Abstract
Background
We have previously reported that expression of the Wnt antagonist genes SFRP1 and SFRP5 is frequently silenced by promoter hypermethylation in breast cancer. SFRP2 is a further Wnt inhibitor whose expression was recently found being downregulated in various malignancies. Here we investigated whether SFRP2 is also implicated in human breast cancer, and if so whether SFRP2 promoter methylation might serve as a potential tumor biomarker.
Methods
We analyzed SFRP2 mRNA expression and SFRP2 promoter methylation in 10 breast cell lines, 199 primary breast carcinomas, 20 matched normal breast tissues and 17 cancer-unrelated normal breast tissues using RT-PCR, realtime PCR, methylation-specific PCR and Pyrosequencing, respectively. SFRP2 protein expression was assessed by immunohistochemistry on a tissue microarray. Proliferation assays after transfection with an SFRP2 expression vector were performed with mammary MCF10A cells. Statistical evaluations were accomplished with SPSS 14.0 software.
Results
Of the cancerous breast cell lines, 7/8 (88%) lacked SFRP2 mRNA expression due to SFRP2 promoter methylation (P < 0.001). SFRP2 expression was substantially restored in most breast cell lines after treatment with 5-aza-2'-deoxycytidine and trichostatin A. In primary breast carcinomas SFRP2 protein expression was strongly reduced in 93 of 125 specimens (74%). SFRP2 promoter methylation was detected in 165/199 primary carcinomas (83%) whereas all cancer-related and unrelated normal breast tissues were not affected by SFRP2 methylation. SFRP2 methylation was not associated with clinicopathological factors or clinical patient outcome. However, loss of SFRP2 protein expression showed a weak association with unfavorable patient overall survival (P = 0.071). Forced expression of SFRP2 in mammary MCF10A cells substantially inhibited proliferation rates (P = 0.045).
Conclusion
The SFRP2 gene is a high-frequent target of epigenetic inactivation in human breast cancer. Its methylation leads to abrogation of SFRP2 expression, conferring a growth advantage to epithelial mammary cells. This altogether supports a tumor suppressive function of SFRP2. Although clinical patient outcome was not associated with SFRP2 methylation, the high frequency of this epimutation and its putative specificity to neoplastic cells may qualify SFRP2 promoter methylation as a potential candidate screening marker helping to improve early breast cancer detection.
Background
Aberrant promoter methylation leading to functional inactivation of tumor suppressor genes is a well recognized mechanism capable of driving carcinogenesis [1,2]. In human breast cancer numerous genes have been identified with abolished expression due to 5'-cytosine methylation within their gene promoter (recently reviewed in [3]). Typically those genes affect important aspects of normal growth control, like cell cycle regulation (p16INK4a) [4], steroid receptor biology (estrogen receptor-α) [5], cell adhesion (E-cadherin) [6], apoptosis (death-associated protein (DAP) kinase-1) [7] or extracellular matrix integrity (ITIH5) [8], all of which confers, in case of expression loss, growth advantages to neoplastic cells. Importantly, the observation that DNA methylation of the same gene may occur both in premalignant lesions, such as atypical hyperplasia of the breast, and in carcinoma [9] suggests that DNA methylation might serve as ideal biomarker for early cancer detection or patient risk assessment in clinical oncology [10]. Thus, identification and validation of epigenetically silenced cancer-related genes is of critical importance in the search of novel tumor biomarkers.
Secreted frizzled-related proteins (SFRPs) constitute a family of extracellular Wnt signaling antagonists, of which five members (SFRP1-5) have been identified to date [11]. SFRPs sequester Wnt molecules at the cell surface membrane [12] and by this are recognized as sensitive regulators of the canonical Wnt signaling pathway [13]. Aberrant activation of Wnt signaling has been associated with the pathogenesis of virtually all human cancers (reviewed in [14]). In breast tumor tissues, activated Wnt signaling has been repeatedly observed as determined by nuclear and cytoplasmic accumulation of β-catenin [15-18], arguing for a disrupted equilibrium between Wnt and SFRP expression in this tumor type. In line with this, previous studies have shown that expression of SFRP genes is commonly silenced by promoter methylation in human cancers [19-26]. In breast cancer, SFRP1 and SFRP5 have been identified as targets of aberrant epigenetic inactivation to date, and either promoter methylation was found to be associated with unfavorable patient prognosis [27,28]. SFRP2 has been previously identified as epigenetic target in other tumor entities, such as colon [29], oesophagus [30], bladder [31], stomach [23,24], liver [25] and lung cancer [32]. Interestingly, in all tumor entities SFRP2 methylation was detected with a high frequency of > 50% of cancer patients, ranging from 52% in lung cancer to 96% in gastric cancer, which suggests that SFRP2 methylation might potentially be useful as a ubiquitous pan-tumor marker in cancerous tissues, and possibly also in body fluids. Consequently, Urakami et al. [22] demonstrated that of all investigated SFRPs only SFRP2 methylation proved to be a valuable independent prediction factor for bladder cancer in urine samples. At the same time, SFRP2 methylation was found to occur high-frequent in colon cancer (83–90%) [33,19], which may have forced the establishment of SFRP2 methylation as a promising sensitive screening marker for the stool-based detection of colorectal cancer and premalignant lesions [34-36].
Very recently, Suzuki et al. [37] reported about SFRP2 methylation in human breast cancer, and their study demonstrated an inhibitory effect of SFRP2 on canonical Wnt signaling in breast cancer cell lines. However, SFRP2 expression analyses in normal and breast carcinoma tissues as well as patient survival analysis in relation to SFRP2 methylation were not addressed. Our approach was to investigate SFRP2 expression and promoter methylation in breast cell lines, primary breast carcinomas and normal breast tissues, followed by comprehensive statistical correlation analysis with clinicopathological factors and patient survival. We also investigated a functional role of SFRP2 in a breast cell line with regard to growth behaviour. In summary, our data confirm that the SFRP2 gene is high-frequently inactivated by promoter methylation in human breast cancer and that loss of SFRP2 expression confers a growth advantage to mammary epithelial cells. In addition, we provide evidence that SFRP2 protein expression is commonly reduced in breast cancer and that SFRP2 methylation might be a potential biomarker useful for early detection of this disease.
Methods
Cryoconserved clinical materials
According to a multi-center study design, 20 matched tumor/macroscopically normal samples of breast cancer patients (median patient age: 67 years; range 48–86 years) and 179 unmatched breast carcinomas (median patient age: 57 years; range 28–96 years) were obtained from patients treated by primary surgery for breast cancer at the Departments of Gynecology at the University Hospitals of Aachen, Jena, Regensburg and Düsseldorf, Germany. None of the patients had received neo-adjuvant chemotherapy. Inclusion criterion for ipsilateral normal breast tissue was a distance of > 2 cm to the carcinoma margin. All patients gave informed consent for retention and analysis of their tissue for research purposes and the Institutional Review Boards of the participating centers approved the study. The selection of cases was based on availability of tissue. Cases were not stratified for any known preoperative or pathological prognostic factor. Tumor histology was determined according to the criteria of the WHO (2003), while disease stage was assessed according to UICC [38]. Tumors were graded according to Bloom and Richardson, as modified by Elston and Ellis [39]. Hormone receptor positivity was defined as an immunoreactivity score (IRS) ≥ 3 [40]. For 136 patients follow-up data were available with a median time of 64 months (range 1–174 months). Tumor material was immediately snap-frozen in liquid nitrogen after surgery. Hematoxylin/Eosin-stained sections were prepared for assessing the percentage of tumor cells; only samples with greater than 70% tumor cells were selected for analysis. Samples were dissolved in lysis buffer followed by DNA isolation using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's recommendations. For patient characteristics see additional file 1.
