Abstract
Background
Toxin complex (Tc) proteins termed TcaABC, TcdAB, and TccABC with insecticidal activity are present in a variety of bacteria including the yersiniae.
Results
The tc gene sequences of thirteen Yersinia strains were compared, revealing a high degree of gene order conservation, but also remarkable differences with respect to pseudogenes, sequence variability and gene duplications. Outside the tc pathogenicity island (tc-PAIYe) of Y. enterocolitica strain W22703, a pseudogene (tccC2'/3') encoding proteins with homology to TccC and similarity to tyrosine phosphatases at its C-terminus was identified. PCR analysis revealed the presence of the tc-PAIYe and of tccC2'/3'-homologues in all biotype 2–5 strains tested, and their absence in most representatives of biotypes 1A and 1B. Phylogenetic analysis of 39 TccC sequences indicates the presence of the tc-PAIYe in an ancestor of Yersinia. Oral uptake experiments with Manduca sexta revealed a higher larvae lethality of Yersinia strains harbouring the tc-PAIYe in comparison to strains lacking this island. Following subcutaneous infection of Galleria mellonella larvae with five non-human pathogenic Yersinia spp. and four Y. enterocolitica strains, we observed a remarkable variability of their insecticidal activity ranging from 20% (Y. kristensenii) to 90% (Y. enterocolitica strain 2594) dead larvae after five days. Strain W22703 and its tcaA deletion mutant did not exhibit a significantly different toxicity towards G. mellonella. These data confirm a role of TcaA upon oral uptake only, and suggest the presence of further insecticidal determinants in Yersinia strains formerly unknown to kill insects.
Conclusion
This study investigated the tc gene distribution among yersiniae and the phylogenetic relationship between TccC proteins, thus contributing novel aspects to the current discussion about the evolution of insecticidal toxins in the genus Yersinia. The toxic potential of several Yersinia spp. towards M. sexta and G. mellonella demonstrated here for the first time points to insects as a natural reservoir for yersiniae.
Background
The toxin complex (Tc) proteins whose insecticidal potential resembles that of the Bacillus thuringiensis Bt-toxin were first purified from Photorhabdus luminescens which lives in symbiosis with nematodes [1]. They have also been identified in other insect-parasitizing bacteria such as Serratia entomophila, Xenorhabdus nematophilus, or Pseudomonas entomophila [2,3]. Homologous insecticidal toxin genes are present in most genomes of Yersinia strains sequenced so far, including Y. mollaretii, seven Yersinia pestis strains and three Y. pseudotuberculosis strains. They have also been found in Y. frederiksenii and in two Y. enterocolitica strains, T83 and W22703, for which a genome sequence is not yet available [4-6]. However, tc genes are absent in Y. bercovieri and in Y. enterocolitica strain 8081 [7]. Interestingly, Tc proteins of three Serratia species and of Y. frederiksenii are plasmid-encoded, indicating that these sepABC-like genes are part of a horizontally mobile region [4].
Little is known about the biological role of the tc genes in Yersinia spp. The genes of the tc operons have been classified into three types according to their homology, namely tcdA/tcaAB/tccAB (type [A]), tcdB/tcaC (type [B]), and tccC (type [C]) [8]. Tc proteins have recently been shown to be secreted in a type III-dependent manner in Y. pestis [9]. Type [A] and [B] Tc proteins are presumably toxins directed against invertebrate and mammalian gut cells, and the variability in terms of Tc composition and Tc sequences may be due to insect- and tissue-specific activity [8,10]. A role of the Tc proteins from Y. enterooclitica strain T83, Y. pseudotuberculosis strain IP32953 and Y. pestis KIM in mice gut colonization and in the actin cytoskeleton rearrangement of human gut cells and mouse fibroblast cells, respectively, has been reported [5,8,11]. The function of TccC remains unknown, but it has been suggested that TccC homologs could contribute to stable biofilm formation in fleas or combatting yet unknown antibacterial effectors in fleas [12], or that they act as universal activator of, or chaperons for, different toxin proteins [13].
Y. enterocolitica was the first member of the Yersinia genus for which insecticidal activity has been experimentally demonstrated, and tcaA encoding a subunit of the toxin complex was identified to be necessary for this activity [6]. The transcription of tcaA in Y. enterocolitica is completely repressed at 37°C, but strongly induced at lower temperatures with a maximum at approximately 10°C to 15°C. In contrast to Y. enterocolitica W22703, tcaABC expression in Y. pseudotuberculosis strain IP32953 was observed at 15°C and at 37°C [14]. Upregulation of tcaA and tcaB, but not tccC, upon temperature shift from 37°C to 26°C have been shown in two Y. pestis strains [15,16]. The IP32953 Tc proteins are toxic against M. sexta larvae when expressed heterologously in E. coli [14]. Temperature-independent, but weak oral toxicity of several Y. pseudotuberculosis to this tobacco hornworm has been reported. Y. pseudotuberculosis, unlike Y. pestis, causes acute oral toxicity to fleas [12]. However, when the tcaAB gene pair from Y. pseudotuberculosis was heterologously expressed in E. coli, the lysates did not cause excess mortality in fleas, and a Y. pseudotuberculosis mutant deleted of the tc genes remained toxic toward the arthropod [8]. This is in line with the finding that two Y. enterocolitica strains containing a tcdB-tccC gene pair (strain CS080) or lacking any tc-like genes (strain 8081) were equally toxic to fleas [12].
The insecticidal potential of a variety of Yersinia spp. has not been tested in an insect infection assay, and the correlation of virulence to the presence or absence of tc operons in yersiniae is unknown. The phylogenetic relationship of the insecticidal toxins is not well understood. Here, we report a genome-based comparison of the tc genes in Y. enterocolitica strain W22703, the phylogenetic analysis of tccC genes in Yersinia species, the tc-PAIYe distribution among six biotypes, and the insecticidal activity of Yersinia spp. towards two model organisms, the greater wax moth G. mellonella and the tobacco hornworm M. sexta.
