Abstract
The origin recognition complex or ORC is a six-subunit protein important for DNA replication and other cell functions. Orc6, the smallest subunit of ORC, is essential for both replication and cytokinesis in Drosophila, and interacts with the septin protein Pnut, which is part of the Drosophila septin complex. In this study, we describe the analysis of the interaction of Orc6 with Pnut and whole Drosophila septin complex. Septin complex was purified from Drosophila embryos and also reconstituted from recombinant proteins. The interaction of Orc6 with the septin complex is dependent on the coiled-coil domain of Pnut. Furthermore, the binding of Orc6 to Pnut increases the intrinsic GTPase activity of the Drosophila septin complex, whereas in the absence of GTP it enhances septin complex filament formation. These results suggest an active role for Orc6 in septin complex function. Orc6 might be a part of a control mechanism directing the cytokinesis machinery during the final steps of mitosis.
INTRODUCTION
The origin recognition complex (ORC) plays a central role in the initiation of DNA replication and the recruitment of essential replication factors to the origins of DNA replication in eukaryotes (Dutta and Bell, 1997; Bell, 2002; Bell and Dutta, 2002; Machida et al., 2005). In addition to initiating DNA replication, ORC is involved in other functions (for reviews, see Bell, 2002; Chesnokov, 2007; Sasaki and Gilbert, 2007). Some of these activities link cell cycle progression to DNA replication, whereas other functions seem distinct from replication. In Drosophila, the smallest subunit of ORC, Orc6, is an essential component of the complex and directly involved in DNA binding (Balasov et al., 2007). Orc6 is also essential for cytokinesis in both Drosophila (Chesnokov et al., 2003) and human cells (Prasanth et al., 2002) because ablation of Orc6 from cells by RNA interference results in cytokinesis defects. In Drosophila, Orc6 colocalizes and interacts with the septin protein Pnut. This interaction is mediated by the C terminus of Orc6 that contains a predicted amphipathic α-helical domain (Chesnokov et al., 2003).
Pnut is a member of the septin family of polymerizing GTPases, which are required for cytokinesis and other processes that involve spatial organization of the cell cortex. Identified originally in yeast, septins are now found in many fungi and animals (Kinoshita, 2003; Pan et al., 2007). Septins are essential for cytokinesis (Hartwell, 1971; Neufeld and Rubin, 1994; Kinoshita et al., 1997); however, they have also been implicated in other processes such as polarity establishment, cell cycle checkpoints, formation of diffusion barrier, spindle alignment, chromosome segregation, vesicle trafficking, and exocytosis (Gladfelter et al., 2001; Kusch et al., 2002; Dobbelaere and Barral, 2004; Martinez et al., 2004; Spiliotis et al., 2005; Spiliotis and Nelson, 2006).
Septins are found as heteromeric complexes that can assemble in larger filaments (Kinoshita, 2006). All septins have in common a GTP binding domain that consists of the binding motifs G1 (P-loop), G3, and G4. Most septins contain a predicted coiled-coil domain at the C terminus (Field and Kellogg, 1999). In many cases, septins seem to serve as a scaffold or matrix for other proteins (Longtine et al., 1998, 2000; Field and Kellogg, 1999; Barral et al., 2000; Takizawa et al., 2000). Therefore, progress in understanding septins' functions will depend in part on identifying the proteins with which the septins interact.
Drosophila melanogaster has at least five septin proteins, named Pnut, Sep1, Sep2, Sep4, and Sep5, whose functions are not yet well understood (Neufeld and Rubin, 1994; Fares et al., 1995; Field et al., 1996; Longtine et al., 1996; Field and Kellogg, 1999; Adam et al., 2000; Kinoshita, 2003). Of these five Drosophila septins, Pnut, Sep1 and Sep2 form a heteromeric six subunit complex consisting of two of each septin subunits (Field et al., 1996). The complex binds and hydrolyzes GTP. This GTPase activity is characterized by a faster GTP-to-GDP conversion but a very slow exchange of bound nucleotide. Pnut and Sep1 colocalize at the cleavage furrows of dividing cells during cytokinesis, to the intercellular bridge that connect postmitotic daughter cells, neurons, the leading edge of epithelial sheaths, and the early embryo cortex (Neufeld and Rubin, 1994; Fares et al., 1995). Larvae, homozygous null for Pnut, die shortly after pupation and pnut− tissues contain multinucleated cells, indicating an involvement for this protein in cytokinesis (Neufeld and Rubin, 1994). Both Pnut (Neufeld and Rubin, 1994) and Orc6 (Chesnokov et al., 2003) are essential for cytokinesis, and their interaction might be critical for their tasks in the cell.
In this study, we describe the role of the interaction of Orc6 protein with functionally active Drosophila septin complex. Our data revealed that the C-terminal coiled-coil domain of Pnut is essential for both septin complex formation and interaction with Orc6. However, different motifs within the C-terminal domain are responsible for these functions. Orc6 increased GTPase activity of wild-type septin complex but not of complex containing coiled-coil domain mutants of Pnut defective for interaction with Orc6. The C-terminal deletion mutant of Orc6, which fails to interact with Pnut, also had no effect on the GTPase activity of the complex. These results indicate that binding of Orc6 via its C-terminal domain to the coiled-coil domain of Pnut is directly related to the effect of Orc6 on septin complex GTPase activity. Orc6, but not Orc6 C-terminal deletion mutant, enhanced filament assembly of recombinant septin complex. In the presence of GTP, the effect of Orc6 on filament formation was significantly decreased or absent. This suggests a dual role for Orc6 in its interaction with the septin complex. In the absence of GTP, Orc6 promotes assembly of septin filaments, whereas in the presence of GTP Orc6 enhances septin complex GTPase activity, resulting in the disassembly of filaments. The data presented here indicate that septins are regulated by additional factors involved in cytokinesis such as Orc6.
MATERIALS AND METHODS
Cloning and Mutagenesis
cDNAs of Sep1 and Sep2 were cloned by polymerase chain reaction (PCR) from a Drosophila embryonic library (MATCHMAKER library; Clontech, Mountain View, CA). C-Terminal deletions of Pnut (Pnut1-508, Pnut1-460, Pnut1-427), the C terminus of Pnut (Pnut427-539), the glutathione transferase (GST)-tag, and the FLAG-tag were generated with standard PCR technique. Triple leucine to alanine substitutions in the coiled-coil domain of Pnut (L463A,L468A,L470A and L481A,L483A,L488A) were introduced with Stratagene's site-directed mutagenesis protocol (Stratagene, La Jolla, CA; http://www.stratagene.com/manuals/200516.pdf). All constructs were analyzed by sequencing. cDNAs were subcloned into desired vectors with standard molecular biology techniques except for the yeast constructs (see below).
Proteins and Antibodies
Purification of Escherichia coli derived recombinant His-tagged wild-type Orc6 and Orc6-200 mutant has been described previously (Balasov et al., 2007). Pnut cDNA was cloned into pET-Duet expression vector (Novagen, Madison, WI) by using the NdeI and KpnI restriction sites. His-tagged Pnut was produced by expression from the plasmid in E. coli strain BL21 DE3. After induction with isopropyl β-d-thiogalactoside (IPTG), His-tagged protein was isolated with nickel-nitrilotriacetic acid (Ni-NTA) beads (QIAGEN, Valencia, CA). Purified antigen was used to generate rabbit polyclonal antibodies (Cocalico Biologicals, Reamstown, PA). Antibodies were purified by affinity chromatography as described previously (Harlow and Lane, 1999). Mouse anti-FLAG (M2 clone) was obtained from Sigma-Aldrich (St. Louis, MO).
RNA Interference (RNAi) Assay
Double-stranded RNA (dsRNA) was obtained by using the Megascript kit from Ambion (Austin, TX). Pnut primers (5′-CGGCCAGTGAATTGTTTAATACGACTCACTATAGGGA ATAGTCCTCGCTCGAACGCG-3′ and 5′-CGGCCAGTGAATTGTTTAATACGACTCACTATAGGGTTAGAACAGACCCT-TCTTTTTC-3′) flanked with T7 promoter were used. The Orc6 primers used have been described previously (Chesnokov et al., 2003). L2 cells were cultured at 27°C in Shields and Sang M3 medium (Sigma-Aldrich) supplemented with 5% fetal bovine serum. For RNAi experiments, 1 × 106 Drosophila L2 cells seeded on a coverslip in a well of a six-well dish were inoculated with 30 μg of dsRNA in 1 ml of serum-free M3 medium. After 1-h incubation, 1 ml of medium supplemented with 10% fetal bovine serum was added to the culture. After 72 h cells were fixed with 2% formaldehyde in PBS. Cells were stained for Pnut and Orc6 and counterstained with 4,6-diamidino-2-phenylindole (DAPI). Coverslips were mounted with 80% glycerol, 20% 1× phosphate-buffered saline (PBS), and 2% N-propyl-gallate and analyzed with fluorescence microscopy. RNAi efficiency was tested by immunoblotting with anti-Pnut antibody.