Formalin-fixed, paraffin-embedded (FFPE) clinical material
A total of 17 archival FFPE normal breast tissues were obtained from the Institute of Pathology, University Hospital of the RWTH Aachen, Germany. These patients had undergone breast reduction surgery without the condition of cancer. The median age in the cancer-unrelated normal breast tissue set was 33 years (range 22–61 years). Per sample, five consecutive sections (each 10 μm) were deparaffinized and rehydrated in a decreasing alcohol series prior to DNA extraction by use of the QIAamp DNA Mini Kit.
SFRP2 protein expression was assessed using a tissue microarray (TMA) consisting of 125 breast carcinomas, four ductal carcinomas in situ (DCIS) and 10 normal breast tissues that have been described previously [41]. The TMA contained one tissue core from non-selected, FFPE primary breast carcinoma specimens diagnosed between 1994 and 2002 at the Institute of Pathology, University of Regensburg, Germany. Histological, all tumors were graded according to Bloom and Richardson, as modified by Elston and Ellis [39]. Clinical follow-up data were available for 124 breast cancer patients with a median follow-up period of 80 months (range 5–114 months). All patients gave informed consent for retention and analysis of their tissue for research purposes and the Institutional Review Board of the participating centers approved the study.
Immunohistochemistry
The TMA was subjected to immunostaining using the K5007 Kit (DAKO, Hamburg, Germany) following the manufacturer's instructions. Antigen retrieval was performed by pretreatment in citrate buffer (pH 7) in a microwave oven (20 min, 200 W). Samples were incubated for 30 min with the primary SFRP2 antibody (rabbit polyclonal IgG; H-140; 1:75; Santa Cruz Biotechnology, Santa Cruz, CA), washed, and incubated for 10 min with the secondary antibody (biotinylated polylink; DAKO). Diaminobenzidin (DAKO) was used for antibody detection. In negative controls the primary antibody was omitted. An experienced breast cancer pathologist (N.B.) scored the immunohistochemical staining according to the scoring system suggested by Remmele and Stegner [40]. Feasibility of the antibody for immunohistochemical analysis of breast tissue has been previously demonstrated e.g. by Lee et al. [42].
Cell lines
The benign cell lines HMEC and MCF10A as well as the cancerous breast cell lines BT20, BT474, Hs578T, MCF7, MDA-MB-231, MDA-MB-453, SKBR3, T47D and ZR75-1 were obtained from the American Type Culture Collection (Rockville, MA) and cultured as recommended by the vendor.
Reverse transcription (RT-) PCR and semi-quantitative realtime PCR
RNA isolation, RT-PCR and SYBR Green I realtime PCR (Roche Diagnostics, Mannheim, Germany) were performed as described elsewhere [27]. Quality of cDNA was checked after each preparation by standard RT-PCR using glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) primers that yield an amplification product of 510 bp. To ensure experiment accuracy, all quantitative measurements were performed in triplicate. Intron-spanning primer sequences and cycle conditions are given in additional file 2.
Sodium bisulfite-modification and methylation-specific PCR (MSP)
Of the genomic DNA, 1 μg was bisulfite-modified using the EZ DNA Methylation Kit (Zymo Research, Orange, CA) according to the manufacturer's recommendations. The final precipitate was eluted in 20 μl TRIS buffer (10 mM). For MSP, one μl of modified DNA was amplified using MSP primers (see additional file 2) that specifically recognized either the unmethylated or methylated gene promoter sequence after bisulfite-conversion. Each primer pair mapped to nine cytosine-phosphate-guanine dinucleotide (CpG) sites in order to specifically discriminate between methylated and non-methylated DNA. Further 11 non-CpG cytosines within the primer pair specific for methylated DNA and 13 non-CpG cytosines within the primer pair specific for non-methylated DNA guaranteed unequivocal amplification of bisulfite-converted DNA. Primers defined an amplicon between +19 and +163 relative to the transcription start site (+1) of the SFRP2 gene. Reaction volumes of 25 μl contained 1 × MSP-buffer [44], 400 nM each of methylation and non-methylation-specific primers, respectively, and 1.25 mM of dNTPs. One drop of mineral oil was added to the reaction tube. The PCR was initiated as "Hot Start" PCR at 94°C and held at 80°C before the addition of 1.25 units Taq DNA polymerase (Promega, Madison, WI). Cycle conditions were: 95°C for 5 min, 35 cycles of 95°C for 30 s, 60°C for 30 s, 72°C for 40 s and a final extension at 72°C for 5 min. Blood lymphocyte DNA from a healthy donor was bisulfite-modified to serve as a control for the unmethylated promoter sequence [45], DNA from the cancerous breast cell line BT20 served as control for methylated alleles. Amplification products were visualized on 3% low range ultra agarose gel (Bio-Rad Laboratories, Hercules, CA) containing ethidium bromide and illuminated under ultraviolet (UV) light.
Pyrosequencing
Quantitative Pyrosequencing of a SFRP2 promoter fragment was performed by use of a Pyromark ID device, PyroGoldSQA Reagent Kit and Pyro Q-CpG software (Biotage, Uppsala, Sweden). Initially, a 291 bp fragment of the SFRP2 promoter (relative position -28 to +263), covering the hybridization sites for MSP primers, was amplified with degenerate primers irrespective of the methylation status, which assures unbiased DNA amplification. To enable single strand preparation the reverse primer was 5'-biotinylated. Reaction volumes of 50 μl contained 1 × GoTaq buffer, 2.5 units GoTaq polymerase (Promega), 2.5 mM of MgCl2, 400 nM of primers, 500 nM of each dNTP, and 3 μl of bisulfite-converted DNA as template. Reactions were initiated as "Hot Start" PCR at 95°C for 3 min and held at 80°C before addition of Taq polymerase. Cycle conditions were: 94°C for 3 min, 50 cycles of 94°C for 15 sec, 58°C for 30 sec, 72°C for 30 sec, and a final extension step at 72°C for 5 min. PCR was carried out in a PTC-200 cycler (Bio-Rad, formerly MJ Research, Hercules, CA). Prior to sequencing, aliquots of the amplificate were analyzed on a 2% agarose gel containing ethidium bromide under UV light. Single strand separation of the remaining amplificate (40 μl) was performed with a PyroMark Vacuum Prep Workstation (Biotage). Amplificate was immobilized to Streptavidin-Sepharose HP beads (Amersham Biosciences, Uppsala, Sweden), washed, denatured and the biotinylated strands were released into 40 μl of annealing buffer containing 400 nM of forward sequencing primer. Sequencing started with position +3 (relative to the TSS) and was continued to position +167, covering a total of 22 sequential CpG sites. The following sequence represents the SFRP2 promoter sequence that was analyzed by pyrosequencing: AYGGTTTATTTTGTTTTTTYGGGTYGGAGT TTTTYGGAGTTGYGYGYGGGTT TGTAGTGTTTYGTTYGYGTTGTTTTTTYGGTGTTTYGTTTTTTYGYGTT TTAGTYGTYGGTTGTT AGTTTTTYGGGGTTTYGAGTYGTATTTAGYGAAGAGAGYGGGTTYGG.
