Abstract
We have identified a novel family of proteins, in which the N-terminal Cystathionine Beta-Synthase (CBS) domain is fused to the C-terminal Zn ribbon domain. Four proteins were over-expressed in E. coli and purified: TA0289 from Thermoplasma acidophilum, TV1335 from Thermoplasma vulcanum, PF1953 from Pyrococcus furiosus, and PH0267 from Pyrococcus horikoshii. The purified proteins had red/purple color in solution and an absorption spectrum typical of rubredoxins. Metal analysis of purified proteins revealed the presence of several metals with iron and zinc being the most abundant metals (2 to 67% of iron and 12 to 74% of zinc). Crystal structures of both mercury- and iron-bound TA0289 (1.5–2.0 Å resolution) revealed a dimeric protein whose inter-subunit contacts are formed exclusively by the α helices of two CBS sub-domains, whereas the C-terminal domain has a classical Zn-ribbon planar architecture. All proteins were reversibly reduced by chemical reductants (ascorbate or dithionite) or by the general rubredoxin reductase NorW from E. coli in the presence of NADH. Reduced TA0289 was found to be able to transfer electrons to cytochrome C from horse heart. Likewise, the purified Zn ribbon protein KTI11 from Saccharomyces cerevisiae had purple color in solution and a rubredoxin-like absorption spectrum, contained both iron and zinc, and was reduced by the rubredoxin reductase NorW from E. coli. Thus, recombinant Zn ribbon domains from archaea and yeast demonstrate a rubredoxin-like electron carrier activity in vitro. We suggest that in vivo some Zn ribbon domains might also bind iron and therefore possess an electron carrier activity adding another physiological role to this large family of important proteins.
INTRODUCTION
The cystathionine-β-synthase (CBS) domain and Zn ribbon domain proteins comprise two large superfamilies of evolutionarily conserved proteins that are present in all kingdoms of life. CBS domains (IPR000644: 7,972 sequences; InterPro database) usually occur as tandem pairs (two or four copies) in stand-alone proteins or fused to different protein domains: cystathionine β-synthase, inosine-5′-monophosphate dehydrogenase (IMPDH), AMP-activated protein kinase (AMPK), and the chloride channels (ClC-0 to ClC-5) 1; 2. Their importance is underlined by the facts that mutations within CBS domains cause several hereditary diseases in humans 2. CBS domains represent a new class of binding module for adenosine derivatives (AMP, ADP, ATP, S-adenosyl methionine) and regulate the activity of the associated protein domain depending upon cellular energy status 2; 3; 4; 5. In addition to the regulatory role of CBS domains, they also seem to be involved in protein oligomerization or protein-protein interactions 6; 7; 8.
3D structures of several characterized CBS domain proteins in a ligand-free state from various organisms have been published 7; 9; 10. In the S. pyogenes IMPDH, two CBS domains form a minibarrel structure with a well-defined cleft between domains that may function as a potential binding site for regulatory molecules 9. Very recently, three structures of eukaryiotic CBS domains (from the human and Saccharomyces pombe AMPK, as well as from the human chloride channel ClC-5) complexed with ATP or AMP were determined 11; 12; 13. Although the ligands bind in equivalent pockets in these structures, their molecules remain surface-exposed and are coordinated by different protein residues. These structures also revealed that the backbone carbonyl and amido groups play an important role in ligand binding by these domains.
Zinc ribbon domains represent the largest fold group of the vast superfamily of zinc fingers 14. In these small protein domains, zinc plays a structural role stabilizing the domain structure through tetrahedral coordination by the side chains of conserved cysteine and histidine residues (2Cys+2His or 4 Cys motifs) 14; 15. Presently, zinc fingers comprise a large group of compact domains stabilized by a “zinc” ion and define one of the largest superfamilies in mammalian genomes 14; 16. These domains function as interaction modules for nucleic acids, proteins, and small molecules and they are present in proteins that perform a broad range of cellular functions, such as replication and repair, transcription and translation 14; 15.
Recent structure-based classification of zinc fingers identified eight fold groups with different main chain conformations and arrangement of secondary structure elements that contribute ligands for zinc 14. In the Zn ribbon fold group, the amino acids for zinc binding (usually four cysteines) are contributed by two zinc-knuckles, which are composed of two β-hairpins forming two structurally similar zinc-binding sub-sites 14. In Zn ribbons, the metal coordinating Cys residues are typically separated by two or four amino acid residues, but this number can vary from 0 to 15 14. The transcription elongation factor TFIIS and the transcription initiation factor TFIIB are typical representatives of this family, which mostly include domains from proteins involved in transcription or translation 14; 17; 18. This family also includes rubredoxins (Rd), small (45–55 amino acids) electron carriers which have red or purple color associated with the presence of a single iron atom, instead of zinc, coordinated in a tetrahedral geometry to four cysteinyl residues. Rds can exist as small monomeric solo proteins or they can be fused to other redox domains (like a bi-nuclear iron center in rubrerythrin, or FMN and di-iron domains in flavorubredoxin) 19; 20. These proteins are involved in oxygen/superoxide detoxification, iron metabolism, alkane hydroxylation, and Photosystem I assembly 21; 22; 23; 24.
The vast majority of CBS and Zn ribbon domain sequences in protein databases represent uncharacterized stand-alone proteins or domains fused to other protein domains. In this work, we report the biochemical and structural characterization of a previously unknown family of CBS domain proteins, which are fused to a C-terminal Zn ribbon-like domain (CBS-Zn ribbon-like proteins). The purified proteins (TA0289, TV1335, PH0267, and PF1953) have absorption spectra typical of Rds, contain both iron and zinc, and demonstrate an electron carrier activity in vitro. The crystal structure of TA0289 reveals that the C-terminal domain binds the metal atom (Hg2+ or Fe3+) with a coordination sphere similar to that of known Zn ribbon proteins whereas the N-terminal tandem CBS domains appear to be involved in protein dimerization. As well, we demonstrated the presence of an electron carrier activity in a known Zn ribbon protein, KTI11 from yeast, which was expressed and purified from E. coli cells grown with the addition of iron. We suggest that in vivo Zn ribbon domains might also bind Fe and exhibit an electron carrier activity.
RESULTS and DISCUSSION
Sequence analysis of the CBS domain proteins containing C-terminal extensions
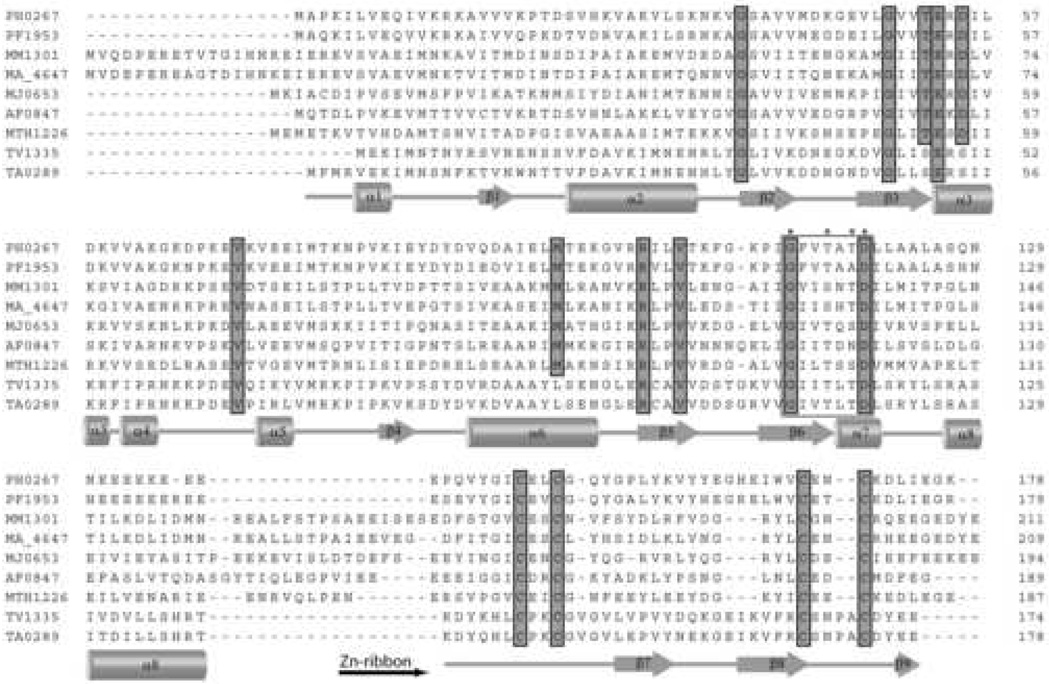
The examination of amino acid sequences of the stand-alone CBS domain proteins available in protein databases revealed the presence of two groups of these proteins with either shorter (130–140 residues) or longer (178–200 residues) amino acid sequences. Over 30 longer proteins have small (~50 amino acids) un-annotated C-terminal extensions containing four cysteine residues arranged in a Cys-X2-Cys-X14-19-Cys-X1-4-Cys motif (Figure 1). Similar cysteine motifs have been described in Zn ribbon domains and in rubredoxins 14; 15; 25.
Figure 1.
Multiple sequence alignment of CBS-Zn ribbon-like proteins from various organisms. Highly conserved residues are highlighted in gray. The secondary structure elements derived from the structure of TA0289 are shown below the sequence alignment. The beginning of the Zn ribbon-like domain is indicated by a horizontal black arrow below the alignment. The residues marked with asterisks (and boxed) represent the predicted ribose-phosphate binding site.
In the Cys motif of most CBS-Zn ribbon-like proteins, there are two amino acid residues separating the conserved cysteines (Figure 1). Two proteins, TA0289 from Thermoplasma acidophilum and TV1335 from Thermoplasma volcanium, have modified the second Cys motif with four amino acid residues between cysteines (Cys-X4-Cys) (Figure 1). This Cys motif was also found in Zn ribbon domains of DNA topoisomerase I and two Rds from Desulfovibrio 22; 26; 27. Another variant of the second Cys motif (Cys-X-Cys) was found in five uncharacterized archaeal proteins: ST2119 from Sulfolobus tokodaii, MsedDRAFT_1422 from Metallosphaera sedula, Saci_0044 and Saci_1167 from Sulfolobus acidocaldarius, and YBP3_ACIAM from Acidianus ambivalens. Using the sequences of TA0289, ST2119, and PF1953 as queries in BLAST searches, we identified over 30 CBS-Zn ribbon-like proteins in sequenced prokaryotic genomes, mostly in thermophilic and methanogenic archaea with just two sequences from thermophilic bacteria (Q3E1R1_CHLAU from Chloroflexus aurantiacus and Q2E2V6_ACICE from Acidothermus cellulolyticus). Most organisms contain one CBS-Zn ribbon-like protein with the exception of Sulfolobus acidocaldarius, which has two (Saci_0044 and Saci_1167; 46% sequence identity). CBS-Zn ribbon-like proteins share less than 30% sequence identity with the exception of TA0289 and TV1335 (82% sequence identity).
The operon prediction method of Yan and Moult, based on the combination of the gene neighbor and gene gap models 28, predicted no operon association of CBS-Zn ribbon-like genes with their neighbor genes in most genomes (TA0289, TV1335, PF1953, PH0267, ST2119, MJ0653, PYRAB02440). In several genomes, genes encoding CBS-Zn ribbon-like proteins are located next to another gene encoding a small CBS domain-like protein (A. fulgidus, Methanopyrus kandleri) or next to a series of four genes encoding small CBS domain-like proteins (MTH1226 from Methanobacterium thermoautotrophicum, MM1301 from Methanosarcina mazei, MA4647 from Methanosarcina acetivorans, Mbar_A0964 from Methanosarcina barkery). These uncharacterized CBS domain-like proteins share low sequence similarity (29% and less sequence identity) suggesting early gene duplication events and/or high rates of mutation/recombination of these genes. Thus, the genes encoding the CBS-Zn ribbon-like proteins are single gene cistrons or associated with unknown or hypothetical genes.