Yeast Two-Hybrid Assays
The Cytotrap yeast two-hybrid system (Stratagene) was used to analyze the interaction between Orc6 and wild-type or mutant Pnut proteins. The system has been described previously (Aronheim et al., 1997). In brief; the system uses the yeast temperature-sensitive mutant strain cdc25H. The cdc25 mutation prevents growth at 37°C but allows growth at the permissive temperature (25°C). The Cytotrap system is based on the ability of the hSos protein to complement the cdc25 defect and to activate the yeast Ras-signaling pathway. Expression of hSos and its subsequent localization to the plasma membrane allows the cdc25H yeast strain to grow at 37°C. The localization of hSos to the plasma membrane occurs through the interaction of its fusion protein encoded by the pSos vector with a myristylated protein encoded by the pMyr vector.
A gateway cloning cassette has been introduced into the pMyr and pSos vectors to introduce the cDNAs of interest from pENTR/D-TOPO plasmids with CL clonase (Invitrogen, Carlsbad, CA) into the yeast expression vectors. PCR products of Orc6 and Pnut cDNAs were cloned into the pENTR/ D-TOPO vector according to protocol. These constructs were used to clone Orc6 and Pnut coding sequences into the pMyr and pSos vectors, respectively. All yeast transformations were performed with the standard lithium acetate method. The expression of pSos-fusion proteins is constitutive in the Cytotrap system with low expression levels. To verify that the pSos-Pnut fusion proteins were properly expressed, yeast lysates were immunoprecipitated with anti-hSos antibody (Santa Cruz Biotechnology, Santa Cruz, CA) bound to protein A-Sepharose 4B beads (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom), and the precipitate was analyzed by SDS-polyacrylamide gel electrophoresis followed by immunoblotting with mouse anti-Pnut antibody.
For the yeast two-hybrid assay, individual colonies expressing both myristylated Orc6 and hSos-Pnut fusion proteins were picked from the plate and resuspended in 35 μl of 10 mM Tris-HCl pH 8.0 1 mM EDTA (TE). 2.5 μl of the suspension was applied on gal/raff plates. Plates were grown at either the permissive (25°C) or restricted (37°C) temperature.
Immunoprecipitation Studies with De Novo-synthesized Proteins
For the in vitro expression of proteins a modified pBluescript KS plasmid was used. A T7 terminator signal that originated from pET15b vector was cloned into the BamHI and HindIII sites of pBluescript KS. Coding sequences of wild-type and mutant Pnut proteins as well as Orc6 were cloned into this vector within the XbaI site downstream from the T7 promoter region. GST-tag was cloned into the SacI and XbaI sites downstream from the T7 promoter region, and Pnut(427-539) was subsequently cloned into the XbaI site to produce a fusion protein. Expression of proteins and immunoprecipitation reactions were performed as described previously (Pak et al., 1997). Septin complexes consisting of Sep1, Sep2 and wild-type or mutant Pnut protein were expressed in one reticulocyte reaction, whereas Orc6 was generated in a separate reaction. Five microliters of a 50-μl reaction was analyzed by SDS-PAGE for synthesis efficiency. Twenty-five microliters of the reactions containing septin proteins, and, if necessary, Orc6 protein were used for interaction studies and subsequent immunoprecipitations. Half of the precipitated material was analyzed by SDS-PAGE followed by autoradiography.
Expression of FLAG-tagged Pnut in L2 Cells
FLAG-tagged Pnut proteins were expressed in L2 cells under control of the Pnut promoter from pCasper vectors containing the 1.7-kb genomic region directly upstream from the start codon of Pnut isolated from Drosophila Canton S strain. L2 cells were cultured at 27°C in Shields and Sang M3 medium (Sigma-Aldrich) supplemented with 5% fetal bovine serum. Drosophila L2 cells (3 × 106) seeded on a coverslip in a well of a six-well dish were transfected with 1 μg of DNA using Insect Genejuice, according to the manufacturer's recommendations (Novagen) in the presence of 5% serum. Twenty-four hours after transfection, medium containing the transfection mix was replaced for regular medium. Seventy-two hours after transfection, cells were fixed with 2% formaldehyde in PBS. Cells were stained for FLAG and counterstained with DAPI. Coverslips were mounted with 80% glycerol, 20% 1× PBS, and 2% N-propyl-gallate, and analyzed with fluorescence microscopy. Expression of FLAG-tagged Pnut proteins was analyzed by immunoblotting with anti-FLAG antibody.
Purification of Native Septin Complex
Cytosolic embryonic extracts were isolated from the wild-type Canton S strain of Drosophila melanogaster by using a modified method initially designed for the isolation of nuclear extracts of Drosophila embryos (Heberlein and Tjian, 1988). Dechorionated 0- to 12-h embryos were homogenized in lysis buffer (15 mM HEPES-KOH, pH 7.6, 10 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 350 mM sucrose, 1 mM dithiothreitol [DTT], 1 mM sodium metabisulfite, 0.2 mM phenylmethylsulfonyl fluoride [PMSF], 3 ml/g of embryo). Nuclei and yolk were removed from the lysate by low-speed centrifugation at 9800 × g in a JA-14 rotor (Beckman Coulter, Fullerton, MA) at 4°C for 15 min. The crude cytosolic lysate was then cleared from any remaining debris by high-speed centrifugation at 142,000 × g in a Ti 45 rotor (Beckman Coulter) at 4°C for 1 h. Proteins in the cleared cytosolic lysate were concentrated by ammonium sulfate precipitation (30%) followed by centrifugation. Subsequently, the precipitated proteins were dissolved in HEM buffer (25 mM HEPES-KOH, pH 7.6, 0.1 mM EDTA, and 12.5 mM MgCl2) containing 100 mM KCl, 1 mM DTT, 0.5 mM sodium metabisulfite, and 0.1 mM PMSF, and dialyzed overnight to remove the ammonium sulfate. After dialysis, the extract was centrifuged to remove precipitated material, glycerol was added to a concentration of 10%, and the cleared extract flash frozen in liquid nitrogen and stored at −80°C until further use. Twenty milliliters of the cleared extract was applied to a 50-ml Heparin-Sepharose column (GE Healthcare) in HEMG (HEM and 5% glycerol) buffer containing 100 mM KCl, 0.005% NP-40, 1 mM DTT, and 0.1 mM PMSF. The flowthrough of this step was concentrated by precipitation with 30% ammonium sulfate, resuspended in 10 ml of HEMG buffer containing 100 mM NaCl, 0.005% NP-40, 1 mM DTT, and 0.1 mM PMSF, and subsequently dialyzed overnight. The dialyzed product was fractionated on a Sephacryl S-300 HR 26/60 column (318 ml; GE Healthcare). Fractions containing the majority of Pnut as detected by Western blot were pooled and applied to a MonoQ10/100 GL column (7.8 ml; GE Healthcare). The flowthrough of this step was diluted to 50 mM NaCl and adjusted to pH 8.0 and applied to a 1-ml MonoQ5/50 GL column (GE Healthcare). Peak fractions containing septin complex were pooled, concentrated with a Centricon YM-100 (100 kDa cut-off; Millipore, Billerica, MA) and loaded on top of a 4-ml 15–35% glycerol gradient in HEM buffer, pH 8.0, with 200 mM NaCl, 0.005% NP-40, 1 mM DTT, and 0.1 mM PMSF. The gradient was centrifuged in a MLS-50 swinging bucket rotor at 227,000 × g at 4°C for 16 h in an Optima MAX-E tabletop ultracentrifuge (Beckman Coulter). Approximately 100-μl fractions were collected and analyzed by SDS-PAGE followed by silver stain or colloidal Coomassie Blue stain. Septin proteins were identified by liquid chromatography-mass spectrometry (LC-MS) as described below.
LC-MS Confirmation of Native Septin Complexes
Individual protein bands (∼70, 50, and 41 kDa) from the glycerol gradient fractions were excised from the gel with a razor blade and subjected to in-gel tryptic digestion (with reduction by 10 mM dithiothreitol and alkylation with iodoacetamide). The tryptic digests were loaded onto a LC-MS system composed of a MicroAS autosampler, LC nanopump (Eksigent, Dublin, CA), and a linear ion trap-Fourier transform ion cyclotron resonance hybrid mass spectrometer (LTQ FT; Thermo Fisher Scientific, San Jose, CA). The pulled tip was packed with Jupiter 5-μm C18 reversed phase beads to give a 100-μm diameter, 11-cm column. A gradient of acetonitrile with 0.1% formic acid was run from 5 to 30% in 60 min at 500 nl min−1. LTQ FT parameters were set as described previously (Renfrow et al., 2007). Drosophila tryptic peptides were identified by use of the TurboSEQUEST algorithm within Bioworks 3.2 (Thermo Fisher Scientific) with a mass accuracy of 2.0 ppm or better. Pnut, Sep2, and Sep1 were each unambiguously identified within their respective excised gel band with >10 unique tryptic peptides.
