Abstract
Comprehensive protein interaction mapping projects are underway for many model species and humans. A key step in these projects is estimating the time, cost, and personnel required for obtaining an accurate and complete map. Here, we model the cost of interaction map completion across a spectrum of experimental designs. We show that current efforts may require up to 20 independent tests covering each protein pair to approach completion. We explore designs for reducing this cost substantially, including prioritization of protein pairs, probability thresholding, and interaction prediction. The best designs lower cost by four-fold overall and >100-fold in early stages of mapping. We demonstrate the best strategy in an ongoing project in Drosophila, in which we map 450 high-confidence interactions using 47 microtiter plates, versus thousands of plates expected using current designs. This study provides a framework for assessing the feasibility of interaction mapping projects and for future efforts to increase their efficiency.
Analysis of molecular networks has exploded in recent years. A wide variety of technologies have been introduced for mapping networks of gene and protein interactions, including yeast two-hybrid assays1–8, affinity purification coupled to mass spectrometry9–11, chromatin immunoprecipitation measurements of transcriptional binding12–14, synthetic-lethal and suppressor networks15,16, expression QTLs17–20, and many others. Using these technologies, network mapping projects are currently underway for many model species2–4,7–13,15, microbial21–23 and viral24,25 pathogens, and humans5,6. As an illustration of how pervasive networks have become, the U.S. National Institutes of Health currently funds 3076 active research grants covering the topic “protein-protein interactions” with 794 of these implementing the technique of “yeast two hybrid system”26.
Mapping a complete gene or protein network evokes similar challenges and considerations as mapping a complete genome sequence. In the case of the human and model genome projects, large-scale sequencing efforts were accompanied by a series of feasibility studies27,28 which used mathematical formulations and pilot projects to explore strategies for genome assembly and the work required for each. In the case of interaction networks, similar feasibility studies are just beginning29–31. Some of the questions to be addressed are: What is the cost of completing an interactome map and what is the best strategy for minimizing that cost? Given practical cost constraints, how can the quality and coverage of interaction data be maximized? How many independent assay types are needed? How should direct pairwise tests for interaction be combined with pooled screening? What is the effect of the test sensitivity on the final cost? How should interaction predictions be incorporated, and what is their effect on the mapping cost? Which specific improvements to experimental and computational methods are likely to have the largest effect?
To approach these questions, we modeled several standard and alternative strategies for using pairwise protein interaction experiments to determine the interactome of the fruit fly Drosophila melanogaster. Our analysis shows that completing the interactome using sequential pairwise or pooled screening is probably too costly to be practical. However, this cost can be reduced substantially using a strategy that combines pooling with prioritized testing and interaction prediction. We carry out several iterations of this strategy to efficiently map 450 new high confidence interactions in Drosophila.
RESULTS
Interactome mapping—problem definition
In contrast to a genome, the interactome has been more difficult to define. Some authors have argued32 that the “True Interactome” should be defined as all possible interactions encoded by a genome— i.e., the set of all pairwise protein interactions that occur in at least one biological condition or cell type. The assumption is that every true interaction will be detectable by some assay, and that given enough independent measurements, most of the interactome could be detected. Many assay types have been described for detecting protein-protein interactions, a few of which have been adapted to large-scale screening1,32–34. On the other hand, some interactions may be immeasurable by any available assay, or will not arise in the conditions surveyed. Therefore, we use the term “Mappable Interactome” for the subset of true pairwise interactions that are reproducibly detectable by any given assay method or combination of methods.
To define appropriate criteria for determining when an interactome map is “complete”, we distinguish between the terms saturation and coverage. Saturation measures the percentage of true interactions that have been experimentally observed at least once. Coverage we define more strictly to mean the percentage of true interactions that have been experimentally validated with high confidence such that the percentage of false interactions (i.e., the False Discovery Rate or FDR) is kept below a predetermined threshold. We treat “completion” as achieving 95% coverage of the Mappable Interactome at 5% FDR, which requires that the map include at least 95% of all true interactions with no more than 5% of the reported interactions being false.
A model of interactome coverage
We simulated a series of mapping strategies implementing a variety of basic and sophisticated features (Fig. 1; Flowcharts of each strategy are provided in Supplementary Fig. 1). All strategies were evaluated using a statistical model based on naïve Bayes to estimate saturation and coverage of the Drosophila interactome as a function of the number of interaction tests. We programmed this model with the assumption that the fly interactome contains approximately 105 interactions overall, along with estimates for the false positive rate (FPR—the probability that a non-interacting protein pair is reported as interacting) and the false negative rate (FNR—the probability that an interacting pair is reported as non-interacting). Although the magnitudes of these errors are still under debate, previous studies2,5,29,35,36 have reported Y2H error rates of FPR < 1% with FNR in the range 50–80% (note that several of these studies erroneously refer to FDR as FPR). Here, we used 0.2% FPR and 66% FNR.