Protein purification and oligomeric state
To experimentally characterize the members of the CBS-Zn ribbon-like family, we cloned four proteins (TA0289, TV1335, PH0267, and PF1953), over-expressed them in E. coli and affinity purified them using a Ni2+-chelate affinity column. These proteins were expressed in soluble form, and their purification produced red- or purple colored proteins suggesting the presence of an iron cofactor. After elution from a Ni2+-column, the red/purple color of two proteins (TA0289 and PH0267) was unstable and quickly (PH0267) or slowly (TA0289) faded suggesting that the red color (probably iron cofactor) is sensitive to the high concentration of imidazole (0.3 M) used for elution. For TA0289, an alternative purification protocol without imidazole was therefore designed using cation exchange chromatography followed by gel-filtration that produced TA0289 preparations with a stable red color. The red/purple color of TV1335 and PF1953 was not sensitive to imidazole.
Analytical gel-filtration experiments with purified proteins demonstrated that in the native state TA0289 had a molecular mass of approximately 38 kDa, TV1335: 36.8 kDa, and PF1953: 62.8 kDa. Since the sequence-based molecular mass of these proteins was 20.3 kDa (TA0289), 19.7 kDa (TV1335), and 20.2 kDa (PF1953), these data indicate that TA0289 and TV1335 exist as a dimer in solution, whereas PF1953 appears to be a trimer.
Absorption spectra
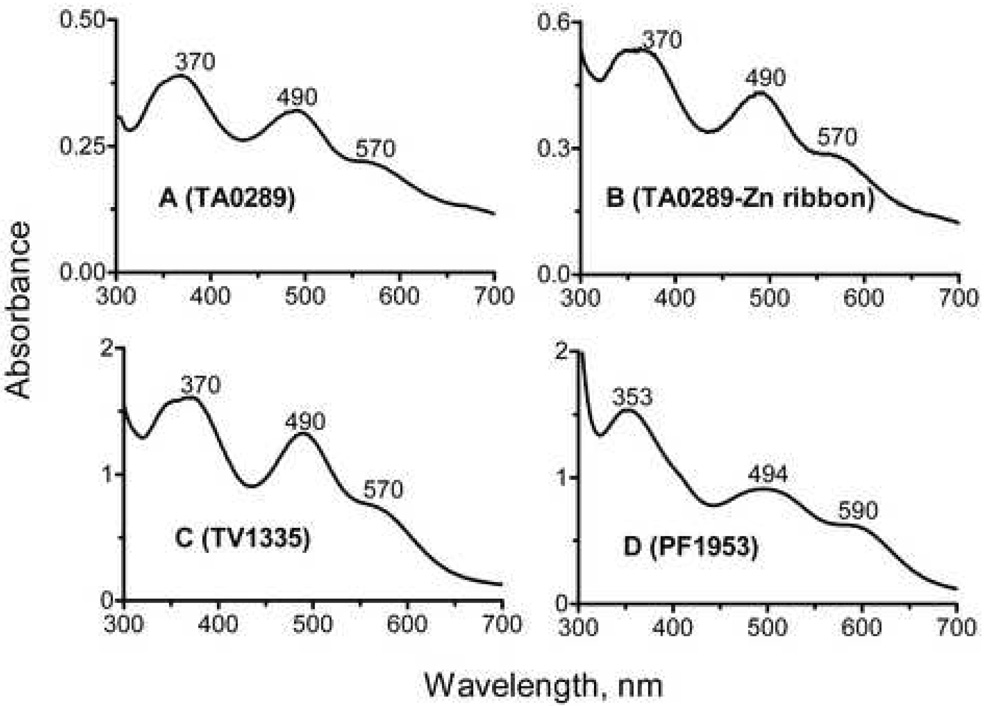
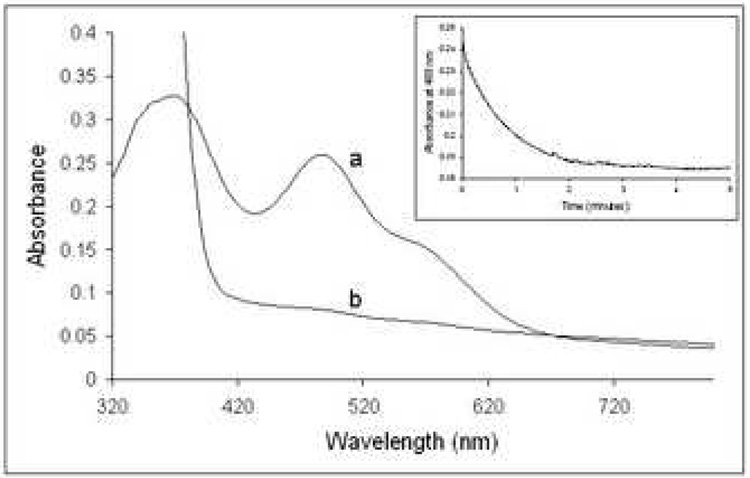
Purified TA0289, TV1335, and PF1953 have absorption spectra typical of Rds (Figure 2). TA0289 and TV1335 have almost identical absorption spectra with prominent peaks at 370 nm and 490 nm and shoulders at 350 nm and 570 nm (Figure 2A, 2C). The absorption spectrum of the individually expressed C-terminal domain of TA0289 (residues 138–178) showed a similar long-wavelength peak (490 nm) with a shoulder at 570 nm, but different short-wavelength peak with a maximum at 341 nm and a shoulder at 370 nm (Figure 2B). Biochemically characterized Rds from clostridia, Desulfovibrio and P. furiosus have similar absorption spectra with two major maxima at 370 nm (with or without a shoulder at 350 nm) and 490 nm and a broad shoulder at 570 nm 26; 29; 30. In the absorption spectrum of PF1953, the 353-nm peak was quite symmetric, and the second peak was shifted to 494 nm with a shoulder at 590 nm (Figure 2D).
Figure 2.
Absorption spectra of purified CBS-Zn ribbon-like proteins. (A), TA0289; (B), Zn ribbon domain of TA0289; (C), TV1335; (D), PF1953.
Metal content
Purified recombinant CBS-Zn ribbon-like proteins were found to contain several metals with Zn, Fe, and Ca being the most abundant metals (25 to 100% occupancy) (Table 1). As expected, the presence of a red/purple color in TA0289, TV1335, and PF1953 correlated with an increased content of Fe (Zn/Fe ratio from 0.25 to 4.1). Like for the P. furiosus Rd and rubrerythrin 31; 32, the iron content of recombinant TA0289 can be increased by growing cells on a minimal medium supplemented with 70 µM FeSO4 (Table 1). From the absorption spectrum of this sample of TA0289, an extinction coefficient for the iron center of this protein (A371 = 0.069) can be calculated to be 10,950 mM−1cm−1. This value is in close agreement with published values of 10,700 mM−1cm−1 and 10,800 mM−1cm−1 for Rds from P. furiosus and C. pasteurianum, respectively 29; 33. Previously, several recombinant Rds were purified as a mixture of Zn- and Fe-containing forms when expressed in E. coli 34; 35; 36. In fact, the metal binding sites of Rds appear to be able to accommodate a range of metals with little disruption to the overall structure of the protein 34; 37. The cadmium, mercury, nickel, zinc, and iron-containing forms of Rd have been investigated by a variety of methods 31; 38. It has been also shown that Zn2+ can directly displace Fe2+ from the C. pasteurianum Rd in vitro without resorting to protein denaturation 39. This displacement is consistent with the higher thiophilicity (affinity to the metal coordinating S atoms) of Zn2+ over Fe2+ 39. Interestingly, Zn-containing Rds have never been observed in the Rd preparations purified from the native microorganisms 32; 35. It is not clear why E. coli mainly inserts zinc rather than iron into the recombinant Rds. It has been suggested that in the native microorganisms most of the Zn2+ is sequestered away from Rd by a special mechanism, whereas Fe2+ is not 39.
Table 1.
Metal content of the recombinant CBS-Zn ribbon-like proteins and KTI11a
| Proteins and growth medium | Metal content (atoms/subunit) | ||||
|---|---|---|---|---|---|
| Fe | Zn | Ni | Cu | Ca | |
| TA0289 (TBb) | 0.18 | 0.74 | n.d.c | 0.09 | n.d. |
| TA0289 (M9b) | 0.67 | 0.17 | n.d. | n.d. | n.d. |
| PH0267 (TB) | 0.02 | 0.12 | 0.01 | 0.01 | 0.15 |
| PF1953 (TB) | 0.16 | 0.35 | 0.09 | 0.02 | 0.18 |
| TV1335 (TB) | 0.13 | 0.25 | 0.02 | 0.01 | 0.04 |
| KTI11 (wt)d (TB) | 0.17 | 0.40 | n.d. | 0.01 | 0.04 |
| KTI11 (wt) (M9+Fe2+) | 0.49 | 0.03 | n.d. | 0.01 | 0.09 |
| KTI11 (wt) (M9+Zn2+) | n.d. | 0.97 | n.d. | n.d. | 0.09 |
| KTI11 (C25S, C27S) (TB) | 0.01 | 0.09 | n.d. | 0.03 | n.d. |
| KTI11 (C47S, C50S) (TB) | 0.03 | 0.03 | 0.01 | n.d. | 0.06 |
| KTI11 (C25S, C27S, C47S, ΔC50) (TB) | 0.01 | 0.02 | 0.02 | n.d. | 0.09 |
Standard deviations for all determinations were 10.9 +/− 3.0%.
TB, terrific broth; M9, minimal medium M9.
n.d., not detected (below the detection limit).
wt, wild type protein.
Electron carrier activity
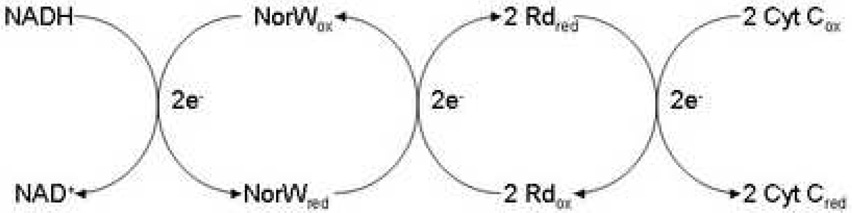
TA0289, TV1335, and PF1953 can be reduced by sodium dithionite (a midpoint redox potential −0.66 mV at pH 7) 40 and slowly reoxidized by air (Figure 3, only TA0289 is shown). These proteins, as well as the individually expressed Zn ribbon-like domain of TA0289 (residues 138–178) can be also reduced by the mild reductant ascorbate (−81 mV at pH 7.0) (data not shown) suggesting that their redox potential is close to that of typical Rds (−57 to +6 mV) 29; 41. To characterize the electron carrier activity of the CBS-Zn ribbon-like proteins we have designed a general Rd assay using the Rd-reductase NorW from E. coli and NADH as an electron donor and cytochrome C from horse heart as the terminal electron acceptor (Figure 4). E.coli NorW is a recently characterized non-specific NADH-dependent Rd-reductase, which is capable of mediating electron transfer from NADH to the E. coli flavo-rubredoxin NorV or to the Desulfovibrio gigas Rd 20; 42.
Figure 3.
Reduction of TA0289 by dithionite and reoxidation by air. 19 µM TA0289 in 50 mM Hepes-Na+ (pH 7.5), 0.2 M NaCl, and 10% glycerol (trace a) was scanned. A 50 mM stock of anaerobic sodium dithionite was then diluted 100-fold into the solution (to a final concentration of 500 µM), resulting in rapid reduction of TA0289 (trace b). After standing for several hours in air, a reoxidized spectrum (trace c) was obtained.
Figure 4.
