Abstract
Capillary electrophoresis with β-CD as a chiral selector has successfully separated the two enantiomers of salsolinol, N-methyl-salsolinol, and 1-benzyl-tetrahydroisoquinoline. The migration times of each enantiomer in capillary electrophoresis reflect the stability of their β-CD inclusion complexes. This paper reports a computational modeling study of the inclusion complexes of β-cyclodextrin (β-CD) with salsolinol, N-methyl-salsolinol, and 1-benzyl-tetrahydroisoquinoline by using PM3 (Parametric Method 3) semi-empirical molecular orbital calculations and the ONIOM hybrid method. Two types of the inclusion complexes, cis- and trans-orientations, are considered for each enantiomer of the guest molecules, salsolinol, N-methyl-salsolinol, and 1-benzyl-tetrahydroisoquinoline. In the cis-orientation, the nitrogen in the salsolinol, N-methyl-salsolinol, and 1-benzyl-tetrahydroisoquinoline points toward the secondary hydroxyls of the β-CD, while in the trans-orientation, the nitrogen in salsolinol, N-methyl-salsolinol, and 1-benzyl-tetrahydroisoquinoline points toward the primary hydroxyls of the β-CD. We found that the stabilization energies of these inclusion complexes from these PM3 and ONIOM different methods correlate very well with the migration order deduced from the study of capillary electrophoretic separation.
Introduction
Native cyclodextrins (CD) are oligosaccharides composed of six or more D-glucopyranose residues attached by α-1,4-linkages in a cyclic array. The hydrophobic character of the central cavity of CDs enables them to form inclusion complexes with a wide variety of compounds, ranging from very polar inorganic ions to non-polar organic molecules. There are no covalent bonds formed or broken during the complex formation process. The driving forces for the complex formations have been attributed to the release of entropy-rich water molecules from the cavity, van der Waals interaction, hydrogen bonding, hydrophobic interactions, release of ring strain in the cyclodextrin molecule, and changes in solvent-surface tension [1-10].
Cyclodextrins are often used in separation methods as receptors that discriminate between enantiomers of various chiral and achiral guest compounds. Cramer and Dietsche [11] reported the pioneering study of chiral recognition by CD, partial optical resolution of mandelic acid derives by β-CD. Since then, a large number of successful applications of CDs to chiral recognition have been carried out [12-21]. The mechanisms underlying the separation in the presence of CD as a selector have not been fully clarified.
Salsolinol (6,7-dihydroxy-1-methyl-1, 2, 3, 4-tetrahydroisoquinoline; Sal), N-methyl-salsolinol (6,7-dihydroxy-1,2-dimethyl-1, 2, 3, 4-tetrahydroisoquinoline; NMSal), and 1-benzyl-tetrahydroisoquinoline (BTIQ) are dopamine (DA)-derived tetrahydroisoquinolines (TIQ). They are thought to be potent dopaminergic neurotoxins, similar to 1-methyl-4-phenyl-1, 2, 3, 6-tetrahydropyridine (MPTP), an exogenous neurotoxin that causes Parkinsonism in humans, monkeys, and various animals [22]. Different neurotoxicological properties were observed for (R)- and (S)-enantiomers of Sal, NMSal, and BTIQ. The chirality of neurotoxic TIQs has been receiving increasing research attention. Many chiral TIQ molecules have been shown to exhibit enantioselective neurotoxicity [23,24]. (R)-Sal was found to be a stronger inhibitor for MAO type-A than (S)-Sal [25]. (S)-Sal was shown to suppress significantly the proopiomelanocotin (POMC) gene expression, but the (R)- enantioner did not exhibit the same effect [26]. N-methyl-(R)-salsolinol (NM(R)Sal) has been proposed to be closely involved in the pathogenesis of Parkinson’s disease. NM(R)Sal is synthesized from dopamine by two enzymes: (R) Salsolinol synthase and a neutral (R) Salsolinol N-methyltransferase. The mechanism of cell death was examined by detection of DNA damage using a single-cell gel electrophoresis (comet) assay in human dopaminergic neuroblastoma SH-SY5Y cells. The (R)-enantiomer of NMSal damaged DNA much more profoundly than the (S)-enantiomer [27]. It was shown that endogenous TIQ formation and TIQ N-methylation involved stereoselective enzymatic processes [28]. Actually, an enzyme, which enantioselectively synthesized (R)-Sal, has been isolated from the rat brain [29]. BTIQ exhibited varying degrees of interaction with β 1-adrenergic receptors.
Studies on the stereoselective neurotoxicity of TIQs have promoted the development of analytical methods for the determination of TIQ enantiomers. Several HPLC procedures [30-33] and a capillary electrochromatographic (CEC) method [34] were reported for the separation of Sal and N-methyl-Sal enantiomers. Gas chromatography-mass spectrometry (GC-MS) has been long used for the quantification of catecholic amines [35-36]. To achieve the separation of TIQ enantiomers, Haber et al. developed a chiral GC-MS method based on a two -step derivatization with N-methyl-N-trimethylsilyltrifluoracetamide and R-(-)-2-phenylbutyrylic acid [37]. This method was later used by Musshoff et al. for the determination of dopamine, (R)-/(S)-Sal, and (R)-/(S)-norsalsolinol in human brain tissue samples [38]. We previously reported a chiral GC-MS method employing a β-cyclodextrin (β-CD) coated capillary column for resolving Sal, NMSal, and BTIQ enantiomers [39-40]. Although a better resolution for Sal enantiomers was obtained by using the GC-MS method, the water-sensitive derivatization procedure involved was not convenient for the assay. Extreme care must be taken to obtain reproducible results.
