Abstract
Appropriate clearance of apoptotic cells (cell corpses) is an important step of programmed cell death. Although genetic and biochemical studies have identified several genes that regulate the engulfment of cell corpses, how these are degraded after being internalized in engulfing cell remains elusive. Here, we show that VPS-18, the Caenorhabditis elegans homologue of yeast Vps18p, is critical to cell corpse degradation. VPS-18 is expressed and functions in engulfing cells. Deletion of vps-18 leads to significant accumulation of cell corpses that are not degraded properly. Furthermore, vps-18 mutation causes strong defects in the biogenesis of endosomes and lysosomes, thus affecting endosomal/lysosomal protein degradation. Importantly, we demonstrate that phagosomes containing internalized cell corpses are unable to fuse with lysosomes in vps-18 mutants. Our findings thus provide direct evidence for the important role of endosomal/lysosomal degradation in proper clearance of apoptotic cells during programmed cell death.
INTRODUCTION
During the development of Caenorhabditis elegans hermaphrodites, three waves of programmed cell death (apoptosis) occur to remove 131 somatic cells and nearly half the germ cells generated in the process of oocyte maturation (Horvitz, 2003; Lettre and Hengartner, 2006). Programmed cell death in C. elegans is essentially controlled by an evolutionarily conserved genetic pathway that regulates different events of cell death, including the decision of cell death, the killing of cells, and the removal of cell debris (Horvitz, 2003). With the identification of more than a dozen of these regulators, the molecular mechanisms underlying several cell death events are becoming evident. It is now known that the genetic and molecular interaction of four factors, EGL-1, CED-9, CED-4, and CED-3, determines the activation of cell-killing process (Horvitz, 2003). After cell killing, apoptotic cells are rapidly engulfed and digested by neighboring cells. The engulfment (phagocytosis) of cell corpse involves two partially redundant genetic pathways, with the genes ced-1, ced-6, ced-7, and dyn-1 functioning in one pathway and the genes ced-2, ced-5, ced-10, ced-12 and psr-1 acting in the other (Wang et al., 2003; Reddien and Horvitz, 2004; Yu et al., 2006). It has been proposed that the ced-1 pathway likely regulates the recognition of cell corpse and transduces engulfing signals. CED-1 clusters at the membrane contact between the engulfing cell and the cell corpse to initiate engulfment, which requires CED-7, a C. elegans homologue of mammalian ATP-binding cassette transporters (Zhou et al., 2001). CED-6 may act downstream of CED-1 to transduce engulfing signals as CED-6 can interact with the intracellular domain of CED-1 through its phosphotyrosine binding domain (Liu and Hengartner, 1998). In the other pathway, PSR-1 may recognize phosphotydylserine, one of the “eat me” signals, and function through CED-5/Dock180 and CED-12/ELMO1, which together with CED-2/CrkII and CED-10/Rac GTPase induces the cytoskeleton reorganization of engulfing cell for cell corpse internalization (Wang et al., 2003).
Although much is known about the genetic and molecular basis of cell corpse engulfment in C. elegans, how cell corpses are degraded after being internalized in engulfing cells remains poorly understood. In mammalian cells, many studies have suggested that lysosome plays pivotal roles in eliminating phagosomal contents such as foreign pathogens after their internalization (Jutras and Desjardins, 2005; Luzio et al., 2007). Lysosomes are single-membrane organelles that are formed by different mechanisms, such as maturation of late endosome/multivesicular body by Rab conversion, fusion into a hybrid organelle, and fission as well as “kiss-and-run” between late endosomes and existing lysosomes (Treusch et al., 2004; Bright et al., 2005; Rink et al., 2005). Lysosomes contain many acidic hydrolases, including proteases, nucleases, glycosidases, sulfatases and lipases, thus serving as the major degradative sites within eukaryotic cells. By dynamically or sequentially fusing with early and late endosomes and ultimately with lysosomes, nascent phagosomes containing foreign pathogens or particles mature into phagolysosomes where the internalized pathogens or particles are degraded (Jutras and Desjardins, 2005). In C. elegans, recent studies on UNC-108/RAB-2 and RAB-7 small GTPases, which are important for lysosome maturation and endocytosis, start to uncover the role of lysosome and its related cellular processes in the removal of cell corpses (Lu et al., 2008; Mangahas et al., 2008; Yu et al., 2008). More recently, it was reported that the C. elegans phosphatidylinositiol 3- kinase VPS-34 and other components of endocytic pathways contribute to the maturation of phagosomes containing cell corpses (Kinchen et al., 2008), but the in-depth mechanisms for these molecules to control cell corpse degradation need further investigation.
In this study, we found that C. elegans VPS-18, a potential ubiquitin ligase important for lysosome maturation, plays a critical role in the clearance of apoptotic cells. VPS-18 is the C. elegans homologue of the yeast class C Vps protein, Vps18p, which forms HOPS complex together with Vps11p, Vps16p, and Vps33p, the other three class C Vps members, and Vps39p and Vps41p, the two class B Vps proteins (Sato et al., 2000; Seals et al., 2000). In yeast, HOPS complex mediates membrane docking and fusion by interacting with Ypt7/Rab7 and soluble N-ethylmaleimide-sensitive factor attachment protein receptor complex, which is required for endosome–lysosome and autophagosome–lysosome docking and fusion (Sato et al., 2000; Seals et al., 2000). It has been reported previously that inactivation of the C. elegans homologues of class C vps genes by RNA interference affected germ cell death (Lackner et al., 2005), but how these genes regulate C. elegans apoptosis is unknown. In addition, the function of HOPS complex in other cellular events in worm remains unexplored. Here, we investigate the role of vps-18 in regulating programmed cell death in C. elegans, with detailed characterization of a vps-18 deletion mutant. We show that inactivation of vps-18 causes severe defects in endosome and lysosome biogenesis in C. elegans. We provide evidence that vps-18 functions in the phase of cell corpse clearance by affecting phagosome–lysosome fusion and thus affecting the degradation of cell corpses. Our results suggest that endosomal/lysosomal degradation activity directly contributes to the removal of apoptotic cells.
MATERIALS AND METHODS
C. elegans Strains and Genetics
The vps-18(tm1125), vps-33(tm327), and vps-39(tm2253) deletion mutants were provided by Dr. Shohei Mitani (Tokyo Women's College, Tokyo, Japan). The vps-11(ok1664) and vps-16(ok719) deletion mutants were provided by C. elegans Genetics Center (CGC, University of Minnesota, Minneapolis, MN). The Bristol strain N2 was used as wild-type. C. elegans cultures, and genetic crosses were performed essentially according to standard procedures (Brenner, 1974). Mutant alleles used in this study are listed by linkage groups: LGI: ced-1(e1735), ced-12(tp2). LGII: vps-11(ok1664), vps-18(tm1125). LGIII: ced-6(n2095), ced-7(n1892), vps-16(ok719), and vps-33(tm327). LGIV: ced-2(n1994), ced-5(n1812). LGV: unc-76(e911), vps-39(tm2253). Deletion strains were outcrossed with N2 strain for at least four times. The strains used to monitor the biogenesis of endosomes and lysosomes as well as chromatin degradation were provided by Dr. Hanna Fares (University of Arizona, Tucson, AZ) and CGC: arIs36 (PhspssGFP), bIs34(RME-8::GFP), bcIs39 (Plim-7CED-1::GFP), cdIs39(GFP::RME-1), cdIs40(GFP::CUP-5), cdEx49(GFP::RAB-5), cdIs66 (GFP::RAB-7), cdIs73(RME-8::mRFP), cdIs97(mCherry::CUP-5), pwIs50 (LMP-1::GFP), and ruIs32(GFP::H2B). Transgenic strain expressing Pced-1LMP-1::mCherry was provided by Dr. Xiaochen Wang (National Institute of Biological Sciences, Beijing, China).
