Abstract
Inhibitor-3 is a potent inhibitor of protein phosphatase-1, with an IC50 in the nanomolar range for the inhibition of the dephosphorylation of phosphorylase a. Human Inhibitor-3 possesses a putative protein phosphatase-1 binding motif, 39KKVEW43. We provide direct evidence that this sequence is involved in PP1 interaction by examining the effects of site-directed mutations of Inhibitor-3 on its ability to inhibit protein phosphatase-1. A second interaction site whose deletion led to loss of inhibitory potency was identified between residues 65–77. The existence of two interaction sites is consistent with the high inhibitory potency of Inhibitor-3, and with current models for other inhibitor and targeting proteins that interact with protein phosphatase-1 with high affinity.
Keywords: protein phosphatase-1, Inhibitor-3, PP1 binding motif, interaction domains
Human Inhibitor-3 (Inh3) is a small protein of 126 amino acids that inhibits protein phosphatase-1 (PP1) activity with an IC50 in the nanomolar range [1]. We identified homologs of Inh3 in C. elegans, S. pombe and S. cerevisiae, so that it is likely to have evolutionarily conserved functions [1]. Inh3 is a nuclear protein, and is localized to the nucleoli and centrosomes and to the mitotic apparatus in mitotic cells [2]. Inh3 has a well-defined nuclear localization sequence located at its N-terminus, as well as a separate nucleolar targeting sequence at its C-terminus [2]. Inh3 may also have a role in apoptosis, as it is a caspase-3 substrate, and is rapidly cleaved during the course of actinomycin-mediated apoptosis [3]. Inh3 demonstrates a specificity for association with the PP1α and PP1γ 1 isoforms, and does not interact with PP1β [3]. Significant fractions of the cellular pools of PP1α and PP1γ 1 are associated with Inh3, suggesting that it may play a role in the modulation of their cellular pools as well as their subcellular localization [3]. The cellular functions of Inh3 are still uncertain, but these are likely to be associated with PP1 regulation in nuclear processes and in the regulation of cell division. Inh3 is essential in yeast and is associated with PP1 as well as with the Sds22 protein, a PP1 regulatory protein that is involved in regulation of mitosis and chromosome segregation [4]. Mammalian Sds22, PP1 and Inh3 are able to form a ternary complex in which PP1 bridges Sds22 and Inh3 [5].
The mammalian PP1 catalytic subunit is a protein of ca. 37 kDa in size that has been extensively studied as a phosphorylase phosphatase [6, 7]. The PP1 catalytic subunit is associated with a large number of regulatory or targeting subunits, which allow PP1 to function in the regulation of a diverse range of cellular processes [8]. These non-catalytic subunits function as targeting proteins, which serve to specify the substrates that are acted on by PP1. In addition to the targeting subunits, there exist inhibitor proteins that inhibit the phosphorylase phosphatase activity of PP1 with IC50’s in the nanomolar range. These were originally identified as heat-stable and trypsin-sensitive proteins [9, 10]. The best known of these inhibitor proteins are Inhibitor-1 [11], its neuronal homolog, DARPP-32 [12], and inhibitor-2 [13–15]. These inhibitors generally share the properties of being hydrophilic proteins that exhibit anomalous behavior on SDS-PAGE and gel filtration, suggesting that they have disordered or highly asymmetric structures. Inh3 is similar to these inhibitors in that it migrates on SDS-PAGE as a 23kDa protein, and elutes on gel filtration with an apparent Mr of 55,000, despite an estimated molecular mass of 14 kDa [1].
PP1 possesses a hydrophobic pocket which binds a peptide motif of the type RVXF that is present in most of the known PP1 targeting/regulatory subunits [16]. This binding site is distal from the catalytic site on PP1. Analysis of the peptide specificity of the hydrophobic pocket of PP1 by random peptide library panning has shown that the bulk of the peptides conformed to the motif [K,R]-[R,H]-V-[H,R,S]-[F,W] [17]. The inhibitor proteins, Inhibitor-1, DARPP-32 and inhibitor-2 also interact with the hydrophobic pocket. Current models for their interaction with PP1 are ones in which they bind to PP1 via the hydrophobic pocket as well as to the PP1 active site through a second interaction domain [15, 18, 19]. A further increase in binding interaction may be provided by the connecting sequence between these two sites which could interact with PP1 along grooves in the PP1 surface [15, 19].
