Abstract
The nervous system coordinates many aspects of body function such as learning, memory, behaviour and locomotion. Therefore, it must develop and maintain an intricate network of differentiated neuronal cells, which communicate efficiently with each other and with non-neuronal target cells. Unlike most somatic cells, differentiated neurons are post-mitotic and characterized by a highly polarized morphology that determines the flow of information. Among other post-translational modifications, the ubiquitination of specific protein substrates was recently shown to have a crucial role in the regulation of neuronal development and differentiation. Here, we review recent findings that illustrate the mechanisms that mediate the temporal and spatial control of neuronal protein turnover by the ubiquitin–proteasome system (UPS), which is crucial for the development and function of the nervous system.
Keywords: neuron, neuronal activity, protein degradation, SCF ligase complex, ubiquitin
Glossary
AMPA α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid
APC/C anaphase-promoting complex/cyclosome
β-TrCP β-transducin repeat-containing protein
BTB broad-complex/tramtrack/brick-a-brac
C4da class IV dendritic arborization
CaMKII calcium/calmodulin-dependent protein kinase II
CDC20 cell division cycle 20 homologue
CDH1 CDC20 homologue 1
DFsn Drosophila homologue of FSN-1
Diap1 Drosophila inhibitor of apoptosis 1
DLK-1 death associated protein kinase-like kinase 1
Dronc Drosophila Nedd2-like caspase
Dunc13 Drosophila unc-13
FMRP fragile X mental retardation protein
FSN-1 F-box synaptic protein 1
GLO-4 gut granule loss 4
GLR-1 glutamate receptor family 1
GluRIIa glutamate receptor IIa
GSK3β glycogen synthase kinase 3β
IgSF immunoglobulin superfamily
KEL-8 Kelch repeat-containing protein 8
Kelch 50-residue motif named after Drosophila Kelch
LIN-23 abnormal cell lineage 23
mRNA messenger RNA
NMDA N-methyl-D-aspartic acid
Phr1 Pam/Highwire/RPM-1
PI(3)K phosphatidyl inositol-3-kinase
PSD postsynaptic density
PSD-95 postsynaptic density protein 95
Rab Ras-related in brain
RIM1 regulating synaptic membrane exocytosis 1
RING really interesting new gene
RPM-1 regulator of presynaptic morphology 1
SAPAP synapse-associated protein 90 (SAP90)/PSD95-associated protein
SCF Skip1, Cullin, F-box
SEL-10 suppressor/enhancer of LIN-12
SHANK SH3 and multiple ankyrin repeat domain-containing protein
Skip1 S-phase kinase-associated protein 1
SKR-1 Skip1-related ubiquitin ligase complex component
SNK serum-inducible kinase
SPAR GTPase-activating protein spine-associated RapGAP
SYG-1 synaptogenesis abnormal
UCHL1 ubiquitin carboxyl-terminal esterase L1
UPS ubiquitin–proteasome system
Regulation of the nervous system by the UPS
A defining characteristic of neurons is their highly polarized morphology, which is typically composed of soma, axons and dendrites. The passage of information between neurons or between neurons and non-neuronal target cells is mediated by chemical synapses. The synapse is a specialized cellular structure that consists of presynaptic boutons at the axon terminal—which are varicosities that contain the machinery for neurotransmitter release—and a postsynaptic dendritic side (Fig 1). The transfer of information typically occurs along an elongated axon towards the presynaptic bouton; at the postsynaptic side, synaptic protrusions that emanate from dendritic shafts, which are known as spines, act as recipients of information for efficient signal transduction. These dendritic spines are composed of a postsynaptic membrane that contains many types of neurotransmitter receptors and an adjacent microdomain beneath the plasma membrane—known as the PSD—that consists of scaffolding and signalling molecules. The abundance and stabilization of neurotransmitter receptors within the plasma membrane is regulated by endocytosis and exocytosis, and controlled through dynamic interactions of the receptors with scaffolding proteins of the PSD. Neuronal stimulation induces the release of specific neurotransmitters from presynaptic vesicles into the synaptic cleft, which subsequently regulate the activity of neurotransmitter receptors at the postsynaptic membrane (Fig 1). The ability of a neuron to respond to differences in signal strength depends on the regulated release of neurotransmitters and on the concentration of receptors at the postsynaptic plasma membrane, which is crucial for information storage and transmission.
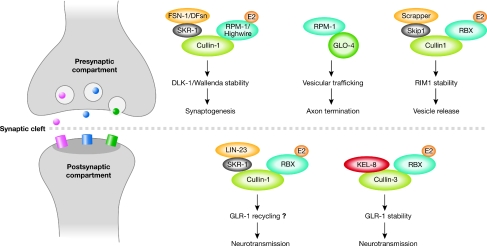
Figure 1.
