nM FRET siPPIB to confirm its efficacy. FRET siPPIB produced robust silencing of its target mRNA (Figure 2D).
Figure 2.
Double-stranded siRNA is detected in P-bodies using FRET technology. (A) HeLa cells were grown on poly-l-lysine–coated coverslips and transfected with 50 nM FRET siPPIB. After 24 h, the cells were fixed and immunostained with human anti-GWB serum. Both siRNA strands colocalized in P-bodies (antisense strand image: Cy3, FRET siPPIB, excitation 543 nm, emission 560–615 nm [red]; sense strand image: Cy5, FRET siPPIB, excitation 643 nm, emission above 660 nm [yellow]; and anti-GWB [green]). Arrows indicate colocalization of siPPIB and P-bodies. (B) Cy3 (donor) and Cy5 (acceptor) are a FRET pair, and the proximity of these dyes on the double-stranded siRNA results in FRET (FRET siPPIB) (images: FRET, FRET siPPIB, excitation 543 nm, emission above 660 nm [yellow]). Colocalization with P-bodies (green) indicated with arrow. Bar, 20 μm. (C) Typical emission wavelength spectrum (lambda spectrum) obtained from a single P-body colocalizing with FRET PPIB (blue). Also included is a spectrum from P-body in cells transfected with control siRNAs (Cy3 siLuc and Cy5 siPPIB) showing little FRET signal (red). Excitation at 543 nm and emission collected every 10 nm after 550 nm. Cy3 emission peak, ∼570 nm; Cy5 emission peak, ∼670 nm. (D) FRET siPPIB is potent at silencing PPIB; Western blot indicating knockdown of PPIB at 72 h after transfection of FRET siPPIB into HeLa cells. Three lanes indicate samples in triplicate. GAPDH used as loading control. (E) Quantification of FRET signal confirms presence of double-stranded siRNA and indicates possible siRNA strand separation. eGFP-tagged hDcp1a was expressed in HeLa cells that were then transfected with either 20 nM FRET siPPIB or 40 nM FRET control (20 nM Cy5-siPPIB and 20 nM Cy3-siLuc). Fa/Fd (Fa is emission from acceptor on excitation of donor; Fd is emission from donor upon excitation of donor) from FRET siPPIB is indicated by light bars, and FRET control by dark bars from ∼20 P-bodies. Up to 48 h, there is a significant difference in Fa/Fd from FRET siPPIB and FRET control (***p < 0.001 as indicated by Student's t test). At 72 h, there is no longer a significant difference. (F) Quantification of target mRNA silencing timeline correlates with decline in FRET signal. HeLa cells were transfected with 50 nM FRET siPPIB. Cyclophilin-B mRNA levels were measured by real-time PCR. The light bars indicate the level of cyclophilin-B normalized to GAPDH, shown as % siLuc control. There is a significant reduction in the levels of cyclophilin-B through 72 h (*p < 0.05, **p < 0.01; Student's t test); however, maximum silencing occurs at 48 h, with levels rising again at 72 h.
To measure the amount of double-stranded siRNA in the P-bodies over time, we measured the FRET signal obtained from siRNA localized to P-bodies in live cells, where P-bodies were visualized with eGFP-hDcp1a (Supplemental Figure 2B). We quantified the FRET signal from the sensitized acceptor intensity relationships, i.e., from the ratio of acceptor Cy5 emission (Fa) to donor Cy3 emission (Fd) upon donor excitation only. This method has been used to quantify FRET signals from double-labeled siRNAs previously, to measure stability of the siRNA as a function of time (Raemdonck et al., 2006). Because microscopically visible P-bodies are structures ∼100–300 nm in diameter (Eystathioy et al., 2002) (a small diameter relative to the distance required between two siRNA strands for a FRET signal), it is possible that a FRET signal may arise from the proximity of the two dyes on individual, separated siRNA strands in the P-body oriented at random such that they are <10 nm apart. Hence, we controlled for this by transfecting equal amounts of two separate siRNAs, siPPIB with 5′ antisense strand Cy5 label and siLuc with 5′ sense strand Cy3 label. Any FRET signal collected from cells transfected with these siRNAs would therefore be the result of the two dyes in the P-body being in sufficient proximity at random.
