Abstract
Conifers are characterized by a large genome size and a rapid decay of linkage disequilibrium, most often within gene limits. Genome scans based on noncoding markers are less likely to detect molecular adaptation linked to genes in these species. In this study, we assessed the effectiveness of a genome-wide single nucleotide polymorphism (SNP) scan focused on expressed genes in detecting local adaptation in a conifer species. Samples were collected from six natural populations of white spruce (Picea glauca) moderately differentiated for several quantitative characters. A total of 534 SNPs representing 345 expressed genes were analysed. Genes potentially under natural selection were identified by estimating the differentiation in SNP frequencies among populations (FST) and identifying outliers, and by estimating local differentiation using a Bayesian approach. Both average expected heterozygosity and population differentiation estimates (HE = 0.270 and FST = 0.006) were comparable to those obtained with other genetic markers. Of all genes, 5.5% were identified as outliers with FST at the 95% confidence level, while 14% were identified as candidates for local adaptation with the Bayesian method. There was some overlap between the two gene sets. More than half of the candidate genes for local adaptation were specific to the warmest population, about 20% to the most arid population, and 15% to the coldest and most humid higher altitude population. These adaptive trends were consistent with the genes’ putative functions and the divergence in quantitative traits noted among the populations. The results suggest that an approach separating the locus and population effects is useful to identify genes potentially under selection. These candidates are worth exploring in more details at the physiological and ecological levels.
Keywords: expressed genes, FST, genome scan, local adaptation, SNP, white spruce
Introduction
Identifying genomic regions involved in local adaptation is a challenging task for plant evolutionary biologists, breeders, and conservationists. It is particularly challenging in forestry because trees such as conifers are among organisms with largest genome size (Wakamiya et al. 1993; Murray 1998) and a rapid decay of linkage disequilibrium (LD), most often within gene limits (Neale & Savolainen 2004). Such factors are likely to reduce the efficiency of genome scans based on intergenic or anonymous markers in detecting adaptive variation linked to coding regions. They also make it hard to adequately disentangle the effects of historical or demographic factors from those of natural selection since a higher marker density would be needed to attain this goal (Storz 2005; Wright & Gaut 2005). Indeed, evidence for discrepancies in population differentiation estimates due to the limited number of loci and their sparse distribution on the genome has been reported (Nybom 2004).
Until recently, most adaptive genome scans in plants have relied on the identification of quantitative trait loci (QTL) by using mapping approaches for inbred or outbred pedigrees (e.g. Frewen et al. 2000; Hurme et al. 2000; Bradshaw & Schemske 2003). They also relied extensively on DNA markers from noncoding or anonymous regions of the genome such as random amplified polymorphic DNA (RAPD), amplified fragment length polymorphism (AFLP), or simple sequence repeats, also known as microsatellites (SSR). Despite its great utility, the QTL approach is difficult to apply in cases where traits cannot be measured before many years of testing in common gardens, or when test crosses cannot be experimentally performed (Storz 2005). It also steps short from elucidating the genetic basis of complex traits when several genes and environmental factors regulate the phenotype under study (e.g. see Howe et al. 2003; Erickson et al. 2004) or when QTLs encompass large genomic regions.
An alternative approach consists in using a multilocus genome scan that identifies outlier loci for population differentiation by screening genomes for genetic polymorphisms directly into natural populations (Luikart et al. 2003; Schlötterer 2003; Storz et al. 2004). This approach has the advantage of not requiring knowledge of genetic divergence in quantitative traits between populations. It also does not require a complete knowledge of the DNA sequence underlying the loci investigated (Storz 2005). Its main assumption is that when an adaptive mutation is driven to fixation by selection at a specific locus, this fixation will lead to a joint fixation of the neutral linked variants (Maynard Smith & Haigh 1974; Kaplan et al. 1989; Stephan et al. 1992), which can be maintained in small fractions in the species genome even when recombination randomizes associations between the selected site and the linked neutral variants (Storz 2005). This process is known as genetic hitch-hiking and can be easily detected since the selected locus and its linked regions exhibit a variation pattern that is distinct from the rest of the genome, mainly a skewed allele frequency distribution, a lower within-population variation, and a higher differentiation for the locally adapted populations (Vasemägi & Primmer 2005).
To date, genome scans for the identification of loci or genes involved in local adaptation have been extensively used in humans (e.g. Altshuler et al. 2000; Akey et al. 2002; Kayser et al. 2003) and for some model organisms (e.g. Cork & Purugganan 2005; Wright et al. 2005), but they are still in their infancy for most species including conifers. In most cases, because of the anonymous or intergenic nature of the markers used, these scans were limited to the identification of outlier loci without a special focus on identifying the underlying genes or possible causes at the molecular level (Scotti-Saintagne et al. 2004; Krutovsky & Neale 2005; Bousquet et al. 2007; Tsumura et al. 2007). In taxa characterized by large genome sizes and low LD such as conifers, these approaches might effectively miss regions of the genome involved in adaptation because coding genes that can be the target of selection are embedded in large recombining noncoding regions. Such a genome configuration necessitates high marker densities to detect the effect of natural selection on linked genomic regions. This configuration becomes even more problematic in species characterized by large genomes. In addition, the accuracy of anonymous or intergenic markers such as RAPDs, AFLPs, and SSRs in estimating allelic variation within or between populations (Hedrick 1999; Isabel et al. 1999; Mariette et al. 2002) and consequently, in detecting natural selection along a species genome, might be decreased by partial DNA digestion or high mutation rates, two potential sources of homoplasious variation.
One way to circumvent these limitations is to rely on polymorphisms located in genes or gene regions closely linked to them. With sequencing becoming more affordable, large-scale sequencing of expressed sequence tags (ESTs) may represent a useful starting point for mining DNA polymorphisms located directly into genes for such species. The detection of DNA polymorphisms in large numbers of expressed genes and their use in population studies has been shown to be feasible, using EST-linked microsatellites (e.g. Vasemägi et al. 2005; Oetjen & Reusch 2007) and single nucleotide polymorphisms (SNP, e.g. Kimura et al. 2007; Zayed & Whitfield 2008).
Recently, Pavy et al. (2006) identified more than 12 000 SNPs from the clustering of about 50 000 ESTs from the white spruce [Picea glauca (Moench) Voss] genome (Pavy et al. 2005). The discovery of such a large number of SNPs provides a useful starting point to perform a genome-wide SNP scan in natural populations of a conifer species in order to identify candidate genes underlying adaptation (Bouck & Vision 2007; Bousquet et al. 2007). Although the characterization of variation in quantitative characters is not an essential condition for the success of such a scan (Storz 2005), one potentially powerful approach would be to link this scan to populations previously characterized for adaptive quantitative characters in common garden studies. In doing so, population sampling could be orientated more effectively based on knowledge of variation in quantitative characters, thus reducing the rate of false–positives. For white spruce, many large-scale replicated provenance tests have already been established for the first-generation breeding cycle and for gene conservation purposes. These tests indicate significant among-population genetic variation for quantitative traits related to growth, phenology, and wood characters (Li et al. 1997; Jaramillo-Correa et al. 2001), but corroborative evidence using a genome-wide scan has not yet been investigated.
In the present study, we took advantage of the new EST resource to assess the effectiveness of a genome-wide SNP scan focused on expressed genes in detecting adaptive polymorphism in six natural populations of white spruce. We also tried to determine whether the adaptive patterns observed at some genes could be associated, at least qualitatively, with the bioclimatic conditions or phenotypic attributes of the populations, as well as with the functional properties of the genes. To identify genes with an adaptive genetic pattern, we used two different approaches: one relying on estimates of overall among-population differentiation (Beaumont & Nichols 1996), and the other based on a Bayesian method aimed at identifying local adaptation (Beaumont & Balding 2004). The latter has the advantage of separating the locus and population effects.
Materials and methods
Gene selection
A total of 656 genes were chosen and resequenced among a sample of 16 500 annotated unigenes derived from the assembly of around 50 000 white spruce ESTs (Pavy et al. 2005). The ESTs were from 5′ and 3′ reads and were generated from 16 cDNA banks. The average length of high-quality reads was 645 nucleotides (Pavy et al. 2005). The present subset of 656 genes represents candidates for regulatory function, wood formation, plant growth, or phenology. Various publicly accessible data banks were used to retrieve annotations and identify functional attributes of the genes including spruceDB (http://biodata.cbri.umn.edu/spruce/), ForestTreeDB (http://foresttree.org:8680/DB/nimbus/project.do), PFAM (http://www.sanger.ac.uk/Software/Pfam/), and Arabidopsis databases (http://arabidopsis.med.ohio-state.edu/AtTFDB/andhttp://datf.cbi.pku.edu.cn/). Because of EST redundancy, high probability in silico SNPs were identified for many of these genes (Pavy et al. 2006). Resequencing was conducted to confirm these SNPs and discover new ones.
SNP discovery
The 656 candidate genes were resequenced for the two white spruce parents of a linkage mapping population in order to identify SNPs enabling gene mapping in a large F1 population (Pavy et al. 2008). The same set of SNPs was used in the present survey. Primers for amplification and resequencing were generally placed in 5′ or 3′ untranslated regions of the genes to increase specificity. Methods for the identification of coding regions and primer design for amplification and resequencing are reported elsewehere (Pavy et al. 2008). Each gene was also resequenced from DNA of a megagametophyte, the tissue surrounding the embryo of a conifer seed, in order to identify and exclude paralogous SNPs (Pavy et al. 2008). Because of the haploid nature of conifer megagametophytes, no polymorphism is expected in haploid DNA sequences. Any exception to the rule indicates polymorphism of paralogous nature that was not considered further. A final screening was conducted to eliminate SNPs with variable flanking regions (e.g. with highly repetitive sequences, palindromes, polymorphism located too close of each other) in order to eliminate SNPs with low probability of genotyping success using the GoldenGate assay (Fan et al. 2003; see below). Of the 656 candidate genes, 487 could be successfully amplified. A subset of 424 genes containing a total of 768 orthologous SNPs were used for the construction of the SNP array.