In recent years, various theoretical studies have been made on CD inclusion complexes with the aim of correlating the experimental results with the mode of the interaction between a CD host and a guest molecule [41-44]. Although molecular mechanics (MM) and molecular dynamics (MD) with various force field approaches are increasingly being used in modern theoretical calculations, semi-empirical methods such as AM1 (Austin Model 1) [45] and PM3 (Parametric Method 3) [46] are more commonly used for structure optimization of very large systems or for reactions involving large molecular systems [44, 47-53]. It has been generally accepted that PM3 has high computational efficiency and permits the modeling of large systems beyond the capacity of ab initio methods [54]. The precision of PM3 is comparable to that of ab initio methods with medium-sized basis sets [55]. As pointed out by Casadesús et al. [56], ab initio methods are prohibitively expensive in treating large systems such as CD inclusion complexes. In addition, PM3 performs better than AM1 in biochemical systems due to its improved description of the interactions between nonbonded atoms such as hydrogen bonding and steric effects [57].
The objective of this work is to use the PM3 semi-empirical molecular orbital method and the ONIOM hybrid method [58] on the inclusion complexes of different enantiomers of Sal, NMMSal, and BTIQ with β-CD. These methods have not previously been applied to the aforementioned complexes. In the experimental part, capillary electrophoresis techniques have been used to measure migration times of the different enantiomers [40].
Three aspects of inclusion complexes were investigated in these studies: (a) PM3 and ab initio density-functional theory (B3LYP/6-31G*) were used to obtain optimized geometries of the guest compounds, Sal, NMSal, and BTIQ; (b) relative stabilization energies of β-CD and isomeric guest inclusion complexes from PM3 and ONIOM (B3LYP/6-31G*:PM3) calculations were obtained; and (c) the relation of the relative stabilization energies of the complexes to the experimentally determined migration order in capillary electrophoretic separation was investigated.
Computational Method
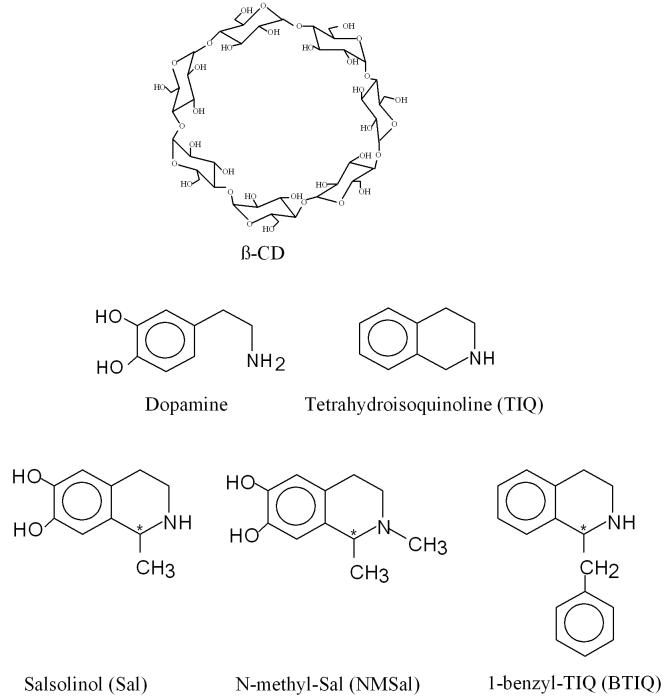
PM3 and ONIOM (B3LYP/6-31G*:PM3) hybrid calculations were performed on Sal, NMSal, BTIQ, β-CD, and β-CD isomeric guest inclusion complexes by using the Gaussian 98 and Gaussian 03 programs [59]. Within the ONIOM procedure, two levels of calculations are defined: density-functional theory with the B3LYP functional and the 6-31G* basis set were performed on the guest compounds, Sal, NMSal, and BTIQ enantiomers, and the semiempirical PM3 method was used for the β-CD. In B3LYP, the Becke exchange functional [60] is coupled with the Lee-Yang-Parr correlation functional [61]. Figure 1 shows the structures of β-CD, Sal, NMSal, and BTIQ. All the compounds were built with the Spartan software program [62]. The size and conformational flexibility of the inclusion complexes make it impossible to study all the possible conformers. In order to search for the lowest energy structures of the inclusion complexes, we have tried several starting points for the PM3 and ONIOM hybrid optimizations, including some determined by molecular mechanics.
Figure 1.
Structures of β-CD and certain compounds involved in this work.