Universal bisulfite-converted polymethylated and unmethylated DNA (Epi Tect Control DNA Set; Qiagen, Hilden, Germany) served as technical controls for SFRP2 methylation and non-methylation, respectively. Pyrosequencing primers are available on request.
5-aza-2'-deoxycytidine (DAC) and trichostatin A (TSA) treatment
We plated cells at 3 × 104 cells/cm2 in a six-well plate on day 0. The demethylating agent DAC (Sigma-Aldrich, Deisenheim, Germany) was added to a final concentration of 1 μM in fresh medium on days 1, 2 and 3. Additionally, 300 nM TSA (Sigma-Aldrich) was added on day 3. Cells were harvested on day 4 for RNA and DNA extraction. Control cells were incubated without the addition of DAC or TSA and fresh medium was also supplied on days 1, 2 and 3.
Transient transfection
Cells were seeded at a density of 3 × 104 cells/cm2 and transfected 24 hours after incubation with 100 ng/cm2 of plasmid DNA in the following manner: 100 ng of empty pCMV-hemagglutinin (HA) vector control (Clontech, Heidelberg, Germany), or 50 ng of pCMV-HA + 50 ng pCMV-HA/SFRP2, or 50 ng of pCMV-HA + 50 ng pcDNA3.1-HisA/WNT1, or 50 ng pCMV-HA/SFRP2 + 50 ng pcDNA3.1-HisA/WNT1 [19] applying the FuGENE 6 transfection system (Roche Diagnostics) and a 3:1 transfection ratio according to the manufacturer's instructions.
Proliferation assays
MCF10A cells were transfected in 96-well plates as described above and an XTT-proliferation assay (Roche Diagnostics) was performed on day 0, 1, 2 and 3 after transfection by determining the optical density of the supernatants at 480 nm minus the optical density of the supernatants at 690 nm. To enhance experimental accuracy, six replicas were seeded. For the colony formation assay cells were transfected accordingly in six-well plates and kept for three weeks under selective force of the antibiotic G418 (700 μg/ml) (Invitrogen, Carlsbad, CA). After incubation, colonies were washed with phosphate-buffered saline, fixed and stained for 30 minutes (0.25% crystal violet in 10% formalin/80% methanol), washed three times with distilled water and photographed.
Statistical methods
Statistical analyses were completed using the software package SPSS, version 14.0 (SPSS Inc., Chicago, IL). Differences were considered significant when P-values were below 0.05. A two-sided non-parametric Mann-Whitney U-test and a paired student's t-test were performed to analyze differences in expression levels. Associations between metrical variables were determined by a linear regression analysis. To study statistical associations between clinicopathological factors and SFRP2 expression or SFRP2 promoter methylation status contingency-tables and two-sided Fisher's exact tests were accomplished. Survival curves were calculated using the Kaplan-Meier method, with significance evaluated by two-sided log-rank statistics. Overall survival (OS) (n = 136 for MSP samples) was measured from the day of surgery until tumor-related death (20.6%, n = 28) and was censored for patients alive at last contact (69.1%, n = 94), in case of death unrelated to the tumor (3.7%, n = 5) or when the death cause was unknown (6.6%, n = 9). Disease-free survival (DFS) (n = 136 for MSP samples) was measured from surgery until local or distant relapse (36.8%, n = 50) and censored for patients alive without evidence of relapse at the last follow-up (63.2%, n = 86).
Results
Expression of SFRP2 mRNA is reduced in breast cancer cell lines
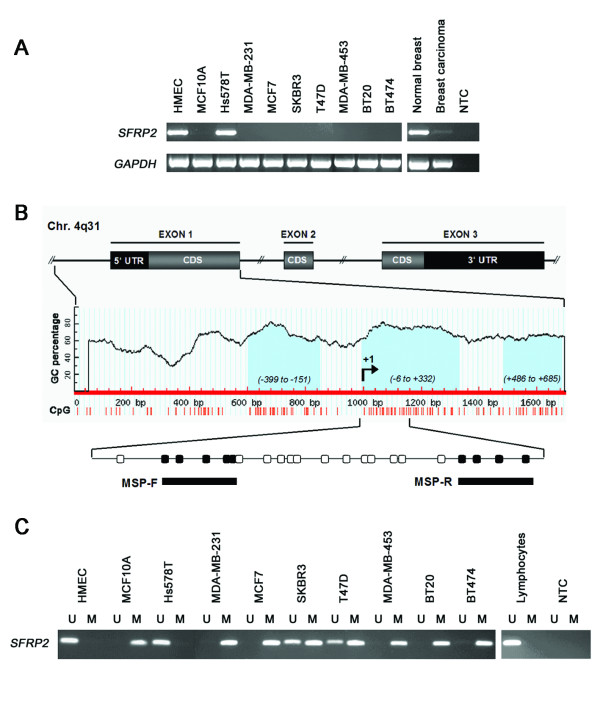
Initiating our analysis, we used the RT-PCR technique to evaluate whether SFRP2 mRNA is differentially expressed in human breast cell lines. SFRP2 mRNA expression was detectable in the benign cell line HMEC, whilst its expression was absent in the cancerous cell lines MDA-MB-231, MCF7, SKBR3, T47D, MDA-MB-453, BT20 and BT474 (Figure 1A). Of the cancerous cell lines only Hs578T exhibited strong SFRP2 expression. However, SFRP2 expression was also absent in the benign cell line MCF10A. Additionally, in a commercially available cDNA of human normal breast tissue (Clontech, Heidelberg, Germany) SFRP2 was strongly expressed while its expression was substantially reduced in Clontech's cDNA of primary breast carcinoma.
Figure 1.
