Abstract
Cell adhesion molecules such as cadherins alternate their expression throughout cranial neural crest (CNC) development, yet our understanding of the role of these molecules during CNC migration remains incomplete. The “mesenchymal” cadherin-11 is expressed in the CNC during migration yet prevents migration when overexpressed in the embryo, suggesting that a defined level of cadherin-11–mediated cell adhesion is required for migration. Here we show that members of the meltrin subfamily of ADAM metalloproteases cleave the extracellular domain of cadherin-11 during CNC migration. We show that a fragment corresponding to the putative shed form of cadherin-11 retains biological activity by promoting CNC migration in vivo, in a non-cell–autonomous manner. Additionally, cleavage of cadherin-11 does not affect binding to β-catenin and downstream signaling events. We propose that ADAM cleavage of cadherin-11 promotes migration by modifying its ability to support cell–cell adhesion while maintaining the membrane-bound pool of β-catenin associated with the cadherin-11 cytoplasmic domain.
INTRODUCTION
The neural crest is a transient population of cells present in all vertebrate embryos. Induced at the border between the neural and nonneural ectoderm, these cells migrate from the dorsal part of the embryo to more ventral locations where they participate in the formation of muscle, cartilage, melanocytes, and ganglia of the peripheral nervous system (PNS; Dupin et al., 2006; Knight and Schilling, 2006; Sandell and Trainor, 2006; Sauka-Spengler and Bronner-Fraser, 2006; Harris and Erickson, 2007). Neural crest cells are separated in two distinct populations depending on their position on the anterior/posterior axis. The most anterior are called cranial neural crest (CNC), responsible for the facial structures, whereas the posterior are the trunk neural crest mostly contributing to the PNS and the melanocytes.
Neural crest cell migration requires tight control over cell adhesion molecules such as integrins and cadherins. To date, there have been four different Cadherin molecules implicated in neural crest migration among the mouse, chick, and Xenopus models (Akitaya and Bronner-Fraser, 1992; Kimura et al., 1995; Nakagawa and Takeichi, 1995; Inoue et al., 1997; Hadeball et al., 1998; Vallin et al., 1998; Borchers et al., 2001; Coles et al., 2007). These four molecules can be divided into two groups in relation to their expression during migration. The first group consisting of N-cadherin and cadherin-6 (also Cad-6A) are both expressed at the beginning of migration, and then their mRNA and protein expression is quickly down-regulated (Akitaya and Bronner-Fraser, 1992; Nakagawa and Takeichi, 1995). The second group comprising cadherin-7 and -11 is continually expressed throughout neural crest cell migration (Kimura et al., 1995; Nakagawa and Takeichi, 1995; Hadeball et al., 1998; Vallin et al., 1998). Not surprisingly, overexpression of any of these four cell adhesion molecules in at least one of the above model organisms blocks neural crest migration (Nakagawa and Takeichi, 1995, 1998; Dufour et al., 1999; Borchers et al., 2001; Coles et al., 2007; Shoval et al., 2007). However, it is likely that there must be unique properties among these cadherins that make one group more conducive to cell migration than the other.
To further understand the role of cadherins in the neural crest, we have examined the regulation of cadherin-11 during CNC migration in Xenopus laevis. In the Xenopus embryo, N-cadherin is replaced by cadherin-11 expression during CNC migration. We suspected that a protease regulates cadherin-11 levels during CNC migration as an extracellular cleavage product of cadherin-11 had been previously detected in tissue culture cells (Kawaguchi et al., 1999).
Among the proteases expressed in the embryo, a member of the ADAM metalloprotease family was a likely candidate for the regulation of cadherin-11 during this process. ADAMs and cadherins have previously been shown to interact in various experimental systems. For example, ADAM15 and VE-cadherin colocalize to adherens junctions and increasing the expression of VE-cadherin results in a corresponding increase in ADAM15 (Ham et al., 2002). Additionally, ADAM10 activity can modify cell adhesion via the cleavage of both N- and E-cadherin (Maretzky et al., 2005). ADAM10 was also found to play a role in the global down-regulation of N-cadherin at the onset of trunk neural crest migration in chick embryos (Shoval et al., 2007). Yet, although ADAM10 is expressed dorsally in Xenopus, it is not enriched in the CNC. On the other hand, another ADAM, ADAM13, is specifically expressed in the Xenopus CNC during migration. Moreover, the proteolytic activity of ADAM13 was previously shown to play a vital role in the migration of this tissue (Alfandari et al., 2001). Our findings show that cadherin-11 is cleaved during Xenopus CNC migration, and that ADAMs from the meltrin subfamily are responsible for this event. We propose that cadherin-11 cleavage is unique when compared with that of other cadherins in the neural crest and provides further insight into the differential roles of cadherins during morphogenesis.
MATERIALS AND METHODS
Eggs and Embryos
Eggs were obtained from X. laevis, fertilized, and cultured as described previously (Alfandari et al., 1997). Embryos were staged according to Nieuwkoop and Faber (1967). UV irradiation and LiCl treatments were performed as described (Pickard and Damjanovski, 2004).
Cell Culture
Cos cells were cultured in RPMI media complemented with Pen/Strep, l-glut, sodium pyruvate, and FBS (10 U/ml, 2 mM; 0.11 mg/ml, 10%; Hyclone, South Logan, UT). Transfections were performed using Fugene 6 reagent (Roche, Basel, Switzerland) following the manufacturer's instructions.
DNA Constructs
The cloning of Xenopus ADAM9, 10, and 13 and the E/A mutants have been previously described (Cai et al., 1998; Alfandari et al., 2001; Smith et al., 2002). Monomeric red fluorescent protein (mRFP) in CS2 was a generous gift from Dr. Jim Smith (Gurdon Institute, Cambridge, United Kingdom). ADAM19 was cloned by homologous PCR using sequences from mouse, chick, and Xenopus tropicalis. 5′ and 3′ ends were obtained by RACE PCR using the generacer kit (Invitrogen, Carlsbad, CA). All full-length ADAM were cloned into the pCS2 vector for expression. The ADAM9-E/A construct was produced using the QuickChange Mutagenesis Kit (Stratagene, La Jolla, CA). The Xenopus full-length cadherin-11 in pcDNA3 was a gift from Dr. Doris Wedlich (Universität Ulm, Ulm, Germany) and was recloned into pCS2. The EC1-3 construct was made by introducing a myc- tag and stop codon between the EC3 and EC4 sequences of cadherin-11. The ΔEC1-3 construct was made by deletion using all around PCR with Pyrococcus furiosus DNA polymerase. All constructs were sequenced and tested for expression using the appropriate antibodies in both Cos-7 cells and embryos.
Morpholino Oligonucleotides
Morpholino oligonucleotides were directed against the 5′ untranslated region of ADAM9, 13, and 19 diluted in water at 5 mg/ml (Gene Tools, Philomath, OR). Ten nanograms of morpholino (MO) was injected into each embryo at the one-cell stage, or 1 ng was injected at the 16-cell stage. The MO sequences directed against the ADAM9, 13, and 19 are listed in Table 1.
Table 1.
MO sequences directed against ADAM9, 13, and 19
| Target | Morpholino sequence |
|---|---|
| xADAM13 | GTCCCAGCCGACCCTCC |
| xADAM9 | GGTGTCCCCTCATCTAC |
| xADAM19 | GAGTCCTGTAGCTCCTT |
Cadherin-11 Antibody Production and Screening
A His-tagged fusion protein (pET 30 vector; Novagen, San Diego, CA) encoding 157 C-terminal amino acids of the cytoplasmic domain of cadherin-11 was purified using standard methods. Fusion protein, 100–300 μg, was combined with Freund's adjuvant and injected intraperitoneally into BALB/c mice. Hybridoma fusion protocol was performed using standard methods (Harlow and Lane, 1988). Hybridomas were screened by ELISA, Western blot, immunofluorescence, and immunoprecipitation to test immunoreactivity to endogenous cadherin-11 and minimal cross-reactivity to N- and C-cadherin. The mAb 1B4 showed a very low affinity for overexpressed N-cadherin and no detectable affinity for C-cadherin. The same fusion protein was used to immunize rabbits from which specific immunoglobulin were purified by affinity on the antigen according to Alfandari et al. (1997).
Antibodies
Rabbit 6615F affinity-purified polyclonal antibody (pAb) to ADAM13 is used at a 0.1 mg/ml concentration in Western blot (Alfandari et al., 1997). The rabbit β-catenin pAb (Abcam, Cambridge, MA) was used at a 1:2000 dilution. Rabbit anti-ADAM9 was described earlier (Cai et al., 1998). Rabbit anti-ADAM19 was produced against a fusion protein to the ADAM19 cytoplasmic domain and affinity-purified before use. As loading controls, antibody to the β1 integrin subunit (mAb 8C8) and PACSIN2 (mAb 3D8) were used (Gawantka et al., 1994; Cousin et al., 2000). To perform Western blot after immunoprecipitation, we biotinylated mAb 1B4 while bound to the antigen using NHS-LC Biotin (Pierce, Rockford, IL). 9E10 mAB and α-mouse-FITC (1:200) were used to detect EC1-3-mt via immunofluorescence. Photographs were taken using a Zeiss Axiovert 200M inverted microscope (Thornwood, NY) equipped with a Hamamatsu Orca camera (Bridgewater, NJ).