Results
The tc-PAIYe in Yersinia spp
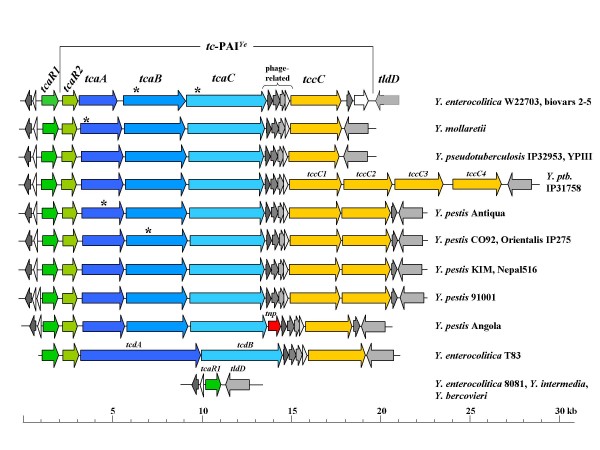
Chromosomal loci encoding Tc proteins have first been sequenced from P. luminescens, S. entomophila and X. nematophilus. In the past few years, the genome sequences of several Yersinia strains became available. Most of them, namely seven Y. pestis strains, three Y. pseudotuberculosis strains and Y. mollaretii, carry a common tc gene cluster termed the tc pathogenicity island of yersiniae (tc-PAIYe). Further DNA fragments encoding insecticidal toxins were detected in the genomes of two Y. enterocolitica strains W22703 and T83 [5,6], and on a plasmid of Y. frederiksenii strain 49 [4]. A comparison of the chromosomal loci of yersiniae containing tc homologues is shown in Fig. 1. The length of the sequences ranges from ~17 kb to ~26 kb. This variation is mainly due to the presence of one to four tccC (1-4) homologues. In all cases, tc-PAIsYe are inserted into an equivalent location with respect to the common Yersinia genome backbone [8], namely between the genes encoding a putative DNA-binding transcriptional regulator and a putative DNA gyrase modulator. These genes are YE3797 (tcaR1) and YE3798 (tldD) with respect to the chromosomal sequence of Y. enterocolitica strain 8081. The overall organisation is similar for all strains, including a second gene, tcaR2 that encodes a homolog of LysR-like regulators, followed by genes belonging to the homology types tcaAB/tcdA, tcaC/tcdB, and tccC. tccC is separated from tcaC/tcdB by two small phage-related genes and two ORFs of unknown function. According to the classification of Waterfield et al. [8] described above, all tc genes belong to tcd operons. Beyond these similarities, several differences were revealed by homology analysis and re-annotation. Y. enterocolitica strain W22703 is characterised by a 2034 bp sequence located between tccC and YE3798 (tldD) harbouring two ORFs of unknown function, one of which is also present in the Y. pestis genomes. A transposon-related sequence (tnp) was identified in front of the first phage-related gene of Y. pestis Angola. Frameshifts (asterisks in Fig. 1) are present in tcaC of strain W22703, in tcaA of strain Antiqua and of Y. mollaretii, and in tcaB of strains W22703, Orientalis IP275 and CO92.
Figure 1.
Comparison of the tc-PAIYe in Yersiniae. Three homology groups are depicted, namely tcaAB/tcdA, tcaC/tcdB, and tccC. tcdA/tcdB homologous are present only in Y. enterocolitica strain T83. tcaR1 (left) encoding a regulator and tldD encoding a putative DNA gyrase modulator (right, checkered) mark the island insertion site common for all Yersinia strains that harbour tc homologues. Identically coloured arrows mark homologous genes. A transposase-like gene (tnp) is present in the genome of Y. pestis Angola (black arrow). The overall gene organisation is similar for all strains harbouring insecticidal determinants, but differences with respect to gene homology, hypothetical ORFs, the presence of transposase-like elements and the number of tccC genes are also visible. Gene lengths and intergenic regions are in scale. Asterisks mark frameshifts. With the exception of tcaC, all frameshifts result in two ORFs. Y. ptb., Y. pseudotuberculosis.
Homologues of tccC located outside tc-PAIYe
By screening a Tn5 luxCDABE reporter library of strain W22703 for genes induced upon low-temperature [17], we identified a transposon insertion located outside the tc-PAIYe. A 4,595 bp sequence encompassing the transposon insertion site was derived, revealing two strain-specific ORFs termed tccC2' (1083 bp) and tccC3' (1680 bp) due to homologies to other Yersinia tccC loci. Obviously, a frameshift had splitted a tccC homologue into two ORFs. Exploring the available genome sequences of Yersinia strains, two additional tccC genes located outside the tc-PAIYe were identified in Y. pestis strains Antiqua, CO92, Nepal516, Orientalis IP275 and 91001, and in Y. pseudotuberculosis IP32953, and one further tccC gene in Y. pestis strains KIM and Angola, and in Y. pseudotuberculosis IP31758. A truncated tccC2' gene with a 1953 bp deletion in comparison to tccC2 of strain IP32953 is present in the genome of Y. pseudotuberculosis YPIII. tccC2' and tccC3' of strain W22703 are located between two genes encoding a lipid A biosynthesis lauroyl acyltransferase, HtrB (YE1612), and a putative membrane protein (YE1611) of strain 8081. In contrast, the non-clustered tccC loci of all other strains are inserted into two equivalent locations on the common Yersinia backbone. These locations are exemplified as between the Y. pestis CO92 genes YPO2379/YPO2381 encoding an N-ethylmaleimide reductase and a lactoylglutathione lyase, and between YPO2311/YPO2313 coding for the insertion element IS1541 and a hypothetical protein. In addition, domain structure of TccC with similarity to a protein tyrosine phosphatase of undefined specificity was identified in the sequence of Y. pestis CO92 TccC2 and, albeit with lower probability, of W22703.
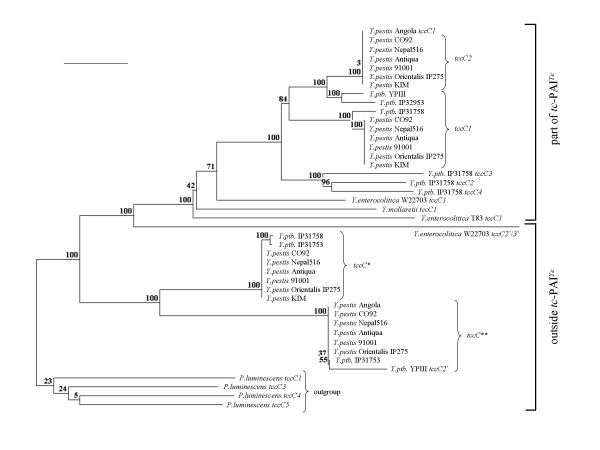
A total of 39 TccC amino acid sequences, derived from yersiniae genes either located within or outside the tc-PAIYe, was compared here by a ClustlX alignment. The cladogram of the yersiniae TccC proteins tc-PAIYe exhibits a significant sequence variability between island- and non island-encoded TccCs (Fig. 2). The TccC proteins expressed from loci outside the tc-PAIYe compose two groups that are characterised by the two common insertion sites as described above. The only exception is TccC2'/3' of strain W22703 that is more closely related to the tc-PAIYe-encoded TccC proteins. TccC sequences derived from genes located within the tc-PAIYe show a more complex relationship. One group of TccCs represents TccC1 proteins, a second group TccC2 proteins, indicating a highly conserved linear order of tccC1 and tccC2 genes in the yersiniae genomes. Interestingly, TccC1 of Y. pestis Angola encoded by a tccC gene located nearby a transposase-like gene (Fig. 1) appears to be more closely related to TccC2 proteins. Two further sublines that show a higher sequence variability are represented by TccC2-4 of Y. pseudotuberculosis IP31758, and by TccC1 of Y. mollaretii and Y. enterocolitica strains W22703 and T83.