The Fa/Fd from the control was significantly lower than Fa/Fd from the double-labeled FRET siPPIB (Figure 2E), indicating the FRET signal we captured was indeed from double-stranded siRNA. We found that the difference in Fa/Fd between the FRET siPPIB and control was highest at 4 h after transfection, reducing over time such that no difference was observed at 72 h. Furthermore, we found that although Fa/Fd from the FRET siPPIB reduced over time, levels from the control experiments showed no significant change.
Two possible reasons for the decline over time in the Fa/Fd of the FRET siPPIB relative to the control are that the siRNA was being actively used for target gene silencing or that it was being degraded. If degradation alone occurred, Fa and Fd would both decrease proportionately over time, but the ratio would remain constant. This is the case for the control. However with FRET siPPIB, Fa reduces disproportionately faster than Fd, which indicates increasing distance between the dyes without degradation leading to a decreasing ratio (possibly indicative of strand separation. We went on to measure the levels of the target mRNA (cyclophilin-B) at 4, 24, 48, and 72 h after transfection of 50 nM siPPIB into HeLa cells, by using real-time RT PCR. We found that maximum silencing occurred at 48 h (cyclophilin-B mRNA <20% of the control) after transfection. At 72 h, silencing of the target mRNA is relieved, with cyclophilin-B mRNA levels reaching ∼45% of control (Figure 2F). This pattern of mRNA silencing correlates closely with the findings in Figure 2E, where the double-stranded siRNA FRET signal in P-bodies was found to decrease with time, to reach a level no different from the control at 72 h.
This pattern is a correlation only and does not show that the siRNA fraction in P-bodies is the only active fraction. Cytoplasmic Ago2 that does not localize to P-bodies has been shown to be involved in silencing (Leung et al., 2006). However, our data does indicate that the siRNA fraction in P-bodies is probably active. One way to test whether this is indeed the case is to see whether Ago2, the main component of RISC, was associated in any way with the siRNA fraction localizing to P-bodies. This Ago2 requirement would indicate that the siRNA fraction localizing to P-bodies does interact with RISC.
Ago2 Is Required for Localization and/or Retention of siRNAs in P-Bodies
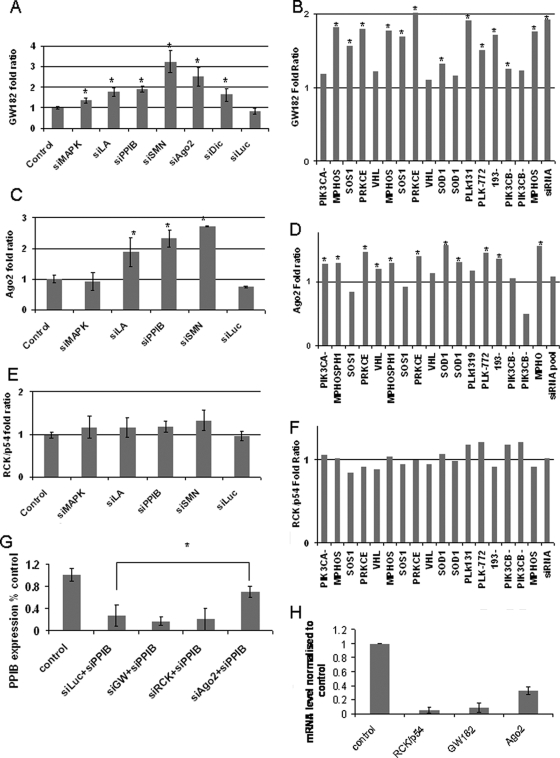
To test the idea that siRNA localization to P-bodies required Ago2, we studied the effect of knocking down Ago2 and other P-body components on both P-body integrity and siRNA localization to P-bodies. We knocked down Ago2, GW182, and RCK/p54 in HeLa cells by using siRNA transfection (Chu and Rana, 2006; Lian et al., 2007), and we found high levels of knockdown in all cases (>80%) (Figure 3A). We confirmed the level of Ago2 protein knockdown at 90% by measuring knockdown on an eGFP reporter (data not shown). It has been reported previously that Ago2 knockdown has no noticeable effect on P-body integrity or number in mammalian cells, whereas knockdown of GW182 and RCK/p54 leads to the disappearance of P-bodies, and our observations confirmed the same (Figure 3B) (Lian et al., 2007). We then transfected Cy3-siPPIB into these cells and observed the localization pattern of siRNAs. In the case of GW182 and RCK/p54 knockdown, we found a diffuse pattern of siRNA localization in the cytoplasm, consistent with the fact that microscopically visible P-body aggregates were no longer present. In the Ago2 knockdown experiments, we found that compared with the control, Cy3-siPPIB in Ago2 knockdown cells showed reduced siRNA colocalization with P-bodies (Figure 3B). This observation was quantified using Costes' randomization-based colocalization and a significant difference was observed in the colocalization coefficient between Ago2 knockdown and control (Figure 3C). We therefore conclude that Ago2 facilitates siRNA localization to and/or retention in P-bodies.
