Abstract
Spinal muscular atrophy (SMA), the leading genetic cause of infant death results from loss of spinal motor neurons causing atrophy of skeletal muscle. SMA is caused by loss of the Survival Motor Neuron 1 (SMN1) gene, however, an identically-coding gene called SMN2 is retained, but is alternatively spliced to produce ~90% truncated protein. Most SMA translational and preclinical drug development has relied on the use of SMA mice to determine changes in SMN protein levels. However, the SMA mouse models are relatively severe and analysis of SMN-inducing compounds is confounded by the early mortality of these animals. An antibody that could detect SMN protein on a Smn background could circumvent this limitation and allow unaffected, heterozygous animals to be examined. Here we describe the generation and characterization of a monoclonal anti-SMN antibody, 4F11, which specifically recognizes human SMN protein. 4F11 detects SMN (human) but not native Smn (mouse) protein in SMN2 transgenic mice and in SMA cell lines. We demonstrate the feasibility of using 4F11 to detect changes in SMN2-derived SMN protein in SMA patient fibroblasts and in healthy SMN2 transgenic mice. This antibody is, therefore, an excellent tool for examining SMN2-inducing therapeutics in patient cells as well as in transgenic mice.
Keywords: spinal muscular atrophy, transgenic mouse, monoclonal antibody, biomarker, motor neuron disease, gem, survival motor neuron
1. Introduction
Proximal spinal muscular atrophy (SMA) is the second most common autosomal recessive disorder and the number one genetic cause of infantile death. It has a carrier frequency of about 1:35 and affects every one in six thousand live births. SMA causes the specific loss of α motor neurons resulting in the atrophy of voluntary muscle groups. There are at least three clinical classifications of SMA dependant upon age of onset and severity of symptoms (Crawford and Pardo, 1996; Munsat and Davies, 1992). Type I and Type II forms are the most severe and are the most common, comprising nearly 80% of all new SMA cases. Type III patients have a normal life-expectancy, but are restricted to a walker or wheelchair. All three types of SMA result from reduced levels of survival of motor neuron (SMN) protein. In humans, there are two nearly identical copies of SMN, SMN1 and SMN2 (Lefebvre et al., 1995). The main difference between SMN1 and SMN2 is a silent C to T transition in exon 7 of SMN2 which results in the majority of SMN2-derived transcripts being alternatively spliced (Lorson et al., 1999; Monani et al., 1999). Importantly, SMN2 has the same coding sequence as SMN1; however, the alternative splicing event results in a truncated, defective, and unstable SMN protein which lacks exon 7. At the genetic level, SMA results from deletion of or mutations in SMN1 and retention of SMN2. There is a relationship between SMN2 copy number and the severity of the disease (Feldkotter et al., 2002; McAndrew et al., 1997) making SMN2 an ideal target for therapeutic development.
SMN is a ubiquitously expressed protein and it is clear that SMN is intimately involved in multiple RNA-related pathways. In all SMA tissues, SMN expression is depressed (Coovert et al., 1997; Lefebvre et al., 1997), but it is unknown why a protein found to operate in every cell would cause the precise death of the alpha motor neurons. Two main hypotheses have emerged (as reviewed by (Monani, 2005)). First, it has been hypothesized that motor neurons require a higher level of snRNP activity and that depletion of SMN makes these cells especially sensitive during development. An alternative hypothesis is that SMN performs a neuron-specific function potentially involving axonal transport of specific mRNAs such as β-actin. The association of SMN with hnRNP-Q and with factors involved in β-actin mRNA transport has been further supported by live cell imaging of SMN granules.
While SMA only affects humans, several animal models of SMA have been developed including those in the nematode (C. elegans), fruit fly, zebrafish and mice (reviewed in (Butchbach, 2004; Monani et al., 2000; Schmid and DiDonato, 2007)). In the SMA mouse models, a complete knock-out of Smn is lethal at the pre-implantation stage (Schrank et al., 1997); however, lethality can be rescued by introduction of the human SMN2 transgene (Monani et al., 2000). A model widely used to test therapeutics is the SMNΔ7 SMA mouse (SMN2+/+;SMNΔ7+/+;Smn−/−) which corresponds to an intermediate phenotype (Le et al., 2005). This mouse model closely mimics the human SMA phenotype in reduced lifespan, body mass, motor behavior and strength (Butchbach et al., 2007a; Le et al., 2005). This model is a tremendous tool for examining various proposed SMA therapeutics such as small molecules or viral vectors which alter SMN2 splicing, increase total SMN2 gene expression or stabilize SMN protein products. Therefore, compound testing can be performed in these SMA animals. In all of these proposed routes of SMA therapy, the primary benefit is being derived from an increase in SMN2-derived SMN protein; therefore, an antibody that specifically recognizes SMN protein in a murine background would be extremely useful.
Here we examine another potential tool for examining SMA therapeutics in the mouse models in the form of a human specific SMN antibody (4F11). Since the SMA mouse models express human transgenes this antibody could be used to specifically examine the changes in SMN protein in response to a given treatment.
2. Methods
2.1. Antibody production
Hybridoma supernatants were produced as previously described (Wolstencroft et al., 2005). 4F11 ascites fluid was produced from BALB/C male mice primed with pristine (2,6,10,14-tetramethylpentadecane) using established protocols (Harlow E, 1988). All experiments were conducted in accordance with the protocols described in the National Institutes of Health Guide for the Care and Use of Animals and were approved by the Ohio State University Institutional Laboratory Animal Care and Use Committee.
2.2. Epitope mapping
GST-tagged exons 1, 2(2a and 2b combined), 2a, 2b, 3 and 4 (respectively) of SMN were expressed in Rosetta pLysS E. coli. Bacteria were lysed in 8 M urea. Cell lyses were run on an 8% SDS-PAGE gel. Anti-GST antibody (Santa Cruz) was used in western blot analysis at a dilution of 1:200 to identify the recombinant SMN exons. The membrane was subsequently stripped (see below), reprobed with mouse monoclonal anti-SMN 4B7, then stripped again (see below) and probed using mouse monoclonal anti-SMN 4F11.
2.3. Cell culture and immunohistochemistry
3813 fibroblasts were plated on coverslips and grown as previously described (Mattis et al., 2006). Immunohistochemistry was performed as previously described (Mattis et al., 2006) using either the mouse anti-SMN antibody 4B7 or 4B11 (as indicated) as the primary antibody.
2.4. Animals and tissue harvesting
5 genotypes of mice representing various mouse models of SMA were used. 1) FVB/N nontransgenic mice were used as a control for distinguishing Smn from SMN. 2) Low copy SMN2 mice were generated from carrier parents of the genotype SMN2+/+;Smn+/− (line 89; FVB.Cg-Tg(SMN2)89Ahmb Smn1tm1Msd (Monani et al., 2000)). 3) High copy SMN2 mice were produced by mating males of the genotype SMN2(89)+/+;SMN2(566)+/+;Smn+/− with females of the genotype Smn+/− (Monani et al., 2000). 4) SMNΔ7 mice were generated from males and females of the genotype SMN2+/+;SmnΔ7+/+;Smn+/− (line 4299; FVB.Cg-Tg(SMN2*delta7)4299Ahmb Tg(SMN2)89Ahmb Smn1tm1Msd (Le et al., 2005)). 5) SMN(A2G) mice were bred from males with the genotype SMN2+/+;SMN(A2G)+/+;Smn+/− (line 2023; FVB.Cg-Tg(SMN1*A2G)2023Ahmb Tg(SMN2)89Ahmb Smn1tm1Msd) and females with the genotype SMN2+/+;Smn+/− (Monani et al., 2003). All of the mice used in these experiments were obtained from an internal (Ohio State University) breeding colony but can be obtained commercially from Jackson. Mouse tissues were harvested from adult carrier mice. The mice were euthanized by rapid decapitation; spinal cords were rapidly dissected and immediately frozen in liquid nitrogen. All tissue samples were stored at −80°C until use. A tail biopsy, which was used for genotyping, was also taken from each mouse. These mice were genotyped using a PCR-based assay on genomic DNA from tail biopsies as described previously (Butchbach et al., 2007b; Monani et al., 2003; Monani et al., 2000).