We have used site-directed and deletion mutagenesis to determine the regions of Inh3 that are required for the inhibition of PP1 activity. These experiments establish the identity of the sequence 39KKVEW43 as an interaction site for PP1 that conforms to the RVXF motif. In addition, we identified a second region between residues 65–77 as being required for PP1 inhibition. Thus, Inh3 has two domains that are required for its interaction with PP1.
Materials and Methods
Proteins
Recombinant PP1α was expressed in E. coil and purified to near-homogeneity as previously described [20]. Inhibitor-3 was expressed in E. coli and purified to near-homogeneity as previously described [1].
Assay of PP1 Activity
PP1 activity was assayed using [32P]-labeled phosphorylase a as the substrate. Assays for inhibition of PP1 activity were performed in triplicate, essentially as described by Zhang et al. [21]. Data were plotted as percentage inhibition against Inh3 concentration and the IC50 values were determined by graphical extrapolation.
Construction of inhibitor-3 mutants
The mutants were constructed using the Altered Site II in vitro Mutagenesis System (Promega). The correctness of the mutant and deletion mutants cDNAs were confirmed by DNA sequencing. The mutant cDNAs were cloned into the pET3a vector [1]. Examples of the primers that were used are as follows: KKVEW/AAAAA, 5′-CACAGTGTCACTTGTCGCTGCTGCGTTGTTCTCTGGCTTCCG-3′; RAF/AAA, 5′-GGAGCTCTCGCCAGCGGCCGCAGGTTTCTCATA-3′; VEW/AAA, 5′-GTGTCACTTGTCGCTGCTTCCTTTTTCTCTGG-3′; D46A, 5′-ATTGTCACAGTGGCACTTGTCCA-3′; W43Y, ‘5′-AGTGTCACTTGTGTATTCTACCTT-3′; W43F, 5′-AGTGTCACTTGTGAATTCTACCTT-3′; W43A:, 5′-AGTGTCACTTGTCGCTTCTACCTTTTTCTC-3′; W43R, 5′-GTCACTTGTCCATCGTACCTTTTTCTC-3′; E42T, 5′-GTCACTTGTCCATGTTACCTTTTTCTC-3′; E42A, 5′-CACTTGTCCATGCTACCTTTTTCTC-3′; V41A, 5′-CTTGTCCATTCTGCCTTTTTCTC-3′. The deletion mutants were constructed by PCR amplification from the wild type Inh3 in the pET3a vector. The 5′-primers were 5′-G-ATCGATATGGCCGAGGCAGGGGCTGGGC 3′ for Inh3(1–47), Inh3(1–64), and Inh3(1–77), respectively. The 3′ primers were 5′-GATGGGATCCTTAAGTGTCACTTGTCCATTCATCCTTTTTC-3′, 5′-GATCGGATCCTTAATAAATACAGCAGCATTTGGATGAGGGGC-3′, 5′-CTTCCTCCTCCTAACTTTGCGTG for Inh3(1–47), Inh3(1–64), and Inh3(1–77), respectively. The primers for Inh3(38–126) were 5′ –GATCCATATGGAGAAAAAGGTAGAATGGACAAGTGACAC-3′ and 5′-GATCGGATCCTTAGTGCTGCATTGGCCCTGG-3′. The PCR conditions used were 94°C/2 min, 42°C/2 min, and 72°C/2 min, for 30 cycles. The PCR products were purified and ligated into the Ned I/BamH I treated pET3a vector. The correctness of the mutations were confirmed by DNA sequencing.
