Abstract
GCC185, a trans-Golgi network-localized protein predicted to assume a long, coiled-coil structure, is required for Rab9-dependent recycling of mannose 6-phosphate receptors (MPRs) to the Golgi and for microtubule nucleation at the Golgi via CLASP proteins. GCC185 localizes to the Golgi by cooperative interaction with Rab6 and Arl1 GTPases at adjacent sites near its C terminus. We show here by yeast two-hybrid and direct biochemical tests that GCC185 contains at least four additional binding sites for as many as 14 different Rab GTPases across its entire length. A central coiled-coil domain contains a specific Rab9 binding site, and functional assays indicate that this domain is important for MPR recycling to the Golgi complex. N-Terminal coiled-coils are also required for GCC185 function as determined by plasmid rescue after GCC185 depletion by using small interfering RNA in cultured cells. Golgi-Rab binding sites may permit GCC185 to contribute to stacking and lateral interactions of Golgi cisternae as well as help it function as a vesicle tether.
INTRODUCTION
The Golgi complex plays a central role in processing and sorting of secreted and membrane proteins. In mammalian cells, it is made up of a set of flattened cisternae that are organized as a stack (Farquhar and Palade, 1998). Within the Golgi, proteins undergo oligosaccharide side chain maturation, and they are sorted to different destinations, including apical or basolateral plasma membranes, prelysosomes, or secretory storage granules, as they exit this compartment.
The Golgi surface is decorated with proteins called Golgins (Short et al., 2005; Sztul and Lupashin, 2006). Golgins are characterized by their predicted, highly extended, coiled-coil structures, and they typically have a high glutamic acid content (Ramirez et al., 2008). Certain Golgins have been implicated as transport vesicle tethers that help capture vesicles and bring them closer to their cognate targets; they are also essential for the maintenance of normal Golgi structure. For example, temperature-sensitive cells lacking GM130 have a disrupted Golgi at the nonpermissive temperature (Vasile et al., 2003); loss of Golgin-84 (Diao et al., 2003), Golgin-245 (Yoshino et al., 2005), TATA modulatory factor (Fridmann-Sirkis et al., 2004), GRASP55 (Feinstein and Linstedt, 2008), GRASP65 (Puthenveedu et al., 2006), and the so-called COG complex (Zolov and Lupashin, 2005) all lead to disruption of the Golgi ribbon. Thus, the Golgi complex is highly sensitive to the loss of any one of a large collection of Golgi-associated proteins.
Most Golgins are peripherally associated with this organelle by virtue of interaction with small GTPases of the Rab and/or Arf (Arl) families; a few contain C-terminal membrane anchors (Short et al., 2005). Four human Golgins contain a so-called GRIP domain at their C termini that enables them to bind the Arl1 GTPase at the trans-Golgi network (TGN); Arl1 is essential for the Golgi targeting of multiple GRIP domain-containing proteins (Lu and Hong, 2003; Panic et al., 2003; Setty et al., 2003; Lu et al., 2006). We study GCC185, a GRIP domain-containing protein that uses cooperative, C-terminal interactions with Rab6 and Arl1 to achieve its TGN localization (Burguete et al., 2008). GCC185 came to our attention when we identified it as a Rab9 GTPase binding partner (Reddy et al., 2006). Using small interfering (siRNA) depletion in live cells and in vitro transport assays, we showed that like Rab9, GCC185 is required for the recycling of mannose 6-phosphate receptors (MPRs) from endosomes to the TGN. Cells lacking GCC185 accumulate MPRs in peripheral vesicles that contain Rab9 GTPase. In addition, the elegant and ordered Golgi ribbon fragments into perinuclear mini-stacks (Reddy et al., 2006; Derby et al., 2007).
The accumulation of transport vesicle intermediates in cells depleted of GCC185 is good evidence that this protein functions in the process of transport vesicle tethering (Reddy et al., 2006). We initially proposed that Rab9 present on transport vesicles (Barbero et al., 2002) could recruit cytosolic GCC185 to the vesicle surface and move along microtubule tracks toward the Golgi, in which Rab9 could pass the tether to Golgi-associated Rab6 and Arl1 proteins (Reddy et al., 2006; Burguete et al., 2008). But the Rab9 binding site we identified was located near the C terminus of GCC185, which also interacts with the TGN surface. This left the rest of the 1684-amino acid-long coiled-coil without a clear role. An obvious alternative possibility was that GCC185 contained additional Rab9 binding sites elsewhere along its length to permit vesicle binding at one site and Golgi association at another. For this reason, we conducted a screen for additional Rab binding sites in GCC185 protein. As we describe herein, this screen was a very fruitful endeavor and simplifies our model for vesicle tethering at the TGN surface.
MATERIALS AND METHODS
Yeast Two-Hybrid Analysis
Yeast two-hybrid analysis was carried out as described previously (Fuchs et al., 2007). Briefly, a library of 57 mutant Rab proteins deficient for guanosine triphosphate (GTP) hydrolysis was cloned into pGBT9 as a Gal4-binding domain fusion library (Bai and Elledge, 1996). Individual GCC185 domains were cloned into pACT2 as Gal4-activation domain hybrids. Cotransformations of these two constructs into the PJ269-4A yeast strain and plating onto synthetic complete agar plates lacking leucine and tryptophan yielded colonies expressing both fusion proteins. Five individual colonies were picked for each Rab/coiled-coil pairing and streaked onto selective plates (synthetic complete media containing 0.4 mg/ml adenine and lacking leucine, tryptophan, and histidine); a Rab/coiled-coil interaction was indicated by the presence of cell growth on this selective media.
Expression Plasmids, Protein Expression, and Purification
Different cDNAs encoding residues 1–50 and 51–1684 of GCC185 were obtained from Kazusa (Kisarasu, Japan) and American Type Culture Collection (Manassas, VA), respectively. Full-length GCC185 cDNA was constructed by three-piece ligation of polymerase chain reaction (PCR)-amplified products into pCDNA3.1(+) (Invitrogen, Carlsbad, CA) modified with an N-terminal myc-tag. ΔCC1 (358-1684), ΔCC12 (752-1684), ΔCC123 (890-1684), and ΔC110 (1-1574) truncation constructs were assembled by PCR and ligated into pCDNA3.1(+)-myc. For the full-length GCC185 siRNA rescue construct, GCC185 was cloned into a myc-tagged pcDNA3.1(+)vector, and eight silent mutations were introduced via QuikChange mutagenesis into the region of CC1 targeted by the GCC185 siRNA; results were verified by sequencing. This plasmid was used to generate GCC185 (Δ862-882). CC1 (34-358), CC2 (394-751), and CC3 (805-889) coiled-coil domains were amplified from the full-length sequence and cloned into both the yeast two-hybrid prey vector pACT2 (Clontech, Mountain View, CA) and the glutathione transferase (GST) fusion vector pGEX4T-1 (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom); CC3 and CC3 L873A I880A were also inserted into pCDNA3.1(+)-myc. The CC3 L873A I880A mutant in pGEX4T-1 was generated via QuikChange and confirmed by sequencing. The ΔCC123 and C343 (1341-1684) domains were cloned separately into the pACT2 prey vector.