In vitro electron carrier assay for Rds and Zn ribbon-like proteins: scheme showing the electron transfer from NADH to cytochrome C via the E. coli NorW and Rd (or other electron carrier).
Under anaerobic conditions and in the presence of NADH, E. coli NorW (1 µM) reduced TA0289 with a first order rate constant of 1.3 ± 0.1 min−1 (at 37°C) (Figure 5; Table 2). This rate was found to be almost independent of NADH concentration in the range of 100 – 300 µM NADH (data not shown). As expected, the individually expressed Zn ribbon-like domain of TA0289 (residues 138–178) was reduced by the E. coli NorW with a rate almost two times higher than the full-size TA0289 (Table 2). Four to six times higher reaction rates were observed for the reduction of TV1335 and PF1953 by NorW (Table 2). The determined rates of reduction of CBS-Zn ribbon-like proteins by the E. coli NorW (at 37°C) are a bit lower than those observed with the physiological electron acceptor NorV (40 min−1) or with the non-physiological mesophilic Rd from Desulfovibrio (20 min−1) 20; 42; 43. This might be explained by the thermophilic character of the studied proteins and/or by lower activity of the NorW Rd-reductase with CBS-Zn ribbon-like proteins. The rate constants for the reduction of small, single domain Rds by the physiological Rd-reductases were found to be in the range from 8 s−1 to 3.6 min−1 44; 45; 46.
Figure 5.
Anaerobic reduction of TA0289 by the E. coli NorW in the presence of NADH. A 24 µM anaerobic sample of TA0289 in a solution containing 50 mM Hepes-Na+ (pH 7.5), 0.2 M NaCl, 10% glycerol and 1 µM NorW was scanned (trace a). Reduction of TA0289 was achieved by the anaerobic addition of 200 µM NADH. The fully reduced spectrum is trace b. INSET: An absorbance trace at 488 nm following the time course for the reduction of 19 µM TA0289 in the presence of 200 µM NADH and 1 µM NorW in the same buffer, obtained from a single wavelength stopped flow experiment. The dotted line represents a fitted first order curve with a rate constant of 1.3 min−1.
Table 2.
First order rate constants of electron transfer reactions observed with CBS-Zn ribbon-like proteins and KTI11
| Protein and reaction | First order rate constants, min−1 |
|---|---|
| TA0289 / reduction by NorW | 1.5 ± 0.1 |
| TA0289 (Zn ribbon-like domain) / reduction by NorW | 2.47 ± 0.01 |
| TA0289 / reduction of cytochrome C | 1.9 ± 0.2 |
| TV1335 / reduction by NorW | 10.96 ± 0.02 |
| PF1953 / reduction by NorW | 11.0 ± 0.1 |
| KTI11 / reduction by NorW | 0.17 ± 0.01 |
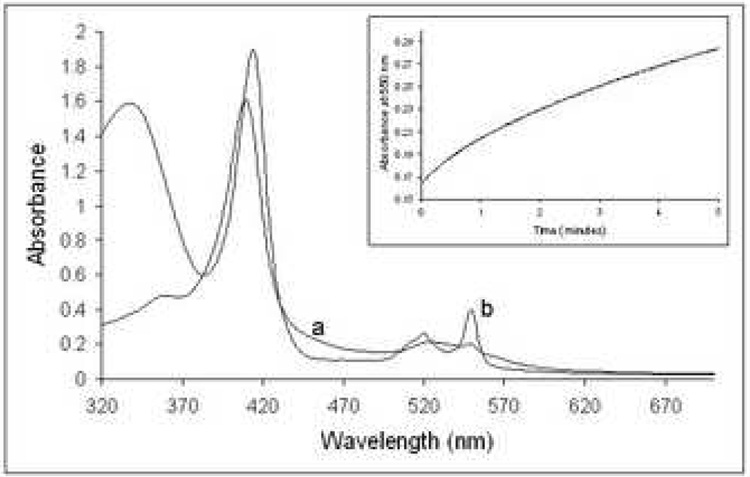
Cytochrome C from horse heart has been shown to be capable to function as a model electron acceptor for several Rds 47; 48; 49. The reported rates of the cytochrome C reduction by the reduced mesophilic Rds from C. pasteurianum 48 or P. oleovorans 47 were 30.7 s−1 and 54.7 s−1, respectively. Like known Rds, TA0289 reduced by the E. coli NorW in the presence of NADH, was able to transfer electrons to cytochrome C under anaerobic conditions (Figure 6). The initial first order reaction has a rate of 1.2 ± 0.3 min−1 at 37°C in the presence of 1 µM NorW, increasing to 1.5 ± 0.1 min−1 at 2 µM NorW, and corresponds to the pre-steady-state reduction of TA0289. When the reaction was carried out with 200 µM NADH, 1 µM NorW, and 1.9 µM TA0289, the steady-state turnover of cytochrome C was measured at 0.9 ± 0.1 min−1 and 1.0 ± 0.2 min−1 for the linear part of the reaction, in the presence of 100 and 200 µg ml−1 cytochrome C, respectively. This rate shows a linear dependence on the concentration of TA0289 (data not shown). Doubling the concentration of NorW to 2 µM at 6.3 µM Fe-TA0289 also results in an approximate doubling of the rate of reduction of cytochrome C to 1.9 ± 0.2 min−1. Therefore, electron transfer between NorW and TA0289 appears to be the rate limiting step of the entire electron transfer chain shown in Figure 4. Thus, our results demonstrate that CBS-Zn ribbon-like proteins are able to function as electron transfer proteins in the manner of Rds.
Figure 6.
Reduction of cytochrome C by TA0289 in the presence of NorW and NADH. A 200 mg ml−1 sample of cytochrome C in 50 mM Hepes-Na (pH 7.5), 0.2 M NaCl, and 10% glycerol in the presence of 1 µM NorW and 1.9 µM TA0289 was scanned (trace a). Reduction of cytochrome C was achieved by the anaerobic addition of 200 µM NADH. The fully reduced spectrum is trace b. INSET: An absorbance trace at 550 nm following the time course for the reduction of 200 µg ml−1 cytochrome C in the presence of 200 µM NADH, 1 µM NorW, and 1.9 µM TA0289 in the same buffer, obtained from a single wavelength stopped flow experiment.
In the T. acidophilum genome, we identified nine uncharacterized proteins (291–451 amino acids), which share 21–30% sequence identity with five experimentally characterized Rd-reductases (Swiss-Prot P37596, Q9AL95, P17052, P42454, Q8U1K9) 20; 46; 48; 50; 51 and might therefore represent the potential physiological reductases for TA0289. Seven genes (TA0837, TA1129, TA1341, TA1435, TA1413, TA1467, and TA0476) were cloned and over-expressed in E. coli. Four clones (TA0837, TA1435, TA1341, and TA1129) produced soluble proteins that were purified as 6His-fusions. Like known Rd-reductases, purified T. acidophilum proteins were yellow in solution and showed absorption spectra typical of a flavin-containing protein with major peaks at 363–369 nm and 447–457 nm and a shoulder at 468–482 nm (not shown). However, they were unable to reduce TA0289 and other CBS-Zn ribbon-like proteins under anaerobic conditions in vitro. Thus, further studies are required to identify the physiological reductase(s) for CBS-Zn ribbon-like proteins.
Crystal structure of TA0289
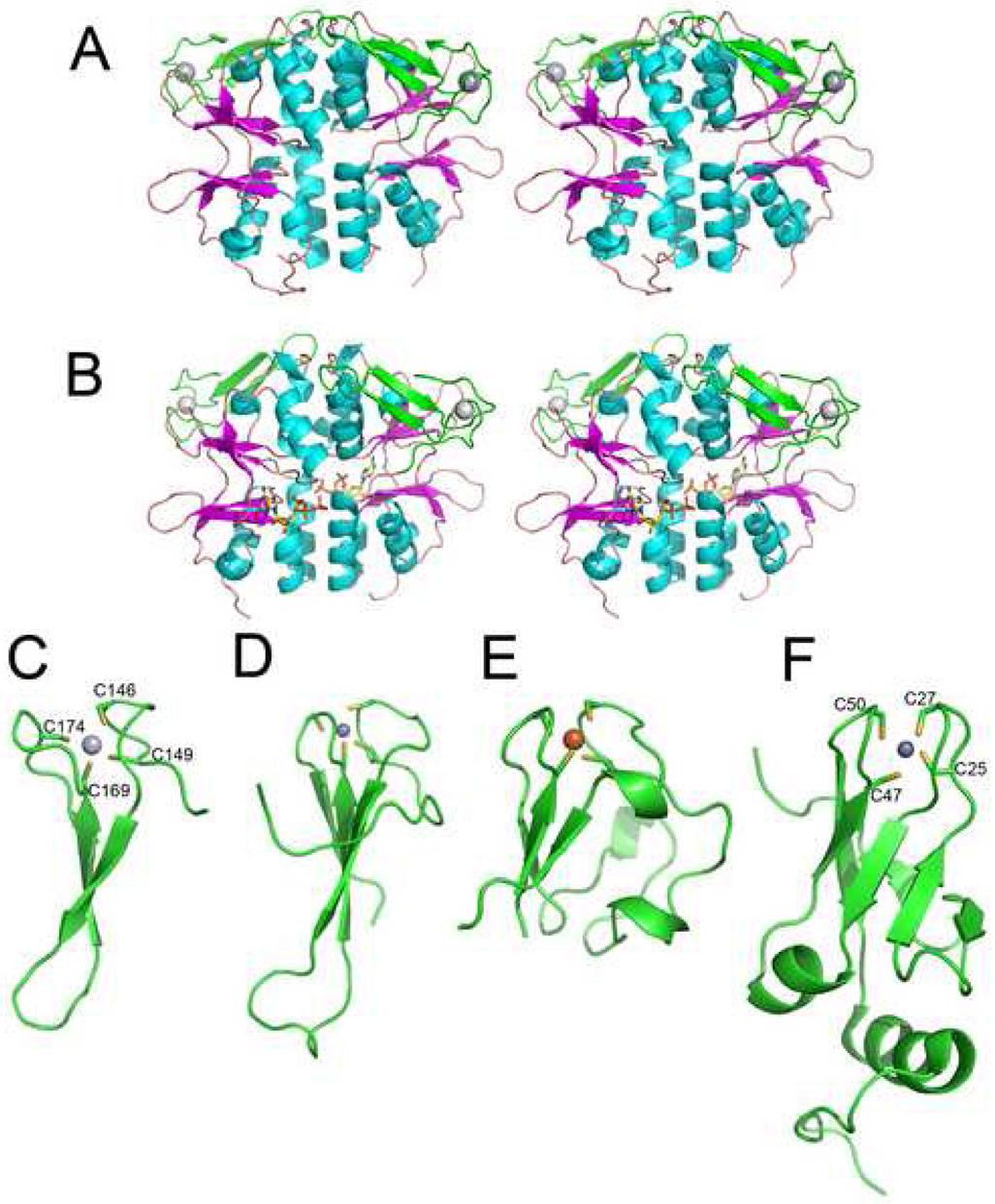
The crystal structure of TA0289 has been determined at a 1.5 Å resolution by the method of single-wavelength anomalous diffraction (SAD) phasing (PDB code 1pvm; Table 3). In agreement with our gel-filtration experiments (see above), the structure revealed that two subunits of TA0289 form a pyramid-shaped dimer with the CBS domains making tight contacts mainly through their α-helices (Figure 7A). In crystal structures of mammalian and bacterial IMP dehydrogenases, the CBS domains are located outside of the protein tetramer, and the intersubunit contacts are made entirely by the catalytic (dehydrogenase) domains 3; 9. However, in most available structures of CBS domain proteins (PDB codes 1vr9, 1o50, 1yav, 1y5h) the subunit-subunit interface is formed entirely by the α-helices of the CBS domains 52. In the TA0289 structure, the extended dimer interface (1,797–1,806 Å) involves at least 30 residues of the helices α2, α3, α4, α7, and α8 of each monomer (Figure 7A, 8). The interface contains both charged and hydrophobic residues and involves 19 hydrogen bonds. Thus, the CBS domains appear to be solely responsible for the dimerization of TA0289.