Figure 2.
Cladogram based on 39 TccC amino acid sequence data. Four TccC sequences of P. luminescens subspecies laumondii strain TT01 served as outgroup. The phylogenetic analysis was performed with the neighbour-joining method and calculated using the two parameter model of Kimura [31]. Values on each branch indicate the occurrence (%) of the branching order in 500 bootstrapped trees. *insertion site between genes coding for the insertion element IS1541 and a hypothetical protein, **insertion site between genes encoding a N-ethylmaleimide reductase and a lactoylglutathione lyase. The frameshift of tccC in strain W22703 was not considered here; hence, one coherent amino acid sequence was used for the alignment. Bar represents 0.1% sequence divergence. Y. ptb., Y. pseudotuberculosis.
Presence of tc genes in Y. enterocolitica strains
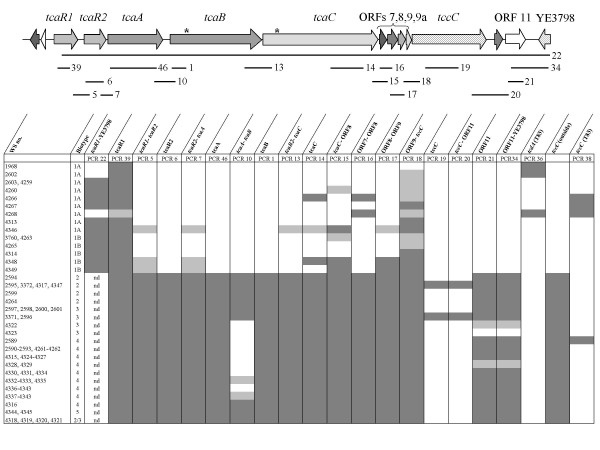
In a previous study, we had studied a restricted number of Y. enterocolitica strains by PCR and Southern Blot for the presence of tcaA, tcaB, and tcaC in representatives of five biotypes [6]. Here, we performed a more thorough investigation for the presence, absence, variability and genetic organisation of tc genes in a total of 68 Y. enterocolitica strains belonging to six biotypes. Chromosomal DNA of all strains was successfully subjected to PCR with oligonucleotides specific for 16S rDNA as control. A series of 22 PCRs designed to amplify intragenic and gene-overlapping fragments was performed (Fig. 3). DNA of strains W22703 and 8081 served as a positive and a negative control. First, tcaR1 was confirmed as part of the common yersiniae genomic backbone. The second regulatory gene, tcaR2, is present in all strains of biotypes 2–5, but absent in biotypes 1A and 1B. Two different primer combinations failed to amplify tcaA-specific fragments from DNA of several strains. Sequencing of four fragments revealed mismatches, and conserved tcaA regions were therefore used to design more appropriate oligonucleotides that showed the presence of tcaA in biotypes 2–5, but not 1A and 1B (PCR 46). Similar patterns were obtained following amplification of intragenic tcaB and tcaC fragments (PCRs 1 and 14), with the exception that tcaC revealed also to be present in one biotype 1A and one biotype 1B strain. Using oligonucleotides homologous to the 3'-end of tcaA and the 5'-end of tcaB, the operon organisation was confirmed for most, but not all, biotype 2–5 strains (PCR 10). Negative results in case of PCR 10 predominantly with DNA of biotype 3 strains might be due to a frameshift in tcaB similar to that in strain W22703, resulting in an unfunctional and possibly degenerated 5'-end of tcaB. Remnants of the phage-related gene cassette which are present in all biotype 2–5 strains could also be amplified from DNA of biotype 1A and 1B strains (ORFs 7,8,9,9a and PCRs 15–18). The results of PCRs 18–20 indicate a highly conserved 5'-region and an otherwise variable tccC sequence. We therefore performed a PCR with primers specific for tccC of strain T83 and found the respective fragment also in three biotype 1 strains and in one biotype 4 strain. To investigate whether tc genes homologous to that of biotype 1A strain T83 are present in other Y. enterocolitica strains, PCRs 36 and 37 (data not shown) with oligonucleotides specific for tcdA of strain T83 were performed. Fragments of the expected length were obtained using DNA of four biotype 1A strains, a finding that correlates well with the negative result of PCR 22 for fragments overlapping the insertion site of the tc-PAIYe. Finally, ORF11 is present in all but one biotype 2–5 strains, but absent in most other Yersinia spp. (Fig. 1). Taken together, the tc operons of strain W22703 are present in all biotype 2–5 representatives tested here. A tc gene cluster homologous to that of strain T83 is probably present in biotype 1A strains 1968, 2602 and 4268. In comparison to all other tc genes, tccC exhibits a higher sequence variability (see chapter above).
Figure 3.
Distribution of the tc-PAIYe in Y. enterocolitica strains. Strains investigated are described in Table 3. The lines below the tc-PAIYe are in scale and mark the fragments amplified by PCR. The PCR numbers correspond to those indicated in Additional file 1. Asterisks mark frameshifts in tcaB and tcaC. Dark grey: fragment amplification, light grey: biased results, white: no amplification; nd, not defined, e.g. no PCR was performed. See text for further details.
Oral infection of M. sexta
Some Yersinia spp. besides the three human pathogenic species have not yet been investigated for their insecticidal activity. Oral infection of first-instar M. sexta neonates was performed by soaking small blocks of an artificial diet with 50 μl aliquots of overnight cultures grown at 15°C (Yersinia spp.) and 37°C (DH5α). Yersinia strains applied were Y. mollaretii, Y. aldovae, Y. ruckeri, and four Y. enterocolitica strains (W22703, W22703-tcaA::Tn5lux, 2594, 4466). Their toxicity towards M. sexta as percentage of dead larvae five days after infection is shown in Table 1. The experimental setting hampered the reproducibility of the assay, resulting in larger standard deviations in comparison to the G. mellonella infection model (see below). The strains Y. enterocolitica 2594 and Y. mollaretii possessing the tc-PAIYe showed the highest killing rates. Strains without tc genes such as Y. ruckeri and Y. aldovae were less toxic towards M. sexta. The only exception was the tc-negative Y. enterocolitica strain 4466 with a lethality rate of 42%, suggesting the presence of further gut-active insecticidal determinants in its genome. The toxicity of strain W22703 (30% dead larvae) in comparison to that of the tcaA knockout mutant W22703-tcaA::Tn5lux (10%) confirms our recent experiments using concentrated protein extracts instead of viable cells. Taken together, the average killing rates of Table 1 show that the tc genes play an important role in the insecticidal activity of Yersinia spp. upon oral uptake.