Results and Discussion
Characterization of the PP1 binding motif of Inh3
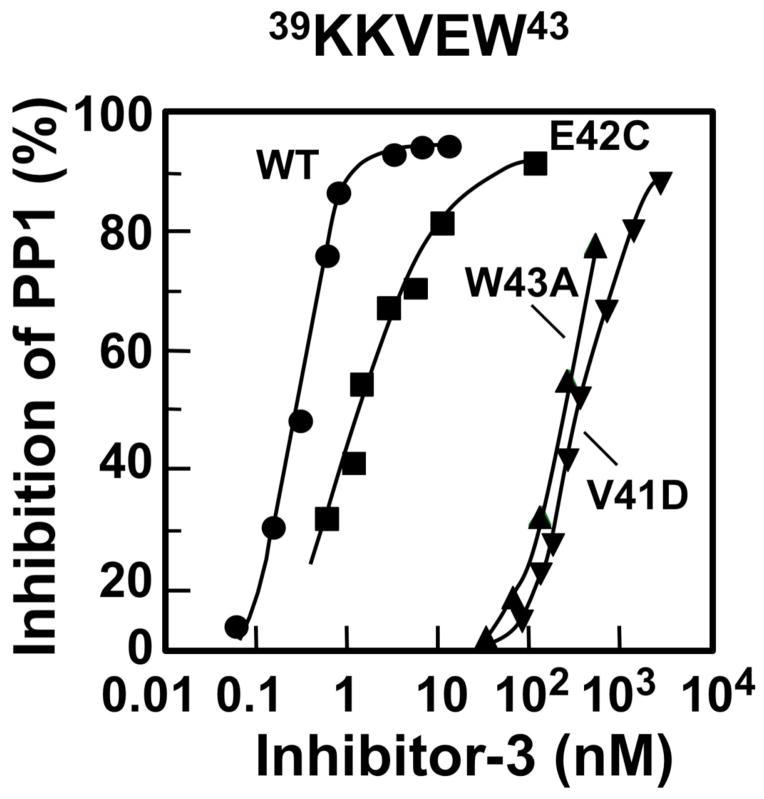
Inspection of the Inh3 sequence shows that it harbors a putative PP1-binding motif, 39KKVEW43 [1]. We examined the inhibitory properties of a series of site-directed mutants of the residues in this motif. Mutants of Inh3 were expressed in E. coli and were purified to homogeneity. They were then tested for their ability to inhibit PP1 using 32P-labeled rabbit muscle phosphorylase a as the substrate. Data were plotted as percentage inhibition against Inh3 concentration on a log scale to obtain the IC50’s, as shown in Fig. 1 for the V41D, W43A and E42C mutants (Fig. 1). The results for the analysis of a series of twelve mutations are shown in Table 1. Mutation of residues 41–43 to alanine (KKAAA) resulted in an increase in IC50 from 0.46 nM to 7.8 μM, i.e., a >104 fold loss in inhibitory potency. This establishes that the KKVEW sequence is a variant of the RVXF type of PP1-binding motif. Further mutations were examined to determine the importance of residues within the motif. The Inh3-V41D and Inh3-W43A mutants had severely reduced abilities to inhibit PP1, as shown by their IC50’s which were 900 and 500 fold greater than that for the wild type Inh3, respectively. Mutation of V41 by a conservative replacement with alanine caused a 50-fold increase in IC50 (Fig. 1, Table 1). Replacement of tryptophan with either phenylalanine or tyrosine was well tolerated as shown by the W43F and W43Y mutants (Table 1). The E42A, E42C, and E42T mutants (Table 1) exhibited only small changes in IC50 (<5 fold). Mutation of the two basic residues (39KK40/NN) resulted in a 47 fold increase in IC50, showing that these residues are important for PP1 interaction (Table 1). Overall, the results are consistent with other studies that show the conservation of the valine and aromatic residues of this motif, as well as with structural studies of PP1 complexes [16, 17].
Figure 1. Effects of mutations of the KKVEW sequence of Inh3 on the inhibition of PP1 activity.
The V41D, E42C, W43A mutants of Inh3 were purified to near-homogeneity and their abilities to inhibit PP1 activity were determined as a function of concentration (Experimental Procedures). Results were plotted as percentage of inhibition of phosphorylase phosphatase activity: wild type Inh3, circles; Inh3-E42C, squares; Inh3-W43A, triangles; Inh3-V41D, inverted triangles.
Table 1.
Inhibition of PP1 by Inh3 Mutantsa
| Mutant | IC50 (nM) | |||
|---|---|---|---|---|
| WT | KKVEW | 0.46 | 1 | |
| 41AAA43 | KKAAA | 7,820 | 17,000 | |
| V41A | KKAEW | 23 | 50 | |
| V41D | KKDEW | 400 | 870 | |
| W43A | KKVEA | 230 | 500 | |
| W43F | KKVEF | 1.5 | 3 | |
| W43Y | KKVEY | 1.5 | 3 | |
| E42A | KKVAW | 1 | 2 | |
| E42C | KKVCW | 2.3 | 5 | |
| E42T | KKVTW | 0.5 | 1 | |
| 42EW43/RF | KKVRF | 0.7 | 2 | |
| 42EW43/HY | KKVHY | 1.4 | 3 | |
| 39KK40/NN | NNVEW | 22 | 47 |
The IC50’s were determined by inhibition of PP1 activity as described in Fig. 1. Mutated residues are shown in bold.