Figure 3.
Knockdown of Ago2 limits localization of siRNA to P-bodies. (A) RCK/p54, GW182, and Ago2 were knocked down by administration of 100 nM siRNA against each target individually in HeLa cells. Knockdown of respective targets was measured using real-time PCR at 72 h after transfection. (B) HeLa cells were grown on poly-l-lysine–coated coverslips and transfected with 100 nM siRNA targeting RCK/p54, GW182, or Ago2. After 48 h, the cells were transfected with Cy3 siPPIB. At 72 h, the cells were fixed and immunostained with human anti-GWB serum. Colocalization of siRNA with P-bodies is observed in the case of control. P-bodies are lost upon knockdown of GW182 and RCK/p54 and no corresponding colocalization of siPPIB and P-bodies is seen. In Ago2 knockdown, marginally reduced localization of siRNA to P-bodies is seen. Bar, 10 μm. (C) Colocalization of siPPIB to P-bodies was statistically quantified using the Jacop program. Six images containing between three and six cells per image were analyzed for both control and Ago2 knockdown, and average Pearson's colocalization coefficient was calculated for both, using Costes' randomization based colocalization. The Pearson's coefficient is significantly lower in Ago2 knockdown (p = 0.035; n = 6).
GW182 Is Up-Regulated upon siRNA Transfection
GW182 is one P-body component that has been shown to directly interact with RISC; an interaction required for silencing of miRNA targets in Drosophila (Behm-Ansmant et al., 2006; Eulalio et al., 2008b). The effect of GW182 knockdown in human cells on the capacity for effective siRNA-mediated silencing has yielded mixed data. However, GW182 has three human paralogues, which may have redundant function making it difficult to assess the importance of GW182 alone in siRNA-mediated silencing. To study whether GW182 (and therefore P-bodies) was associated with siRNA-mediated RNAi, we investigated if siRNA transfection had any effect on GW182 (trinucleotide repeat containing 6A, TNRC6A) expression. Transfection of six different siRNAs with complementary targets all caused an up-regulation of TNRC6A in 72 h (Figure 4A) compared with a control siRNA against luciferase (lacking a complementary target) that did not. To see whether this finding is applicable to all siRNAs, we interrogated publicly available microarray data from the NCBI GEO website. Eighteen sets of arrays were chosen using the following criteria: comparable microarray platform, 100 nM siRNA used on HeLa cells and RNA extracted 24 h after transfection. These constituted arrays from the following studies (Jackson et al., 2006a,b; Schwarz et al., 2006). See Supplemental Table 1 for list of target genes/siRNAs. Analysis of the entire data set showed highly significant up-regulation of TNRC6A by 70% (p < 0.0001). Because this analysis was across all arrays, it indicates this up-regulation occurs in response to transfection of most siRNAs. Individually, of the 18 array sets, 13 showed significant up-regulation of TNRC6A (Figure 4B), confirming our experimental findings in Figure 4A and showing that up-regulation of TNRC6A upon siRNA transfection is a general phenomenon. From our study, we found a smaller but significant up-regulation of Ago2 (eukaryotic initiation factor 2C, EIF2C2) (50%; p < 0.001) (Figure 4C), which we confirmed by quantifying the level of Ago2 mRNA in samples transfected with four different siRNAs. Three of the four showed significant up-regulation of Ago2, and this required the presence of a target mRNA (Figure 4D). The microarray data were taken at a 24-h time point, and PCR analysis was conducted at a 72-h time point. At 72 h, a greater degree of up-regulation can be observed both in the case of GW182 and Ago2. We also investigated the expression levels of other characterized P-body components by using the microarray data sets and real-time PCR, but we found no significant changes; the case of RCK/p54 (Figure 4, E and F) is given as an example.