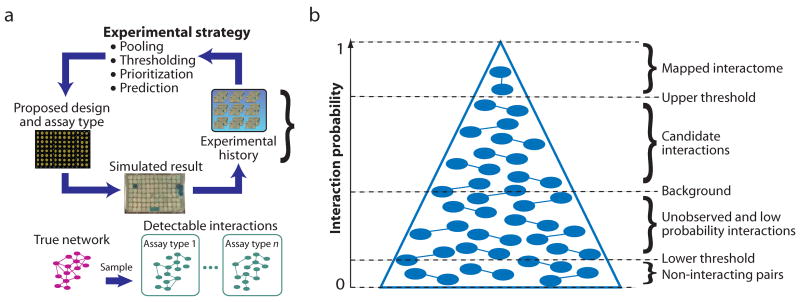
Figure 1. Simulating an interaction mapping project.
(a) At any given point in the project, every pair of proteins is assigned an interaction probability based on its experimental history (initially these probabilities are set to background or informed by predictions). The interaction probabilities and experimental history are used to design a 96-well plate Y2H experiment according to one of the strategies. The result of this experiment is simulated based on the detectability of the tested interactions (given the assay type) and the pooling sensitivity. The new experimental results are recorded in the history and also (b) used to update the interaction probabilities of the relevant protein pairs. The pyramid represents the ordered list of protein pairs ranked by probability. It is wider at the bottom than at the top to reflect that most pairs are negative—i.e., most pairs will have low probability and only a few pairs will percolate to the top of the list with high probability. Interactions with probability above an upper threshold are added to the mapped interactome, which is compared to the simulated “True Network” at intervals of 1,000 plates for reporting coverage and FDR.
Due to the high FNR of a particular assay, it becomes clear that multiple assay types will likely be needed to achieve complete coverage, and that these assays should be independent or at least only partially dependent. Although the precise correlations between different assay types have not been well studied, complementarity between assays has been widely assumed and occasionally demonstrated: For instance, protein interactions have been shown to be of substantially higher confidence if they are detected in different orientations (bait-prey vs. prey-bait)2; in different Y2H screens3,8,35; by different types of Y2H system such as LexA-based vs. Gal4-based36; or by both Y2H and co-affinity purification29.
Basic mapping strategies in current use
We first simulated a “Basic serial” strategy, in which all pairs of proteins are tested for interaction sequentially. Under this formulation, achieving a saturation of 95% required eight comprehensive screens, in which each protein pair was tested by eight independent assays, equivalent to ~7×108 pairwise tests assuming a total of 13,600 protein-encoding genes in fly37 (Table 1 and Fig. 2a). Moreover, 93% of all observed interactions in this network were false positives (including 99% of interactions observed exactly once and 21% of interactions observed twice—Fig. 2b). The false-positives predominate because, although the 0.2% FPR seems low, the number of non-interacting protein pairs is far in excess of the number of true interactions.
Table 1.
Summary of the features and performance of the different strategies*
| Fly Interactome | Human Interactome | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Strategy | Figure(s) | Pooling | Repeated screens | Pooling sensitivity (%) | Thresholding and Prioritization | Prediction | Intermediate (50%) coverage cost | Complete (95%) coverage cost | Intermediate (50%) coverage cost | Complete (95%) coverage cost | |
| 1 | Basic serial | Fig. 2c | ✘ | — | — | ✘ | ✘ | 7.5M | 19.9M | 18.9M | 51.8M |
|
| |||||||||||
| 2.1 | Fig. 3a | ✓ | 4 | 40 | ✘ | ✘ | 1.6M | 4.4M | 4.2M | 11.3M | |
| 2.2 | Pooling | Fig. 3a | ✓ | 1 | 20 | ✘ | ✘ | 1.6M | 4.9M | 3.9M | 12.3M |
| 2.3 | Fig. 3a | ✓ | 1 | 40 | ✘ | ✘ | 1.4M | 4.1M | 3.5M | 10.4M | |
| 2.4 | Fig. 3a | ✓ | 1 | 100 | ✘ | ✘ | 1.4M | 3.7M | 3.5M | 9.5M | |
|
| |||||||||||
| 3.1 | Fig. 3a | ✓ | 1 | 20 | ✓ | ✘ | 443K | 1.9M | 1.1M | 4.8M | |
| 3.2 | Thresholding | Fig. 3a–b | ✓ | 1 | 40 | ✓ | ✘ | 391K | 1.7M | 969K | 4.3M |
| 3.3 | Fig. 3b | ✓ | 1 | 100 | ✓ | ✘ | 354K | 1.5M | 916K | 3.9M | |
|
| |||||||||||
| Predictions: FPR, FNR, FDR (%) | |||||||||||
| 4.1 | 10, 20, 99.2 | Fig. 3b | ✓ | 1 | 40 | ✓ | ✓ | 249K | 1.6M | 611K | 4.1M |
| 4.2 | 1, 50, 95.0 | Fig. 3b | ✓ | 1 | 40 | ✓ | ✓ | 111K | 1.4M | 293K | 3.6M |
| 4.3 | 5, 20, 98.3 | Fig. 3b | ✓ | 1 | 40 | ✓ | ✓ | 126K | 1.3M | 313K | 3.3M |
| 4.4 | 1, 20, 92.2 | Fig. 3b | ✓ | 1 | 40 | ✓ | ✓ | 28K | 925K | 69K | 2.3M |
Interaction costs are given in units of total number of plates (K = Thousands, M = Millions) required for 50% or 95% coverage. When 95% coverage is achieved more than once, the greatest cost is presented.