SFRP2 expression and promoter methylation in breast cancer cell lines. (A) SFRP2 mRNA expression in benign and malignant cell lines was determined by RT-PCR. All but one malignant cell line (Hs578T) completely lacked SFRP2 mRNA expression. Of the two benign breast cell lines (MCF10A and HMEC) only HMEC cells were found to express abundant SFRP2 mRNA. In a commercially available human normal breast tissue cDNA SFRP2 expression was clearly detectable, while expression was strongly reduced in a corresponding cDNA of human malignant breast tissue. (B) Genomic structure of the human SFRP2 gene on chromosome 4q31. Bioinformatic analysis revealed three CGIs (light blue), located within the SFRP2 promoter (left CGI), the 5' untranslated region (UTR; central CGI) and the coding sequence (CDS; right CGI). Methylation of the central CGI was explored by MSP. Circles indicate CpG sites; filled circles represent MSP forward (MSP-F) and reverse (MSP-R) primer hybridization sites. Indicated positions are related to the transcription start site (+1) initiating the 5'-UTR. (C) MSP was performed with bisulfite-treated DNA from the same breast cell lines as in A. DNA bands in lanes labeled with U indicate PCR products amplified with primers recognizing unmethylated SFRP2 promoter sequence. DNA bands in lanes labeled with M represent amplification products with methylation-specific primers. Five out of eight mammary tumor cell lines exhibit complete promoter methylation (MDA-MB-231, MCF7, MDA-MB-453, BT20 and BT474), two cell lines show partial SFRP2 methylation (SKBR3 and T47D). In Hs578T, only unmethylated SFRP2 promoter sequence could be detected, like it was also found in benign HMEC cells. In addition, lymphocyte DNA from a healthy donor did not reveal SFRP2 methylation. GAPDH served as cDNA loading control; NTC represents the no template control.
Methylation of the SFRP2 promoter in breast cancer cell lines
Analysis of the SFRP2 gene promoter on chromosome 4q31 [46] using the genomic DNA information contained in Ensembl contig ENSG00000145423 [47] revealed a CpG island (CGI) between base position -818 to +743 relative to the expected transcription start site (+1), according to the CGI definition of Takai and Jones [48]. Further exploration of this CGI using Methprimer software [49] identified three regions of particularly high CpG density (-399 to -151, -6 to +332, and +486 to +685) (Figure 1B). Since sequence integrity of the first SFRP2 exon was demonstrated to be most essential for efficient RNA transcription in a luciferase promoter assay [24] we chose the central CGI (-6 to +332) for subsequent methylation analysis by application of the highly specific MSP primers described by Suzuki et al. [29] and others [24,37]. First, we assessed SFRP2 promoter methylation in eight cancerous and two non-cancerous cell lines. Six of the analyzed cell lines (MCF10A, MDA-MB-231, MCF7, MDA-MB-453, BT20 and BT474) exhibited a methylated SFRP2 promoter sequence in the analyzed region (Figure 1C). Two cell lines (SKBR3 and T47D) showed partial promoter methylation, since a mixture of unmethylated and methylated DNA sequence could be detected in the same sample. One malignant cell line (Hs578T) and benign HMEC cells revealed solely unmethylated SFRP2 promoter sequence. This result correlates with the above described finding that Hs578T and HMEC cells exhibited strong SFRP2 mRNA expression whereas in all cell lines with aberrant methylation SFRP2 mRNA expression was absent. Interestingly, SFRP2 mRNA was not expressed at detectable levels from unmethyated alleles in SKBR3 and T47D, which may be due to repressing mechanisms that are codominant to the effect of promoter methylation in these cells.
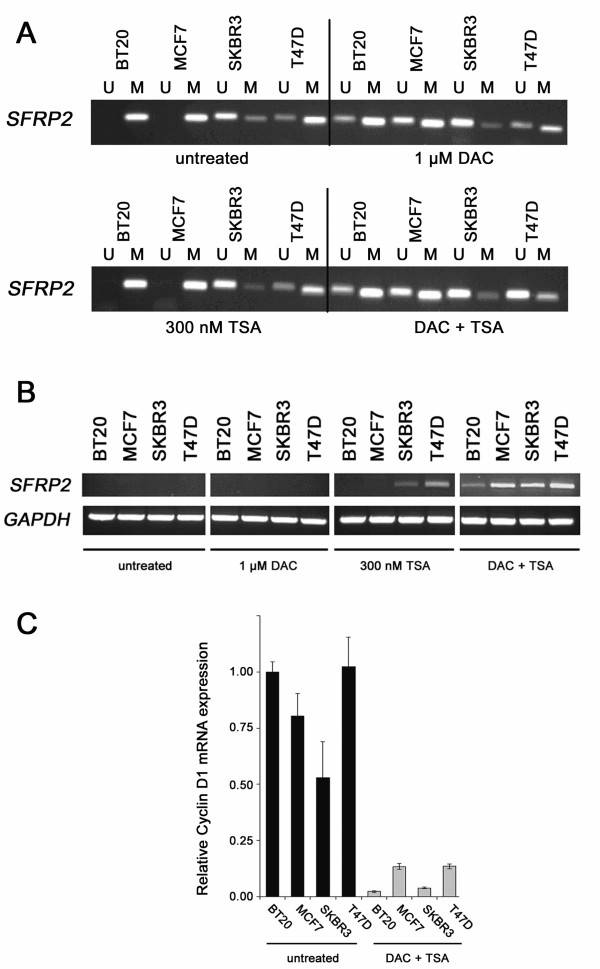
Re-expression of SFRP2 mRNA after in vitro DNA demethylation
We next asked whether SFRP2 promoter methylation is functionally associated with loss of SFRP2 mRNA expression in breast cell lines. To address this question we treated four representative breast cancer cell lines for four days with 1 μM of the methyltransferase inhibitor DAC and one day with 300 nM of the histone deacetylase inhibitor TSA. As seen in Figure 2A, in three cell lines (BT20, MCF7 and T47D) the DAC/TSA treatment resulted in a clear increase of unmethylated SFRP2 promoter sequence, only in SKBR3 cells there was no visible difference detectable. Interestingly, BT20 and MCF7 showed demethylation after the addition of DAC alone, whereas in T47D cells such effect was only achieved by the combination of DAC and TSA. Before the treatment, none of the cell lines showed detectable SFRP2 mRNA expression (Figure 2B). The treatment with 1 μM DAC alone did not reverse SFRP2 gene repression in these cells. TSA, in contrast, was able to induce some SFRP2 expression in SKBR3 and T47D cells. Importantly, only the combination of both drugs substantially restored SFRP2 mRNA expression in all analyzed cell lines.
Figure 2.
Global demethylation and histone acetylation restores SFRP2 expression. (A) MSP of four malignant cell lines was performed with DNA from either untreated cells, or after treatment with 1 μM DAC, or after treatment with 300 nM TSA, or after a combined treatment applying both drugs. In three cell lines (BT20, MCF7, T47D) a promoter demethylating effect could be visually detected, since signals indicative of unmethylated SFRP2 promoter arise (BT20, MCF7) or become enhanced (T47D) after the combined treatment. In T47D, DAC alone had no detectable demethylating effect on the SFRP2 promoter. (B) Expression of SFRP2 mRNA before treatment, or after treatment with 1 μM DAC, or after treatment with 300 nM TSA, or after a combined treatment applying both drugs. Treatment with DAC alone was not able to induce SFRP2 expression in all cell lines, in contrast to TSA which induced expression in two out of four cell lines (SKBR3 and T47D) previously showing partial SFRP2 methylation. However, only combined promoter demethylation and histone reacetylation leads to strong induction of SFRP2 mRNA expression in all cell lines. GAPDH served as cDNA loading control. (C) Suppression of Cyclin D1 mRNA expression after global DNA demethylation of breast cancer cell lines as determined by realtime PCR. Untreated tumor cells (black bars) and cells treated with DAC/TSA (grey bars) show significantly different expression levels of Cyclin D1 mRNA (P = 0.029, two-sided Mann-Whitney U-test). Expression level of each sample is normalized to its GAPDH expression and related to untreated BT20 cells (set to 1).