Table 1.
Oral infection of M. sexta
| strain | tc-PAIYe | total number | dead | alive | dead [%] ± sd |
| Y. enterocolitica 2594 | present2) | 18 | 15 | 3 | 83 ± 3 |
| Y. mollaretii | present1) | 25 | 12 | 13 | 48 ± 9 |
| Y. enterocolitica 4466 | absent2) | 36 | 15 | 21 | 42 ± 15 |
| W22703 | present2) | 33 | 10 | 23 | 30 ± 14 |
| Y. ruckeri | absent2) | 19 | 4 | 15 | 21 ± 26 |
| W22703-tcaA::Tn5 lux | present, but tcaA knockout | 21 | 2 | 19 | 10 ± 11 |
| Y. aldovae | absent2) | 23 | 2 | 21 | 9 ± 2 |
| control | |||||
| DH5α | absent1) | 21 | 1 | 20 | 5 ± 7 |
Yersinia strains were orally fed to first-instar M. sexta neonates. Each experiment was independently performed at least three times with a minimum of six larvae, and the average dead rates are shown. Y. enterocolitica W22703: biotype 2, serotype O:9; Y. enterocolitica 2594: biotype 2, serotype O:9; Y. enterocolitica 4466: biotype 1B, serotype O:21. 1) according to the genome sequence, 2) according to PCR analysis performed in this study.
Subcutaneous infection of G. mellonella
For an additional bioassay, we chose the model organism G. mellonella that is also used to determine the toxicity of P. luminescens and X. nematophilus. We infected larvae of this greater wax moth subcutaneously with 1:10 and 1:100 diluted aliquots of Y. mollaretii, Y. kristensenii, Y. bercovieri, Y. aldovae, Y. ruckeri, and four Y. enterocolitica strains (W22703, W22703-tcaA::Tn5lux, 2594, 4466) incubated at 15°C until they reached stationary phase. After five days, only 10% of the larvae had survived an infection of strains 2594 or 4466. In total, four strains showed high killing rates of 81–90% (Table 2). To determine whether the toxicity towards the insect larvae depends on the presence of Tc proteins, we analysed chromosomal DNA of Y. ruckeri, Y. aldovae, Y. kristensenii, and Y. enterocolitica strain 4466 with primers described in Additional file 1 and shown in Fig. 3. No fragments were obtained with primers specific to tcaR2 (PCR 6), tcaA (PCR 46), tcaB (PCR 1), tcaB-tcaC (PCR 13), tcaC (PCR 14), ORF11-YE3798 (PCR 34), and tccC of Y. enterocolitica strain T83 (PCR 38). As a control, amplification of a tcaR1 fragment which is part of the common Yersinia backbone sequence (PCR 39) succeeded with DNA of Y. aldovae and Y. enterocolitica strain 4466. Therefore, we conclude that the tc-PAIYe is absent in these strains (Table 2). Thus, only two of the strains exhibiting a killing rate of at least 81%, namely Y. mollaretii, and the biotype 2 strain 2594, harbour the tc-PAIYe, while it is absent in Y. enterocolitica strain 4466 and Y. bercovieri. On the other hand, Y. ruckeri and Y. aldovae killed G. mellonella larvae in approximately 40–50% of all infection experiments, and the insecticidal activity of Y. kristensenii is only slightly higher than that of the control strain S. enterica serovar Typhimurium. These three strains do not carry tc genes, too. No significant difference (p = 0.21) in the insecticidal activity between W22703 and its tcaA knockout mutant could be observed. Plasmid-encoded sep-like genes of Y. frederiksenii isolate 49 could also not be detected using primers specific for tcYF1, tcYF2 or tcYF3 [see Additional file 1] in bacterial lysates of Y. enterocolitica strain 4466, Y. ruckeri, Y. aldovae, or Y. kristensenii. According to BLAST analysis, homologous of type III secretion system (T3SS) genes are present in the genomes of W22703 and the other strains from Table 2 sequenced so far, but the role of T3SS and insect virulence as shown for Y. pestis remains to be determined for these strains. Taken together, this data strongly suggests that insecticidal activity of Yersinia spp. towards G. mellonella upon subcutaneous infection is not caused by Tc proteins, and that yet unknown determinants contribute to the insecticidal activity of Yersinia strains towards the insect larvae. The similar lethality of strain W22703 and its tcaA-negative mutant not only confirms the assumption of Tc-independent killing, but also of a gut-related TcaA activity following oral infection as demonstrated above and recently suggested [6,8,10].
Table 2.
Subcutaneous infection of G. mellonella
| 1:10 | 1:100 | total | |||||||
| strain | tc-PAIYe | total number | dead | alive | dead | alive | dead | alive | dead [%] ± sd |
| Y. enterocolitica 2594 | present2) | 79 | 36 | 4 | 34 | 5 | 70 | 9 | 90 ± 9 |
| Y. enterocolitica 4466 | absent2) | 96 | 47 | 1 | 34 | 14 | 81 | 15 | 88 ± 11 |
| Y. mollaretii | present1) | 64 | 30 | 2 | 24 | 8 | 54 | 10 | 84 ± 5 |
| Y. bercovieri | absent1) | 64 | 28 | 8 | 23 | 5 | 51 | 13 | 81 ± 11 |
| Y. ruckeri | absent2) | 85 | 33 | 11 | 20 | 21 | 53 | 32 | 53 ± 21 |
| W22703-tcaA::Tn5 lux | present, but tcaA knockout | 93 | 30 | 25 | 18 | 20 | 48 | 45 | 51 ± 13 |
| W22703 | present2) | 114 | 32 | 25 | 20 | 37 | 52 | 62 | 41 ± 17 |
| Y. aldovae | absent2) | 68 | 23 | 13 | 4 | 28 | 27 | 41 | 41 ± 6 |
| Y. kristensenii | absent2) | 88 | 19 | 27 | 1 | 41 | 20 | 68 | 20 ± 12 |
| controls | |||||||||
| S. typhimurium | absent1) | 88 | 10 | 33 | 10 | 35 | 20 | 68 | 18 ± 9 |
| DH5α | absent1) | 63 | 5 | 34 | 2 | 22 | 7 | 56 | 13 ± 6 |
| LB | 64 | 3 | 61 | 5 ± 0 | |||||
G. mellonella larvae were subcutaneously infected with 5 μl of diluted overnight cultures. Strain W22703 and its tcaA knockout mutant confer similar toxicity towards the larvae (depicted in italics). Each experiment was independently performed at least three times, and the average dead rates are shown. Y. enterocolitica W22703: biotype 2, serotype O:9; Y. enterocolitica 2594: biotype 2, serotype O:9; Y. enterocolitica 4466: biotype 1B, serotype O:21. 1) according to the genome sequence, 2) according to PCR analysis performed in this study.