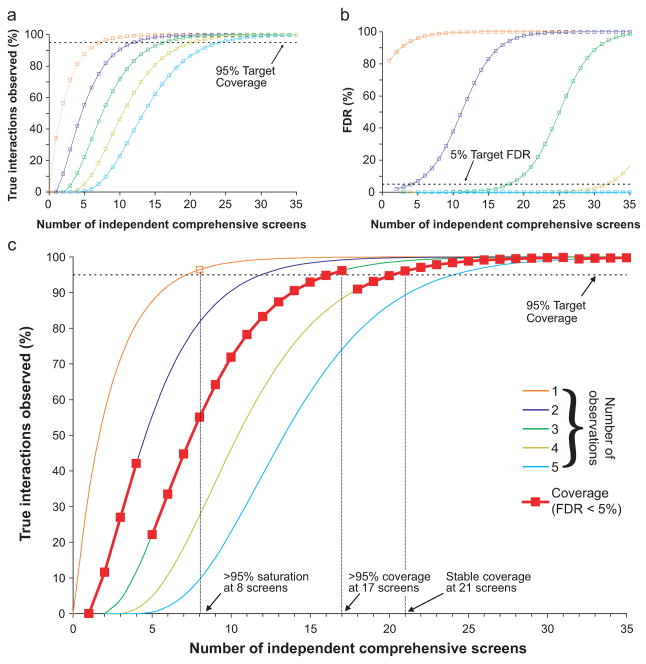
Figure 2. Analysis of the coverage and saturation of the fly interactome as a function of the number of independent screens.
(a) The percentage of true interactions that are observed >1 (orange), >2 (purple), >3 (green), >4 (yellow), and >5 (cyan) times as a function of the number of times they are tested with independent assays. Increasing the threshold of independent observations increases the number of independent assays needed to reach the 95% coverage target. (b) The false discovery rate (FDR) of interactions that are observed exactly once (orange), twice (purple), thrice (green), four times (yellow), and five times (cyan) as a function of the number of times they are tested with independent assays. To achieve FDR < 5% interactions should be observed at least twice when tested with < 5 independent assays, and at least three times when tested with 5–17 assays. (c) The effective coverage level at FDR < 5% is shown (red curve) by embedding the observation threshold from (b) into the curves of (a). While saturation is achieved after 8 screens, 21 screens are required for 95% coverage at FDR < 5%.
To ensure an overall FDR < 5%, we found that every interaction must be reported by at least three independent assays. After eight screens 55% of the interactome was covered under these conditions. The coverage goal of 95% was achieved only after 21 comprehensive pair-wise screens (Fig. 2c). This overall outcome—that the number of experiments required to reach full coverage is two to three times that required to reach saturation—was observed over a range of error parameters (Supplementary Table 1). Clearly, completing the interactome map under these conditions is impractical, as it would require testing 92 million protein pairs 21 separate times.
To reduce the number of tests, assays such as Y2H typically use pooled screens in which a single protein “bait” is tested for interaction against pools of protein “preys” (phase I)38. For pools that test positive, pairwise tests are immediately conducted between the bait and each prey in the pool (phase II—this second phase can also be conducted by sequencing3,5). The benefit of pooling is that large numbers of potential interactions can be sampled at relatively low cost. This comes at the expense of FNR, as the chance a true interaction is missed in the pool is higher than the chance it would be missed by direct pairwise tests38. Through simulation, we found that basic two-phase pooling (Pooling strategy) does indeed achieve a four- to five-fold reduction in coverage cost over pairwise testing (~4 million plates for Pooling compared to ~20 million plates for Basic-serial, Table 1). However, assuming the rate of interaction screening of a typical laboratory (e.g., ~2400 plate-matings per person per year), pooled screens would still require approximately 1700 person-years to achieve completion of the Drosophila protein network.