Table 3.
Summary of X-ray data collection and refinement statistics
| Data collection | Ta0289 (Hg2+ form, 1pvm) | Ta0289 (Fe2+ form, 2qh1) |
|---|---|---|
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | a=58.74, b=62.84, c=97.28 | a= 58.81, b=63.03, c=97.47 |
| α, β, γ (°) | α=β=γ=90° | α=β=γ=90° |
| Wavelength | 1.0081 | 1.54178 |
| Resolution (Å) | 50 - 1.50 | 50 – 2.0 |
| Rsyma | 0.056 (0.271) | 0.081 (0.340) |
| I/σI | 44.1 (5.0) | 52.7 (13.6) |
| Completeness (%) | 97.0 | 98.8 (97.4) |
| Redundancy | 10.5 | 10.1 |
| Overall figure of merit for SAD fasing at 1.5 Å resolution | ||
| Acentric | 0.282 | |
| Centric | 0 | |
| Overall | 0.270 | |
| Refinement | ||
| Resolution (Å) | 50 – 1.5 | 20-2.0 |
| Number of reflections | 105550 | 25113 |
| Rwork / Rfreeb,c | 19.4/22.8 | 20.4/23.7 |
| Number of atoms | ||
| Protein | 2887 | 2902 |
| Major ligand | 2 (Hg2+) | 2 (Fe2+) |
| Solvent | 501 | 345 |
| B-factors | 17.3 | 27.2 |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.005 | 0.006 |
| Bond angles (°) | 1.3 | 1.3 |
Values in parentheses are for the highest-resolution shell.
Rsym = Σ|I - <I>|/Σ I
Rwork = 100 X Σ |Fo – Fc| / Σ Fo, where Fo and Fc are defined as the observed and calculated structure factors, respectively.
Rfree is calculated as defined by Rwork using test set reflections, representing 5% of the total.
Figure 7.
Structures of TA0289 and other Zn ribbon domain proteins. (A), overall structure of the TA0289 dimer (stereo view). CBS domains are shown in orange (β-strands) and cyan (α-helices), Zn ribbon domains are shown in green, and metal atoms are shown as grey spheres. (B), stereo view of the TA0289 dimer shown with docked ATP from in silico analysis. (C), structure of the TA0289 Zn ribbon-like domain showing the tetrahedral coordination of the metal atom (Hg2+) by conserved cysteines (labeled). (D), structure of the Zn ribbon domain of the human transcriptional elongation factor TFIIS (1tfi). (E), structure of the P. furiosus rubredoxin PF1282 (1brf). (F), structure of KTI11 (1yop, 1yws). Metals are shown as: dark gray spheres (zinc; D and F), light gray sphere (mercury; C), and orange sphere (iron; E).
Figure 8.
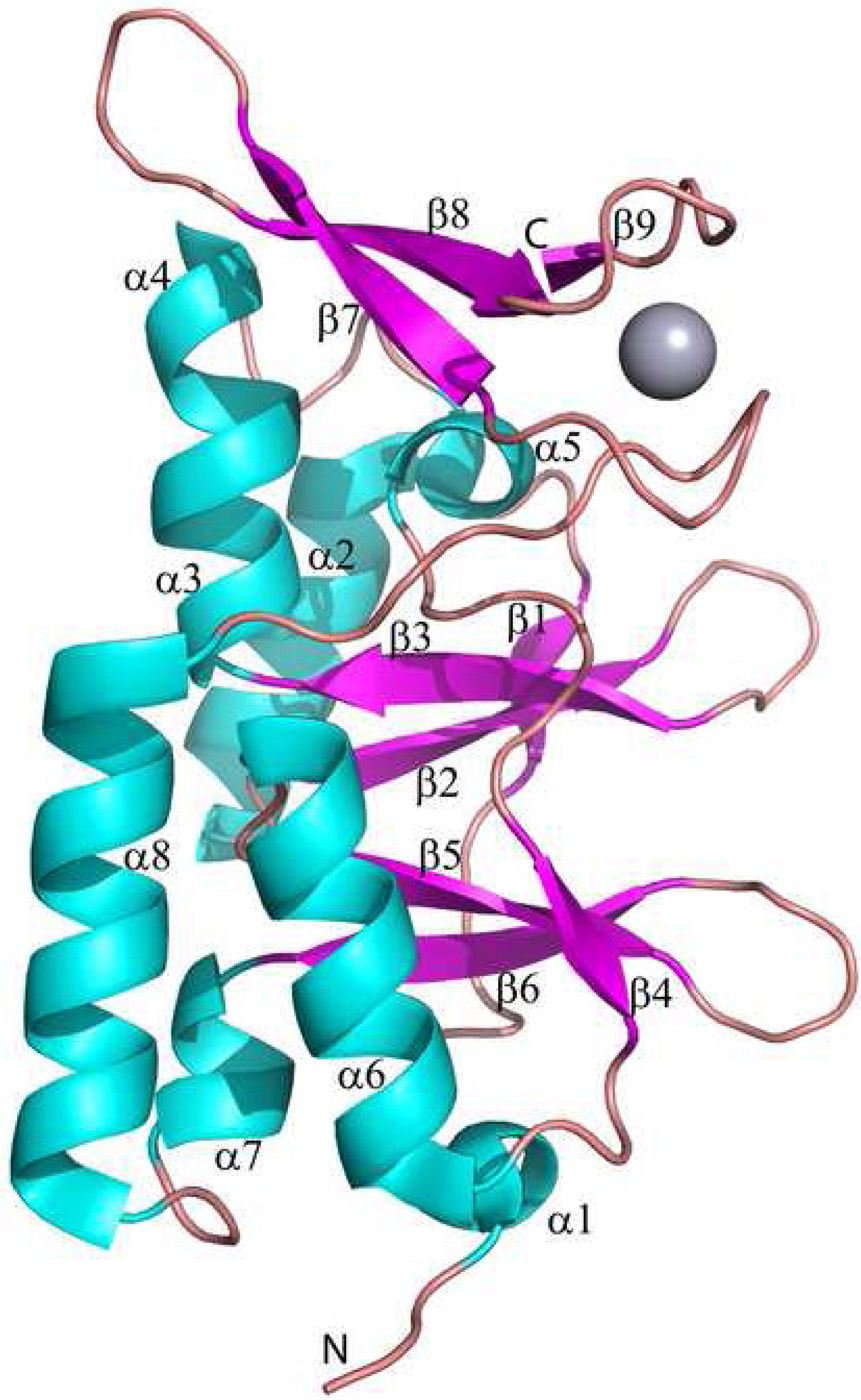
Structure of the TA0289 subunit. The secondary structure elements are numbered and shown in different colors (α-helices – cyan, β-strands – magenta, and loops – salmon). The position of the bound metal (Hg2+) is shown as a gray sphere.
Structure of the CBS domain
The TA0289 CBS domain comprises two sub-domains with similar structure and sequence: the CBS-1 sub-domain (residues 1–64; α1, β1, α2, β2, β3, α3, and α4) and CBS-2 sub-domain (residues 65–126; α5, β4, α6, β5, β6, and α7) (Figure 8). The CBS-2 sub-domain is connected to the Zn ribbon-like domain by the long α8 helix and a flexible loop (Figure 8). Both CBS sub-domains have hydrophobic cores formed exclusively by aliphatic residues: Val16, Ala25, Val39, Val47, Leu50, Ile55, and Ile71 (for CBS-1 sub-domain), and Val5, Val82, Val91, Leu95, Val105, Val106, Val116, Leu118, Leu121, and Ile133 (for CBS-2 sub-domain). CBS domains from various proteins and organisms show little sequence conservation but have been structurally conserved. A DALI search for structural homologues of TA0289 identified several uncharacterized CBS domain proteins: MTH1622 (1pbj, Z-score 7.6, rmsd 1.76 Å), Rv2626c from Mycobacterium tuberculosis (1xkf and 1y5h, Z-score 6.5 – 6.8, rmsd 1.77 – 1.84 Å), TM0935 (1o50, Z-score 6.4, rmsd 1.89 Å), and TM0892 (1vr9, Z-score 5.9, rmsd 2.23 Å).
In the TA0289 monomer, there is a deep cleft between CBS-1 and CBS-2 sub-domains formed by two β-sheets and occupied by both hydrophobic and charged residues (Figure 7A). In three recent structures of the eukaryotic CBS domains complexed with ATP or AMP 11; 12; 13, this cleft binds the ligand with the adenine ring on the bottom sandwiched between the hydrophobic side chains of four non-conserved residues (Ile, Val, Pro, Phe, or Tyr). These structures suggested that four semi-conserved residues (Gly-h-x-Ser/Thr-x-Ser/Thr-Asp) might represent a novel recognition motif for the ribose-phosphate moiety 12. This motif is also recognizable in the sequence of CBS-Rd-like proteins (Figure 1), and in the TA0289 structure these residues (Gly114, Thr117, Thr119, and Asp120) are located in or close to the deep cleft (β3-α3 and β6-α7) (Figure 8) suggesting that CBS domains in these proteins might also be involved in AMP/ATP binding. In silico docking experiments with the structure of TA0289 (Figure 7B) demonstrated that two ATP molecules can be docked to the TA0289 dimer in a binding mode similar to that in the human ClC-5:ATP complex (2j9l). However, our thermal shift experiments with circular dichroism as a detection method produced no evidence for the binding of ATP, ADP, AMP or their cytidine, guanosine and thymidine analogs to TA0289 (data not shown). In addition, TA0289 crystallized quite easily in the presence of adenosine, AMP, ADP, or ATP, but the analysis of electron density maps of these crystals revealed no ligand bound to the protein.
Three 3D structures of prokaryotic CBS domain proteins revealed the presence of a bound ligand. The structure of MTH1622 (1pbj) showed a Mg2+ ion coordinated by Glu72 located on the side opposite the long cavity. However, in the TA0289 structure the access to the homologous Glu68 is blocked by the Zn ribbon-like domain. The structure of the hypothetical protein Rv2626c from Mycobacterium tuberculosus (1xkf) revealed the presence of two bound Zn2+ ions, one of which is coordinated by the His122 located in the long cavity. In TA0289, this residue is replaced by the conserved Asp120 (Figure 1). In the structure of YkuL from Bacillus subtilis (1yav), the SO42− anion is bound to the long cavity and is coordinated by Arg133, which occupies a position similar to that of Arg102 in TA0289. However, it is not clear presently if these ligand-CBS domain complexes of prokaryotic proteins are physiologically relevant.
Structure of the C-terminal domain
The C-terminal domain of TA0289 is formed by the final 36 amino acids (Gln143-Glu178). Three short β-strands (β7, β8, and β9) form a compact anti-parallel β-sheet that packs against the CBS domain (Figure 7A). The overall structure of the TA0289 C-terminal domain (Figure 7C) is more similar to the planar architecture of classical Zn-ribbon domains (1tfi, 1pft) (Figure 7D) and Rd-like domains of rubrerythrins (1yux), whereas typical stand-alone Rds (1brf) have also a globular hydrophobic core (Figure 7E). To date, more than 50 Rd structures have been deposited in the PDB with several of them having a resolution better than 1.0 Å (e.g. 1brf, 2dsx, 1rb9, 1yk4). The best studied Rd, PF1282 from P. furiosus, has been characterized using several forms of this protein containing different metals: cadmium, mercury, nickel, zinc, and iron 31; 38; 53; 54. The first structure of TA0289 (1pvm) was solved using mercury-soaked crystals obtained with the colorless (iron-free) form of the protein. In this structure, the sulfur atoms of the four cysteine residues bind the mercury atom with nearly equal bond lengths, creating an almost tetrahedral coordination sphere with an average Hg-S distance of 2.55 ± 0.04 Å and an S-S distance of 4.18 ± 0.2 Å (Figure 7C). As in Rds, two Cys residues of the TA0289 Zn ribbon-like domain (Cys149 and Cys174) are located close to the domain surface, whereas the other two cysteines (Cys146 and Cys169) are buried in the domain core.