Microinjection Experiments
Transcription reactions and injections were performed as previously described in (Cousin et al., 2000). The injection volume was determined by capillary calibration of the injection needle. We injected 5 nl at the one- to eight-cell stages and 2.5 nl at 16- or 32-cell stages.
Whole Mount In Situ Hybridization
Whole mount in situ hybridization was performed as previously described (Harland, 1991). Diogoxigenin-rUTP–labeled transcripts were synthesized in vitro from Xenopus Sox10 and Twist plasmids. Synthetic mRNA encoding β-galactosidase was also included in the microinjections of embryos that were analyzed via in situ hybridization. The x-gal reaction was performed as in Smith and Harland (1991) to indicate the site of injection. Embryos that were expressing β-galactosidase in the posterior region were excluded from our statistical analysis. Images were recorded using a Nikon D50 camera on a Nikon SMZ1500 dissecting scope (Melville, NY).
Protein Extraction and Analysis
For direct Western blot analysis of transfected Cos cells, each well of a six-well plate was extracted with 200 μl of reducing Laemmli buffer, and 10% of the extract was applied to a SDS-PAGE gel. Immunoprecipitation were carried out exactly as described in Alfandari et al. (1995) using protein G beads (Roche, Indianapolis, IN) and 10 μg of mAb-1B4. Western blot protocol was followed as previously reported (Cousin et al., 2000). Embryo extraction and analysis was performed similar to above but 1× Modified Barth's Saline (MBS) was used instead of 1× TBS in the extraction buffer and washes. Extraction buffer, 20 μl, was used per embryo. Total embryo number for each experiment is noted in the figure legends. Glycoproteins were purified from total protein extract using concanavalin-A agarose beads (Vector Laboratories, Burlingame, CA) as previously described (Alfandari et al., 1997).
Quantitative PCR Analysis
RNA from stage-21 embryos was purified using guanidine isothiocyanate as described in Alfandari et al. (1995). Reverse transcription reactions were performed as in Alfandari et al. (1997). Sequences for xActin, Sox8, xTwist, cyclin-Dl, and c-myc probes are listed in the Table 2. Quantitative PCR (qPCR) reactions and data generation were performed using CYBR Green Premix Ex Taq (Takara, Kyoto, Japan) and the LightCycler system 1.5 (Roche). The 2(−ΔΔCT) method was used for target quantification (Livak and Schmittgen, 2001), where actin was used to normalize for total cDNA quantities.
Table 2.
Sequences for xActin, Sox8, xTwist, cyclin-Dl, and c-myc probes
| Probe | Sequence 5′ |
|---|---|
| Cyclin D1 sense | ATCCCACTGACCGCA |
| Cyclin D1 anti-sense | TCTGATGAAGCGTTGT |
| c-myc sense | ACTGAACGACAGCAT |
| c-myc anto-sense | TGTGCGTCTTCCTCTT |
| xTwist sense | GCCATGTCAGGGAGC |
| xTwist anti-sense | GATTTGGCGAACCTA |
| Sox8 sense | AAGGTCTCTGGTGGC |
| Sox8 anti-sense | CACCGCCACATTTCA |
| Actin sense | AACAGAGAAAAGATC |
| Actin anti-sense | CAAAGTCAAGAGCAA |
RESULTS
Endogenous Cadherin-11 Is Cleaved In Vivo during CNC Migration
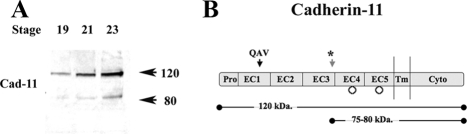
Because a shed form of cadherin-11 had been observed in tissue culture, we predicted that migrating CNC cells could regulate cadherin-11 surface levels by an extracellular cleavage event (Kawaguchi et al., 1999). To investigate this possibility, we produced a mAb directed against the cytoplasmic domain of cadherin-11 and studied its expression and changes in molecular weight during CNC migration in X. laevis embryos. The Western blot analysis in Figure 1A depicts the expression of endogenous cadherin-11 at the beginning (stage 19) and during CNC migration (stages 21 and 23). In these embryos, the amount of total cadherin-11 increases as the crest progresses through migration. Furthermore, we can detect the presence of a cadherin-11 cleavage product of ∼75–80 kDa, which also increases during migration. This cleavage product corresponds in size to the cytoplasmic and transmembrane domains, as well as a portion of the extracellular domain, and retains at least one glycosylation site because it can be purified on concanavalin A-beads. Using the primary amino acid sequence and the putative N-glycosylation sites, we estimate the cleavage site to be between the EC3 and EC4 domain of the cadherin-11 protein (Figure 1B). The timing and sizes of the cadherin-11 fragments suggest that the homophylic binding site in the first Cadherin domain (EC1) is removed during CNC migration, thus decreasing cell–cell interactions.
Figure 1.
Cadherin-11 is cleaved in vivo during cranial neural crest migration. (A) Wild-type embryos were extracted at stage 19, stage 21, and stage 23 representative of the different phases of CNC migration (20 embryos/lane). Western blot analysis for Cad-11 shows an increase in full-length protein (120 kDa) as migration proceeds, as well as the appearance of one 80-kDa cleavage product. (B) Schematic representation of full-length Cad-11. The EC1 domain contains a QAV homophilic binding motif consistent with type II cadherins (Hadeball et al., 1998). The cleavage site (*) is determined by calculating the relative molecular mass of the C-terminal fragment taking into account the N-glycosylation sites. The cytoplasmic region of Cad-11 can bind to β-catenin (Kawaguchi et al., 1999).
Cadherin-11 Can Be Cleaved In Vitro by ADAM9 and 13
Our next objective was to find which protease is responsible for cleaving Cadherin-11 during CNC migration. Because ADAM10 was previously shown to cleave members of the cadherin superfamily, we first investigated if cadherin-11 could also be processed by an ADAM. Cos-7 cells were transfected with cadherin-11 and various ADAM constructs. We selected ADAMs that have been previously shown to be expressed in an overlapping pattern with cadherin-11, namely ADAM9, 10, 13, and 19. Western blot analysis on the cell extract revealed the presence of the 80-kDa cadherin-11 fragments in the ADAM9 and 13 cotransfected cells, but not with their proteolytic inactive E/A mutants (Supplemental Figure S1). This fragment was not present in the ADAM10 and 19 cotransfections.
Binding of Endogenous ADAM13 and Cadherin-11 Occurs during CNC Migration and Corresponds to Cadherin-11 Cleavage
As described above, both ADAM9 and 13 were shown to cleave cadherin-11 in tissue culture. However, we pursued ADAM13 as the protease most likely responsible for cadherin-11 cleavage because of its highly specific expression in the CNC and its previously established role in CNC migration (Alfandari et al., 2001). To further investigate if ADAM13 is directly interacting with cadherin-11, we tested the ability of the two proteins to coprecipitate. Indeed, ADAM13 coprecipitates with cadherin-11 in extracts from transfected Cos cells, as well as from embryos overexpressing these two proteins (Supplemental Figure S2). Interestingly, although overexpressed cadherin-11 binds to both the pro and mature forms of ADAM13, endogenous cadherin-11 only coimmunoprecipitates the overexpressed mature ADAM13, suggesting that cadherin-11 preferentially binds with this form in embryos (Supplemental Figure S2).
To determine when the interaction between endogenous ADAM13 and cadherin-11 occurs during early development, we performed another coimmunoprecipitation experiment using noninjected embryos at four different stages of development (Figure 2). We used blastula (stage 7) embryos as negative control because neither ADAM13 nor cadherin-11 is expressed at that stage. We also used gastrula stage embryos (stage 10.5) because both proteins are expressed but the CNC has not yet been induced. Finally we used neurula stage embryos (stage 19), where the CNC have just begun migration and tailbud stage (stage 23), when the CNC migration is nearly completed. The results show that ADAM13 coprecipitates with cadherin-11 during the migration of CNC cells, but not at blastula or gastrula stages. In addition, only the mature form of endogenous ADAM13 (M) is bound to endogenous cadherin-11 (Figure 2A). At stage 23 a 50-kDa band also coprecipitate with cadherin-11. A similar size band was previously described for ADAM13 (Alfandari et al., 1997) and could correspond to the protein lacking both the pro and metalloprotease domain. Surprisingly this form is not significantly enriched when ADAM13 is purified by affinity to concanavalin-A (Figure 2C). As expected, the levels of the 80-kDa cadherin-11 cleavage fragment increase as the CNC is migrating (Figure 2B). We find that cadherin-11 cleavage is also occurring at gastrula stage, whereas no detectable level of ADAM13 is associated, suggesting that another ADAM, possibly ADAM9, may also cleave cadherin-11 during gastrulation in vivo. Although the mRNA for cadherin-11 is also expressed in the somites, Western blotting experiments suggest that during CNC migration the vast majority of the protein is restricted to CNC in the head of the embryo, and not in the trunk where the somites are (Supplemental Figure S3).