Advanced mapping strategies
We next considered an interaction mapping strategy that, rather than treat all protein pairs equally, maintains a rank-ordered list of pairs according to their probabilities of interaction (Thresholding strategy, Table 1). All probabilities start at the background frequency of interaction for random protein pairs (as for Basic-serial and Pooling). Protein interactions are initially tested using pooled screening, and after each two-phase pooled experiment the probabilities increase for interactions that are observed and decrease for interactions that are tested but not observed. Unlike previous strategies, however, protein pairs with probability greater than an upper threshold (i.e., 95%) are declared to be definite “interactors” and are removed from subsequent testing (Fig. 1b). Likewise, interactions with probability beneath a lower threshold are declared to be “non-interactors” and are also removed from further consideration. The motivation for thresholding is to more quickly exclude the overwhelming number of non-interacting protein pairs. Finally, candidate interactions are defined as those with probabilities between the upper threshold and background. When candidates are available they are always tested immediately using pairwise assays, before resorting to pooling, until their probabilities are pushed above the upper threshold or below background. The motivation for prioritizing candidate interactions is to more quickly cover the interactions likely to be positive. Overall, Thresholding resulted in more than a two-fold cost reduction compared to Pooling (Table 1 and Fig. 3a).
Figure 3. Fly and Human Interactome coverage costs for different experimental strategies.
(a) Comparison of the Pooling strategy (numbers 2.1–2.4) with a Thresholding strategy (numbers 3.1–3.3) which combines pooling with direct experiments based on thresholding and prioritization. (a INSET) Zoomed view showing the number of plates required to add the first 450 interactions to the map using Pooling. (b) Performance of the Prediction strategy (numbers 4.1–4.4) over a range of FPR, FNR, and FDR of the predictions. (b INSET) Zoomed view including an experimental proof-of-principle based on predictions from network conservation (cFwd, cRev) or multiple types of evidence (mFwd, mRev). Fwd and Rev denote the series of experiments performed in the forward then reverse Y2H orientations, respectively. X-axis units of INSETs are absolute number of plates, not millions of plates as for the parent figures (a) and (b).
Lastly, we considered whether computational prediction of interactions, based on prior knowledge and data, could hasten the time to interactome completion. A variety of prediction methods have been proposed based on evolutionary conservation39–41—i.e., transfer of interaction measurements from one species to another—or based on integrating conservation with additional features such as co-expression and co-annotation42–47. Such predictions impact the experimental design by setting the prior probabilities of interaction for each protein pair in lieu of background probabilities. In the Prediction strategy, we explored the effect of setting these prior probabilities using theoretical prediction methods simulated over a range of predetermined prediction accuracies (a range of different values for FPR, FNR, and corresponding FDR of the predictions). We found that even predictors with very high FDRs could have a major impact on the mapping cost (Table 1 and Fig. 3b). For example, a predictor with 92.2% FDR gave a four-fold reduction in cost over Pooling, with a >50-fold reduction in cost to achieve 50% coverage and a savings of hundreds of fold in the early stages of mapping. Moreover, the 92.2% FDR means that even a predictor that makes 12 false predictions for every true one can lead to a major reduction in the cost of interactome completion. The best prediction method required approximately 385 person-years to achieve 95% coverage of the Drosophila protein network and 12 person-years to achieve 50% coverage. Thus, while obtaining full coverage of an interactome map may still be some years away, a draft scaffold providing half coverage might be feasibly achieved by a team of ~12 technicians working over a period of one year.
From theory to practice: An experimental proof-of-concept
Given the high performance of the Prediction strategy in simulations, we explored an experimental implementation in which Drosophila protein interactions were predicted using the cross-species method of Sharan et al.39 (Fig. 4a). According to this method, existing protein interaction networks in yeast, worm, and fly are aligned based on sequence similarity to identify conserved interaction clusters, and these alignments are used to transfer interactions that have been observed in some species but not yet in others (Fig. 4b). A total of 1,294 interactions were predicted using this method, each of which was prioritized as a candidate with high prior probability (92.4%) based on the FDR reported by Sharan et al.39 (7.6%).