Multimeric cullin-based E3 ligases control neuronal development and activity. Cullin complexes direct protein turnover in the presynaptic and postsynaptic compartments. A Cullin-1 complex containing the F-box molecule FSN-1 (Caenorhabditis elegans)/DFsn (Drosophila) and the RING-finger protein RPM-1 (C. elegans)/Highwire (Drosophila) degrades DLK-1 (C. elegans)/Wallenda (Drosophila) at the presynaptic compartment to control synaptogenesis. In C. elegans, RPM-1 also functions in an FSN-1-independent pathway during axon termination. In this case, it interacts with the guanine nucleotide-exchange factor GLO-4 independently of its RING-finger domain and promotes vesicular trafficking through a Rab GTPase pathway. The F-box protein Scrapper forms a cullin-based complex to degrade the vesicle-priming factor RIM1, thereby controlling vesicle release from the presynaptic side. At the postsynaptic compartment, a Cullin-1 complex including the F-box protein LIN-23 influences the recycling of the AMPA-type receptor GLR-1. The stability of GLR-1 itself is also regulated by the Cullin-3 complex through its substrate-recognition factor KEL-8. AMPA, α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid; DFsn, Drosophila homologue of FSN-1; DLK-1, death associated protein kinase-like kinase 1; E2, E2 ligase; FSN-1, F-box synaptic protein 1; GLO-4, gut granule loss 4; GLR-1, glutamate receptor family 1; KEL-8, Kelch repeat-containing protein 8; LIN-23, abnormal cell lineage 23; Rab, Ras-related in brain; RBX, Ring box molecule; RIM1, regulating synaptic membrane exocytosis 1; RING, really interesting new gene; RPM-1, regulator of presynaptic morphology 1; Skip1, S-phase kinase-associated protein 1; SKR-1, Skip1-related ubiquitin ligase complex component.
Therefore, it is not surprising that the establishment, maintenance and modulation of axons, dendrites and synapses involve several post-translational regulation pathways such as protein phosphorylation and ubiquitination. Among other protein modifications, the spatial and temporal control of ubiquitination have a crucial role during neuronal development and differentiation (Hegde & Upadhya, 2007; Yi & Ehlers, 2007). The degradation of proteins conjugated to ubiquitin allows the neuron to adapt its excitability rapidly at defined regions such as the postsynaptic side (Fig 1). The fact that synaptic transmission depends on the composition of a highly dynamic multiprotein structure, consisting of membrane-bound receptors, ion channels and adhesion molecules, makes it susceptible to proteolytic regulation. For example, once neurotransmitter receptors are synthesized, their density on the postsynaptic membrane is controlled through the concerted and balanced action of ligand-induced endocytosis, protein recycling to the plasma membrane or lysosomal proteolysis. In contrast to proteasomal degradation, which mainly affects intracellular substrates that are conjugated to a chain of several ubiquitin molecules, the endocytosis of membrane-bound receptors—which are subsequently either sorted into recycling endosomes or degraded by acidic hydrolases in the lysosome—occurs mainly after mono-ubiquitination (Hicke & Dunn, 2003).
Recent data have revealed a crucial role of the UPS in the spatial and temporal control of protein turnover in the nervous system, which regulates the development and maintenance of specialized neuronal structures and, consequently, neuronal transmission. The involvement of the UPS during these processes is well established and has been previously reviewed (Hegde, 2004; Hegde & Upadhya, 2007; Yi & Ehlers, 2007). However, the precise mechanisms that underlie the local formation of enzyme–substrate complexes and the potential influence of neuronal activity on local protein degradation often remain to be identified. The idea that local degradation of proteins might be a central process underlying synaptic plasticity has emerged recently (Hegde, 2004). Here, we discuss spatially controlled ubiquitin-dependent degradation pathways and highlight the regulatory principles that govern these processes, thereby providing new insights into neuronal development and function.
Ubiquitin-dependent protein degradation
The UPS constitutes the main eukaryotic protein degradation system and its correct function is required in diverse cellular processes such as cell-cycle progression, signal transduction, development and protein quality control. The multi-ubiquitination of a substrate to target it for proteasomal degradation is usually mediated by a cascade that includes ubiquitin-activating E1 enzymes, ubiquitin-conjugating E2 enzymes and E3 ubiquitin ligases (Kerscher et al, 2006; Pickart, 2001). However, multi-ubiquitin chains can also be disassembled by deubiquitination enzymes, which cleave off ubiquitin molecules from the chain, thereby antagonizing substrate degradation.
In contrast to E1 and E2 enzymes, E3 ligases are highly diverse. For example, RING finger-type E3 enzymes can function as monomers or as multimeric E3 complexes, which contain a RING-finger enzyme associated with a core protein that binds to the substrate through alternative adaptor molecules. Therefore, the RING-finger ligase, together with an E2 enzyme, is able to promote substrate ubiquitination. The core protein defines two groups of RING-finger E3 complexes: the cullin-based E3s and the APC/C-based E3s (Sumara et al, 2008). Among the cullin-based E3s, SCF complexes contain the scaffold protein Cullin1, the adaptor Skip1 and various F-box proteins for substrate recognition, whereas the substrate-recognition molecules of Cullin3-containing E3s have a BTB domain, and directly bridge the interaction between the substrate and Cullin3 (Fig 1; Sumara et al, 2008). However, the composition of APC/C-based E3s is more complex. In vertebrates, it consists of at least 12 subunits, which include the cullin-like scaffold protein APC2, the RING finger-type protein APC11, and the activating cofactors CDC20 and CDH1 that mediate substrate recognition (Peters, 2006). Hence, the variability of subunit combination results in many different E3 ligase complexes, which allows for a broad spectrum of substrate selectivity (Fig 1). Moreover, the subcellular and developmental availability of ubiquitination enzymes confers an additional layer of spatial and temporal control of protein degradation (Pines & Lindon, 2005).