2.5. Western blot analysis
U20S, 3814, 3813 or A9 cells were harvested from a 60 mm dish by scraping. Cells were lysed in 8M Urea and sonicated. Total protein was measured using the BioRad Protein Assay according to the manufacturer’s directions. Equal amounts of total protein were then loaded into a SDS-PAGE gel. Mouse monoclonal anti-SMN antibody 4F11 was used in western blot analysis at a dilution of 1:25 to identify SMN in cellular extracts. The membrane was subsequently stripped with β-mercaptoethanol [0.625% β-mercaptoethanol, 0.02% SDS, 6.25% 1M Tris pH 6.8, 45 min at 50°C] and reprobed with an ti-SMN (BD Biosciences; clone 8) at a dilution of 1:2000. The HRP was then inactivated using 1:1 H2O2:PBS for 45 min at room temperature and reprobed using an anti-actin polyclonal rabbit antibody at 1:300 dilution (Sigma). Probes were visualized by chemiluminescence using the Pierce SuperSignal Pico reagents.
For mouse tissues, spinal cords were homogenized in lysis buffer (0.1% Triton X-100, complete protease inhibitor cocktail in PBS, pH 7.4) containing 4 M urea. Tissue extracts (100 µg) were resolved through a 12% PA gel containing 0.1% SDS and electrotransfered onto a PVDF membrane. This blot was first blocked with 3% BSA in PBST (0.2% Tween-20 in PBS) for 60 min at room temperature and then incubated with 4F11 ascites fluid (1:5,000 in 0.3% BSA in PBST) overnight at 4°C. After thorough rinsing with PBST, the blot was incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (1:10,000 (Rockland Immunochemicals for Research) in 0.3% BSA in PBST) for 60 min at room temperature. This blot was then rinsed with PBST and the probe was visualized by enhanced chemiluminescence using the SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific). The blot was then stripped and reprobed with either mouse anti-SMN mAb (1:5000; BD Biosciences, clone 8) or mouse anti-actin mAb (1:10,000; Sigma-Aldrich, clone AC-40).
2.6. Drug Treatment of SMA Fibroblasts
Treatment of SMA patient fibroblast (3813) cultures with drug compounds was accomplished as described previously (Novoyatleva et al 2008; Thurmond et al 2008). 3813 cells were exposed to sodium valproate (VPA, 5 mM), hydroxyurea (HU, 65.7 µM) or vehicle (PBS) for 3 days with fresh compound added each day. These cells were processed for SMN immunostaining as described in (Novoyatleva et al., 2008; Thurmond et al., 2008) except that 4F11 supernatant (dilution 1:100) was used as the primary antibody. Confocal images were obtained with a Zeiss LSM510 fluorescent microscope as described previously (Novoyatleva et al., 2008; Thurmond et al., 2008). For gem counting, immunostained cells were examined with a Nikon fluorescent microscope and the following measures were obtained: the number of cells containing gems, the number of gems in 100 randomly selected nuclei and the number of cells containing multiple gems.
2.7. Drug Treatment of SMN2 Transgenic Mice
Adult severe SMA carrier (SMN2+/+;Smn+/−) mice (Monani et al. 2000) were used to examine the effects of HU and VPA on human SMN protein expression in vivo. Mice received either HU (5 mg/kg), VPA (200 mg/kg) or vehicle (PBS) via intraperitoneal (i.p.) injection at a dose volume of 1 µL/g. The treated mice were euthanized 6 hr after dosing and spinal cords were dissected and rapidly frozen in N2(l). Tissues were stored at −80°C until use.
2.8. Statistical analysis
All parametric data were expressed as mean ± standard error. Comparisons between data were made using one-way ANOVA and a Bonferonni post hoc test. All statistical analyses were performed using SPSS v.16.
3. Results
3.1. Production and characterization of anti-SMN antibodies
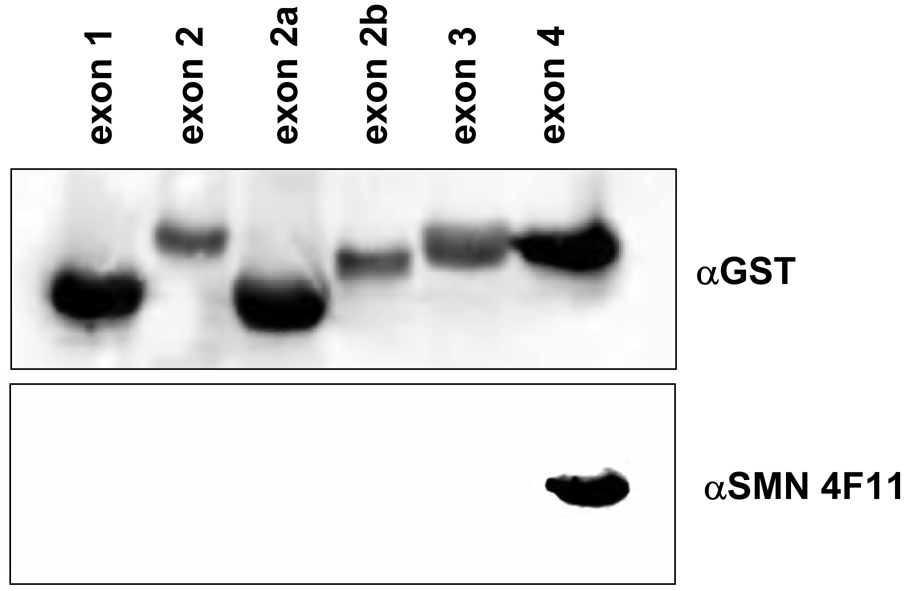
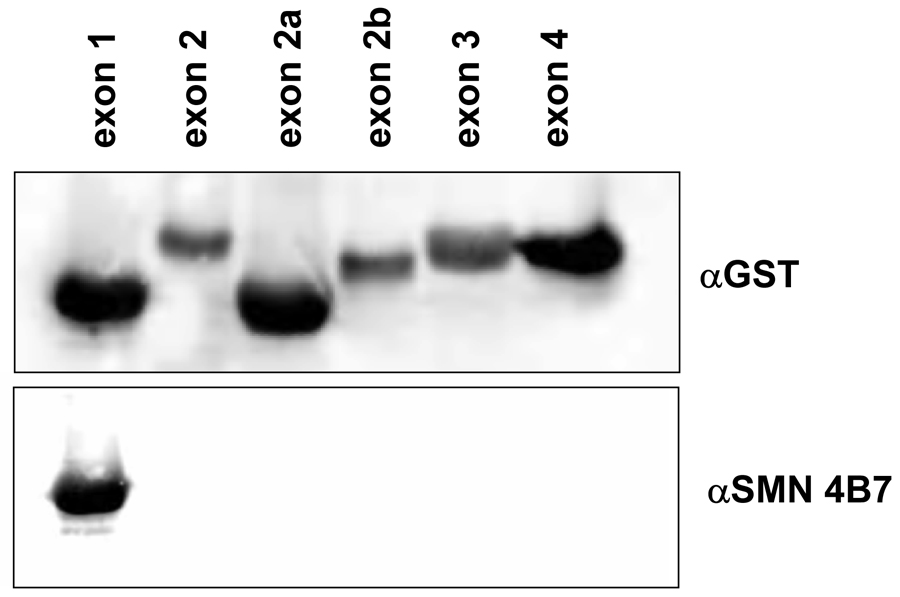
Monoclonal anti-SMN antibodies, clones 4B7 and 4F11, were produced in a mouse by injecting purified His-tagged full-length SMN protein. To identify the epitope recognized by these novel antibodies, epitope mapping experiments were performed using purified recombinant proteins encoding specific SMN domains fused to GST. Each of the SMN exon-encoded peptides was resolved on a denaturing polyacrylamide gel and expression was confirmed by blotting with an anti-GST antibody (Fig. 1A and 1B). Blots were stripped and re-probed with anti-SMN 4F11. The detection of a single reactive species identifies SMN exon 4 peptide as the specific epitope for 4F11 (Fig. 1A). To confirm the specificity of the reaction, anti-SMN 4B7 was reacted similarly and shown to recognize SMN exon 1 peptide (Fig. 1B).