Plasmid Construction
To make Pvps-18vps-18::gfp construct, the genomic DNA containing the entire coding region and 2 kb of the 5′ upstream promoter region of vps-18 gene was amplified by polymerase chain reaction (PCR) and subsequently cloned into the vector pPD95.77 between the PstI and XbaI sites. The expression vector for VPS-18(C859A, H861A) mutant protein was generated by using a PCR-based assay with Pvps-18vps-18::gfp expression vector as template. To generate Pvps-33mVps18 construct, a 2-kb DNA fragment of the 5′ promoter region of vps-33 gene was amplified by PCR and cloned into pPD95.77 between the SphI and XbaI sites, and the mVps18 cDNA with a stop codon was amplified by reverse transcription-PCR and subsequently cloned into the above vector between the BamHI and XhoI sites. To make Plim-7vps-18::gfp construct, a DNA fragment of 4129 base pairs containing the lim-7 promoter and a DNA region encoding the first 61 amino acids of LIM-7 were first cloned into pPD95.77 and then the cDNA of vps-18 was inserted in-frame between the lim-7 promoter and the green fluorescent protein (GFP) coding region.
Acridine Orange (AO) Staining of Cell Corpses
To stain the germ cell corpses with AO, adult worms were incubated in M9 medium containing AO (100 μg/ml) and OP50 bacteria in dark for 2 h at room temperature. Worms were then transferred to nematode growth medium plates for recovery for 1 h to decrease background gut fluorescence. The stained worms were examined under fluorescence microscopy to score the AO-positive germ cell corpses from >150 germ cell corpses of each examined strain.
Quantification of Cell Corpses
Cell corpses were scored by using Nomarski optics. For embryonic cell corpses, only those in the head region of embryos were scored. Germ cell corpses in the germline meiotic region of one gonad arm were scored 12, 24, 36, 48, and 60 h after L4 larval stage.
Four-dimensional Analysis of Cell Corpse Duration
To record the duration of embryonic cell corpses, early embryos (2-cell stage) were put in egg salt buffer (118 mM NaCl and 48 mM KCl) and mounted on slides with agar pads. The slides were sealed with beeswax and petroleum jelly (1:1). Images in a 28 Z-serial sections (1 μm/section) were captured every minute for 400 min by using an Axioimager M1 coupled with AxioCam monochrome digital camera and Axiovision release 4.6 software (Carl Zeiss, Jena, Germany). To record the duration of germ cell corpses, animals were mounted in M9 buffer containing 2 mM levamisole and observed under Nomarski optics at 20°C. The gonadal region was recorded every minute at 1 μm/section for 30 Z-sections. Animals were constantly examined for viability during recording.
In Vivo Protein Uptake and Degradation Assay
We performed the in vivo protein uptake and degradation assay using the transgenic strain arIs36(PhspssGFP) and vps-18(tm1125);arIs36. To follow the uptake and degradation of secreted soluble (ss)GFP in coelomocytes, worms were heat shocked for 30 min at 33°C and continued to grow at 20°C. The signal of ssGFP in body cavity and coelomocytes was observed by using fluorescence microscopy at indicated times, and pictures were taken with equal exposure time.
RESULTS
vps-18 Affects the Clearance of Cell Corpses in C. elegans
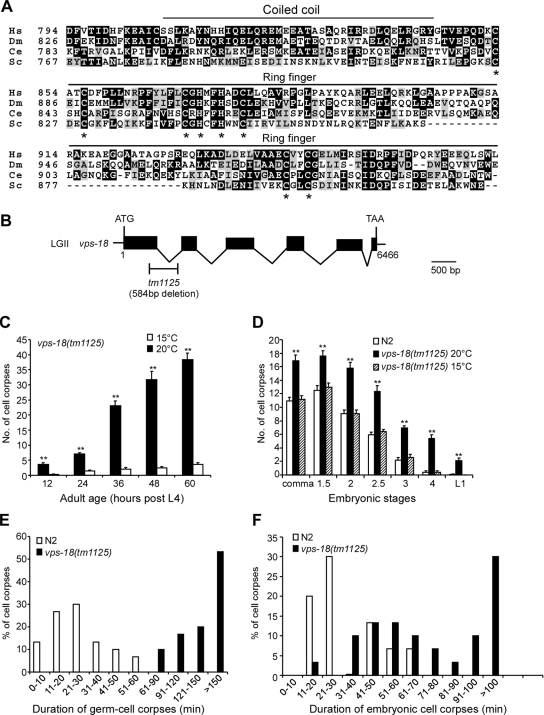
C. elegans vps-18 gene is defined by the open reading frame W06B4.3 located on the linkage group II and encodes a protein of 962 amino acids that shares homology with yeast, Drosophila, and human Vps18p proteins. In the C-terminal region, C. elegans VPS-18 contains a predicted coiled-coil domain and a RING-finger motif (C3H2C3 type), which are implicated in protein–protein interaction and protein ubiquitination, respectively (Figure 1A). In humans, hVps18p was shown to possess ubiquitin ligase activity that monoubiquitinates the adaptor protein GGA3 involved in endocytosis (Yogosawa et al., 2006). But whether hVps18p is involved in apoptosis is not known.
Figure 1.
C. elegans vps-18 affects cell corpse clearance. (A) Sequence alignment of the C-terminal region of VPS proteins of human (Hs), Drosophila (Dm), C. elegans (Ce), and yeast (Sc). The identical amino acids are shaded in black and similar amino acids are shaded in gray. The top lines indicate the regions for coiled-coil domain and RING-finger motif. Asterisks indicate the residues critical for RING-finger motif. (B) Schematic representation of vps-18(tm1125) deletion. Solid boxes indicate exons and waved lines indicate introns. The fragment below the gene indicates the region and size of the deletion mutation of vps-18. The scale bar represents 500 base pairs. (C) The profile of germ cell death in vps-18(tm1125) mutants. Germ cell corpses in one gonad arm of each animal were scored every 12 h after L4 stage. The error bars represent SEM. Fifteen animals were scored for each time point. Comparisons were performed with unpaired t test. Single asterisks indicate p < 0.05 and double asterisks indicate p < 0.001. (D) The profile of embryonic cell death in vps-18(tm1125) mutants. Cell corpses of 15 embryos at each embryonic stage were scored. Comparisons were performed as described in C. (E and F) Four-dimensional microscopy analysis of durations of persistence of germ cell corpses (E) and embryonic cell corpses (F) in N2 and vps-18(tm1125) animals. The duration of 30 germ cell corpses and 30 embryonic cell corpses in N2 and vps-18(tm1125) animals were recorded, respectively.
In an effort to identify potential ubiquitin ligases affecting both programmed cell death and lysosome biogenesis, we found that inactivation of C. elegans vps-18 by RNA interference (RNAi) caused an increase of germ cell corpses in an age-dependent manner (data not shown). To investigate further how vps-18 affects programmed cell death in C. elegans, we analyzed a deletion mutant vps-18(tm1125) that removes 584 base pairs between the first two exons, which results in an early stop codon and likely represents a strong loss-of-function mutation of vps-18 gene (Figure 1B). vps-18(tm1125) animals seem moderately dumpy and exhibit temperature-dependent embryonic lethality. At 15 and 20°C, vps-18(tm1125) embryos develop properly to adults; but eggs do not hatch at 25°C. vps-18(tm1125) adult animals possess normal germline, and their brood size is slightly smaller than that of wild-type animals at 20°C (data not shown).