The role of the UPS during the cell cycle of actively dividing cells is well established, whereas its regulatory function in non-dividing cells—such as differentiated neurons—is rather unclear. Primary evidence for a neuronal function of the UPS was shown during learning and memory in the marine mollusc Aplysia (Chain et al, 1999; Hegde et al, 1993, 1997). Moreover, a mutation in the Drosophila E2 enzyme bendless results in an altered jump response caused by impaired synaptic connectivity between the giant fibre neuron and its muscle motor neuron (Muralidhar & Thomas, 1993). Intriguingly, mutations in the E3 ligase gene parkin and the human deubiquitination enzyme UCHL1 cause loss of dopaminergic neurons in patients affected by familial forms of Parkinson disease (Leroy et al, 1998; Shimura et al, 2000). Together, these initial observations indicate a crucial role of the UPS in correct neuronal development and function. Recent studies in various model organisms have provided additional mechanistic insights into how the local and temporal regulation of ubiquitin-mediated protein turnover contributes to the establishment and maintenance of the neuronal circuit.
Protein turnover during neuronal development
The formation of the nervous system is a complex and highly dynamic event, during which neuronal processes and connections are established and integrated with the development of the entire organism. During larval metamorphosis in Drosophila, a set of peripheral sensory neurons, known as C4da neurons, undergo severe dendritic remodelling. This specific remodelling of dendrites has been proposed to require a two-step regulatory pathway that involves ubiquitin-dependent degradation and apoptotic protein degradation (Kuo et al, 2005). In fact, ubiquitin-mediated turnover of the E3 ligase Diap1 results in the stabilization and enhanced activation of the pro-apoptotic caspase Dronc specifically in dendrites (Kuo et al, 2006). Therefore, the study by Kuo and colleagues represents an interesting mechanistic example of how neuronal remodelling can be regulated at the level of ubiquitin conjugation. It is tempting to speculate that the spatial control of E3 ligase activity might be a common principle to adjust neuronal degradation pathways to developmental stages.
The UPS is also linked to the formation and maintenance of neuronal polarity. Rat hippocampal neurons that are grown in cell culture undergo extensive remodelling before their final polarization is established. The soma of these neurons extends several processes, known as neurites, only one of which forms a single axon, whereas the others develop into dendrites (Craig & Banker, 1994). The establishment and maintenance of neuronal polarity depend on the activity of the PI(3)K pathway (Arimura & Kaibuchi, 2005). PI(3)K activates the serine/threonine kinase AKT at the tip of the axon, which, in turn, inactivates GSK3β, thereby promoting axon formation. Interestingly, axon growth and maintenance are determined by AKT protein levels, which are stable in neurites that develop into axons, but are reduced in a ubiquitin-dependent manner in neurites that differentiate into dendrites (Yan et al, 2006). Therefore, the development of neuronal polarity in cell culture is established by the locally coordinated degradation of the serine/threonine kinase AKT.
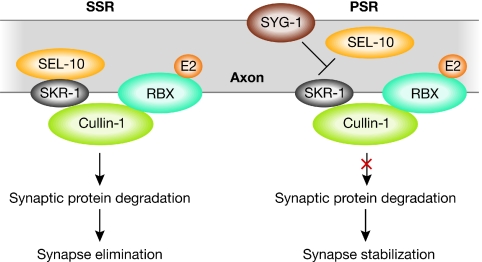
Recently, another developmental degradation pathway that regulates the local elimination of synapses was identified in Caenorhabditis elegans (Ding et al, 2007; Shen & Bargmann, 2003). In adult hermaphrodite worms, the motor neuron that regulates egg-laying behaviour connects to its target muscle cells in the primary synapse region (PSR). During larval development, this motor neuron assembles an additional secondary synpase region (SSR) that is eliminated when worms become adult. In a screen for molecules that regulate the elimination of the SSR, Ding and colleagues identified the IgSF protein SYG-1, which controls the formation of a multimeric SCF E3 ligase complex. Intriguingly, SYG-1 is exclusively localized to the PSR rather than the SSR. At the PSR, SYG-1 inhibits the formation of a functional SCF ligase complex by preventing the interaction of the SKR-1 adaptor with the substrate-recognition subunit SEL-10. The authors suggested that the amount of functional SCFSEL-10 complex is limiting in the motor neuron, where it functions to degrade synaptic proteins. A high concentration of the SCFSEL-10 inhibitor SYG-1 helps to stabilize the PSR, whereas the functional SCFSEL-10 ligase complex eliminates the SSR (Fig 2). Hence, local inhibition of an E3 ligase complex controls the survival of synapses, thereby regulating the formation of a specific neuronal circuit during C. elegans development. To confirm the local degradation pathway indicated by this study, the identification of SCFSEL-10 substrates is eagerly awaited.
Figure 2.