Fig. 1.
Epitope mapping of two mouse anti-SMN monoclonal antibodies. GST-tagged SMN exons were individually expressed in bacterial cells. Immunoblots were performed against the GST tag, the blot was then stripped and reprobed with either anti-SMN 4F11 (A) or 4B7 (B). The 4F11 antibody maps to exon 4 of SMN while 4B7 maps to exon 1 of SMN.
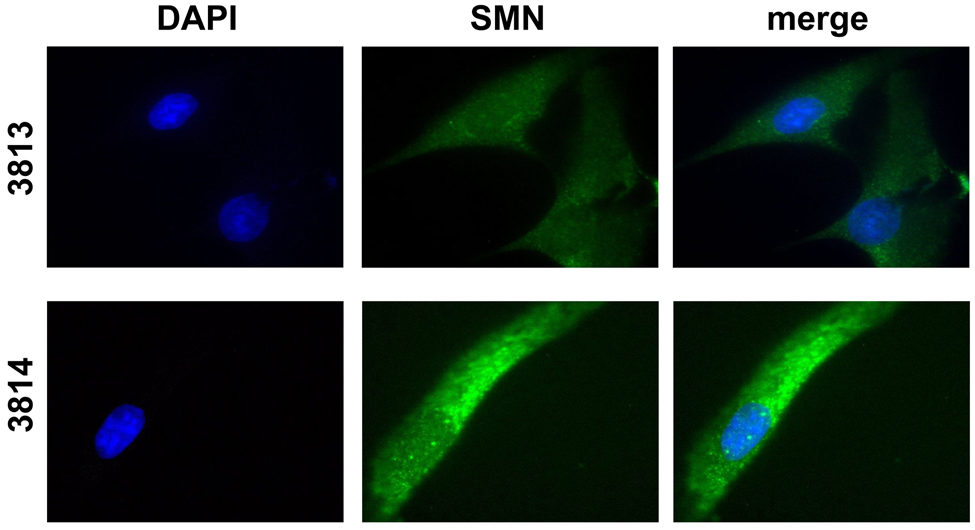
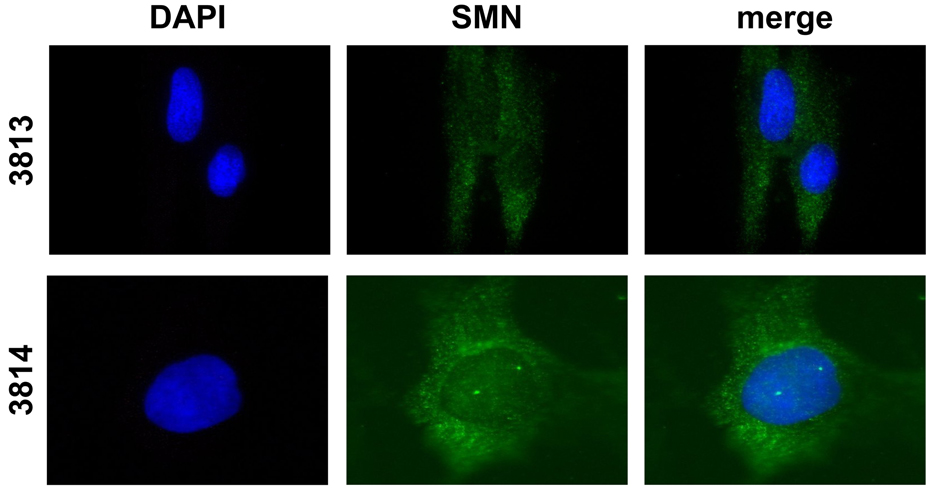
SMN localizes to nuclear structures referred to as “gems.” Gems have become a useful biomarker for total SMN levels and frequently serve as a tool in the screening of SMN-inducing compounds. To determine whether 4F11 was capable of detecting SMN in gems, immunofluorescence was performed using either anti-SMN 4F11 or anti-SMN 4B7 on type 1 SMA patient fibroblasts (3813) and carrier fibroblasts (3814) (Fig. 2A,B). Both antibodies resulted in the expected SMN staining in 3814 cells: diffuse cytoplasmic staining and punctuate nuclear structures (gems). Similarly, both antibodies display proper localization in the 3813 cells: low, but diffuse cytoplasmic staining and few to no gems.
Fig. 2.
Localization of endogenous SMN in human fibroblast cultures. Skin fibroblasts from a Type 1 SMA patient (3813) or from the carrier parent (3814) were immunolabeled with either anti-SMN 4F11 (A) or 4B7 (B). SMN staining is shown in green while the blue, DAPI staining outlines the nucleus.