Figure 2.
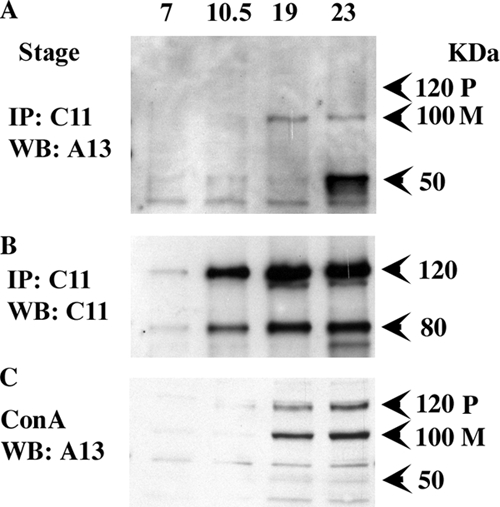
Binding of endogenous ADAM13 and cadherin-11 occurs during CNC migration and corresponds with cadherin-11 cleavage. Wild-type embryos (50 embryos/stage) were extracted at stages 7, 10.5, 19, and 23. Embryo extracts were immunoprecipitated for Cad-11 and detected by Western blot for ADAM13 (A) and Cad-11 (B). (C) Glycoproteins from five embryos were purified using Con-A agarose, separated by SDS PAGE, and blotted using ADAM13 antibodies. Both pro- (P) and mature (M) forms are detected.
Overexpression of ADAM9 or 13 But Not ADAM19 Rescues Migration of CNC Cells Blocked by the Overexpression of Cadherin-11
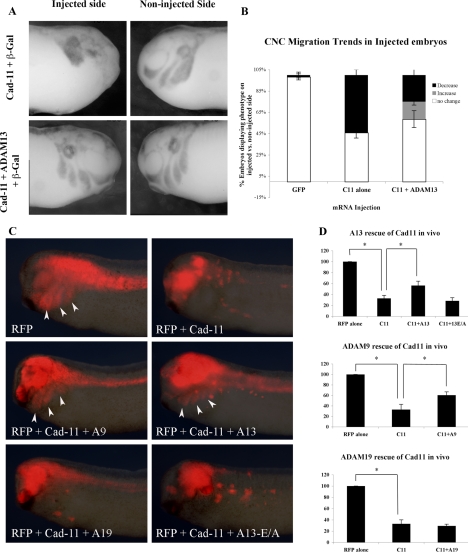
Previous work has shown that overexpression of cadherin-11 results in the inhibition of CNC migration in Xenopus embryos (Borchers et al., 2001). Our results indicate that the cadherin-11 level is regulated by proteolytic cleavage during CNC migration. Our hypothesis is that overexpression of cadherin-11 reaches protein levels that can no longer be regulated by endogenous ADAM13. To investigate this possibility, we tested whether the overexpression of ADAM13 could rescue CNC migration in embryos overexpressing cadherin-11. Embryos were injected into one blastomere at the two-cell stage with synthetic mRNA for cadherin-11 alone, or in combination with ADAM13 (Figure 3). Synthetic mRNA for β-galactosidase was also included to identify the injected side of the embryos. The noninjected sides of these embryos serve as a stage-match control for embryo development and were used in each case to quantify the extent of migration. At stage 25 embryos were fixed and processed for whole mount in situ hybridization using a mix of RNA probes for Sox10 and Twist to label CNC. Sox10 was used in combination with Twist because previous work had shown that Twist could be down-regulated in CNC overexpressing cadherin-11 (Borchers et al., 2001). Our results confirm that overexpression of cadherin-11 severely disrupted the CNC migration on the injected side (Figure 3A). In contrast, expression of both cadherin-11 and ADAM13 was able to rescue CNC migration in a large fraction of the injected embryos (Figure 3, A and B; p < 0.05).
Figure 3.
ADAM9 and 13 rescue CNC migration in cells overexpressing cadherin-11. (A) In situ hybridization was performed using a combination of CNC markers xTwist and Sox10. Embryos were injected into one blastomere at the two-cell stage with synthetic mRNA for either Cad-11 alone (top left) or in combination with ADAM13 (bottom left). The site of injection was determined by coinjecting mRNA for β-galactosidase. The right panels correspond to the noninjected side of each embryo. Disruption of CNC migration was determined by comparing the distance migrated on the injected side (left panels) versus the noninjected side (right panels) of the same embryo. (B) Quantification of three independent rescue experiments. n = 30 for GFP-injected embryos, n = 79 for Cad-11–injected embryos, and n = 80 for Cad-11– and ADAM13-injected embryos. (C) Visualization of CNC cell migration in vivo using RFP as a lineage tracer. One dorsal animal cell at the eight-cell stage was injected with mRNA encoding RFP and cadherin-11 to inhibit CNC migration. Synthetic RNA (0.25 ng) encoding ADAM9, ADAM13, ADAM19, and ADAM13-E/A were each coinjected with cadherin-11 to determine their ability to rescue migration. (D) Histograms representing the percentage of embryos in which the RFP-labeled cells migrated. Significance was determined by students t test (p <0.05). The number of embryo analyzed was as follows: RFP = 51, Cad-11 = 61, Cad-11+ADAM9 = 67, Cad-11+ADAM13 = 59, Cad-11+ADAM13-E/A = 70, and Cad-11+ADAM19 = 56.
Because ADAM9 but not ADAM19 can cleave cadherin-11 in vitro, we analyzed their ability to rescue migration of CNC cells overexpressing cadherin-11 by targeted injection at the 16-cell stage. The cell targeted at the 16-cell stage is defined as D1-2 and contributes to a large fraction of the CNC cell population (Moody, 1987). To follow the ability of CNC cells to migrate, the various mRNA were coinjected with mRNA for RFP (Figure 3C). ADAM13 was used in this assay as a positive control and the ADAM13-E/A mutant as a negative control. These experiments showed that ADAM9 can rescue migration with the same efficiency as ADAM13, whereas ADAM19 or the ADAM13-E/A do not (Figure 3D). This experiment shows that the ability of ADAM to rescue CNC migration blocked by an excess of cadherin-11 depends on the presence of the active proteolytic site and is specific of a subset of ADAM metalloproteases.
Inhibition of ADAM Activity Blocks Cadherin-11 Cleavage In Vivo
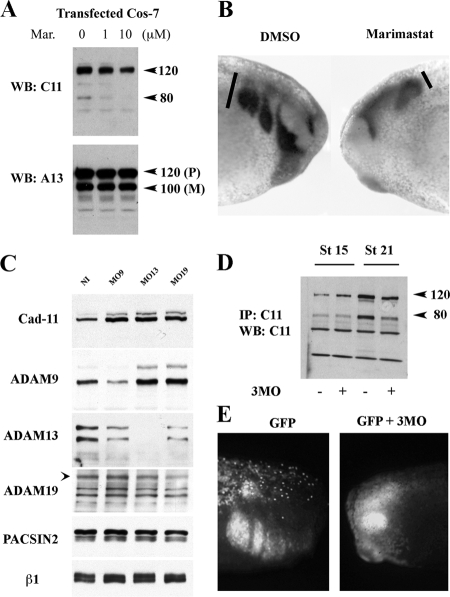
We have previously shown using a dominant negative approach that ADAM13 is critical for CNC migration in vivo (Alfandari et al., 2001). To resolve the importance of cadherin-11 cleavage by ADAMs during CNC migration, we further investigated the effect of blocking ADAM function on this process. We first used a hydroxamate-based inhibitor marimastat that inhibits a wide range of metalloprotease function including ADAMs (Orth et al., 2004). Cos-7 cells transfected with ADAM13 and cadherin-11 were treated with various concentrations of marimastat. Western blot analysis shows that marimastat inhibits ADAM13 cleavage of cadherin-11 in a dose-dependent manner (Figure 4A).
Figure 4.