Wild-type, untagged Rab1A and Rab9A were purified according to Aivazian et al. (2006); His-tagged Rab6A was purified as described previously (Ganley et al., 2008). Untagged Rab9B in pet14b was transformed into BL21(DE3) cells, and cultures grown at 37°C (OD600 = 0.6) were induced with 0.4 mM isopropyl β-d-thiogalactoside (IPTG) for 3 h at 37°C. Cell pellets were resuspended in Rab lysis buffer (50 mM Tris, pH 8.0, 100 mM NaCl, 8 mM MgCl2, 2 mM EDTA, 0.5 mM dithiothreitol [DTT], and 10 μM GDP) and lysed by two passes at 20,000 psi through an EmulsiFlex-C5 apparatus (Avestin, Ottawa, ON, Canada). Clarified lysates (20,000 rpm for 20 min in a JA 20 rotor; Beckman Coulter, Fullerton, CA) were applied to a Q Sepharose ion exchange column and eluted with a 100–400 mM NaCl gradient. Fractions containing Rab9B were concentrated via ammonium sulfate precipitation (at 50% saturation) and loaded onto a Superdex 200 16/60 gel filtration column equilibrated in Rab storage buffer (20 mM HEPES pH 7.4, 200 mM NaCl, 8 mM MgCl2, 2 mM EDTA, 10 μM GDP, and 0.5 mM DTT). Fractions containing Rab9B were pooled and stored at −80°C in 10% glycerol. His-tagged Rab33B Q92A was also cloned into the pET14b expression vector; expression and cell lysis was performed as for Rab9B. Clarified cell lysate was loaded onto a Q Sepharose column and washed with 10 column volumes of Rab lysis buffer. The His-Rab33b enriched column wash was concentrated by ammonium sulfate precipitation (at 50% saturation) and applied to a gel filtration column equilibrated in Rab storage buffer. His-Rab33b fractions were pooled and stored at −80°C in 10% glycerol.
GST-coiled-coil constructs were transformed into BL21(DE3) Rosetta II cells, and cultures (OD600 = 0.7) were induced with 0.5 mM IPTG for 2.5 h at 37°C. Cells were resuspended in 50 mM HEPES, pH 7.4, 250 mM KCl containing 1 mM phenylmethylsulfonyl fluoride, 1 mM DTT and lysed by emulsification at 15,000 psi in a EmulsiFlex-C5 apparatus (Avestin); lysates were centrifuged at 20,000 rpm for 30 min in a JA 20 rotor (Beckman Coulter). Clarified lysates were incubated with glutathione 4B-Sepharose beads (GE Healthcare) for 2 h at 4°C, eluted with 50 mM HEPES, pH 7.4, 250 mM KCl, and 20 mM glutathione, and dialyzed overnight against 50 mM HEPES, pH 7.4, 250 mM KCl. Purified proteins were stored on ice at 4°C to prevent freezing-induced aggregation. The C110 I1588A L1595A mutant was obtained as described previously (Burguete at al., 2008).
Binding Assays
Binding reactions were carried out similarly to that described previously (Aivazian et al., 2006). Purified Rab proteins were loaded with radioactive [35S]guanosine 5′-O-(3-thio)triphosphate (GTPγS) (PerkinElmer Life and Analytical Sciences, Boston, MA) in loading buffer (50 mM HEPES pH 7.4, 150 mM KCl, 2 mM EDTA, 1 mM MgCl2, 0.1 mg/ml bovine serum albumin (BSA), 7 μM GTPγS, and 2.7 μM Rab) for 2 h (Rab9A and Rab9B) or 3 h (Rab6A, Rab1A, and Rab33B) at room temperature (Rab9A and Rab9B) or 37°C (Rab6A, Rab1A, and Rab33B). Rabs were then desalted on a PD-10 column (GE Healthcare) into binding buffer (50 mM HEPES pH 7.4, 150 mM KCl, 5 mM MgCl2, and 0.1 mg/ml BSA). Binding reactions were assembled in a 250-μl reaction containing 0.1 μM Rab and 3 μM (for single point binding) or varying (for concentration curve) amounts of GST-coiled-coil in binding buffer. Reactions proceeded at room temperature for 1.5 h. Glutathione-Sepharose beads (15 μl) equilibrated in binding buffer were added to each reaction and incubated for 30 min at room temperature with rotation. Beads were then washed with 10 column volumes of binding buffer, and the amount of retained Rab protein was detected through liquid scintillation counting of the beads.
Cell Culture
HeLa cells from American Type Culture Collection were cultured at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 7.5% fetal calf serum, penicillin, and streptomycin. For RNA interference (RNAi), HeLa cells were transfected at 50% confluence with fluorescein labeled duplex RNA (Dharmacon RNA Technologies, Lafayette, CO) by using Oligofectamine (Invitrogen) according to the manufacturer's instructions. The GCC185 siRNA sequence was located in the first coiled-coil domain and targeted the sequence GGAGTTGGAACAATCACAT. RNAi depletion was carried out for 72 h posttransfection and followed by fixation and staining. For rescue experiments, plasmids containing GCC185 constructs were transfected 48 h after initial siRNA treatment (24-h rescue) by using FuGENE6 (Roche Diagnostics, Indianapolis, IN) according to the manufacturer's instructions. Transfection of GCC185 constructs in the absence of RNAi depletion was carried out by treatment of cells by using FuGENE6 24 h before fixation.
Immunofluorescence Microscopy
Twenty-four hours before fixation, cells were split onto 22- × 22-mm coverslips in a six-well plate. Cells were washed twice in phosphate-buffered saline (PBS) and fixed for 20 min in 3.7% formaldehyde in 200 mM HEPES, pH 7.4 (Warren et al., 1984). After fixation, cells were washed twice and incubated for 15 min in DMEM and 10 mM HEPES, pH 7.4, to quench. Cells were permeabilized for 5 min with 0.2% Triton X-100 in PBS, followed by two washes and a 15-min incubation with 1% BSA in PBS. Cells were incubated with primary antibody, diluted 1:500 (anti GM130), 1:750 (anti myc), or 1:1000 (anti-β-COP or anti-GCC185) in BSA/PBS for 30 min. This was followed by three 5-min washes and a 30-min incubation in secondary antibody diluted 1:1000 in BSA/PBS. The following antibodies were used in this study: rabbit anti-GCC185 residues 1301–1684 (Reddy et al., 2006); mouse anti-GM130 (BD Biosciences, San Jose, CA), chicken anti-myc (Bethyl Laboratories, Montgomery, TX), Cy3-goat anti chicken (Jackson ImmunoResearch Laboratories, West Grove, PA), and Cy5-goat anti mouse/rabbit (Invitrogen). A rabbit anti-β-COP polyclonal antibody was generated according to Pepperkok et al. (1995). Micrographs were acquired using a microscope (Eclipse 80i; Nikon, Tokyo, Japan) fitted with a 60×/numerical aperture (NA) 1.40 plan apochromat objective lens and a charge-coupled device camera (CoolSnapHQ; Photometrics, Tucson, AZ) and controlled by MetaMorph Imaging software (Molecular Devices, Sunnyvale, CA). Pictures were analyzed using MetaMorph and Adobe Photoshop (Adobe Systems, Mountain View, CA) software. Confocal microscopy was performed with a Leica TCS SP2 SE confocal scanner in conjunction with a Leica DM6000 B upright scope (with attached Leica HCX PL apochromatic 63×/NA 1.4 objective) and a Leica CTR 6000 confocal control box. This setup was controlled by Leica Control software (Leica Microsystems, Heidelberg, Germany). Images taken represent single slices of a confocal Z-stack at 63×.