The crystals of the Fe-containing (red form) TA0289 (2qh1) produced very similar structure with the Fe center located at the periphery of the molecule, but with different metal-S distances: from 2.02 Å to 3.0 Å with an average of 2.41 ± 0.43 Å (not shown). Since the Fe atom (1.17Å) has a smaller radius than the Hg atom (1.49Å), the Fe-containing TA0289 possesses a more compact Fe center (an average S-S distance of 3.95 ± 0.15 Å), which is close to the size of the Fe center of the P. furiosus Rd (3.81 ± 0.014 Å; 1brf). The TA0289 Fe center is stabilized by an interaction between the side chains of Lys148 and Asp175 (2.67 Å), as well as by H-bonds between the side chain of Ser170 and the main chain NH-group of Val154 (3.01 Å) and between the Lys148 side chain and the main chain carbonyl oxygen of Asp175 (2.94 Å). Structural studies of mesophilic and thermophilic Rds identified three main components essential for the integrity of the metalloprotein structure: the hydrogen bond network, the hydrophobic protein core, and the electrostatic interactions between surface residues 55; 56. Like typical Zn ribbon domains (1tfi, 1pft), the C-terminal domain of TA0289 has no hydrophobic core and is stabilized mainly by the intra-domain H-bonds between its β7 and β8 strands, as well as by the electrostatic interactions between the side chains of Glu163/Lys161 (3.5 Å) and Arg168/Glu177 (3.1 Å). Additional stabilization of the TA0289 Zn ribbon-like domain is achieved through the interaction of its residues with the CBS domain: Glu163/Asn63 (2.6 Å), Glu178/Lys77 (3.2 Å), as well as by H-bonds between the main chain carbonyl oxygens of Pro61 and Glu178 with the NH group of Ile164 and the Arg62 side chain, respectively.
In redox proteins, electrons can travel up to 14 Å between redox centers through the protein medium, whereas the transfer over longer distances always involves a chain of redox cofactors 57. In the Desulfovibrio gigas desulforedoxin, a small homo-dimeric (2×36 amino acids) Rd-like protein, the two Fe atoms are 16 Å apart suggesting rapid electron exchange between the Fe centers in the half-reduced species 58. However, the two Fe atoms within the TA0289 dimer are 53 Å apart suggesting that no electron exchange takes place between the Fe centers in the half-reduced species and that these Fe atoms are likely to function as independent redox centers.
Experiments with the Zn ribbon protein KTI11 from yeast
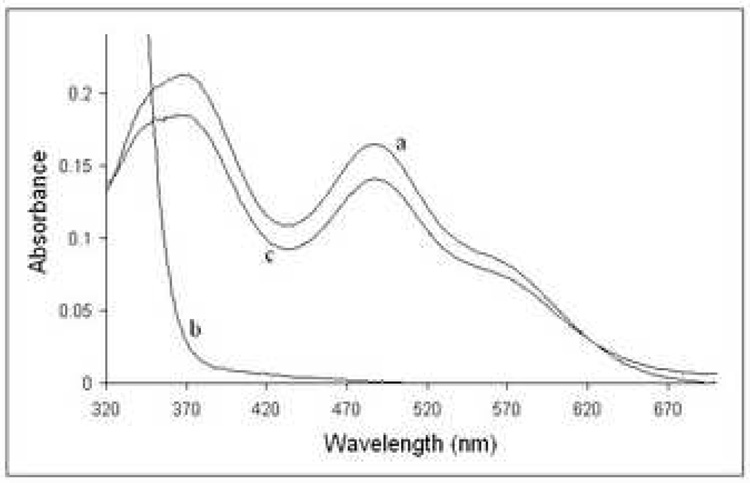
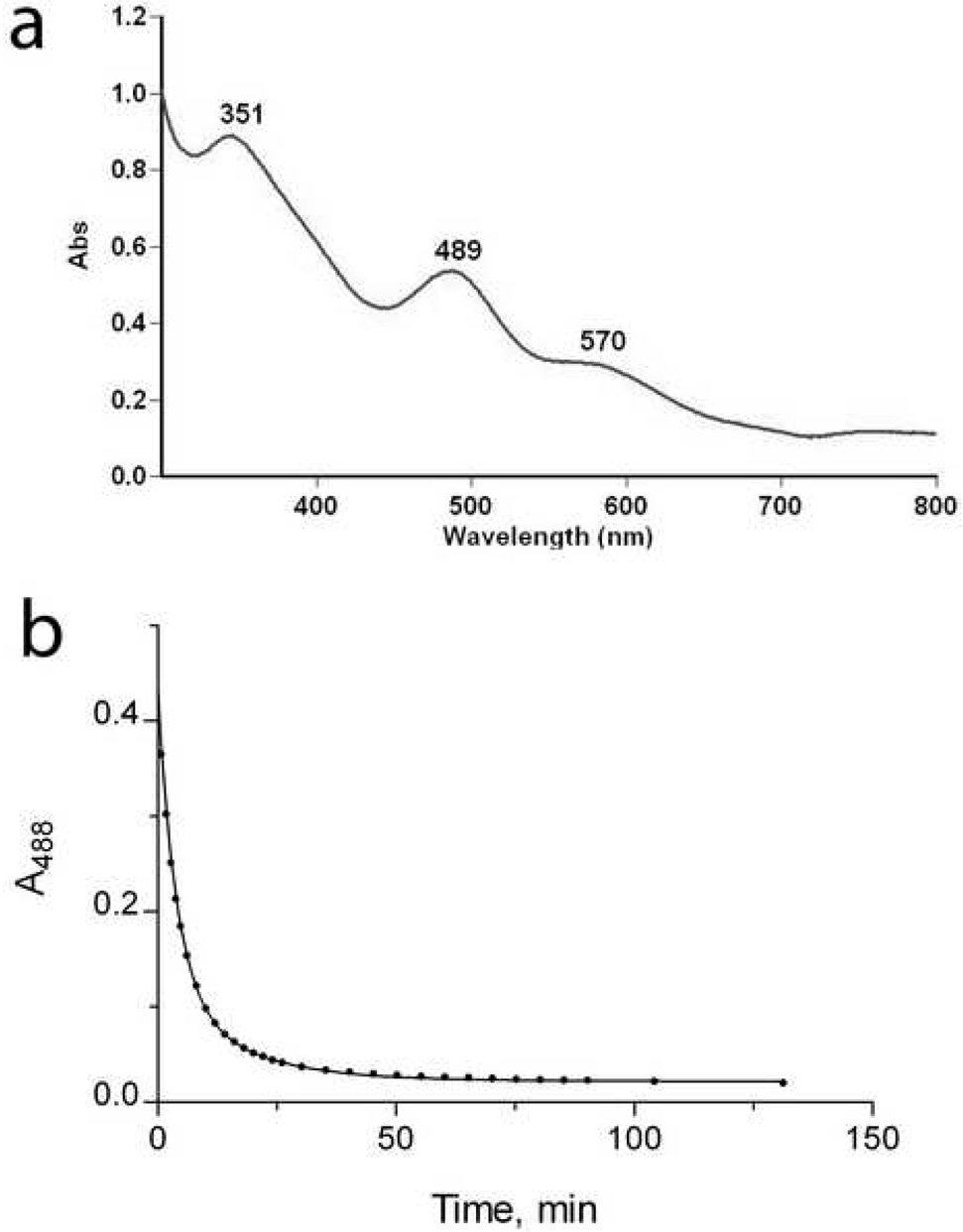
To produce additional evidence that Zn ribbon proteins can bind iron and exhibit an electron carrier activity, we over-expressed in E. coli and purified the recently characterized Zn ribbon protein KTI11 from yeast. This protein (249 residues) contains one Cys-X-Cys…Cys-X2-Cys motif and is involved in the biosynthesis of diphthamide on the translation elongation factor eEF-2 59. Recently, another laboratory (1yop) 59 and ours (1yws) solved structures of KTI11 purified from E. coli cells grown on the minimal medium M9. These structures revealed the presence of a typical Zn ribbon fold with one Zn atom tetrahedrically coordinated by four cysteines of the Cys motif (Figure 7F). However, purification experiments produced the purple colored KTI11 when they were performed using E. coli cells grown on rich medium (Terrific Broth). The purple colored preparations of KTI11 had a classical Rd-like absorption spectrum with two major peaks at 351 nm and 489 nm and a shoulder at 570 nm (Figure 9A). Metal analysis of these preparations demonstrated that 17% of protein molecules contained Fe and 40% - Zn (Table 1). E. coli cells grown on the minimal medium supplemented with Fe2+ (70 µM) also produced the purple-colored KTI11 containing mainly Fe (49%) and little Zn (3%), whereas cells grown on the Zn2+-supplemented M9 medium produced only colorless, Zn-containing KTI11 (Table 1). Therefore, like in Rds, the metal content of the recombinant Zn ribbon protein KTI11 depends on the presence of Fe in the growth medium.
Figure 9.
Biochemical characterization of KTI11. (a), absorption spectrum of KTI11 purified from iron-supplemented cells. (b), anaerobic reduction of KTI11 (180 µM) by the E. coli NorW (20 nM) in the presence of NADH (0.2 mM). An absorbance trace at 488 nm following the time course for the reduction KTI11.
In Rds, amino acid replacement mutagenesis of a single metal-coordinating cysteine produced iron-containing (colored) proteins, although with changed redox properties 60; 61. When two cysteines of KTI11 were simultaneously changed to serines (C25S/C27S and C47S/C50S), the proteins were still expressed in colored form, but they quickly lost the color upon elution from the Ni2+-chelate column by 0.25 M imidazole. We also designed a KTI11 protein in which all four metal-coordinating cysteines were replaced by serine (C25, C27, and C47) or deleted (C50). This protein was expressed in colorless form indicating that it completely lost the ability to bind metal. This was confirmed by metal analysis of the purified mutant KTI11 proteins which demonstrated 5 – 20 times lower content of both Fe and Zn (Table 1) indicating that these cysteines coordinate both Fe and Zn.
Like CBS-Zn ribbon-like proteins, the Fe-containing KTI11 is redox active and can be reversibly reduced by the Rd-reductase NorW in the presence of NADH and transfer electrons to cytochrome C from horse heart (Figure 9B). Compared to TA0289, the rates of KTI11 reduction by NorW (Table 2) and cytochrome C reduction by the reduced KTI11 (not shown) were somewhat lower. The redox potential of the Fe-containing KTI11 was determined using xanthine oxidase and indigo carmine (as an internal reference) and was found to be 6 mV, which is close to the redox potential of Rds (−57 mV – 6 mV) 29; 41; 62. Thus, the recombinantly expressed Zn ribbon protein KTI11 incorporates both Fe and Zn into its molecules and demonstrates an electron carrier activity in vitro.