Developmental control of synaptogenesis of the Caenorhabditis elegans egg-laying motor neuron. During early post-embryonic development, two synaptic structures are assembled at the SSR and PSR. The Cullin-1 E3 ligase is present at both sites, but is active only at the SSR where it degrades synaptic proteins. The function of Cullin E3 is inhibited at the PSR by SYG-1, thereby preventing the binding of the F-box protein SEL-10 to the Cullin complex. This spatial control of SCF complex formation leads to the elimination of the SSR, while maintaining the PSR during adulthood. E2, E2 ligase; PSR, primary synapse region; RBX, Ring box molecule; SCF, Skip1, Cullin, F-box; SEL-10, suppressor/enhancer of LIN-12; Skip1, S-phase kinase-associated protein 1; SKR-1, Skip1-related ubiquitin ligase complex component; SSR, secondary synapse region; SYG-1, synaptogenesis abnormal.
Presynaptic ubiquitin-dependent protein turnover
A main function of the presynaptic compartment is the release of specific neurotransmitters from presynaptic vesicles into the synaptic cleft upon neuronal activation. Intriguingly, several recent studies provide evidence that presynaptic processes are also under the control of local ubiquitin-dependent regulation pathways. For example, enzymes of the UPS localize to presynaptic boutons in Drosophila, and the inhibition of the 26S proteasome elevates synaptic transmission (Speese et al, 2003). One known presynaptic target of the UPS is Dunc13, which is important for synaptic vesicle fusion (Speese et al, 2003). However, the E3 ligase required for Dunc13 ubiquitination remains to be identified and it is likely that additional presynaptic substrates exist. Similarly, the APC/C controls synaptic size through the regulation of Liprin-α protein levels at the presynaptic compartments of Drosophila neuromuscular junctions (NMJs; van Roessel et al, 2004).
Furthermore, in mouse hippocampal neurons, synaptic plasticity is modulated by the F-box protein Scrapper. This protein is part of an SCF ligase complex and mediates substrate recognition. Local enrichment of Scrapper controls the presynaptic concentration of RIM1, which is a Ca2+-dependent vesicle-priming factor that regulates neurotransmitter release (Yao et al, 2007). Intriguingly, a different SCF E3 ligase complex was reported to control the formation of synapses in C. elegans and Drosophila, suggesting that conserved regulatory degradation mechanisms exist. The C. elegans protein RPM-1 forms an SCF complex with the Skip1 homologue SKR-1, Cullin-1 and the F-box protein FSN-1 (Liao et al, 2004), which regulates the stability of the kinase DLK-1 (Nakata et al, 2005). Loss of RPM-1 results in a reduction of NMJs. Similarly, the RPM-1 and FSN-1 homologues Highwire and DFsn in Drosophila control the abundance of the DLK-1 homologue Wallenda in the synapses of NMJs (Collins et al, 2006; Wu et al, 2007). Interestingly, mutations in Highwire and DFsn cause increased numbers of synaptic boutons (Wan et al, 2000; Wu et al, 2007), which has also been reported for a conditional mouse mutant affecting the RPM-1/Highwire homologue Phr1 (Bloom et al, 2007).
Clearly, several E3 ligase complexes exist that control different aspects of presynaptic function through ubiquitination, such as synapse formation and size, as well as neurotransmitter release. It is tempting to speculate that the assembly of various SCF complexes might be under developmental and spatial control, possibly as a result of the limited abundance of certain subunits. It is interesting to note that, depending on the nerve-cell type, C. elegans RPM-1 also functions independently of ubiquitin. As mentioned above, RPM-1-mediated ubiquitination is important to regulate synaptogenesis in motor neurons; however, in mechanosensory neurons, RPM-1 controls axon termination through the control of vesicle trafficking through the guanine nucleotide-exchange factor GLO-4 (Fig 1; Grill et al, 2007). Therefore, the function of RPM-1 seems to be modulated in a cell type-specific manner, which emphasizes the idea that SCF subunits could be differentially combined or activated in distinct neurons.
Postsynaptic ubiquitin-dependent protein turnover
In contrast to the presynaptic compartment, the postsynaptic side receives information by the binding of neurotransmitters to their receptors and, for example, subsequent receptor activation, which leads to the opening of ligand-gated ion channels, thereby transducing the chemical information into ion flux. The balance between endocytosis and exocytosis is usually crucial for regulating the amount of neurotransmitter receptors present at the postsynaptic membrane and defines the synaptic conductivity. Notably, receptor endocytosis is induced by ubiquitination, whereas receptor degradation typically takes place in the lysosome and is not mediated by the 26S proteasome (Hicke & Dunn, 2003).