We observed that a large number of germ cell corpses were present in the germline of vps-18(tm1125) animals. A time course analysis indicated that the increase of germ cell corpses in vps-18(tm1125) mutants occurred in an age-dependent manner (Figure 1C). Intriguingly, the amount of germ cell corpses was significantly reduced at 15°C, indicating that the increase of germ cell corpses in vps-18(tm1125) was temperature sensitive (Figure 1C). Furthermore, in the double mutants vps-18(tm1125);ced-3(n717) and vps-18(tm1125);ced-4(n1162), the germ cell corpses were completely suppressed (data not shown), suggesting that the germ cell corpses in vps-18(tm1125) animals are resulted from physiological cell death. We also examined the embryonic cell death profiles in the mutant animals. Consistent with the observation on germ cell corpses, vps-18(tm1125) worms exhibited significantly higher number of embryonic cell corpses than wild-type animals at every examined developmental stage at 20°C, but they seemed the same as wild-type animals at 15°C (Figure 1D).
The increase of both embryonic and germ cell corpses in vps-18(tm1125) could be caused by either excessive apoptosis or defects in cell corpse clearance. To distinguish between these two possibilities, we first examined whether there were extra “undead cells” or “missing cells” caused by insufficient or ectopic cell death in the anterior pharyngeal region or in the ventral cord Pn.aap cells (Reddien et al., 2001), but we did not find either undead cells or missing cells in these tissues. We also monitored the occurrence of cell death in the U-turn region in the germline of both wild-type and mutant animals. In wild type (n = 10), an average of 3.9 ± 0.7 germ cells die within 200 min in the U-turn region observed. A comparable number (4.3 ± 0.5) of cell death events were observed in the same germline region within the same period in vps-18(tm1125) mutants (n = 10), indicating that the mutation of vps-18 does not result in a significant increase of germ cell apoptosis.
We next carried out four-dimensional microcopy analysis to measure the duration of persistence of both embryonic and germ cell corpses. In wild-type animals, the average duration of germ cell corpse was 23.6 ± 2.5 min, and we seldom observed germ cell corpses persisting >60 min. In vps-18(tm1125) mutant animals, however, the average persistence time of germ cell corpses were greatly expanded such that >50% of examined cell corpses persisted >150 min; and we did not observe any germ cell corpses existing >60 min (Figure 1E). In embryos, the average duration of embryonic cell corpses in wild-type animals was 32.6 ± 2.6 min, whereas 50% of cell corpses persisted >70 min in vps-18(tm1125) animals (Figure 1F). These findings suggest that the increase of cell corpses in vps-18(tm1125) animals is likely caused by defects in the removal of cell corpses.
In addition, we also examined the deletion mutants of other C. elegans homologues of yeast HOPS complex, including vps-11(ok1664), vps-16(ok719), vps-33(tm327), and vps-39(tm327), and observed a large number of persistent germ cell corpses in these mutants (Supplemental Figure S1). RNAi depletion of these genes in wild-type animals also gave rise to an increase of germ cell corpses. However, RNAi of these genes in vps-18(tm1125) did not enhance the defects in cell corpse clearance in vps-18(tm1125) mutants (Supplemental Figure S1C), suggesting that these C. elegans homologues of HOPS components function together with VPS-18 to regulate cell corpse clearance.
vps-18 Does Not Act within a Specific Engulfment Pathway
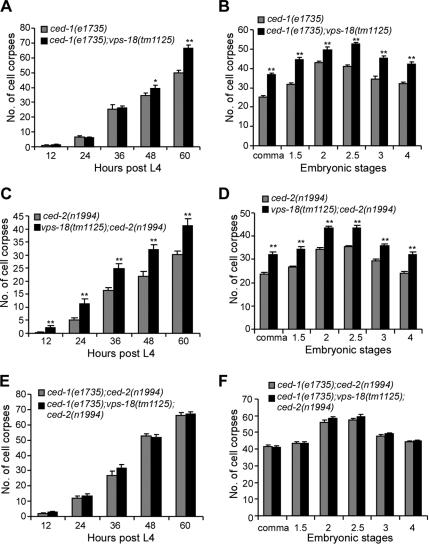
Because vps-18 affects the clearance of cell corpses, we wondered whether it functions together with the engulfment genes to regulate cell corpse engulfment. In C. elegans, the engulfment of cell corpses is regulated by two parallel and partially redundant pathways of which nine genes have been identified, with the genes ced-1, -6, -7, and dyn-1 functioning in one pathway and the genes ced-2, -5, -10, -12, and psr-1 in the other pathway. To test which pathway vps-18 may act in, we constructed and analyzed double mutants between vps-18(tm1125) and loss-of-function alleles of genes from each pathway. Our results indicated that vps-18(tm1125) significantly enhanced the defects in both embryonic and germ cell corpse engulfment at late adult stages in ced-1(e1735) and ced-7(n1892) animals (Figure 2, A and B; data not shown). Intriguingly, we found that vps-18(tm1125) also enhanced the engulfment defects of both embryonic and germ cell corpses in the mutants ced-2(n1994) and ced-5(n1812), which function in the other genetic pathway to regulate cell corpse engulfment (Figure 2, C and D; data not shown). These findings suggest that vps-18 does not act specifically within either pathway to regulate cell corpse removal. To further test whether vps-18 functions in parallel to the two pathways of cell corpse engulfment, we constructed ced-1(e1735); vps-18(tm1125);ced-2(n1994) triple mutants. However, we found that neither embryonic cell corpses nor germ cell corpses were increased in the triple mutants compared with that in the ced-1(e1735);ced-2(n1994) double mutants (Figure 2, E and F). In addition, our further analysis indicated that the persistent cell corpses in vps-18(tm1125) animals are internalized (see below). Thus, vps-18 likely functions downstream of engulfment genes to affect the removal of cell corpses.
Figure 2.
vps-18(tm1125) mutation enhances engulfment defects in the mutants of both engulfment pathways. Comparisons were performed with unpaired t test as described above. (A, C, and E) Germ cell corpses of one gonad arm from indicated animals were scored every 12 h post-L4 stage. Fifteen animals were scored for each time point. (B, D, and F) Embryonic cell corpses of 15 embryos of indicated animals at each embryonic stage were scored. The error bars represent SEM.
The Persistent Cell Corpses in vps-18(tm1125) Are Internalized but Not Digested
To investigate how vps-18 is involved in the clearance of cell corpses, we first examined whether vps-18 affected the clustering of CED-1::GFP around cell corpses by crossing vps-18(tm1125) mutant with the transgenic strain bcIs39 that expressed CED-1::GFP fusion protein (Plim-7ced-1::gfp) in gonadal sheath cells (Zhou et al., 2001). In wild-type animals, germ cell corpses were surrounded by CED-1::GFP (Supplemental Figure S2), which were internalized into phagosome-like structures (Zhou et al., 2001). In vps-18(tm1125) mutant animals, the majority of the persisting germ cell corpses were also surrounded by CED-1::GFP produced by germline sheath cells, indicating that vps-18 does not affect the clustering of CED-1 around germ cell corpses (Supplemental Figure S2).