Figure 4.
Transfection of siRNA induces up-regulation of GW182. (A) Seven different siRNAs (100 nM) were transfected into HeLa cells, and the level of GW182 was measured after 72 h by using real-time PCR. Significant up-regulation of GW182 was seen in all targeting siRNAs (between 35 and 222%; average 106%; *p < 0.05). siLuc, which did not have a complementary target, did not induce any increase. (B) From the NCBI GEO website, 18 microarray data sets were chosen based on satisfying the following conditions: 1) 100 nM siRNA against any target. 2) Transfected into HeLa cells. 3) RNA harvested at 24 h. 4) Microarrays done in duplicates in dye reversal. 5) Microarrays done on either of two platforms, GPL3991 or GPL3992, for easy comparison between samples. 6) p values available to indicate significance in ratios. Of the 18, 13 showed significant up-regulation of GW182, *p < 0.05. (C) In a similar experiment as A, levels of Ago2 were measured using real-time PCR, and results indicate three of four siRNAs cause a significant up-regulation of Ago2 * = p < 0.05. (D) Ago2 up-regulation as indicated by the same microarray analysis as (B), * = p < 0.05. (E) In a similar experiment as (A), levels of RCK/p54 were measured using real-time PCR, results indicate none of the siRNAs tested cause a significant up-regulation of RCK/p54. (F) Confirmation of lack of RCK/p54 up-regulation using the same microarray analysis as described in B. (G) Silencing of GW182 does not affect siRNA-mediated silencing. HeLa cells were transfected with 100 nM siRCK, siGW, or siAgo2 and after 48 h with siPPIB. Levels of siPPIB mRNA were measured after 24 h by using real time PCR. Knockdown of RCK/p54 and GW182 did not affect PPIB silencing, whereas knockdown of Ago2 significantly relieved silencing. (H) Knockdown of RCK/p54, GW182, and Ago2 at the same time point as PPIB measurement in G.
To investigate whether these components were required for siRNA-mediated silencing, we knocked down GW182, Ago2, and RCK/p54, and 48 h later we transfected the cells with 100 nM siPPIB. After 24 h, levels of PPIB knockdown were measured using real-time PCR. We found that knockdown of GW182 and PCK/p54 had no effect on PPIB silencing, whereas knockdown of Ago2 relieved PPIB silencing (Figure 4G), thereby confirming a previous report on the requirement of Ago2 for siRNA-mediated silencing (Lian et al., 2007). The lack of an effect on silencing after GW182 knockdown can be either due to redundancy of GW182 paralogues or/and because it may share silencing function with Ago2 (Behm-Ansmant et al., 2006). The presence of nonfunctional siRNA alone is not sufficient to cause this effect, as seen with the case of siLuc, which lacks a complementary target.
DISCUSSION
We have shown that siRNAs rapidly localize to P-bodies in double-stranded form. Furthermore, effective localization requires Ago2 and transfection of functional siRNAs in general causes an up-regulation of the key P-body component GW182 and the important RNAi effector Ago2. Together, our results suggest an important role for P-bodies in siRNA-mediated RNAi as cytoplasmic microenvironments that facilitate the interaction of double-stranded siRNAs with other RNAi elements. It is known that a functional RNAi pathway is required for the formation of P-bodies and furthermore, that Ago2 is concentrated in P-bodies even in the absence of miRNAs, as seen in miRNA-deficient dicer knockout cell lines (Leung et al., 2006). This indicates that RNAi and P-bodies are intimately linked; here, we show that this relationship extends to siRNA-mediated RNAi.