Discussion
Two basic methods have been used here to determine the insecticidal potential of Yersinia spp., namely the oral application of viable cells and the subcutaneous injection of protein extracts or living bacterial cells. Upon oral application of W22703 and W22703-tcaA::Tn5lux protein extract to M. sexta larvae, we could recently demonstrate the role of TcaA in Y. enterocolitica toxicity towards insects [6]. Further five Yersinia strains were tested here for the first time with respect to their oral toxicity in the M. sexta model (Table 1). The presence of the tc-PAIYe correlates with a higher toxicity of yersiniae towards larvae of the tobacco hornworm, while strains such as Y. ruckeri or Y. aldovae lacking the tc genes are less insecticidal in this assay. The variable insecticidal activity of strains with tc genes might be the result of sequence variations, or the presence of further insecticidal components. Due to a higher toxin concentration, the feeding of protein extracts led to higher lethality rates using strain W22703 [8]. In contrast, subcutaneous infection of G. mellonella does not result in a significantly different toxicity of these strains (Table 2). In comparison to P. luminescens that causes death of G. mellonella larvae within 24 hours following injection of several thousand cells [18], approximately 5 × 105 Y. enterocolitica strain W22730 cells are required to kill G. mellonella within five days. The most surprising result of the injection study performed here was the high variability of the insecticidal potential among Yersinia strains that is probably caused by tc-independent determinants. Examples for factors required for full virulence towards insect larvae are the hemolysin XhlA of X. nematophila or the gene mcf of P. luminescens. [13,19]. In Y. enterocolitica, XaxAB, an apoptotic AB toxin, and the putative macrophage toxin MT have been identified as candidates with potential insecticidal activity, but their biological role still remains to be uncovered [20]. The overall results of the Galleria bioassay correlate with the finding that among 147 Yersinia isolates from the environment, 15.6% were Y. enterocolitica, but only 0.7% belonged to Y. kristensenii [21].
Although the biological role of Tc proteins has still to be experimentally defined, sequence analysis already revealed several interesting aspects. Regions of significant sequence similarities have been identified in all TcdA-like elements characterized so far [14]. Especially, TcaC is well conserved within the Yersinia genus, but TcaB and TcaA show significant sequence variability [8]. When the TccC sequences derived from the tc-PAIYe of yersiniae were aligned, a high degree of sequence conservation was obtained at amino acids 1–680, followed by a remarkably high sequence diversity [14] as is confirmed by the TccC cladogram (Fig. 2). Some Tc sequences show evidence of undergoing degradation with frameshifts that often result in the splitting of tc genes into two separate ORFs (Fig. 1). Frameshifts in Y. pestis, especially in tcaB of CO92, are discussed as a critical step in the recent evolution of flea-borne transmission in the genus Yersinia due to the loss of one or more of those insect gut toxins [12,14]. This data indicates that the tc genes of yersiniae may be under diversifying selection [8] which might result in insecticidal proteins with host-specific activity and with varying insecticidal potential.
It has been suggested that the genomes of different strains have taken up different tc genes after strain separation [22]. However, the data presented here point to a common Yersinia ancestor that has aquired the tc-PAIYe. The plasmid-encoded Tc proteins in Y. frederiksenii and a transposon-like element downstream of Y. pestis Angola tcaC hint to putative mechanisms that might have played a role during horizontal transfer of insecticidal toxin genes (Fig. 1). This hypothesis is strongly supported by the common insertion site of the tc-PAIYe that indicates one horizontal gene transfer (HGT) event, by the highly conserved phage-related genes within the tc-PAIYe, and by a similar gene order including tccC1 and tccC2 in all islands investigated. Moreover, the cladogram derived from a comprehensive alignment of TccC protein sequences (Fig. 2) essentially reflects the phylogeny of Yersinia based on 16S rDNA sequences, including the clonal diversity among Y. enterocolitica strains [23]. As an additional insecticidal determinant, tccC genes located outside the tc-PAIYe might have been acquired by a further HGT event following the separation of Y. pseudotuberculosis and Y. enterocolitica, because all available genomes of the Y. pseudotuberculosis and Y. pestis subline share two tccC insertion sites. Thus, reductive evolution by genetic drift might explain the lack of tc-PAIYe in several Yersinia species and strains (Table 2) as examplified by the identification of rudimentary tc genes in biotypes 1A and 1B (Fig. 3).
Conclusion
The prevalence of the tc-PAIYe in many genomes, its proven functionality in Y. enterocolitica and Y. pseudotuberculosis, as well as the common insecticidal potential of Yersinia spp. towards M. sexta and G. mellonella, hints to insects as yet unknown host organisms of yersiniae. This is in line with the hypothesis that environmental predators such as nematodes or insect larvae play a role in the evolution of pathogens [22,24]. The tc-PAIYe has probably been acquired by an ancestral Yersinia strain before the separation of Y. pestis, Y. pseudotuberculosis, Y. enterocolitica, and others. This ancestor strain could then have evolved the ability to exploit invertebrates by the acquisition of further genetic determinants required for the interaction of yersiniae with those hosts [20]. Distinct sequence variation, and reductive evolution especially within the genomes of Y. pestis serovars, might have allowed yersiniae to occupy specific ecological niches [22]. The role of the tc genes and other insecticidal determinants in proliferation and transmission of the three human pathogenic Yersinia species remains to be elucidated in more detail.
Methods
Bacterial strains and growth conditions
Y. enterocolitica strains used in this study are listed in Table 3. Y. mollaretii (CIP 103324), Y. ruckeri (CIP 82.80), Y. bercovieri (CIP 103323), Y. aldovae (CIP 103162) and Y. kristensenii (CIP 80.30) were obtained from the Collection Institute Pasteur (Paris, France). Strain W22703-tcaA::Tn5lux is a tcaA knockout mutant [6].Salmonella enterica serovar Typhimurium is the ATCC strain 14028. All cultures were grown in Luria-Bertani (LB) broth (10 g l-1 tryptone, 5 g l-1 yeast extract, and 5 g l-1 NaCl) or on LB agar (LB broth supplemented with 1.5% w/v agar). E. coli was grown at 37°C and Yersinia strains at 15°C or 30°C. If appropriate, the media were supplemented with the following antibiotics: 50 μg ml-1 kanamycin and 20 μg ml-1 nalidixic acid.
Table 3.