Results and Discussion
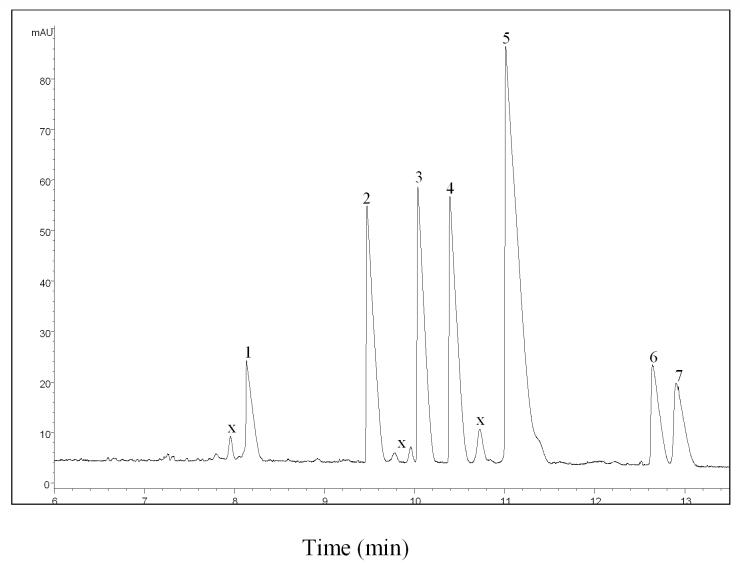
As mentioned in our previous paper [40], various CE conditions were used to achieve the optimum enantiomeric separation efficiency. A 75 μm i.d. capillary was selected for better detection sensitivity and reproducibility. The maximal voltage output available with the power supply (30 kV) was used for a shortened running time, and β-CD was selected for the experiments. As can be seen from Figures 2 and 3, TIQ, DA, (R/S)-Sal, (R)-NMSal and (R/S)-BTIQ were well separated using a running buffer consisting of 1.5 M urea, 12 mM β-CD, and 50 mM phosphate. In that study, three pairs of TIQ enantiomers, i.e. (R/S)-BTIQ, (R/S)-Sal, and (R/S)-NMSal were tested. All enantiomers were baseline resolved under the CE conditions described above. All (R)-enantiomers had a longer migration time than their optical antipodes.
Figure 2.
Electropherogram from separating mixtures of TIQ derivatives. Peak identification. 1. TIQ; 2. dopamine; 3. (S)-Sal; 4. (R)-Sal; 5. (R)-N-methyl-Sal; 6. (S)-1-benzyl-TIQ; 7. (R)-1-benzyl-TIQ. “x” stands for unknown identity. The concentrations of chemicals were: 0.0125 mg/ml TIQ, 0.018 mg/ml dopamine, (R)-Sal and (S)-Sal, 0.08 mg/ml (R)-NMSal and 0.009 mg/ml (R)- and (S)- 1-benzyl-TIQ. The running buffer contained 12 mM -CD, 1.5 M Urea and 50 mM phosphate (pH 3.0).
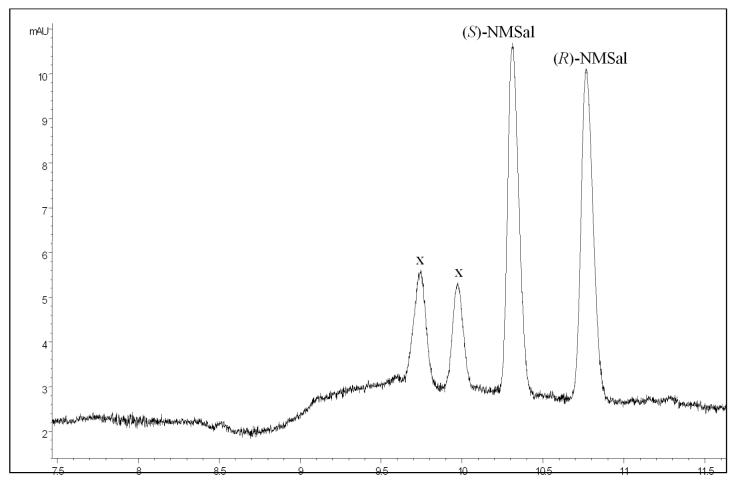
Figure 3.
Electropherogram obtained from a mixture of racemic NMSal, “x” stands for unknown identity.
As also mentioned in our previous paper [40], to gain insight into the separation mechanism, consider the geometry of a Sal molecule. The benzene ring and the atoms directly attached to the ring are rigid and in a single plane. In the presence of β-CD, Sal enantiomers form inclusion complexes with β-CD. In these complexes, the aromatic moiety in a Sal molecule is accommodated in the hydrophobic cavity of a β-CD molecule. Due to the chirality effects of β-CD molecules, the stability of the inclusion complexes can be different for the two Sal enantiomers. From the CE separation results, it has been shown that the inclusion complex formed by (R)-Sal and β-CD is more stable than that formed from (S)-Sal and β-CD, which causes (R)-Sal to take more time to migrate out than (S)-Sal. Further, the stability difference for NMSal enantiomers is greater than that for Sal enantiomers, which is in accordance with the fact that racemic NMSal was better resolved (i.e. a larger RS value was obtained for NMSal enantiomers) than racemic Sal.