To further prove that the demethylating treatment did not result in unspecific upregulation of gene expression, we determined the expression of the growth promoting gene cyclin D1, which is a direct read-out gene of active Wnt signaling [50] and whose expression is commonly elevated in breast cancer [51]. Using realtime PCR we observed that cyclin D1 mRNA expression was significantly reduced (P = 0.029, two-tailed Mann-Whitney U-test) after the demethylation treatment (43-fold in BT20, 14-fold in SKBR3, 8-fold in T47D and 6-fold in MCF7, Figure 2C), suggesting that inhibitors of proliferation, such as SFRPs which are downregulated in breast cancer cell lines, have been reactivated and were able to block Wnt signaling in these cells.
Methylation of the SFRP2 promoter in primary breast carcinoma and normal breast tissue
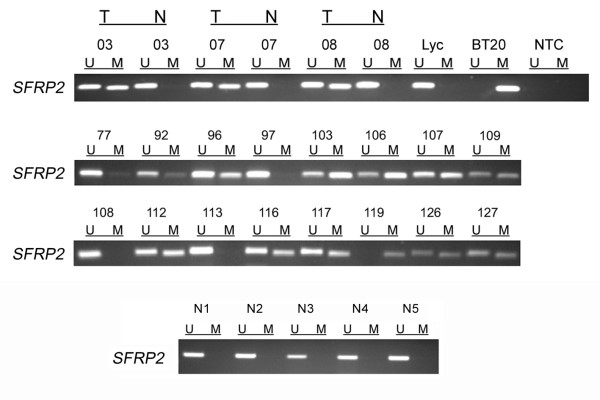
In order to answer the question whether SFRP2 promoter methylation occurs in primary breast carcinoma as well we analyzed 199 mammary tumor samples by MSP. For 20 breast tumors corresponding tissues of histological normal breast epithelium were available and analyzed in parallel. Representative results are shown in Figure 3. In total, 165 of 199 tumor samples (83%) showed SFRP2 promoter methylation as a PCR product could be amplified with methylation-specific primers (e.g. #103 in Figure 3), and 34 of 199 tumors (17%) showed no evidence of promoter methylation since exclusive amplification signals were obtained with primers specific to unmethylated DNA (e.g. #108 in Figure 3). None of the 20 matched normal breast tissue samples showed a methylation signal (e.g. #03 in Figure 3). Tumor samples generally exhibited also unmethylated promoter sequences due to possible contamination with small amounts of stromal and endothelial cells, as has also been described by Suzuki et al. [19]. To further confirm that SFRP2 promoter methylation in breast cancer is restricted to malignant tissue, we analyzed 17 cancer-unrelated normal breast samples. Again, none of these normal breast tissues (e.g. Figure 3) harbored detectable SFRP2 methylation.
Figure 3.
SFRP2 methylation analysis of primary breast cancer specimens. MSP was performed on bisulfite-treated DNA from primary invasive breast cancer tissues. MSP results from 19 representative patient samples are shown. DNA bands in lanes labeled with U indicate PCR products amplified with primers recognizing the unmethylated SFRP2 promoter sequence. DNA bands in lanes labeled with M represent amplified products with methylation-specific primers. In addition, five representative normal cancer-unrelated breast tissues (N1 – N5) are shown. DNA from the breast cancer cell line BT20 and lymphocyte DNA from a healthy donor (Lyc) served as positive controls for MSP. NTC designates the no template control; T indicates tumor tissue; N indicates normal breast tissue.
Correlation of SFRP2 promoter methylation and SFRP2 mRNA expression in breast cancer cell lines
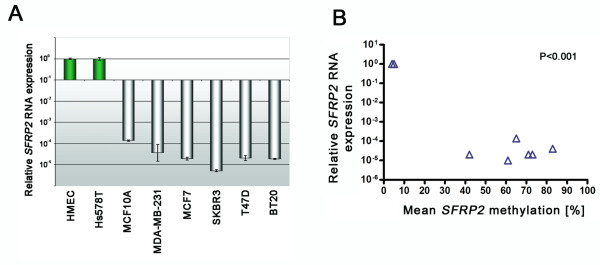
In order to investigate the association of SFRP2 promoter methylation with transcriptional silencing in a quantitative manner we assessed in parallel SFRP2 mRNA expression by realtime PCR and SFRP2 methylation by quantitative Pyrosequencing in breast cell lines. The technical controls for Pyrosequencing revealed median methylation of 4% (unmethylated control) and 90% (methylated control) in the 22 sequential CpG sites, by this defining the detection limits of this assay. HMEC cells exhibited abundant SFRP2 mRNA expression (Δcycle threshold (CT) GAPDH:SFRP2 = 5.1) (Figure 4A) together with median SFRP2 methylation of 4%, in contrast to e.g. BT20 cells which exhibited mean methylation of 83% together with absence of SFRP2 mRNA expression (ΔCT GAPDH:SFRP2 = 19.9). Median SFRP2 methylation for Hs578T, MCF10A, MCF7, MDA-MB-231, SKBR3 and T47D was 5%, 73%, 73%, 71%, 61% and 42%, respectively. A direct comparison of SFRP2 mRNA expression and SFRP2 promoter methylation indicates that in breast cell lines SFRP2 methylation is correlated with loss of SFRP2 mRNA expression (P < 0.001; Figure 4B). Thus, when applying an empiric cut-off of > 5% to discriminate between SFRP2 methylation and non-methylation, all semi-quantitative results for SFRP2 methylation in breast cell lines obtained by MSP could be confirmed by the quantitative Pyrosequencing assay.
Figure 4.
Correlation of SFRP2 promoter methylation and loss of SFRP2 mRNA expression in breast cell lines. (A) Realtime PCR of eight breast cell lines revealed abundant SFRP2 mRNA expression in benign HMEC and malignant Hs578T cells (green bars), whereas in all other investigated malignant breast cell lines SFRP2 mRNA expression was substantially reduced (grey bars). (B) Plotting of SFRP2 mRNA expression from the same cell lines (Y-axis) against each mean SFRP2 promoter methylation value (X-axis) reveals a significant correlation between loss of RNA expression and SFRP2 promoter methylation (P = 0.001; Pearson's correlation coefficient r = -0.9241).