The Inh3 motif conforms to the RVXF motif, but exhibits two uncommon features. The first is the presence of tryptophan rather than phenylalanine as is found in most naturally occurring PP1 binding motifs [16, 17]. Analysis of the sequence specificity of PP1 by panning of a random peptide library has shown that tryptophan occurs as frequently as phenylalanine [17]. We tested two Inh3 mutants containing the VRF and VHY sequences that were identified by peptide library panning [17]. These exhibited inhibitory potencies similar that of wild type Inh3 (Table 1), showing that these motifs were functional in the context of their location in a protein. PP1-binding motifs containing tryptophan (KSVTW or KSVSW) have been found in p99 [22] and in four members of the metabotropic glutamate receptor family [23].
The second feature of note is that Inh3 contains a negatively charged glutamate residue (KKVEW). This is unusual, in that phosphorylation of PP1-binding proteins with the RVSF motif have been shown to lead to their dissociation; structural analysis of the RVSF-PP1 complex indicates that the introduction of a phosphoserine could cause steric hindrance and charge repulsion [16]. Although this suggests that a charged residue such as glutamate might not be observed in a PP1 binding peptide, our data indicate otherwise. Since the side chain of glutamate is more extended than that of serine, it may be able to adopt a conformation that would not interfere with binding.
Identification of a second Inh3 domain that is required for inhibition of PP1
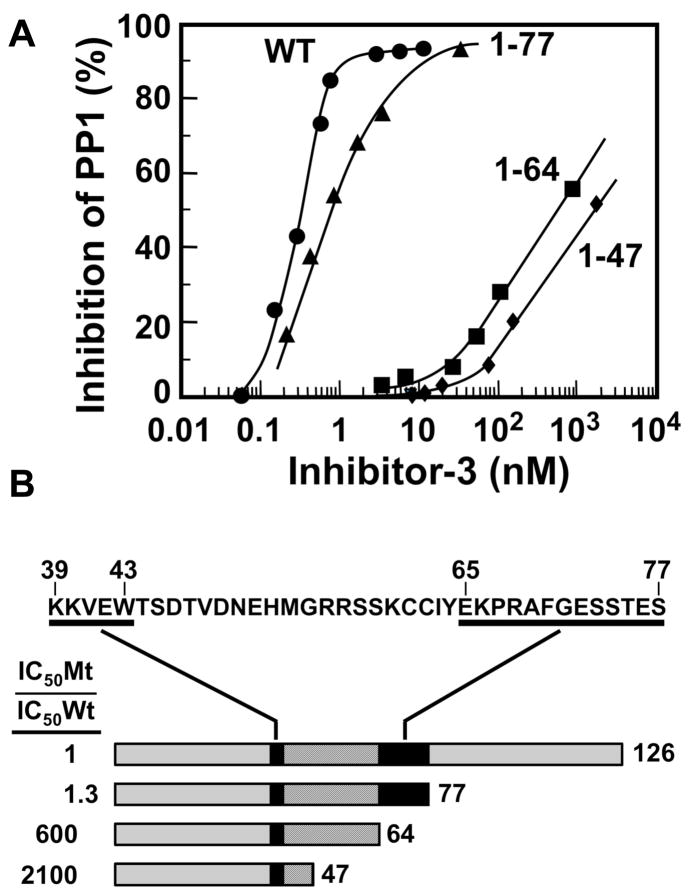
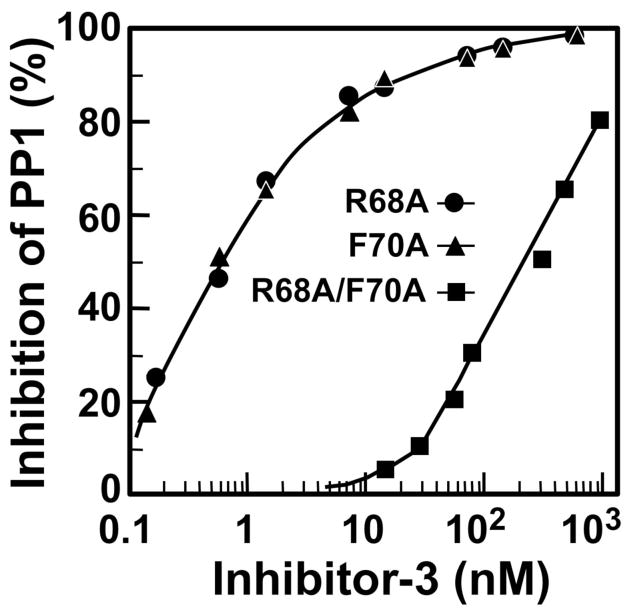
We analyzed a series of deletion mutants of Inh3 by determination of their IC50’s for the inhibition of PP1 (Fig. 2A, Fig. 2B). This analysis focused on the region between residues 39 to 77, which is very highly conserved in the C. elegans and yeast homologs of Inh3 [1]. This analysis revealed the existence of a second PP1 interacting domain (Fig. 2A). Inh3(1–77), in which the C-terminal 49 residues were deleted, inhibited PP1 as effectively as wild type Inh3. The Inh3(1–64) mutant in which an additional thirteen residues