Each of the guest structures of Sal, NMSal, and BTIQ has one chiral center, so there are two enantiomers for each of the guests. The energies for the PM3 and B3LYP/6-31G* optimized structures of the guest compounds (Sal, NMSal, and BTIQ) and β-CD with the PM3 method are shown in Table 1.
Table 1.
Energies (in units of a.u.) for Sal, NMSal, and BTIQ from PM3 and B3LYP/6-31G* methods and for β-cyclodextrin from PM3 methods.
| PM3 | B3LYP/6-31G* | |
|---|---|---|
| β-CD | -2.31056 | |
| Sal | -0.12371 | -594.07690 |
| NMSal | -0.12538 | -633.38493 |
| BTIQ | 0.06264 | -674.69503 |
The inclusion complexes have two types of orientations, namely cis- and trans-orientations that are possibly formed with each isomer of Sal, NMSal, and BTIQ. Figure 4 shows the two orientations for Sal and BTIQ guests inside the β-CD. In the cis-orientation the nitrogen in the Sal, NMSal, and BTIQ points toward the secondary hydroxyls of the β-CD, and in the trans-orientation, the nitrogen in Sal, NMSal, and BTIQ points toward the primary hydroxyls of the β-CD.
Figure 4.

The two types of orientation of the inclusion complexes for Sal and BTIQ.
The PM3 and ONIOM (B3LYP/6-31G*:PM3) energies, stabilization energies, and ΔHtotal for the inclusion complexes of β-CD with Sal, NMSal, and BTIQ are shown in Table 2. The stabilization energies (enthalpies) of complexation were calculated as follows:
and the total stabilization energy is one-half of the sum of the stabilization energies of the cis and trans orientations, i.e.,
Table 2.
The PM3 and ONIOM energies (ΔH in units of a.u.), stabilization energies (ΔΔH in units of kcal/mol) and ΔHtotal (in units of kcal/mol) of the β-CD inclusion complexes of Sal, NMSal, and BTIQ.
| CD inclusion complex |
Orientation | ΔH (a.u.) | ΔΔH (kcal/mol) | ΔHtotal (kcal/mol) |
|---|---|---|---|---|
| PM3 | ||||
| Sal-(R) | cis | -2.46338 | -18.27 | -19.94 |
| trans | -2.46870 | -21.60 | ||
| Sal-(S) | cis | -2.45842 | -15.15 | -16.66 |
| trans | -2.46322 | -18.17 | ||
| NMSal-(R) | cis | -2.46897 | -20.72 | -20.79 |
| trans | -2.46919 | -20.86 | ||
| NMSal-(S) | cis | -2.45971 | -14.92 | -17.41 |
| trans | -2.46766 | -19.90 | ||
| BTIQ-(R) | cis | -2.27626 | -17.78 | -20.07 |
| trans | -2.28356 | -22.36 | ||
| BTIQ-(S) | cis | -2.27256 | -15.45 | -17.81 |
| trans | -2.28007 | -20.17 | ||
| ONIOM | ||||
| Sal-(R) | cis | -596.41600 | -17.91 | -19.01 |
| trans | -596.41949 | -20.10 | ||
| Sal-(S) | cis | -596.41182 | -15.29 | -18.01 |
| trans | -596.42048 | -20.72 | ||
| NMSal-(R) | cis | -635.72722 | -19.91 | -20.04 |
| trans | -635.72761 | -20.16 | ||
| NMSal-(S) | cis | -635.71747 | -13.79 | -15.64 |
| trans | -635.72337 | -17.49 | ||
| BTIQ-(R) | cis | -677.03719 | -19.83 | -22.38 |
| trans | -677.04533 | -24.94 | ||
| BTIQ-(S) | cis | -677.03319 | -17.32 | -21.67 |
| trans | -677.04704 | -26.01 | ||
From Table 2, the total stabilization energies from PM3 calculations for the Sal-(R), Sal-(S), NMSal-(R), NMSal-(S), BTIQ-(R), and BTIQ-(S) are -19.94 kcal/mol, - 17.01 kcal/mol, -20.79 kcal/mol, -17.41 kcal/mol, -20.07 kcal/mol, and -17.81 kcal/mol, respectively. The more stable the inclusion complex is, the longer time for the inclusion complex to come out during the separation. The total stabilization energies show that the inclusion complexes formed by (R)-isomers are more stable than those formed by (S)-isomers, which coincides with the CE results. As mentioned, from the CE separation results [40], it has been shown that the inclusion complex formed by (R)-Sal and β-CD is more stable than that formed from (S)-Sal and β-CD, which causes (R)-Sal to take more time to migrate out than (S)-Sal. Further, the stability difference for NMSal enantiomers was greater than that for Sal enantiomers, which is in accordance with the fact that racemic NMSal was better resolved (i.e. a larger RS value was obtained for NMSal enantiomers) than for racemic Sal [40]. The resolution is defined by RS= 2(R2-R1)/(W1+W2), where (R2-R1) represents the difference of migration times for two analytes, and W1 and W2 are the widths for the corresponding peaks. The resolution values from Figures 2 and 3 are 2.01, 3.04, and 1.18 for Sal, NMSal, and BTIQ enantiomers, respectively. The greater the resolution value is, the better the separation is, i.e. the greater the differences in stability are. The total stabilization energy differences between the inclusion complexes of the (R)- and (S)- isomers are 3.28 kcal/mol, 3.38 kcal/mole and 2.26 kcal/mol for Sal, NMSal, and BTIQ respectively. Our theoretical calculations show that the stabilization energy differences between the inclusion complexes of the (R)- and (S)- isomers for NMSal are much larger than those for Sal, which agrees with the experimental data very well.