Population sampling and DNA isolation for the population scan
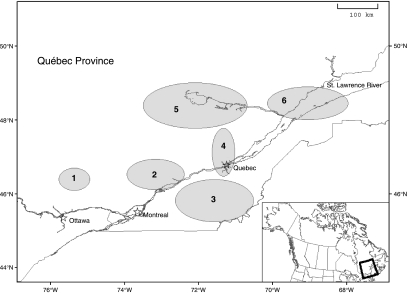
Trees were sampled from natural populations of white spruce distributed in different ecological regions in Québec. They extended from the temperate hardwood to the boreal conifer forest (Fig. 1). In total, six broad populations representative of as many distinct ecological regions were analysed in the present study. Previous studies showed that these six populations were in drift–migration equilibrium, exhibited no significant genetic differentiation at neutral loci, but were significantly differentiated in quantitative traits related to wood density, phenology, and growth, as determined in common garden studies and by QST differentiation estimates (Jaramillo-Correa et al. 2001). Trees were represented by ramets maintained in a clonal bank and open-pollinated families grown in common garden tests previously established in 1979 and 1980 (Li et al. 1993, 1997). Bioclimatic data and phenotypic attributes of these populations are presented in Table 1. From 20 to 34 trees per population could be sampled, for a total of 158 trees. For each tree, DNA was extracted from dormant buds using a DNeasy Plant mini kit according to the manufacturer's instructions (QIAGEN).
Fig. 1.
Geographical distribution of the six populations (ecoregions) analysed.
Table 1.
Average climatic parameters and quantitative traits for the six sampled populations (ecoregions) of white spruce*
| Population | ||||||
|---|---|---|---|---|---|---|
| Climatic parameters and traits | No. 1 | No. 2 | No. 3 | No. 4 | No. 5 | No. 6 |
| Elevation (metres) | 269 | 211 | 205 | 771 | 334 | 198 |
| Annual number of degree-days†‡ | 1570 | 1633 | 1789 | 970 | 1305 | 1353 |
| Precipitation from June to August (millimetres)† | 305 | 319 | 348 | 430 | 345 | 292 |
| Aridity index† | 4.1 | 4.0 | 3.8 | 0.9 | 2.5 | 4.2 |
| Total annual precipitation (millimetres)† | 1058 | 1111 | 1127 | 1599 | 1090 | 1079 |
| Annual number of days without frost† | 176 | 181 | 194 | 139 | 161 | 171 |
| Annual average temperature (°C)† | 3.3 | 3.6 | 4.8 | –0.3 | 1.1 | 2.2 |
| Maximum temperature (°C)† | 33.0 | 32.8 | 32.7 | 29.2 | 31.7 | 31.6 |
| Minimum temperature (°C)† | –36.8 | –34.9 | –32.1 | –37.7 | –38.4 | –33.4 |
| Height (centimetres)§ | 516 (4) | 494 (4) | 447 (4) | 466 (5) | 493 (3) | 449 (5) |
| Diameter at breast height (centimetres)§ | 7.1 (0.1) | 6.9 (0.1) | 5.9 (0.1) | 6.5 (0.1) | 6.7 (0.1) | 6.0 (0.1) |
| Wood density (g/cm3)§ | 0.361 (0.002) | 0.369 (0.002) | 0.382 (0.002) | 0.368 (0.003) | 0.367 (0.002) | 0.387 (0.001) |
| Date of budset (julian days)§¶ | 230 (2) | 236 (1) | 236 (1) | 229 (1) | 231 (1) | 235 (1) |
| Date of budburst (julian days)§¶ | 143 (< 1) | 143 (< 1) | 143 (< 1) | 144 (< 1) | 143 (< 1) | 143 (< 1) |
| Duration of growing season (days)§ | 87 (2) | 93 (1) | 94 (1) | 84 (1) | 87 (1) | 92 (1) |
Traits were measured in a common garden study (see Materials and Methods);
average estimates are for the reference period 1971–2000 and were obtained by using the biosim climate simulator (Régnière 1996);
the number of degree-days corresponds to the cumulative number of degrees above 5 °C;
quantitative traits were measured at age 22 on half-sib progenies raised in a common garden in three replicated sites for height, diameter at breast height, and wood density, and at age 3 at a nursery site with replications for phenological traits; numbers in parentheses indicate the standard errors;
the number of julian days corresponds to the number of days elapsed since noon Greenwich Mean Time on January 1st.
SNP genotyping
SNP genotyping of the 158 sampled individuals was performed by using the Illumina SNP bead array platform (Illumina, San Francisco, California) and the GoldenGate allele-specific extension assay, a highly multiplexed genotyping assay (Fan et al. 2003; Shen et al. 2005). It was carried in 96-well plates using 2 µg of DNA extract normalized at 50 ng/µL for each sample. Briefly, the GoldenGate assay consists in genotyping genomic DNA directly without the need for polymerase chain reaction (PCR) amplification by hybridizing two allele-specific (ASO) and one locus-specific oligos (LSO) with each DNA sample in the array matrix. It allows highly multiplex genotyping, up to 1536 SNPs in the GoldenGate genotyping assay. Further details about this technique can be found in Shen et al. (2005).
GenCall and GenTrain scores were used to evaluate the accuracy and efficiency of SNP genotyping. These scores reflect the degree of separation between homozygote and heterozygote clusters for each SNP (Fan et al. 2003). The lowest acceptable score was set at 0.25, which represents a stringent criterion used in human genetic studies (http://www.illumina.com/; Fan et al. 2003). In the present study, this threshold corresponded to SNPs with accurate scoring for at least 95% of the individuals, with most successful SNPs scored for over 99% of the individuals analysed.
Data analysis
Genetic diversity was estimated for each population by calculating the percentage of polymorphic SNPs (PO), the mean number of alleles per SNP (A), observed (HO) and expected (HE) heterozygosities (unbiased, Nei 1978), and the within-population fixation index (F). The deviation of fixation indices from zero was tested by 10 000 permutations of alleles between individuals. Population departure from Hardy–Weinberg equilibrium was also tested. Among-population differentiation estimates (FST) were calculated and their deviation from zero was tested by 10 000 allele permutations. These various parameters were calculated with arlequin version 2.3 software (http://anthro.unige.ch/arlequin). Detection of genes carrying the signature of natural selection or ‘outliers’ was first performed with the fdist2 program that uses the summary-statistic approach described in Beaumont & Nichols (1996) and further developed in Beaumont & Balding (2004). This program is available at http://www.rubic.reading.ac.uk/~mab/software/fdist2.zip. It first calculates FST for each sampled locus with the Weir & Cockerham (1984) formula (#10, page 1364). It then uses coalescent simulations to generate a null distribution of FSTvalues based on an infinite island model for the populations and an infinite allele model for polymorphism (Beaumont & Nichols 1996). Loci with an unusually high or low FST value conditional on heterozygosity are considered as potentially under selection. In this study, we simulated the neutral distribution of FST with 60 000 iterations at the 95% confidence level.
The program newfst developed by Beaumont & Balding (2004) was also used to identify genes subject to selection. This program relies on a Bayesian model to generate FST values by implementing a Metropolis Hastings Markov chain Monte Carlo (MCMC) algorithm based on the likelihood of allele counts. It has the advantage of disentangling the locus effect (αi), the population effect (βj), and the interaction between the locus and the population effects (γij). In general, a large positive αi indicates the presence of a positive selection on the studied gene, while a large positive γij indicates an important locus–population interaction, thus a potentially advantageous mutation that would be locally adapted to a particular population (Beaumont & Balding 2004). The probability densities for FST values were obtained with the assumption of independent, lognormal (1, 1.8, 0.5) prior distributions for the αi, βi, and γij. For this, Gaussian kernel density estimation was used based on 160 000 iterations of the Metropolis algorithm with a thinning interval of 320. The convergence of the 10 000 parameters series αi, βi, and γij generated by the MCMC algorithm was tested with the Heidelberger & Welch (1981) convergence diagnostic using the coda package of r version 2.2.1. r is an open-source statistical software available at http://www.r-project.org/. In a first step, we set the confidence level at 95% to identify large positive values of αi and γij. Genes identified with a positive αi value were called outliers, while those identified with a large positive γijvalue were termed candidate for local adaptation throughout the paper. Although only two of them reached the 95% confidence level of the simulations (see Results), those with high positive γijvalues (above 0.10) were retained for further investigation as they possibly reflected true adaptive trends. To compare these results to those obtained with the summary-statistic method (fdist 2), we adjusted the confidence levels to 90% and 99% for newfst and fdist 2, respectively. This 10-fold difference in confidence level was suggested by Beaumont & Balding (2004) to make the two methods comparable for their own data by maintaining the same rate of false-positives.