The abundance of the C. elegans AMPA-type glutamate receptor GLR-1 at the postsynaptic side is regulated by LIN-23—the F-box subunit of an SCF complex—and the APC/C. Given that the amount of ubiquitinated GLR-1 is not altered in worms lacking LIN-23 or APC/C subunits, different concentrations at the postsynaptic membrane might be a consequence of GLR-1 recycling effects rather than its ubiquitin-dependent degradation (Dreier et al, 2005; Juo & Kaplan, 2004). By contrast, the BTB-Kelch protein KEL-8 is involved in GLR-1 turnover, as GLR-1–ubiquitin conjugates are stabilized in C. elegans kel-8 loss-of-function mutants. Moreover, KEL-8 localizes adjacent to GLR-1 in postsynaptic clusters of the ventral cord (Schaefer & Rongo, 2006), although the direct ubiquitination of GLR-1 by KEL-8 remains to be shown. Similarly, mutations in the APC/C gene Apc2 in Drosophila result in elevated levels of the glutamate receptor GluRIIa (van Roessel et al, 2004) and, in mammals, inhibition of the 26S proteasome increases the amount of ubiquitinated AMPA-type and NMDA-type ionotropic glutamate receptors (Kato et al, 2005; Rezvani et al, 2007). Moreover, the endocytosis of AMPA-type receptors is influenced indirectly by the ubiquitination of PSD-95, which acts as a scaffold molecule for receptors and signalling molecules at the PSD (Colledge et al, 2003; Patrick et al, 2003). Hence, the endocytosis of glutamate receptors seems to involve their ubiquitination, as well as the UPS-mediated degradation of PSD scaffolding molecules. Therefore, ubiquitin-dependent protein degradation also has a significant role in signalling processes at the postsynaptic side. Several different E3 ligase complexes are involved in regulating the abundance of the GLR-1 receptor at the postsynaptic surface and it will be important to identify the mechanisms that determine the preferential use of one ubiquitination pathway over another, which might be defined by the combination of alternative E3 ligase subunits that are expressed at a particular developmental stage. The ubiquitin-dependent turnover of other neurotransmitter receptors might also be important to modulate synaptic transmission. In light of GLR-1 regulation, future studies on neuronal receptor turnover will need to distinguish between endocytotic and proteasomal degradation pathways.
Protein degradation modulated by neuronal activity
The primary function of a typical neuron is to receive and transmit information. In this context, an interesting aspect is how protein degradation is integrated with neuronal activity. Using an in vivo substrate as a reporter for ubiquitin-mediated proteolysis, Bingol and Schuman showed that neuronal activation results in increased proteasomal degradation in the dendritic spines of rat hippocampal neurons. Additional localization studies revealed that the exit rate of the proteasome from dendritic spines is reduced, probably owing to its sequestration by the actin cytoskeleton (Bingol & Schuman, 2006). Along this line, the relationship between ubiquitin-dependent proteolysis and neuronal activity was previously shown for various substrates (Ehlers, 2003; Pak & Sheng, 2003). In fact, an increase in synaptic activity enhances the ubiquitination and degradation of several synaptic scaffolding proteins, including SHANK and SAPAP, which are crucial components of the PSD and are involved in postsynaptic remodelling. Importantly, the activity-dependent decrease in protein levels could be reversed by proteasomal inhibition (Ehlers, 2003). Although the long time scale (24 h) used in these experiments does not allow us to conclude whether the effects observed are direct, it suggests an activity-induced postsynaptic function of the UPS that is important for synaptic remodelling. Indeed, the ubiquitin-dependent degradation of the spine-associated protein SPAR controls the local morphogenesis of dendritic spines. The kinase SNK phosphorylates SPAR and promotes its degradation in activated hippocampal neurons. SPAR ubiquitination is mediated by an SCF ligase complex that contains the F-box protein β-TrCP. This protein—the mammalian orthologue of C. elegans LIN-23, which is implicated in the postsynaptic turnover of glutamate receptors (see above)—recognizes SPAR through a canonical phosphodegron sequence and promotes its ubiquitin-dependent degradation in hippocampal neurons (Ang et al, 2008). As SPAR is essential for spine maintenance, its degradation causes a loss of mature spines. The ubiquitin-mediated control of SPAR turnover therefore represents an elegant mechanism that illustrates how synaptic transmission can be adjusted in response to neuronal activity (Pak & Sheng, 2003; Pak et al, 2001).
Conclusions
Recent studies have shown that the formation and maintenance of specialized neuronal structures and neuronal activity involve local protein degradation controlled by ubiquitin conjugation. However, the way in which spatially restricted proteolysis is mechanistically achieved in non-dividing neurons has remained unclear (Sidebar A). Conceivably, this could be initiated by the selective phosphorylation of a substrate, as shown for AKT and SPAR. Alternatively, the UPS itself could be regulated locally by the subcellular recruitment of the proteasome (Bingol & Schuman, 2006), the inhibition of E3 ligase activity or the degradation of ubiquitination enzymes (Kuo et al, 2006).
Local degradation of ubiquitinated substrates might be achieved by the differential activation of the 26S proteasome, depending on the subcellular context. In line with this hypothesis, proteasomal function was shown to be increased in synaptosomal compartments, in comparison to nuclear compartments, in Aplysia and mouse neurons (Upadhya et al, 2006). In this case, proteasomal activity seems to be regulated either by differential phosphorylation of the 26S proteasome subunits or by differential assembly of cofactors. It will be important to identify the precise mechanism underlying this spatial modulation of proteasomal activity.