Y. enterocolitica strains used in this study
| WS no. | Biotype | Serotype | Strain | Geographic origin | Biological origin |
| 1968 | 1A | n. d. | MZ0124a) | n. d. | Concentrate of whey |
| 4346 | 1A | O:5 | Y755c) | France | Pony |
| 2602 | 1A | O:5 | H79/83b) | Germany | Man |
| 2603 | 1A | O:5 | H1527/93b) | Germany | Man |
| 4259 | 1A | O:41,43 | SZ593/04b) | Germany | Baby food |
| 4260 | 1A | O:41,43 | SZ554/04b) | Germany | Food |
| 4266 | 1A | O:4,33 | SZ1167/04b) | Germany | Man |
| 4267 | 1A | O:10 | SZ671/04b) | Germany | Man |
| 4268 | 1A | O:41,43 | SZ634/04b) | Germany | Man |
| 4313 | 1A | O:5 | NFOc) | New Foundland | Man |
| 4346 | 1A | O:5 | Y755c) | France | Pony |
| 3760 | 1B | O:8 | 8081g) | USA | Man |
| 4263 | 1B | O:8 | SZ506/04b) | Germany | Man |
| 4265 | 1B | O:8 | SZ375/04b) | Germany | Man |
| 4314 | 1B | O:8 | WA-314c) | USA | Man |
| 4348 | 1B | O:8 | Y286d) | USA | n. d. |
| 4349 | 1B | O:13 | Y293d) | n. d. | n. d. |
| 4466 | 1B | O:21 | 209–36/84b) | Germany | Man |
| 2594 | 2 | O:9 | H692/94b) | Germany | n. d. |
| 2595 | 2 | O:9 | H621/87b) | Germany | Man |
| 2599 | 2 | O:5,27 | H280/83b) | Germany | n. d. |
| 3372 | 2 | O:9 | W22703 h) | n. d. | n. d. |
| 4264 | 2 | O:5,27 | SZ1249/0b)4 | Germany | Man |
| 4317 | 2 | O:9 | Y738c) | France | Man |
| 4347 | 2 | O:9 | Y127d) | n. d. | n. d. |
| 2596 | 3 | O:9 | H324/78b) | n. d. | Pig |
| 2597 | 3 | O:9 | H7580/93b) | n. d. | n. d. |
| 2598 | 3 | O:9 | H7692/93b) | n. d. | n. d. |
| 2600 | 3 | O:5,27 | H230/89b) | Germany | Man |
| 2601 | 3 | O:5,27 | H582/87b) | n. d. | Man |
| 3371 | 3 | O:1 | NCTC 10460f) | Denmark | Chinchilla |
| 4322 | 3 | O:3 | Y745 c) | Japan | Man |
| 4323 | 3 | O:3 | Y746c) | Japan | Man |
| 2589 | 4 | O:3 | H270/78b) | n. d. | Dog feces |
| 2590 | 4 | O:3 | H31/80b) | n. d. | Pig |
| 2591 | 4 | O:3 | H608/87b) | n. d. | Man |
| 2592 | 4 | O:3 | H450/87b) | n. d. | Man |
| 2593 | 4 | O:3 | H469/87b) | n. d. | Pig |
| 4261 | 4 | O:3 | SZ425/04 b) | Germany | Pig tongue |
| 4262 | 4 | O:3 | SZ687/04b) | Germany | Dog feces |
| 4315 | 4 | O:3 | Y-108c) | Germany | Man |
| 4324 | 4 | O:3 | Y747c) | Sweden | Man |
| 4325 | 4 | O:3 | Y750c) | China | Man |
| 4326 | 4 | O:3 | Y751c) | Great Britain | Man |
| 4327 | 4 | O:3 | Y752c) | Brazil | Man |
| 4328 | 4 | O:3 | Y753c) | New Caledonia | Man |
| 4329 | 4 | O:3 | Y754c) | New Caledonia | Man |
| 4330 | 4 | O:3 | Y755c) | South Africa | Man |
| 4331 | 4 | O:3 | Y756c) | South Africa | Man |
| 4332 | 4 | O:3 | Y757c) | Hungary | Man |
| 4333 | 4 | O:3 | Y758c) | Hungary | Man |
| 4334 | 4 | O:3 | Y759c) | Canada | Man |
| 4335 | 4 | O:3 | Y763c) | Canada | Man |
| 4336 | 4 | O:3 | Y764c) | Canada | Man |
| 4337 | 4 | O:3 | Y765c) | Australia | Man |
| 4338 | 4 | O:3 | Y766c) | Australia | Man |
| 4339 | 4 | O:3 | Y767c) | Australia | Man |
| 4340 | 4 | O:3 | Y768c) | Australia | Man |
| 4341 | 4 | O:3 | Y769c) | New Zealand | Man |
| 4342 | 4 | O:3 | Y770c) | New Zealand | Man |
| 4343 | 4 | O:3 | Y771c) | New Zealand | Man |
| 4316 | 4 | O:3 | Y11 d) | n. d. | n. d. |
| 4344 | 5 | O:2a,2b,3 | Y772c) | France | Hare |
| 4345 | 5 | O:2a,2b,3 | Y773 c) | France | Hare |
| 4318 | 2/3 | O:5,27 | 237 c) | USA | n. d. |
| 4319 | 2/3 | O:5,27 | 238 c) | Great Britain | n. d. |
a) own collection, b) Institut für Hygiene und Umwelt, Hamburg, Germany; c) Max von Pettenkofer-Institut, München, Germany; d) Institut für Mikrobiologie der Bundeswehr, München, Germany; e) Robert Koch-Institut, Berlin, Germany; f) NCTC, London, UK; g) Virginia Miller, St. Louis, USA; h) Roos Goverde, Utrecht, Netherlands. WS, Weihenstephaner Sammlung. N. d., not defined.
General molecular techniques
DNA and RNA manipulation was performed according to standard procedures [25]. To isolate chromosomal DNA, 1.5 ml of a bacterial culture was centrifuged, and the sediment was resuspended in 400 μl of lysis buffer (100 mM Tris pH 8.0, 5 mM EDTA, 200 mM NaCl). After incubation for 15 min on ice, 10 μl of 10% SDS and 5 μl of proteinase K (10 mg/ml) were added, and the sample was incubated overnight at 55°C. The chromosomal DNA was then precipitated with 500 μl of isopropanol, washed in ethanol, dried, and dissolved in 500 μl of TE buffer (10 mM Tris-HCl, 1 mM Na2 EDTA, pH 7.4) containing 1 μl of RNase (10 mg/ml). Polymerase chain reactions (PCR) were carried out with Taq polymerase (Fermentas, Vilnius, Lithunia) and the following programme: one cycle at 95°C for 2 min; 30 cycles at 95°C for 10 sec, at the appropriate annealing temperature for 30 sec, at 72°C for 45 sec to 180 sec depending on the expected fragment length; one cycle at 72°C for 10 min. All primers used are listed in Additional file 1. 4 μl of chromosomal DNA (100 ng ml-1) was used as template for PCR amplification, and the GeneRuler DNA mix (Fermentas) served as DNA ladder.