In an attempt to find corroborative evidence for the genetic patterns observed, we qualitatively assessed the presence of possible relationships between the genes identified as candidate for local adaptation and the bioclimatic and phenotypic attributes of each population. The bioclimatic parameters included the annual number of degree-days, the aridity index, the total precipitation during summer, the total annual precipitation, the annual number of days without frost, the annual average temperature, and the maximum and minimum temperatures (Table 1). The quantitative traits were estimated from common garden studies (see Li et al. 1993, 1997 for study design) and included tree height, diameter at breast height (d.b.h.), and wood density at age 22, as well as the date of budburst, the date of budset, and the duration of the growing season at age 3. We also looked at the current functional annotation and classification of the candidate genes for local adaptation as described above. However, the present effort of linking statistical and functional inferences has to be considered as preliminary since many of the genes used in this study are still not well characterized at the functional or transcriptional level.
Results
Genotyping success
Among the 768 SNPs analysed, 166 failed to provide an acceptable GenTrain quality score, 68 had an acceptable quality score but were monomorphic in all samples, and the remaining 534 (70%) were polymorphic with a high quality score in more than 95% of the sampled individuals (Table 2). The 534 successful SNPs were located on 345 expressed genes including 260 (75%) that were regulatory. Regulatory genes were identified by comparing spruce sequences to Arabidopsis transcription factors (http://arabidopsis.med.ohio-state.edu/AtTFDB/ and http://datf.cbi.pku.edu.cn/) and to PFAM domains (http://www.sanger.ac.uk/Software/Pfam/) (Pavy et al. 2005). Given that the complete sequence of the Picea genome is not available to fully validate SNPs before spotting them on the SNP-bead array, the proportion of SNPs successfully genotyped in the populations (70%) compares well with that for human SNPs using the same technology (80%) (Shen et al. 2005). The 534 successful SNPs and their 345 genes were distributed along the 12 linkage groups of white spruce (Pelgas et al. 2006) at a rate of 19–31 gene locus per linkage group (Pavy et al. 2008).
Table 2.
Genotyping success of SNP markers and genes using the Illumina GoldenGate multiplex assay
| Category | Number of SNPs | Percentage of SNPs | Number of genes | Percentage of genes |
|---|---|---|---|---|
| Failed SNPs (Gentrain score < 0.25)* | 166 | 21.6 | 20 | 4.7 |
| Successful SNPs but monomorphic | 68 | 8.9 | 27 | 6.4 |
| Successful SNPs and polymorphic | 534 | 69.5 | 345 | 81.4 |
| Total | 768 | 100 | 424 | 100 |
The GenTrain score reflects the degree of separation between homozygote and heterozygote clusters for each SNP and the placement of the individual call within each cluster (Shen et al. 2005).
Genetic diversity
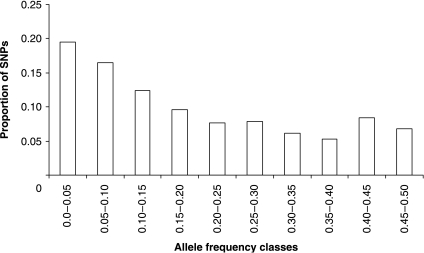
No more than two nucleotides could be detected per SNP. Accordingly, the mean number of alleles per polymorphic locus (A) per population ranged from 1.90 to 1.94 with a grand mean of 1.92. About 65% of all SNPs had an overall frequency equal to or larger than 0.10 (Fig. 2) and were considered common SNPs. Out of 3204 tests performed, only two departed from Hardy–Weinberg equilibrium (at α = 0.05). Overall genetic diversity (i.e. considering all polymorphic SNPs together) expressed by the average observed heterozygosity (HO) ranged from 0.263 to 0.293 among populations with a grand mean of 0.276 (SD = ±0.011). Average unbiased expected heterozygosity (HE) was generally slightly lower than HO and ranged from 0.266 to 0.274 per population with a grand mean of 0.270 (SD = ±0.003). The average within-population fixation index F (averaged over all loci in each population) showed a significant excess of heterozygotes in population nos 1, 2, and 4 (Table 3). The largest number of SNPs with a significant excess of homozygotes was in population nos 3 (14 SNPs for 14 genes) and 6 (15 SNPs for 14 genes).
Fig. 2.
Allele frequency distribution at 534 SNP loci.
Table 3.
Genetic parameters of the six sampled populations (ecoregions) of white spruce*
| Population | Number of trees sampled | A | HO | HE | F |
|---|---|---|---|---|---|
| No. 1 | 25 | 1.93 | 0.293 | 0.274 | –0.073† |
| No. 2 | 31 | 1.94 | 0.277 | 0.273 | –0.019† |
| No. 3 | 25 | 1.91 | 0.263 | 0.266 | 0.011 |
| No. 4 | 20 | 1.90 | 0.283 | 0.268 | –0.059† |
| No. 5 | 34 | 1.93 | 0.269 | 0.269 | –0.001 |
| No. 6 | 23 | 1.92 | 0.271 | 0.269 | –0.008 |
| Mean | 26.3 | 1.92 | 0.276 | 0.270 | –0.025 |
| SD | 5.2 | 0.01 | 0.011 | 0.003 | 0.033 |
A is the mean number of alleles per SNP, HOis the average observed heterozygosity; HEis the average unbiased expected heterozygosity (Nei 1978); F is the average fixation index; SD is the standard deviation;
P ≤ 0.01 based on 10 000 permutations between individuals within the same population.
Genetic differentiation among populations and detection of genes targeted by selection
The average level of genetic differentiation among populations was extremely low (FST = 0.006) but significantly different from zero at the 95% confidence level. FST values per SNP ranged from –0.019 to 0.137, with 73 SNPs (14%) having a value significantly different from zero at the 95% confidence level. In this study, negative FST values were considered equal to zero because they do not bear any biological interpretation. In general, FST values followed an L-shaped distribution with 223 SNPs (42%) having an FST value below or equal to zero, and 21 SNPs (4%) in the tail of the distribution with a value higher than 0.04.
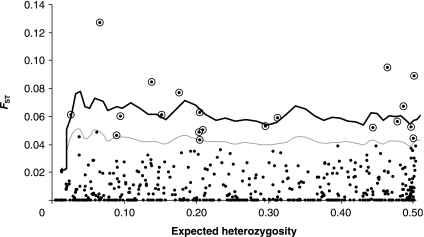
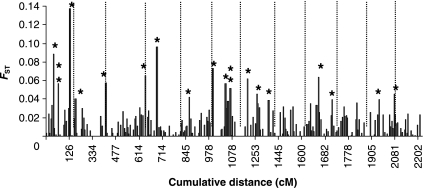
fdist 2, which is based on the summary-statistic method, identified 20 SNPs (3.7%) located on 19 genes (5.5%) with an FSTvalue above the 95% confidence level. Most of the outlier SNPs had an empirical FST higher than 0.04 (Fig. 3) and were distributed over the 12 linkage groups (Fig. 4). As expected, newfst, which is based on a Bayesian method that isolates the locus and population effects, provided a much lower figure at the same 95% confidence level: only two SNPs (0.4%) located on two different genes (0.6%) located on two different linkage groups carried the signature of positive selection when the locus effect parameter (αi) alone was considered, while none carried the signature of local adaptation when considering the locus × population parameter (γij). However, 49 SNPs (9.2%) located on 47 genes (13.6% including the two with a high positive αi value) exhibited a relatively large positive mean γijvalue (> 0.10) in one or two populations while maintaining negative or close to zero mean γijvalues in the other populations (Fig. 5b–e). This can be indicative of a possible adaptive trend in some populations. These SNPs also had a significant average FSTafter permutations (Fig. 5a) and were therefore considered as candidate for local adaptation. Conversely, no SNP had a significantly negative αivalue, indicating the absence of balancing selection. Most outliers identified with fdist 2 were also identified as candidate for local adaptation by newfst. These 49 SNPs were distributed over the 12 linkage groups of white spruce and included 21 SNPs (20 genes) located on exons and 28 SNPs (27 genes) on introns or the 5′UTR and 3′UTR regions (χ2 = 0.02, d.f. = 1, P = 0.88). Those located on exons included 10 synonymous and 11 nonsynonymous SNPs (χ2 = 0.99, d.f. = 1, P = 0.32), and the number of those with low allele frequency classes (below 10%) was not significantly different from the number of those considered as common (χ2 = 0.12, d.f. = 1, P = 0.73). When adjusting the confidence levels to 99% and 90% for fdist 2 and newfst, respectively (Beaumont & Balding 2004), the results became much similar: nine SNPs (1.7%) located on eight different genes (2.3%) could be declared outliers with fdist 2, compared to five SNPs (0.9%) located on five genes (1.4%) with newfst.
Fig. 3.
Outlier detection and distribution of empirical FST values as a function of expected heterozygosity. The solid line indicates the 99% upper and lower confidence levels and the grey line indicates the 95% upper and lower confidence levels, as estimated using the summary-statistic method of Beaumont & Nichols (1996). The identification of 20 outlier SNPs (above the 95% confidence level, fdist 2) is indicated by circled dots.
Fig. 4.
Distribution of empirical FST values over the 12 linkage groups of white spruce with linkage groups (LG) order following Pelgas et al. (2006). The vertical dotted lines indicate approximate boundaries between consecutive linkage groups. The identification of 20 outlier SNPs (above the 95% confidence level, fdist 2, Fig. 3) is indicated by asterisks above vertical solid lines. cM is centimorgans.