were removed exhibited a sharp (604-fold) increase in IC50. A further truncation to the Inh3(1–47) produced a further 3.5 fold increase in IC50. This indicates that the sequence between residues 65 and 77 is also involved in Inh3 inhibition of PP1. Thus, Inh3 possess two domains that are required for interaction with PP1 (Fig. 2B). We mutated residues R68 and F70 to alanines; this did not result in a significant effect on the IC50 (Fig. 3). However, the double mutation R68A/F70A caused a ca. 500 fold increase in IC50 compared to wild type Inh3 (Fig. 2A, Table 1). These results demonstrate that there is a second region between residues 65 and 77 that is required for potent inhibition of PP1 activity. The existence of two interaction sites would greatly increase the binding affinity between Inh3 and PP1, and is consistent with the nanomolar efficiency of inhibition of PP1 by Inh3.
Figure 2. Identification of a second domain required for Inh3 inhibition of PP1 activity.
A. Deletion mutants of Inh3 containing residues 1–77, 1–64 and 1–47 were assayed for their abilities to inhibit PP1 activity as described for Fig. 1. Data for wild type Inh3 are shown as circles; Inh3(1–77), triangles; Inh3(1–64), squares, Inh3(1–47), diamonds. B. B. Model for the interaction domains of Inh3. The sequence of the region between residues 39 and 77 is shown at the top. The underlined sequences are residues 39–43 and 65–77. The shaded bars below represent wild type Inh3, Inh3(1–77), Inh3(1–64) and Inh3(1–47) mutants drawn to approximate scale. The binding regions are shown as the black bars, and the connector regions as the striped bars. The ratios for the increases in IC50, expressed as the IC50 of the mutant divided by the IC50 of wild type Inh3, are shown at the left of each mutant.
Figure 3. Effects of mutation of R68 and F70 on the inhibition of PP1 activity by Inh3.
The inhibition of PP1 activity by the R68A, F70A and the R68A/F70A mutants of Inh3 was determined as described in Fig. 1. Data for Inh3-R68A are shown as circles; Inh3-F70A, triangles; R68A/F70A, squares. The IC50’s for R68A, F70A and R68A/F70A were 0.6, 0.7 and 240 nM respectively.