From Table 2, the ONIOM (B3LYP/6-31G*:PM3) results follow the same trend as the PM3 data. The total stabilization energies from ONIOM (B3LYP/6-31G*:PM3) calculations for the Sal-(R), Sal-(S), NMSal-(R), NMSal-(S), BTIQ-(R), and BTIQ-(S) are -19.01 kcal/mol, -18.01 kcal/mol, -20.04 kcal/mol, -15.64 kcal/mol, -22.38 kcal/mol, and -21.67 kcal/mol, respectively. The total stabilization energies for the inclusion complexes formed by (R)-isomers are more stable than those formed by (S)-isomers. The total stabilization energy differences from ONIOM (B3LYP/6-31G*:PM3) for the inclusion complexes of the (R)- and (S)- isomers are 1.00 kcal/mol, 4.40 kcal/mol and 0.71 kcal/mol for Sal, NMSal, and BTIQ, respectively. ONIOM calculations also indicate that the total stabilization energy differences for NMSal enantiomers are greater than those for Sal enantiomers. These results are in accordance with the experimental fact that racemic NMSal was better resolved than racemic Sal.
From Table 2, we also noticed that the trans orientations for the inclusion complexes have higher stabilization energies than the cis orientations. Acomparison of the number of hydrogen bonds in the inclusion complexes with the number in β-CD molecule is shown in Table 3. The hydrogen bond here is defined as an O-H · · · O interaction in which the H · · · O distance is less than or equal to 3.00 Å and the angle at H is greater than 145°. By this definition, there are three intramolecular hydrogen bonds in β-CD itself. For the Sal-(R) and β-CD inclusion complexes in the cis orientation, one of the β-CD intramolecular hydrogen bonds is broken, and another intramolecular hydrogen bond is formed in β-CD. For the same inclusion complex in the trans orientation, one of the β-CD intramolecular hydrogen bonds is broken, and another four intramolecular hydrogen bonds are formed in β-CD. In general, the total number of hydrogen bonds in the inclusion complexes correlates with the stabilization energies except for the NMSal-(R) and β-CD inclusion complexes with PM3 and Sal-(S) and NMSal-(R) and β-CD inclusion complexes with ONIOM; those have the same number of hydrogen bonds. From PM3 calculations on NMSal-(R) and β-CD inclusion complexes with cis orientation is no intramolecular hydrogen bonds are broken and two new intramolecular hydrogen bonds are formed in β-CD. For the same inclusion complexes with trans orientation, PM3 calculations indicate that one intramolecular hydrogen bond is broken and three new intramolecular hydrogen bonds are formed in β-CD. ONIOM calculations on Sal-(S) and β-CD inclusion complexes with cis orientation suggest one intramolecular hydrogen bond is broken, while two new intramolecular hydrogen bonds are formed in β-CD. Thus, one more intermolecular hydrogen bond is formed between the Sal-(S) and β-CD. For the same inclusion complexes with trans orientation, ONIOM calculations indicate that two intramolecular hydrogen bonds broken, but three new intramolecular hydrogen bonds are formed in β-CD, i.e. one more intermolecular hydrogen bond is formed between the Sal-(S) and β-CD. For NMSal-(R) and β-CD inclusion complexes with cis orientation, ONIOM calculations indicate no intramolecular hydrogen bonds are broken while two new intramolecular hydrogen bonds are formed in β-CD. For the same inclusion complexes with trans orientation, ONIOM calculations imply one intramolecular hydrogen bond is broken and three new intramolecular hydrogen bonds are formed in β-CD. For the above three exceptional cases, calculations show that the broken intramolecular hydrogen bonds have been replaced by forming new intramolecular hydrogen bonds. As mentioned in one of our previous papers [52], the hydrogen-bonding attribution alone cannot explain the stabilization energies of the inclusion complexes.
Table 3.
Comparison of the number of hydrogen bonds in inclusion complexes with the number in β-CD.