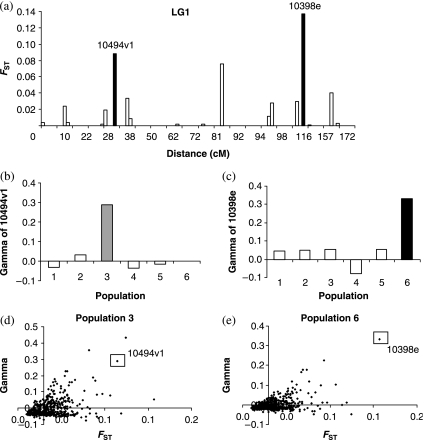
Fig. 5.
(a) Distribution of empirical FST values over linkage group 1 (LG1) with focus on two outlier SNPs (above the 99% confidence level, fdist 2) in grey and black. cM is centimorgans. (b) and (c) Distribution of the mean gamma values (γij) of the two outlier SNPs highlighted in (a) and indicative of local adaptation among the six populations studied. Gamma values were estimated with a Bayesian method and reflect the interaction between the locus and the population effects. (d) and (e) Distribution of gamma values (γij) for all SNPs in populations no. 3 and no. 6 relative to their overall FST values estimated with the summary-statistic method (fdist 2).
Geographical and functional trends
The number of candidate SNPs for local adaptation was significantly different among the six populations (χ2 = 39.10, d.f. = 5, P < 0.01), with the largest number being in population no. 3, then population nos 6 and 4 (Table 4). These three populations harboured 84% and 88% of the outlier SNPs identifed by fdist 2 at the 95% and 99% confidence levels, respectively. Population no. 3 that harboured the largest number of candidate SNPs for local adaptation had the warmest environmental conditions with the annual average temperature, number of degree-days and days without frost at the high end of the spectrum observed among the six populations (Table 1). Interestingly, growth parameters were at the low end. Population no. 6 that came second in terms of the number of candidate SNPs for local adaptation was on the high end of the aridity spectrum and its trees had the highest wood density (Table 1). Population no. 4 with the third largest number of candidate SNPs for local adaptation was the most elevated in altitude. It was also the most humid and the coldest with an annual average temperature of –0.3 °C (Table 1). Its trees had the shortest duration of growing season with the latest budburst and earliest budset (Table 1).
Table 4.
List and properties of the 49 candidate SNPs for local adaptation representative of 47 genes*
| SNP† | Locus‡ | Gene family | tblastx (e-value) | HE§ | FST (fdist 2)¶ | Population with mean γij > 0.10** | Putative biological function†† |
|---|---|---|---|---|---|---|---|
| 01424a | AT1G50640.1 | AP2 | 3E-22 | 0.090 | 0.009 | No. 3 | GR, RE, WF, BS, AS |
| 01471a | AT3G04730.1 | Auxin | 6E-28 | 0.480 | 0.004 | No. 3 | GR, RE, WF |
| 01687n | AT1G62360.1 | Homeobox | 7E-68 | 0.446 | 0.067 | No. 4 | GR |
| 02603e | AT1G25280.2 | Tubby | 3E-46 | 0.415 | 0.028 | No. 5 | GR |
| 02892v3 | AT3G23920.1 | Beta amylase | 3E-90 | 0.088 | 0.060 | No. 5 | GR, AS |
| 03056e | AT3G12130.1 | C3H RING finger | 3E-60 | 0.413 | 0.095 | No. 3 | GR, RE |
| 03579f | AT1G76880.1 | Trihelix | 3E-46 | 0.470 | 0.044 | No. 4 | PH, RE |
| 03579 g | AT AT1G76880.1 | Trihelix | 3E-46 | 0.475 | 0.0401 | No. 4 | PH, RE |
| 03693f | AT2G37630.1 | Myb | 2E-89 | 0.333 | 0.009 | No. 6 | GR, WF, AS |
| 03713 m | AT1G66230.1 | Myb | 4E-72 | 0.426 | 0.007 | No. 3 | GR, WF, AS |
| 04148e | AT1G69780.1 | Homeobox | 5E-33 | 0.482 | 0.015 | No. 1 and No. 3 | GR |
| 04190t1 | AT5G02810.1 | C2C2-colike | 6E-23 | 0.012 | 0 | No. 1 | PH, RE |
| 04514 g | AT3G20770.1 | Pseudouridine synthase A | 2E-22 | 0.190 | 0.048 | No. 4 | GR, RE, AS |
| 04885a | AT1G50640.1 | AP2 | 3E-26 | 0.105 | 0.005 | No. 3 | GR, RE, WF, BS, AS |
| 05090v1 | AT2G19940.1 | Gamma-glutamyl- phosphate reductase | 1E-136 | 0.462 | 0.052 | No. 4 | AS |
| 05937v1 | AT5G49580.1 | Chaperone protein DnaJ | 4E-24 | 0.126 | 0.032 | No. 3 | AS |
| 05953 g | AT5G17600.1 | C3H RING finger | 3E-47 | 0.188 | 0.063 | No. 6 | GR, RE, AS |
| 06620 g | AT4G04885.1 | C2H2 zinc finger | 1E-24 | 0.414 | 0.032 | No. 6 | GR, AS |
| 06684v1 | AT1G70320.1 | E3 ubiquitin protein ligase | 4E-56 | 0.058 | 0.048 | No. 1 | GR, RE, AS |
| 07106p1 | AT1G04140.2 | WD40 | 4E-85 | 0.289 | 0.059 | No. 3 | GR, RE, PH |
| 07248e | AT3G16830.1 | WD40 | 1E-142 | 0.448 | 0.089 | No. 4 | GR, RE, PH |
| 07393f | AT1G69440.1 | Argonaute | 7E-56 | 0.195 | 0.050 | No. 1 and No. 5 | GR |
| 07506a | AT1G60710.1 | Oxidoreductase | 1E-152 | 0.482 | 0 | No. 3 | GR, BS |
| 07604v4 | AT5G26680.1 | Flap endonuclease-1 | 1E-41 | 0.192 | 0.043 | No. 6 | GR, WF |
| 07977f | AT2G47900.2 | Tubby | 1E-82 | 0.367 | 0.003 | No. 3 | GR |
| 08080 m | AT4G38620.1 | Myb | 1E-81 | 0.084 | 0.046 | No. 3 and No. 6 | GR, WF, AS |
| 08177v1 | AT4G11790.1 | Ran binding protein | 2E-44 | 0.403 | 0.016 | No. 3 | GR |
| 08349a | AT1G25280.2 | Tubby | 9E-76 | 0.264 | 0.015 | No. 3 | GR |
| 08438b | AT3G55770.2 | LIM | 4E-84 | 0.328 | 0 | No. 2 and No. 6 | WF |
| 08987p1 | AT1G22190.1 | AP2 | 1E-26 | 0.130 | 0.029 | No. 5 | GR, RE, WF, BS, AS |
| 09562a | AT2G19810.1 | C3H RING finger | 6E-83 | 0.109 | 0.011 | No. 3 | GR, RE, AS |
| 09644v2 | AT1G60420.1 | Peroxidase | 6E-71 | 0.326 | 0.024 | No. 3 | WF, AS |
| 09863a | AT2G01570.1 | GRAS | 1E-140 | 0.231 | 0.011 | No. 3 | GR, WF |
| 09889b | AT3G12390.1 | NAC | 6E-48 | 0.241 | 0.025 | No. 3 | GR, BS |
| 09982e | AT1G10200.1 | LIM | 1E-73 | 0.177 | 0 | No. 5 | WF |
| 10016v1 | AT5G06720.1 | Peroxidase | 7E-91 | 0.443 | 0.056 | No. 3 | WF, AS |
| 10125n | AT3G15610.1 | WD40 | 1E-128 | 0.469 | 0.037 | No. 6 | GR, RE, PH |
| 10398e | AT1G08830.2 | Superoxide dismutase | 3E-49 | 0.056 | 0.127 | No. 6 | PH, AS, BS |
| 10494v1 | AT1G55670.1 | Photosystem I reaction centre subunit V | 6E-53 | 0.124 | 0.084 | No. 3 | PH |
| 10583v1 | AT2G21490.1 | Dehydrin | 1E-21 | 0.473 | 0.035 | No. 4 | AS |
| 10614t2 | PTU09554 | AGP | 3E-85 | 0.338 | 0.013 | No. 3 | WF |
| 11176a | AT4G24020.1 | Nin-like | 2E-58 | 0.141 | 0.061 | No. 4 | BS |
| 11176b | AT4G24020.1 | Nin-like | 2E-58 | 0.160 | 0.077 | No. 1 and No. 6 | BS |
| 12347a | AT1G16070.2 | Tubby | 2E-31 | 0.105 | 0.008 | No. 3 | GR |
| 13634e | AT3G13040.2 | Myb | 7E-34 | 0.486 | 0.006 | No. 3 | GR, WF, AS, BS |
| 14328e | AT4G32600.1 | C3H RING finger | 4E-76 | 0.143 | 0.023 | No. 1 | GR, RE, AS |
| 14745f | AT1G78070.2 | WD40 | 5E-70 | 0.394 | 0.028 | No. 3 | GR, RE, PH |
| 15115e | AT5G47390.1 | Myb | 4E-58 | 0.413 | 0.052 | No. 3 | GR, WF, AS, BS |
| 90002e | AT5G47670.2 | CCAAT box-binding factor E22 | 1E-54 | 0.220 | 0.036 | No. 3 | RE, PH |
Candidate SNPs for local adaptation are those exhibiting a mean locus–population interaction parameter (γij) higher than 0.10 based on the Bayesian method;
SNP annotations and nomenclature are detailed in Pavy et al. (2008); related transcript sequences can be found in the spruce gene database at http://biodata.cbri.umn.edu/spruce/;
Sequences were paired to an Arabidopsis locus based on sequence highest homology through tblastx searches against Arabidopsis blastsets database (ftp://ftp.arabidopsis.org/home/tair/Sequences/blast_datasets/TAIR7_blastsets/TAIR7_seq_20070320);
Expected heterozygosity;
FST was calculated as an estimate of Weir & Cockerham's (1984) β statistics as described in Beaumont & Nichols (1996);
The γij parameter is the mean locus–population interaction parameter as defined by Beaumont & Balding (2004);
PH, phenology; GR, growth; RE, reproduction; AS, abiotic stress; BS, biotic stress; WF, wood formation.