We propose that Inh3 may conform to current models for the interaction of PP1 with its inhibitor/regulatory proteins, viz. that it does so via the binding of the KKVEW sequence to the hydrophobic groove of PP1, while the region between residues 65–77 harbors an inhibitory domain that binds at or in the vicinity of the active site (Fig. 2B). This model is one that has been proposed for Inhibitor-1, DARPP-32, Inhibitor-2, and the MYPT1 subunit of myosin phosphatase [15, 18, 19]. While the structure of an Inhibitor-1-PP1 complex has not been determined, Barford et al. [18] have modeled a two site binding interaction between Inhibitor-1 and PP1. In this model, binding of the 9KIQF12 sequence of Inhibitor-1 to the PP1 hydrophobic pocket that interacts with RVxF motifs serves as one point of attachment, while phospho-Thr35 interacts with the active site. The connector sequence of 22 residues between the two sites was modeled to take a path along the acidic groove of PP1 [18]. The latter is one of two major grooves that intersect with the active site groove [18]. Thus, it is noteworthy that the distance between the 39KKVEW43 motif of Inh3 to the inhibitory site between residues 65–77 is of similar length. The first structural evidence for a two-site interaction for PP1 came from the structure of the N-terminal 299 residues of the myosin phosphatase targeting subunit MYPT1 complexed to PP1 [19]. The binding of MYP1 is anchored by binding of a RVXF motif to the hydrophobic pocket, and by a region that interacts with the active site region to augment substrate specificity. The latter is located 34 residues N-terminal to the RVXF motif, and the connector sequence follows a path that follows the C-terminal groove of PP1, providing additional protein-protein interactions. It was also noted that several other proteins, NIPP1, neurabin and spinophilin, also have N-terminal sequences that interact with PP1, and are separated from their RVXF motifs by similar spacings of ca. 34 residues; it was suggested that the connector sequences traverse a common route over the surface of PP1 [19]. In the model of Barford et al. [18] for Inhibitor-1, the connector sequence takes an alternative route across the surface of PP1 that accommodates the shorter connector sequence of 22 residues. The recently solved crystal structure of Inhibitor-2 in complex with PP1 has revealed a more complex set of interactions, with three sites of interaction. Nevertheless, Inhibitor-2 interacts with PP1 with the common elements of a site that binds to the hydrophobic pocket by a non-canonical motif, and a second site that inserts into active site and which has an adjacent sequence that interacts with the acid groove of PP1 [15].
Acknowledgments
This work was supported by a grant from the National Institutes of Health (DK18512) to EYCL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References cited
- 1.Zhang J, Zhang L, Zhao S, Lee EYC. Identification and characterization of the human HCG V gene product as a novel inhibitor of protein phosphatase-1. Biochemistry. 1998;37:16728–16734. doi: 10.1021/bi981169g. [DOI] [PubMed] [Google Scholar]
- 2.Huang HS, Pozarowski P, Gao Y, Darzynkiewicz Z, Lee EYC. Protein phosphatase-1 inhibitor-3 is co-localized to the nucleoli and centrosomes with PP1gamma1 and PP1alpha, respectively. Arch Biochem Biophys. 2005;443:33–44. doi: 10.1016/j.abb.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 3.Huang HS, Lee EYC. Protein phosphatase-1 inhibitor-3 is an in vivo target of caspase-3 and participates in the apoptotic response. J Biol Chem. 2008;283:18135–18146. doi: 10.1074/jbc.M709735200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pedelini L, Marquina M, Arino J, Casamayor A, Sanz L, Bollen M, Sanz P, Garcia-Gimeno MA. YPI1 and SDS22 proteins regulate the nuclear localization and function of yeast type 1 phosphatase Glc7. J Biol Chem. 2007;282:3282–3292. doi: 10.1074/jbc.M607171200. [DOI] [PubMed] [Google Scholar]
- 5.Lesage B, Beullens M, Pedelini L, Garcia-Gimeno MA, Waelkens E, Sanz P, Bollen M. A complex of catalytically inactive protein phosphatase-1 sandwiched between Sds22 and inhibitor-3. Biochemistry. 2007;46:8909–8919. doi: 10.1021/bi7003119. [DOI] [PubMed] [Google Scholar]