Inverse PCR and DNA sequencing
Identification of the transposon insertion site in mutant W22703-tccC(405)::Tn5lux was performed as described previously [17]. Briefly, 400 ng chromosomal DNA of the transposon mutant was completely digested with ClaI, HindIII or SspI (Fermentas), enzymes were heat-inactivated, and fragments were treated with T4 DNA ligase (Invitrogen, Carlsbad, USA) to allow self-ligation resulting in circular molecules. Inverse PCR [26] was then performed using transposon-specific primers [17], and the resulting fragments were sequenced with primers hybridizing to transposon regions near the O-end and the I-end. Sequencing of the strain-specific DNA was performed following inverse PCR using the restriction enzymes HaeIII (USB, Cleveland, USA), HhaI, HindIII, HpaI, MspI, MunI, RsaI, SspI and VspI (Fermentas), and primers derived from the sequence already obtained. Sequencing was done by 4 base lab (Reutlingen, Germany) and by MWG-Biotech (Ebersberg, Germany).
Bioinformatics
Mapping of the mini-Tn5 luxCDABE insertion was performed using the Y. enterocolitica Blast Server from the Sanger Institute http://www.sanger.ac.uk/cgi-bin/blast/submitblast/y_enterocolitica. The reference genome sequence was that of Y. enterocolitica 8081 (accession numbers AM286415 and AM286416). Sequence assembly was done with Vector NTI Advance™ (Invitrogen, Carlsbad, USA). The resulting sequence was annotated using the NCBI ORF-Finder http://www.ncbi.nlm.nih.gov/gorf/gorf.html. Homology searches of predicted proteins were performed by BLAST analysis http://www.ncbi.nlm.nih.gov/BLAST/. Genome sequences of Yersinia strains were obtained from the NCBI database and compared using the homepage http://www.microbesonline.org/. Protein sequence alignment was done with the ClustalW program [27], and cladogram was constructed with TREECON [28]. Promoter sequences located upstream of the identified genes were deduced with BPROM http://www.softberry.com/. The accession number of the W22703 tccC2' and tccC3' sequence is AM941739.
Bioassays
M. sexta were reared as described recently [29]. For oral bioassays, bacteria were grown at 15°C (Yersinia strains) or 37°C (DH5α) until stationary phase. 50 μl of a culture was applied to 4-mm3 disks of an agar-based artificial diet [30]. The liquid was allowed to soak into the agar block which was then dried under a laminar flow. First-instar M. sexta neonate larvae were then placed on the disk and incubated at 22°C. The application of bacterial culture aliquots was repeated after three days, and the larvae mortality was recorded after 5 days.
Larvae of the greater wax moth, G. mellonella, were obtained from the Zoo-Fachmarkt (München, Germany), and stored for less than one week at room temperature. Bacterial strains were grown to stationary phase at 15°C (Yersinia spp.) or 37°C (S. enterica serovar Typhimurium and DH5α) and then diluted 1:10 and 1:100. 5–7.5 μl of each dilution corresponding to approximately 5–7.5 × 105 and 5–7.5 × 104 viable cells were subcutaneously injected into larvae of 2–3 cm length and of 90–140 mg weight using a sterilized micro syringe (Hamilton 1702 RN, 25 μl). Infected larvae were then incubated for five days at 15°C, and the numbers of killed and living larvae were enumerated.
Authors' contributions
TMF compared the Yersinia genomes sequences, performed the bioassays, supervised the study, and drafted the manuscript. GB identified the novel tccC gene and derived the distribution pattern. LM performed the PCR analysis. JS supported the bioassays. SS contributed to the conception and revised the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Oligonucleotides used in this study.
Acknowledgments
Acknowledgements
We thank Peter Roggentin, Alexander Rakin, Jürgen Heesemann, Henry Derschum and Herbert Nattermann for providing Y. enterocolitica strains, and Patrick Schiwek for technical assistance. This work was supported by a grant of the Hochschul- und Wissenschaftsprogramm (HWPII): Fachprogramm "Chancengleichheit für Frauen in Forschung und Lehre" to G. B.
Contributor Information
Thilo M Fuchs, Email: thilo.fuchs@wzw.tum.de.
Geraldine Bresolin, Email: geraldine@bresolin.de.
Lisa Marcinowski, Email: lisa.marcinowski@gmx.de.
Joachim Schachtner, Email: schachtj@staff.uni-marburg.de.
Siegfried Scherer, Email: siegfried.scherer@wzw.tum.de.
References
- Bowen DJ, Ensign JC. Purification and characterization of a high-molecular-weight insecticidal protein complex produced by the entomopathogenic bacterium Photorhabdus luminescens. Appl Environ Microbiol. 1998;64:3029–3035. doi: 10.1128/aem.64.8.3029-3035.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterfield NR, Bowen DJ, Fetherston JD, Perry RD, ffrench-Constant RH. The tc genes of Photorhabdus: a growing family. Trends Microbiol. 2001;9:185–191. doi: 10.1016/s0966-842x(01)01978-3. [DOI] [PubMed] [Google Scholar]
- Vodovar N, Vallenet D, Cruveiller S, Rouy Z, Barbe V, Acosta C, Cattolico L, Jubin C, Lajus A, Segurens B, Vacherie B, Wincker P, Weissenbach J, Lemaitre B, Medigue C, Boccard F. Complete genome sequence of the entomopathogenic and metabolically versatile soil bacterium Pseudomonas entomophila. Nat Biotechnol. 2006;24:673–679. doi: 10.1038/nbt1212. [DOI] [PubMed] [Google Scholar]
- Dodd SJ, Hurst MR, Glare TR, O'Callaghan M, Ronson CW. Occurrence of sep insecticidal toxin complex genes in Serratia spp. and Yersinia frederiksenii. Appl Environ Microbiol. 2006;72:6584–6592. doi: 10.1128/AEM.00954-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennant SM, Skinner NA, Joe A, Robins-Browne RM. Homologues of insecticidal toxin complex genes in Yersinia enterocolitica biotype 1A and their contribution to virulence. Infect Immun. 2005;73:6860–6867. doi: 10.1128/IAI.73.10.6860-6867.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresolin G, Morgan JA, Ilgen D, Scherer S, Fuchs TM. Low temperature-induced insecticidal activity of Yersinia enterocolitica. Mol Microbiol. 2006;59:503–512. doi: 10.1111/j.1365-2958.2005.04916.x. [DOI] [PubMed] [Google Scholar]