Differential expression of SFRP2 protein in primary breast cancer
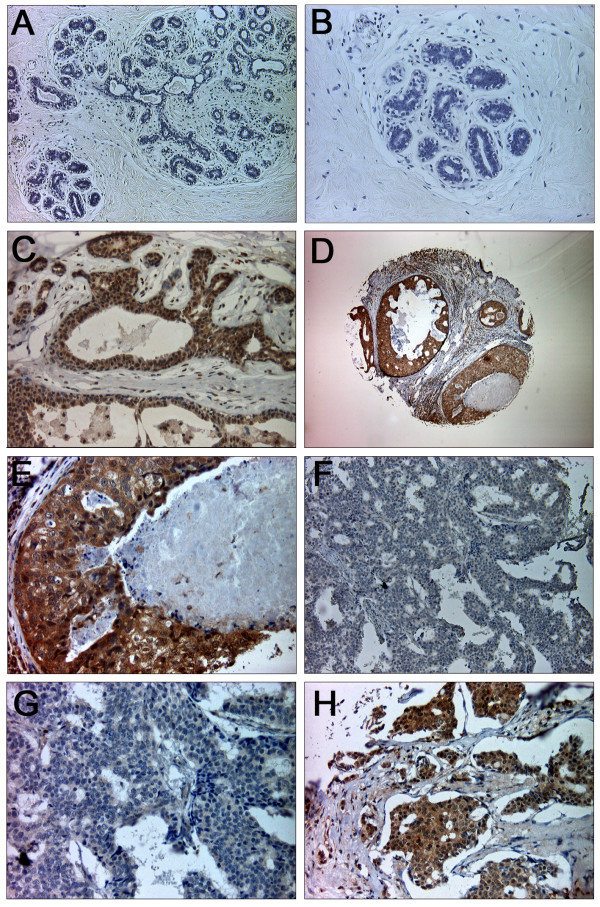
Immunohistochemical analysis was used to investigate SFRP2 protein expression in normal and malignant breast tissue. A total of 125 informative breast cancer cases, four DCIS and 10 normal breast tissues were analyzed. Intensity of immunohistochemical staining was evaluated using a semi-quantitative IRS [39]. SFRP2 protein was clearly detectable in 90% (9/10) of normal breast tissue samples analyzed, as defined by an IRS ≥ 8 (Figure 5C). The mean expression was determined to be IRS = 7.5 (range 3–8; standard deviation (SD) ± 1.6), and median expression to be IRS = 8. Expression was predominantly localized in luminal and basal epithelial cells of the normal breast while weak expression was detectable in adjacent stromal cells. In four DCIS, SFRP2 expression was slightly reduced (mean IRS = 6.5; range 4–8; SD ± 1.9; median IRS = 7; Figure 5D, E) with one sample showing strong reduction (25%, IRS = 4). However, the mean expression in invasive breast carcinomas was determined to be IRS = 4.6 (range 0–8; SD ± 2.1) and median expression to be IRS = 4. Invasive breast carcinomas showed strongly reduced or complete loss (IRS ≤ 4) of SFRP2 expression in 74% (93/125) of cases (Figure 5F, G). The SFRP2 expression difference between tumors and normal breast tissues was statistically significant (P = 0.001).
Figure 5.
SFRP2 protein expression in normal and malignant breast tissues. (A) In the negative control showing normal breast tissue the primary SFRP2 antibody was omitted. (B) Scale-up of negative control shown in A. (C) Strong SFRP2 expression in ducts and lobules of normal breast tissue (IRS = 12). SFRP2 expression is abundant in epithelial cells, while there is only weak expression in stromal cells. (D) Very abundant SFRP2 expression in ductal carcinoma in situ of the breast. (E) Scale-up of specimen shown in D. (F) High-grade tumor exhibiting substantial loss of SFRP2 expression (IRS = 0). (G) Scale-up of specimen shown in F. (H) High-grade breast carcinoma with intensive cytoplasmic SFRP2 staining (IRS = 12). Original magnifications: A, F = 100×; B, C, G, H = 200×; D = 40×; E = 400×.
Correlation of SFRP2 expression and SFRP2 methylation with clinicopathological parameters and patient survival
Clinicopathological characteristics of breast cancer patients were first correlated with SFRP2 protein expression for descriptive data analysis (Table 1). Loss of SFRP2 expression in tumor tissue (IRS ≤ 4) was not associated with age at diagnosis, tumor size, histological grade, histological type, estrogen/progesterone receptor status, Her2 status or expression of p53. A prevalence of more abundant SFRP2 expression was detected in node negative breast tumors (P = 0.033). In univariate survival analysis using log-rank test, loss of SFRP2 protein expression was not associated with DFS (P = 0.237), but a weak trend was detected towards reduced OS (P = 0.071) (Figure 6). SFRP2 promoter methylation in breast carcinomas was not associated with age at diagnosis, tumor size, lymph node status, histological grade, histological type, or estrogen/progesterone receptor status (Table 2). In univariate survival analysis, lymph node status, histological grade and histological type were significantly associated with DFS; lymph node status and histological grade were significantly associated with OS (Table 3). However, SFRP2 methylation was neither associated with DFS (P = 0.192) nor with OS intervals (P = 0.686).
Table 1.
Clinicopathological and immunohistochemical factors in relation to SFRP2 immunoreactivity
| SFRP2 immunoreactivity | |||||
| Variable | na | IRS 0 – 4 (%) | IRS > 4 (%) | Rd | Pe |
| Total | 125 | 93 (74) | 32 (26) | ||
| Clinicopathological factors | |||||
| Age at diagnosis (median: 57 years; range 29 – 82 years) | |||||
| < 60 years | 69 | 49 (71) | 20 (29) | -0.103 | 0.255 |
| ≥ 60 years | 55 | 44 (80) | 11 (20) | ||
| Tumor sizeb | |||||
| pT1 | 42 | 29 (69) | 13 (31) | -0.110 | 0.226 |
| pT2 – 4 | 81 | 64 (79) | 17 (21) | ||
| Lymph node statusb | |||||
| pN0 | 62 | 41 (66) | 21 (34) | -0.194 | 0.033 |
| pN1 – 3 | 59 | 49 (83) | 10 (17) | ||
| Histological grade | |||||
| G1 – G2 | 67 | 49 (73) | 18 (27) | -0.047 | 0.606 |
| G3 | 57 | 44 (77) | 13 (23) | ||
| Histological type | |||||
| ductal | 98 | 73 (75) | 25 (26) | 0.024 | 0.796 |
| lobular | 9 | 9 (100) | 0 (0) | ||
| other | 14 | 9 (64) | 5 (36) | ||
| Immunohistochemistry | |||||
| Estrogen receptor status | |||||
| negative (IRSc 0 – 2) | 28 | 20 (71) | 8 (29) | -0.079 | 0.424 |
| positive (IRS 3 – 12) | 76 | 60 (79) | 16 (21) | ||
| Progesterone receptor status | |||||
| negative (IRSc 0 – 2) | 72 | 55 (76) | 17 (24) | 0.048 | 0.611 |
| positive (IRS 3 – 12) | 43 | 31 (72) | 12 (28) | ||
| Her2 | |||||
| negative (0, 1+) | 91 | 70 (77) | 21 (23) | 0.110 | 0.238 |
| positive (2+, 3+) | 26 | 17 (65) | 9 (35) | ||
| p53 | |||||
| negative (< 5%) | 70 | 53 (76) | 17 (24) | 0.033 | 0.724 |
| positive (≥ 5%) | 44 | 32 (73) | 12 (27) | ||
aOnly female patients with primary, unilateral, invasive breast cancer were included. bAccording to UICC: TNM Classification of Malignant Tumours [38]. cIRS = immunoreactivity score according to Remmele and Stegner [40]. dPearson's correlation coefficient. eFisher's exact test (two-sided). Significant P-values are marked in bold face. Percentages may not sum to 100 due to rounding.