RESULTS
Rab9A, Rab6A, and Arl1 GTPases interact with the C-terminal 110 amino acids of GCC185 (Reddy et al., 2006; Burguete et al., 2008). We used a yeast two-hybrid approach to determine whether Rab proteins might also interact with other domains within the full-length, 1684-residue GCC185 protein. Secondary structure prediction algorithms revealed five distinct regions with high coiled-coil probability along the length of GCC185 (Figure 1A). This analysis guided the construction of shortened GCC185 constructs spanning the length of the protein (CC1, CC2, CC3, ΔCC123, and C343). A library of 57 human Rab proteins (all GTP hydrolysis deficient) representing ∼88% of the full complement of human Rabs (∼65 excluding nonprenylated Ran related members; Colicelli, 2004) was screened against each of the GCC185 constructs in a yeast two-hybrid assay. As summarized in Figure 1A, as many as 14 different Rab GTPases showed binding to different regions of GCC185 by this method (Supplemental Figure 1). The screen for binding to GCC185 fragment C343 is shown in Figure 1B. As expected, both Rab6A and Rab9A showed robust interaction with the C343 construct, in agreement with previous work that identified Rab6 and Rab9 binding sites within the C-terminal most 110 amino acids (Figure 1B). In addition, C343 interacted with four additional Golgi-associated Rabs: Rab6B, Rab9B (Yoshimura et al., 2007), Rab30, and Rab33B (Table 1). This, coupled with the discrete and distinct Rab interactions for each coiled-coil truncation construct, suggested that the observed interactions were likely significant.
Figure 1.
Yeast two-hybrid analysis of Rab GTPase binding to distinct coiled-coils in GCC185. (A) Top, secondary structure prediction for GCC185 using Paircoils software. Residue number is shown at bottom. Middle, schematic representation of predicted coiled-coils: CC1, CC2, CC3, and fragments representing the remainder of the protein (ΔCC123, C343, and C110). Bottom, summary of Rab binding to each of the domains as determined by yeast two-hybrid analysis. Rab9A is asterisked because it was variably detected by two hybrid; biochemical experiments shown below confirm its binding to CC3. (B) Yeast two-hybrid data for the construct C343. Each box shows five colonies streaked on selective media; numbers represent activated versions of the human Rab proteins.
Table 1.
GCC185 binding partners: localization and function
| Rab protein | Localization | Function | References |
|---|---|---|---|
| Rab1A, Rab1B | Golgi (see Yoshimura et al., 2007) | ER-to-Golgi and intra-Golgi transport; Golgi maintenance | Allan et al., (2000); Moyer et al., (2001); Haas et al., (2007) |
| Rab2A, Rab2B | Golgi (see Yoshimura et al., 2007) | ER-to-Golgi transport and Golgi structure | Short et al., (2001); Tisdale et al., (1992) |
| Rab6A, Rab6B | Golgi | Retrograde transport to the ER and GCC185 localization at the TGN | Opdam et al., (2000); Echard et al., (2000); Mallard et al., (2002); Burguete et al., (2008) |
| Rab9A | Perinuclear late endosomes, transport vesicles | Late endosome-to-TGN transport | Lombardi et al., (1993); Barbero et al., (2002) |
| Rab9B | Golgi | Unknown | Yoshimura et al., (2007); our unpublished data |
| Rab15 | Perinuclear recycling endosomes | Not precisely known; traffic modulation | Strick and Elferink, (2005); Zuk and Elferink, (2000) |
| Rab27B | Golgi (see Yoshimura et al., 2007) | Regulated secretion | Zhao et al., (2002); Tolmachova et al., (2007); Gomi et al., (2007) |
| Rab30 | Golgi | Unknown | Yoshimura et al., (2007) |
| Rab33B | Golgi | Retrograde transport to the ER | Zheng et al., (1998); Valsdottir et al., (2001) |
| Rab35 | Plasma membrane, perinuclear endosomes | Not precisely known; traffic modulation, cytokinesis | Kouranti et al., (2006); Yoshimura et al., (2007) |
| Rab36 | Unknown | Unknown |
It is of interest to note that yeast two-hybrid screens of full-length GCC185 yielded strong interaction with Rabs1A, 1B, and 35; weak binding was detected for Rabs9B and 6B (data not shown). Most of these Rabs interact with GCC185's first coiled-coil (Figure 1A). We consider it likely that the GAL4 activation domain at GCC185's N terminus cannot be brought close enough to the Gal4 DNA binding domain, when Rab partners interact at too great a distance from GCC185's N terminus. Thus, screens of individual coiled-coils provided additional information regarding possible Rab interactions.
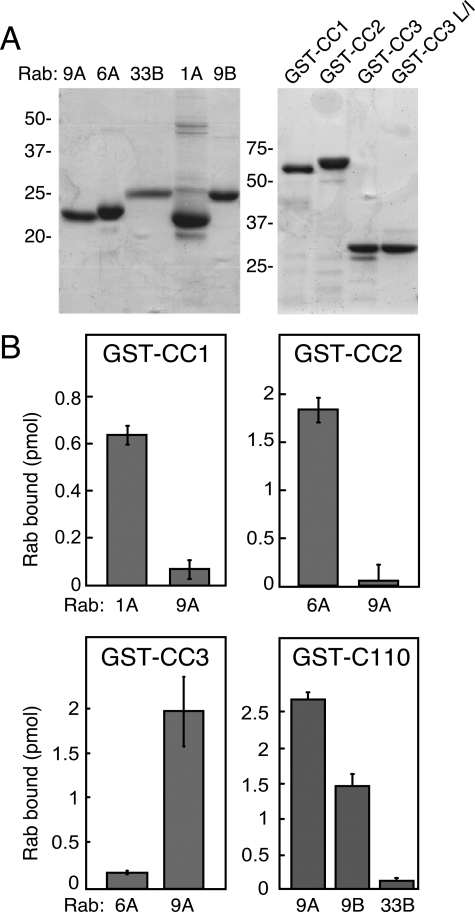
Several of the Rab interactions discovered by yeast two hybrid (Figure 1) were verified independently using purified proteins and an in vitro binding assay. For this purpose, we purified untagged Rab9A, Rab9B, Rab1A, and His-tagged Rab6A and Rab33B, as well as GST fused to GCC185 CC1, CC2, CC3, and C110 (Figure 2A). Rab proteins were preloaded with [35S]GTPγS, and binding of Rabs to glutathione Sepharose-immobilized GCC185 constructs was tested. As shown in Figure 2B, almost all of the yeast two-hybrid interactions that we tested could be verified using purified components. For example, CC1 showed significant binding to Rab1A but poor binding to Rab9A; CC2 bound Rab6A but not Rab9A; in contrast, CC3 bound Rab9A in strong preference to Rab6A. Finally, the C-terminal 110 residue fragment bound Rab9A as we have reported previously, in addition to Rab9B, but not Rab33B. Because Rab33B was unable to bind C110, we infer that its binding site lies within the region upstream of C110, but downstream of the C343 N terminus. Unfortunately, it was not possible to obtain soluble C343 protein for in vitro binding studies.