Reduction of ADAM function decreases cadherin-11 cleavage and CNC migration. (A) Cos-7 cells overexpressing Cad-11 and ADAM13 were treated with 0 μM, 1 μM, or 10 μM of marimastat. Cad-11 cleavage was determined by Western blot analysis of Cad-11 (top panel). ADAM13 levels were also detected by Western blot (bottom panel). M, mature-form ADAM13; P, proform of ADAM13. (B) Lateral view of tailbud stage embryos treated by whole-mount in situ hybridization using slug to label neural crest cells. Embryos at stage 17 were injected under the epidermis with 10 nl of 10% DMSO (left) or the same amount of 1 mM marimastat in 10% DMSO (right). At tailbud stage the CNC in control embryos have migrated in the hyoid, branchial, and mandibular segments (100%, n = 24). In contrast, 87.5% of the embryos injected with the marimastat inhibitor have severe inhibition of CNC migration (n = 24). (C) Western blot analysis detecting ADAM and Cad-11 expression in control noninjected embryos (NI) or injected with morpholinos directed against ADAM9 (MO9), ADAM13 (MO13), or ADAM19 (MO19). Each lane represents the glycoproteins from five embryos equivalent. PACSIN2 and the β1-integrin protein levels are unaffected by MO injection. In contrast, the uncleaved cadherin-11 protein level is increased twofold with each MO. (D) ADAM9, 13, and 19 protein expression was knocked down using a cocktail of all three specific MO. Embryos were extracted at stage 15 (premigration) or at stage 21(mid-migration), and were immunoprecipitated for Cad-11. Cad-11 was then detected by Western blot (20 embryos/lane). At stage 21, the cadherin-11 cleavage fragments are reduced in embryos injected with the 3MO. (E) In vivo migration analysis of embryos injected at the 16-cell stage with mRNA encoding GFP alone (0.5 ng/injection) or combined with 1 ng of the 3MO cocktail (0.33 ng of each MO/injection). The CNC in GFP mRNA injected embryos migrated in 21 of 21 embryos. The CNC in GFP mRNA combined with 3MOs migrated in only eight of 33 embryos (24%).
We then investigated the effect marimastat treatment has on CNC migration in vivo by injecting the inhibitor in the pathway of the migrating cells. At stage 22 the CNC cells of the embryo injected with the carrier solution containing 10% DMSO (10 nl) migrated in the hyoid, branchial, and mandibular segments (Figure 4B). In contrast, injection of the inhibitor blocked CNC migration in vivo in a similar but more robust manner as the ADAM13 DN (Figure 4B; Alfandari et al., 2001). These results suggest that at least one metalloprotease inhibited by marimastat, possibly ADAM13, is essential for releasing cadherin-mediated cell–cell adhesion during CNC migration.
To further investigate this hypothesis, we knocked down individual ADAM metalloproteases via morpholino injection (Figure 4C). The embryos used in this study were injected with MO oligonucleotides to ADAM9, 13, and 19 and then were raised to tailbud stage (stage 24) before the analysis. The total proteins were then extracted and the glycoproteins purified by affinity to concanavalin-A. Western blot using antibodies to each ADAM, cadherin-11, PACSIN2, and the β1-integrin subunit were performed. The results show that MOs directed against ADAM9, 13, and 19 decreased the translation of their corresponding proteins. Western blot analysis also revealed that the level of uncleaved cadherin-11 at 120 kDa is increased by about twofold in embryos with each of the ADAM MO, suggesting that ADAM9, 13, and 19 may all participate, directly or indirectly, in the cleavage of cadherin-11 in vivo. As a control we tested the cadherin-11 mRNA level using real-time qPCR and found no increase in expression of the gene (Supplemental Figure S4), confirming that the increase in cadherin-11 protein level is due to “stabilization” of the protein and not increased gene expression. In support of this hypothesis, injection of an ADAM9, 13, and 19 MO cocktail significantly decreases the amount of cleaved cadherin-11 at stage 21 (Figure 4D). Additionally, injection of the MO cocktail (3MO) also blocks CNC migration in vivo (Figure 4E).
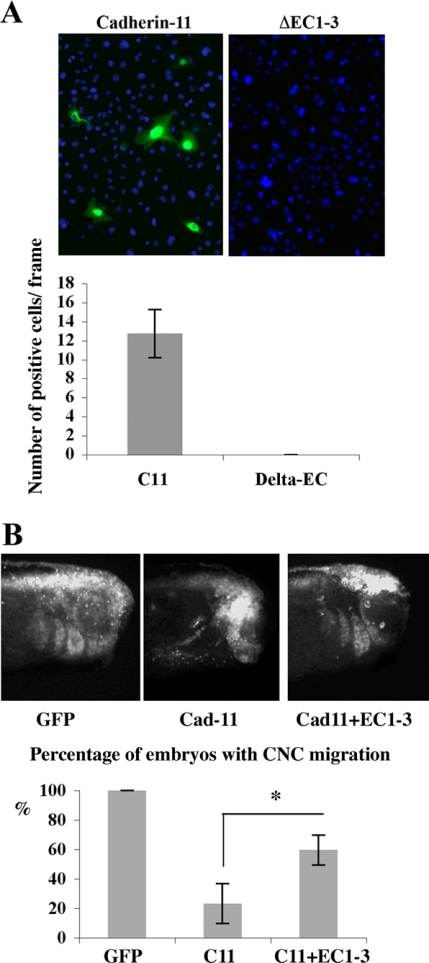
The Extracellular Cleavage Fragment Binds to Full-Length Cadherin-11 Molecules and Promotes CNC Cell Migration
Thus far we have provided evidence that ADAM cleavage of cadherin-11 produces a 80-kDa fragment that remains in the plasma membrane. Consequently, it is likely that the extracellular fragments containing the homophylic binding site are released by the shedding events and may interfere with cadherin-11 function in cell adhesion. To determine if the extracellular fragment may participate in the migration, we made a construct designed to mimic the cadherin-11 extracellular cleavage fragment (EC1-3). To test whether the fragment can bind to full-length cadherin-11, we applied the media from EC1-3–transfected cells onto live Cos-7 cells overexpressing either cadherin-11 or a cadherin-11 mutant missing the homophilic binding site (ΔEC1-3). Immunofluorescence detected the EC1-3 fragment only on cadherin-11–transfected cells, suggesting that the extracellular cleavage fragment can bind to full-length cadherin-11 (Figure 5A). Because EC1-3 can bind to cells expressing full-length cadherin-11, we predicted that this fragment might also help promote CNC cell migration. To further explore this hypothesis we overexpressed both of these proteins with green fluorescent protein (GFP) to follow CNC migration in vivo. Although overexpression of cadherin-11 alone expectedly blocks CNC migration, coexpression of EC1-3 rescues this phenotype (Figure 5B, p < 0.05). This result suggests that the cleavage fragment may compete with full-length cadherin-11 molecules for cell–cell adhesion and that CNC cells require a defined ratio of cleaved to uncleaved cadherin-11 for migration.
Figure 5.
The Cad-11 extracellular cleavage fragment binds to full-length cadherin-11 and promotes CNC cell migration. (A) EC1-3 binding experiment performed in cell culture. Conditioned media from EC1-3-mt–transfected cells was incubated with live cells transfected with full-length Cad-11 (left) or an extracellular truncated form (ΔEC1-3, right). After 20 min, the cells were washed, fixed, and processed for immunofluorescence using mAb 9E10 (myc). The green fluorescence represent EC1-3-mt bound to cells, whereas DAPI was used to stain all cell nuclei. Bars represent the average number of cells positive for EC1-3 that were counted in 19 frames. We found no fluorescence associated with cells expressing the cadherin-11 lacking the EC1-3 domain. (B) Lateral view of embryos (St 26) that were injected in one CNC precursor cell at the 16-cell stage with synthetic mRNA for GFP alone, GFP (top left) and Cad-11 (top right), or GFP, Cad-11 and EC1-3 (bottom). GFP mRNA, 0.5 ng, 1 ng of Cad-11 mRNA, and 1 ng of EC1-3 mRNA was used. The extent of CNC migration was determined by GFP fluorescence, and results from three independent experiments are plotted. Bars represent the percentage of embryos in which CNC migration was observed. The total number of embryos counted was GFP alone (n = 40), GFP+Cad-11 (n = 75), or GFP+Cad-11+EC1-3 (n = 82). *Statistically significant at p < 0.05.
EC1-3 Rescues CNC Migration in Embryos with Reduced ADAM13 Expression
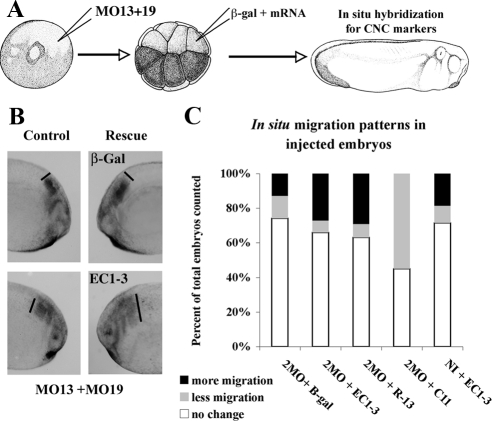
To test if the EC1-3 fragment could promote migration in CNC cells with reduced level of ADAM protein, we chose two complementary approaches (Figures 6 and 7). The first one consists of injecting the MO at the one-cell stage, to produce embryos with reduced level of ADAMs and then injecting either a lineage tracer alone or with the EC1-3 fragment at the 16-cell stage D1-2 (Figure 6). In that case we can compare using in situ hybridization the position of CNC cells that express the lineage tracer to the ones that do not. The second approach is to inject the MO with the lineage tracer at the 16-cell stage in D1-2 and follow the position of the injected cells in live embryos, thus directly assessing the capacity of the injected cells to migrate (Figure 7).