In terms of gene ontology based on biological processes and in concordance with the physiological patterns described above, population no. 3 was, among the six populations surveyed, the one with the largest number of candidate adaptive genes involved in growth (19 genes) followed by abiotic stress (10 genes), reproduction, and wood formation (nine genes each). In population no. 6 characterized by the most arid conditions, candidate adaptive genes affecting wood formation were the second most abundant after growth related ones. This pattern was not seen for the other populations. For population no. 4 characterized by a lower annual average temperature and a shorter growing season, a disproportionate number of candidate genes related to phenology, reproduction, and stress were observed, compared to the other populations (Table 4). Interestingly, most of the candidate SNPs for local adaptation in population nos 4 and 6 were located in introns or in 5′UTR or 3′UTR regions, and half of them were located in exonic regions in population no. 3, almost equally divided between synonymous and nonsynonymous SNPs. No significant difference could be observed in the number of candidates for local adaptation between regulatory and nonregulatory gene categories (χ2 = 0.55, d.f. = 1, P = 0.46). The gene family with the largest number of candidate genes for local adaptation was that for the Myb genes, which play an important role in plant growth and wood and lignin formation (Bedon et al. 2007). However, this family was over-represented in the gene sampling, which makes this trend only indicative.
Discussion
Genome scans in conifer species
To the best of our knowledge, this is the first report on a genome-wide SNP scan of expressed genes in a nonmodel species. It is also the first one to be conducted in natural conifer populations for which significant genetic differentiation in quantitative traits has been demonstrated from common garden studies (Jaramillo-Correa et al. 2001). The average among-population genetic differentiation estimated by FST was very low (0.006 or 0.60%), but comparable to that previously reported with allozymes and expressed sequence tag polymorphisms (ESTP) (mostly indels) in the same populations (GST = 1.4% and 2.0%, respectively; Jaramillo-Correa et al. 2001). It was also comparable to that observed in other white spruce populations using nuclear molecular markers (GST = 1.6%; Godt et al. 2001) and in black spruce populations in the same region (Isabel et al. 1995; Perry & Bousquet 2001), a species reproductively well isolated from white spruce but similar in terms of demography and population genetic parameters. As expected, white spruce harboured little population structure at the nuclear DNA level in the region surveyed.
When compared with other SNP studies in expressed gene regions, the average observed heterozygosity (HO = 0.276) fell in the range of that observed in other species such as humans (Moffatt et al. 2000; Beaty et al. 2005), maize (Ching et al. 2002), and Arabidopsis (Ostrowski et al. 2006). However, direct comparisons of heterozygosity estimates to those obtained with other markers in conifers (allozymes, RAPDs, AFLPs, SSRs, ESTPs) can be misleading. This is because SNPs are diallelic and have relatively low mutation rates (10−8–10−9; Brumfield et al. 2003). Also, plant coding regions generally have lower nucleotide diversity than noncoding or untranslated regions (e.g. Zhu et al. 2003; Guillet-Claude et al. 2004), which may further bias the comparison with heterozygosity estimates derived from anonymous markers or DNA polymorphisms in nongenic regions.
Detection of candidate SNPs for adaptation
At the 95% confidence level, the observed proportion of outlier gene loci (5.5%) was slightly higher than most rates reported with AFLPs such as the 2.6% to 3.3% in Norway spruce (Acheréet al. 2005), the 1.4% and 3.2% in lake whitefish ecotypes (Campbell & Bernatchez 2004), and the 1.3% to 3.6% in common frog (Bonin et al. 2006). It was also slightly higher than the 3.0% (Akey et al. 2002), 5.5% (Wang et al. 2005), and 2.6% (Kelley et al. 2006) reported with SNPs from coding and noncoding regions in the human genome. However, it fell well below the 9.5% outlier rate reported in salmon populations by using EST-linked microsatellites and relying on the union of results from three different statistical approaches (Vasemägi et al. 2005). Apparently, the proportion of outlier genes recorded in the present study could only be temptatively compared to other studies that used summary-statistic methods. This is because all our markers were located within expressed gene regions, while most outlier studies from the literature were based on anonymous markers or markers falling outside genic regions. Moreover, caution needs to be exercised in such comparisons as the results may also depend on the statistical procedure and associated parameters used to detect outliers. For instance, our estimated proportions of outliers became lower when we adjusted the confidence levels of fdist 2 and newfst to maintain the same rate of false-positives with both programs. We expect a similar pattern in the previously published studies.
With the Bayesian method, we did not detect strong local adaptation (no positive γij at the 95% or the 99% confidence levels), but 49 SNPs showed a trend towards local adaptation (γijvalue > 0.10). This is not surprising because strong local adaptive responses are more likely to be detected with this method when selective forces are important (Beaumont & Balding 2004). In the present case, the study covers only a small part of the natural range of white spruce, which extends from Alaska to Newfoundland. Also, no clear-cut bioclimatic or biogeographical differences exist between the six populations surveyed (with the exception of population no. 4 located at a higher altitude in the Laurentians), which could have facilitated gene flow among them and thus limited the efficiency of natural selection in promoting strong adaptive responses (Jaramillo-Correa et al. 2001). In support of this observation, a significant differentiation in adaptive traits was detected among the populations surveyed, mainly for phenological characters and wood traits, but it was moderate and QST values remained below 0.30 (Jaramillo-Correa et al. 2001).
Another plausible explanation is that multiple gene loci may be responsible for regulating the expression of a given trait (Brem et al. 2002). The present gene sample, although containing many genes coding for transcription factors involved in growth, phenology, stress response and wood formation, most probably covers only a part of the gene sets controlling these traits. Even if a strong emphasis was put on sampling transcription factors in the present study (260 out of the 345 genes genotyped were of regulatory nature), the proportion of outlier SNPs was not higher for regulatory than for nonregulatory genes and only a fraction of the regulatory gene space could be tested. On the other hand, if some adaptive traits are controlled by few loci as suggested in annual plants for certain characters such as apical dominance, flowering time, or photoperiod sensitivity (Glazier et al. 2002), then a large number of genes needs to be surveyed to identify outliers in absence of detailed gene expression data. In such a situation, we might have missed many key adaptive genes. Lastly, the high recombination rate in conifer genomes (Brown et al. 2004) may have reversed the effect of selective sweeps in purging variations at linked loci (Lynch 2006). Given the long generation time and the recent Holocene history of spruce populations in eastern Canada, longer periods may in fact be needed to observe a stronger adaptive response for the genes under natural selection.
One concern related to the ability to detect outliers is the power of detection. Positive selective sweeps, usually eliciting high genetic differentiation at selected linked loci, are expected to remain for only a transient phase because recombination breaks the linkage with genes targeted by selection (Przeworski 2002). This is especially true in natural conifer populations, which harbour a rapid decay of linkage disequilibrium usually within gene limits (e.g. Brown et al. 2004; Neale & Savolainen 2004; Bousquet et al. 2007). As a result, a limited population sample size, as that used in ours and most previous studies, may reduce the efficiency of detecting low-frequency variants responsible for adaptation. Nonetheless, many association studies have shown that common alleles (c. 10% and more in frequency) are responsible for a large proportion of the variance in multigenic complex traits (Miller et al. 2000; Thumma et al. 2005; Broderick et al. 2007). In our study, the rate of candidate SNPs for adaptation belonging to the low allele frequency classes (frequency lower than 10%) was not significantly different than that found in the higher allele frequency classes (10% and more). Another possible concern with the identification of outliers is that the model assumptions and derivations used for the Bayesian simulations might not reflect the real population demography and structure. However, previous simulations demonstrated the robustness of the Bayesian model to population demography (Beaumont & Balding 2004). Moreover, the weak amount of population differentiation in white spruce because of allogamy and extensive gene flow (e.g. Jaramillo-Correa et al. 2001) provides little support for this concern.
The fact that a large proportion of the candidate SNPs for local adaptation were located in introns and in 5′- and 3′UTRs does not imply that these SNPs should be considered de facto as false-positives. Natural selection targeting intronic regions was reported in humans and chimpanzee and was shown to play an important role in species divergence (e.g. Keightley & Gaffney 2003; Drake et al. 2005; Gazave et al. 2007). Also, introns and other regions outside exons possess splicing control elements or transcriptional regulatory elements that can have multiple effects on gene expression (Majewski & Ott 2002; Carmel et al. 2007). Similarly, the numerous synonymous candidate SNPs for local adaptation identified in the present study should not be simply dismissed as false-positives. This is because natural selection may affect synonymous codon usage in some genes, leading to codon usage bias (Williams & Hurst 2000; Chamary & Hurst 2004). Furthermore, mutation bias or transcription related mutation/repair mechanisms may translate into unequal substitutions on each of the two DNA strands (Frank & Lobry 1999). In many cases, both mutation bias and selective pressure from functional effects on mRNA are reported to be at the origin of the asymmetry between nucleotides at a synoymous site (Frank & Lobry 1999). Conversely, genetic hitch-hiking that results from the linkage between positions along the gene can not explain these patterns. This is because linkage disequilibrium decays within a few hundreds of base pairs in gene regions in white spruce natural populations (M.-C. Namroud, C. Guillet-Claude, J. Mackay, N. Isabel & J. Bousquet, Arborea, unpublished results) as is the case in other conifers (Brown et al. 2004; Neale & Savolainen 2004).