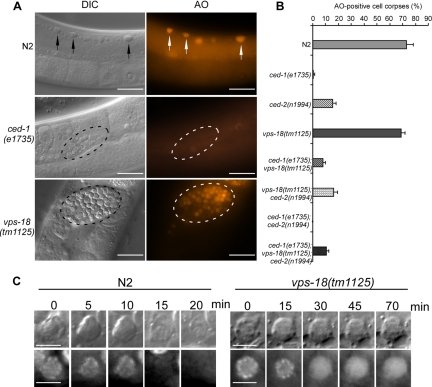
We next used AO staining to determine whether the persistent cell corpses in vps-18(tm1125) animals were internalized. AO is an acidophilic fluorescent dye that preferentially stains acidic cellular compartments and thus labels the cell corpses internalized in living cells (Gumienny et al., 1999; Lettre et al., 2004; Lu et al., 2008). In the germline of wild-type animals, cell corpses were rapidly engulfed of which 70% were AO positive (Figure 3, A and B). In contrast, only a small amount of germ cell corpses in ced-1(e1735) and ced-2(n1994) animals were labeled by AO because most cell corpses were not phagocytosed in these mutants (Figure 3, A and B). However, a majority of germ cell corpses (68.7%) in vps-18(tm1125) animals were stained by AO, which was comparable with that in wild-type animals (Figure 3, A and B), indicating that these persistent germ cell corpses were indeed internalized in engulfing cells. Consistently, we found that the embryonic cell corpses in vps-18(tm1125) were labeled by AO as in N2 animals (data not shown). Furthermore, the number of AO-positive germ cell corpses were greatly reduced in the double mutants ced-1(e1735);vps-18(tm1125) and vps-18(tm1125);ced-2(n1994) as well as in the triple mutants ced-1(e1735);vps-18(tm1125);ced-2(n1994), confirming that vps-18 functions downstream of engulfing genes (Figure 3B) and likely affects the degradation of internalized cell corpses.
Figure 3.
Cell corpses are internalized in vps-18(tm1125) mutants. (A) AO staining of germ cell corpses in indicated animals. Arrows and fragmented lines indicate cell corpses in N2, ced-1(e1735) and vps-18(tm1125) worms, respectively. Bars, 20 μm. (B) Quantification of AO-positive germ cell corpses in wild type and indicated single, double, and triple mutants. Error bars represent SEM. (C) Time-lapse recording of chromatin degradation of the cell corpses of both wild-type and vps-18(tm1125) animals. Bars, 5 μm.
To directly monitor the degradation process of cell corpses, we introduced in vps-18(tm1125) mutants a transgenic marker GFP::H2B which is specifically expressed in germline to label chromatin (Praitis et al., 2001). In wild type, the chromatin in early germ cell corpses was condensed and disappeared within 30 min (28.7 ± 5.8 min; n = 8) (Figure 3C). In vps-18(tm1125) mutants, although the chromatin in early germ cell corpses was also condensed, it became diffusive in the later stage and the GFP signal remained for >70 min (n = 8), indicating that the degradation of the chromatin is greatly delayed (Figure 3C). Thus, these findings demonstrate that the accumulation of cell corpses caused by vps-18 mutation is due to the defects in cell corpse digestion in engulfing cells.
VPS-18 Activity in Engulfing Cells Is Important for Cell Corpse Clearance
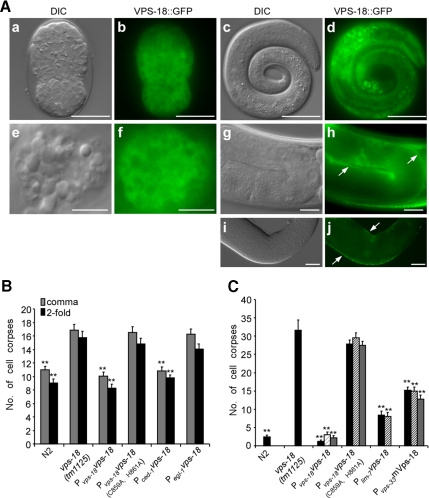
To determine the subcellular localization and expression pattern of VPS-18, we analyzed a VPS-18::GFP fusion protein under the control of the vps-18 promoter (Pvps-18vps-18::gfp). VPS-18::GFP was ubiquitously expressed in early embryo and seemed cytoplasmic (Figure 4A, a and b). In early larvae, VPS-18::GFP was mainly seen in hypodermal cells, seam cells and some body wall muscle cells (Figure 4A, c and d; data not shown). In adult hermaphrodite, VPS-18::GFP was detected in coelomocytes (Figure 4A, e and f), the C. elegans scavenger cells functioning similarly to mammalian macrophages for endocytosing fluid from pseudocoelom (Fares and Greenwald, 2001a). VPS-18::GFP signal was also present in gonadal sheath cells (Figure 4A, g–j). In addition, VPS-18::GFP was expressed in intestine and spermatheca (data not shown).
Figure 4.
Expression and rescuing activity of VPS-18. (A) VPS-18::GFP expression in embryo (a and b), larva (c and d), adult coelomocyte (e and f), and germline (g and h) as well as the dissected-out germline (i and j). Arrows in (h and j) indicate germline sheath cells. The scale bars represent 20 μm (a–d, i, and j), 5 μm (e and f), and 10 μm (g and h). (B) Rescuing activity of Pvps-18vps-18::gfp, Pvps-18 vps-18(C859A, H861A)::gfp, Pced-1vps-18 and Pegl-1vps-18 in vps-18(tm1125) embryos. Embryonic cell corpses from 15 animals were scored for comma- (gray bar) and twofold stage (black bar). Comparisons were performed between vps-18(tm1125) and N2 animals and between vps-18(tm1125) and transgenic animals by using unpaired t test. For transgenes, only one transgenic line from a total of three lines that exhibit similar rescuing activity was shown. (C) Rescuing activity of Pvps-18vps-18::gfp, Pvps-18 vps-18(C859A, H861A)::gfp, Pvps-33mVps-18, and Plim-7vps-18::gfp in vps-18(tm1125) germline. Germ cell corpses in one gonad arm of 15 animals were scored 48 h post-L4 stage for two to three independent transgenic lines as indicated by black, waved, and gray columns. Comparisons were performed between vps-18(tm1125) and all transgenic lines by using unpaired t test. Error bars represent SEM, and double asterisks (**) indicate p < 0.001.
Hypodermal and sheath cells are the major engulfing cells for the engulfment of embryonic and germ cell corpses, respectively. Because we detected the expression of VPS-18 in these cells, we tested whether expression of VPS-18 in engulfing cells was sufficient to rescue the cell corpse clearance defects in vps-18(tm1125) mutants. We used the promoter of ced-1 that is only expressed in engulfing cells to drive the expression of vps-18 (Pced-1vps-18) and found that it fully rescued the defects in cell corpse clearance in vps-18(tm1125) embryos (Figure 4B). In comparison, we also used the promoter of egl-1 that was only expressed in dying cells to drive the expression of vps-18 (Pegl-1vps-18) but found it did not rescue the cell corpse clearance defects in vps-18(tm1125) embryos (Figure 4B). In germline, VPS-18::GFP under the control of the vps-18 promoter, which was seen in gonadal sheath cells, fully rescued the defects in germ cell corpse clearance in vps-18(tm1125) animals (Figure 4C). Moreover, we expressed VPS-18::GFP under the control of the promoter of the lim-7 gene which is expressed in gonadal sheath cells (Plim-7vps-18::gfp). We found that VPS-18::GFP driven by the lim-7 promoter also strongly reduced the accumulation of germ cell corpses in vps-18(tm1125) mutants, albeit to a less extent than that of Pvps-18vps-18::gfp (Figure 4C). Together, these findings demonstrate that vps-18 activity in engulfing cells is important for the proper degradation of internalized cell corpses.