3.2. 4F11 is specific to human SMN
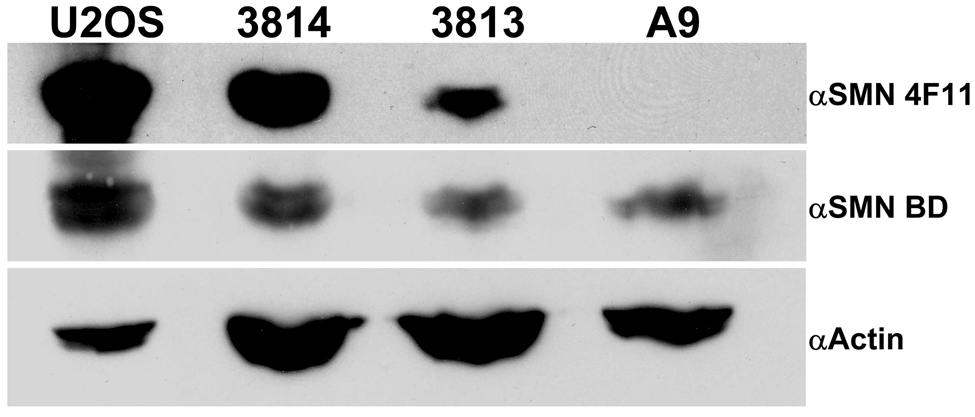
Since anti-SMN 4F11 specifically recognizes SMN exon 4, which only shares 57% amino acid identity with it’s murine ortholog, Smn, (Fig. 3A) and even less so with other animals further evolutionarily divergent, we wanted to determine if this antibody would recognize other SMN species. To determine whether 4F11 recognized Smn, extracts from human (U2OS, 3814 and 3813) and mouse (A9) tissue culture cells were resolved on a SDS-PAGE gel and analyzed by Western blotting. 4F11 clearly identifies the human SMN protein, but fails to react with the murine Smn protein (Fig. 3B). As a confirmatory measure to demonstrate that Smn was present on the blot, a commercially available antibody known to recognize a highly conserved Smn epitope was used to re-probe the membrane (Fig. 3B). Collectively, these results identify 4F11 as a human SMN specific antibody that recognizes the exon 4 peptide.
Fig. 3.
4F11 detect SMN in human cells but not in mouse cells. (A) The amino acid sequence of exon 4 of human SMN were aligned to orthologues in mouse (Mus musculus, 57% identity), rat (Rattus norvegicus, 57%), bull (Bos taurus, 75%), dog (Canis familiaris, 82%), cat (Felis catus, 75%) and zebrafish (Danio rerio, 28%). Bold/color sequence indicates an identical amino acid to human homolog; normal sequence indicates amino acid change. Green sequences are amide amino acids, red sequences are Acidic amino acids, pink sequences are nucleophilic amino acids, yellow sequences are hydrophobic amino acids, and blue sequences are Basic amino acids. Sequences and identities found on NCBI by protein-protein blast of exon 4 amino acid sequence and aligned using STRAP project. (B) Blots of extracts from human cell lines (HeLa, 3814 and 3813) and the A9 mouse cell line were probed with 4F11. 4F11 recognizes SMN but not Smn. The blot was then stripped and reprobed with a commercially available anti-SMN antibody (clone 8, BD Biosciences) that recognizes both human and mouse SMN. Actin was used to show that equal amounts of total protein were loaded in each lane.
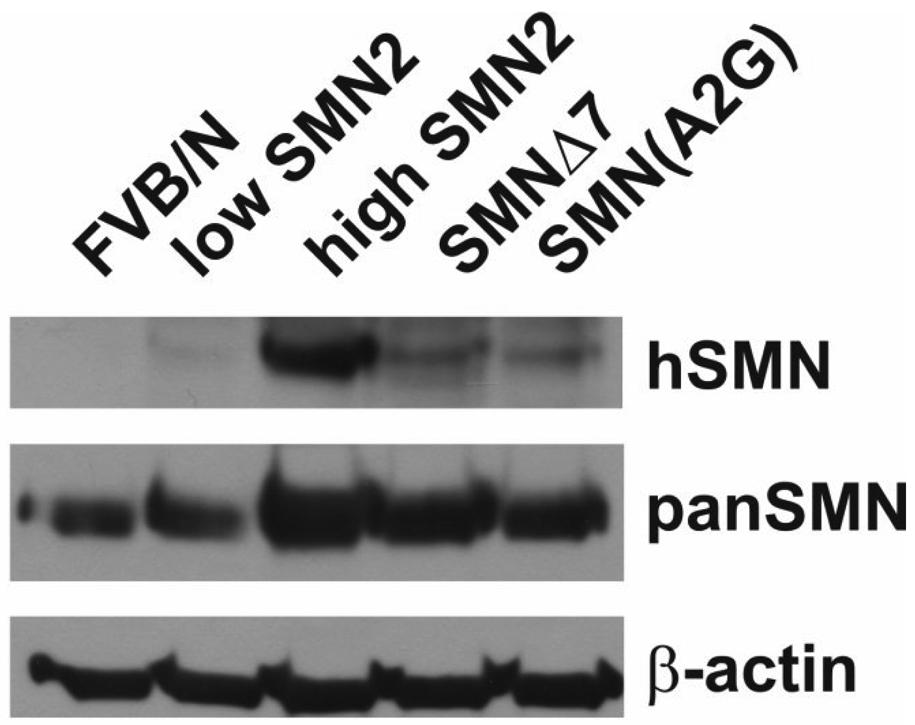
3.3. 4F11 detects human SMN in SMN2 transgenic mice
We wanted to determine if 4F11 could detect SMN in a native Smn background. Spinal cord extracts from adult mice containing various human SMN transgenes were probed with either 4F11 or a pan-specific SMN antibody. The following transgenic lines were tested: 1) low copy SMN2 (line 89) carrier mice (2 copies of SMN2; SMN2+/+;Smn+/− (Monani et al., 2000)), 2) high copy SMN2 (line 566) carrier mice (9 copies of SMN2; SMN2(89)+/−;SMN2(566)+/−;Smn+/− (Monani et al., 2000)), 3) SMNΔ7 carrier mice (SMN2+/+;SmnΔ7+/+;Smn+/− (Le et al., 2005)) and 4) SMN(A2G) carrier mice (SMN2+/+;SMN(A2G)+/−;Smn+/− (Monani et al., 2003)). As a control, spinal cord extracts from non-transgenic FVB/N mice were used. As shown in figure 4, we can detect SMN with 4F11 in transgenic mouse samples but not in FVB/N controls. The addition of 4 M urea to the extraction buffer greatly facilitates detection of SMN in mouse samples (data not shown). The SMN band intensity relates to the copy number of SMN2 transgenes and to the presence of either SMNΔ7 or SMN(A2G) transgenes. Reprobing with a pan-specific anti-SMN antibody showed that Smn was present in every sample but the band intensity was proportional to the amount of SMN present in each transgenic line.
Fig. 4.
Detection of human SMN protein in SMA mouse models. Spinal cord extracts from adult nontransgenic FVB/N mice, low SMN2 (SMN2(89)+/+;Smn+/−) mice, high SMN2 (SMN2(89)+/− ;SMN2(566)+/−;Smn+/−) mice, SMNΔ7 (SMN2+/+;SMNΔ7+/+;Smn+/−) mice and SMN(A2G) (SMN2+/+;SMN(A2G)+/−;Smn+/−) mice were analyzed for human SMN protein levels by immunoblot using 4F11 and total SMN protein using a pan-specific anti-SMN antibody (clone 8, BD Biosciences). β-Actin bands show equal loading of protein in each lane.