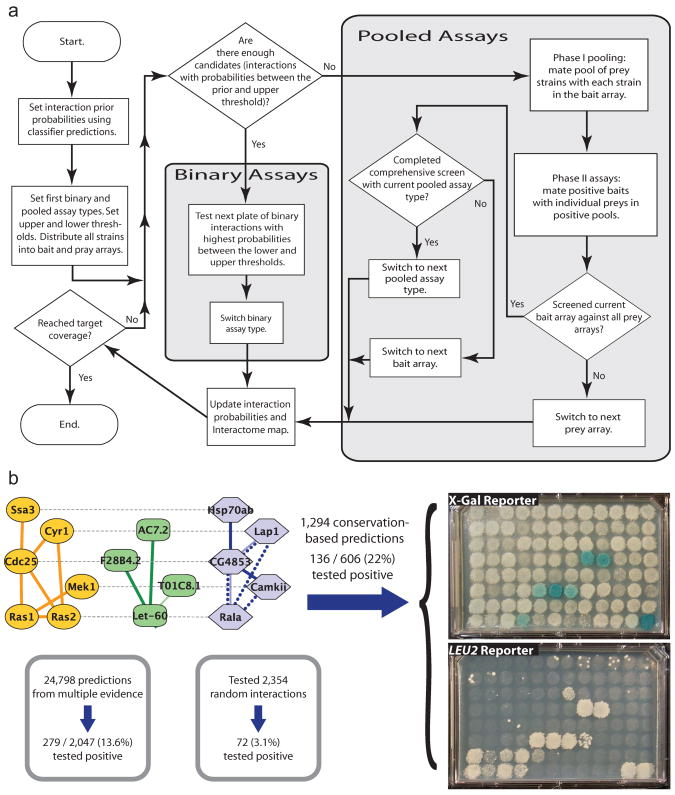
Figure 4. Design and implementation of the Prediction strategy for mapping the Interactome.
(a) State diagram of the Prediction strategy. This strategy combines interaction predictions with direct and pooled experiments to reduce the intermediate and total costs of Interactome mapping. (b) Application of the first steps of the Prediction strategy to the Drosophila interactome using conservation-based predictions from Sharan et al.39 or predictions made by integrating multiple evidence types (this study). Representative plates are shown for tests of conservation-based predictions using the X-Gal (top) or LEU2 (bottom) reporters.
Since this prior was much greater than the background probability of other protein pairs (0.1%), we began by using the pairwise Lex-A Y2H assay48 to test all 606 predictions for which sequence-verified clones were available. Of these, 136 tested positive and 470 negative. After each 96-well plate of tests (seven plates total), the interaction probabilities were updated resulting in an increase to >99.9% for pairs testing positive and a decrease to 90.5% for pairs testing negative. Since the 136 positives now had probability greater than the upper threshold (95%), all of these could be added to the interactome map and removed from further testing.
Although the remaining 470 predictions had tested negative once, their high probability (90.5%) still prioritized them as candidate interactions. Therefore, as dictated by the Prediction strategy these pairs were tested again immediately using a second assay type.
For this second assay, Lex-A Y2H was run in a “reverse” orientation in which the two proteins cloned as bait and prey, respectively, were exchanged as prey and bait. We tested 251 of the 470 predictions for which sequence-verified clones were available in the opposite orientation. This resulted in 35 positives, elevating these interactions to probability >99.9% and adding them to the map. The pairs that tested negative in the reverse orientation were downgraded to 88.1% probability. Overall, after performing Y2H in both forward and reverse orientations, 171 new interactions were identified out of 606 protein pairs for a success rate of 28%. Although we ended our experimentation at this point, the Prediction strategy could be continued by next testing the “double negatives” (pairs testing negative in both orientations of Lex-A Y2H) using a third type of assay such as Gal4-based Y2H.
A means of predicting additional protein interactions is to probabilistically integrate many different lines of evidence into a single classifier42–47. Along these lines, we applied a machine-learning-based classifier for predicting interactions that combined many relevant features including gene expression, domain-domain interactions, conserved protein-protein interactions, genetic interactions, and shared gene annotations (Supplementary Methods). We used this approach to generate 24,798 high confidence predictions. We randomly selected 2,047 of these for testing using forward-orientation Y2H and, as above, retested the negative pairs using reverse-orientation Y2H (for which clones were available). In total, this procedure added 279 new high-confidence interactions to the map for a 13.6% success rate. Combined over both conservation-based and multiple-evidence-based predictions, 450 new protein-protein interactions were added to the Drosophila map using 47 96-well plates (Fig. 3a,b). To establish the background rate of interaction, we also tested 2,354 randomly chosen pairs, 72 of which were positive yielding a 3% background rate (Fig. 4b). These results show that both types of prediction are highly enriched for true interactions. Note that even if all predicted interactions were true, the expected confirmation rate would be limited by the false negative rate of the Y2H assay, equal to 1–FNR =33% in our model.