| CD inclusion complex |
Orientation | Number of hydrogen bonds |
|
|---|---|---|---|
| β-CD | 3 | ||
| PM3 | |||
| Sal-(R) | cis | 3 | -1 + 1 (intra) |
| trans | 6 | -1 + 4 (intra) | |
| Sal-(S) | cis | 5 | -1 + 2 (intra) + 1 (inter) |
| trans | 6 | -0 + 3 (intra) | |
| NMSal-(R) | cis | 5 | -0 + 2 (intra) |
| trans | 5 | -1 + 3 (intra) | |
| NMSal-(S) | cis | 5 | -1 + 3 (intra) |
| trans | 6 | -1 + 3 (intra) + 1 (inter) | |
| BTIQ-(R) | cis | 3 | -1 + 1 (intra) |
| trans | 4 | -1 + 2 (intra) | |
| BTIQ-(S) | cis | 3 | -2 + 2 (intra) |
| trans | 4 | -0 + 1 (intra) | |
| ONIOM | |||
| Sal-(R) | cis | 3 | -1 + 1 (intra) |
| trans | 6 | -1 + 4 (intra) | |
| Sal-(S) | cis | 5 | -1 + 2 (intra) + 1 (inter) |
| trans | 5 | -2 + 3 (intra) + 1 (inter) | |
| NMSal-(R) | cis | 5 | -0 + 2 (intra) |
| trans | 5 | -1 + 3 (intra) | |
| NMSal-(S) | cis | 5 | -1 + 3 (intra) |
| trans | 6 | -1 + 2 (intra) + 2 (inter) | |
| BTIQ-(R) | cis | 4 | -1 + 2 (intra) |
| trans | 7 | -0 + 4 (intra) | |
| BTIQ-(S) | cis | 3 | -0 + 0 (intra) |
| trans | 7 | -0 + 4 (intra) | |
In conclusion, a theoretical study of inclusion complexes of β-CD with Sal, NMSal, and BTIQ enantiomers has been performed with PM3 semi-empirical molecular orbital calculations and ONIOM hybrid calculations. These two different sets of calculations show that the total stabilization energies for the inclusion complexes formed by (R)-isomers are more stable than those formed by (S)-isomers and the total stabilization energy differences for NMSal enantiomers are greater than those for Sal enantiomers, which correlates very well with the migration order observed in the capillary electrophoretic separation study. From the stabilization energies and hydrogen bonding studies of the inclusion complexes, we found that the principal driving forces for the formation of the inclusion complexes are from the van der Waals interactions.
Acknowledgements
The work was supported in part by National Institutes of Health grants (S06 GM08047 for M.-J. H. and NS44177 for Y.-M. L.), by the National Science Foundation (NSF) CREST grant (HRD0318519 for M.-J. H.), and by the NSF-EPSCoR grant (NSF440900-362427-02 for M.-J. H.). M.-J. H. thanks the Army High-Performance Computing Research Center (AHPCRC) for providing generous computing facilities. The content of this publication does not necessarily reflect the position or policy of the Government, and no official endorsement should be inferred. We thank Dr. John Watts for reading the manuscript.
References
- (1).VanEtten RL, Sebastain JF, Clowes GA, Bender ML. J. Am. Chem. Soc. 1967;89:3242. [Google Scholar]
- (2).Cramer F. Angew. Chem. 1956;68:115. [Google Scholar]
- (3).Nishijo J, Nagai M. J. Pharm. Sci. 1991;80:58. doi: 10.1002/jps.2600800115. [DOI] [PubMed] [Google Scholar]
- (4).Cohen J, Lach JL. J. Pharm. Sci. 1963;52:132. doi: 10.1002/jps.2600520206. [DOI] [PubMed] [Google Scholar]
- (5).Jones SP, Grant DJW, Hadgraft J, Parr GD. Acta Pharm. Technol. 1984;30:213. [Google Scholar]
- (6).Tong W-Q, Lach JL, Chin T-F, Guillory JK. Pharm. Res. 1991;8:951. doi: 10.1023/a:1015880218535. [DOI] [PubMed] [Google Scholar]
- (7).Tabushi I, Kiyosuke Y, Sugiomoto T, Yamamura K. J. Am. Chem. Soc. 1978;100:916. [Google Scholar]
- (8).Orstan A, Ross JBA. J. Phys. Chem. 1987;91:2735. [Google Scholar]
- (9).Aki H, Niiya T, Iwase Y, Yamamoto M. J. Pharm. Sci. 2001;90:1186–1197. doi: 10.1002/jps.1072. [DOI] [PubMed] [Google Scholar]
- (10).Connors KA. Chem. Rev. 1997;97:1325–1357. doi: 10.1021/cr960371r. [DOI] [PubMed] [Google Scholar]
- (11).Cramer F, Dietsche W. Chem. Ber. 1959;92:278–385. [Google Scholar]
- (12).Wan H, Blomberg LG. J. Chromatogr. A. 1995;704:179–193. [Google Scholar]
- (13).Pumera M, Felegel M, Lepša L, Jelinek I. Electrophoresis. 2002;23:2449–2456. doi: 10.1002/1522-2683(200208)23:15<2449::AID-ELPS2449>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- (14).Zbrožek J, Pumera M, Felegel M. J. Chrom. Sci. 2002;40:505–508. doi: 10.1093/chromsci/40.9.505. [DOI] [PubMed] [Google Scholar]