3.4. Effect of valproic acid and hydroxyurea on SMN localization in patient fibroblasts
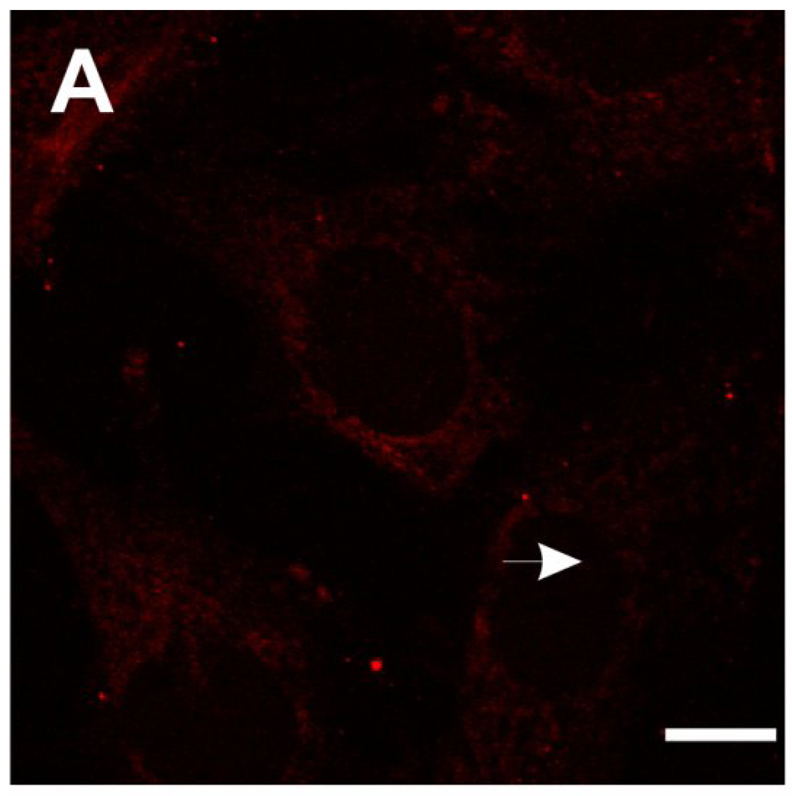
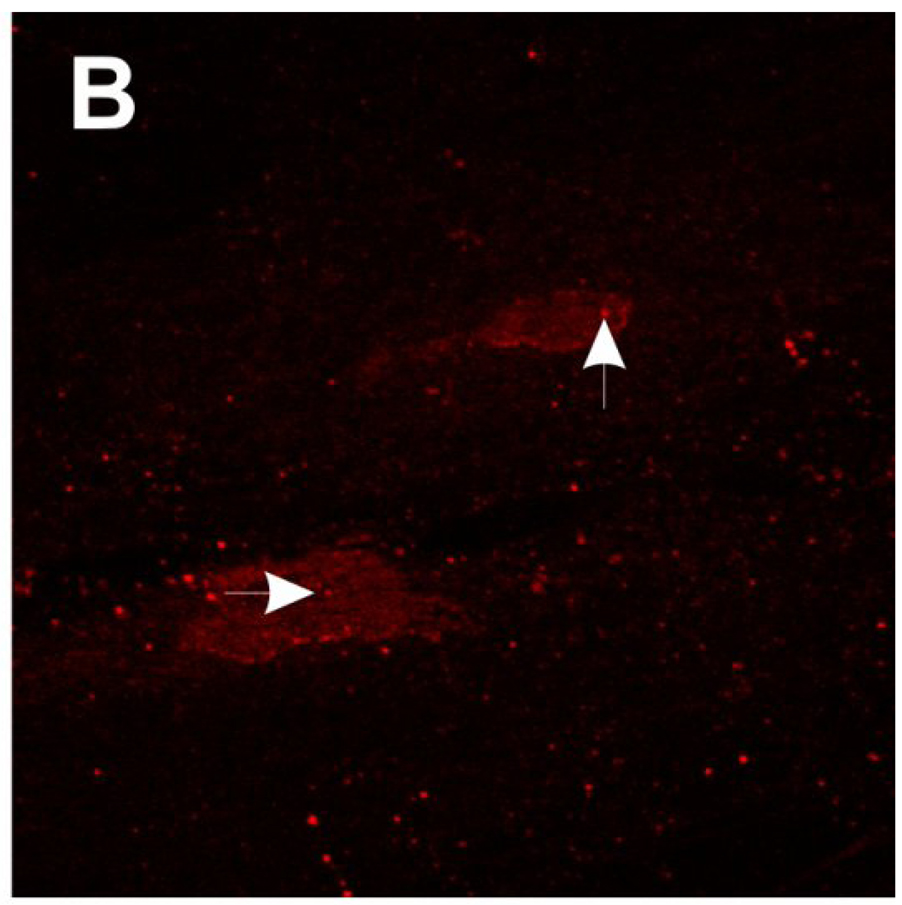
SMA patient fibroblasts (3813) were treated with valproic acid (VPA, 5 mM) or hydroxyurea (HU, 65.7 µM) and then stained for SMN using 4F11. These drugs have been previously shown to increase SMN protein in SMA patient cultured cells (Brichta et al., 2003; Grzeschik et al., 2005; Sumner et al., 2003). Carrier fibroblasts (3814) were also immunolabeled with 4F11 as a control. As shown in the confocal images in Fig. 5, SMN staining in SMA fibroblasts (Fig. 5A) is much weaker than that observed in carrier fibroblasts (Fig. 5B). Treatment of 3813 cells with either VPA (Fig. 5C) or HU (Fig. 5D) increases SMN immunoreactivity in both the nucleus and the cytosol.
Fig. 5.
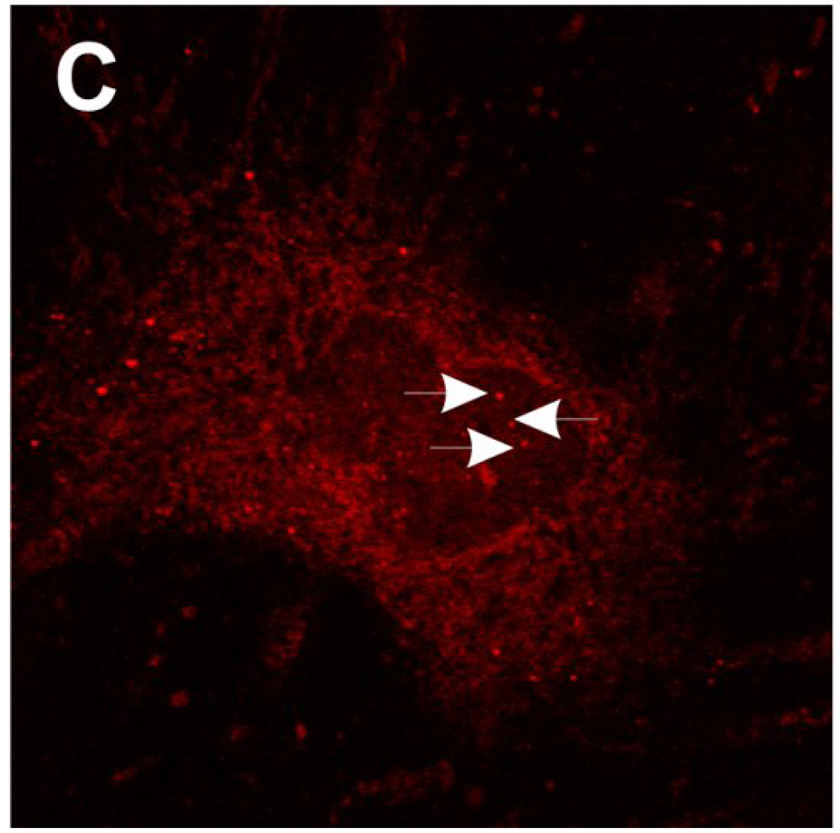
Immunofluorescent localization of SMN in SMA fibroblasts treated with valproic acid (VPA) or hydroxyurea (HU). SMA patient fibroblasts (3813) were treated for 3 days with either 5 mM VPA (C), 65.7 µM HU (D) or vehicle (ddH2O; A). Fibroblasts were then labeled for SMN protein using 4F11. As a control, SMA carrier fibroblasts (3814; B) were also stained for SMN localization with 4F11. VPA and HU increase the amount of SMN in the nucleus and cytosol of patient fibroblasts. Arrowheads show SMN-positive gems in the nuclei. Scale bar, 10 µm.