Reconstitution and Purification of Recombinant Drosophila Septin Complex
cDNA encoding for Pnut was cloned into the pET-Duet expression vector to generate a His-tagged protein. cDNAs encoding Sep1 and Sep2 were cloned into the pCDF-Duet expression vector resulting in full-length (untagged) proteins. Plasmids were transformed into E. coli strain BL21 DE3. Colonies were grown in Miller's LB Broth with 50 μg/ml ampicillin and 50 μg/ml streptomycin to an OD600 of 0.6, after which protein expression was induced with 1 mM IPTG for 3 h. Cells were lysed in a French press after which protein was isolated with Ni-NTA according to the manufacturer's recommendations (QIAGEN). The eluate fractions containing recombinant septin complex were diluted to 50 mM NaCl with dilution buffer (25 mM HEPES-NaOH, pH 8.0, and 5% glycerol) and applied to a 5-ml HP Q column (GE Healthcare). Protein was eluted from the column using a gradient of 50 mM to 1 M NaCl in 25 mM HEPES-NaOH, pH 8.0, 5% glycerol, and 1 mM DTT. Fractions containing septin complex were further purified by centrifugation on a 15–35% glycerol gradient in 25 mM HEPES-NaOH pH 8.0, 200 mM NaCl, and 1 mM DTT. Centrifugation was carried out as mentioned above.
Recombinant baculoviruses were generated using the Bac-to-Bac expression system (Invitrogen). Viruses carrying Sep1 and Sep2 were mixed with either wild-type or mutant N-terminally His-tagged Pnut baculovirus constructs. P3 viral stocks were used to express protein in High Five cells. Cells were infected with the viral stocks of all three septins. Seventy-two to-90 h after transfection, cells were isolated and lysed with a Dounce homogenizer in lysis buffer (10 mM HEPES-KOH, pH 7.6, 15 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 2 mM β-mercaptoethanol, and the protease inhibitors leupeptin, aprotinin, and pepstatin). NaCl was added to a final concentration of 300 mM, and nuclei were precipitated by low-speed centrifugation at 1800 × g in a JS-4.2 rotor with a JH-4B centrifuge (Beckman Coulter) at 4°C for 10 min. Supernatant was adjusted to 10 mM imidazole and added to Ni-NTA beads (QIAGEN). Bead suspension was incubated at 4°C for 2 h. Subsequently, beads were washed with 50 volumes of wash buffer (10 mM HEPES-KOH pH 7.6, 300 mM NaCl, 2 mM MgCl2, 0.1 mM EDTA, 2 mM β-mercaptoethanol, and 20 mM imidazole). Proteins were eluted with elution buffer (10 mM HEPES-NaOH, pH 8.0, 300 mM NaCl, 2 mM MgCl2, 0.1 mM EDTA, 2 mM β-mercaptoethanol, 250 mM imidazole, and 5% glycerol). Eluate fractions containing septin complex were diluted to 50 mM NaCl and further purified with anion chromatography on a 1-ml MonoQ 5/5 or 1-ml HiTrap HP Q column using a gradient of 50 mM to 1 M NaCl in 25 mM HEPES, pH 8.0, 12.5 mM MgCl2, 0.1 mM EDTA, 5% glycerol, and 1 mM DTT. Septin complex eluted at 200 mM NaCl.
Electron Microscopy
Septin filament formation was analyzed essentially as described previously (Field et al., 1996). Purified septin complex was applied to nickel Formvar carbon-coated grids and incubated at room temperature for 4 min. Excess solution was removed from the grids and proteins were fixed with 1% uranyl acetate in 30% ethanol for 30 s. Fixative was removed, and grids were dried after which images were taken on a Philips FEI Tecnai Spirit transmission electron microscope at 60 kV.
GTPase Assays
GTP hydrolysis assays were based on the method described by Field et al. (1996). The septin complex (2.5 μg) was incubated with affinity-purified rabbit-anti-Pnut antibody and 5 μl of protein A-Sepharose 4B beads in a total volume of 100 μl of GTPase buffer (20 mM HEPES-KOH, pH 7.6, 3 mM MgCl2, 1 mM EGTA, 50 mM KCl, and 1 mM DTT) at 4°C for 30 min. Beads were isolated, washed with GTPase buffer and finally resuspended in 50 μl of GTPase buffer. Then, 0.5 μCi of [α-32P]GTP (3000 Ci/mmol; GE Healthcare) and cold GTP were added to a final concentration of 2 μM. Next, 0.5 μg of purified recombinant Orc6 protein was included in the mixture when indicated. After incubation at 21–22°C with continuous agitation, beads were washed four times with 0.5 ml of cold GTPase buffer. Nucleotides bound to the septin complex were eluted with 10 μl of 8 M urea, 5 mM EDTA, and 20 mM Tris-HCl, pH 7.5. Two microliters of the eluate was analyzed for total bound nucleotide by liquid scintillation counting. Four microliters of the eluate was analyzed for nucleotide by thin layer chromatography (TLC) on polyetheleneimine-cellulose plates (Selecto Scientific, Suwanee, GA) developed with 0.85 M KH2PO4 pH 3.4. Radioactive GTP and GDP spots were quantified by phosphorimaging.
RESULTS
Pnut Depletion by RNAi Results in Loss of Cytoplasmic Orc6 Localization
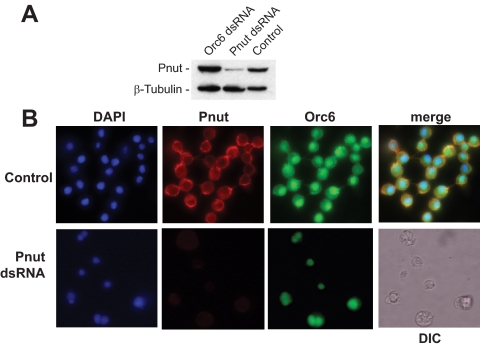
Pnut mutations in Drosophila result in cytokinesis defects (Neufeld and Rubin, 1994). To address the potential role of Pnut in Orc6 localization, L2 cells were depleted of Pnut by RNAi, and Orc6 localization was analyzed by immunohistochemistry. Immunoblot analysis of Pnut dsRNA-treated L2 cells revealed that the level of Pnut was greatly reduced by 72 h, whereas cells transfected with Orc6 dsRNA showed the same levels of Pnut as control, untransfected cells (Figure 1A). Analysis of cells treated with Pnut dsRNA for 72 h revealed that, depending on the experiment, up to 35% of the cells showed a binucleated phenotype as measured among a population of 100–300 cells per experiment. Figure 1B shows that in cells lacking Pnut due to the dsRNA treatment the majority of Orc6 was confined to the nucleus, with some residual cytoplasmic staining, whereas in untreated cells Orc6 and Pnut colocalized at the plasma membrane and at cleavage furrows of dividing cells. These results suggest that Pnut is important for recruiting Orc6 to the plasma membrane of Drosophila cells.
Figure 1.
Silencing of Pnut in Drosophila L2 cells by dsRNA causes multinucleation and Orc6 mislocalization. (A) Immunoblotting with anti-Pnut antibody of whole cell extract Drosophila L2 cells transfected with either Orc6 dsRNA or Pnut dsRNA for 72 h, or nontransfected (control). Tubulin was used as a loading control. (B) Drosophila L2 cells, either control or transfected with Pnut dsRNA were fixed 72 h after transfection, and stained as described in Materials and Methods. For the Pnut dsRNA-treated cells, the differential interference contrast (DIC) is also shown.
The Predicted Coiled-Coil Domain of Pnut Is Essential for Its Interaction with Orc6
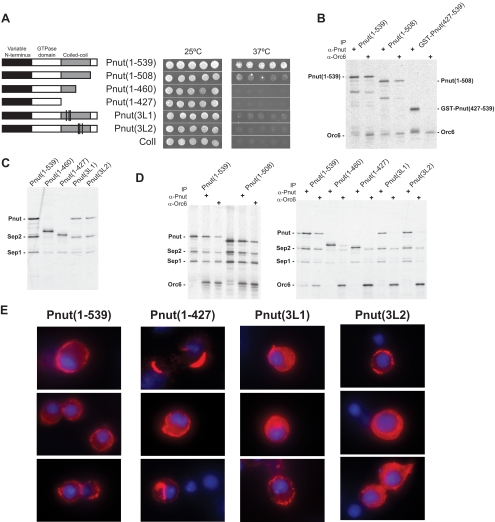
Pnut contains a predicted coiled-coil domain at the C terminus. The importance of the coiled-coil domain of Pnut for its interaction with Orc6 was first tested in a yeast two-hybrid assay. C-Terminal deletions that disrupt the coiled-coil domain of Pnut were analyzed. The importance of regularly spaced leucine residues that often form the hydrophobic core of the coiled-coil domain were also examined. Figure 2A shows that the coiled-coil domain of Pnut is essential for its interaction with Orc6. The Pnut1-508 mutant lacks the last C-terminal 31 amino acids but leaves the predicted coiled-coil domain intact. Although significantly reduced compared with wild type (Pnut1-539), yeast growth was still supported at the restrictive temperature, indicating that Orc6 did interact with this Pnut deletion mutant. This was confirmed by direct interaction between Orc6 and the Pnut(1-508) deletion mutant from in vitro transcription–translation reactions as shown in Figure 2B. Yeast two-hybrid assay further revealed that the removal of half of the coiled-coil domain of Pnut (Pnut1-460) or the whole coiled-coil domain of Pnut (Pnut1-427) resulted in abolishment of growth at the restrictive temperature. Pnut proteins containing triple alanine substitutions for leucine residues at positions L463A, L468A, L470A [Pnut(3L1)] and L481A, L483A, L488A [Pnut(3L2)] in the coiled-coil domain were severely impaired in their ability to interact with Orc6. The yeast two-hybrid results were confirmed by direct interaction between Orc6 and Pnut deletion and substitution mutant proteins from in vitro transcription–translation reactions (data not shown). Our data suggest that the coiled-coil domain of Pnut and the leucine residues herein are important for the interaction with Orc6. Direct interaction between the coiled-coil region of Pnut (Pnut427-539) and wild-type Orc6 could not be confirmed with proteins from in vitro transcription–translation reactions by either immunoprecipitation (Figure 2B) or the use of glutathione or chitin beads with GST- or chitin binding domain-fusion proteins, respectively (data not shown). This indicates that although essential the coiled-coil domain of Pnut is not sufficient for the interaction of this protein with Orc6.