Evidence of adaptation from gene functions and patterns of population variation
The significant imbalance between populations for the number of candidate SNPs and genes for local adaptation is likely an indication of potential adaptation because an even distribution across populations would be expected with false-positives alone. Moreover, the known functions for many of these genes identified as candidate for local adaptation reflect an inclination towards maintaining biological processes that are vital for growth and survival under stressful environmental conditions. This second observation is not surprising, given the bias in the present study in selecting candidate genes for wood formation, growth, phenology, and stress response. However, only a modest proportion of candidate genes turned out to be candidates for local adaptation. In particular, one of the five genes showing the highest level of putative local adaptation belonged to the zinc finger family, which is known to control the flowering time and reproductive success (Quesada et al. 2005). The second one with the highest putative local adaptation belonged to the zf-B_box (superoxide dismutase) family, which plays an important role in the response to oxidative stress (Hassett & Cohen 1989), while the third one belonged to the Nin_like (ribosomal protein) family usually associated with the control of nitrogen uptake and nutrition (Schieble et al. 2004). These findings are in agreement with those reported by Ford (2002), who compiled more than 119 cases of plant genes or gene groups subject to positive selection in host–parasite interactions, sexual reproduction, and energy metabolism.
One interesting finding that emerges from our results and worth exploring in future physiological and ecological investigations pertains to the potential associations between the biological processes of the candidate genes for local adaptation in certain populations on one hand, and the bioclimatic and phenotypic attributes of the populations on the other hand. Although these associations are made a posteriori and are only qualitative in nature, they do provide insights about possible relationships between genetic, environmental, and quantitative trait attributes. For instance, population no. 3 had a disproportionate number of genes involved in growth in its list of candidate genes for local adaptation. At the same time, it had the warmest climate and the lowest growth (Table 1), perhaps reflecting misadaptation at the southern edge of the species natural range (Andalo et al. 2005). Similarly, a disproportionate number of candidate genes for local adaptation in population no. 6 were involved in wood and lignin formation, which suggests a relationship with the higher level of aridity and higher wood density observed for this population (Table 1). Higher wood density in trees is usually related to a higher proportion of latewood as an adaptation to higher aridity (Corcuera et al. 2004). In population no. 4 characterized by the highest altitude, coldest climate, and lowest growth potential (Table 1), there was a disproportionate representation of genes involved in phenology and stresses in the candidate genes for local adaptation, which could reflect multilocus genetic adaptation to colder climatic conditions (Table 4). Cold adaptation has been mapped on the genome of a number of forest tree species such as Douglas-fir Pseudotsuga menziesii (Mirb.), loblolly pine (Pinus taeda L.) and Populus spp., and it is thought to be controlled by multiple genes with small effects (Frewen et al. 2000; Howe et al. 2003). Similar QTL studies are underway for white spruce (B. Pelgas, P.G. Meirmans, C. Dhont, J. Cooke, J. Bousquet & N. Isabel, Arborea, and K. Ritland, Treenomix, personal communication).
Study limitations and future perspectives
In this study, we used a genome-wide scan to identify genes potentially involved in local adaptation in a nonmodel undomesticated plant species. The proportion of outliers varied with the model and confidence levels used, but an approach focusing on expressed genes and taking into consideration the population effect appears promising in functional population genomic studies at the exploratory stage. This would be especially true for species similar to white spruce, with large genome sizes and a rapid decay of linkage disequilibrium in natural populations.
However, much work remains to be carried out to overcome some limitations. One of these is related to the ascertainment bias associated with the discovery of SNPs. In general, SNPs are identified in a limited discovery panel and those with common allele frequencies have more chance to be detected than rare alleles. In such cases and regardless of the ability of rare or common alleles to best account for the observed quantitative variation, rare alleles potentially involved in directional selection might be under-represented among our 534 SNPs, thus reducing the power to detect natural selection (Morin et al. 2004).
Another limitation pertains to the lack of information about the genes’ physiological roles. Because genotypes observed at locally adapted genes could not be directly linked to fitness-related phenotypes, genes identified as being candidate for local adaptation should be further validated and investigated using complementary approaches such as association genetic studies. Studies based on segregating unstructured populations for a number of key adaptive traits as well as QTL studies in genetically structured spruce populations are underway (N. Isabel, J. Beaulieu, J. Mackay & J. Bousquet, Arborea, personal communication). Studies are also underway to characterize patterns of variation of the candidate SNPs for local adaptation at the rangewide level across Canada. Moreover, expression profiling of these genes (J. Mackay, J. Cooke & B. Boyle, Arborea, personal communication) is expected to shed more light on the genes’ specific physiological roles, and help validate which candidate genes for local adaptation represent true positives.
New promising mass-paralleled DNA sequencing technologies (e.g. Huse et al. 2007) that translate into tumbling costs for sequencing and genotyping recently appeared on the market. They will likely make genome scan approaches for the search of adaptive polymorphisms more accessible than ever for nonmodel or undomesticated species. Major efforts devoted to sequencing ESTs in various species are also multiplying (e.g. Kirst et al. 2003; Li et al. 2003; Pavy et al. 2005; Vasemägi & Primmer 2005). With sufficient sequencing depth and appropriate statistical filtering, these EST collections might represent useful sources of common SNPs (Pavy et al. 2006). As a result, genome-wide SNP scans aimed at identifying outlier gene loci in population surveys should become a standard exploratory approach to detect genes under potential selection, especially for nonmodel and undomesticated species.
Acknowledgments
We are grateful to F. Gagnon, S. Beauseigle, D. Plourde, S. Gerardi, S. Senneville, and P. Marchand for assistance at various stages of the study. We also thank B. Boyle, N. Pavy, H. Maaroufi, P. Laplante, and M. Deslauriers (Arborea, Université Laval and Canadian Forest Service) for assistance with the search for gene annotations and Prof D. Balding (Imperial College, London) for his helpful advices with the NEWFST software. We also thank anonymous reviewers for their helpful comments. This work was supported by grants from NSERC of Canada and from Genome Canada, Génome Québec and the Canadian Biotechnology Strategy, through the white spruce genome project Arborea II led by J. Mackay and J. Bousquet.
Footnotes
This collaborative project reflects the interests of the authors to identify genetic variation of adaptive nature in undomesticated species using population genomics approaches. Marie-Claire Namroud is a postdoctoral associate interested in the use of genomic information to design evolutionary applications. Jean Beaulieu is a research scientist interested in quantitative genetics and problems related to the biodiversity of tree species. Nicolas Juge is a mathematician involved in computational biology. Jérôme Laroche is a research biologist interested in the analysis of diversity from a population and phylogenomic perspective. Jean Bousquet is a professor involved in studying the evolutionary biology of plant and tree species from a molecular and genomic perspective.