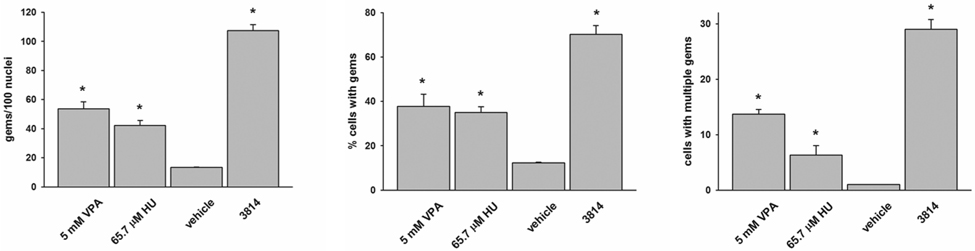
As a quantitative marker for SMN protein induction in cultured cells, the number of SMN-positive nuclear gems was counted in 3813 cells treated with VPA, HU or vehicle. Treatment of SMA fibroblasts with VPA increased the number of gems by 3-fold (p<0.001) and HU increased the number of gems by 2.2-fold (p=0.003) relative to vehicle-treated cells (Fig. 6A). The number of cells that contain gems also increased significantly in VPA (p=0.003)- and HU (p=0.013)-treated SMA fibroblasts (Fig. 6B). The number of cells containing multiple gems per nucleus increased by almost 13-fold in VPA-treated cells (p=0.001) and by 5-fold in HU-treated cells (p=0.023) relative to vehicle-treated SMA fibroblasts (Fig. 6C). Interestingly, none of gem-based measures examined in SMA fibroblasts treated with either VPA or HU approached those values seen in carrier (3814) fibroblasts.
Fig. 6.
Analysis of gems in SMA fibroblasts treated with valproic acid (VPA) or hydroxyurea (HU). SMA patient fibroblasts (3813) were treated for 3 days with either 5 mM VPA, 65.7 µM HU or vehicle (ddH2O). As a control, SMA carrier fibroblasts (3814) were also used. Treated cells were immunostained for SMN with 4F11 and the number of gems per 100 nuclei (A), the number of cells with gems (B) and the number of cells with multiple gems (C) were counted (n=3/group). In agreement with previously published results, VPA and HU increase the number of gems in SMA patient fibroblasts. *p<0.05 (one-way ANOVA, Bonferroni post hoc test).
3.5. Effects of Hydroxyurea on Human SMN in SMN2 Transgenic Mice
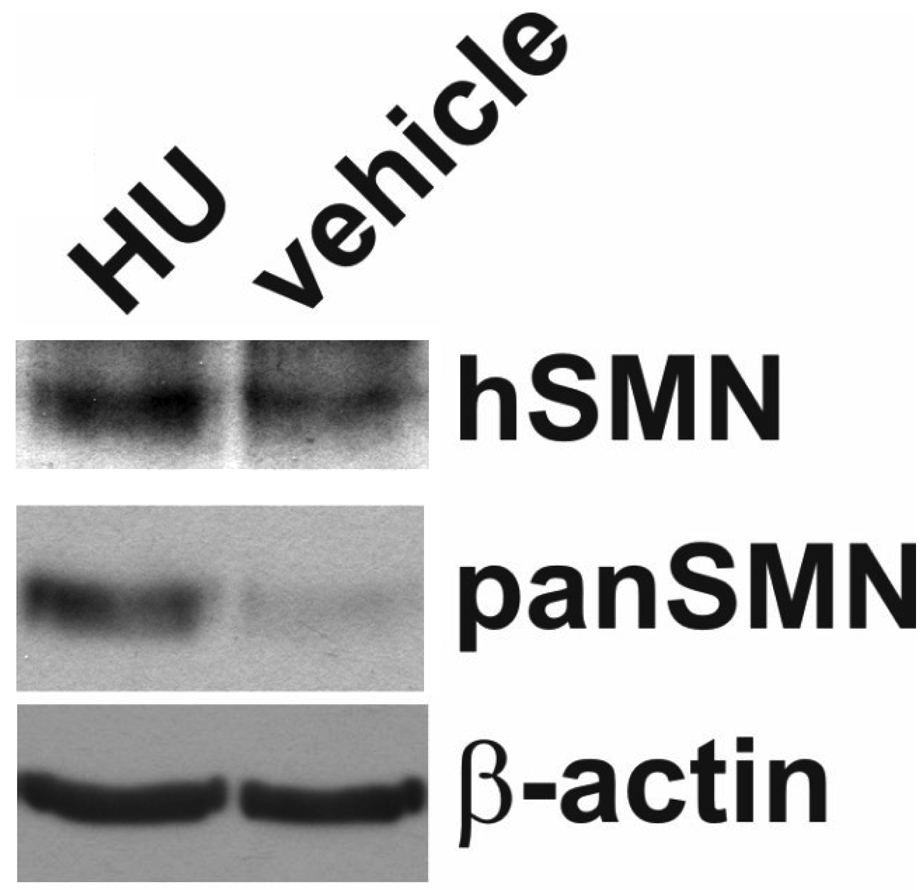
To determine whether 4F11 can be used to detect changes in SMA mice treated with previously identified SMN-inducing agents, severe SMA carrier mice (SMN2+/+;Smn+/−) received single intraperitoneal injections (n=3/group) of hydroxyurea (HU; 5 mg/kg), VPA (200 mg/kg) or vehicle (PBS). Spinal cords were harvested from euthanized mice at 6 hours after injection and examined for SMN protein levels. As anticipated, HU increased SMN protein, as visualized with 4F11, relative to vehicle treated spinal cord extracts (Fig. 7). Similar observations were seen when the blot was reprobed with an anti-SMN antibody that recognized both SMN and Smn (panSMN). Equal loading of protein in each lane was confirmed by the β-actin band. VPA administration, however, did not increase SMN protein levels in the spinal cord in vivo (data not shown).
Fig. 7.
Hydroxyurea (HU) increases human SMN protein in SMN2 transgenic mouse spinal cords. SMN2+/+;Smn+/− transgenic mice received a single intraperitoneal (i.p.) injection of HU (5 mg/kg) or vehicle (PBS). Six hours after injection, spinal cords were harvested and analyzed for human SMN protein using 4F11. Total SMN protein levels were shown using a pan-specific anti-SMN antibody. HU increases SMN2-derived SMN protein levels in the spinal cord. β-actin shows equal loading of protein in each lane.
4. Discussion
We describe in this study the generation and characterization of a human-specific SMN monoclonal antibody, 4F11. This antibody detects SMN in human cells, but not Smn in mouse cells; furthermore, 4F11 can detect SMN protein in various transgenic mouse models that express SMN2. This antibody is, therefore, an excellent tool for examining potential therapeutics in SMA mouse models. Additionally, since 4F11 recognizes exon 4, it should be able to recognize both splice isoforms of SMN.