Figure 2.
Interaction of Orc6 with Pnut and septin complex. (A and B) The coiled-coil domain of Pnut is essential for its interaction with Orc6. (A) Results from the Cytotrap yeast two-hybrid system are presented. Individual colonies expressing both myristylated Orc6 and hSos-Pnut fusion proteins were picked and resuspended in 35 μl of TE. Then, 2.5 μl of the suspension was applied on gal/raff plates, which were incubated at either the permissive (25°C) or restricted (37°C) temperature. hSos-ColI is a fusion protein that serves as a negative control. A schematic representation is given for each mutant of Pnut tested. The lines for the representations of Pnut(3L1) and Pnut(3L2) represent the relative locations of the mutated leucines, which are Pnut(L463A,L468A,L470A) and Pnut(L481A,L483A,L488A), respectively. (B) Direct interaction studies of in vitro-synthesized Pnut and Orc6 proteins. Proteins were synthesized individually in an in vitro transcription translation reaction containing [35S]methionine. Reactions containing wild-type or mutant Pnut protein were mixed with Orc6. Proteins were immunoprecipitated with either anti-Pnut antibody or anti-Orc6 antibody and further analyzed by SDS-PAGE followed by autoradiography. The GST-Pnut(427-539) synthesis reaction contained a degradation product of slightly bigger size than Orc6 that was pulled down with anti-Pnut antibody only. (C) The coiled-coil domain of Pnut is essential for septin complex formation. Sep1, Sep2, and wild-type or mutant Pnut protein were simultaneously synthesized in an in vitro transcription translation reaction containing [35S]methionine. Proteins were immunoprecipitated with anti-Pnut antibody and further analyzed by SDS-PAGE followed by autoradiography. Wild-type (Pnut1-539) and deletion mutants (Pnut1-460, Pnut 1-427) as well as the triple leucine mutants within the coiled coil domain Pnut(3L1) and Pnut(3L2) are shown. (D) Orc6 interacts with the septin complex via Pnut. Sep1, Sep2, and wild-type or mutant Pnut protein were simultaneously synthesized in an in vitro transcription translation reaction containing [35S]methionine, whereas Orc6 was synthesized individually. Reactions were mixed and proteins immunoprecipitated with either anti-Pnut antibody or anti-Orc6 antibody. Immunoprecipitates were analyzed by SDS-PAGE followed by autoradiography. (E) Expression of Pnut mutants under control of native promoter in Drosophila tissue culture cells. Drosophila L2 cells were transfected with pCasper constructs expressing FLAG-tagged Pnut under control of the Pnut promoter. Cells were fixed 72 h after transfection, stained with anti-FLAG antibody, and counterstained with DAPI. Columns of three representative images for wild-type Pnut and Pnut mutants are shown.
The Coiled-Coil Domain of Pnut Is Important in Septin Complex Formation
The C-terminal regions of most septins contain a predicted coiled-coil domain, which is important for protein–protein interaction (Mason and Arndt, 2004). To test whether this domain of Pnut is important for septin complex assembly, we analyzed the ability of C-terminal deletion Pnut mutants to form septin complex with Sep1 and Sep2. Sep1, Sep2, and wild-type or mutant Pnut proteins were expressed simultaneously in vitro by using a transcription–translation assay. Pnut was immunoprecipitated with a polyclonal anti-Pnut antibody, and pulled down material was analyzed by SDS-PAGE. Figure 2C shows that the synthesized proteins Sep1 and Sep2 formed a complex with wild-type Pnut. However, in the presence of a Pnut mutant lacking half of the coiled coil domain (Pnut1-460), the pulled down material contained little to no Sep2 and low amounts of Sep1, resulting in nonstoichiometric ratios of Sep1:Sep2:Pnut. This indicates that complex formation was significantly impaired. The deletion of the whole coiled-coil domain (Pnut1-427) produced the same results. Moreover, Pnut mutants lacking the coiled-coil domain were not able to reconstitute into septin complex in baculovirus expression system (data not shown). Interestingly, the triple leucine mutants of Pnut (3L1 and 3L2), which were unable to interact with Orc6, did not inhibit the formation of septin complex (Figure 2C). Figure 2D shows that the C-terminal mutant Pnut(1-508), which contains the whole predicted coiled-coil domain, was able to assemble into a stoichiometric septin complex, indicating that within the C terminus of Pnut specifically the coiled-coil region is important for septin complex formation.
In Vivo Localization of Pnut Mutants
To investigate the effect of the mutations on Pnut localization FLAG-tagged Pnut proteins were expressed in L2 cells. Overexpression of Pnut with inducible promoters was not successful as it resulted in aggregated rods and spirals of the expressed protein (Supplemental Figure S4). To obtain lower expression levels of wild-type and mutant FLAG-tagged Pnut proteins the native Pnut promoter was used. Overall, the expression of FLAG-tagged Pnut proteins was modest but detectable as immunoblot analysis confirmed expression of correct size proteins (data not shown). Localization of the expressed proteins was analyzed by immunohistochemistry. Figure 2E shows that wild-type FLAG-tagged Pnut localized to the plasma membrane of L2 cells similar to endogenous Pnut. The FLAG-tagged coiled-coil deletion mutant of Pnut, Pnut(1-427), which failed to assemble into septin complex in vitro, had the tendency to accumulate into crescent shaped aggregates, although plasma membrane localization was observed in rare cases. Expressed FLAG-tagged triple leucine mutants of Pnut, Pnut3L1 and Pnut3L2, displayed mainly diffuse cytoplasmic staining but also some plasma membrane localization.
Interaction of Orc6 with the Septin Complex
Immunofluorescence studies on septins imply that they form a complex in vivo (Fares et al., 1995). Orc6 and Pnut interact and colocalize at cleavage furrows of dividing cells (Chesnokov et al., 2003). These results suggest that in vivo Orc6 interacts with Pnut as part of the septin complex. To investigate this possibility, immunoprecipitation experiments were set up with in vitro synthesized septin complex and Orc6. Proteins were immunoprecipitated with either anti-Pnut antibody or anti-Orc6 antibody. Figure 2D shows autoradiograms of the precipitated proteins after separation by SDS-PAGE. Again, coiled-coil deletion mutants of Pnut were not able to incorporate into septin complex, suggesting an important role for the coiled-coil domain of Pnut in complex formation. Furthermore, Orc6 did not interact with Pnut mutants lacking part of the coiled-coil domain. The triple leucine mutants of Pnut, Pnut(3L1), and Pnut(3L2), were able to integrate into septin complex, but little to none Orc6 precipitated with the complex. The anti-Orc6 antibody pull-down experiments show that indeed Orc6 interacted with the complex when wild-type Pnut was present. However, in the case of the Pnut triple leucine mutants Orc6 was not able to pull down significant amounts of complex. Interestingly, immunoprecipitation reactions with anti-Orc6 antibody also pulled down a significant amount of Sep2. This was also confirmed by direct interaction between the two proteins (data not shown). This result suggests that Orc6 can interact with a subset of Sep2 not integrated into the septin complex.
Purification, Reconstitution, and Characterization of Drosophila Septin Complex
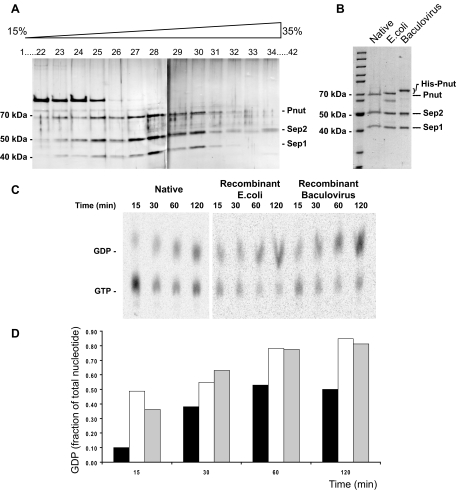
To study the interaction of Orc6 with the septin complex in more detail septin complex was purified from Drosophila embryonic extracts. A new purification method was developed that avoids the previously reported immunoaffinity technique (Field et al., 1996) and uses only chromatography steps as described in Materials and Methods. The procedure is not detrimental to the septin complex and has the advantage that it could be used to isolate native complex from other organisms for which no (epitope specific) antibodies against subunits of the septin complex are available. The protein fractions containing Pnut protein were determined by Western blotting using anti-Pnut antibodies. At the final stage of purification, samples containing the septin complex were subjected to a 15–35% glycerol gradient centrifugation. Figure 3A shows a silver stained gel of glycerol gradient fractions containing Drosophila septin complex. The proteins in this fractions consisted of Pnut (∼70 kDa), Sep2 (∼50 kDa), and Sep1 (∼41 kDa). The protein identities were verified by Western blotting and reversed phase C18 LC-MS as described previously (Renfrow et al., 2007). With complete cDNAs for each of the Drosophila septin complex subunits available, we wanted to determine whether coexpression of the genes would be sufficient for complex formation. The reconstitution of the septin complex would also allow the purification of the complex in high amounts for subsequent biochemical assays. Both E. coli and baculovirus expression systems were used for reconstitution of the Drosophila septin complex. On coexpression of all vectors carrying Pnut, Sep1, and Sep2, all proteins remained soluble and readily formed a complex. His-tagged Pnut facilitated isolation of the recombinant septin complexes with Ni-NTA beads. Complex was further purified as described in Materials and Methods. The protein pattern observed in peak fractions is shown in Figure 3B for both E. coli and baculovirus-derived complexes. His-tagged Pnut runs slightly higher than native Pnut during electrophoresis.