Figure 6.
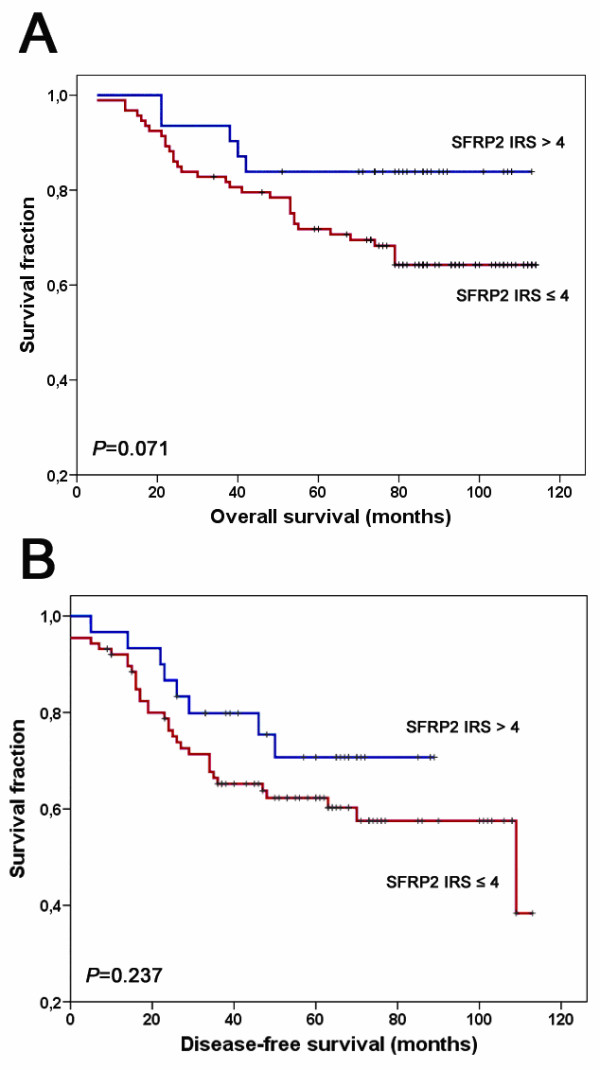
Loss of SFRP2 protein expression in correlation to patient survival. (A) Univariate Kaplan-Meier analysis of patient overall survival stratified between SFRP2 expresser (IRS > 4, blue line) and SFRP2 non-expresser (IRS ≤ 4, red line). A weak association of loss of SFRP2 protein expression with unfavorable outcome could be detected (P = 0.071, two-sided log-rank test). (B) Univariate Kaplan-Meier analysis regarding disease-free patient survival. The visual impression of a clinical impact of SFRP2 protein expression loss on breast tumor recurrence was statistically not significant (P = 0.237, two-sided log-rank test). Vertical tick marks represent censored patients.
Table 2.
Correlation analysis of SFRP2 promoter methylation with clinicopathological and immunohistochemical patient characteristics
| SFRP2 promoter | |||||
| Variable | na | Unmethylated (%) | Methylated (%) | Rb | Pc |
| Total | 199 | 34 (17) | 165 (83) | ||
| Clinicopathological factors | |||||
| Age at diagnosis | |||||
| < 60 years | 114 | 24 (21) | 90 (79) | 0.122 | 0.091 |
| ≥ 60 years | 85 | 10 (12) | 75 (88) | ||
| Tumor sized | |||||
| pT1 | 70 | 11 (16) | 59 (84) | -0.013 | 1.000 |
| pT2 – 4 | 114 | 19 (17) | 95 (83) | ||
| Lymph node statusd | |||||
| pN0 | 89 | 19 (21) | 70 (79) | 0.129 | 0.106 |
| pN1 – 3 | 85 | 10 (12) | 75 (88) | ||
| Histological grade | |||||
| G1 – G2 | 109 | 13 (12) | 96 (88) | -0.139 | 0.069 |
| G3 | 76 | 17 (22) | 59 (78) | ||
| Histological type | |||||
| ductal | 155 | 26 (17) | 129 (83) | -0.057 | 0.434 |
| lobular | 24 | 1 (4) | 23 (96) | ||
| other | 14 | 5 (36) | 9 (64) | ||
| Immunohistochemistry | |||||
| Estrogen receptor | |||||
| negative (IRSe 0 – 2) | 64 | 13 (20) | 51 (80) | 0.084 | 0.295 |
| positive (IRS 3 – 12) | 123 | 17 (14) | 106 (86) | ||
| Progesterone receptor | |||||
| negative (IRSe 0 – 2) | 70 | 14 (20) | 56 (80) | 0.083 | 0.304 |
| positive (IRS 3 – 12) | 117 | 16 (14) | 101 (86) | ||
Table 3.