The RING-Finger Motif Is Important for VPS-18 Proper Function
C. elegans VPS-18 contains a RING-finger domain in its C terminus, implying that VPS-18 is potentially involved in ubiquitin-related cellular processes. To provide further mechanistic insight into how VPS-18 functions, we mutated two amino acid residues (C859A and H861A) that are important for the RING-finger motif. We then tested whether the mutated VPS-18(C859A, H861A)::GFP fusion protein could rescue the defects in cell corpse clearance in vps-18(tm1125) mutants. Our results indicated that the RING-finger motif was important for the proper function of VPS-18. In contrast to the wild-type VPS-18::GFP fusion protein that fully rescued the defects of cell corpse degradation in both embryo and germline in vps-18(tm1125) animals, the mutated VPS-18(C859A, H861A)::GFP fusion protein showing the same expression pattern as that of wild-type VPS-18 failed to rescue the cell corpse clearance defects in either embryo or germline in vps-18(tm1125) animals (Figure 4, B and C).
Because sequence analysis revealed that VPS-18 is evolutionarily conserved across diverse species (Figure 1), we wanted to know whether it was also functionally conserved. To answer this, we tried to use the promoter of vps-18 to drive the expression of mouse Vps18 (mVps18) in C. elegans, but we found that the transcriptional fusion of vps-18 promoter region and gfp (Pvps-18gfp) was not expressed. We thus expressed mVps18 under the control of the promoter of vps-33 gene (Pvps-33mVps18::gfp). C. elegans vps-33 encodes another HOPS complex component and exhibits similar expression pattern to that of VPS-18 (our unpublished data). Expression of mVps18 in vps-18(tm1125) animals significantly reduced the number of germ cell corpses, indicating that mVps18 partially rescued the cell corpse clearance defects in C. elegans caused by vps-18 mutation (Figure 4C). Thus, the function of VPS-18 in regulating cell corpse removal is evolutionarily conserved from nematode to mammals.
vps-18(tm1125) Affects the Biogenesis of Endosomes and Lysosomes in Coelomocyte
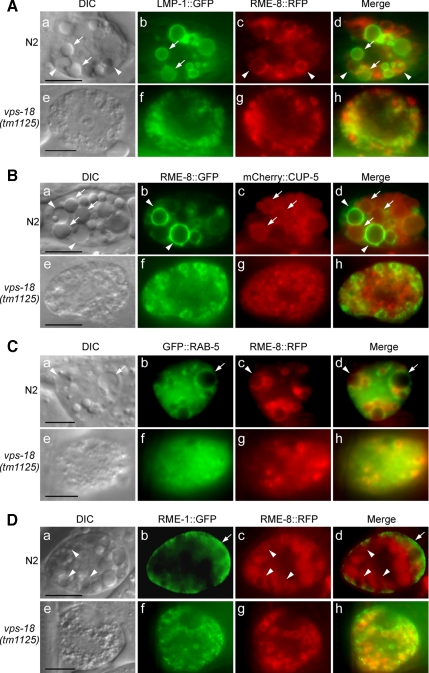
In yeast and mouse, Vps18p is critical to late endosome/lysosome docking and fusion (Rieder and Emr, 1997; Poupon et al., 2003). To determine whether C. elegans VPS-18 functions similarly to yeast and mouse Vps18p, we analyzed in detail whether vps-18 affects the formation of endosomes and lysosomes in coelomocytes in which the biogenesis of endocytic vesicles can be easily monitored (Fares and Greenwald, 2001a,b; Fares and Grant, 2002). In wild-type animals, the coelomocytes of adult hermaphrodite contain vacuoles of different sizes under differential interference contrast (DIC) optics. In contrast, the vps-18 mutant coelomocytes contain only punctate granules (Figure 5, see DIC images). To identify the origins of these granules, we crossed vps-18(tm1125) mutant with several transgenic strains expressing lysosomal and endosomal markers in coelomocytes, including LMP-1, CUP-5, RME-8, RAB-5, and RME-1. In wild-type coelomocytes, some vacuoles of different sizes were labeled by LMP-1::GFP and mCherry ::CUP-5 (Figure 5, A, a and b; and B, a and c), the two lysosome membrane-associated proteins (Fares and Greenwald, 2001b; Treusch et al., 2004). In comparison, red fluorescent protein (RFP)- and GFP-tagged RME-8 label early and late endosomes of different sizes (Figures 5A, a and c; and 5B, a and b) (Zhang et al., 2001; Poteryaev et al., 2007), which do not overlap with LMP-1– and CUP-5–positive vacuoles (Figure 5, A, d; and B, d). In vps-18(tm1125) animals, however, the sizes of LMP-1–, CUP-5–, and RME-8–positive organelles are dramatically reduced. Moreover, neither LMP-1- nor CUP-5–positive organelles with reduced sizes overlap with RME-8–positive organelles in vps-18(tm1125) mutants (Figure 5, A, e–h; and B, e–h). These results indicate that vps-18 mutation severely impaired the maturation of both endosomes and lysosomes.
Figure 5.
vps-18(tm1125) mutation affects the biogenesis of endosomes and lysosomes. DIC and fluorescence images are shown for both N2 (a–d) and vps-18(tm1125) (e–h) coelomocytes in the following panels. (A and B) vps-18(tm1125) reduces the sizes of lysosomes and endosomes. Lysosomes are labeled by LMP-1::GFP (A) or mCherry:: CUP-5 (B) and indicated by arrows. Endosomes (early and late) are labeled by RME-8::RFP (A) or RME-8::GFP (B) and indicated by arrowheads. (C) vps-18(tm1125) affects the sizes of vesicles labeled by GFP::RAB-5 and RME-8::RFP. Vesicles that are only labeled by GFP::RAB-5 are indicated by arrows and vesicles that are only labeled by RME-8::RFP are indicated by arrowheads. (D) vps-18(tm1125) affects recycling endosomes. Recycling endosomes are labeled by RME-1::GFP and endosomes are labeled by RME-8::RFP. Arrows indicate RME-1::GFP in the peripheral membrane region and arrowheads indicate RME-8::RFP-positive vesicles. Bars, 5 μm.
Because we observed that the formation of endosomes and lysosomes are severely affected in vps-18(tm1125) colomocytes, we wanted to know whether vps-18(tm1125) affects other endocytic vesicles. Thus we examined the early endosomes and recycling endsosomes. In wild-type coelomocyte, GFP::RAB-5 labels early endosomes of different sizes (Poteryaev et al., 2007), of which some are also marked by RME-8::RFP (Figure 5C, a–d), in the meantime, GFP::RAB-5 and RME-8::RFP also label some distinct vesicles (Figure 5C, a–d, indicated by arrows and arrowheads, respectively). In vps-18(tm1125) coelomocyte, by contrast, the sizes of GFP::RAB-5-marked early endosomes are greatly reduced that overlap with those small RME-8–positive organelles, suggesting that vps-18 mutation impeded the early endsosome formation (Figure 5C, e–h). Similarly, we examined recycling endosomes by checking RME-1, an EH-domain-containing ATPase that is associated with recycling endosomes (Grant et al., 2001; Poteryaev et al., 2007). In wild-type coelomocytes, RME-1::GFP was enriched in proximity to plasma membrane which is distinct from RME-8–labeled vesicles (Figure 5D, a–d) but displayed a more evenly distributed pattern in vps-18 mutant coelomocytes (Figure 5D, e–h), suggesting that vps-18 mutation also strongly affected recycling endosomes.