Figure 3.
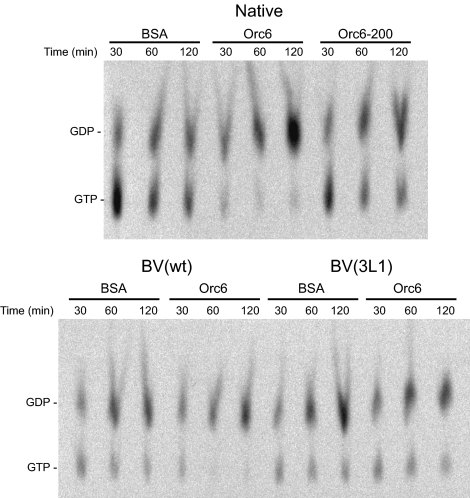
Purification of native and recombinant Drosophila septin complexes. (A) SDS-PAGE gel stained with silver of glycerol gradient fractions. In this last step in the purification protocol of septin complex from Drosophila embryos, samples enriched for septin proteins were fractionated by glycerol gradient centrifugation. Fractions of the gradient were analyzed by SDS-PAGE followed by silver stain for the presence of septin proteins as depicted for a typical purification. (B) SDS-PAGE gel stained with colloidal Coomassie depicts purified reconstituted septin complexes next to purified native complex for comparison. His-tagged Pnut runs slightly higher than native Pnut. The E. coli-derived complex preparation contains some degraded Pnut protein. (C) Phosphorimaging picture of TLC plate containing separated nucleotides eluted from native and recombinant septin complex incubated with GTP at 21°C for the time points indicated. The conversion of GTP to GDP was traced with [α-32P]GTP. (D) GTPase activity of purified septin complex. Graph depicts the amount of GDP generated as a fraction of the total of GTP and GDP measured on the TLC plate. The origin of the septin complex is as follows: black bars, native; white bars, E. coli; and gray bars, baculovirus.
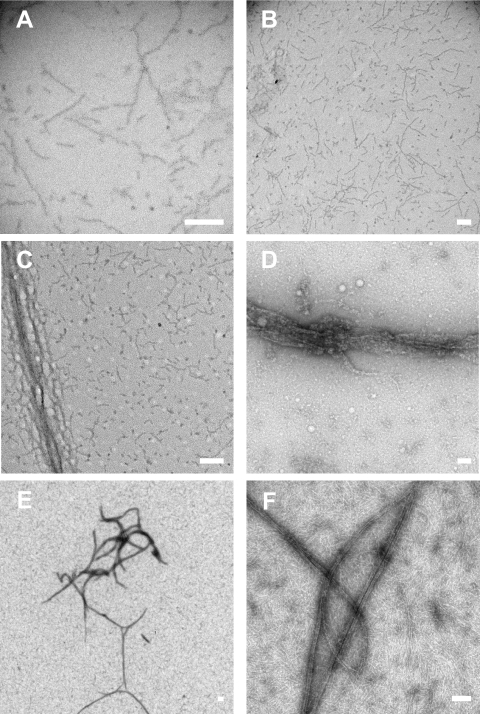
GTPase activities of native as well as recombinant septin complexes were assayed essentially as described by Field et al. (1996). Purified complex was bound to protein A beads via anti-Pnut antibody and used in a GTPase assay. A control experiment in which pulled down material was analyzed for Pnut by Western blot confirmed that despite different sources of Drosophila septin complex equal amounts of complex were precipitated and used throughout the GTPase assay (Supplemental Figure S1A). The hydrolysis of GTP to GDP was analyzed by TLC. Figure 3C shows representative phosphorimaging pictures of TLC plates with separated nucleotides eluted from both native and recombinant septin complexes. Graphic representation of the data are shown in Figure 3D. For native septin complex, ∼50–80% of newly bound nucleotide was retrieved as GDP after a 2 h incubation in individual experiments, depending on the batch of isolated protein. For the recombinant septin complexes this number was ∼75–80%. This is in good agreement with the results reported previously (Field et al., 1996). Recombinant septin complex seems to have a higher initial hydrolysis rate. Potential posttranslational modifications might be the cause of the lower GTPase activity displayed by the purified native septin complex. Septins are polymerizing proteins and purified septin complexes are known to form filaments in vitro (Field et al., 1996). Preparations of both native and recombinant (baculovirus) complexes were analyzed with negative-stain electron microscopy (EM) for filament formation. Figure 4, A–D, shows filaments formed by native septin complex. Because the concentration of the proteins is relatively low in the glycerol gradient fractions large aggregates were not observed (Figure 4, A and B). The addition of 1.5% polyethylene glycol (PEG) to the sample resulted in aggregation of the filaments (Figure 4C). Concentration of the proteins in the glycerol gradient samples by using a Centricon concentrator also resulted in the formation of larger filaments (Figure 4D). The concentration dependency of septin complex solutions to form larger aggregates has also been described previously (Field et al., 1996). This process involves a lateral association of filaments. The samples containing a highly concentrated, baculovirus produced reconstituted septin complex formed many larger cable-like aggregates as shown in Figure 4, E and F. Overall, recombinant septin protein complex behaved in a similar manner as purified native complex.
Figure 4.
Negative-stain electron microscopy of filaments of native (A–D) and recombinant septin complex (E and F). Native complex shows smaller filaments due to the lower concentration of protein (A and B). The addition of 1.5% PEG 8000 (C) or concentrating the sample (D) induces larger cable-like aggregates. Cable-like aggregates are also present in the concentrated recombinant protein complex (E and F). Images taken at different magnifications. Bars, 100 nm.
Orc6 Increases the GTP Hydrolysis Rate of the Septin Complex
The GTPase activities of native and recombinant, baculovirus-derived septin complex were measured in the presence of purified wild type and C-terminal mutant Orc6 proteins. Proteins were prepared as described previously (Balasov et al., 2007). Figure 5 shows the TLC analysis of a typical experiment with native and recombinant (baculovirus) septin complexes. Quantification of TLC data for all septin complexes used is summarized in Table 1. Both for native and recombinant septin complex the addition of Orc6 increased the GDP over GTP ratio of bound nucleotide significantly compared with the control experiment with BSA. An Orc6-200 mutant that is unable to interact with Pnut (Chesnokov et al., 2003) did not have an additional effect on GTPase activity of the septin complex. In fact the addition of Orc6-200 or BSA resulted in similar GDP to GTP ratios of the complex. Overall, amount of newly bound nucleotide was in the same range for all septin complexes and resulted in ∼0.05–0.1 mol nucleotide/mol Pnut after a 2-h incubation, in good agreement with previously reported data (Field et al., 1996). The increased GTPase activity is not detrimental to the integrity of the septin complex. SDS-PAGE analysis followed by silver stain revealed that after a 2-h incubation of septin complex with Orc6 and GTP a stoichiometric septin complex with bound Orc6 could be retrieved (Supplemental Figure S1B). Table 1 further shows that Orc6 did not have an effect on the GTPase activity of purified baculovirus recombinant septin complex containing triple leucine mutants of Pnut, which are not able to interact with Orc6 (Figure 2D). Septin complex containing wild-type Pnut, either native or reconstituted, showed a 1.5- to 2-fold higher bound GDP over GTP ratio for wild-type Orc6 than for the Orc6-200 mutant protein. For both wild-type and C-terminal deletion mutant of Orc6, equal ratios were observed with septin complex containing a triple leucine mutant of Pnut. These data demonstrate that the effect of Orc6 on septin GTPase activity is via its direct interaction with Pnut.
Figure 5.
Orc6 increases GTP hydrolysis of septin complex. Phosphorimaging picture of TLC plate containing separated nucleotides eluted from either native or recombinant baculovirus septin complex incubated with GTP at 21°C in the presence of BSA, wild-type Orc6, and the Orc6-200 mutant for the time points indicated. BV(wt), baculovirus-derived recombinant septin complex containing wild-type Pnut. BV(3L1), baculovirus derived recombinant septin complex containing 3L1 triple leucine mutant of Pnut.
Table 1.