Testing the conditional independence between assay types
An underlying assumption of our simulations is that different assay types are conditionally independent—i.e., given that a tested protein pair is known to be positive or negative, the result of one assay is uncorrelated with that of another. To examine the extent to which this assumption holds, we compared Y2H data for protein pairs tested in both forward and reverse orientations—the two assay types used in our study. Overall, we obtained Y2H tests in both orientations for 309 conservation-based predictions (including data reported above combined with additional tests; Supplementary Data). Of these, we observed 58 positives in the forward orientation and 50 positives in the reverse orientation, for an average positive rate of 17% [(58 + 50)/(309 * 2)]. Fifteen positives were found in both orientations, representing 4.9% of the tests. Assuming all predictions are true interactions, this percentage is very close to that predicted by conditional independence, for which 3.1% of tests are expected to be positive in both orientations [17% ^ 2]. If some predictions are not true as expected, the percentages come into even better agreement—e.g., a prediction FDR of 20% predicts that 4.8% positives would arise in both orientations. A similar analysis was performed on a set of 1,572 combined-evidence predictions that were tested in both orientations, leading to similar agreement with the conditional independence assumption.
DISCUSSION
The interactions predicted by cross-species conservation were at least as accurate as we had assumed in our simulations. On the other hand, their power to prioritize interactions is dependent on the network coverage in other species, and the long-term viability of this approach will depend on obtaining greater numbers of predictions than the 1,294 that are currently available. As interactome maps progress across an ever-widening array of species, these maps might be dynamically cross-compared to continually generate sufficient numbers of candidate interactions for testing. The second set of predictions, made by integrating various lines of evidence, had a lower success rate than the predictions based solely on conservation. Their potential utility is higher, however, since the number of available predictions is nearly 20 times that of the conservation-based predictions and could be increased further by including lower confidence predictions. Even with a lower success rate, the performance of the integrated classifier was superior to the best theoretical predictor we simulated.
Predictions lead to a lower interactome mapping cost for two reasons. First, predicted protein pairs are much more likely than arbitrary pairs to be true. Second, protein pairs with high prior probabilities do not require repeated positive measurements to confirm them as true interactions. Both effects underlie the finding that 450 new predicted interactions could be added to the interaction map using just 47 microtiter plates. In contrast, the Pooling strategy would require nearly 105 plates to add this number of interactions to the map.
One might intuitively object that, rather than test predicted interactions, a better strategy would focus on the “novel” areas of the interactome that have never before been suggested by any species or data set. The problem with such an approach is that it would very quickly produce an interactome map with a very high error rate. Conversely, the rationale behind the Thresholding and Prediction strategies is that one should first clean up the map by validating predicted interactions using real experiments, and only then resort to testing random protein pairs in pools.
A second objection might be that prioritizing candidate interactions requires the corresponding Y2H baits and preys to be rearrayed in microtiter plates in different orders over the course of an interaction mapping project. While the cost of rearraying was not included in our analysis, in our lab (Finley) these costs are greatly alleviated through robotic transfer systems. Certainly, failure to rearray leads to a ~4-fold increase in cost and a ~10-fold increase in the early stages of mapping (compare Pooling versus Prediction in Table 1).
Regardless, mapping the Interactome remains a daunting task. Our study makes it clear that achieving 95% coverage of an interactome requires many more screens than one pass through all pools or over all protein pairs. If complete coverage is to be obtained in the near future, it will be necessary to invoke better strategies for experimental design, technologies reporting fewer false negatives, or both. In terms of experimental design, we have shown that the cost of completion is reduced substantially by careful ordering of pooled screens. In terms of technology, our study underscores the importance of decreasing the FNR or of different assays that provide independent samples of a protein pair. Even if the error rates are lower than assumed here, advanced mapping strategies are still likely to be worthwhile (Suppl. Table 1). Here we have used two types of Y2H assay, forward and reverse orientations, to obtain multiple samples which appear largely independent. If the assays were partially dependent, multiple tests might still be worth the cost as long as they were not perfectly correlated (and the dependence could be handled quantitatively using a statistical model). In the present study, the conditional independence assumption leads to a “best-case scenario” or lower-bound on the number of interaction tests that will likely be required to achieve full coverage of an interactome. Further work will be needed to better characterize the relative dependencies among the wide range of other interaction assays that are currently available— if the current assays are highly dependent, then the required number of tests will be greater than was estimated here.
METHODS
Simulation procedure
“True” reference interactomes for fly and human were generated by random sampling of interactions from the set of all possible pairs of proteins using the interaction probabilities in the String database46. Protein pairs not included in the String database were sampled using a low background probability, such that the total number of interactions in the sampled interactomes agreed with current estimates of interactome sizes30 (~100,000 fly interactions and ~260,000 human interactions). The detectability of each protein pair was independently sampled for each new assay type (representing a new type of measurement technology or new bait/prey orientation) using a 66% FNR for true interactions and 0.2% FPR for false interactions (corresponding to 82% FDR). Once an interaction was defined as “Detectable/Undetectable”, direct pairwise experiments were assumed to be 100% reproducible for a given protein pair and assay. For pooled assays, each detectable interaction in the sample space of a pool was assumed to be observed in the pool with probability equal to the pooling sensitivity (Table 1). Pools with at least one observed interaction were declared positive. For each strategy, after every 1000 experiments the mapped interactomes were compared to the “true” interactomes and the coverage and FDR were recorded.