Endosomal/Lysosomal Degradation Activity Is Reduced in vps-18(tm1125) Mutants
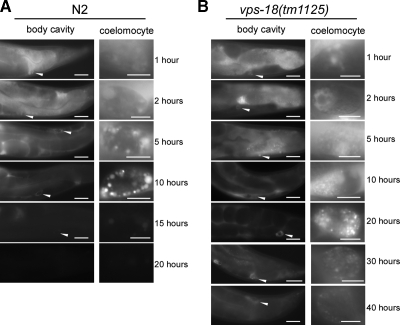
Because vps-18 mutation caused defects in endosome and lysosome biogenesis in coelomocytes, we further investigated whether the endosomal/lysosomal degradation activity was affected in vps-18(tm1125) mutants. To do so, we introduced a transgene into vps-18(tm1125) animals that expressed ssGFP under the control of a heat-shock-inducible promoter (Fares and Greenwald, 2001b). We then monitored the uptake and degradation of ssGFP by coelomocytes in a time course manner. In wild-type worms, ssGFP started to accumulate in body cavity (pseudocoelom) and was taken into coelomocytes within 1 h after heat-shock treatment (Figure 6A). Within 10 h after heat shock, ssGFP signal in body cavity gradually decreased, which correlated with an increase of GFP signal in coelomocytes. At 20 h after heat shock, ssGFP both in body cavity and in coelomocytes had disappeared (Figure 6A), indicating that ssGFP was taken in and degraded by coelomocytes. In vps-18 mutant animals, ssGFP reached coelomocytes within 1 h after heat shock as in wild type, suggesting that vps-18 mutation does not obviously affect the initial uptake of ssGFP into coelomocytes. However, ssGFP was observed both in body cavity and in coelomocytes even 40 h after heat shock in mutant animals (Figure 6B), indicating that endosomal/lysosomal protein degradation was strongly impeded in the mutant coelomocytes. Thus, the abnormal biogenesis of endosomes and lysosomes severely reduced the endosomal/lysosomal degradation activity in vps-18(tm1125) animals.
Figure 6.
vps-18(tm1125) mutation affects ssGFP degradation. (A) Wild-type animals expressing ssGFP controlled by a heat-shock promoter were heat shocked for 30 min at 33°C, and the uptake and degradation of ssGFP in coelomoctyes were monitored at indicated time points. Left, accumulation of ssGFP in body cavity (400×), and the arrowheads indicate coelomocytes that are enlarged in the right pictures (1000×). Bars, 20 μm (left) and 5 μm (right). (B) vps-18(tm1125) animals expressing ssGFP were treated and monitored as in described in A.
vps-18(tm1125) Affects Phagosome–Lysosome Fusion
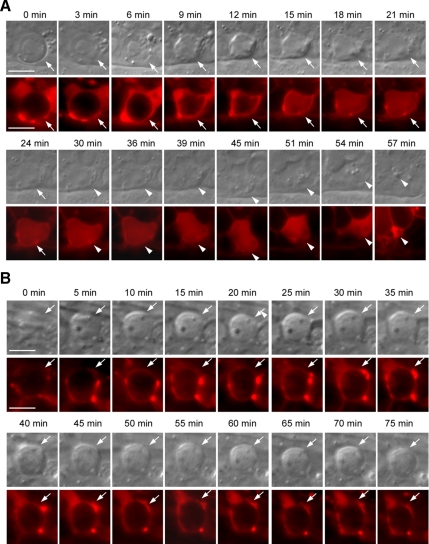
The observations that the endosome and lysosome maturation were severely compromised in vps-18(tm1125) coelomocytes suggest that the abnormal biogenesis of endosomes and lysosomes is likely the direct cause for the persistent cell corpses in vps-18(tm1125) mutants. To test this possibility, we examined whether the internalized germ cell corpses (phagosomes) were fused with lysosomes in both wild-type and vps-18 mutant animals by using a transgenic marker LMP-1::mCherry expressed in gonadal sheath cells under the control of ced-1 promoter (Pced-1lmp-1::mCherry). In wild-type animals, a time-lapse chase of the fusion between the phagosome containing germ cell corpse and LMP-1::mCherry-positive organelles indicated that LMP-1::mCherry started to cluster around the phagosome when the cell corpse was just about to be morphologically seen under DIC optics (Figure 7A, 0 min). Within a very short time (Figure 7A, 3–9 min), the cell corpse become more condensed and LMP-1::mCherry was further enriched around the cell corpse. Then LMP-1::mCherry quickly fused with the cell corpse and displayed an even distribution until the cell corpse disappeared under DIC optics (Figure 7A, 15–24 min). On average, the fusion between LMP-1::mCherry and the cell corpse took place within 15.5 ± 1.9 min (n = 10), indicating that lysosomes are rapidly recruited to the surface of the phagosome and they further fused into phagolysosome in wild-type animals. Interestingly, the evenly distributed LMP-1::mCherry still remained for a certain time after the disappearance of cell corpse (Figure 7A, 30–39 min) before it gradually condensed (Figure 7A, 45–57 min), which may indicate a continuing degradation of phagosomal contents and the recycle of lysosomes. In vps-18(tm1125) mutants, however, the fusion between LMP-1::mCherry and the germ cell corpse-containing phagosome was severely impaired. As shown in Figure 7B, although LMP-1::mCherry was found to cluster around the internalized germ cell corpse, it seemed a punctate and less smooth distribution around the cell corpse all the way through the whole time-lapse monitoring period. Moreover, we did not observe further fusion between LMP-1::mCherry and the cell corpse as seen in wild type even with longer time-lapse chasing (>70 min; n = 10) (Figure 7B). Thus, in vps-18(tm1125) mutants, although LMP-1::mCherry-positive organelles, which are likely aberrant lysosomes, could be recruited to the cell corpse-containing phagosome, they were not able to fuse further into phagolysosome, which may lead to the accumulation of undigested cell corpses.
Figure 7.
vps-18 is important for phagosome–lysosome fusion. (A) Time-lapse chasing of the fusion between germ cell corpses and LMP-1::mCherry in wild-type animals expressing LMP-1::mCherry. DIC (top row) and fluorescence (bottom row) pictures were taken every 3 min. Arrows indicate the position of cell corpses. Arrowheads indicate the continuing existence of LMP-1::mCherry in the locations where cell corpses are degraded. (B) Time-lapse chasing of the fusion between germ cell corpses and LMP-1::mCherry in vps-18(tm1125) animals. Pictures were taken every 5 min and shown as in described in A. Bars, 5 μm.
DISCUSSION
VPS-18 and HOPS Complex Are Important for Clearance of Apoptotic Cells
Although two partially redundant pathways consisting of nine genes have been identified in regulating the engulfment of cell corpses in C. elegans, our understanding of the cellular events after the internalization of cell corpses by engulfing cells is still very limited. In this study, we have investigated the role of C. elegans VPS-18, a potential ubiquitin ligase, in regulating cell corpse clearance during programmed cell death. We found that genetic inactivation vps-18 gave rise to strong increase of both embryonic and germ cell corpses in C. elegans. Our time-lapse analysis revealed that the cell corpses in mutant animals persisted much longer than in wild type, indicating that they were caused by defects in cell corpse clearance, but not excessive apoptosis. Our further genetic analysis indicated that vps-18 does not act specifically within either pathway for cell corpse engulfment but likely functions downstream of engulfment genes. This was supported by that the persistent cell corpses in vps-18 mutants were internalized but not digested promptly, because these cell corpses overlapped with acidic compartments in engulfing cells but failed to be degraded. Consistently, we found that VPS-18 was expressed in engulfing cells and its activity in engulfing cells but not dying cells was required for fulfilling its role in cell corpse removal. Together, these findings established that VPS-18 plays a critical role in the clearance of apoptotic cells by affecting the degradation of cell corpses.