- Thomson NR, Howard S, Wren BW, Holden MT, Crossman L, Challis GL, Churcher C, Mungall K, Brooks K, Chillingworth T, Feltwell T, Abdellah Z, Hauser H, Jagels K, Maddison M, Moule S, Sanders M, Whitehead S, Quail MA, Dougan G, Parkhill J, Prentice MB. The complete genome sequence and comparative genome analysis of the high pathogenicity Yersinia enterocolitica strain 8081. PLoS Genet. 2006;2:e206. doi: 10.1371/journal.pgen.0020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterfield N, Hares M, Hinchliffe S, Wren B, ffrench-Constant R. The insect toxin complex of Yersinia. Adv Exp Med Biol. 2007;603:247–257. doi: 10.1007/978-0-387-72124-8_22. [DOI] [PubMed] [Google Scholar]
- Gendlina I, Held KG, Bartra SS, Gallis BM, Doneanu CE, Goodlett DR, Plano GV, Collins CM. Identification and type III-dependent secretion of the Yersinia pestis insecticidal-like proteins. Mol Microbiol. 2007;64:1214–1227. doi: 10.1111/j.1365-2958.2007.05729.x. [DOI] [PubMed] [Google Scholar]
- Silva CP, Waterfield NR, Daborn PJ, Dean P, Chilver T, Au CP, Sharma S, Potter U, Reynolds SE, ffrench-Constant RH. Bacterial infection of a model insect: Photorhabdus luminescens and Manduca sexta. Cell Microbiol. 2002;4:329–339. doi: 10.1046/j.1462-5822.2002.00194.x. [DOI] [PubMed] [Google Scholar]
- Hares MC, Hinchliffe SJ, Strong PC, Eleftherianos I, Dowling AJ, ffrench-Constant RH, Waterfield N. The Yersinia pseudotuberculosis and Yersinia pestis toxin complex is active against cultured mammalian cells. Microbiology. 2008;154:3503–3517. doi: 10.1099/mic.0.2008/018440-0. [DOI] [PubMed] [Google Scholar]
- Erickson DL, Waterfield NR, Vadyvaloo V, Long D, Fischer ER, ffrench-Constant R, Hinnebusch BJ. Acute oral toxicity of Yersinia pseudotuberculosis to fleas: implications for the evolution of vector-borne transmission of plague. Cell Microbiol. 2007;9:2658–2666. doi: 10.1111/j.1462-5822.2007.00986.x. [DOI] [PubMed] [Google Scholar]
- ffrench-Constant R, Waterfield N, Daborn P, Joyce S, Bennett H, Au C, Dowling A, Boundy S, Reynolds S, Clarke D. Photorhabdus: towards a functional genomic analysis of a symbiont and pathogen. FEMS Microbiol Rev. 2003;26:433–456. doi: 10.1111/j.1574-6976.2003.tb00625.x. [DOI] [PubMed] [Google Scholar]
- Pinheiro VB, Ellar DJ. Expression and insecticidal activity of Yersinia pseudotuberculosis and Photorhabdus luminescens toxin complex proteins. Cell Microbiol. 2007;9:2372–2380. doi: 10.1111/j.1462-5822.2007.00966.x. [DOI] [PubMed] [Google Scholar]
- Motin VL, Georgescu AM, Fitch JP, Gu PP, Nelson DO, Mabery SL, Garnham JB, Sokhansanj BA, Ott LL, Coleman MA, Elliott JM, Kegelmeyer LM, Wyrobek AJ, Slezak TR, Brubaker RR, Garcia E. Temporal global changes in gene expression during temperature transition in Yersinia pestis. J Bacteriol. 2004;186:6298–6305. doi: 10.1128/JB.186.18.6298-6305.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Zhou D, Pang X, Song Y, Zhang L, Bao J, Tong Z, Wang J, Guo Z, Zhai J, Du Z, Wang X, Zhang X, Wang J, Huang P, Yang R. Microarray analysis of temperature-induced transcriptome of Yersinia pestis. Microbiol Immunol. 2004;48:791–805. doi: 10.1111/j.1348-0421.2004.tb03605.x. [DOI] [PubMed] [Google Scholar]
- Bresolin G, Neuhaus K, Scherer S, Fuchs TM. Transcriptional analysis of long-term adaptation of Yersinia enterocolitica to low-temperature growth. J Bacteriol. 2006;188:2945–2958. doi: 10.1128/JB.188.8.2945-2958.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen DJ, Ensign JC. Isolation and characterization of intracellular protein inclusions produced by the entomopathogenic bacterium Photorhabdus luminescens. Appl Environ Microbiol. 2001;67:4834–4841. doi: 10.1128/AEM.67.10.4834-4841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert EE, Goodrich-Blair H. Friend and foe: the two faces of Xenorhabdus nematophila. Nat Rev Microbiol. 2007;5:634–646. doi: 10.1038/nrmicro1706. [DOI] [PubMed] [Google Scholar]
- Heermann R, Fuchs TM. Comparative analysis of the Photorhabdus luminescens and the Yersinia enterocolitica genomes: uncovering candidate genes involved in insect pathogenicity. BMC Genomics. 2008;9:40. doi: 10.1186/1471-2164-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shayegani M, DeForge I, McGlynn DM, Root T. Characteristics of Yersinia enterocolitica and related species isolated from human, animal, and environmental sources. J Clin Microbiol. 1981;14:304–312. doi: 10.1128/jcm.14.3.304-312.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterfield NR, Wren BW, ffrench-Constant RH. Invertebrates as a source of emerging human pathogens. Nat Rev Microbiol. 2004;2:833–841. doi: 10.1038/nrmicro1008. [DOI] [PubMed] [Google Scholar]
- Ibrahim A, Goebel BM, Liesack W, Griffiths M, Stackebrandt E. The phylogeny of the genus Yersinia based on 16S rDNA sequences. FEMS Microbiol Lett. 1993;114:173–177. doi: 10.1111/j.1574-6968.1993.tb06569.x. [DOI] [PubMed] [Google Scholar]
- Hilbi H, Weber SS, Ragaz C, Nyfeler Y, Urwyler S. Environmental predators as models for bacterial pathogenesis. Environ Microbiol. 2007;9:563–575. doi: 10.1111/j.1462-2920.2007.01238.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3. Cold Spring Harbor Laboratory, Cold Spring Harbor, N. Y; 2001. [Google Scholar]
- Ochman H, Ajioka JW, Garza D, Hartl DL. Inverse polymerase chain reaction. Biotechnology (NY) 1990;8:759–760. doi: 10.1038/nbt0890-759. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer Y Van de, De Wachter R. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- Schachtner J, Huetteroth W, Nighorn A, Honegger HW. Copper/zinc superoxide dismutase-like immunoreactivity in the metamorphosing brain of the sphinx moth Manduca sexta. J Comp Neurol. 2004;469:141–152. doi: 10.1002/cne.10992. [DOI] [PubMed] [Google Scholar]
- David WAL, Gardiner BOC. Rearing Pieris brassicae larvae on a semi-synthetic diet. Nature. 1965;207:882–883. [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oligonucleotides used in this study.