- (15).Pumera M, Jelinek I, Jindrich J. Fresenius J. Ana. Chem. 2001;369:666–669. doi: 10.1007/s002160100721. [DOI] [PubMed] [Google Scholar]
- (16).Pumera M, Jelinek I, Jindrich J. J. Chromatogr A. 2000;891:201–206. doi: 10.1016/s0021-9673(00)00628-2. [DOI] [PubMed] [Google Scholar]
- (17).Pumera M, Jelinek I, Jindrich J, Benada O. J. Liq. Chrom. Relat. Technol. 2002;25:2473–2484. [Google Scholar]
- (18).Kano K, Hasegawa H. J. Am. Chem. Soc. 2001;123:10616–10627. doi: 10.1021/ja0112644. [DOI] [PubMed] [Google Scholar]
- (19).Kano K. In: In Bioorganic Chemistry Frontiers. Dugas H, Schmidtchen FP, editors. Vol. 3. Spring-Verlag; Berlin, Heidelberg: 1993. pp. 1–23. [Google Scholar]
- (20).Kano K. J. Phys. Org. Chem. 1997;10:286–291. [Google Scholar]
- (21).Rekharsky M, Inoue Y. J. Am. Chem. Soc. 2000;122:4418–4435. [Google Scholar]
- (22).Heikkila RE, Hess A, Devoisin RC. Science. 1984;224:1451. doi: 10.1126/science.6610213. [DOI] [PubMed] [Google Scholar]
- (23).Abe K, Taguchi K, Wasai T, Ren J, Utsunomiya I, Shinohara T, Miyatake T, Sano T. Brain Res Bull. 2001;56(1):55–60. doi: 10.1016/s0361-9230(01)00603-7. [DOI] [PubMed] [Google Scholar]
- (24).Naoi M, Maruyama W, Akao Y, Yi H. Neurotoxicol Teratol. 2002;24(5):579–591. doi: 10.1016/s0892-0362(02)00211-8. [DOI] [PubMed] [Google Scholar]
- (25).Minami M, Maruyama W, Dostert P, Nagatsu T, Naoi M. J. Neural Transm. 1993;92:125. doi: 10.1007/BF01244872. [DOI] [PubMed] [Google Scholar]
- (26).Putscher I, Haber H, Winkler A, Fickel J, Melzig MF. Alcohol. 1995;12:447. doi: 10.1016/0741-8329(95)00028-p. [DOI] [PubMed] [Google Scholar]
- (27).Maruyama W, Naoi N, Kasamatsu T, Hashizume Y, Takahashi T, Kohda K, Dostert P. J. Neurochem. 1997;69:322. doi: 10.1046/j.1471-4159.1997.69010322.x. [DOI] [PubMed] [Google Scholar]
- (28).Muller Th., Baum SS, Haussermann D, Woitalla D, Rommelspacher H, Przuntek H, Kuhn WJ. Neural Transm. 1998;105:239–245. doi: 10.1007/s007020050052. [DOI] [PubMed] [Google Scholar]
- (29).Naoi M, Maruyama W, Dostert P, Kohda K, Kaiya T. Neurosci. Lett. 1996;103:183–190. doi: 10.1016/0304-3940(96)12807-x. [DOI] [PubMed] [Google Scholar]
- (30).Baum SS, Rommelspacher H. J. Chromatogr. 1994;660B:235–241. doi: 10.1016/0378-4347(94)00300-9. [DOI] [PubMed] [Google Scholar]
- (31).Deng Y, Maruyama W, Kawai M, Dostert P, Yamamura H, Takahashi T, Naoi M. J. Chromatogr. B Biomed. Appl. 1997;689:313–319. doi: 10.1016/s0378-4347(96)00359-3. [DOI] [PubMed] [Google Scholar]
- (32).McMurtrey K, Strawbridge C, McCoy JG. Enantiomers. 2000;5:377–383. [PubMed] [Google Scholar]
- (33).Sällström-Baum S. J. Chromatogr. B. 1994;660:235. [Google Scholar]
- (34).Deng Y, Zhang J, Tsuda T, Yu PH, Boulton AA, Cassidy RM. Anal. Chem. 1998;70:4586–4593. doi: 10.1021/ac980366i. [DOI] [PubMed] [Google Scholar]
- (35).Niwa T, Maruyama W, Nakahara D, Takeda N, Yoshimizu H, Tatematsu A, Takahashi A, Dostart P, Naoi M, Nagatsu T. J. Chromatogr. 1992;578:109–115. doi: 10.1016/0378-4347(92)80231-e. [DOI] [PubMed] [Google Scholar]
- (36).Haber H, Haber HM, Melzig MF. Anal. Biochem. 1995;224:256–262. doi: 10.1006/abio.1995.1038. [DOI] [PubMed] [Google Scholar]
- (37).Haber H, Henklein P, Georgi M, Melzig MF. J. Chromatogr. 1995;672B:179–187. doi: 10.1016/0378-4347(95)00213-3. [DOI] [PubMed] [Google Scholar]
- (38).Musshoff F, Schmidt P, Dettmeyer R, Priemer F, Jachau K, Madea B. Forensic Sci Int. 2000;113(1-3):359–366. doi: 10.1016/s0379-0738(00)00225-5. [DOI] [PubMed] [Google Scholar]
- (39).Liu Y-M, Gordon P, Green S, Sweedler J. Anal. Chim. Acta. 2000;420:81–88. [Google Scholar]
- (40).Quan Z, Song Y, Peters G, Shenwu M, Sheng YH, Hwang HM, Liu YM. Anal. Sci. 2005;21(2):115–119. doi: 10.2116/analsci.21.115. [DOI] [PubMed] [Google Scholar]