C. elegans possesses the homologues of the components of yeast HOPS complex consisting of four members of class C Vps proteins (Vps11p, 16p, 18p, and 33p) and two members of class B VPS proteins (Vps39p and Vps41p). It was reported previously that RNAi knockdown of these C. elegans homologues of HOPS complex components gave rise to an increase of germ cell corpses (Lackner et al., 2005), but the underlying mechanisms were not understood. Our findings excluded the likelihood that such increase of germ cell corpses was resulted from excessive apoptosis because the germ cell death rate in vps-18(tm1125) mutants was not significantly different from that in wild-type animals. RNA interference of other HOPS complex genes in the vps-18(tm1125) mutants revealed that these double mutants exhibited cell corpse clearance defects indistinguishable from vps-18(tm1125) mutants alone. Thus, C. elegans VPS-18 functions together with the other HOPS complex proteins to regulate the degradation of cell corpses.
VPS-18 Regulates Lysosome-mediated Degradation in C. elegans
VPS-18 is evolutionarily conserved across diverse species ranging from yeast to mammals. In yeast, Vps18 mediates the docking and fusion of transport vesicles and late endosomes at the surface of vacuoles/lysosomes (Sato et al., 2000). Yeast vps18tsf allele accumulated multivesicular bodies, autophagosomes and other membrane compartments representing blocked transport intermediates (Rieder and Emr, 1997). In Drosophila, mutations of deep orange (dor), the Drosophila homologue of Vps18p, led to failure of Golgi-derived vesicles to fuse with Rab7-positive endosomes and caused defects in endosomal degradation (Sevrioukov et al., 1999; Sriram et al., 2003), suggesting a role of dor similar to that of yeast Vps18p in regulating membrane docking and fusion. In mouse, overexpression of mVps18p recruited other HOPS complex proteins and induced clustering of late endosomes and lysosomes that were surrounded by actin cytoskeleton, whereas knockdown of mVps18p by RNAi caused lysosomes to disperse away from their juxtanuclear location (Poupon et al., 2003). Thus mVps18p also functions as a tethering and docking factor to promote aggregation and fusion of late endosome and lysosomes (Poupon et al., 2003).
In C. elegans, our data showed that inactivation of vps-18 caused severe defects in lysosomes and endosomes (early and late) in that they were unable to develop into matured lysosomes. Loss-of-function of vps-18 also led to defects in recycling endosomes. These findings clearly indicate that VPS-18 is critical to endocytic pathways in C. elegans. In agreement with this notion, we found that inactivation of vps-18 strongly decreased the endosomal/lysosomal degradation activity as supported by that the degradation of ssGFP in coelomocytes was strongly delayed in vps-18(tm1125) mutants although the initial uptake of ssGFP from body cavity into coelomocytes was not affected. The reduced lysosomal degradation activity may contribute to the accumulation of cell corpses in vps-18(tm1125) mutants. Our results thus demonstrate that C. elegans VPS-18, as its counterparts in other organisms, mediates multiple steps of endocytic vesicle tethering and fusion and plays an essential role in the biogenesis of lysosomes of normal functions in C. elegans.
VPS-18 Mediates Phagosome–Lysosome Fusion
Our findings that inactivation of vps-18 caused accumulation of persistent cell corpses and that vps-18 affected the biogenesis of endosomes and lysosomes, suggest that the endosome–lysosome degradation pathway is responsible for the degradation of cell corpses in C. elegans. In support of this conclusion, we found that the internalized cell corpses, or phagosomes, rapidly recruited lysosomes to their surface in wild-type worms, which further fused into phagolysosomes in which the cell corpses were quickly degraded. When vps-18 was mutated, although the phagosomes containing internalized cell corpses could recruit LMP-1–positive organelles that are likely aberrant lysosomes to their surface, the further fusion between these aberrant lysosomes and cell corpses was blocked, leading to a failure of prompt degradation of the internalized cell corpses. In addition, the reduced degradation activity of the aberrant lysosomes caused by vps-18 mutation may also contribute to the defects in cell corpse degradation. These findings thus provide direct evidence that the fusion of phagosomes containing cell corpses with lysosomes is crucial to the adequate degradation of cell corpses.
The detailed molecular mechanisms for VPS-18 to mediate the phagosome–lysosome fusion are to be uncovered. VPS-18 contains two distinct motifs in its C-terminal region: a coiled-coil domain and a RING-finger motif, which are implicated in protein-protein interactions. More interestingly, the RING-finger motif is also implicated in protein ubiquitination. In humans, hVps18p was shown to possess an E3-ubiquitin ligase activity, which interacted with and ubiquitinated serum-inducible kinase in HeLa cells (Yogosawa et al., 2005). It was also found that hVps18p interacted with and monoubiquitinated GGA3, an adaptor protein that regulates the delivery of clathrin-coated vesicles from trans-Golgi network to endosomes (Yogosawa et al., 2006), implying that hVps18p may regulate endocytic vesicle trafficking and endosome/lysosome fusion through ubiquitination of other factors involved in membrane docking and fusion. In our study, we found that point mutations (C859A and H861A) in the RING-finger motif of VPS-18 abrogated its ability to rescue the defects in both cell corpse clearance and endosome/lysosome biogenesis in vps-18(tm1125) mutants, suggesting that a possible ubiquitin ligase activity is essential for the proper function of VPS-18 in C. elegans. Our results are consistent with previous reports that mutations in the RING-finger motifs of yeast and Drosophila Vps18p caused abnormal biogenesis of late endocytic organelles (Emr and Malhotra, 1997; Sevrioukov et al., 1999).
In conclusion, we have demonstrated that VPS-18 plays a crucial role in the clearance of apoptotic cells during programmed cell death. Moreover, our findings suggest that abnormal biogenesis of endosomes/lysosomes is responsible for inappropriate degradation of cell corpses in C. elegans.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Shohei Mitani, Hanna Fares, and Xiaochen Wang as well as CGC for providing C. elegans deletion and marker strains; Qun Lu and Weida Li for providing reagents; and Dr. Xiaochen Wang for helpful suggestions and critical reading of the manuscript. C. Y. is supported by the CAS 100-Talents Program. This research was supported by grants 2007CB947201 and 2006CB504100 from the National Basic Research Program of China and grants KSCX2-YW-R-125 and KSC1-YW-R-70 from the Chinese Academy of Sciences.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-04-0441) on October 15, 2008.