Figure 2.
Rab GTPase binding to purified GCC185 coiled-coils. (A) Coomassie stained SDS-polyacrylamide gel electrophoresis of untagged Rab proteins and GST-coiled-coil domains used in this study. Molecular mass is shown to the left. (B) Solution binding of [35S]GTPγS-loaded Rab proteins to purified coiled-coil constructs. Reactions contained 3 μM coiled-coil and 0.1 μM active, nucleotide-loaded Rab protein. Products were collected on glutathione Sepharose and determined by scintillation counting. Binding to GST alone was subtracted; error bars represent SE of the mean for three independent experiments.
The levels of binding observed in our in vitro binding assays (∼1–2 pmol) were comparable with the levels of binding detected for proteins known to interact with C110 at 2–4 μM KD under similar conditions (Burguete et al., 2008). This is a typical affinity for Rab-effector binding. It should also be noted that at higher concentrations of GST-CC3 (>25 μM), some binding of Rab6A was detected, although it was substantially lower than that of Rab9A (data not shown).
Interaction of Rab9A and Rab6A with the C110 region of GCC185 requires a surface-exposed leucine/isoleucine pair that are found separated by one helix turn, on the same face of the C110 coiled-coil (Burguete et al., 2008). A helical wheel plot of GCC185 CC3 revealed a similar leucine/isoleucine motif near the C terminus of this peptide (Figure 3A). We therefore engineered and purified a GST-CC3 mutant protein containing leucine-to-alanine and isoleucine-to-alanine mutations to test the role of these residues in Rab9A binding. As shown in Figure 3B, the predicted, surface-exposed leucine 873 and isoleucine 880 residues were important for Rab9 interaction with CC3: KD estimates for the interaction were ∼8 μM for the wild-type CC3 and ∼29 μM for the mutant CC3. This suggests that Rab9A interacts with two distinct coiled-coils by a similar surface interaction.
Figure 3.

(A) Helical wheel plot of residues near the C terminus of GCC185 CC3 predicts a solvent exposed leucine/isoleucine pair. (B) GCC185 I880A L873A are important for Rab9A binding to coiled-coil 3 (CC3). Binding was carried out as in Figure 2 by using the indicated concentrations of CC3 and 0.1 μM Rab9A.
Rab9A is localized to late endosomes (Lombardi et al., 1993) and to the transport vesicles that carry MPRs from endosomes to the Golgi complex (Barbero et al., 2002). Because GCC185 seems to be a tether for this transport process (Reddy et al., 2006), it made little sense that GCC185 would both attach to transport vesicles (via Rab9) and to its Golgi target (via Rab6 and Arl1) by using only its C terminus. The presence of a second Rab9A binding site near the middle of the protein, in GCC185 CC3, provided a solution to this conundrum and suggested that CC3 might be especially important for MPR transport.
To test this, we used an in vitro transport assay that measures the retrograde transport of cation-dependent (CD) MPRs from late endosomes to the TGN (Itin et al., 1997). Figure 4shows that purified CC3 inhibited transport in vitro. The ability of CC3 to inhibit transport was concentration dependent (Figure 4B) and with an apparent Ki value similar to the affinity of interaction of Rabs with this fragment. As expected, a control, mutated fragment of GCC185 that contains a coiled-coil but cannot bind Rab9A or Rab6A (C110 I/L → A/A) failed to inhibit in vitro transport (Figure 4A). We have shown previously that the wild-type C110 fragment inhibits in vitro transport >60% (Reddy et al., 2006). In contrast, the CC3 L/I → A/A mutant was still able to inhibit transport (although perhaps slightly less well than wild type), implicating other regions of the CC3 domain in this transport step. Alternatively, Rab9A may bind sufficiently well to the mutant protein (Figure 3B) to still result in inhibited transport under these conditions. Nevertheless, together, these data support the importance of GCC185's CC3 in MPR recycling.
Figure 4.

Coiled-coil 3 inhibits in vitro transport of mannose 6-phosphate receptors from late endosomes to the trans-Golgi network. (A) Ninety minute reactions were carried out as described in Reddy et al. (2006) and contained 10 μg of membranes, 0.5 mg/ml cytosol, and where indicated, 4 μM GST-tagged GCC185 coiled-coil domains. Reactions were corrected to the higher salt buffer of the coiled-coil preps added. Shown is a representative of three experiments carried out in triplicate; error bars represent SE of the mean. (B) In vitro transport in reactions containing GST-CC3. Values represent means of at least triplicates; shown are aggregate data from seven independent experiments.
In an attempt to characterize further the functional importance of CC3, we overexpressed this protein fragment and tested whether it interfered with MPR trafficking as monitored by an increase in lysosomal enzyme secretion. Although the protein was observed to be entirely cytosolic upon expression in HeLa, human embryonic kidney 293, or COS-1 cells, we were not able to obtain high enough expression to detect a change in hexosaminidase secretion (data not shown).
Functional Analysis of GCC185 Coiled-Coil Domains 1 and 2
GCC185's TGN localization requires Rab6A and Arl1 GTPases and specifically, Rab6A's precise binding site (I1588, L1595) within the C110 coiled-coil (Burguete et al., 2008). Consistent with these previous observations, a set of GCC185 deletion constructs lacking the first, second, and third coiled-coil domains (ΔCC1, ΔCC12, and ΔCC123) localized normally to the Golgi (Figure 5). As expected, a GCC185 construct lacking the C-terminal 110 amino acids, ΔC110, failed to associate with the Golgi and was completely cytosolic (Figure 5). This confirmed that the first three coiled-coil domains are dispensable for GCC185 Golgi localization. In addition, at least one of the individual coiled-coil domains (CC2) was unable to localize to the Golgi and was predominantly cytosolic (data not shown). Moreover, association of full-length GCC185 with purified Golgi membranes was Rab dependent: removal of Rab proteins from Golgi membranes by pretreatment with GDP dissociation inhibitor protein decreased GCC185's Golgi binding (Supplemental Figure 2). Thus, these data suggest that the multiple, non–C-terminal, Rab binding sites located across the GCC185 protein are not sufficient to mediate stable Golgi association. In addition, the ability of ΔCC1, ΔCC12, and ΔCC123 to show normal localization indicates that these proteins are not grossly misfolded.
Figure 5.