Interestingly, local protein degradation might be coupled with the local translation of neuronal proteins. The necessity of a concerted balance of protein synthesis and proteasomal degradation during synaptic plasticity has been shown in mammals and Aplysia (Fonseca et al, 2006; Hegde & DiAntonio, 2002). Moreover, a recent study in mice supports a direct role for the proteasome in stabilizing locally translated proteins in dendrites (Dong et al, 2008). In this regard, the translation of the PSD protein SHANK seems to be spatially restricted because its mRNA was found specifically in the distal dendrites of hippocampal and cerebellar neurons (Bockers et al, 2004). In addition, neuronal activity causes SHANK degradation by the UPS (Ehlers, 2003). These findings suggest an interplay between mRNA targeting and protein degradation, which is integrated with neuronal activity. Moreover, the UPS might modulate local protein synthesis directly because the translational regulator FMRP is degraded in a ubiquitin-dependent manner, thereby affecting the translation of FMRP target mRNAs in the soma and dendrites of mouse hippocampal neurons (Hou et al, 2006). Similarly, the proteasomal substrate Armitage regulates the translation of CaMKII mRNA in Drosophila synapses (Ashraf et al, 2006). It is attractive to speculate that such a function of the UPS in gene expression might also be involved in the local synthesis of ubiquitination enzymes, thereby constituting an autoregulatory feedback loop.
Notably, one of the main regulatory principles for spatially restricted protein turnover in neurons seems to be based on the combination of alternative E3 ligase subunits. As described above, different E3 ligase complexes target specific substrates for degradation in defined neuronal compartments (Table 1). Furthermore, the activity of an E3 ligase could be under local control depending on the presence of regulatory cofactors (Figs 1, 2). Therefore, the temporal and spatial control of E3 ligase activity seems to provide neurons with an additional mechanism to rapidly regulate protein levels in subcellular compartments. It will be necessary to establish new biochemical purification strategies and novel in vivo UPS reporter assays in multicellular model organisms to elucidate further the connection between ubiquitination, neuronal development and activation.
Table 1.
Neuronal ubiquitin-dependent degradation factors
| Ligase | Target | Localized function | Neuronal function | Organism | References |
|---|---|---|---|---|---|
| Unknown | Dunc13 | Presynaptic | Synaptic transmission | Drosophila | Speese et al, 2003 |
| SCFScrapper | RIM1 | Presynaptic | Synaptic transmission | Mouse | Yao et al, 2007 |
| RPM-1 | DLK-1 | Presynaptic | Synaptogenesis | C. elegans | Nakata et al, 2005 |
| Phr1 | Unknown DLK | Synapse Axon | Synaptogenesis Axon termination | Mouse | Bloom et al, 2007; Lewcock et al, 2007 |
| Highwire | Wallenda | Presynaptic | Synaptic growth | Drosophila | Collins et al, 2006 |
| KEL-8 | GLR-1 | Postsynaptic | Synaptic transmission | C. elegans | Schaefer & Rongo, 2006 |
| Unknown | AKT | Neurites | Neuron polarization | Rat | Yan et al, 2006 |
| Unknown | Diap1 | Postsynaptic Dronc activation | Dendritic pruning | Drosophila | Kuo et al, 2006 |
| SCFSEL-10 | Unknown | Axonal | Synapse elimination | C. elegans | Ding et al, 2007 |
| SCFβ-TrCP | SPAR | Postsynaptic | Dendritic spine morphogenesis | Rat | Pak & Sheng, 2003; Ang et al, 2008 |
Diap1, Drosophila inhibitor of apoptosis 1; DLK-1, death associated protein kinase-like kinase; Dunc13, Drosophila unc-13; GLR-1, glutamate receptor family 1; KEL-8, Kelch repeat-containing protein; Phr1, Pam/Highwire/RPM-1; RIM1, regulating synaptic membrane exocytosis 1; RPM-1, regulator of presynaptic morphology 1; SCF, Skip1, Cullin, F-box; SPAR, GTPase-activating protein spine-associated RapGAP.
Sidebar A | In need of answers.
Are proteasomal subunit composition and activity differentially regulated in neuronal compartments or specialized nervous tissues?
What are the E3 ligase complexes and their substrates that act in neuronal subcompartments or neuronal tissue types?
How does neuronal development and activity influence the local function of ubiquitin-proteasome system (UPS) components?
Is there a link between messenger RNA trafficking and local synthesis of UPS components?
Does proteasomal degradation locally control translational repressors of the UPS in an autoregulatory feedback loop?

Alexandra Segref

Thorsten Hoppe
Acknowledgments
We are grateful for critical comments on the manuscript from members of the Hoppe laboratory and colleagues at the Research Unit FOR 885. Work in the laboratory of T.H. is funded by the Deutsche Forschungsgemeinschaft (especially the Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases and the Research Unit FOR 885), the European Molecular Biology Organization Young Investigator Programme and the Rubicon European Union Network of Excellence. The authors apologize to all colleagues whose work could not be cited owing to space limitations.