Amount of bound nucleotide converted to GDP on septin complex incubated with the protein indicated
| Septin complex | Incubation time (min) | Protein added to septin complex |
||
|---|---|---|---|---|
| BSAa | Orc6-200 | Orc6 (wt)b,c | ||
| Native | n = 5 | n = 4 | ||
| 30 | 1.00 | 1.04 ± 0.21 | 2.48 ± 0.79d | |
| 60 | 1.00 | 1.03 ± 0.11 | 1.66 ± 0.31d | |
| 120 | 1.00 | 1.06 ± 0.14 | 1.39 ± 0.35 | |
| Baculovirus | n = 4 | n = 3 | ||
| 30 | 1.00 | 0.97 ± 0.22 | 2.99 ± 1.30e | |
| 60 | 1.00 | 1.03 ± 0.07 | 1.93 ± 0.21d | |
| 120 | 1.00 | 0.97 ± 0.09 | 1.40 ± 0.40 | |
| 3L1 | n = 3 | n = 3 | ||
| 30 | 1.00 | 1.00 ± 0.04 | 1.03 ± 0.02 | |
| 60 | 1.00 | 1.03 ± 0.02 | 1.01 ± 0.02 | |
| 120 | 1.00 | 1.01 ± 0.01 | 1.04 ± 0.05 | |
| 3L2 | n = 3 | n = 3 | ||
| 30 | 1.00 | 0.94 ± 0.02 | 0.89 ± 0.14 | |
| 60 | 1.00 | 1.00 ± 0.02 | 0.92 ± 0.15 | |
| 120 | 1.00 | 1.00 ± 0.01 | 1.04 ± 0.01 | |
Values are means ± SD.
a Data is normalized to the amount of GDP associated with septin complex incubated with BSA.
b Wild-type Orc6.
c Student's t test probabilities for wild type Orc6 compared with Orc6-200 assuming the null hypothesis are d p < 0.01 and e p < 0.05.
Orc6 Enhances Filament Assembly of the Septin Complex
GTP binding has been reported to induce filament formation of the recombinant expressed Xenopus septin protein Sept2 (Mendoza et al., 2002). GTP hydrolysis was not important for filament formation and even seemed to be inhibitory because incubating the septin protein with nonhydrolyzable guanosine 5′-O-(3-thio)triphosphate (GTPγS) instead of GTP resulted in a higher degree of polymerization. These results were obtained for the single Xenopus septin protein. Filament formation of septin complexes isolated from yeast (Frazier et al., 1998), Drosophila (Field et al., 1996), or from reconstituted human septins (Kinoshita et al., 2002) did not show a dependency for GTP. It was suggested that the GTPase activity of septin complexes might promote disassembly of the heteromeric complex (Mitchison and Field, 2002). In our experiments, a 2-h incubation of wild type recombinant septin complex in the presence of GTP and Orc6 did not seem to induce complex disassembly in vitro (Supplemental Figure S1B).
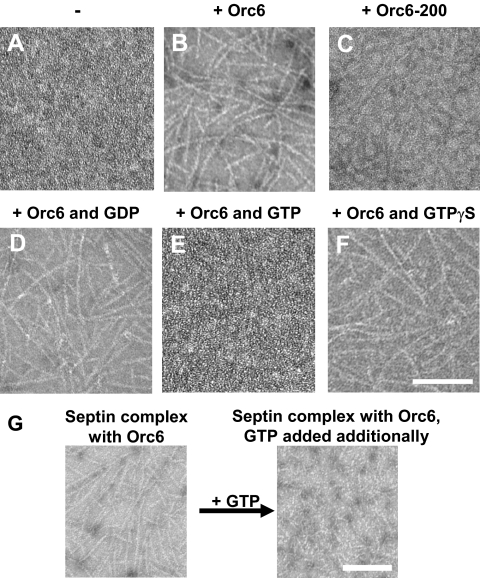
To investigate the effect of Orc6 on septin filaments, EM was used to analyze septin filament formation under the conditions of the GTPase assay. Recombinant (baculovirus) septin complex was incubated with or without Orc6 in GTPase buffer for 2 h at room temperature and analyzed for filaments. The effect of wild-type Orc6 and Orc6-200 deletion mutant protein on septin filament assembly/disassembly was tested in the presence or absence of nucleotide. When diluted in GTPase buffer septin complex did not exhibit large filaments as is shown in Figure 6A. Very small filaments or individual complexes were not detected under the conditions used for EM. The addition of either 2 μM GDP, GTP, or GTPγS had no detectable effect on filament formation (Supplemental Figure S2). The addition of Orc6 significantly increased filament length as shown in Figure 6B. However, in the presence of GTP this effect of Orc6 on septin filament formation was greatly reduced (Figure 6E). On the other hand, incubation of Orc6 with septin complex in the presence of either GDP (Figure 6D) or GTPγS (Figure 6F) still resulted in the formation of long filaments. One could argue that the presence of GTP merely prevented assembly of long filaments by Orc6 rather than aiding in their disassembly. However, in a follow-up experiment the filaments formed by the incubation of Orc6 with septin complex were significantly reduced in length when the incubation was continued after the addition of 2 μM GTP (Figure 6G). This result suggests that GTP hydrolysis leads to filament disassembly. The Orc6-200 mutant was not able to enhance filament formation compared with wild-type Orc6 (Figure 6C). No filament formation was detected in GTPase buffer containing Orc6 but no septin complex (data not shown). Furthermore, although septin complexes containing the triple leucine mutants of Pnut are able to form higher order filaments in concentrated samples (Supplemental Figure S3A), filament formation of these complexes was not dramatically affected by the addition of Orc6 under GTPase assay conditions (Supplemental Figure S3B). This indicates that the larger filaments are formed from the septin complex and that the assembly is induced/enhanced by its interaction with Orc6.
Figure 6.
Orc6 enhances septin filament assembly. Recombinant baculovirus derived wild-type septin complex was diluted in GTPase buffer to 80 ng Pnut/μl, and incubated with 80 ng/μl Orc6 protein and 2 μM nucleotide where indicated. Samples were incubated at room temperature for 2 h after which they were prepared for EM. Proteins incubated are recombinant septin complex (A), recombinant septin complex and Orc6 (B), recombinant septin complex and Orc6-200 (C), recombinant septin complex and Orc6 in the presence of GDP (D), recombinant septin complex and Orc6 in the presence of GTP (E), and recombinant septin complex and Orc6 in the presence of GTPγS (F). (G) Recombinant baculovirus derived wild-type septin complex was diluted in GTPase buffer to 80 ng Pnut/μl and incubated with 80 ng/μl wild-type Orc6 at room temperature for 2 h. At this point, half the sample was used for EM analysis, confirming filament formation. Subsequently, 2 μM GTP was added to the remaining sample after which incubation at room temperature was continued for an additional 2 h. Sample was processed and analyzed with EM. Bars, 100 nm.
DISCUSSION
Septin proteins have been discovered throughout the Animal Kingdom as well as in fungi. The Drosophila septins are essential for cytokinesis and are likely involved in other functions as well (Adam et al., 2000). Studies in yeast suggested that a primary function of the septins is to serve as a matrix or scaffold for the organization of other proteins at the cell surface (Field and Kellogg, 1999; Versele and Thorner, 2005). Therefore, the identification of the septin-interacting proteins should be critical to the elucidation of septin functions. Previously, we found that the smallest subunit of Drosophila ORC complex, Orc6, is important for cytokinesis and interacts with the septin protein Pnut (Chesnokov et al., 2003).
Both in Drosophila (Chesnokov et al., 2001) and human cells (Prasanth et al., 2002), a considerable pool of Orc6 is cytoplasmic, and the protein is either associated with or proximal to the plasma membrane and cleavage furrows of dividing cells. In Drosophila, Orc6 and Pnut colocalize in vivo at cell membranes and cleavage furrows of dividing cells, and during cellularization in Drosophila early embryos (Chesnokov et al., 2001). The C-terminal domain of Orc6 is necessary for this colocalization with Pnut. Moreover, Orc6 RNAi results in cytokinesis defects in Drosophila tissue culture cells (Chesnokov et al., 2003), whereas Pnut RNAi disrupts the localization of Orc6 to the plasma membrane (Figure 1B).
Analysis of the cells treated with Pnut dsRNA revealed an elevated number of binucleated cells (5- to 30-fold increase, depending on the experiment). This is in contrast with previously reported data (Somma et al., 2002) in which no elevated numbers of binucleated cells were detected in cultures treated with Pnut dsRNA. Differences in culture conditions, amount of dsRNA used, as well as cell preparation protocols for analysis might have contributed to the discrepancies between the two studies.
Deletion of part of the predicted coiled-coil domain of Pnut impaired its ability to form a complex with both Sep1 and Sep2 together, but it was still able to interact with Sep1 (Figure 2C). A previous study (Adam et al., 2000) proposed that different septin complexes may exist within Drosophila. Our data suggests that Pnut and Sep1 might form a precomplex that joins with Sep2 to form the complete septin complex. The interaction of Orc6 with Pnut is also disrupted in C-terminal deletion mutants of Pnut, suggesting that the coiled-coil domain of Pnut is important for both binding with Orc6 and for the formation of the septin complex. However, leucine to alanine substitutions within the coiled-coil domain of Pnut prevent Orc6 binding but do not inhibit complex assembly, indicating that these protein interactions are based on different structural moieties within the C terminus. Furthermore, direct interaction studies (Figure 2B) revealed that the coil-coil domain of Pnut is not sufficient for the interaction of this septin with Orc6 and that other structural features may also be important for the interaction between the two proteins.