Figure 6.
The cadherin-11 extracellular cleavage fragment rescues CNC migration in embryos with reduced ADAM13 expression. (A) Schematic representation of the experimental method. Embryos were injected at the one-cell stage with MO13 and MO19 (5 ng each) and then again at the 16-cell stage, with a lineage tracer and mRNA encoding the various constructs, in D1.2 to target CNC. In this case we are testing the ability of the mRNA to rescue CNC migration. (B) In situ hybridization using Twist and Sox10 to label the CNC. The left panels represent the control side where migration was inhibited by the MO. The right panels represent the experimental side injected either with β-Gal or the EC1-3 mRNA. The black lines represent the extent of migration of the most posterior segment. After injection of the EC1-3 migration is rescued. (C) Quantification of three individual experiments described above (2MO is MO13 + 19). The total number of embryos for each injection set was n = 77 (2MO + β-gal), n = 96 (2MO + β-gal + EC1-3), n = 82 (2MO + β-gal + R13), n = 66 (2MO + β-gal + C11), and n = 91 (noninjected + β-gal + EC1-3).
Figure 7.
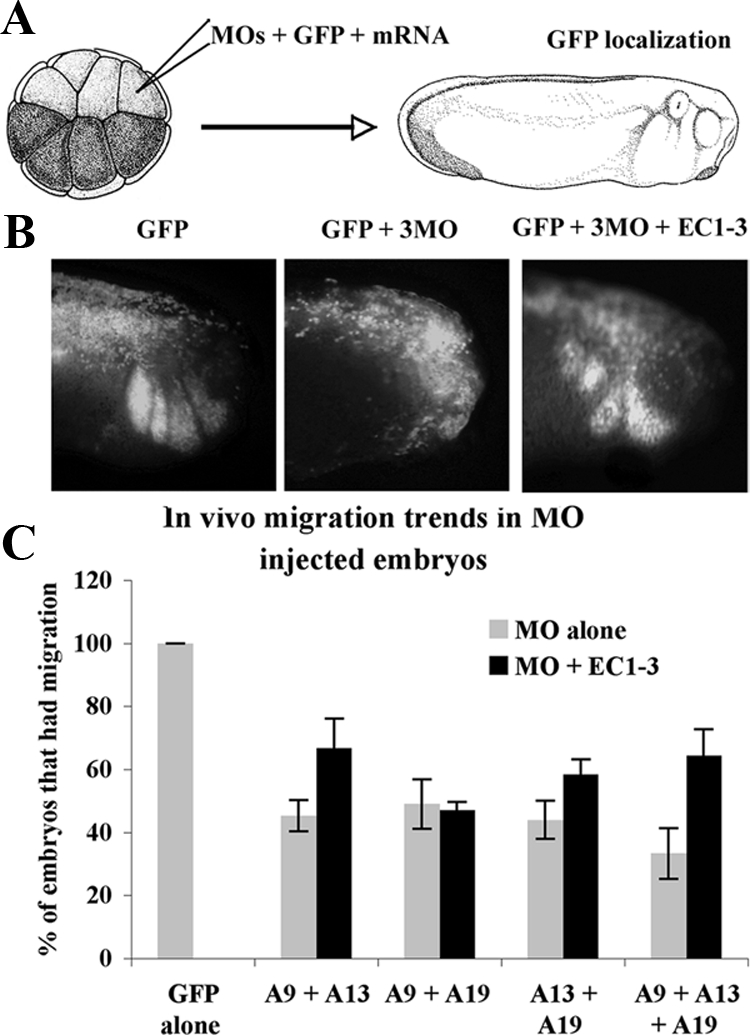
Combinations of MO to the different meltrins result in a reduction of CNC migration. (A) Embryos were injected at the 16-cell stage in D1-2 with the MO (0.33 ng each) and GFP as a lineage tracer to test their ability to prevent CNC migration. (B) Lateral views of representative embryos at tailbud stage. Migration was determined by the presence of GFP-labeled cells in the CNC pathways as evident in the GFP control. (C) Histogram representing the percentage of migration in embryos injected with the various MO (0.5 ng of each A9, A13, and A19) with or without the EC1-3 mRNA.
The first approach shows that the EC1-3 cadherin-11 fragment can rescue CNC positioning in embryos with reduced level of ADAM13 and 19 with the same efficiency as the injection of an ADAM13 mRNA lacking the MO target sequence (R13; Figure 6). Using the second approach, we find that combination of MO oligonucleotides to ADAM9, 13, and 19 can all decrease CNC migration in vivo (Figure 7). The most efficient inhibition was found using all three MOs (66% inhibition). CNC migration in all MO combinations containing the ADAM13 MO was rescued by the expression of the EC1-3 domain of cadherin-11. However, this fragment had no effect on CNC inhibited by the ADAM9 and 19 MO combination. These results suggest that meltrin ADAMs may all participate in CNC migration or may compensate for each other in vivo. Because the cadherin-11 extracellular domain could only rescue migration in embryos that had decreased ADAM13 (MO13 + 9, MO13 + 19 or all 3MO), but not in embryos lacking both ADAM9 and 19, it is likely that ADAM13 is the principal ADAM responsible for cadherin-11 cleavage during CNC migration.
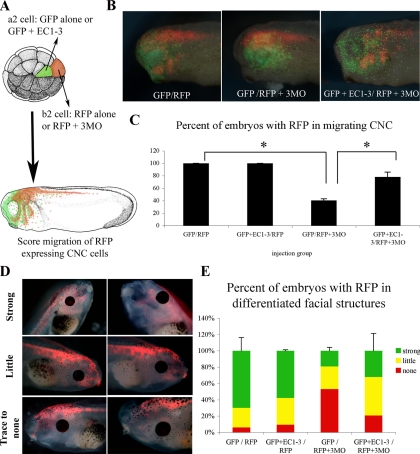
EC1-3 Promotes Migration in a Non-Cell-Autonomous Manner
The studies described in Figures 5, 6, and 7 show that EC1-3 can rescue CNC migration when coexpressed in cells that are either overexpressing full-length cadherin-11 or have knocked down ADAM expression through the use of MOs. To determine if this rescue is cell-autonomous or not, we injected embryos at the 32-cell stage with the EC-1–3 and GFP mRNAs in the a2 cell while we injected the ADAM MO cocktail with RFP mRNA in b2. Both a2 and b2 contribute to the CNC. The result show that expression of the EC1-3 domain rescued CNC migration of RFP expressing cells lacking ADAM proteins, whereas GFP alone did not, demonstrating that the cadherin-11 extracellular domain can act in a non-cell-autonomous manner.
Surprisingly, the RFP we expressed in the cells with knocked-down ADAM expression (to visualize CNC migration in vivo) remained stable throughout the later stages of CNC cell differentiation (stages 45–47). This unexpected feature made it possible to analyze if the rescued ADAM KD cells also differentiated into the craniofacial structures (Figure 8, D and E). Indeed, embryos that were mosaic for EC1-3 and ADAM knockdown (KD) cells had a tendency to have more ADAM KD cells in the developing facial cartilages and muscles than those embryos not expressing EC1-3 (Figure 8E). These results show that EC1-3 can rescue migration noncell autonomously and suggest that ADAM KD cells rescued by EC1-3 retain the ability to differentiate into craniofacial structures.
Figure 8.
The cadherin-11 extracellular fragment (EC1-3) is not cell-autonomous. (A) Schematic representation of the experimental design. The EC1-3 mRNA was coinjected with GFP mRNA at the 32-cell stage in the a2 cell. The 3MO cocktail (0.5 ng ADAM9, 13 and 19) was injected with RFP mRNA in the b2 cell of the same embryo (all mRNA were at 0.25 ng). Embryos were grown to stage 26 before imaging the GFP and RFP fluorescence (B). The percentage of embryos with migrating CNC cells expressing RFP was then counted and is presented in C. Asterisks indicate statistical significance as determined by Student's t test (p < 0.05). (D) Late stage (stages 45–47) analysis of RFP localization in differentiated facial structures in the dual injected embryos from above. Embryos were scored for having strong, little, or trace to no expression in the developing facial cartilage. (E) Histogram representing scoring data from late stage embryo analysis.