References
- Ang XL, Seeburg DP, Sheng M, Harper JW (2008) Regulation of postsynaptic rapgap spar by polo-like kinase 2 and the SCFβ-TRCP ubiquitin ligase in hippocampal neurons. J Biol Chem 283: 29424–29432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura N, Kaibuchi K (2005) Key regulators in neuronal polarity. Neuron 48: 881–884 [DOI] [PubMed] [Google Scholar]
- Ashraf SI, McLoon AL, Sclarsic SM, Kunes S (2006) Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell 124: 191–205 [DOI] [PubMed] [Google Scholar]
- Bingol B, Schuman EM (2006) Activity-dependent dynamics and sequestration of proteasomes in dendritic spines. Nature 441: 1144–1148 [DOI] [PubMed] [Google Scholar]
- Bloom AJ, Miller BR, Sanes JR, DiAntonio A (2007) The requirement for Phr1 in CNS axon tract formation reveals the corticostriatal boundary as a choice point for cortical axons. Genes Dev 21: 2593–2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockers TM, Segger-Junius M, Iglauer P, Bockmann J, Gundelfinger ED, Kreutz MR, Richter D, Kindler S, Kreienkamp HJ (2004) Differential expression and dendritic transcript localization of Shank family members: identification of a dendritic targeting element in the 3′ untranslated region of Shank1 mRNA. Mol Cell Neurosci 26: 182–190 [DOI] [PubMed] [Google Scholar]
- Chain DG, Casadio A, Schacher S, Hegde AN, Valbrun M, Yamamoto N, Goldberg AL, Bartsch D, Kandel ER, Schwartz JH (1999) Mechanisms for generating the autonomous cAMP-dependent protein kinase required for long-term facilitation in Aplysia. Neuron 22: 147–156 [DOI] [PubMed] [Google Scholar]
- Colledge M, Snyder EM, Crozier RA, Soderling JA, Jin Y, Langeberg LK, Lu H, Bear MF, Scott JD (2003) Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron 40: 595–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CA, Wairkar YP, Johnson SL, DiAntonio A (2006) Highwire restrains synaptic growth by attenuating a MAP kinase signal. Neuron 51: 57–69 [DOI] [PubMed] [Google Scholar]
- Craig AM, Banker G (1994) Neuronal polarity. Annu Rev Neurosci 17: 267–310 [DOI] [PubMed] [Google Scholar]
- Ding M, Chao D, Wang G, Shen K (2007) Spatial regulation of an E3 ubiquitin ligase directs selective synapse elimination. Science 317: 947–951 [DOI] [PubMed] [Google Scholar]
- Dong C, Upadhya SC, Ding L, Smith TK, Hegde AN (2008) Proteasome inhibition enhances the induction and impairs the maintenance of late-phase long-term potentiation. Learn Mem 15: 335–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreier L, Burbea M, Kaplan JM (2005) LIN-23-mediated degradation of β-catenin regulates the abundance of GLR-1 glutamate receptors in the ventral nerve cord of C. elegans. Neuron 46: 51–64 [DOI] [PubMed] [Google Scholar]
- Ehlers MD (2003) Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci 6: 231–242 [DOI] [PubMed] [Google Scholar]
- Fonseca R, Vabulas RM, Hartl FU, Bonhoeffer T, Nagerl UV (2006) A balance of protein synthesis and proteasome-dependent degradation determines the maintenance of LTP. Neuron 52: 239–245 [DOI] [PubMed] [Google Scholar]
- Grill B, Bienvenut WV, Brown HM, Ackley BD, Quadroni M, Jin Y (2007) C. elegans RPM-1 regulates axon termination and synaptogenesis through the Rab GEF GLO-4 and the Rab GTPase GLO-1. Neuron 55: 587–601 [DOI] [PubMed] [Google Scholar]
- Hegde AN (2004) Ubiquitin-proteasome-mediated local protein degradation and synaptic plasticity. Prog Neurobiol 73: 311–357 [DOI] [PubMed] [Google Scholar]
- Hegde AN, DiAntonio A (2002) Ubiquitin and the synapse. Nat Rev Neurosci 3: 854–861 [DOI] [PubMed] [Google Scholar]
- Hegde AN, Upadhya SC (2007) The ubiquitin-proteasome pathway in health and disease of the nervous system. Trends Neurosci 30: 587–595 [DOI] [PubMed] [Google Scholar]
- Hegde AN, Goldberg AL, Schwartz JH (1993) Regulatory subunits of cAMP-dependent protein kinases are degraded after conjugation to ubiquitin: a molecular mechanism underlying long-term synaptic plasticity. Proc Natl Acad Sci USA 90: 7436–7440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde AN, Inokuchi K, Pei W, Casadio A, Ghirardi M, Chain DG, Martin KC, Kandel ER, Schwartz JH (1997) Ubiquitin C-terminal hydrolase is an immediate-early gene essential for long-term facilitation in Aplysia. Cell 89: 115–126 [DOI] [PubMed] [Google Scholar]
- Hicke L, Dunn R (2003) Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol 19: 141–172 [DOI] [PubMed] [Google Scholar]
- Hou L, Antion MD, Hu D, Spencer CM, Paylor R, Klann E (2006) Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron 51: 441–454 [DOI] [PubMed] [Google Scholar]
- Juo P, Kaplan JM (2004) The anaphase-promoting complex regulates the abundance of GLR-1 glutamate receptors in the ventral nerve cord of C. elegans. Curr Biol 14: 2057–2062 [DOI] [PubMed] [Google Scholar]