- 6.Lee EYC, Zhang L, Zhao S, Wei Q, Zhang J, Qi ZQ, Belmonte ER. Phosphorylase phosphatase: new horizons for an old enzyme. Front Biosci. 1999;4:D270–85. doi: 10.2741/lee. [DOI] [PubMed] [Google Scholar]
- 7.Cohen P. The structure and regulation of protein phosphatases. Annu Rev Biochem. 1989;58:453–508. doi: 10.1146/annurev.bi.58.070189.002321. [DOI] [PubMed] [Google Scholar]
- 8.Ceulemans H, Bollen M. Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol Rev. 2004;84:1–39. doi: 10.1152/physrev.00013.2003. [DOI] [PubMed] [Google Scholar]
- 9.Brandt H, Killilea SD, Lee EYC. Activation of phosphorylase phosphatase by a novel procedure: evidence for a regulatory mechanism involving the release of a catalytic subunit from enxyme-inhibitor complex(es) of higher molecular weight. Biochem Biophys Res Commun. 1974;61:598–604. doi: 10.1016/0006-291x(74)90999-1. [DOI] [PubMed] [Google Scholar]
- 10.Brandt H, Capulong ZL, Lee EYC. Purification and properties of rabbit liver phosphorylase phosphatase. J Biol Chem. 1975;250:8038–8044. [PubMed] [Google Scholar]
- 11.Connor JH, Quan H, Oliver C, Shenolikar S. Inhibitor-1, a regulator of protein phosphatase 1 function. Methods Mol Biol. 1998;93:41–58. doi: 10.1385/0-89603-468-2:41. [DOI] [PubMed] [Google Scholar]
- 12.Svenningsson P, Nishi A, Fisone G, Girault JA, Nairn AC, Greengard P. DARPP-32: an integrator of neurotransmission. Annu Rev Pharmacol Toxicol. 2004;44:269–296. doi: 10.1146/annurev.pharmtox.44.101802.121415. [DOI] [PubMed] [Google Scholar]
- 13.Bollen M, Stalmans W. The structure, role, and regulation of type 1 protein phosphatases. Crit Rev Biochem Mol Biol. 1992;27:227–281. doi: 10.3109/10409239209082564. [DOI] [PubMed] [Google Scholar]
- 14.Eto M, Leach C, Tountas NA, Brautigan DL. Phosphoprotein inhibitors of protein phosphatase-1. Methods Enzymol. 2003;366:243–260. doi: 10.1016/s0076-6879(03)66019-2. [DOI] [PubMed] [Google Scholar]
- 15.Hurley TD, Yang J, Zhang L, Goodwin KD, Zou Q, Cortese M, Dunker AK, Depaoli-Roach AA. Structural basis for regulation of protein phosphatase 1 by inhibitor-2. J Biol Chem. 2007;282:28874–28883. doi: 10.1074/jbc.M703472200. [DOI] [PubMed] [Google Scholar]
- 16.Egloff MP, Johnson DF, Moorhead G, Cohen PT, Cohen P, Barford D. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. Embo J. 1997;16:1876–1887. doi: 10.1093/emboj/16.8.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao S, Lee EYC. A protein phosphatase-1-binding motif identified by the panning of a random peptide display library. J Biol Chem. 1997;272:28368–28372. doi: 10.1074/jbc.272.45.28368. [DOI] [PubMed] [Google Scholar]
- 18.Barford D, Das AK, Egloff MP. The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu Rev Biophys Biomol Struct. 1998;27:133–164. doi: 10.1146/annurev.biophys.27.1.133. [DOI] [PubMed] [Google Scholar]
- 19.Terrak M, Kerff F, Langsetmo K, Tao T, Dominguez R. Structural basis of protein phosphatase 1 regulation. Nature. 2004;429:780–784. doi: 10.1038/nature02582. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Z, Bai G, Shima M, Zhao S, Nagao M, Lee EYC. Expression and characterization of rat protein phosphatases-1 alpha, -1 gamma 1, -1 gamma 2, and -1 delta. Arch Biochem Biophys. 1993;303:402–406. doi: 10.1006/abbi.1993.1301. [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, Zhang Z, Long F, Lee EYC. Tyrosine-272 is involved in the inhibition of protein phosphatase-1 by multiple toxins. Biochemistry. 1996;35:1606–1611. doi: 10.1021/bi9521396. [DOI] [PubMed] [Google Scholar]
- 22.Kreivi JP, Trinkle-Mulcahy L, Lyon CE, Morrice NA, Cohen P, Lamond AI. Purification and characterisation of p99, a nuclear modulator of protein phosphatase 1 activity. FEBS Lett. 1997;420:57–62. doi: 10.1016/s0014-5793(97)01485-3. [DOI] [PubMed] [Google Scholar]
- 23.Croci C, Sticht H, Brandstatter JH, Enz R. Group I metabotropic glutamate receptors bind to protein phosphatase 1C. Mapping and modeling of interacting sequences. J Biol Chem. 2003;278:50682–50690. doi: 10.1074/jbc.M305764200. [DOI] [PubMed] [Google Scholar]