GCC185 coiled-coil deletion constructs containing an intact C terminus are correctly localized to the Golgi complex. Left column, myc-tagged constructs lacking the indicated domains (green); right column, Golgi localization as determined using rabbit anti-βCOP antibodies (red). Alexa488-goat anti mouse and Alexa594 goat anti-rabbit antibodies were used for detection. Bar, 10 μm.
What role do the non–C-terminal, Rab binding domains play for GCC185? To explore whether these domains contribute to Golgi organization, we made use of the striking Golgi fragmentation phenotype observed when GCC185 is depleted from cells by using siRNA (Reddy et al., 2006). GCC185 was depleted from cells by 72-h treatment with fluorescein conjugated siRNA. We then attempted to rescue Golgi fragmentation with plasmids containing different regions of GCC185. These plasmids were constructed so that they would not be targets of the siRNA used (see Materials and Methods) and were expressed at very similar levels as determined by total pixel intensity.
In Figure 6, cells that contained fluorescent, GCC185 siRNA are indicated with an asterisk. As expected, asterisked cells that displayed only green (GM130) fluorescence (and had not received the complementing GCC185 plasmid) showed Golgi fragmentation compared with nonasterisked (nondepleted), green labeled cells (bottom row). Expression of Myc-tagged, full-length GCC185 (red; top row) restored normal Golgi morphology (Figure 6A, top row). In contrast, neither ΔCC1 nor ΔCC12 constructs were capable of reversing the fragmented Golgi phenotype upon siRNA treatment (Figure 6A, middle and bottom rows). Quantitation of these data confirmed the importance of at least CC1 (and possibly also CC2) in contributing to Golgi organization. Similar experiments were carried out using full-length GCC185 that lacked 20 residues in the middle of CC3 as the rescue plasmid. In these cells, Golgi structure seemed restored at the same frequency as seen with wild-type plasmid rescue (data not shown). Therefore, full-length GCC185 is required for proper maintenance of Golgi structure; CC3 residues that contribute to Rab9 binding seem to be dispensable for this aspect of GCC185 function.
Figure 6.
GCC185 coiled-coils one and two are required to rescue Golgi disruption caused by GCC185 depletion. (A) Cells were treated with fluorescein-labeled GCC185 siRNA for 72 h. After 48 h of siRNA treatment, cells were transfected with plasmids encoding the indicated myc-tagged proteins. Golgi localization was determined using mouse anti-GM130 and Cy5-goat anti-mouse antibodies (green); GCC185 rescue construct expression was detected using chicken anti-myc and Cy3-goat anti-chicken antibodies (red). Cells containing siRNA (as determined by detection of fluorescein) are indicated with asterisks. (B) Quantitation of rescue experiments. Golgi fragmentation was scored visually; data represent the average of two experiments in which a total of ≥40 cells were counted for each condition. Error bars represent SE of the mean of two independent experiments. Bar, 10 μm.
Does GCC185 reach across the entire Golgi stack? If GCC185 exists as a fully extended coiled-coil, it would achieve a length of ≈240 nm (1.4 Å/residue), a length that is almost sufficient to stretch across the entire Golgi stack in several cell types (200–300 nm; Rambourg et al., 1979; Clermont et al., 1995; Ladinsky et al., 1999) (Figure 7A). In addition, the complement of GCC185-interacting Rabs includes those localized to the trans (Rab6)-, medial (Rab6, Rab33B), and cis/medial (Rab1)-Golgi cisternae. Although these interactions are not required for localization of GCC185 to the TGN (Figure 5; Burguete et al., 2008), they could provide contacts for maintaining Golgi structure with contacts across the height of the stack (Figure 7C). It is intriguing to note that the binding site for the Rab protein that may be located the farthest away from the TGN (Rab1) is located the farthest away from GCC185's TGN anchoring sites near its C terminus (Figure 1).
Figure 7.
Model for GCC185 function as a tether and Golgi organizer. (A) Dimensions of GCC185 if fully extended, transport vesicles and Golgi stacks. (B) A model for GCC185 in vesicle tethering. In this model, cytosolic GCC185 would first associate with Rab9-positive transport vesicles via GCC185 coiled-coil 3. GCC185 on vesicles brought close to the Golgi by cytoplasmic dynein could then engage Rab6 and Arl1 GTPases at the target membrane. SNARES would then drive fusion. (C) Although GCC185 is potentially long enough to reach across the stack, most molecules do not seem to; for details, see Figure 8 and text.
To explore this possibility further, we carried out confocal microscopy experiments in which we could distinguish the localizations of GCC185's N and C termini, independently. Specifically, we compared the distributions of N-terminally myc-tagged, full-length GCC185, and N-terminally myc-tagged, C-terminal C-110 fragment, in relation to the cis-Golgi markers GM130 (Figure 8) and p115 (data not shown). In both cases, very low levels of overlap were observed in terms of the staining of anti-myc antibodies with the cis-Golgi markers.
Figure 8.
Neither the N or C terminus of GCC185 colocalizes with the cis-Golgi marker GM130. (A) Confocal micrographs of cells either nontransfected (left column) or expressing N-terminally myc-tagged GCC185 (center column) or N-terminally myc tagged C110 (C-terminal) fragment (right column) were labeled for GM130 (green), endogenous GCC185 C terminus (left column only), or myc localization (center and right columns) as described in Materials and Methods. Bar, 10 μm. (B) Manders colocalization coefficients for each data set.
Overlap was quantified in at least 50 different images for each marker by using a colocalization coefficient according to Manders (R; Manders et al., 1993; Zinchuk and Zinchuk, 2008). Visually apparent colocalization yields Manders R values of 0.6–1.0 (Zinchuk and Zinchuk, 2008). As shown in Figure 8B, in the cells studied, labeling of the endogenous cis-Golgi marker GM130 and the TGN marker GCC185,yielded an R value of 0.55, which represents a baseline for two adjacent, distinct compartments in these experiments. The myc-tagged, C-terminal C110 fragment yielded an R value of 0.53; N-terminally myc-tagged, full-length GCC185 yielded an R value of 0.58. Thus, although the trend was slightly upward, we did not detect a significantly greater degree of colocalization of the N terminus of the full-length protein with cis-Golgi markers than that seen with the much shorter, C-terminal construct.
From these data, we conclude that the majority of GCC185 molecules do not extend fully toward the cis-Golgi; rather, they are likely to reach laterally across the stack to stabilize this dynamic compartment and outward to help in vesicle tethering. This does not preclude a small subset of the proteins from reaching all the way from the TGN to the cis-Golgi. Given the pancake stack-structure of the Golgi (Rambourg et al., 1979; Clermont et al., 1995; Ladinsky et al., 1999), our findings are consistent with most GCC185 molecules facing outward from the bottom of the stack, rather than being localized to the rims of the stack, where they would have the potential to reach upward.