- Kato A, Rouach N, Nicoll RA, Bredt DS (2005) Activity-dependent NMDA receptor degradation mediated by retrotranslocation and ubiquitination. Proc Natl Acad Sci USA 102: 5600–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol 22: 159–180 [DOI] [PubMed] [Google Scholar]
- Kuo CT, Jan LY, Jan YN (2005) Dendrite-specific remodeling of Drosophila sensory neurons requires matrix metalloproteases, ubiquitin-proteasome, and ecdysone signaling. Proc Natl Acad Sci USA 102: 15230–15235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CT, Zhu S, Younger S, Jan LY, Jan YN (2006) Identification of E2/E3 ubiquitinating enzymes and caspase activity regulating Drosophila sensory neuron dendrite pruning. Neuron 51: 283–290 [DOI] [PubMed] [Google Scholar]
- Leroy E et al. (1998) The ubiquitin pathway in Parkinson's disease. Nature 395: 451–452 [DOI] [PubMed] [Google Scholar]
- Lewcock JW, Genoud N, Lettieri K, Pfaff SL (2007) The ubiquitin ligase Phr1 regulates axon outgrowth through modulation of microtubule dynamics. Neuron 56: 604–620 [DOI] [PubMed] [Google Scholar]
- Liao EH, Hung W, Abrams B, Zhen M (2004) An SCF-like ubiquitin ligase complex that controls presynaptic differentiation. Nature 430: 345–350 [DOI] [PubMed] [Google Scholar]
- Muralidhar MG, Thomas JB (1993) The Drosophila bendless gene encodes a neural protein related to ubiquitin-conjugating enzymes. Neuron 11: 253–266 [DOI] [PubMed] [Google Scholar]
- Nakata K, Abrams B, Grill B, Goncharov A, Huang X, Chisholm AD, Jin Y (2005) Regulation of a DLK-1 and p38 MAP kinase pathway by the ubiquitin ligase RPM-1 is required for presynaptic development. Cell 120: 407–420 [DOI] [PubMed] [Google Scholar]
- Pak DT, Sheng M (2003) Targeted protein degradation and synapse remodeling by an inducible protein kinase. Science 302: 1368–1373 [DOI] [PubMed] [Google Scholar]
- Pak DT, Yang S, Rudolph-Correia S, Kim E, Sheng M (2001) Regulation of dendritic spine morphology by SPAR, a PSD-95-associated RapGAP. Neuron 31: 289–303 [DOI] [PubMed] [Google Scholar]
- Patrick GN, Bingol B, Weld HA, Schuman EM (2003) Ubiquitin-mediated proteasome activity is required for agonist-induced endocytosis of GluRs. Curr Biol 13: 2073–2081 [DOI] [PubMed] [Google Scholar]
- Peters JM (2006) The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol 7: 644–656 [DOI] [PubMed] [Google Scholar]
- Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70: 503–533 [DOI] [PubMed] [Google Scholar]
- Pines J, Lindon C (2005) Proteolysis: anytime, any place, anywhere? Nat Cell Biol 7: 731–735 [DOI] [PubMed] [Google Scholar]
- Rezvani K, Teng Y, Shim D, De Biasi M (2007) Nicotine regulates multiple synaptic proteins by inhibiting proteasomal activity. J Neurosci 27: 10508–10519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer H, Rongo C (2006) KEL-8 is a substrate receptor for CUL3-dependent ubiquitin ligase that regulates synaptic glutamate receptor turnover. Mol Biol Cell 17: 1250–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, Bargmann CI (2003) The immunoglobulin superfamily protein SYG-1 determines the location of specific synapses in C. elegans. Cell 112: 619–630 [DOI] [PubMed] [Google Scholar]
- Shimura H et al. (2000) Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet 25: 302–305 [DOI] [PubMed] [Google Scholar]
- Speese SD, Trotta N, Rodesch CK, Aravamudan B, Broadie K (2003) The ubiquitin proteasome system acutely regulates presynaptic protein turnover and synaptic efficacy. Curr Biol 13: 899–910 [DOI] [PubMed] [Google Scholar]
- Sumara I, Maerki S, Peter M (2008) E3 ubiquitin ligases and mitosis: embracing the complexity. Trends Cell Biol 18: 84–94 [DOI] [PubMed] [Google Scholar]
- Upadhya SC, Ding L, Smith TK, Hegde AN (2006) Differential regulation of proteasome activity in the nucleus and the synaptic terminals. Neurochem Int 48: 296–305 [DOI] [PubMed] [Google Scholar]
- van Roessel P, Elliott DA, Robinson IM, Prokop A, Brand AH (2004) Independent regulation of synaptic size and activity by the anaphase-promoting complex. Cell 119: 707–718 [DOI] [PubMed] [Google Scholar]
- Wan HI, DiAntonio A, Fetter RD, Bergstrom K, Strauss R, Goodman CS (2000) Highwire regulates synaptic growth in Drosophila. Neuron 26: 313–329 [DOI] [PubMed] [Google Scholar]
- Wu C, Daniels RW, Diantonio A (2007) DFsn collaborates with Highwire to down-regulate the Wallenda/DLK kinase and restrain synaptic terminal growth. Neural Develop 2: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Guo L, Wang Y (2006) Requirement of dendritic Akt degradation by the ubiquitin-proteasome system for neuronal polarity. J Cell Biol 174: 415–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao I et al. (2007) SCRAPPER-dependent ubiquitination of active zone protein RIM1 regulates synaptic vesicle release. Cell 130: 943–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi JJ, Ehlers MD (2007) Emerging roles for ubiquitin and protein degradation in neuronal function. Pharmacol Rev 59: 14–39 [DOI] [PubMed] [Google Scholar]