Our results indicate that siRNAs rapidly localize to P-bodies in double-stranded conformation and interact with core P-body components. However knockdown of GW182 does not impair the silencing of PPIB by siRNAs; therefore, we conclude siRNAs do not require the presence of microscopically visible P-bodies for efficient gene silencing to occur. The complete structure of P-bodies is yet to be defined; moreover, we do not yet know whether the P-body population displays structural and/or functional heterogeneity. Together, our data suggest that there may be a minimum P-body composition for effective siRNA-mediated RNAi. Thus, submicroscopic aggregates or “minimal” P-bodies produced as a result of knockdown of P-body components such as Lsm or RCK/p54 may retain the minimal P-body components necessary for siRNA-mediated RNAi. That knockdown of these two proteins does not prevent the reassembly of visible P-bodies formed due to siRNA function lends support to this idea (Lian et al., 2007). Our data from FRET imaging of double-stranded siRNA in P-bodies show an intriguing pattern of decay, whereby there is a significant difference between the rates at which the signal decays from the FRET siRNA compared with the control. This indicates a greater loss of FRET than that which occurs simply due to decay, indicating strand separation may contribute to this. If this is the case, P-bodies may be centers where double-stranded siRNA interacts with the RNAi machinery and processed to single-stranded form.
Importantly, both our experimental work and interrogation of publically available microarray data show that up-regulation of GW182 and Ago2 is a general response to siRNA function. In both cases, there are few instances where statistically significant up-regulation is not seen. This could be due to any of the following reasons: experimental variation and noise, certain siRNAs causing a higher up-regulation than other siRNAs, or off-target effects of the siRNA and further downstream effects that affect the levels of these proteins. That Ago2 is up-regulated in response to the presence of functional siRNA is not surprising, because it is the main effector of siRNA-mediated silencing. The up-regulation of GW182 is more intriguing, because GW182's exact function is yet to be determined. In Drosophila, Ago1–GW182 complexes are essential for miRNA-mediated silencing (Eulalio et al., 2008b), although how this complex achieves silencing is still debated (Eulalio et al., 2008a; Wu and Belasco, 2008). Tethering of GW182 to target mRNA shows that GW182 may facilitate silencing through a mode that bypasses the requirement for Ago2 (Behm-Ansmant et al., 2006). Other work has shown that the GW/WG repeat domains in GW182 form evolutionarily and functionally conserved binding platforms for RNAi machinery (El-Shami et al., 2007). Our results show that a general phenomenon upon transfecting siRNA is that both GW182 and Ago2 are up-regulated. Increases in their levels may be the cellular system's response to large increases in RNAi activity. It is possible that these increases ensure that endogenous RNAi function is maintained. siRNA against luciferase that lacks a complementary target does not cause this effect, indicating that it is not simply the presence of siRNA but rather siRNA function against a complementary target mRNA that induces up-regulation. These results are consistent with previous studies that showed siRNA-mediated silencing induced increases in P-body size and number in a target-dependent manner (Lian et al., 2007) and that GW182 is an important player in the RNAi pathway and is required for silencing (Liu et al., 2005a; Eulalio et al., 2008b). Our data, in the light of these studies, confirms that siRNAs do interact with P-bodies and their RNAi-related components.
In summary, our results show that P-bodies are cytoplasmic sites where siRNAs aggregate, in addition to being centers where nontranslating mRNA and RNAi components are found. Moreover, siRNA transfection induces the up-regulation of two important P-body components involved in RNAi, Ago2 and GW182. This suggests that although microscopically visible P-bodies are not absolutely required for siRNA silencing, the interaction of siRNAs with core P-body components, whether in visible P-bodies or in submicroscopic minimal P-bodies, provides a microenvironment that facilitates siRNA-mediated silencing (Jakymiw et al., 2005).
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Sridhar Vasudevan, Prof. Edward Chan, Dr. Andrew Jakymiw, Dr. Grant Churchill, Dr. Raman Parkesh, and Dr. Shankar Srinivas for technical assistance and helpful discussions. We also thank Drs. Lykke-Andersen and Marvin Fritzler for providing reagents. This work was supported by grants from the UK Medical Research Council (to M. W.) and a Clarendon Scholarship (to A. J.).
Abbreviations used:
- Ago2
Argonaute 2
- P-body
processing body.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-08-0796) on October 22, 2008.