To study the effect of Pnut mutations in vivo in Drosophila tissue culture cells, we used various expression systems, including heat shock and metallothionein promoters. In all cases, expression of GFP-Pnut in L2 cells resulted in rod- and spiral-like structures present throughout the cytosol (Supplemental Figure S4). Expression of either N-terminal or C-terminal GFP fusions to Pnut, or a FLAG-Pnut protein also resulted in the same aberrant structures, compromising the in vivo analysis of Pnut mutants. However, when under control of the native Pnut promoter, proteins could be expressed in L2 cells at lower levels as is shown in Figure 2E. This figure further shows that the coiled-coil domain of Pnut, which is important for Drosophila septin complex assembly, also is essential for the in vivo localization of Pnut, because FLAG-Pnut(1-427), lacking the coiled-coil domain, had the tendency to accumulate into crescent shaped aggregates. The FLAG-tagged triple leucine mutants of Pnut exhibited mainly diffuse cytoplasmic staining when expressed in L2 cells, although some plasma membrane staining was observed. It is possible that due to the mutations in the coiled-coil domain these Pnut mutants do not interact properly with other proteins (as shown for Orc6), resulting in a release from the plasma membrane at specific cell stages.
Native septin complex as well as reconstituted septin complexes exhibit the characteristic properties of filament formation and GTPase activity, indicating that they are functional complexes. Because insect cell lines are closely related to Drosophila, we used baculovirus-derived reconstituted septin complex for further biochemical studies.
The human SEPT2–SEPT6–SEPT7 complex can be formed from recombinant proteins all lacking their predicted coiled-coil domains, suggesting that their C termini are dispensable for complex formation (Sirajuddin et al., 2007). Structural analysis of crystals of the human septin complex revealed that the filaments consist of an assembly of GTP binding domains. However, the coiled-coil domains of SEPT6 ands SEPT7 do interact directly with each other (Low and Macara, 2006), suggesting that although not required for the human septin complex, coiled-coils may further stabilize filament formation (Sirajuddin et al., 2007). The GTP binding domains of human septin proteins can also interact with coiled-coil structures within the multiple subunit complex (Low and Macara, 2006). This might also occur with the Drosophila septins when they assemble into complex. However, the interaction of Orc6 with the septin complex seems strongly dependent on the coiled-coil domain of Pnut.
One possible role of the interaction of Orc6 with the septin complex could be the regulation of the GTPase activity of the complex during cytokinesis. Orc6 reproducibly increased the GDP-to-GTP ratio of bound nucleotide of the whole septin complex, but no significant increase of total nucleotide bound to complex was detected. It has been hypothesized that GTP hydrolysis might promote disassembly of the septin complex (Mitchison and Field, 2002). However, purified recombinant septin complex was retrieved intact with bound Orc6 after 2-h incubation in the presence of GTP, although potentially the disassembly of a small amount of complex might have occurred. We observed that many larger filaments present in concentrated recombinant septin complex samples were not detected under GTPase assay conditions (Figure 6A), most likely due to the dilution of concentrated sample. No differences in filament size were observed for septin complexes incubated either in the presence or absence of GTP. However, due to limitations of our EM setup, subtle changes in small filament size could not be detected.
Although GTP hydrolysis by septin complex was accelerated by Orc6 binding (because the presence of a nonbinding mutant of Orc6 had no additional affect on hydrolysis), no significant changes could be detected in turnover rate. The higher turnover rates reported for individual Xenopus (Mendoza et al., 2002), mouse (Kinoshita et al., 1997), and human (Sheffield et al., 2003; Huang et al., 2006) recombinant septin proteins do not exclude a regulatory function for these subunits in vivo when not assembled in complex. A structural rather than regulatory role for septin complex-bound GTP and GDP was proposed from the results obtained with yeast septins (Vrabioiu et al., 2004). No turnover of yeast septin-bound GTP and GDP could be detected during a cell cycle in vivo. Furthermore, in vitro experiments revealed that GTP hydrolysis of yeast septin complex was limited by its slow binding or exchange activity, similar to the properties described initially for the Drosophila septin complex (Field et al., 1996). The role of Orc6 in GTP hydrolysis and filament disassembly of septin complex also suggests that in the case of Drosophila the guanine nucleotides bound to septins may contribute to the structural properties of the complex. Additionally, the importance of the GTP binding domains for the assembly of the human septin complex and potentially filament formation (Sirajuddin et al., 2007) also indicates a role for guanine nucleotide in septin complex structure.
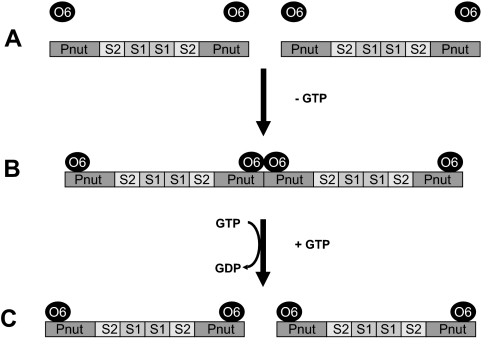
The addition of Orc6 to septin complex in the absence of GTP, in contrast, greatly induced filament formation, whereas in the presence of GTP the effect was not observed. This indicates that Orc6 exhibits two opposite effects in its interactions with the septin complex, which are described in the model outlined in Figure 7. Based on the sequence homologies between human or Drosophila septins (Pan et al., 2007), hexamers are depicted as a linear protein similar to the crystal structure of the human septin complex (Sirajuddin et al., 2007) with Pnut at either end of the complex, as shown in Figure 7A. In the absence (or low concentration) of GTP Orc6 binding to Pnut enhances linear filament assembly (Figure 7B), potentially due to conformational changes in either Pnut or other septin subunits. Orc6 stabilizes the formation of the filaments by protein–protein interactions. It is interesting to note that purified recombinant Orc6 protein behaves as a dimer in biochemical assays (Chesnokov, unpublished). In the presence of GTP, Orc6 increases the GTPase activity of the septin complex, at the same time resulting in filament disassembly (Figure 7C). Increased GTPase activity may lead to conformational changes in the septin complex (data not shown), causing disassembly of septin filaments. Our results suggest that Orc6 may regulate either assembly or disassembly of septin filaments. Whether Orc6 actively induces filament disassembly in the presence of GTP or this process is a result of the increased GTPase activity of the septin complex remains to be investigated.
Figure 7.
Model for the interaction of Orc6 with the septin complex. Pnut subunits within the linear complex are marked. Sep1 is marked as S1, Sep2 is marked as S2, and Orc6 is marked as O6. For details, see text.
The septins are important for cytokinesis but molecular mechanisms of their functions in this process are not completely understood. Our data on interactions between Drosophila Orc6 and the septin complex reveal some new aspects for these proteins. Orc6 has an effect on both GTPase activity and filament formation of the septin complex, suggesting that Orc6 might have a direct role in septin complex functions during the last stage of mitosis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Leigh Millican and Melissa Chimento from the High Resolution Imaging Facility at University of Alabama for producing the EM images; Olga Chesnokova for providing the pMYR and pSOS plasmids with gateway cassettes, and Kirill Popov for comments and suggestions on the manuscript. This work was supported by National Institutes of Health grant GM-69681 (to I. C.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-07-0754) on November 5, 2008.