DISCUSSION
We have shown here that GCC185 contains at least five and likely six distinct binding sites for multiple Rab proteins that are distributed along its entire length. Yeast two-hybrid screening revealed the ability of 14 different Rabs to bind to these sites; direct, in vitro binding studies confirmed their existence and documented binding for Rabs1A, 6A, 9A, and 9B. In previous work, we showed that Rab6 binds to a C-terminal coiled-coil via a pair of surface exposed, leucine, and isoleucine residues (Burguete et al., 2008). A similar mode of interaction was revealed for Rab9A binding to GCC185's coiled-coil 3: mutation of this motif resulted in significantly lowered Rab9A affinity. GCC185's coiled-coil 3 inhibited transport of MPRs from late endosomes to the TGN in vitro, consistent with its importance for GCC185 function. The importance of GCC185's first and second, N-terminal coiled-coil domains was revealed in experiments that tested the ability of truncation constructs to rescue the Golgi fragmentation phenotype that is seen upon depletion of GCC185. Although full-length GCC185 was able to fully rescue Golgi fragmentation in GCC185-depleted cells, constructs lacking the first or first and second coiled-coil domains failed to promote reestablishment of a normal Golgi ribbon. Thus, length matters—and despite a sequence that at first glance, seems to be just a long coiled-coil, GCC185's N-terminal domains play important and unexpected functional roles for this protein in live cells.
As summarized in Table 1, the overwhelming majority of Rab proteins identified in this study as GCC185 binding partners are Golgi-localized. Some of the Rabs are cell type specific: for example, Rab27B is present is regulated secretory cells, but localizes to the Golgi when expressed in cultured fibroblasts (Yoshimura et al., 2007). Why might GCC185 interact with so many Golgi-localized Rab proteins? We favor a model in which GCC185 binding to multiple Rab proteins contributes to its role in maintaining normal Golgi stack morphology. GCC185 is specifically localized to the TGN (Derby et al., 2004). It is hard to imagine why its depletion would lead to the generation of mini-stacks (Reddy et al., 2006; Derby et al., 2007), unless GCC185 could link ministacks together laterally, or contribute to stack stability, vertically. Simple de-polymerization of microtubules is sufficient to generate Golgi mini-stacks, which suggests that the steady state Golgi structure is normally subject to dynamic, lateral fusion and fission, with the fusion process requiring intact microtubules. Under normal conditions, proteins such as GCC185 could reach across laterally, from one mini-stack domain to another, to promote Golgi ribbon stability. Indeed, GCC185's connection to the microtubule based cytoskeleton via CLASP proteins (Efimov et al., 2007) may provide cytoskeletal coupling for such linking events.
Given GCC185's role in tethering at the TGN, it is important that the protein be specifically localized to that compartment. It is thus not surprising that the C terminus dictated TGN localization (Luke et al., 2003; Burguete et al., 2008) (Figure 5) and that distal Rab binding domains were not sufficient to localize the ΔC110 protein elsewhere on the Golgi stack. If GCC185 could bind tightly to Rabs located on other Golgi compartments, those interactions might interfere with its ability to mediate vesicle docking at the TGN. Instead, multiple, weak interactions let GCC185 interact reversibly with the Golgi surface, and stabilize this dynamic compartment. Multiple weak interactions could also retain transport vesicles destined for distinct Golgi compartments within the vicinity of the Golgi stack.
A few Rab proteins that bound GCC185 by yeast two hybrid are not Golgi residents. This includes Rab9A, which is present on inbound transport vesicles (Barbero et al., 2002), and Rab15 and Rab35 that also seem to be present on perinuclear endosomal compartments (Strick and Elferink, 2005; Kouranti et al., 2006). It is possible that these Rabs participate in a retrograde transport step that also uses GCC185. Alternatively, Rab35 is most closely related to Golgi-bound Rabs 1A and 1B; binding in the two-hybrid system may reflect interaction with homologous portions of the Rab protein. We are most confident of the interactions that could be recapitulated with purified proteins and include Rabs 1A, 6A, 9A, and 9B, a collection that spans the entire Golgi stack. GCC185 is not the first Golgin shown to bind to multiple Rabs. For example, GM130 binds Rabs 1, 2 and 33B (Short et al., 2005). But, the ability of GCC185 to potentially reach out to incoming transport vesicles and to earlier Golgi compartments seems to be a novel feature of this GRIP domain family member.
A Model for Vesicle Tethering at the TGN
In addition to contributing to Golgi structure, GCC185 is also required for vesicle docking at the Golgi (Reddy et al., 2006; Derby et al., 2007). Figure 7B presents our most current working model for this process. GCC185 exists in the cytosol and at the TGN (Luke et al., 2003). We propose that Rab9, present on late endosome-derived transport vesicles, binds cytosolic GCC185 via CC3 in the middle of GCC185 (and/or C110). Transport vesicles carrying Rab9-GCC185 then move along microtubules toward their targets (Itin et al., 1999; Barbero et al., 2002). Once close to the TGN, GCC185's C terminus could bind TGN-localized Rab6, triggering concomitant Arl1 binding (Figure 7B, step 1). The transport vesicle would then be brought into proximity to the TGN membrane surface, either by dynamic mobility of a stably anchored GCC185, a GCC185 conformational change, or directed transport of the vesicle down the length of the GCC185 molecule (Figure 7B, step 2). Fusion of the Rab9-containing transport vesicle would then follow.
Validation of these steps in our model will require determining the length of TGN-associated GCC185 protein, and characterizing in greater detail each of its Rab binding sites. In the future, single vesicle analyses of vesicle tethering at the TGN should enable us to elucidate the mechanisms by which Rab9-vesicles find and engage their fusion targets.
Supplementary Material
ACKNOWLEDGMENTS
We thank James Godfrey and John King for help with two-hybrid screens and Benedikt Brommer for plasmid construction. This work was supported by grant R01 GM-79322 (to S.R.P.) and fellowships to G.L.H. (NIH T32 GM-007276 and the National Science Foundation) and F.C.B. (NIH T90 DK-070090).
Abbreviations used:
- MPR
mannose 6-phosphate receptor
- TGN
trans-Golgi network.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-07-0740) on October 22, 2008.