REFERENCES
- Behm-Ansmant I., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR 4, NOT deadenylase and DCP 1, DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S., Cordelieres F. P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- Chu C. Y., Rana T. M. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costes S. V., Daelemans D., Cho E. H., Dobbin Z., Pavlakis G., Lockett S. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys. J. 2004;86:3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N., Babajko S., Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Shami M., Pontier D., Lahmy S., Braun L., Picart C., Vega D., Hakimi M. A., Jacobsen S. E., Cooke R., Lagrange T. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 2007;21:2539–2544. doi: 10.1101/gad.451207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 2007a;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Schweizer D., Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 2007b;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Huntzinger E., Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008a;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- Eulalio A., Huntzinger E., Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat. Struct. Mol. Biol. 2008b;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- Eystathioy T., Chan E. K., Tenenbaum S. A., Keene J. D., Griffith K., Fritzler M. J. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol. Biol. Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige J. N., Sage D., Wahli W., Desvergne B., Gelman L. PixFRET, an ImageJ plug-in for FRET calculation that can accommodate variations in spectral bleed-throughs. Microsc. Res. Tech. 2005;68:51–58. doi: 10.1002/jemt.20215. [DOI] [PubMed] [Google Scholar]
- Hammond S. M. Dicing and slicing–the core machinery of the RNA interference pathway. FEBS Lett. 2005;579:5822–5829. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- Hammond S. M., Bernstein E., Beach D., Hannon G. J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Ingelfinger D., Arndt-Jovin D. J., Luhrmann R., Achsel T. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2002;8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- Jackson A. L., et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006a;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A. L., Burchard J., Schelter J., Chau B. N., Cleary M., Lim L., Linsley P. S. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006b;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakymiw A., Lian S., Eystathioy T., Li S., Satoh M., Hamel J. C., Fritzler M. J., Chan E. K. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- Jarve A., et al. Surveillance of siRNA integrity by FRET imaging. Nucleic Acids Res. 2007;35:e124. doi: 10.1093/nar/gkm694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A. K., Calabrese J. M., Sharp P. A. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc. Natl. Acad. Sci. USA. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian S., Fritzler M. J., Katz J., Hamazaki T., Terada N., Satoh M., Chan E. K. Small interfering RNA-mediated silencing induces target-dependent assembly of GW/P bodies. Mol. Biol. Cell. 2007;18:3375–3387. doi: 10.1091/mbc.E07-01-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Rivas F. V., Wohlschlegel J., Yates J. R., 3rd, Parker R., Hannon G. J. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 2005a;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Valencia-Sanchez M. A., Hannon G. J., Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005b;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J., Patkaniowska A., Urlaub H., Luhrmann R., Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- Massey M., Algar W. R., Krull U. J. Fluorescence resonance energy transfer (FRET) for DNA biosensors: FRET pairs and Forster distances for various dye-DNA conjugates. Anal. Chim. Acta. 2006;568:181–189. doi: 10.1016/j.aca.2005.12.050. [DOI] [PubMed] [Google Scholar]
- Pauley K. M., Eystathioy T., Jakymiw A., Hamel J. C., Fritzler M. J., Chan E. K. Formation of GW bodies is a consequence of microRNA genesis. EMBO Rep. 2006;7:904–910. doi: 10.1038/sj.embor.7400783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai R. S., Bhattacharyya S. N., Artus C. G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Raemdonck K., Remaut K., Lucas B., Sanders N. N., Demeester J., De Smedt S. C. In situ analysis of single-stranded and duplex siRNA integrity in living cells. Biochemistry. 2006;45:10614–10623. doi: 10.1021/bi060351b. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J., Behm-Ansmant I., Gatfield D., Izaurralde E. A crucial role for GW182 and the DCP 1, DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz D. S., Ding H., Kennington L., Moore J. T., Schelter J., Burchard J., Linsley P. S., Aronin N., Xu Z., Zamore P. D. Designing siRNA that distinguish between genes that differ by a single nucleotide. PLoS Genet. 2006;2:e140. doi: 10.1371/journal.pgen.0020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen G. L., Blau H. M. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- Serman A., Le Roy F., Aigueperse C., Kress M., Dautry F., Weil D. GW body disassembly triggered by siRNAs independently of their silencing activity. Nucleic Acids Res. 2007;35:4715–4727. doi: 10.1093/nar/gkm491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U., Parker R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125:1095–1109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk E., Cougot N., Meyer S., Babajko S., Wahle E., Seraphin B. Human Dcp 2, a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S., Steitz J. A. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Belasco J. G. Let me count the ways: mechanisms of gene regulation by miRNAs and siRNAs. Mol. Cell. 2008;29:1–7. doi: 10.1016/j.molcel.2007.12.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.