REFERENCES
- Adam J. C., Pringle J. R., Peifer M. Evidence for functional differentiation among Drosophila septins in cytokinesis and cellularization. Mol. Biol. Cell. 2000;11:3123–3135. doi: 10.1091/mbc.11.9.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronheim A., Zandi E., Hennemann H., Elledge S. J., Karin M. Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Mol. Cell Biol. 1997;17:3094–3102. doi: 10.1128/mcb.17.6.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasov M., Huijbregts R. P., Chesnokov I. Role of the Orc6 protein in origin recognition complex-dependent DNA binding and replication in Drosophila melanogaster. Mol. Cell Biol. 2007;27:3143–3153. doi: 10.1128/MCB.02382-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral Y., Mermall V., Mooseker M. S., Snyder M. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol. Cell. 2000;5:841–851. doi: 10.1016/s1097-2765(00)80324-x. [DOI] [PubMed] [Google Scholar]
- Bell S. P. The origin recognition complex: from simple origins to complex functions. Genes Dev. 2002;16:659–672. doi: 10.1101/gad.969602. [DOI] [PubMed] [Google Scholar]
- Bell S. P., Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Chesnokov I., Remus D., Botchan M. Functional analysis of mutant and wild-type Drosophila origin recognition complex. Proc. Natl. Acad. Sci. USA. 2001;98:11997–12002. doi: 10.1073/pnas.211342798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnokov I. N. Multiple functions of the origin recognition complex. Int. Rev. Cytol. 2007;256:69–109. doi: 10.1016/S0074-7696(07)56003-1. [DOI] [PubMed] [Google Scholar]
- Chesnokov I. N., Chesnokova O. N., Botchan M. A cytokinetic function of Drosophila ORC6 protein resides in a domain distinct from its replication activity. Proc. Natl. Acad. Sci. USA. 2003;100:9150–9155. doi: 10.1073/pnas.1633580100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere J., Barral Y. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science. 2004;305:393–396. doi: 10.1126/science.1099892. [DOI] [PubMed] [Google Scholar]
- Dutta A., Bell S. P. Initiation of DNA replication in eukaryotic cells. Annu. Rev. Cell. Dev. Biol. 1997;13:293–332. doi: 10.1146/annurev.cellbio.13.1.293. [DOI] [PubMed] [Google Scholar]
- Fares H., Peifer M., Pringle J. R. Localization and possible functions of Drosophila septins. Mol. Biol. Cell. 1995;6:1843–1859. doi: 10.1091/mbc.6.12.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field C. M., al-Awar O., Rosenblatt J., Wong M. L., Alberts B., Mitchison T. J. A purified Drosophila septin complex forms filaments and exhibits GTPase activity. J. Cell Biol. 1996;133:605–616. doi: 10.1083/jcb.133.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field C. M., Kellogg D. Septins: cytoskeletal polymers or signalling GTPases? Trends Cell Biol. 1999;9:387–394. doi: 10.1016/s0962-8924(99)01632-3. [DOI] [PubMed] [Google Scholar]
- Frazier J. A., Wong M. L., Longtine M. S., Pringle J. R., Mann M., Mitchison T. J., Field C. Polymerization of purified yeast septins: evidence that organized filament arrays may not be required for septin function. J. Cell Biol. 1998;143:737–749. doi: 10.1083/jcb.143.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter A. S., Pringle J. R., Lew D. J. The septin cortex at the yeast mother-bud neck. Curr. Opin. Microbiol. 2001;4:681–689. doi: 10.1016/s1369-5274(01)00269-7. [DOI] [PubMed] [Google Scholar]
- Harlow E., Lane M. E., editors. Using Antibodies. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. [Google Scholar]
- Hartwell L. H. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp. Cell Res. 1971;69:265–276. doi: 10.1016/0014-4827(71)90223-0. [DOI] [PubMed] [Google Scholar]
- Heberlein U., Tjian R. Temporal pattern of alcohol dehydrogenase gene transcription reproduced by Drosophila stage-specific embryonic extracts. Nature. 1988;331:410–415. doi: 10.1038/331410a0. [DOI] [PubMed] [Google Scholar]
- Huang Y. W., Surka M. C., Reynaud D., Pace-Asciak C., Trimble W. S. GTP binding and hydrolysis kinetics of human septin 2. FEBS J. 2006;273:3248–3260. doi: 10.1111/j.1742-4658.2006.05333.x. [DOI] [PubMed] [Google Scholar]
- Kinoshita M. The septins. Genome Biol. 2003;4:236. doi: 10.1186/gb-2003-4-11-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita M. Diversity of septin scaffolds. Curr. Opin. Cell Biol. 2006;18:54–60. doi: 10.1016/j.ceb.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Kinoshita M., Field C. M., Coughlin M. L., Straight A. F., Mitchison T. J. Self- and actin-templated assembly of mammalian septins. Dev. Cell. 2002;3:791–802. doi: 10.1016/s1534-5807(02)00366-0. [DOI] [PubMed] [Google Scholar]
- Kinoshita M., Kumar S., Mizoguchi A., Ide C., Kinoshita A., Haraguchi T., Hiraoka Y., Noda M. Nedd5, a mammalian septin, is a novel cytoskeletal component interacting with actin-based structures. Genes Dev. 1997;11:1535–1547. doi: 10.1101/gad.11.12.1535. [DOI] [PubMed] [Google Scholar]
- Kusch J., Meyer A., Snyder M. P., Barral Y. Microtubule capture by the cleavage apparatus is required for proper spindle positioning in yeast. Genes Dev. 2002;16:1627–1639. doi: 10.1101/gad.222602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., DeMarini D. J., Valencik M. L., Al-Awar O. S., Fares H., De Virgilio C., Pringle J. R. The septins: roles in cytokinesis and other processes. Curr. Opin. Cell Biol. 1996;8:106–119. doi: 10.1016/s0955-0674(96)80054-8. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., Fares H., Pringle J. R. Role of the yeast Gin4p protein kinase in septin assembly and the relationship between septin assembly and septin function. J. Cell Biol. 1998;143:719–736. doi: 10.1083/jcb.143.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., Theesfeld C. L., McMillan J. N., Weaver E., Pringle J. R., Lew D. J. Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae. Mol. Cell Biol. 2000;20:4049–4061. doi: 10.1128/mcb.20.11.4049-4061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low C., Macara I. G. Structural analysis of septin 2, 6, and 7 complexes. J. Biol. Chem. 2006;281:30697–30706. doi: 10.1074/jbc.M605179200. [DOI] [PubMed] [Google Scholar]
- Machida Y. J., Hamlin J. L., Dutta A. Right place, right time, and only once: replication initiation in metazoans. Cell. 2005;123:13–24. doi: 10.1016/j.cell.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Martinez C., Sanjuan M. A., Dent J. A., Karlsson L., Ware J. Human septin-septin interactions as a prerequisite for targeting septin complexes in the cytosol. Biochem. J. 2004;382:783–791. doi: 10.1042/BJ20040372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason J. M., Arndt K. M. Coiled coil domains: stability, specificity, and biological implications. Chembiochem. 2004;5:170–176. doi: 10.1002/cbic.200300781. [DOI] [PubMed] [Google Scholar]
- Mendoza M., Hyman A. A., Glotzer M. GTP binding induces filament assembly of a recombinant septin. Curr. Biol. 2002;12:1858–1863. doi: 10.1016/s0960-9822(02)01258-7. [DOI] [PubMed] [Google Scholar]
- Mitchison T. J., Field C. M. Cytoskeleton: what does GTP do for septins? Curr. Biol. 2002;12:R788–R790. doi: 10.1016/s0960-9822(02)01295-2. [DOI] [PubMed] [Google Scholar]
- Neufeld T. P., Rubin G. M. The Drosophila peanut gene is required for cytokinesis and encodes a protein similar to yeast putative bud neck filament proteins. Cell. 1994;77:371–379. doi: 10.1016/0092-8674(94)90152-x. [DOI] [PubMed] [Google Scholar]
- Pak D. T., Pflumm M., Chesnokov I., Huang D. W., Kellum R., Marr J., Romanowski P., Botchan M. R. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell. 1997;91:311–323. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- Pan F., Malmberg R. L., Momany M. Analysis of septins across kingdoms reveals orthology and new motifs. BMC Evol. Biol. 2007;7:103. doi: 10.1186/1471-2148-7-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth S. G., Prasanth K. V., Stillman B. Orc6 involved in DNA replication, chromosome segregation, and cytokinesis. Science. 2002;297:1026–1031. doi: 10.1126/science.1072802. [DOI] [PubMed] [Google Scholar]
- Renfrow M. B., Mackay C. L., Chalmers M. J., Julian B. A., Mestecky J., Kilian M., Poulsen K., Emmett M. R., Marshall A. G., Novak J. Analysis of O-glycan heterogeneity in IgA1 myeloma proteins by Fourier transform ion cyclotron resonance mass spectrometry: implications for IgA nephropathy. Anal. Bioanal. Chem. 2007;389:1397–1407. doi: 10.1007/s00216-007-1500-z. [DOI] [PubMed] [Google Scholar]
- Sasaki T., Gilbert D. M. The many faces of the origin recognition complex. Curr. Opin. Cell Biol. 2007;19:337–343. doi: 10.1016/j.ceb.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Sheffield P. J., Oliver C. J., Kremer B. E., Sheng S., Shao Z., Macara I. G. Borg/septin interactions and the assembly of mammalian septin heterodimers, trimers, and filaments. J. Biol. Chem. 2003;278:3483–3488. doi: 10.1074/jbc.M209701200. [DOI] [PubMed] [Google Scholar]
- Sirajuddin M., Farkasovsky M., Hauer F., Kuhlmann D., Macara I. G., Weyand M., Stark H., Wittinghofer A. Structural insight into filament formation by mammalian septins. Nature. 2007;449:311–315. doi: 10.1038/nature06052. [DOI] [PubMed] [Google Scholar]
- Somma M. P., Fasulo B., Cenci G., Cundari E., Gatti M. Molecular dissection of cytokinesis by RNA interference in Drosophila cultured cells. Mol. Biol. Cell. 2002;13:2448–2460. doi: 10.1091/mbc.01-12-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiliotis E. T., Kinoshita M., Nelson W. J. A mitotic septin scaffold required for mammalian chromosome congression and segregation. Science. 2005;307:1781–1785. doi: 10.1126/science.1106823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiliotis E. T., Nelson W. J. Here come the septins: novel polymers that coordinate intracellular functions and organization. J. Cell Sci. 2006;119:4–10. doi: 10.1242/jcs.02746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa P. A., DeRisi J. L., Wilhelm J. E., Vale R. D. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 2000;290:341–344. doi: 10.1126/science.290.5490.341. [DOI] [PubMed] [Google Scholar]
- Versele M., Thorner J. Some assembly required: yeast septins provide the instruction manual. Trends Cell Biol. 2005;15:414–424. doi: 10.1016/j.tcb.2005.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrabioiu A. M., Gerber S. A., Gygi S. P., Field C. M., Mitchison T. J. The majority of the 0 septin complexes do not exchange guanine nucleotides. J. Biol. Chem. 2004;279:3111–3118. doi: 10.1074/jbc.M310941200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.