Screens of cell lines have be instrumental in identifying many compounds that can increase the expression of SMN2 and the inclusion of exon 7 in SMN2 transcripts. These compounds include interferons-β and -γ (Baron-Delage et al., 2000), forskolin (Majumder et al., 2004), ortho-vanadate (Zhang et al., 2001), cantharidin (Novoyatleva et al., 2008), tautomycin (Novoyatleva et al., 2008), aclarubicin (Andreassi et al., 2001), butyrate (BA; (Chang et al., 2001)), 4-phenylbutyrate (4-PBA; (Andreassi et al., 2004)), valproic acid (Brichta et al., 2003; Sumner et al., 2003), hydroxyurea (Grzeschik et al., 2005), aminoglycosides (Mattis et al., 2006; Wolstencroft et al., 2005), resveratrol (Sakla and Lorson, 2008), suberoylanilide hydroxamic acid (SAHA; (Hahnen et al., 2006)) and M344 (Riessland et al., 2006). High-throughput screening of compounds that induce the expression of SMN2 from two independent groups yielded indoprofen (Lunn et al., 2004) and a family of quinazolines (Jarecki et al., 2005) as potential therapeutic agents for SMA.
SMA mouse models have been used to analyze potential SMA therapeutics, not only to validate the induction of SMN but also to examine phenotypic changes (Avila et al., 2007; Butchbach MER, 2005). Since these mice lack native Smn, many of the commercially available SMN antibodies can be used to examine SMN induction. However, SMA pups are very frail and typically represent ~25% of each litter. Now that we have identified a monoclonal antibody that specifically detects SMN, unaffected heterozygous Smn+/− mice carrying the human SMN2 transgene can be used to identify SMN2-inducing agents. This alternative could serve as an important preliminary in vivo screen of SMN induction before examining SMN induction in a relatively severe model of disease. These healthy SMN2 carrier mice can provide the same information about the effectiveness of a SMN-inducing agent as using SMA mice, while being a greater percentage of the litter and having increased viability. We have demonstrated that compounds identified in cultured cells as SMN inducers, such as HU (this study) and cantharidin (Novoyatleva et al., 2008), can increase human SMN protein levels in the spinal cords of SMN2 carrier mice (SMN2+/+;Smn+/−). Thus, therapeutic agents that increase SMN2-derived SMN protein in the spinal cord can be more readily identified using healthy mice expressing SMN2 transgenes and 4F11, a monoclonal antibody that detects SMN protein in the context of native Smn protein.
Acknowledgments
We would like to thank the Campus Microscopy & Imaging Facility (Ohio State University) for providing access to the LSM510 microscope. This project was supported in part by grants from Families of SMA (M.E.R.B.). V.B.M. is supported by the Clinical BioDectives Program (NIH T90 DK070105). This work was also funded by grants from FightSMA (C.L.L.), the Muscular Dystrophy Association (C.L.L.) and the National Institutes of Health (C.L.L, R01 NS41584; R01 HD054413).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Virginia B. Mattis, Email: Vbm7aa@mizzou.edu.
Christian L. Lorson, Email: LorsonC@missouri.edu.
References
- Andreassi C, Angelozzi C, Tiziano FD, Vitali T, De Vincenzi E, Boninsegna A, Villanova M, Bertini E, Pini A, Neri G, Brahe C. Phenylbutyrate increases SMN expression in vitro: relevance for treatment of spinal muscular atrophy. Eur J Hum Genet. 2004;12:59–65. doi: 10.1038/sj.ejhg.5201102. [DOI] [PubMed] [Google Scholar]
- Andreassi C, Jarecki J, Zhou J, Coovert DD, Monani UR, Chen X, Whitney M, Pollok B, Zhang M, Androphy E, Burghes AH. Aclarubicin treatment restores SMN levels to cells derived from type I spinal muscular atrophy patients. Hum Mol Genet. 2001;10:2841–2849. doi: 10.1093/hmg/10.24.2841. [DOI] [PubMed] [Google Scholar]
- Avila AM, Burnett BG, Taye AA, Gabanella F, Knight MA, Hartenstein P, Cizman Z, Di Prospero NA, Pellizzoni L, Fischbeck KH, Sumner CJ. Trichostatin A increases SMN expression and survival in a mouse model of spinal muscular atrophy. J Clin Invest. 2007;117:659–671. doi: 10.1172/JCI29562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron-Delage S, Abadie A, Echaniz-Laguna A, Melki J, Beretta L. Interferons and IRF-1 induce expression of the survival motor neuron (SMN) genes. Mol Med. 2000;6:957–968. [PMC free article] [PubMed] [Google Scholar]
- Brichta L, Hofmann Y, Hahnen E, Siebzehnrubl FA, Raschke H, Blumcke I, Eyupoglu IY, Wirth B. Valproic acid increases the SMN2 protein level: a well-known drug as a potential therapy for spinal muscular atrophy. Hum Mol Genet. 2003;12:2481–2489. doi: 10.1093/hmg/ddg256. [DOI] [PubMed] [Google Scholar]
- AHM. Butchbach MaB. Perspectives on models of spinal muscular atrophy for drug discovery. Drug Disc Today Disease Models. 2004;1:151–156. [Google Scholar]
- Butchbach ME, Edwards JD, Burghes AH. Abnormal motor phenotype in the SMNDelta7 mouse model of spinal muscular atrophy. Neurobiol Dis. 2007;27:207–219. doi: 10.1016/j.nbd.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butchbach ME, Edwards JD, Schussler KR, Burghes AH. A novel method for oral delivery of drug compounds to the neonatal SMNDelta7 mouse model of spinal muscular atrophy. J Neurosci Methods. 2007;161:285–290. doi: 10.1016/j.jneumeth.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butchbach MER, SLaBA SMN-independent amelioration of mouse spinal muscular atrophy by butyrate prodrugs. Society for Neuroscience 2005. 2005 Abstract Viewer/Itinerary Planner. [Google Scholar]
- Chang JG, Hsieh-Li HM, Jong YJ, Wang NM, Tsai CH, Li H. Treatment of spinal muscular atrophy by sodium butyrate. Proc Natl Acad Sci U S A. 2001;98:9808–9813. doi: 10.1073/pnas.171105098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coovert DD, Le TT, McAndrew PE, Strasswimmer J, Crawford TO, Mendell JR, Coulson SE, Androphy EJ, Prior TW, Burghes AH. The survival motor neuron protein in spinal muscular atrophy. Hum Mol Genet. 1997;6:1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- Crawford TO, Pardo CA. The neurobiology of childhood spinal muscular atrophy. Neurobiol Dis. 1996;3:97–110. doi: 10.1006/nbdi.1996.0010. [DOI] [PubMed] [Google Scholar]
- Feldkotter M, Schwarzer V, Wirth R, Wienker TF, Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet. 2002;70:358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzeschik SM, Ganta M, Prior TW, Heavlin WD, Wang CH. Hydroxyurea enhances SMN2 gene expression in spinal muscular atrophy cells. Ann Neurol. 2005;58:194–202. doi: 10.1002/ana.20548. [DOI] [PubMed] [Google Scholar]