Conclusion
In this work, we have structurally and biochemically characterized several members of a novel family of CBS-domain proteins, which are fused to a Zn ribbon-like domain. We demonstrated that the Zn ribbon-like domain of these proteins, as well as the Zn ribbon protein KTI11 from yeast, can also bind iron and function in the manner of a rubredoxin in vitro. In addition, the recent work with the anaerobic class III ribonucleotide reductase revealed the presence of both Fe and Zn in the crystals of its Zn ribbon domain raising the question whether the true active center contains Fe or Zn 63. Although Zn ribbon domains are generally believed to bind zinc in vivo, this opinion is based on the in vitro Zn2+ binding experiments with purified proteins 64; 65. These experiments also demonstrated that Co2+, Cu2+, and Cd2+ (but not Mg2+ or Mn2+) compete with Zn2+ for binding to Zn ribbon proteins, however the effect of Fe2+ was not tested 64; 65. Nevertheless, these results revealed that in vitro Zn ribbons are not strictly specific to zinc and can also incorporate other metal ions. Since Zn ribbon domains are structurally similar to Rds, which can incorporate both Fe and Zn, we can suggest that some Zn ribbons might be also capable of coordinating Fe in vivo. In this case, the Fe center would preserve a structural role and add the additional, electron transfer function to these domains. Given the presence of Zn ribbon domains in a broad range of important enzymes and their ability to interact with nucleic acids and proteins, the presence of electron transfer activity in some Zn ribbons would bring another level of complexity to the processes they are involved in. Future experiments should be focused on the identification of the physiological metal ion in Zn ribbon domains and their functions.
MATERIALS and METHODS
Protein Expression and Purification
Cloning of the genes encoding CBS-Zn ribbon-like proteins (TA0289, TV1335, PF1953, and PH0267), the Zn ribbon domain of TA0289 (residues 138–178), potential Rd reductases (TA0837, TA1129, TA1341, TA1435, TA1413, TA1467, and TA0476), the E. coli NorW, and the yeast KTI11 into the modified pET15b vector was carried out as previously described 66. These proteins were expressed as a fusion with an N-terminal 6His-tag in Escherichia coli strain BL21(DE3) cells upon induction with 0.2 mM isopropyl-β-D-thiogalactopyranoside (IPTG). Cultures were either grown in the rich medium Terrific Broth, or in a minimal medium based on the method of Neidhardt et. al. with the zinc salt omitted from the culture 67. Cells were grown in Terrific Broth at 37 °C up to an OD600 of approximately 1.0. The temperature was then lowered to 16 °C and the expression of cloned proteins was induced with 0.2 mM IPTG. After induction the cells were grown overnight at 16 °C prior to harvesting by centrifugation. The cell pellets were flash frozen in liquid N2 and stored at −20 °C. For manipulation of the metal content of recombinant proteins, a minimal medium 67 was used for the growth of BL21 (DE3) cultures, which was supplemented with either 70 µM FeSO4 or 70 µM ZnCl2 immediately prior to induction of expression with 0.2 mM IPTG. Cultures were then grown overnight at 30 °C overnight prior to harvesting by centrifugation.
Purification of His6-tagged proteins was carried out using metal-chelate affinity chromatography on nickel affinity resin (Qiagen) as previously described 66. Frozen cell pellets were thawed and resuspended in binding buffer: 0.5 M NaCl, 5 % glycerol, 50 mM Na+Hepes (pH 7.5), 5 mM imidazole, and 0.1 mM phenylmethylsulfonyl fluoride (PMSF). The cells were lysed by sonication and the lysate was clarified by centrifugation (78,000 g for 30 minutes) prior to being applied to a metal chelate affinity column charged with Ni2+ which had been pre-equilibrated with binding buffer. Bound protein was washed with wash buffer: 0.5 M NaCl, 5 % glycerol, 50 mM Na+Hepes (pH 7.5), 30 mM imidazole, and then eluted with elution buffer:0.5 M NaCl, 5 % glycerol, 50 mM Na+Hepes (pH 7.5), and 250 mM imidazole.
Since TA0289 released Fe in the presence of the high imidazole concentrations (0.25 M) used for the elution of proteins bound to the nickel resin, an alternative purification protocol was developed for this protein, which was based on anion exchange chromatography on SP-Sepharose (Amersham Biosciences). Bacteria were thawed and resuspended in 50 mM MES-Na (pH 6.5) containing 0.1 mM PMSF. Approximately 30 g cells were lysed by sonication and the cell debris was pelleted by centrifugation at 78,000 g for 45 minutes. The supernatant was applied to a 12 cm × 2.6 cm SP Sepharose Fastflow column (Amersham Biosciences) which had been previously equilibrated with 50 mM MES-Na (pH 6.5), and then washed with five column volumes of the same buffer. The protein was then eluted from the column with a linear gradient of 0–1 M NaCl in the same buffer. The partially purified protein was concentrated to 4 ml, clarified by centrifugation at 78,000 g for 30 minutes, filtered through a 0.22 micron filter, and applied to a Superdex 200 26/60 column (Amersham Biosciences), which had previously been equilibrated with 50 mM Hepes-Na (pH 7.5) and 200 mM NaCl, at 1 ml min−1 for purification by gel filtration. The resulting protein was at least 95 % pure as indicated by SDS PAGE. The purified protein was flash frozen in liquid N2 and stored at −80°C prior to use.
Analytical gel filtration
The molecular weight of the CBS Zn-ribbon-like proteins in solution was estimated using a Superdex 75 10/300 GL gel filtration column (Amersham Biosciences) equilibrated with 50 mM Na+-Hepes (pH 7.5), 0.2M NaCl, and 0.5 mM Tris-2-carboxyethylphosphine-HCl (TCEP), attached to an AKTA FPLC system (Amersham Biosciences). 0.3 mg of protein in a volume of 100 µl was loaded onto the column and run at a flow rate of 0.5 ml min−1. A calibration run was carried out with the following standards: ribonuclease A (13,700 Da), chymotrypsinogen (25,000 Da), ovalbumin (43,000 Da), albumin (67,000 Da) and blue dextran (2,000,000 Da) in the same buffer system.
Metal analysis
The metal content of the purified and dialyzed proteins was determined using inductively coupled plasma-atomic emission spectrometry (ICP-AES) at the Department of Chemistry of the University of Toronto as described previously 68. Metal analysis was performed using proteins containing a His6-tag, since previous control experiments demonstrated no effect of the His6-tag on the protein’s metal content 68.
Anaerobic reduction of Zn ribbon proteins and cytochrome C
The reduction of the Zn ribbon proteins by E. coli NorW, and the reduction of cytochrome c (Sigma, from horse heart) by Zn ribbons and E. coli NorW in the presence of NADH as an electron donor, were performed under anaerobic conditions. Glycerol (10% v/v) was included in all buffers in as this was found to increase the stability of proteins in solution. For the reduction of Zn ribbon proteins, 19–180 µM protein in 50 mM Hepes-Na buffer (pH 7.5), 0.2 M NaCl in the presence of 1 µM NorW was made anaerobic by treatment with alternate cycles of evacuation and re-equilibration with oxygen-free argon in a tonometer. NADH solutions in the same buffer were made anaerobic by bubbling with oxygen-free argon in a glass syringe. The two solutions were mixed rapidly with an RX.2000 Stopped Flow Mixing Accessory (Applied Photophysics), which had been scrubbed of oxygen by overnight incubation with a 5 mM solution of sodium dithionite and then rinsed thoroughly with anaerobic buffer. Reduction of Zn ribbon proteins was followed at 488 nm using a Cary 50 spectrophotometer (Varian). Concentrations of TA0289 used in the present study have been calculated using ε371 = 10,950 mM−1cm−1. For the reduction of cytochrome C, a mixture of cytochrome C (Sigma, from horse heart), Zn ribbon protein (TA0289 or KTI11), and NorW at varying concentrations in 50 mM Hepes-Na (pH 7.5) containing 0.2 M NaCl was made anaerobic and mixed rapidly with an equal volume of 400 µM NADH as described above. Reduction of cytochrome C was followed at 550 nm.
Ligand binding experiments with TA0289
A minimum at 220 nm in the CD spectrum of TA0289 arises from the considerable amount of α-helix in the CBS domains. The temperature dependence of the CD signal of TA0289 at 220 nm in the absence and presence of potential ligand was determined. The CD signal at 220 nm was measured at 25°C and at 1 °C increments up to 95 °C. The melting temperature of the protein was then estimated from a plot of CD signal strength at 220 nm versus temperature. Five samples each contained 1 mg ml−1 TA0289 and a mixture of either no ligand, or 2 mM adenosine, AMP, ADP and ATP, or the equivalent cytidine, guanosine, or thymidine analogs, in 25 mM Hepes-Na (pH 7.5) and 100 mM NaCl.
Site-directed mutagenesis of KTI11
Site-directed mutagenesis was performed using the QuikChange™ site-directed mutagenesis kit (Stratagene) according to the manufacturer’s protocol. Because the targeted cysteines are located close to each other in the KTI11 sequence (Cys-X-Cys and Cys-X2-Cys), double Cys mutants (C25S/C27S and C47S/C50S) were prepared using mutagenic primers containing two mutations converting these cysteines to serine. For some unexplained reason, our attempts to prepare a quadruple cysteine mutant of KTI11 (C25S/C27S/C47S/C50S) repeatedly produced a plasmid with deleted Cys50, which was therefore used in the experiments (C25S/C27S/C47S/ΔC50).
Measurement of the redox potential of KTI11
KTI11 was slowly reduced under anaerobic conditions with xanthine, xanthine oxidase and benzyl viologen in the presence of indigo carmine as an internal reference. A 2.9 ml sample of 25 µM KTI11 containing 19 nM bovine xanthine oxidase, 5 µM benzyl viologen, and 10 µM indigo carmine was treated with alternate cycles of evacuation and re-equilibration with oxygen-free argon in an anaerobic cuvette. Reduction was initiated by the addition of 90 µl of 10 mM xanthine from a side arm (a final concentration of 300 µM) and the spectra were recorded from 320 to 750 nm. The extent of KTI11 reduction was estimated from the absorbance at 488 nm, where no absorbance change for the reduction of indigo carmine is observed. The extent of reduction of indigo carmine was estimated from the absorbance change at 605 nm after the small contribution from the reduction of KTI11 at this wavelength was subtracted. The difference in redox potential between KTI11 and indigo carmine was calculated from a plot of log(ox/red) indigo carmine versus log(ox/red) KTI11 according to the method of Minneart 69.
Protein crystallization and data collection
Crystals of TA0289 selected for diffraction studies were grown in 0.1 M Tris HCl (pH 7.8), 0.2 M ammonium formate, 20% PEG 3350, and 8% glycerol. The Hg2+-bound crystal was obtained by overnight soaking at room temperature of an iron-free TA0289 crystal in mother liquor solution containing 1.6 mM HgSO4. Diffraction data were collected at 100K at the 19ID beamline of the Structural Biology Center at the Advanced Photon Source, Argonne National Laboratory. The one-wavelength inverse-beam SAD data set (peak: 12.284 keV (1.0081 Å), was collected from a Hg labeled protein crystal. One crystal (0.2 × 0.2 × 0.2 mm) was used to collect all SAD data to 1.50 Å resolution, with 3 sec. exposure/1°/frame using a 160 mm crystal to the detector distance. The total oscillation range was 160 degrees as predicted using the strategy module within the HKL2000 suite 70. The space group was P212121 with cell dimensions of a=58.74, b=62.84, c=97.28, α=β=γ= 90°. The data were processed and scaled with HKL2000 (Table 3) to an Rmerge of 5.6%. To demonstrate the presence of Fe+2 in the native crystal, a single dataset on a second (Hg-free) crystal was collected using a crystal to detector distance of 150 mm, an oscillation range of 267.5 degrees and 0.5 degrees per oscillation. Data was collected on a Rigaku FR-E rotating anode generator at a wavelength of 1.54178 Å. The space group for this crystal was also P212121 with cell dimensions of a=58.81, b=63.03, c=97.47, α=β=γ= 90°.