Cadherin-11 Cleavage Does Not Affect Canonical Wnt Signaling In Vivo
Cadherin-11, like many other cadherin proteins, can bind to β-catenin via its cytoplasmic domain. Cleavage of N-cadherin by ADAM10 decreases its ability to bind β-catenin, increasing the cytoplasmic pools, and resulting in the stimulation of Wnt downstream markers c-myc, cyclin-D1, and c-jun in tissue culture (Reiss et al., 2005). Furthermore, overexpression of cadherin-11 in Xenopus decreased the expression of Twist, a CNC marker that is also downstream of canonical Wnt signaling. This effect is caused by cadherin-11–sequestering β-catenin at the cell surface, because coexpression of β-catenin rescues Twist expression (Borchers et al., 2001). In light of these findings, we considered two possible ways ADAM processing of cadherin-11 could affect β-catenin. Either cleavage of cadherin-11 destabilizes its interaction with β-catenin, increasing the cytoplasmic pool and possibly promoting nuclear signaling, or ADAM cleavage of cadherin-11 only affects its adhesive properties and not its ability to bind β-catenin. To test how cadherin-11 cleavage affects its interaction with β-catenin, we overexpressed cadherin-11 in embryos either alone or with ADAM13. Cadherin-11 was then immunoprecipitated and the association with β-catenin was tested by Western blot (Supplemental Figure S5A). Here, the level of associated β-catenin was not affected by the coexpression of ADAM13, suggesting that the 80-kDa fragment is still capable of binding to β-catenin. This was further confirmed by coimmunoprecipitation experiments using a truncated form of cadherin-11 lacking the EC1-3 domains expressed in Cos cells (Supplemental Figure S5B). This truncated form associated with the endogenous β-catenin with efficiency similar to that of the wild-type cadherin-11. Finally, we performed real-time qPCR analysis on the expression of canonical Wnt target genes xTwist, cyclin-D1, and c-myc using cDNA from stage-21 embryos injected with the same mRNA combination as described above (Supplemental Figure S5C). Coexpression of ADAM13 with cadherin-11 did not stimulate the expression of any of the markers. Cumulatively, these results indicate that cadherin-11 cleavage by ADAM13 does not affect Wnt signaling through β-catenin.
DISCUSSION
The Differential Role of Cadherins in the Neural Crest
One objective of the studies described in this manuscript was to further understand the role of cadherins during neural crest migration. Among the different model organisms, both cadherin-11 and -7 are expressed in neural crest cells throughout migration (Kimura et al., 1995; Nakagawa and Takeichi, 1995; Hadeball et al., 1998; Nakagawa and Takeichi, 1998; Vallin et al., 1998). N-Cadherin and cadherin-6B are also expressed in neural crest cells but are down-regulated shortly after the onset of migration (Akitaya and Bronner-Fraser, 1992; Nakagawa and Takeichi, 1995; Inoue et al., 1997). Although N-cadherin and cadherin-6 are expressed at the beginning of migration, it is speculated that they may have an inhibitory function in this process (Coles et al., 2007; Shoval et al., 2007). This hypothesis is supported by the observation that down regulating cadherin-6B in the chick neural crest results in premature migration of these cells (Coles et al., 2007).
One intriguing question is how these cadherins may either promote (Cad-7 and -11) or prevent (N-Cad and Cad-6B) cell migration since they all share similar domain organization, intracellular binding partners, and the ability to support cell–cell adhesion. Of course each cadherin family member has unique adhesive properties such as homophilic binding tendencies and exclusive dissociation constants that may make one more useful to migrating cells than the other (Bayas et al., 2006; Patel et al., 2006). However, we suspect that their adhesive properties are more strongly influenced by regulatory proteins during the process of migration. In line with this premise we have shown that cadherin-11 is continuously regulated by ADAM13 via an extracellular cleavage event during CNC migration in Xenopus. Similarly, cadherin-7 was shown to have a rapid turnover rate in migrating neural crest cells when compared with N-cadherin (Dufour et al., 1999). This turnover is likely due to proteolysis as shown in cell culture experiments, but the enzyme responsible for this proteolysis remains to be identified (Kawano et al., 2002). Thus, the pairing of selected ADAM metalloproteases with cadherins may provide them with unique properties such as promoting cellular migration.
Promigratory Function of the Cleaved Extracellular Domain
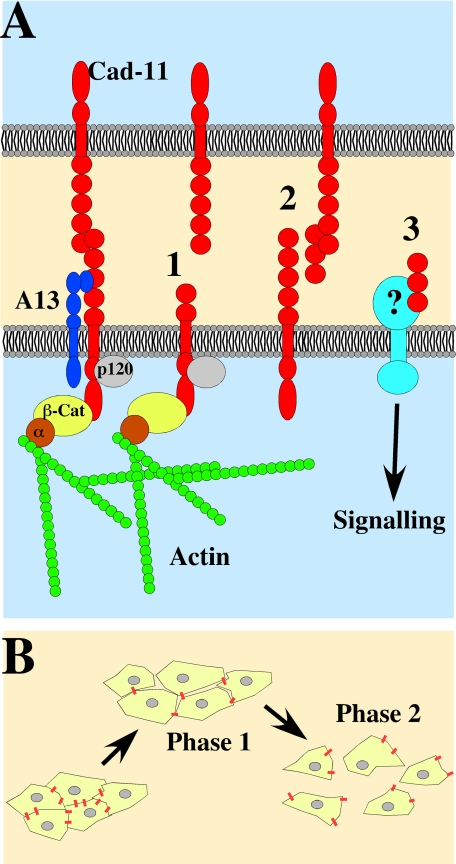
We have also discovered that the cleavage of cadherin-11 produces a fragment that has promigratory activity in vivo. This extracellular fragment can rescue CNC migration when there is an overabundance of full-length cadherin-11, either via overexpression of cadherin-11 mRNA or by blocking the cleavage of endogenous cadherin-11 through MOs directed against ADAMs (Figures 5–7). This cleavage fragment can also rescue the migration of ADAM KD CNC cells via a non-cell–autonomous mechanism (Figure 8). Our results show that the fragment can bind to full-length cadherin-11 molecules (Figure 5), suggesting that in vivo it could act as a competitor and prevent cadherin-11–mediated interactions among cells (Figure 9). Some invasive cancers may use a similar mechanism to promote cell migration via the expression of an alternatively spliced cadherin-11 product. This variant encodes a secreted form of cadherin-11 and has been found in aggressive cancer cell lines (Pishvaian et al., 1999; Feltes et al., 2002). Although competition with cadherin-11 is a likely hypothesis another possibility is that the cleaved cadherin-11 extracellular fragments may bind to an unrelated protein acting as a receptor similar to what has been shown for the L1 adhesion molecule (Figure 9). In the case of L1, the cleaved domain binds to the αVβ5 integrins and stimulate haptotactic migration (Mechtersheimer et al., 2001).
Figure 9.
Why cleave cadherin-11? (A) Cadherin-11 (red) is expressed at the surface of CNC throughout their migration and is associated with ADAM13 (blue). Cleavage of cadherin-11 could promote CNC cell migration via three possible molecular mechanisms. (1) Removal of the cadherin-11 extracellular adhesive (HAV) sequence prevents its ability to bind to full-length cadherin-11 molecules on neighboring cells. (2) The extracellular cleavage fragment retains the adhesive domain and could act as a competitive inhibitor by binding to full-length cadherin-11 and preventing its interaction with other full-length cadherin-11 molecules. (3) The extracellular cleavage fragment may also interact with unknown receptors and activate promigratory signaling cascade. We have also shown that the cleaved cadherin-11 fragment that remains in the plasma membrane can still interact with cytoplasmic protein such as β-catenin and possibly p120 maintaining cytoskeletal organization and controlling β-catenin signaling. (B) The decrease in cell adhesion may increase the fluidity of the CNC tissue allowing for the migration of a cohesive sheet of cells during the first phase of CNC migration. Further decrease of cell adhesion would promote the separation of CNC and the migration of single cells during phase 2.
Although the extracellular cleavage fragment of other cadherin molecules has also been shown to retain biological activity, it appears that no generalizations about their function can be made. For example, although the extracellular fragment of E-cadherin was shown to decrease cell–cell adhesion in vitro, the extracellular fragment of N-cadherin promotes neural cell–cell adhesion in chick embryos (Paradies and Grunwald, 1993; Noe et al., 2001). Thus, the expression of specific cadherins along with the generation of their cleavage fragments seems to play an active role in mediating a specific cellular response, such as cell migration.
Which Meltrin Cleaves Cadherin-11 during CNC Migration?
Meltrin family members ADAM9, 13, and 19 are all expressed in CNC cells during migration. Because all three of these proteins are active proteases, they could all potentially cleave cadherin-11 during this process. However, our studies in vitro reveal that ADAM9 and 13 can cleave cadherin-11, whereas ADAM19 cannot (Supplemental Figure S1). Additionally, both ADAM9 and 13, but not ADAM19, are capable of rescuing the migration of CNC expressing an excess of cadherin-11 in vivo (Figure 3, C and D). Yet, resolving which meltrin predominantly cleaves cadherin-11 during CNC migration was complicated by the ability of the ADAMs to compensate for each other's function. We have shown further evidence of the compensation among the meltrins in vivo by detecting an increase in ADAM9 expression when either ADAM13 or 19 expression is knocked down via MO injection (Figure 4C). To prevent the effects of compensation in our experiments, we used combinations of MOs to knock down at least two meltrins at one time. We have shown that injection of a combination of any two ADAM MOs blocks CNC migration in about half of the embryos screened (Figure 7). This phenotype can be rescued by the expression of the cadherin-11 extracellular domain when ADAM13 MO is included in the injection. However, the cadherin-11 extracellular domain does not rescue embryos with double MO knockdown of ADAM9 and ADAM19. This distinction suggests that ADAM13 is the main enzyme responsible for cadherin-11 cleavage during CNC migration.