Yeast two-hybrid test of predicted interactions
We used the LexA-based yeast two-hybrid mating assay48 using sequence-verified clones as previously described36 (Supplementary Methods). All new protein interactions have been submitted to the IMEx consortium (http://imex.sf.net) through IntAct49 and assigned the identifier IM-9552. The data are also available at DroID (www.droidb.org).
Additional Methods
Detailed descriptions of the interaction probability model, the combined-evidence method for interaction prediction, the computation of thresholds, and the yeast two-hybrid test protocol appear in the Supplementary Methods.
Supplementary Material
Supplementary Data Results from protein-protein interaction predictions in Drosophila.
Supplementary Figure 1 Flow charts detailing the different interactome mapping strategies.
Supplementary Table 1 Interactome completion times for a range of error parameters.
Supplementary Table 2 Results of yeast two-hybrid test of predicted interactions.
Supplementary Methods
Acknowledgments
We thank S. Bandyopadhyay for critical reading of the manuscript and I. Bronner, K. Gulyas, B. Mangiola, and H. Zhang for expert technical assistance with the two-hybrid assays. We thank R. Karp and R. Sharan for discussions on earlier versions of this work. This work was supported by National Institutes of Health grants RR018627, GM070743, and HG001536.
Footnotes
AOP
Different experimental designs for protein interaction mapping were modeled to compare their efficiency in completing an interactome map. The strategy that minimized the final cost was tested in an ongoing Drosophila melanogastor interactome project where it found 450 high-confidence interactions using only 47 microtiter plates.
ISSUE
Different experimental designs for protein interaction mapping were modeled to compare their efficiency in completing an interactome map. The strategy that minimized the final cost was tested in an ongoing Drosophila melanogastor interactome project where it found 450 high-confidence interactions using only 47 microtiter plates.
References
- 1.Fields S. High-throughput two-hybrid analysis. The promise and the peril. Febs J. 2005;272:5391–9. doi: 10.1111/j.1742-4658.2005.04973.x. [DOI] [PubMed] [Google Scholar]
- 2.Giot L, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–36. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 3.Ito T, et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569–74. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li S, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–3. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rual JF, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–8. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 6.Stelzl U, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–68. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki H, et al. Protein-protein interaction panel using mouse full-length cDNAs. Genome Res. 2001;11:1758–65. doi: 10.1101/gr.180101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uetz P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–7. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 9.Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–6. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 10.Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–7. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 11.Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–43. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 12.Harbison CT, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pokholok DK, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–27. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 14.Ren B, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–9. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 15.Tong AH, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–8. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 16.Collins SR, Schuldiner M, Krogan NJ, Weissman JS. A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome Biol. 2006;7:R63. doi: 10.1186/gb-2006-7-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bao L, et al. Combining gene expression QTL mapping and phenotypic spectrum analysis to uncover gene regulatory relationships. Mamm Genome. 2006;17:575–83. doi: 10.1007/s00335-005-0172-2. [DOI] [PubMed] [Google Scholar]
- 18.Chesler EJ, Lu L, Wang J, Williams RW, Manly KF. WebQTL: rapid exploratory analysis of gene expression and genetic networks for brain and behavior. Nat Neurosci. 2004;7:485–6. doi: 10.1038/nn0504-485. [DOI] [PubMed] [Google Scholar]
- 19.Petretto E, et al. Heritability and tissue specificity of expression quantitative trait loci. PLoS Genet. 2006;2:e172. doi: 10.1371/journal.pgen.0020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schadt EE, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 21.Rain JC, et al. The protein-protein interaction map of Helicobacter pylori. Nature. 2001;409:211–5. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- 22.Parrish JR, et al. A proteome-wide protein interaction map for Campylobacter jejuni. Genome Biol. 2007;8:R130. doi: 10.1186/gb-2007-8-7-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LaCount DJ, et al. A protein interaction network of the malaria parasite Plasmodium falciparum. Nature. 2005;438:103–7. doi: 10.1038/nature04104. [DOI] [PubMed] [Google Scholar]