REFERENCES
- Aivazian D., Serrano R. L., Pfeffer S. TIP47 is a key effector for Rab9 localization. J. Cell Biol. 2006;173:917–926. doi: 10.1083/jcb.200510010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan B. B., Moyer B. D., Balch W. E. Rab1 recruitment of p115 into a cis-SNARE complex: programming budding COPII vesicles for fusion. Science. 2000;289:444–448. doi: 10.1126/science.289.5478.444. [DOI] [PubMed] [Google Scholar]
- Bai C., Elledge S. J. Gene identification using the yeast two-hybrid system. Methods Enzymol. 1996;273:331–347. doi: 10.1016/s0076-6879(96)73029-x. [DOI] [PubMed] [Google Scholar]
- Barbero P., Bittova L., Pfeffer S. R. Visualization of Rab9-mediated vesicle transport from endosomes to the trans Golgi in living cells. J. Cell Biol. 2002;156:511–518. doi: 10.1083/jcb.200109030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burguete A. S., Fenn T. D., Brunger A. T., Pfeffer S. R. Rab and Arl GTPase family members cooperate in the localization of the golgin GCC185. Cell. 2008;132:286–298. doi: 10.1016/j.cell.2007.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clermont Y., Rambourg A., Hermo L. Trans-Golgi network (TGN) of different cell types: three-dimensional structural characteristics and variability. Anat. Rec. 1995;242:289–301. doi: 10.1002/ar.1092420302. [DOI] [PubMed] [Google Scholar]
- Colicelli J. Human RAS superfamily proteins and related GTPases. Sci. STKE. 2004;250:RE13. doi: 10.1126/stke.2502004re13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derby M. C., Lieu Z. Z., Brown D., Stow J. L., Goud B., Gleeson P. A. The trans-Golgi network Golgin, GCC185, is required for endosome-to-Golgi transport and maintenance of Golgi structure. Traffic. 2007;8:758–773. doi: 10.1111/j.1600-0854.2007.00563.x. [DOI] [PubMed] [Google Scholar]
- Derby M. C., van Vliet C., Brown D., Luke M. R., Lu L., Hong W., Stow J. L., Gleeson P. A. Mammalian GRIP domain proteins differ in their membrane binding properties and are recruited to distinct domains of the TGN. J. Cell Sci. 2004;117:5865–5874. doi: 10.1242/jcs.01497. [DOI] [PubMed] [Google Scholar]
- Diao A., Rahman D., Pappin D. J., Lucocq J., Lowe M. The coiled-coil membrane protein Golgin-84 is a novel rab effector required for Golgi ribbon formation. J. Cell Biol. 2003;160:201–212. doi: 10.1083/jcb.200207045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echard A., Opdam F. J., de Leeuw H. J., Jollivet F., Savelkoul P., Hendriks W., Voorberg J., Goud B., Fransen J. A. Alternative splicing of the human Rab6A gene generates two close but functionally different isoforms. Mol. Biol. Cell. 2000;11:3819–3833. doi: 10.1091/mbc.11.11.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov A., et al. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev. Cell. 2007;12:917–930. doi: 10.1016/j.devcel.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar M. G., Palade G. E. The Golgi apparatus—100 years of progress and controversy. Trends Cell Biol. 1998;8:2–10. doi: 10.1016/S0962-8924(97)01187-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein T. N., Linstedt A. D. GRASP55 regulates Golgi ribbon formation. Mol. Biol. Cell. 2008;19:2696–2707. doi: 10.1091/mbc.E07-11-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridmann-Sirkis Y., Siniossoglou S., Pelham H.R.B. TMF is a golgin that binds Rab6 and influences Golgi morphology. BMC Cell Biol. 2004;5:18–27. doi: 10.1186/1471-2121-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E., Haas A. K., Spooner R. A., Yoshimura S., Lord J. M., Barr F. A. Specific Rab GTPase-activating proteins define the Shiga toxin and epidermal growth factor uptake pathways. J. Cell Biol. 2007;177:1133–1143. doi: 10.1083/jcb.200612068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley I. G., Espinosa E., Pfeffer S. R. A syntaxin 10-SNARE complex distinguishes two distinct transport routes from endosomes to the trans-Golgi in human cells. J. Cell Biol. 2008;180:159–172. doi: 10.1083/jcb.200707136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomi H., Mori K., Itohara S., Izumi T. Rab27b is expressed in a wide range of exocytic cells and involved in the delivery of secretory granules near the plasma membrane. Mol. Biol. Cell. 2007;18:4377–4386. doi: 10.1091/mbc.E07-05-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas A. K., Yoshimura S.-I., Stephens D. J., Preisinger C., Fuchs E., Barr F. A. Analysis of GTPase-activating proteins: Rab1 and Rab43 are key Rabs required to maintain a functional Golgi complex in human cells. J. Cell Sci. 2007;120:2997–3010. doi: 10.1242/jcs.014225. [DOI] [PubMed] [Google Scholar]
- Itin C., Rancaño C., Nakajima Y., Pfeffer S. R. A novel assay reveals a role for alpha-SNAP in mannose 6-phosphate receptor transport from endosomes to the TGN. J. Biol. Chem. 1997;272:27737–27744. doi: 10.1074/jbc.272.44.27737. [DOI] [PubMed] [Google Scholar]
- Itin C., Ulitzur N., Muhlbauer B., Pfeffer S. R. Mapmodulin, cytoplasmic dynein, and microtubules enhance the transport of mannose 6-phosphate receptors from endosomes to the transgolgi network. Mol. Biol. Cell. 1999;10:2191–2197. doi: 10.1091/mbc.10.7.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranti I., Sachse M., Arouche N., Goud B., Echard A. Rab35 regulates an endocytic recycling pathway essential for the terminal steps of cytokinesis. Curr. Biol. 2006;16:1719–1725. doi: 10.1016/j.cub.2006.07.020. [DOI] [PubMed] [Google Scholar]
- Ladinsky M. S., Mastronarde D. N., McIntosh J. R., Howell K. E., Staehelin L. A. Golgi structure in three dimensions: functional insights from the normal rat kidney cell. J. Cell Biol. 1999;144:1135–1149. doi: 10.1083/jcb.144.6.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi D., Soldati T., Riederer M. A., Goda Y., Zerial M., Pfeffer S. R. Rab9 functions in transport between late endosomes and the trans Golgi network in vitro. EMBO J. 1993;12:677–682. doi: 10.1002/j.1460-2075.1993.tb05701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Hong W. Interaction of Arl1-GTP with GRIP domains recruits auto- antigens Golgin-97 and Golgin-245/p230 onto the Golgi. Mol. Biol. Cell. 2003;14:3767–3781. doi: 10.1091/mbc.E03-01-0864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Tai G., Wu M., Song H., Hong W. Multilayer interactions determine the Golgi localization of GRIP golgins. Traffic. 2006;7:1399–1407. doi: 10.1111/j.1600-0854.2006.00473.x. [DOI] [PubMed] [Google Scholar]
- Luke M. R., Kjer-Nielsen L., Brown D. L., Stow J. L., Gleeson P. A. GRIP domain-mediated targeting of two new coiled-coil proteins, GCC88 and GCC185, to subcompartments of the trans-Golgi network. J. Biol. Chem. 2003;278:4216–4226. doi: 10.1074/jbc.M210387200. [DOI] [PubMed] [Google Scholar]