On the other hand, the above observation does not explain why MOs for ADAM9 and 19 can also block CNC migration. In this regard, we observed a decrease in ADAM13 expression in both ADAM9 and ADAM19 MO injected embryos (Figure 4C), suggesting that there is cross-talk among these meltrins. Thus the CNC migration phenotype (Figure 7), and the increase in uncleaved cadherin-11 levels (Figure 4C) in ADAM9 and 19 knocked down embryos could be at least partially attributed to this secondary effect on the ADAM13 protein level. Additionally, loss of either ADAM9 or 19 may affect CNC induction. For example, conditional knockout of ADAM19 in the mouse neural crest does not prevent migration, but interferes with the specification of cardiac neural crest cells and the proper morphogenesis of the heart (Komatsu et al., 2007). We have also observed that ADAM19 KD interferes with CNC specification in Xenopus, (Neuner and Alfandari, unpublished results), and this could contribute to the partial inhibition of CNC migration observed here.
We propose that ADAM13 is responsible for the cleavage of cadherin-11 during CNC migration and that other meltrins, such as ADAM9 can compensate for this function when ADAM13 protein expression decreases. Our results also suggest that another protein cleaved by the meltrins may be important in the specification and/or migration of the CNC.
ADAM10 Cleavage of N-cadherin versus ADAM13 Cleavage of Cadherin-11 in the Neural Crest
Although ADAM10 cleavage of N-cadherin was shown to play an important role in trunk neural crest delamination in chick, there are fundamental differences with the ADAM13 cleavage of cadherin-11 in the Xenopus CNC (Shoval et al., 2007). First, as described previously, cleavage of cadherin-11 occurs continuously during migration, whereas cleavage of N-cadherin is part of the global down-regulation of this protein required at the onset of migration (Akitaya and Bronner-Fraser, 1992; Shoval et al., 2007). Second, in the avian neural crest, cleavage of N-cadherin releases β-catenin that relocalizes to the nucleus to activate the transcription of promigratory genes such as cyclin-D1 (Shoval et al., 2007). Cyclin-D1 in addition to its role in controlling cell division also controls cell motility by inhibiting ROCK (Rho-associated protein kinase) signaling and TSP-1 expression (Li et al., 2006). Conversely, we have shown that cleavage of cadherin-11 does not affect its interaction with β-catenin.
Although β-catenin signaling appears to be important for the initial delamination of the neural crest, it seems to play a different role during neural crest cell migration (de Melker et al., 2004; Shoval et al., 2007). Some signaling through β-catenin is important for the expression of neural crest markers, such as Twist, during migration (Borchers et al., 2001). However, conditional knock out of β-catenin in mouse embryos showed that β-catenin signaling is not required for neural crest cell migration (Brault et al., 2001). In fact, exogenous stimulation of β-catenin via LiCl treatment will stop the migration of avian neural crest ex vivo (de Melker et al., 2004). It is possible that cadherin-11 helps to control the “intensity” of β-catenin signaling by sequestering a pool of this molecule at the cell membrane during CNC migration. On the other hand, it has been suggested that signaling through β-catenin is involved in the differentiation of neural crest cells once they reach their target locations (Hari et al., 2002; Paratore et al., 2002). Because neural crest differentiation occurs mostly after migration ceases, cadherin-11's interaction with β-catenin at the cell membrane may also play a role in the maintenance of an undifferentiated state while the cells are still moving.
In summary, we propose that the differential role of cadherins in the neural crest is in part mediated by ADAM cleavage. Here we show that ADAM regulates cadherin-11 throughout CNC migration. The continuous cleavage of cadherin-11 could promote migration by removing its adhesive domain, and by producing an extracellular fragment that retains biological activity. The decrease in cell adhesion could be important to increase the fluidity of the CNC tissue as well as promote the dispersion into single cells during the second phase of CNC migration (Figure 9).
Supplementary Material
ACKNOWLEDGMENTS
The author thanks Melissa Morell-Smith for her help in generating ADAM9 constructs and Dr. Doris Wedlich for helpful comments and discussion. The author also thanks Dr. Sewald and Dr. Jenssen (both of the University of Bielefeld, Bielefeld, Germany) for their generous gift of the marimastat inhibitor supported by a grant from the Deutsche Forschungsgemeinschaft (SFB 549) to Dr. Sewald and Dr. Jenssen. This work was supported by a Grant DE016289 from the U.S. Public Health Service to Dr. Alfandari.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-05-0535) on October 22, 2008.
REFERENCES
- Akitaya T., Bronner-Fraser M. Expression of cell adhesion molecules during initiation and cessation of neural crest cell migration. Dev. Dyn. 1992;194:12–20. doi: 10.1002/aja.1001940103. [DOI] [PubMed] [Google Scholar]
- Alfandari D., Cousin H., Gaultier A., Smith K., White J. M., Darribere T., DeSimone D. W. Xenopus ADAM 13 is a metalloprotease required for cranial neural crest-cell migration. Curr. Biol. 2001;11:918–930. doi: 10.1016/s0960-9822(01)00263-9. [DOI] [PubMed] [Google Scholar]
- Alfandari D., Whittaker C. A., DeSimone D. W., Darribere T. Integrin alpha v subunit is expressed on mesodermal cell surfaces during amphibian gastrulation. Dev. Biol. 1995;170:249–261. doi: 10.1006/dbio.1995.1212. [DOI] [PubMed] [Google Scholar]
- Alfandari D., Wolfsberg T. G., White J. M., DeSimone D. W. ADAM13, a novel ADAM expressed in somitic mesoderm and neural crest cells during Xenopus laevis development. Dev. Biol. 1997;182:314–330. doi: 10.1006/dbio.1996.8458. [DOI] [PubMed] [Google Scholar]
- Bayas M. V., Leung A., Evans E., Leckband D. Lifetime measurements reveal kinetic differences between homophilic cadherin bonds. Biophys. J. 2006;90:1385–1395. doi: 10.1529/biophysj.105.069583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchers A., David R., Wedlich D. Xenopus cadherin-11 restrains cranial neural crest migration and influences neural crest specification. Development. 2001;128:3049–3060. doi: 10.1242/dev.128.16.3049. [DOI] [PubMed] [Google Scholar]
- Brault V., Moore R., Kutsch S., Ishibashi M., Rowitch D. H., McMahon A. P., Sommer L., Boussadia O., Kemler R. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- Cai H., Kratzschmar J., Alfandari D., Hunnicutt G., Blobel C. P. Neural crest-specific and general expression of distinct metalloprotease-disintegrins in early Xenopus laevis development. Dev. Biol. 1998;204:508–524. doi: 10.1006/dbio.1998.9017. [DOI] [PubMed] [Google Scholar]
- Coles E. G., Taneyhill L. A., Bronner-Fraser M. A critical role for Cadherin6B in regulating avian neural crest emigration. Dev. Biol. 2007;312:533–544. doi: 10.1016/j.ydbio.2007.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousin H., Gaultier A., Bleux C., Darribere T., Alfandari D. PACSIN2 is a regulator of the metalloprotease/disintegrin ADAM13. Dev. Biol. 2000;227:197–210. doi: 10.1006/dbio.2000.9871. [DOI] [PubMed] [Google Scholar]
- de Melker A. A., Desban N., Duband J. L. Cellular localization and signaling activity of beta-catenin in migrating neural crest cells. Dev. Dyn. 2004;230:708–726. doi: 10.1002/dvdy.20091. [DOI] [PubMed] [Google Scholar]
- Dufour S., Beauvais-Jouneau A., Delouvee A., Thiery J. P. Differential function of N-cadherin and cadherin-7 in the control of embryonic cell motility. J. Cell Biol. 1999;146:501–516. doi: 10.1083/jcb.146.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupin E., Creuzet S., Le Douarin N. M. The contribution of the neural crest to the vertebrate body. Adv. Exp. Med. Biol. 2006;589:96–119. doi: 10.1007/978-0-387-46954-6_6. [DOI] [PubMed] [Google Scholar]