- 24.Uetz P, et al. Herpesviral protein networks and their interaction with the human proteome. Science. 2006;311:239–42. doi: 10.1126/science.1116804. [DOI] [PubMed] [Google Scholar]
- 25.von Brunn A, et al. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS ONE. 2007;2:e459. doi: 10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.CRISP-- Computer Retrieval of Information on Scientific Projects. 2008. (National Institutes of Health, http://crisp.cit.nih.gov/
- 27.Lander ES, Waterman MS. Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics. 1988;2:231–9. doi: 10.1016/0888-7543(88)90007-9. [DOI] [PubMed] [Google Scholar]
- 28.Weber JL, Myers EW. Human whole-genome shotgun sequencing. Genome Res. 1997;7:401–9. doi: 10.1101/gr.7.5.401. [DOI] [PubMed] [Google Scholar]
- 29.von Mering C, et al. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 30.Hart GT, Ramani AK, Marcotte EM. How complete are current yeast and human protein-interaction networks? Genome Biol. 2006;7:120. doi: 10.1186/gb-2006-7-11-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lappe M, Holm L. Unraveling protein interaction networks with near-optimal efficiency. Nat Biotechnol. 2004;22:98–103. doi: 10.1038/nbt921. [DOI] [PubMed] [Google Scholar]
- 32.Cusick ME, Klitgord N, Vidal M, Hill DE. Interactome: gateway into systems biology. Hum Mol Genet. 2005;14:R171–81. doi: 10.1093/hmg/ddi335. Spec No 2. [DOI] [PubMed] [Google Scholar]
- 33.Kocher T, Superti-Furga G. Mass spectrometry-based functional proteomics: from molecular machines to protein networks. Nat Methods. 2007;4:807–15. doi: 10.1038/nmeth1093. [DOI] [PubMed] [Google Scholar]
- 34.Parrish JR, Gulyas KD, Finley RL., Jr Yeast two-hybrid contributions to interactome mapping. Curr Opin Biotechnol. 2006;17:387–93. doi: 10.1016/j.copbio.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 35.Deane CM, Salwinski L, Xenarios I, Eisenberg D. Protein interactions: two methods for assessment of the reliability of high throughput observations. Mol Cell Proteomics. 2002;1:349–56. doi: 10.1074/mcp.m100037-mcp200. [DOI] [PubMed] [Google Scholar]
- 36.Stanyon CA, et al. A Drosophila protein-interaction map centered on cell-cycle regulators. Genome Biol. 2004;5:R96. doi: 10.1186/gb-2004-5-12-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–95. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 38.Zhong J, Zhang H, Stanyon CA, Tromp G, Finley RL., Jr A strategy for constructing large protein interaction maps using the yeast two-hybrid system: regulated expression arrays and two-phase mating. Genome Res. 2003;13:2691–9. doi: 10.1101/gr.1134603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharan R, et al. Conserved patterns of protein interaction in multiple species. Proc Natl Acad Sci U S A. 2005;102:1974–9. doi: 10.1073/pnas.0409522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matthews LR, et al. Identification of potential interaction networks using sequence-based searches for conserved protein-protein interactions or “interologs”. Genome Res. 2001;11:2120–6. doi: 10.1101/gr.205301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boulton SJ, et al. Combined functional genomic maps of the C. elegans DNA damage response. Science. 2002;295:127–31. doi: 10.1126/science.1065986. [DOI] [PubMed] [Google Scholar]
- 42.Ben-Hur A, Noble WS. Kernel methods for predicting protein-protein interactions. Bioinformatics. 2005;21 (Suppl 1):i38–46. doi: 10.1093/bioinformatics/bti1016. [DOI] [PubMed] [Google Scholar]
- 43.Jansen R, et al. A Bayesian networks approach for predicting protein-protein interactions from genomic data. Science. 2003;302:449–53. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- 44.Lee I, Date SV, Adai AT, Marcotte EM. A probabilistic functional network of yeast genes. Science. 2004;306:1555–8. doi: 10.1126/science.1099511. [DOI] [PubMed] [Google Scholar]
- 45.Lu LJ, Xia Y, Paccanaro A, Yu H, Gerstein M. Assessing the limits of genomic data integration for predicting protein networks. Genome Res. 2005;15:945–53. doi: 10.1101/gr.3610305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Mering C, et al. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–61. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu H, Paccanaro A, Trifonov V, Gerstein M. Predicting interactions in protein networks by completing defective cliques. Bioinformatics. 2006;22:823–9. doi: 10.1093/bioinformatics/btl014. [DOI] [PubMed] [Google Scholar]
- 48.Finley RL, Jr, Brent R. Interaction mating reveals binary and ternary connections between Drosophila cell cycle regulators. Proc Natl Acad Sci U S A. 1994;91:12980–4. doi: 10.1073/pnas.91.26.12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kerrien S, et al. IntAct--open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–5. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data Results from protein-protein interaction predictions in Drosophila.
Supplementary Figure 1 Flow charts detailing the different interactome mapping strategies.
Supplementary Table 1 Interactome completion times for a range of error parameters.
Supplementary Table 2 Results of yeast two-hybrid test of predicted interactions.
Supplementary Methods