References
- Acheré V, Favre JM, Besnard G, Jeandroz S. Genomic organization of molecular differentiation in Norway spruce (Picea abies. Molecular Ecology. 2005;14:3191–3201. doi: 10.1111/j.1365-294X.2005.02646.x. [DOI] [PubMed] [Google Scholar]
- Akey JM, Zhang G, Zhang K, Jin L, Shriver MD. Interrogating a high-density SNP map for signatures of natural selection. Genome Research. 2002;12:1805–1814. doi: 10.1101/gr.631202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler D, Pollara VJ, Cowles CR, et al. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature. 2000;407:513–516. doi: 10.1038/35035083. [DOI] [PubMed] [Google Scholar]
- Andalo C, Beaulieu J, Bousquet J. The impact of climate change on growth of local white spruce populations in Québec, Canada. Forest Ecology and Management. 2005;205:169–182. [Google Scholar]
- Beaty TH, Fallin MD, Hetmanski JB, et al. Haplotype diversity in 11 candidate genes across four populations. Genetics. 2005;171:259–267. doi: 10.1534/genetics.105.043075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont MA, Balding DJ. Identifying adaptive genetic divergence among populations from genome scans. Molecular Ecology. 2004;13:969–980. doi: 10.1111/j.1365-294x.2004.02125.x. [DOI] [PubMed] [Google Scholar]
- Beaumont MA, Nichols RA. Evaluating loci for use in the genetic analysis of population structure. Proceedings of the Royal Scoiety B: Biological Sciences. 1996;263:1619–1626. [Google Scholar]
- Bedon F, Grima-Pettenati J, Mackay J. Conifer R2R3-MYB transcription factors: sequence analyses and gene expression in wood-forming tissues of white spruce (Picea glauca. BMC Plant Biology. 2007;7:17. doi: 10.1186/1471-2229-7-17. (17p.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonin A, Taberlet P, Miaud C, Pompanon F. Explorative genome scan to detect candidate loci for adaptation along a gradient in the common frog (Rana temporaria. Molecular Biology and Evolution. 2006;23:773–783. doi: 10.1093/molbev/msj087. [DOI] [PubMed] [Google Scholar]
- Bouck A, Vision T. The molecular ecologist's guide to expressed sequence tag. Molecular Ecology. 2007;16:909–924. doi: 10.1111/j.1365-294X.2006.03195.x. [DOI] [PubMed] [Google Scholar]
- Bousquet J, Isabel N, Pelgas B. Spruce. In: Kole C, et al., editors. Forest Trees. Vol. 7. Berlin Heidelberg, Germany: Springer-Verlag; 2007. pp. 93–114. (Genome Mapping and Molecular Breeding in Plants). [Google Scholar]
- Bradshaw HDJ, Schemske DW. Allele substitution at a flower colour locus produces a pollinator shift in monkey flowers. Nature. 2003;426:176–178. doi: 10.1038/nature02106. [DOI] [PubMed] [Google Scholar]
- Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- Broderick P, Carvajal-Carmona L, Webb E, et al. A genome–wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nature Genetics. 2007;39:1315–1317. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- Brown GR, Gill GP, Kuntz RJ, Langley CH, Neale D. Nucleotide diversity and linkage disequilibrium in loblolly pine. Proceedings of the National Academy of Sciences, USA. 2004;101:15255–15260. doi: 10.1073/pnas.0404231101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumfield RT, Beerli P, Nickerson DA, Edwards SV. The utility of single nucleotide polymorphisms in inferences of population history. Trends in Ecology and Evolution. 2003;18:249–256. [Google Scholar]
- Campbell D, Bernatchez L. Generic scan using AFLP markers as a means to assess the role of directional selection in the divergence of sympatric whitefish ecotypes. Molecular Biology and Evolution. 2004;21:945–956. doi: 10.1093/molbev/msh101. [DOI] [PubMed] [Google Scholar]
- Carmel L, Wolf YI, Rogozin IB, Koonin EV. Three distinct modes of intron dynamics in the evolution of eukaryotes. Genome Research. 2007;17:1034–1044. doi: 10.1101/gr.6438607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamary JV, Hurst LD. Similar rates but different modes of sequence evolution in introns and at exonic silent sites in rodents: evidence for selectively driven codon usage. Molecular Biology and Evolution. 2004;21:1014–1023. doi: 10.1093/molbev/msh087. [DOI] [PubMed] [Google Scholar]
- Ching A, Caldwell K, Jung M, et al. SNP frequency, haplotype structure and linkage disequilibrium in elite maize inbred lines. BMC Genetics. 2002;3:19. doi: 10.1186/1471-2156-3-19. (14p.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcuera L, Camarero JJ, Gil-Pelegrín E. Effects of a severe drought on growth and wood anatomical properties of Quercus Faginea. IAWA Journal. 2004;25:185–204. [Google Scholar]
- Cork JM, Purugganan MD. High-diversity genes in the Arabidopsis genome. Genetics. 2005;170:1897–1911. doi: 10.1534/genetics.104.036152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JA, Bird C, Nemesh J, et al. Conserved noncoding sequences are selectively constrained and not mutation cold spots. Nature Genetics. 2005;38:223–227. doi: 10.1038/ng1710. [DOI] [PubMed] [Google Scholar]
- Erickson DL, Fenster CB, Stenoien HK, Price D. Quantitative trait locus analyses and the study of evolutionary process. Molecular Ecology. 2004;13:2505–2522. doi: 10.1111/j.1365-294X.2004.02254.x. [DOI] [PubMed] [Google Scholar]
- Fan JB, Oliphant A, Shen R, et al. Cold Spring Harbor Symposia on Quantitative Biology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2003. Highly parallel SNP genotyping; pp. 69–78. [DOI] [PubMed] [Google Scholar]
- Ford MJ. Applications of selective neutrality tests to molecular ecology. Molecular Ecology. 2002;11:1245–1262. doi: 10.1046/j.1365-294X.2002.01536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank AC, Lobry JR. Asymmetric substitution patterns: a review of possible underlying mutational or selective mechanisms. Gene. 1999;238:65–77. doi: 10.1016/s0378-1119(99)00297-8. [DOI] [PubMed] [Google Scholar]
- Frewen BE, Chen THH, Howe GT, et al. Quantitative trait loci and candidate gene mapping of bud set and bud flush in Populus. Genetics. 2000;154:837–845. doi: 10.1093/genetics/154.2.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazave E, Marqués-Bonet T, Fernando O, Charlesworth B, Navarro A. Patterns and rates of intron divergence between humans and chimpanzees. Genome Biology. 2007;8:R21. doi: 10.1186/gb-2007-8-2-r21. (13p.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazier AM, Nadeau JH, Aitman TJ. Finding genes that underlie complex traits. Science. 2002;298:2345–2349. doi: 10.1126/science.1076641. [DOI] [PubMed] [Google Scholar]
- Godt MJW, Hamrick MA, Edwards-Burke MA, Williams JH. Comparisons of genetic diversity in white spruce (Picea glauca) and jack pine (Pinus banksiana) seed orchards with natural populations. Canadian Journal of Forest Research. 2001;31:943–949. [Google Scholar]
- Guillet-Claude C, Birolleau-Touchard C, Manicacci D, et al. Nucleotide diversity of the ZmPox3 maize peroxidase gene: relationships between a MITE insertion in exon 2 and variation in forage maize digestibility. BMC Genetics. 2004;5:19. doi: 10.1186/1471-2156-5-19. (11p.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassett DJ, Cohen MS. Bacterial adaptation to oxidative stress: implications for pathogenesis and interaction with phagocytic cells. The Federation of American Societies for Experimental Biology. 1989;3:2574–2582. doi: 10.1096/fasebj.3.14.2556311. [DOI] [PubMed] [Google Scholar]
- Hedrick PW. Perspective: highly variable loci and their interpretation in evolution and conservation. Evolution. 1999;53:313–318. doi: 10.1111/j.1558-5646.1999.tb03767.x. [DOI] [PubMed] [Google Scholar]
- Heidelberger P, Welch P. Spectral method for confidence interval generation and run length control in simulations. Communications of the ACM. 1981;24:233–245. [Google Scholar]
- Howe GT, Aitken SN, Neale DB, et al. From genotype to phenotype: unravelling the complexities of cold adaptation in forest trees. Canadian Journal of Botany. 2003;81:1247–1266. [Google Scholar]
- Hurme P, Sillanpaa MJ, Arjas E, Repo T, Savolainen O. Genetic basis of climatic adaptation in Scots pine by Bayesian quantitative trait locus analysis. Genetics. 2000;156:1309–1322. doi: 10.1093/genetics/156.3.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biology. 2007;8:R143. doi: 10.1186/gb-2007-8-7-r143. (19p.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabel N, Beaulieu J, Bousquet J. Complete congruence between gene diversity estimates derived from genotypic data at enzyme and RAPD loci in black spruce. Proceedings of the National Academy of Sciences, USA. 1995;92:6369–6373. doi: 10.1073/pnas.92.14.6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabel N, Beaulieu J, Theriault P, Bousquet J. Direct evidence for biased gene diversity estimates from dominant random amplified polymorphic DNA (RAPD) fingerprints. Molecular Ecology. 1999;8:477–483. [Google Scholar]
- Jaramillo-Correa JP, Beaulieu J, Bousquet J. Contrasting evolutionary forces driving population structure at expressed sequence tag polymorphisms, allozymes and quantitative traits in white spruce. Molecular Ecology. 2001;10:2729–2740. doi: 10.1046/j.0962-1083.2001.01386.x. [DOI] [PubMed] [Google Scholar]
- Kaplan NL, Hudson RR, Langley CH. The ‘hitchhiking effect′ revisited. Genetics. 1989;123:887–899. doi: 10.1093/genetics/123.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M, Brauer S, Stoneking M. A genome scan to detect candidate regions influenced by local natural selection in human populations. Molecular Biology and Evolution. 2003;20:893–900. doi: 10.1093/molbev/msg092. [DOI] [PubMed] [Google Scholar]