- Feltes C. M., Kudo A., Blaschuk O., Byers S. W. An alternatively spliced cadherin-11 enhances human breast cancer cell invasion. Cancer Res. 2002;62:6688–6697. [PubMed] [Google Scholar]
- Gawantka V., Joos T. O., Hausen P. A beta 1-integrin associated alpha-chain is differentially expressed during Xenopus embryogenesis. Mech. Dev. 1994;47:199–211. doi: 10.1016/0925-4773(94)90039-6. [DOI] [PubMed] [Google Scholar]
- Hadeball B., Borchers A., Wedlich D. Xenopus cadherin-11 (Xcadherin-11) expression requires the Wg/Wnt signal. Mech. Dev. 1998;72:101–113. doi: 10.1016/s0925-4773(98)00022-7. [DOI] [PubMed] [Google Scholar]
- Ham C., Levkau B., Raines E. W., Herren B. ADAM15 is an adherens junction molecule whose surface expression can be driven by VE-cadherin. Exp. Cell Res. 2002;279:239–247. doi: 10.1006/excr.2002.5606. [DOI] [PubMed] [Google Scholar]
- Hari L., Brault V., Kleber M., Lee H. Y., Ille F., Leimeroth R., Paratore C., Suter U., Kemler R., Sommer L. Lineage-specific requirements of beta-catenin in neural crest development. J. Cell Biol. 2002;159:867–880. doi: 10.1083/jcb.200209039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland R. M. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Harlow E., Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Harris M. L., Erickson C. A. Lineage specification in neural crest cell pathfinding. Dev. Dyn. 2007;236:1–19. doi: 10.1002/dvdy.20919. [DOI] [PubMed] [Google Scholar]
- Inoue T., Chisaka O., Matsunami H., Takeichi M. Cadherin-6 expression transiently delineates specific rhombomeres, other neural tube subdivisions, and neural crest subpopulations in mouse embryos. Dev. Biol. 1997;183:183–194. doi: 10.1006/dbio.1996.8501. [DOI] [PubMed] [Google Scholar]
- Kawaguchi J., Takeshita S., Kashima T., Imai T., Machinami R., Kudo A. Expression and function of the splice variant of the human cadherin-11 gene in subordination to intact cadherin-11. J. Bone Miner. Res. 1999;14:764–775. doi: 10.1359/jbmr.1999.14.5.764. [DOI] [PubMed] [Google Scholar]
- Kawano R., Matsuo N., Tanaka H., Nasu M., Yoshioka H., Shirabe K. Identification and characterization of a soluble cadherin-7 isoform produced by alternative splicing. J. Biol. Chem. 2002;277:47679–47685. doi: 10.1074/jbc.M205328200. [DOI] [PubMed] [Google Scholar]
- Kimura Y., Matsunami H., Inoue T., Shimamura K., Uchida N., Ueno T., Miyazaki T., Takeichi M. Cadherin-11 expressed in association with mesenchymal morphogenesis in the head, somite, and limb bud of early mouse embryos. Dev. Biol. 1995;169:347–358. doi: 10.1006/dbio.1995.1149. [DOI] [PubMed] [Google Scholar]
- Knight R. D., Schilling T. F. Cranial neural crest and development of the head skeleton. Adv. Exp. Med. Biol. 2006;589:120–133. doi: 10.1007/978-0-387-46954-6_7. [DOI] [PubMed] [Google Scholar]
- Komatsu K., Wakatsuki S., Yamada S., Yamamura K., Miyazaki J., Sehara-Fujisawa A. Meltrin beta expressed in cardiac neural crest cells is required for ventricular septum formation of the heart. Dev. Biol. 2007;303:82–92. doi: 10.1016/j.ydbio.2006.10.037. [DOI] [PubMed] [Google Scholar]
- Li Z., et al. Cyclin D1 regulates cellular migration through the inhibition of thrombospondin 1 and ROCK signaling. Mol. Cell. Biol. 2006;26:4240–4256. doi: 10.1128/MCB.02124-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Maretzky T., Reiss K., Ludwig A., Buchholz J., Scholz F., Proksch E., de Strooper B., Hartmann D., Saftig P. ADAM10 mediates E-cadherin shedding and regulates epithelial cell-cell adhesion, migration, and beta-catenin translocation. Proc. Natl. Acad. Sci. USA. 2005;102:9182–9187. doi: 10.1073/pnas.0500918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechtersheimer S., et al. Ectodomain shedding of L1 adhesion molecule promotes cell migration by autocrine binding to integrins. J. Cell Biol. 2001;155:661–673. doi: 10.1083/jcb.200101099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody S. A. Fates of the blastomeres of the 16-cell stage Xenopus embryo. Dev. Biol. 1987;119:560–578. doi: 10.1016/0012-1606(87)90059-5. [DOI] [PubMed] [Google Scholar]
- Nakagawa S., Takeichi M. Neural crest cell-cell adhesion controlled by sequential and subpopulation-specific expression of novel cadherins. Development. 1995;121:1321–1332. doi: 10.1242/dev.121.5.1321. [DOI] [PubMed] [Google Scholar]
- Nakagawa S., Takeichi M. Neural crest emigration from the neural tube depends on regulated cadherin expression. Development. 1998;125:2963–2971. doi: 10.1242/dev.125.15.2963. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop P. D., Faber J. 2nd ed. Amsterdam: North-Holland; 1967. Normal table of Xenopus laevis (Daudin) [Google Scholar]
- Noe V., Fingleton B., Jacobs K., Crawford H. C., Vermeulen S., Steelant W., Bruyneel E., Matrisian L. M., Mareel M. Release of an invasion promoter E-cadherin fragment by matrilysin and stromelysin-1. J. Cell Sci. 2001;114:111–118. doi: 10.1242/jcs.114.1.111. [DOI] [PubMed] [Google Scholar]
- Orth P., et al. Crystal structure of the catalytic domain of human ADAM33. J. Mol. Biol. 2004;335:129–137. doi: 10.1016/j.jmb.2003.10.037. [DOI] [PubMed] [Google Scholar]
- Paradies N. E., Grunwald G. B. Purification and characterization of NCAD90, a soluble endogenous form of N-cadherin, which is generated by proteolysis during retinal development and retains adhesive and neurite-promoting function. J. Neurosci. Res. 1993;36:33–45. doi: 10.1002/jnr.490360105. [DOI] [PubMed] [Google Scholar]
- Paratore C., Hagedorn L., Floris J., Hari L., Kleber M., Suter U., Sommer L. Cell-intrinsic and cell-extrinsic cues regulating lineage decisions in multipotent neural crest-derived progenitor cells. Int. J. Dev. Biol. 2002;46:193–200. [PubMed] [Google Scholar]
- Patel S. D., et al. Type II cadherin ectodomain structures: implications for classical cadherin specificity. Cell. 2006;124:1255–1268. doi: 10.1016/j.cell.2005.12.046. [DOI] [PubMed] [Google Scholar]
- Pickard B., Damjanovski S. Overexpression of the tissue inhibitor of metalloproteinase-3 during Xenopus embryogenesis affects head and axial tissue formation. Cell Res. 2004;14:389–399. doi: 10.1038/sj.cr.7290239. [DOI] [PubMed] [Google Scholar]
- Pishvaian M. J., Feltes C. M., Thompson P., Bussemakers M. J., Schalken J. A., Byers S. W. Cadherin-11 is expressed in invasive breast cancer cell lines. Cancer Res. 1999;59:947–952. [PubMed] [Google Scholar]
- Reiss K., Maretzky T., Ludwig A., Tousseyn T., de Strooper B., Hartmann D., Saftig P. ADAM10 cleavage of N-cadherin and regulation of cell-cell adhesion and beta-catenin nuclear signalling. EMBO J. 2005;24:742–752. doi: 10.1038/sj.emboj.7600548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell L. L., Trainor P. A. Neural crest cell plasticity. size matters. Adv. Exp. Med. Biol. 2006;589:78–95. doi: 10.1007/978-0-387-46954-6_5. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T., Bronner-Fraser M. Development and evolution of the migratory neural crest: a gene regulatory perspective. Curr. Opin. Genet. Dev. 2006;16:360–366. doi: 10.1016/j.gde.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Shoval I., Ludwig A., Kalcheim C. Antagonistic roles of full-length N-cadherin and its soluble BMP cleavage product in neural crest delamination. Development. 2007;134:491–501. doi: 10.1242/dev.02742. [DOI] [PubMed] [Google Scholar]
- Smith K. M., Gaultier A., Cousin H., Alfandari D., White J. M., DeSimone D. W. The cysteine-rich domain regulates ADAM protease function in vivo. J. Cell Biol. 2002;159:893–902. doi: 10.1083/jcb.200206023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith W. C., Harland R. M. Injected Xwnt-8 RNA acts early in Xenopus embryos to promote formation of a vegetal dorsalizing center. Cell. 1991;67:753–765. doi: 10.1016/0092-8674(91)90070-f. [DOI] [PubMed] [Google Scholar]
- Vallin J., Girault J. M., Thiery J. P., Broders F. Xenopus cadherin-11 is expressed in different populations of migrating neural crest cells. Mech. Dev. 1998;75:171–174. doi: 10.1016/s0925-4773(98)00099-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.