Structure determination and refinement
The structure of TA0289 was determined by SAD phasing using CNS 71. The initial model was built using ARP/wARP 72, and the final model was finished manually using QUANTA 73. The model was refined against SAD peak data to 1.5 Å using CNS (HLML target). The final R was 0.194 with a free R of 0.228 with zero σ cut-off (Table 3). The stereochemistry of the structure was checked with PROCHECK 74, and the Ramachandran plot. The main chain torsion angles for all residues are in allowed regions. The initial structure solution for the second (iron-bound) crystal was performed by rigid-body refinement in CNS 71 using the protein coordinates from the Hg2+-coordinated structure since the space group and cell dimensions between the two crystals were very similar (see Table 3). Following rigid body refinement, difference density was observed in the same positions of the Hg2+ ion; Fe2+ ions were substituted in place of Hg2+, and refinement using CNS was performed as usual to a R value and R-free value of 0.204 and 0.237, respectively. As with the Hg2+ structure, all main chain torsions were in allowed regions of the Ramachandran plot. The atomic coordinates and structure factors for both Hg2+- and Fe2+-containing TA0289 have been deposited in the Protein Data Bank with the accession codes 1pvm (Hg2+-bound) and 2qhi (Fe2+-bound).
ACKNOWLEDGMENTS
We thank all members of the Ontario Centre for Structural Proteomics for help in conducting experiments. Deborah Zamble is thanked for the help with metal analysis. This work was supported in part by Genome Canada (through the Ontario Genomics Institute) and the Protein Structure Initiative of the National Institutes of Health (Midwest Center for Structural Genomics) Grant GM62414.
Abbreviations used
- AMPK
AMP-activated protein kinase
- CBS
cystathionine beta-synthase
- ICP-AES
inductively coupled plasma-atomic emission spectrometry
- IMPDH
inosine 5′-monophosphate dehydrogenase
- IPTG
isopropyl-beta-D-thiogalactopyranoside
- PMSF
phenylmethylsulfonyl fluoride
- Rd
rubredoxin
- SAD
single-wavelength anomalous diffraction
- TCEP
tris-2-carboxyethylphosphine-HCl
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Bateman A. The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem Sci. 1997;22:12–13. doi: 10.1016/s0968-0004(96)30046-7. [DOI] [PubMed] [Google Scholar]
- 2.Ignoul S, Eggermont J. CBS domains: structure, function, and pathology in human proteins. Am J Physiol Cell Physiol. 2005;289:C1369–C1378. doi: 10.1152/ajpcell.00282.2005. [DOI] [PubMed] [Google Scholar]
- 3.Scott JW, Hawley SA, Green KA, Anis M, Stewart G, Scullion GA, Norman DG, Hardie DG. CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J Clin Invest. 2004;113:274–284. doi: 10.1172/JCI19874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennetts B, Rychkov GY, Ng HL, Morton CJ, Stapleton D, Parker MW, Cromer BA. Cytoplasmic ATP-sensing domains regulate gating of skeletal muscle ClC-1 chloride channels. J Biol Chem. 2005;280:32452–32458. doi: 10.1074/jbc.M502890200. [DOI] [PubMed] [Google Scholar]
- 5.Biemans-Oldehinkel E, Mahmood NA, Poolman B. A sensor for intracellular ionic strength. Proc Natl Acad Sci U S A. 2006;103:10624–10629. doi: 10.1073/pnas.0603871103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller MD, Schwarzenbacher R, von Delft F, Abdubek P, Ambing E, Biorac T, Brinen LS, Canaves JM, Cambell J, Chiu HJ, Dai X, Deacon AM, DiDonato M, Elsliger MA, Eshagi S, Floyd R, Godzik A, Grittini C, Grzechnik SK, Hampton E, Jaroszewski L, Karlak C, Klock HE, Koesema E, Kovarik JS, Kreusch A, Kuhn P, Lesley SA, Levin I, McMullan D, McPhillips TM, Morse A, Moy K, Ouyang J, Page R, Quijano K, Robb A, Spraggon G, Stevens RC, van den Bedem H, Velasquez J, Vincent J, Wang X, West B, Wolf G, Xu Q, Hodgson KO, Wooley J, Wilson IA. Crystal structure of a tandem cystathionine-beta-synthase (CBS) domain protein (TM0935) from Thermotoga maritima at 1.87 A resolution. Proteins. 2004;57:213–217. doi: 10.1002/prot.20024. [DOI] [PubMed] [Google Scholar]
- 7.Meyer S, Dutzler R. Crystal structure of the cytoplasmic domain of the chloride channel ClC-0. Structure. 2006;14:299–307. doi: 10.1016/j.str.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 8.Jhee KH, McPhie P, Miles EW. Domain architecture of the heme-independent yeast cystathionine beta-synthase provides insights into mechanisms of catalysis and regulation. Biochemistry. 2000;39:10548–10556. doi: 10.1021/bi001020g. [DOI] [PubMed] [Google Scholar]
- 9.Zhang R, Evans G, Rotella FJ, Westbrook EM, Beno D, Huberman E, Joachimiak A, Collart FR. Characteristics and crystal structure of bacterial inosine-5′-monophosphate dehydrogenase. Biochemistry. 1999;38:4691–4700. doi: 10.1021/bi982858v. [DOI] [PubMed] [Google Scholar]
- 10.Rudolph MJ, Amodeo GA, Iram SH, Hong SP, Pirino G, Carlson M, Tong L. Structure of the Bateman2 domain of yeast Snf4: dimeric association and relevance for AMP binding. Structure. 2007;15:65–74. doi: 10.1016/j.str.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 11.Meyer S, Savaresi S, Forster IC, Dutzler R. Nucleotide recognition by the cytoplasmic domain of the human chloride transporter ClC-5. Nat Struct Mol Biol. 2007;14:60–67. doi: 10.1038/nsmb1188. [DOI] [PubMed] [Google Scholar]
- 12.Day P, Sharff A, Parra L, Cleasby A, Williams M, Horer S, Nar H, Redemann N, Tickle I, Yon J. Structure of a CBS-domain pair from the regulatory gamma1 subunit of human AMPK in complex with AMP and ZMP. Acta Crystallogr D Biol Crystallogr. 2007;63:587–596. doi: 10.1107/S0907444907009110. [DOI] [PubMed] [Google Scholar]
- 13.Townley R, Shapiro L. Crystal structures of the adenylate sensor from fission yeast AMP-activated protein kinase. Science. 2007;315:1726–1729. doi: 10.1126/science.1137503. [DOI] [PubMed] [Google Scholar]
- 14.Krishna SS, Majumdar I, Grishin NV. Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 2003;31:532–550. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laity JH, Lee BM, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol. 2001;11:39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- 16.Ravasi T, Huber T, Zavolan M, Forrest A, Gaasterland T, Grimmond S, Hume DA. Systematic characterization of the zinc-finger-containing proteins in the mouse transcriptome. Genome Res. 2003;13:1430–1442. doi: 10.1101/gr.949803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu W, Zeng Q, Colangelo CM, Lewis M, Summers MF, Scott RA. The N-terminal domain of TFIIB from Pyrococcus furiosus forms a zinc ribbon. Nat Struct Biol. 1996;3:122–124. doi: 10.1038/nsb0296-122. [DOI] [PubMed] [Google Scholar]
- 18.Qian X, Gozani SN, Yoon H, Jeon CJ, Agarwal K, Weiss MA. Novel zinc finger motif in the basal transcriptional machinery: three-dimensional NMR studies of the nucleic acid binding domain of transcriptional elongation factor TFIIS. Biochemistry. 1993;32:9944–9959. doi: 10.1021/bi00089a010. [DOI] [PubMed] [Google Scholar]
- 19.deMare F, Kurtz DM, Jr, Nordlund P. The structure of Desulfovibrio vulgaris rubrerythrin reveals a unique combination of rubredoxin-like FeS4 and ferritin-like diiron domains. Nat Struct Biol. 1996;3:539–546. doi: 10.1038/nsb0696-539. [DOI] [PubMed] [Google Scholar]
- 20.Gomes CM, Vicente JB, Wasserfallen A, Teixeira M. Spectroscopic studies and characterization of a novel electron-transfer chain from Escherichia coli involving a flavorubredoxin and its flavoprotein reductase partner. Biochemistry. 2000;39:16230–16237. doi: 10.1021/bi001844y. [DOI] [PubMed] [Google Scholar]
- 21.Gomes CM, Silva G, Oliveira S, LeGall J, Liu MY, Xavier AV, Rodrigues-Pousada C, Teixeira M. Studies on the redox centers of the terminal oxidase from Desulfovibrio gigas and evidence for its interaction with rubredoxin. J Biol Chem. 1997;272:22502–22508. doi: 10.1074/jbc.272.36.22502. [DOI] [PubMed] [Google Scholar]
- 22.da Costa PN, Romao CV, LeGall J, Xavier AV, Melo E, Teixeira M, Saraiva LM. The genetic organization of Desulfovibrio desulphuricans ATCC 27774 bacterioferritin and rubredoxin-2 genes: involvement of rubredoxin in iron metabolism. Mol Microbiol. 2001;41:217–227. doi: 10.1046/j.1365-2958.2001.02509.x. [DOI] [PubMed] [Google Scholar]
- 23.McKenna EJ, Coon MJ. Enzymatic omega-oxidation. IV. Purification and properties of the omega-hydroxylase of Pseudomonas oleovorans. J Biol Chem. 1970;245:3882–3889. [PubMed] [Google Scholar]
- 24.Shen G, Antonkine ML, van der Est A, Vassiliev IR, Brettel K, Bittl R, Zech SG, Zhao J, Stehlik D, Bryant DA, Golbeck JH. Assembly of photosystem I. II. Rubredoxin is required for the in vivo assembly of F(X) in Synechococcus sp. PCC 7002 as shown by optical and EPR spectroscopy. J Biol Chem. 2002;277:20355–20366. doi: 10.1074/jbc.M201104200. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Lee HS, Sugar FJ, Jenney FE, Jr, Adams MW, Prestegard JH. PF0610, a novel winged helix-turn-helix variant possessing a rubredoxin-like Zn ribbon motif from the hyperthermophilic archaeon, Pyrococcus furiosus. Biochemistry. 2007;46:752–761. doi: 10.1021/bi061870h. [DOI] [PubMed] [Google Scholar]
- 26.LeGall J, Liu MY, Gomes CM, Braga V, Pacheco I, Regalla M, Xavier AV, Teixeira M. Characterisation of a new rubredoxin isolated from Desulfovibrio desulfuricans 27774: definition of a new family of rubredoxins. FEBS Lett. 1998;429:295–298. doi: 10.1016/s0014-5793(98)00610-3. [DOI] [PubMed] [Google Scholar]
- 27.Grishin NV. C-terminal domains of Escherichia coli topoisomerase I belong to the zinc-ribbon superfamily. J Mol Biol. 2000;299:1165–1177. doi: 10.1006/jmbi.2000.3841. [DOI] [PubMed] [Google Scholar]
- 28.Yan Y, Moult J. Detection of operons. Proteins. 2006;64:615–628. doi: 10.1002/prot.21021. [DOI] [PubMed] [Google Scholar]
- 29.Lovenberg W, Sobel BE. Rubredoxin: a new electron transfer protein from Clostridium pasteurianum. Proc Natl Acad Sci U S A. 1965;54:193–199. doi: 10.1073/pnas.54.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ragsdale SW, Ljungdahl LG. Characterization of ferredoxin, flavodoxin, and rubredoxin from Clostridium formicoaceticum grown in media with high and low iron contents. J Bacteriol. 1984;157:1–6. doi: 10.1128/jb.157.1.1-6.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenney FE, Jr, Adams MW. Rubredoxin from Pyrococcus furiosus. Methods Enzymol. 2001;334:45–55. doi: 10.1016/s0076-6879(01)34457-9. [DOI] [PubMed] [Google Scholar]