- Mallard F., Tang B. L., Galli T., Tenza D., Saint-Pol A., Yue X., Antony C., Hong W., Goud B., Johannes L. Early/recycling endosomes-to-TGN transport involves two SNARE complexes and a Rab6 isoform. J. Cell Biol. 2002;156:653–664. doi: 10.1083/jcb.200110081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manders E.M.M., Verbeek F. J., Aten J. A. Measurement of co-localization of objects in dual-colour confocal images. J. Microsc. 1993;169:375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- Moyer B. D., Allan B. B., Balch W. E. Rab1 interaction with a GM130 effector complex regulates COPII vesicle cis-Golgi tethering. Traffic. 2001;2:268–276. doi: 10.1034/j.1600-0854.2001.1o007.x. [DOI] [PubMed] [Google Scholar]
- Opdam F. J., Echard A., Croes H. J., van den Hurk J. A., van de Vorstenbosch R. A., Ginsel L. A., Goud B., Fransen J. A. The small GTPase Rab6B, a novel Rab6 subfamily member, is cell-type specifically expressed and localised to the Golgi apparatus. J. Cell Sci. 2000;113:2725–2735. doi: 10.1242/jcs.113.15.2725. [DOI] [PubMed] [Google Scholar]
- Panic B., Whyte J. R., Munro S. The ARF-like GTPases Arl1p and Arl3p act in a pathway that interacts with vesicle-tethering factors at the Golgi apparatus. Curr. Biol. 2003;13:405–410. doi: 10.1016/s0960-9822(03)00091-5. [DOI] [PubMed] [Google Scholar]
- Pepperkok R., Scheel J., Horstmann H., Hauri H. P., Griffiths G., Kreis T. E. Beta-COP is essential for biosynthetic membrane transport from the endoplasmic reticulum to the Golgi complex in vivo. Cell. 1993;74:71–82. doi: 10.1016/0092-8674(93)90295-2. [DOI] [PubMed] [Google Scholar]
- Puthenveedu M. A., Bachert C., Puri S., Lanni F., Linstedt A. D. GM130 and GRASP65-dependent lateral cisternal fusion confers uniform Golgi enzyme distribution. Nat. Cell Biol. 2006;8:238–248. doi: 10.1038/ncb1366. [DOI] [PubMed] [Google Scholar]
- Rambourg A., Clermont Y., Hermo L. Three dimensional architecture of the Golgi apparatus in Sertoli cells of the rat. Am. J. Anat. 1979;154:455–476. doi: 10.1002/aja.1001540402. [DOI] [PubMed] [Google Scholar]
- Ramirez I. B., de Graffenried C. L., Ebersberger I., Yelinek J., He C. Y., Price A., Warren G. TbG63, a golgin involved in Golgi architecture in Trypanosoma brucei. J. Cell Sci. 2008;121:1538–1546. doi: 10.1242/jcs.014324. [DOI] [PubMed] [Google Scholar]
- Reddy J. V., Burguete A. S., Sridevi K., Ganley I. G., Nottingham R. M., Pfeffer S. R. A functional role for the GCC185 golgin in mannose 6-phosphate receptor trafficking. Mol. Biol. Cell. 2006;17:4353–4363. doi: 10.1091/mbc.E06-02-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty S. R., Shin M. E., Yoshino A., Marks M. S., Burd C. G. Golgi recruitment of GRIP domain proteins by Arf-like GTPase 1 is regulated by Arf-like GTPase 3. Curr. Biol. 2003;13:401–404. doi: 10.1016/s0960-9822(03)00089-7. [DOI] [PubMed] [Google Scholar]
- Short B., Haas A., Barr F. A. Golgins and GTPases, giving identity and structure to the Golgi apparatus. Biochim. Biophys. Acta. 2005;1744:383–395. doi: 10.1016/j.bbamcr.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Short B., Preisinger C., Körner R., Kopajtich R., Byron O., Barr F. A. A GRASP55-rab2 effector complex linking Golgi structure to membrane traffic. J. Cell Biol. 2001;155:877–883. doi: 10.1083/jcb.200108079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strick D. J., Elferink L. A. Rab15 effector protein: a novel protein for receptor recycling from the endocytic recycling compartment. Mol. Biol. Cell. 2005;16:5699–5709. doi: 10.1091/mbc.E05-03-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztul E., Lupashin V. Role of tethering factors in secretory membrane traffic. Am. J. Physiol. Cell Physiol. 2006;290:C11–C26. doi: 10.1152/ajpcell.00293.2005. [DOI] [PubMed] [Google Scholar]
- Tisdale E. J., Bourne J. R., Khosravi-Far R., Der C. J., Balch W. E. GTP-binding mutants of rab1 and rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the Golgi complex. J. Cell Biol. 1992;119:749–761. doi: 10.1083/jcb.119.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolmachova T., Abrink M., Futter C. E., Authi K. S., Seabra M. C. Rab27b regulates number and secretion of platelet dense granules. Proc. Natl. Acad. Sci. USA. 2007;104:5872–5877. doi: 10.1073/pnas.0609879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valsdottir R., Hashimoto H., Ashman K., Koda T., Storrie B., Nilsson T. Identification of rabaptin-5, rabex-5, and GM130 as putative effectors of rab33b, a regulator of retrograde traffic between the Golgi apparatus and ER. FEBS Lett. 2001;508:201–209. doi: 10.1016/s0014-5793(01)02993-3. [DOI] [PubMed] [Google Scholar]
- Vasile E., Perez T., Nakamura N., Krieger M. Structural integrity of the Golgi is temperature sensitive in conditional-lethal mutants with no detectable GM130. Traffic. 2003;4:254–272. doi: 10.1034/j.1600-0854.2003.00080.x. [DOI] [PubMed] [Google Scholar]
- Warren G., Davoust J., Cockcroft A. Recycling of transferrin receptors in A431 cells is inhibited during mitosis. EMBO J. 1984;3:2217–2225. doi: 10.1002/j.1460-2075.1984.tb02119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S., Egerer J., Fuchs E., Haas A. K., Barr F. A. Functional dissection of Rab GTPases involved in primary cilium formation. J. Cell Biol. 2007;178:363–369. doi: 10.1083/jcb.200703047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino A., Setty S. R., Poynton C., Whiteman E. L., Saint-Pol A., Burd C. G., Johannes L., Holzbaur E. L., Koval M., McCaffery J. M., Marks M. S. tGolgin-1 (p230, golgin-245) modulates Shiga-toxin transport to the Golgi and Golgi motility towards the microtubule-organizing centre. J. Cell Sci. 2005;118:2279–2293. doi: 10.1242/jcs.02358. [DOI] [PubMed] [Google Scholar]
- Zhao S., Torii S., Yokota-Hashimoto H., Takeuchi T., Izumi T. Involvement of Rab27b in the regulated secretion of pituitary hormones. Endocrinology. 2002;143:1817–1824. doi: 10.1210/endo.143.5.8823. [DOI] [PubMed] [Google Scholar]
- Zheng J. Y., Koda T., Fujiwara T., Kishi M., Ikehara Y., Kakinuma M. A novel Rab GTPase, Rab33B, is ubiquitously expressed and localized to the medial Golgi cisternae. J. Cell Sci. 1998;111:1061–1069. doi: 10.1242/jcs.111.8.1061. [DOI] [PubMed] [Google Scholar]
- Zinchuk V., Zinchuk O. Quantitative Colocalization Analysis of Confocal Fluorescence Microscopy Images. Curr. Protoc. Cell Biol. 2008;39:4.19.1–4.19.16. doi: 10.1002/0471143030.cb0419s39. [DOI] [PubMed] [Google Scholar]
- Zolov S. N., Lupashin V. V. Cog3p depletion blocks vesicle-mediated Golgi retrograde trafficking in HeLa cells. J. Cell Biol. 2005;168:747–759. doi: 10.1083/jcb.200412003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk P. A., Elferink L. A. Rab15 differentially regulates early endocytic trafficking. J. Biol. Chem. 2000;275:26754–26764. doi: 10.1074/jbc.M000344200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.