- Keightley PD, Gaffney DJ. Functional constraints and frequency of deleterious mutations in noncoding DNA of rodents. Proceedings of the National Academy of Sciences, USA. 2003;100:13402–13406. doi: 10.1073/pnas.2233252100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley JL, Madeoy J, Calhoun JC, Swanson W, Akey JM. Genomic signatures of positive selection in humans and the limits of outlier approaches. Genome Research. 2006;16:980–989. doi: 10.1101/gr.5157306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura R, Fujimoto A, Tokunaga K, Ohashi J. A practical genome scan for population-specific strong selective sweeps that have reached fixation. PLoS ONE. 2007;2:E286. doi: 10.1371/journal.pone.0000286. doi: 10.1371/journal.pone.0000286 (10p.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirst M, Johnson AF, Baucom C, et al. Apparent homology of expressed genes from wood-forming tissues of loblolly pine (Pinus taeda L.) with Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 2003;100:7383–7388. doi: 10.1073/pnas.1132171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutovsky KV, Neale DB. Nucleotide diversity and linkage disequilibrium in cold-hardiness and wood quality-related candidate genes in Douglas fir. Genetics. 2005;171:2029–2041. doi: 10.1534/genetics.105.044420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Beaulieu J, Corriveau A, Bousquet J. Genetic variation in juvenile growth and phenology in a white spruce provenance-progeny test. Silvae Genetica. 1993;42:52–59. [Google Scholar]
- Li P, Beaulieu J, Bousquet J. Genetic structure and patterns of genetic variation among populations in eastern white spruce (Picea glauca. Canadian Journal of Forest Research. 1997;27:189–198. [Google Scholar]
- Li L, Brunk BP, Kissinger JC, et al. Gene discovery in the Apicomplexa as revealed by EST sequencing and assembly of a comparative gene database. Genome Research. 2003;13:443–454. doi: 10.1101/gr.693203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart G, England P, Tallmon D, Jordan S, Taberlet P. The power and promise of population genomics: from genotyping to genome typing. Nature Reviews of Genetics. 2003;4:981–994. doi: 10.1038/nrg1226. [DOI] [PubMed] [Google Scholar]
- Lynch M. The origins of eukaryotic gene structure. Molecular Biology and Evolution. 2006;23:450–468. doi: 10.1093/molbev/msj050. [DOI] [PubMed] [Google Scholar]
- Majewski J, Ott J. Distribution and characterization of regulatory elements in the human genome. Genome Research. 2002;12:1827–1836. doi: 10.1101/gr.606402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariette S, Le Corre V, Austerlitz F, Kremer A. Sampling within the genome for measuring within-population diversity: trade-offs between markers. Molecular Ecology. 2002;11:1145–1156. doi: 10.1046/j.1365-294x.2002.01519.x. [DOI] [PubMed] [Google Scholar]
- Maynard Smith J, Haigh J. The hitch-hiking effect of a favorable gene. Genetic Research. 1974;23:23–35. [PubMed] [Google Scholar]
- Miller WG, Englen MD, Kathariou S, et al. Single nucleotide polymorphism and linkage disequilibrium within the TCR α/δ. Human Molecular Genetics. 2000;9:1011–1019. doi: 10.1093/hmg/9.7.1011. [DOI] [PubMed] [Google Scholar]
- Moffatt MF, Traherne JA, Abecasis JR, Cookson WOCM. Single nucleotide polymorphism and linkage disequilibrium within the TCR α/δ locus. Human Molecular Genetics. 2000;9:1011–1019. doi: 10.1093/hmg/9.7.1011. [DOI] [PubMed] [Google Scholar]
- Morin PA, Luikart G, Wayne RK, the SNP workshop Group SNPs in ecology, evolution and conservation. Trends in Ecology and Evolution. 2004;19:208–216. [Google Scholar]
- Murray BG. Nuclear DNA amounts in gymnosperms. Annals of Botany. 1998;82:3–15. [Google Scholar]
- Neale DB, Savolainen O. Association genetics of complex traits in conifers. Trends in Plant Science. 2004;9:325–330. doi: 10.1016/j.tplants.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybom H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Molecular Ecology. 2004;13:1143–1155. doi: 10.1111/j.1365-294X.2004.02141.x. [DOI] [PubMed] [Google Scholar]
- Oetjen K, Reusch TBH. Identification and characterization of 14 polymorphic EST-derived microsatellites in eelgrass (Zostera marina. Molecular Ecology Notes. 2007;7:777–780. [Google Scholar]
- Ostrowski M-F, David J, Santoni S, et al. Evidence for a large-scale population structure among accessions of Arabidopsis thaliana: possible causes and consequences for the distribution of linkage disequilibrium. Molecular Ecology. 2006;15:1507–1517. doi: 10.1111/j.1365-294X.2006.02865.x. [DOI] [PubMed] [Google Scholar]
- Pavy N, Paule C, Parsons L, et al. Generation, annotation, analysis and database integration of 16 500 white spruce EST clusters. BMC Genomics. 2005;6:144. doi: 10.1186/1471-2164-6-144. (19p.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavy N, Parsons L, Paule C, Mackay J, Bousquet J. Automated SNP detection from a large collection of white spruce expressed sequences: contributing factors and approaches for the categorization of SNPs. BMC Genomics. 2006;7:174. doi: 10.1186/1471-2164-7-174. (14p.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavy N, Pelgas B, Beauseigle S, et al. Enhancing genetic mapping of complex genomes through the design of highly-multiplexed SNP arrays: application to the large and unsequenced genomes of white spruce and black spruce. BMC Genomics. 2008;9:21. doi: 10.1186/1471-2164-9-21. (17p.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelgas B, Beauseigle S, Acheré V, et al. Comparative genome mapping among Picea glauca, P. abies and P. mariana X rubens, and correspondance with other Pinaceae. Theoretical and Applied Genetics. 2006;113:1371–1393. doi: 10.1007/s00122-006-0354-7. [DOI] [PubMed] [Google Scholar]
- Perry DJ, Bousquet J. Genetic diversity and mating system of post-fire and post-harvest black spruce: an investigation using codominant sequence-tagged site (STS) markers. Canadian Journal of Forest Research. 2001;31:32–40. [Google Scholar]
- Przeworski M. The signature of positive selection at randomly chosen loci. Genetics. 2002;160:1179–1189. doi: 10.1093/genetics/160.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Dean C, Simpson GG. Regulating RNA processing in the control of Arabidospsis flowering. International Journal of Developmental Biology. 2005;49:773–780. doi: 10.1387/ijdb.051995vq. [DOI] [PubMed] [Google Scholar]
- Régnière J. A generalized approach to landscape-wide seasonal forecasting with temperature-driven simulation models. Environmental Entomology. 1996;25:869–881. [Google Scholar]
- Schieble WR, Morcuende R, Czechowski T, et al. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to Nitrogen. Plant Physiology. 2004;136:2483–2499. doi: 10.1104/pp.104.047019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlötterer C. Hitchhiking mapping — functional genomics from the population genetics perspective. Trends in Genetics. 2003;19:32–38. doi: 10.1016/s0168-9525(02)00012-4. [DOI] [PubMed] [Google Scholar]
- Scotti-Saintagne C, Mariette S, Porth I, et al. Genome scanning for interspecific differentiation between two closely related oak species [Quercus robur L. & Quercus petraea (Matt.) Liebl.] Genetics. 2004;168:1615–1626. doi: 10.1534/genetics.104.026849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen R, Fan JB, Campbell D, et al. High-throughput SNP genotyping on universal bead arrays. Mutation Research. 2005;573:70–82. doi: 10.1016/j.mrfmmm.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Stephan W, Wiehe THE, Lenz MW. The effect of strongly selected substitutions on neutral polymorphism: analytical results based on diffusion theory. Theoretical Population Biology. 1992;41:237–254. [Google Scholar]
- Storz JF. Using genome scans of DNA polymorphism to infer adaptive population divergence. Molecular Ecology. 2005;14:671–688. doi: 10.1111/j.1365-294X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- Storz JF, Payseur BA, Nachman MW. Genome scans of DNA variability in humans reveal evidence for selective sweeps outside of Africa. Molecular Biology and Evolution. 2004;21:1800–1811. doi: 10.1093/molbev/msh192. [DOI] [PubMed] [Google Scholar]
- Thumma BR, Nolan MF, Evans R, Moran GF. Polymorphisms in Cinnamoyl CoA Redustase (CCR) are associated with variation in microfibril angle in Eucalyptus spp. Genetics. 2005;171:1257–1265. doi: 10.1534/genetics.105.042028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumura Y, Kado T, Takahashi T, et al. Genome scan to detect genetic structure and adaptive genes of natural populations of Cryptomeria japonica. Genetics. 2007;176:2393–2403. doi: 10.1534/genetics.107.072652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasemägi A, Primmer CR. Challenges for identifying functionally important genetic variation: the promise of combining complementary research strategies. Molecular Ecology. 2005;14:3623–3642. doi: 10.1111/j.1365-294X.2005.02690.x. [DOI] [PubMed] [Google Scholar]
- Vasemägi A, Nilsson J, Primmer CR. Expressed sequence tag-linked microsatellites as a source of gene-associated polymorphisms for detecting signatures of divergent selection in Atlantic Salmon (Salmo salar L.) Molecular Biology and Evolution. 2005;22:1067–1076. doi: 10.1093/molbev/msi093. [DOI] [PubMed] [Google Scholar]
- Wakamiya I, Newton RJ, Johnston JS, Price HJ. Genome size and environmental factors in the genus Pinus. American Journal of Botany. 1993;80:1235–1241. [Google Scholar]
- Wang KS, Liu M, Paterson A. Evaluating outlier loci and their effect on the identification of pedigree errors. BMC Genetics. 2005;6:S155. doi: 10.1186/1471-2156-6-S1-S155. (5p.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BC, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1137. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Williams EJ, Hurst LD. The proteins of linked genes evolve at similar rates. Nature. 2000;407:900–903. doi: 10.1038/35038066. [DOI] [PubMed] [Google Scholar]
- Wright SI, Gaut BS. Molecular population genetics and the search for adaptive evolution in plants. Molecular Biology and Evolution. 2005;22:506–519. doi: 10.1093/molbev/msi035. [DOI] [PubMed] [Google Scholar]
- Wright SI, Bi IV, Schroeder SG, Yamasaki M, Doebley JF. The effects of artificial selection on the maize genome. Science. 2005;308:1310–1314. doi: 10.1126/science.1107891. [DOI] [PubMed] [Google Scholar]
- Zayed A, Whitfield CW. A genome-wide signature of positive selection in ancient and recent invasive expansions of the honey bee Apis mellifera. Proceedings of the National Academy of Sciences, USA. 2008;105:3421–3426. doi: 10.1073/pnas.0800107105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YL, Song QJ, Hyten DL, et al. Single-nucleotide polymorphisms in soybean. Genetics. 2003;163:1123–1134. doi: 10.1093/genetics/163.3.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]