- 32.Weinberg MV, Jenney FE, Jr, Cui X, Adams MW. Rubrerythrin from the hyperthermophilic archaeon Pyrococcus furiosus is a rubredoxin-dependent, iron-containing peroxidase. J Bacteriol. 2004;186:7888–7895. doi: 10.1128/JB.186.23.7888-7895.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blake PR, Park JB, Bryant FO, Aono S, Magnuson JK, Eccleston E, Howard JB, Summers MF, Adams MW. Determinants of protein hyperthermostability: purification and amino acid sequence of rubredoxin from the hyperthermophilic archaebacterium Pyrococcus furiosus and secondary structure of the zinc adduct by NMR. Biochemistry. 1991;30:10885–10895. doi: 10.1021/bi00109a012. [DOI] [PubMed] [Google Scholar]
- 34.Dauter Z, Wilson KS, Sieker LC, Moulis JM, Meyer J. Zinc- and iron-rubredoxins from Clostridium pasteurianum at atomic resolution: a high-precision model of a ZnS4 coordination unit in a protein. Proc Natl Acad Sci U S A. 1996;93:8836–8840. doi: 10.1073/pnas.93.17.8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eidsness MK, O'Dell SE, Kurtz DM, Jr, Robson RL, Scott RA. Expression of a synthetic gene coding for the amino acid sequence of Clostridium pasteurianum rubredoxin. Protein Eng. 1992;5:367–371. doi: 10.1093/protein/5.4.367. [DOI] [PubMed] [Google Scholar]
- 36.Mathieu I, Meyer J, Moulis JM. Cloning, sequencing and expression in Escherichia coli of the rubredoxin gene from Clostridium pasteurianum. Biochem J. 1992;285(Pt 1):255–262. doi: 10.1042/bj2850255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maher M, Cross M, Wilce MC, Guss JM, Wedd AG. Metal-substituted derivatives of the rubredoxin from Clostridium pasteurianum. Acta Crystallogr D Biol Crystallogr. 2004;60:298–303. doi: 10.1107/S090744490302794X. [DOI] [PubMed] [Google Scholar]
- 38.Blake PR, Day MW, Hsu BT, Joshua-Tor L, Park JB, Hare DR, Adams MW, Rees DC, Summers MF. Comparison of the X-ray structure of native rubredoxin from Pyrococcus furiosus with the NMR structure of the zinc-substituted protein. Protein Sci. 1992;1:1522–1525. doi: 10.1002/pro.5560011113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonomi F, Iametti S, Kurtz DM, Ragg EM, Richie KA. Direct metal ion substitution at the [M(SCys)(4)](2−) site of rubredoxin. Journal of Biological Inorganic Chemistry. 1998;3:595–605. [Google Scholar]
- 40.Mayhew SG. The redox potential of dithionite and SO-2 from equilibrium reactions with flavodoxins, methyl viologen and hydrogen plus hydrogenase. Eur J Biochem. 1978;85:535–547. doi: 10.1111/j.1432-1033.1978.tb12269.x. [DOI] [PubMed] [Google Scholar]
- 41.Moura I, Moura JJ, Santos MH, Xavier AV, Le Gall J. Redox studies on rubredoxins from sulphate and sulphur reducing bacteria. FEBS Lett. 1979;107:419–421. doi: 10.1016/0014-5793(79)80421-4. [DOI] [PubMed] [Google Scholar]
- 42.Gomes CM, Giuffre A, Forte E, Vicente JB, Saraiva LM, Brunori M, Teixeira M. A novel type of nitric-oxide reductase. Escherichia coli flavorubredoxin. J Biol Chem. 2002;277:25273–25276. doi: 10.1074/jbc.M203886200. [DOI] [PubMed] [Google Scholar]
- 43.Gardner AM, Gessner CR, Gardner PR. Regulation of the nitric oxide reduction operon (norRVW) in Escherichia coli. Role of NorR and sigma54 in the nitric oxide stress response. J Biol Chem. 2003;278:10081–10086. doi: 10.1074/jbc.M212462200. [DOI] [PubMed] [Google Scholar]
- 44.Lode ET, Coon MJ. Enzymatic omega-oxidation. V. Forms of Pseudomonas oleovorans rubredoxin containing one or two iron atoms: structure and function in omega-hydroxylation. J Biol Chem. 1971;246:791–802. [PubMed] [Google Scholar]
- 45.Chen L, Liu MY, LeGall J, Fareleira P, Santos H, Xavier AV. Rubredoxin oxidase, a new flavo-hemo-protein, is the site of oxygen reduction to water by the "strict anaerobe" Desulfovibrio gigas. Biochem Biophys Res Commun. 1993;193:100–105. doi: 10.1006/bbrc.1993.1595. [DOI] [PubMed] [Google Scholar]
- 46.Lee HJ, Basran J, Scrutton NS. Electron transfer from flavin to iron in the Pseudomonas oleovorans rubredoxin reductase-rubredoxin electron transfer complex. Biochemistry. 1998;37:15513–15522. doi: 10.1021/bi981853v. [DOI] [PubMed] [Google Scholar]
- 47.Ueda T, Lode ET, Coon MJ. Enzymatic Omega-Oxidation. 6. Isolation of Homogeneous Reduced Diphosphopyridine Nucleotide-Rubredoxin Reductase. Journal of Biological Chemistry. 1972;247:2109-&. [PubMed] [Google Scholar]
- 48.Petitdemange H, Marczak R, Blusson H, Gay R. Isolation and properties of reduced nicotinamide adenine dinucleotiderubredoxin oxidoreductase of Clostridium acetobutylicum. Biochem Biophys Res Commun. 1979;91:1258–1265. doi: 10.1016/0006-291x(79)91202-6. [DOI] [PubMed] [Google Scholar]
- 49.Perry A, Lian LY, Scrutton NS. Two-iron rubredoxin of Pseudomonas oleovorans: production, stability and characterization of the individual iron-binding domains by optical, CD and NMR spectroscopies. Biochem J. 2001;354:89–98. doi: 10.1042/0264-6021:3540089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Claus R, Asperger O, Kleber HP. Properties of rubredoxin reductase from the alkane-assimilating bacterium Acinetobacter calcoaceticus. Z Allg Mikrobiol. 1979;19:695–704. [PubMed] [Google Scholar]
- 51.Grunden AM, Jenney FE, Jr, Ma K, Ji M, Weinberg MV, Adams MW. In vitro reconstitution of an NADPH-dependent superoxide reduction pathway from Pyrococcus furiosus. Appl Environ Microbiol. 2005;71:1522–1530. doi: 10.1128/AEM.71.3.1522-1530.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meier M, Janosik M, Kery V, Kraus JP, Burkhard P. Structure of human cystathionine beta-synthase: a unique pyridoxal 5′-phosphate-dependent heme protein. Embo J. 2001;20:3910–3916. doi: 10.1093/emboj/20.15.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blake PR, Park JB, Zhou ZH, Hare DR, Adams MW, Summers MF. Solution-state structure by NMR of zinc-substituted rubredoxin from the marine hyperthermophilic archaebacterium Pyrococcus furiosus. Protein Sci. 1992;1:1508–1521. doi: 10.1002/pro.5560011112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blake PR, Lee B, Summers MF, Adams MW, Park JB, Zhou ZH, Bax A. Quantitative measurement of small through-hydrogen-bond and 'through-space' 1H-113Cd and 1H-199Hg J couplings in metal-substituted rubredoxin from Pyrococcus furiosus. J Biomol NMR. 1992;2:527–533. doi: 10.1007/BF02192814. [DOI] [PubMed] [Google Scholar]
- 55.Day MW, Hsu BT, Joshua-Tor L, Park JB, Zhou ZH, Adams MW, Rees DC. X-ray crystal structures of the oxidized and reduced forms of the rubredoxin from the marine hyperthermophilic archaebacterium Pyrococcus furiosus. Protein Sci. 1992;1:1494–1507. doi: 10.1002/pro.5560011111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bau R, Rees DC, Kurtz DM, Scott RA, Huang HS, Adams MWW, Eidsness MK. Crystal structure of rubredoxin from Pyrococcus furiosus at 0.95 angstrom resolution, and the structures of N-terminal methionine and formylmethionine variants of Pf Rd. Contributions of N-terminal interactions to thermostability. Journal of Biological Inorganic Chemistry. 1998;3:484–493. [Google Scholar]
- 57.Page CC, Moser CC, Chen X, Dutton PL. Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature. 1999;402:47–52. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]
- 58.Archer M, Huber R, Tavares P, Moura I, Moura JJ, Carrondo MA, Sieker LC, LeGall J, Romao MJ. Crystal structure of desulforedoxin from Desulfovibrio gigas determined at 1.8 A resolution: a novel non-heme iron protein structure. J Mol Biol. 1995;251:690–702. doi: 10.1006/jmbi.1995.0465. [DOI] [PubMed] [Google Scholar]
- 59.Liu S, Leppla SH. Retroviral insertional mutagenesis identifies a small protein required for synthesis of diphthamide, the target of bacterial ADP-ribosylating toxins. Mol Cell. 2003;12:603–613. doi: 10.1016/j.molcel.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 60.Kummerle R, Zhuang-Jackson H, Gaillard J, Moulis JM. Site-directed mutagenesis of rubredoxin reveals the molecular basis of its electron transfer properties. Biochemistry. 1997;36:15983–15991. doi: 10.1021/bi971636e. [DOI] [PubMed] [Google Scholar]
- 61.Cross M, Xiao Z, Maes EM, Czernuszewicz RS, Drew SC, Pilbrow JR, George GN, Wedd AG. Removal of a cysteine ligand from rubredoxin: assembly of Fe(2)S(2) and Fe(S-Cys)(3)(OH) centres. J Biol Inorg Chem. 2002;7:781–790. doi: 10.1007/s00775-002-0355-1. [DOI] [PubMed] [Google Scholar]
- 62.Sieker LC, Stenkamp RE, LeGall J. Rubredoxin in crystalline state. Methods Enzymol. 1994;243:203–216. doi: 10.1016/0076-6879(94)43016-0. [DOI] [PubMed] [Google Scholar]
- 63.Logan DT, Mulliez E, Larsson KM, Bodevin S, Atta M, Garnaud PE, Sjoberg BM, Fontecave M. A metal-binding site in the catalytic subunit of anaerobic ribonucleotide reductase. Proc Natl Acad Sci U S A. 2003;100:3826–3831. doi: 10.1073/pnas.0736456100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Treich I, Riva M, Sentenac A. Zinc-binding subunits of yeast RNA polymerases. J Biol Chem. 1991;266:21971–21976. [PubMed] [Google Scholar]
- 65.Carles C, Treich I, Bouet F, Riva M, Sentenac A. Two additional common subunits, ABC10 alpha and ABC10 beta, are shared by yeast RNA polymerases. J Biol Chem. 1991;266:24092–24096. [PubMed] [Google Scholar]
- 66.Zhang RG, Skarina T, Katz JE, Beasley S, Khachatryan A, Vyas S, Arrowsmith CH, Clarke S, Edwards A, Joachimiak A, Savchenko A. Structure of Thermotoga maritima stationary phase survival protein SurE: a novel acid phosphatase. Structure. 2001;9:1095–1106. doi: 10.1016/s0969-2126(01)00675-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hogbom M, Ericsson UB, Lam R, Bakali HM, Kuznetsova E, Nordlund P, Zamble DB. A high throughput method for the detection of metalloproteins on a microgram scale. Mol Cell Proteomics. 2005;4:827–834. doi: 10.1074/mcp.T400023-MCP200. [DOI] [PubMed] [Google Scholar]
- 69.Minnaert K. Measurement of the equilibrium constant of the reaction between cytochrome c and cytochrome a. Biochim Biophys Acta. 1965;110:42–56. doi: 10.1016/s0926-6593(65)80093-5. [DOI] [PubMed] [Google Scholar]
- 70.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 71.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 72.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 73.Oldfield TJ, Hubbard RE. Analysis of C alpha geometry in protein structures. Proteins. 1994;18:324–337. doi: 10.1002/prot.340180404. [DOI] [PubMed] [Google Scholar]
- 74.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]