- (41).Carofiglio T, Fornasier R, Jicsinszky L, Saielli G, Tonellato U, Vetta R. Eur. J. Org. Chem. 2002;7:1191–1196. [Google Scholar]
- (42).Santos H. F. Dos, Duarte HA, Sinisterra RD, De Melo Mattos SV, De Oliveria LC, De Almeida WB. Chem. Phys. Lett. 2000;319:569–575. [Google Scholar]
- (43).Cao YJ, Xiao XH, Ji SF, Lu RH, Guo QX. Spectrochim Acta Part A. 2004;60:815–820. doi: 10.1016/S1386-1425(03)00306-8. [DOI] [PubMed] [Google Scholar]
- (44).Deng Y, Huang M-J. Int. J. Quant. Chem. 2004;100:746–752. [Google Scholar]
- (45).Dewar MJS, Zoebisch EG, Healy EF, Stewart JJP. J. Am. Chem. Soc. 1985;107:3902. [Google Scholar]
- (46).Stewart JJP. J. Comp. Chem. 1989;10:209. [Google Scholar]; Stewart JJP. J. Comp. Chem. 1989;10:221. [Google Scholar]
- (47).Brewster ME, Braunstein AJ, Bartruff MSM, Kibbey C, Huang M-J, Pop E, Bodor N. Supramol. Chem. 1994;4:69–76. [Google Scholar]
- (48).Bodor NS, Huang M-J, Watts JD. J. Pharm. Sci. 1995;84(3):330–336. doi: 10.1002/jps.2600840313. [DOI] [PubMed] [Google Scholar]
- (49).Bodor N, Huang M-J, Watts JD. Proceedings of the Eighth International Symposium on Cyclodextrins Symposium, Budapest (Hungary): Mar. 30 to Apr. 2, 1996.1996. pp. 209–214. [Google Scholar]; J. of Inclusion Phenomena and Molecular Recognition in Chemistry. 1996;25:97–102. [Google Scholar]
- (50).Huang M-J, Watts JD, Bodor N. Int. J. Quantum Chem. 1997;64:711–719. [Google Scholar]
- (51).Huang M-J, Watts JD, Bodor N. Int. J. Quantum Chem. 1997;65:1135–1152. [Google Scholar]
- (52).Huang M-J, Yi M. Int. J. Quantum Chem. 2004;100:771–778. [Google Scholar]
- (53).Pumera M, Rulišek L. J. Mod. Model. 2006;12:799–803. doi: 10.1007/s00894-005-0082-y. [DOI] [PubMed] [Google Scholar]
- (54).Murrell JN. J. Mol. Struct. (Theochem) 1998;424:93. [Google Scholar]
- (55).Boyd DB. J. Mol. Struct. (Theochem) 1997;401:219. [Google Scholar]
- (56).Casadesús R, Moreno M, González-Lafont À, Lluch JM, Repasky MP. J. Comput. Chem. 2004;25:99–105. doi: 10.1002/jcc.10371. [DOI] [PubMed] [Google Scholar]
- (57).Zhang Y-J, Merz KM., Jr. J. Comput. Chem. 1992;13:1151. [Google Scholar]
- (58).Maseras F, Morokuma K. J. Comp. Chem. 1995;16:1170. [Google Scholar]; Humbel S, Sieber S, Morokuma K. J. Chem. Phys. 1996;105:1959. [Google Scholar]; Matsubara T, Sieber S, Morokuma K. Int. J. Quant. Chem. 1996;60:1101. [Google Scholar]; Svensson M, Humbel S, Froese RDJ, Matsubara T, Sieber S, Morokuma K. J. Phys. Chem. 1996;100:19357. [Google Scholar]; Svensson M, Humbel S, Morokuma K. J. Chem. Phys. 1996;105:3654. [Google Scholar]; Dapprich S, Komáromi I, Byun KS, Morokuma K, Frisch MJ. J. Mol. Struct. (Theochem) 1999;462:1. [Google Scholar]; Vreven T, Morokuma K. J. Comp. Chem. 2000;21:1419. [Google Scholar]
- (59).Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Zakrzewski VG, Montgomery JA, Jr., Stratmann RE, Burant JC, Dapprich S, Millam JM, Daniels AD, Kudin KN, Strain MC, Farkas O, Tomasi J, Barone V, Cossi M, Cammi R, Mennucci B, Pomelli C, Adamo C, Clifford S, Ochterski J, Petersson GA, Ayala PY, Cui Q, Morokuma K, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Cioslowski J, Ortiz JV, Baboul AG, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Gomperts R, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Gonzalez C, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Andres JL, Gonzalez C, Head-Gordon M, Replogle ES, Pople JA. GAUSSIAN 98; Revisions A.6 and A.7, 1998Gaussian,Inc.Pittsburgh, PA Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Jr., Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian 03, Revision B.03, 2003Gaussian, Inc.Pittsburgh PA
- (60).Becke AD. Phys. Rev. 1988;A38:3098. doi: 10.1103/physreva.38.3098. [DOI] [PubMed] [Google Scholar]
- (61).Lee C, Yang W, Parr RG. Phys. Rev. B. 1988;37:785. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- (62).SPARTAN 5.2; 2001Wavefunction, Inc.Irvine, CA [Google Scholar]