- Hahnen E, Eyupoglu IY, Brichta L, Haastert K, Trankle C, Siebzehnrubl FA, Riessland M, Holker I, Claus P, Romstock J, Buslei R, Wirth B, Blumcke I. In vitro and ex vivo evaluation of second-generation histone deacetylase inhibitors for the treatment of spinal muscular atrophy. J Neurochem. 2006;98:193–202. doi: 10.1111/j.1471-4159.2006.03868.x. [DOI] [PubMed] [Google Scholar]
- Jarecki J, Chen X, Bernardino A, Coovert DD, Whitney M, Burghes A, Stack J, Pollok BA. Diverse small-molecule modulators of SMN expression found by high-throughput compound screening: early leads towards a therapeutic for spinal muscular atrophy. Hum Mol Genet. 2005;14:2003–2018. doi: 10.1093/hmg/ddi205. [DOI] [PubMed] [Google Scholar]
- Le TT, Pham LT, Butchbach ME, Zhang HL, Monani UR, Coovert DD, Gavrilina TO, Xing L, Bassell GJ, Burghes AH. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum Mol Genet. 2005;14:845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, Munnich A, Dreyfuss G, Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci U S A. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn MR, Root DE, Martino AM, Flaherty SP, Kelley BP, Coovert DD, Burghes AH, Man NT, Morris GE, Zhou J, Androphy EJ, Sumner CJ, Stockwell BR. Indoprofen upregulates the survival motor neuron protein through a cyclooxygenase-independent mechanism. Chem Biol. 2004;11:1489–1493. doi: 10.1016/j.chembiol.2004.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder S, Varadharaj S, Ghoshal K, Monani U, Burghes AH, Jacob ST. Identification of a novel cyclic AMP-response element (CRE-II) and the role of CREB-1 in the cAMP-induced expression of the survival motor neuron (SMN) gene. J Biol Chem. 2004;279:14803–14811. doi: 10.1074/jbc.M308225200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mattis VB, Rai R, Wang J, Chang CW, Coady T, Lorson CL. Novel aminoglycosides increase SMN levels in spinal muscular atrophy fibroblasts. Hum Genet. 2006 doi: 10.1007/s00439-006-0245-7. [DOI] [PubMed] [Google Scholar]
- McAndrew PE, Parsons DW, Simard LR, Rochette C, Ray PN, Mendell JR, Prior TW, Burghes AH. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am J Hum Genet. 1997;60:1411–1422. doi: 10.1086/515465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monani UR. Spinal muscular atrophy: a deficiency in a ubiquitous protein; a motor neuron-specific disease. Neuron. 2005;48:885–896. doi: 10.1016/j.neuron.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Monani UR, Coovert DD, Burghes AH. Animal models of spinal muscular atrophy. Hum Mol Genet. 2000;9:2451–2457. doi: 10.1093/hmg/9.16.2451. [DOI] [PubMed] [Google Scholar]
- Monani UR, Lorson CL, Parsons DW, Prior TW, Androphya EJ, Burghes AH, McPherson JD. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet. 1999;8:1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- Monani UR, Pastore MT, Gavrilina TO, Jablonka S, Le TT, Andreassi C, DiCocco JM, Lorson C, Androphy EJ, Sendtner M, Podell M, Burghes AH. A transgene carrying an A2G missense mutation in the SMN gene modulates phenotypic severity in mice with severe (type I) spinal muscular atrophy. J Cell Biol. 2003;160:41–52. doi: 10.1083/jcb.200208079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monani UR, Sendtner M, Coovert DD, Parsons DW, Andreassi C, Le TT, Jablonka S, Schrank B, Rossol W, Prior TW, Morris GE, Burghes AH. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(−/−) mice and results in a mouse with spinal muscular atrophy. Hum Mol Genet. 2000;9:333–339. doi: 10.1093/hmg/9.3.333. [DOI] [PubMed] [Google Scholar]
- Munsat TL, Davies KE. International SMA consortium meeting. (26–28 June 1992, Bonn, Germany) Neuromuscul Disord. 1992;2:423–428. doi: 10.1016/s0960-8966(06)80015-5. [DOI] [PubMed] [Google Scholar]
- Novoyatleva T, Heinrich B, Tang Y, Benderska N, Butchbach ME, Lorson CL, Lorson MA, Ben-Dov C, Fehlbaum P, Bracco L, Burghes AH, Bollen M, Stamm S. Protein phosphatase 1 binds to the RNA recognition motif of several splicing factors and regulates alternative pre-mRNA processing. Hum Mol Genet. 2008;17:52–70. doi: 10.1093/hmg/ddm284. [DOI] [PubMed] [Google Scholar]
- Riessland M, Brichta L, Hahnen E, Wirth B. The benzamide M344, a novel histone deacetylase inhibitor, significantly increases SMN2 RNA/protein levels in spinal muscular atrophy cells. Hum Genet. 2006;120:101–110. doi: 10.1007/s00439-006-0186-1. [DOI] [PubMed] [Google Scholar]
- Sakla MS, Lorson CL. Induction of full-length survival motor neuron by polyphenol botanical compounds. Hum Genet. 2008;122:635–643. doi: 10.1007/s00439-007-0441-0. [DOI] [PubMed] [Google Scholar]
- Schmid A, DiDonato CJ. Animal models of spinal muscular atrophy. J Child Neurol. 2007;22:1004–1012. doi: 10.1177/0883073807305667. [DOI] [PubMed] [Google Scholar]
- Schrank B, Gotz R, Gunnersen JM, Ure JM, Toyka KV, Smith AG, Sendtner M. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc Natl Acad Sci U S A. 1997;94:9920–9925. doi: 10.1073/pnas.94.18.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner CJ, Huynh TN, Markowitz JA, Perhac JS, Hill B, Coovert DD, Schussler K, Chen X, Jarecki J, Burghes AH, Taylor JP, Fischbeck KH. Valproic acid increases SMN levels in spinal muscular atrophy patient cells. Ann Neurol. 2003;54:647–654. doi: 10.1002/ana.10743. [DOI] [PubMed] [Google Scholar]
- Thurmond J, Butchbach ME, Palomo M, Pease B, Rao M, Bedell L, Keyvan M, Pai G, Mishra R, Haraldsson M, Andresson T, Bragason G, Thosteinsdottir M, Bjornsson JM, Coovert DD, Burghes AH, Gurney ME, Singh J. Synthesis and biological evaluation of novel 2,4-diaminoquinazoline derivatives as SMN2 promoter activators for the potential treatment of spinal muscular atrophy. J Med Chem. 2008;51:449–469. doi: 10.1021/jm061475p. [DOI] [PubMed] [Google Scholar]
- Wolstencroft EC, Mattis V, Bajer AA, Young PJ, Lorson CL. A non-sequence-specific requirement for SMN protein activity: the role of aminoglycosides in inducing elevated SMN protein levels. Hum Mol Genet. 2005;14:1199–1210. doi: 10.1093/hmg/ddi131. [DOI] [PubMed] [Google Scholar]
- Zhang ML, Lorson CL, Androphy EJ, Zhou J. An in vivo reporter system for measuring increased inclusion of exon 7 in SMN2 mRNA: potential therapy of SMA. Gene Ther. 2001;8:1532–1538. doi: 10.1038/sj.gt.3301550. [DOI] [PubMed] [Google Scholar]