REFERENCES
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright N. A., Gratian M. J., Luzio J. P. Endocytic delivery to lysosomes mediated by concurrent fusion and kissing events in living cells. Curr. Biol. 2005;15:360–365. doi: 10.1016/j.cub.2005.01.049. [DOI] [PubMed] [Google Scholar]
- Emr S. D., Malhotra V. V. Membranes and sorting. Curr. Opin. Cell Biol. 1997;9:475–476. doi: 10.1016/s0955-0674(97)80021-x. [DOI] [PubMed] [Google Scholar]
- Fares H., Grant B. Deciphering endocytosis in Caenorhabditis elegans. Traffic. 2002;3:11–19. doi: 10.1034/j.1600-0854.2002.30103.x. [DOI] [PubMed] [Google Scholar]
- Fares H., Greenwald I. Genetic analysis of endocytosis in Caenorhabditis elegans: coelomocyte uptake defective mutants. Genetics. 2001a;159:133–145. doi: 10.1093/genetics/159.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares H., Greenwald I. Regulation of endocytosis by CUP-5, the Caenorhabditis elegans mucolipin-1 homolog. Nat. Genet. 2001b;28:64–68. doi: 10.1038/ng0501-64. [DOI] [PubMed] [Google Scholar]
- Grant B., Zhang Y., Paupard M. C., Lin S. X., Hall D. H., Hirsh D. Evidence that RME-1, a conserved C. elegans EH-domain protein, functions in endocytic recycling. Nat. Cell Biol. 2001;3:573–579. doi: 10.1038/35078549. [DOI] [PubMed] [Google Scholar]
- Gumienny T. L., Lambie E., Hartwieg E., Horvitz H. R., Hengartner M. O. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development. 1999;126:1011–1022. doi: 10.1242/dev.126.5.1011. [DOI] [PubMed] [Google Scholar]
- Horvitz H. R. Nobel lecture. Worms, life and death. Biosci. Rep. 2003;23:239–303. doi: 10.1023/b:bire.0000019187.19019.e6. [DOI] [PubMed] [Google Scholar]
- Jutras I., Desjardins M. Phagocytosis: at the crossroads of innate and adaptive immunity. Annu. Rev. Cell Dev. Biol. 2005;21:511–527. doi: 10.1146/annurev.cellbio.20.010403.102755. [DOI] [PubMed] [Google Scholar]
- Kinchen J. M., Doukoumetzidis K., Almendinger J., Stergiou L., Tosello-Trampont A., Sifri C. D., Hengartner M. O., Ravichandran K. S. A pathway for phagosome maturation during engulfment of apoptotic cells. Nat. Cell Biol. 2008;10:556–566. doi: 10.1038/ncb1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner M. R., et al. Chemical genetics identifies Rab geranylgeranyl transferase as an apoptotic target of farnesyl transferase inhibitors. Cancer Cell. 2005;7:325–336. doi: 10.1016/j.ccr.2005.03.024. [DOI] [PubMed] [Google Scholar]
- Lettre G., Hengartner M. O. Developmental apoptosis in C. elegans: a complex CEDnario. Nat. Rev. Mol. Cell Biol. 2006;7:97–108. doi: 10.1038/nrm1836. [DOI] [PubMed] [Google Scholar]
- Lettre G., Kritikou E. A., Jaeggi M., Calixto A., Fraser A. G., Kamath R. S., Ahringer J., Hengartner M. O. Genome-wide RNAi identifies p53-dependent and -independent regulators of germ cell apoptosis in C. elegans. Cell Death Differ. 2004;11:1198–1203. doi: 10.1038/sj.cdd.4401488. [DOI] [PubMed] [Google Scholar]
- Liu Q. A., Hengartner M. O. Candidate adaptor protein CED-6 promotes the engulfment of apoptotic cells in C. elegans. Cell. 1998;93:961–972. doi: 10.1016/s0092-8674(00)81202-7. [DOI] [PubMed] [Google Scholar]
- Lu Q., Zhang Y., Hu T., Guo P., Li W., Wang X. C. elegans Rab GTPase 2 is required for the degradation of apoptotic cells. Development. 2008;135:1069–1080. doi: 10.1242/dev.016063. [DOI] [PubMed] [Google Scholar]
- Luzio J. P., Pryor P. R., Bright N. A. Lysosomes: fusion and function. Nat. Rev. Mol. Cell Biol. 2007;8:622–632. doi: 10.1038/nrm2217. [DOI] [PubMed] [Google Scholar]
- Mangahas P. M., Yu X., Miller K. G., Zhou Z. The small GTPase Rab2 functions in the removal of apoptotic cells in Caenorhabditis elegans. J. Cell Biol. 2008;180:357–373. doi: 10.1083/jcb.200708130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteryaev D., Fares H., Bowerman B., Spang A. Caenorhabditis elegans SAND-1 is essential for RAB-7 function in endosomal traffic. EMBO J. 2007;26:301–312. doi: 10.1038/sj.emboj.7601498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poupon V., Stewart A., Gray S. R., Piper R. C., Luzio J. P. The role of mVps18p in clustering, fusion, and intracellular localization of late endocytic organelles. Mol. Biol. Cell. 2003;14:4015–4027. doi: 10.1091/mbc.E03-01-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D., Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien P. W., Cameron S., Horvitz H. R. Phagocytosis promotes programmed cell death in C. elegans. Nature. 2001;412:198–202. doi: 10.1038/35084096. [DOI] [PubMed] [Google Scholar]
- Reddien P. W., Horvitz H. R. The engulfment process of programmed cell death in Caenorhabditis elegans. Annu. Rev. Cell Dev. Biol. 2004 doi: 10.1146/annurev.cellbio.20.022003.114619. [DOI] [PubMed] [Google Scholar]
- Rieder S. E., Emr S. D. A novel RING finger protein complex essential for a late step in protein transport to the yeast vacuole. Mol. Biol. Cell. 1997;8:2307–2327. doi: 10.1091/mbc.8.11.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink J., Ghigo E., Kalaidzidis Y., Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- Sato T. K., Rehling P., Peterson M. R., Emr S. D. Class C Vps protein complex regulates vacuolar SNARE pairing and is required for vesicle docking/fusion. Mol. Cell. 2000;6:661–671. doi: 10.1016/s1097-2765(00)00064-2. [DOI] [PubMed] [Google Scholar]
- Seals D. F., Eitzen G., Margolis N., Wickner W. T., Price A. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc. Natl. Acad. Sci. USA. 2000;97:9402–9407. doi: 10.1073/pnas.97.17.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevrioukov E. A., He J. P., Moghrabi N., Sunio A., Kramer H. A role for the deep orange and carnation eye color genes in lysosomal delivery in Drosophila. Mol. Cell. 1999;4:479–486. doi: 10.1016/s1097-2765(00)80199-9. [DOI] [PubMed] [Google Scholar]
- Sriram V., Krishnan K. S., Mayor S. deep-orange and carnation define distinct stages in late endosomal biogenesis in Drosophila melanogaster. J. Cell Biol. 2003;161:593–607. doi: 10.1083/jcb.200210166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treusch S., Knuth S., Slaugenhaupt S. A., Goldin E., Grant B. D., Fares H. Caenorhabditis elegans functional orthologue of human protein h-mucolipin-1 is required for lysosome biogenesis. Proc. Natl. Acad. Sci. USA. 2004;101:4483–4488. doi: 10.1073/pnas.0400709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., et al. Cell corpse engulfment mediated by C. elegans phosphatidylserine receptor through CED-5 and CED-12. Science. 2003;302:1563–1566. doi: 10.1126/science.1087641. [DOI] [PubMed] [Google Scholar]
- Yogosawa S., Hatakeyama S., Nakayama K. I., Miyoshi H., Kohsaka S., Akazawa C. Ubiquitylation and degradation of serum-inducible kinase by hVPS18, a RING-H2 type ubiquitin ligase. J. Biol. Chem. 2005;280:41619–41627. doi: 10.1074/jbc.M508397200. [DOI] [PubMed] [Google Scholar]
- Yogosawa S., Kawasaki M., Wakatsuki S., Kominami E., Shiba Y., Nakayama K., Kohsaka S., Akazawa C. Monoubiquitylation of GGA3 by hVPS18 regulates its ubiquitin-binding ability. Biochem. Biophys. Res. Commun. 2006;350:82–90. doi: 10.1016/j.bbrc.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Yu X., Lu N., Zhou Z. Phagocytic receptor CED-1 initiates a signaling pathway for degrading engulfed apoptotic cells. PLoS Biol. 2008;6:e61. doi: 10.1371/journal.pbio.0060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Odera S., Chuang C. H., Lu N., Zhou Z. C. elegans Dynamin mediates the signaling of phagocytic receptor CED-1 for the engulfment and degradation of apoptotic cells. Dev. Cell. 2006;10:743–757. doi: 10.1016/j.devcel.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Grant B., Hirsh D. RME-8, a conserved J-domain protein, is required for endocytosis in Caenorhabditis elegans. Mol. Biol. Cell. 2001;12:2011–2021. doi: 10.1091/mbc.12.7.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Hartwieg E., Horvitz H. R. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell. 2001;104:43–56. doi: 10.1016/s0092-8674(01)00190-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.