Univariate survival analysis of clinicopathological and immunohistochemical parameters with SFRP2 promoter methylation
| Disease-free survival (DFS) | Overall survival (OS) | |||||
| Variable | na | Events | Pb | na | Events | Pb |
| Clinicopathological factors | ||||||
| Age at diagnosis | ||||||
| < 60 years | 84 | 33 | 0.391 | 83 | 15 | 0.414 |
| ≥ 60 years | 52 | 18 | 53 | 13 | ||
| Tumor sizec | ||||||
| pT1 | 51 | 16 | 0.059 | 51 | 8 | 0.165 |
| pT2 – 4 | 82 | 35 | 82 | 20 | ||
| Lymph node statusc | ||||||
| pN0 | 59 | 15 | 0.008 | 59 | 7 | 0.026 |
| pN1 – 3 | 66 | 31 | 66 | 18 | ||
| Histological grade | ||||||
| G1 – G2 | 72 | 19 | 0.003 | 73 | 7 | 0.001 |
| G3 | 61 | 32 | 61 | 21 | ||
| Histological type | ||||||
| ductal | 109 | 35 | 0.009 | 109 | 25 | 0.622 |
| lobular | 18 | 9 | 18 | 2 | ||
| other | 9 | 7 | 9 | 1 | ||
| Immunohistochemistry | ||||||
| Estrogen receptor | ||||||
| negative (IRSd 0 – 2) | 47 | 16 | 0.644 | 46 | 12 | 0.118 |
| positive (IRS 3 – 12) | 84 | 35 | 85 | 16 | ||
| Progesterone receptor | ||||||
| negative (IRSd 0 – 2) | 44 | 19 | 0.318 | 45 | 12 | 0.093 |
| positive (IRS 3 – 12) | 87 | 32 | 86 | 16 | ||
| SFRP2 promoter | ||||||
| unmethylated | 23 | 5 | 0.192 | 23 | 5 | 0.686 |
| methylated | 113 | 45 | 113 | 23 | ||
SFRP2 inhibits proliferation in breast cell lines
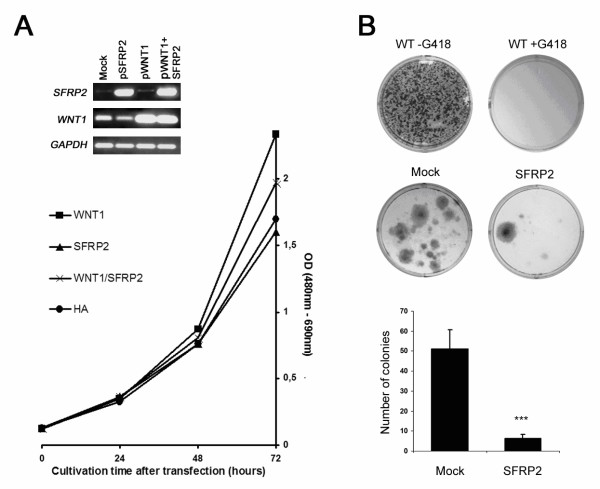
Finally, we asked whether SFRP2 influences proliferation rates in breast cell lines. For gain-of-function experiments we chose mammary MCF10A cells since these cells were found to lack SFRP2 mRNA expression. As shown in Figure 7A, WNT1 overexpressing MCF10A cells notably increased proliferation compared to cells mock-transfected with empty vector. The equimolar co-transfection with SFRP2resulted in a decreased proliferation rate as compared to WNT1 alone. Furthermore, SFRP2-transfectants revealed a slightly reduced proliferation rate compared to cells containing empty vector. In order to support these findings we performed a colony formation assay and selected transfected clones for three weeks by the antibiotic G418. Representative results are shown in Figure 7B. Controls assured the feasibility of this assay, showing that wild type cells without G418 abundantly form colonies, whereas in the presence of G418 wild type cells fail to survive. SFRP2-transfected cells exhibit a reduced number of colonies as compared to mock-transfected cells. The difference in colony numbers from three independent experiments was statistically significant (P = 0.045, two-tailed Mann-Whitney U-test).
Figure 7.
SFRP2 inhibits proliferation in breast epithelial cells. (A) MCF10A cells transfected with empty vector (Mock), SFRP2-expression vector, WNT1-expression vector or a combination of SFRP2+WNT1-expression vectors were subjected to XTT proliferation assays. At three subsequent time points, optical densities (OD) of the supernatants were measured. RNA Expression of the respective genes was surveyed in parallel 4 days post transfection. GAPDH served as cDNA loading control. SFRP2 was able to reduce a proliferative effect mediated by WNT1 overexpression, whereas the inhibiting effect of SFRP2 on cells not stimulated by WNT1 was only marginal (HA). (B) Long-term inhibiting effects of SFRP2 on the ability to form colonies in MCF10A cells. Cells were either transfected with empty vector (Mock) or with SFRP2-expression vector, and after 3 weeks of selective pressure (antibiotic G418) fixed, stained and photographed. Non-transfected wild type cells (WT) are shown as controls with (+) and without (-) the application of G418. The difference in colony numbers between mock-transfected and SFRP2-transfected cells is significant (P = 0.045; two-tailed Mann-Whitney U-test).
Discussion
Aberrant methylation of CpG islands in gene promoters has been ascertained as a primary mechanism for the inactivation of tumor suppressor genes in human malignancies, including colon and breast cancer (for review see [3]). Clinically, the identification of genes that are prone to abnormal methylation and consequently become downregulated is of critical importance since this is considered to provide a good source of novel tumor biomarkers [52] and potential targets for chemotherapeutics [53,54]. The family of SFRP genes, functionally acting as Wnt signaling inhibitors, was recently shown to be a common target of promoter hypermethylation in numerous tumor entities [19-26]. In human breast cancer, we have previously shown that the SFRP1 and SFRP5 promoter is epigenetically silenced in 61% and 73% of invasive breast carcinomas, respectively, each of which was associated with unfavorable patient prognosis [27,28]. We here demonstrate that promoter methylation of SFRP2 is a further tumor-related alteration in human breast cancer occurring with even higher incidence.
Initiating our study, we found that many breast cancer cell lines revealed abolished SFRP2 expression presumably due to methylation of the SFRP2 promoter, since those cell lines lacking SFRP2 methylation abundantly expressed SFRP2 mRNA, whereas all cell lines lacking SFRP2 expression harbored SFRP2 promoter methylation. A direct coherence between promoter methylation and loss of RNA expression was shown by a combined DAC/TSA treatment of breast cancer cell lines, demonstrating that the SFRP2 gene was effectively demethylated and re-expressed after the treatment. Furthermore, those cell lines revealed a significant reduction of cyclin D1 expression, suggesting reactivation of anti-proliferative genes, of which SFRP2 is supposed to be a member [19,23,24]. Interestingly, in partially methylated SKBR3 and T47D cells the sole inhibition of histone deacetylases by TSA led to restoration of some SFRP2 expression, indicating that besides DNA methylation in these cells further reversible chromatin repressing histone modifications may exist.
Since cell lines may acquire de novo genetic and epigenetic lesions during cultivation [55,56] it is mandatory to investigate such aberrations in primary tissues as well. To this end we analyzed SFRP2 promoter methylation in 199 infiltrating breast carcinomas by MSP. We found a high-frequent incidence of SFRP2 methylation in the tumors (83%), confirming the recent results from Suzuki et al. [37], who reported of SFRP2 methylation in 60 of 78 (77%) primary breast carcinomas. Importantly, SFRP2 methylation was independent of relevant clinicopathological factors, thus being unlikely related to disease stage or a molecular breast cancer subtype. SFRP2 methylation was equally prevalent in small sized (pT1) and in larger sized (pT2-4) breast carcinomas, suggesting it occurs as early epigenetic aberration in breast tumorigenesis with no further increase in methylation frequency during tumor progression. Whether SFRP2 methylation is already present in benign and earlier premalignant lesions such as atypical hyperplasia and carcinoma in situ, like it was recently reported for the 14-3-3-σ gene [9], will be of particular importance in regard of early breast tumor detection. Yet, this remains to be determined in a future study.
Interestingly, Suzuki et al. [37] reported that a certain number of cancer-related normal breast tissues also showed weak SFRP2 methylation in their study, whereas in our study none of the normal breast tissues harbored a methylated SFRP2 promoter, irrespective of whether the tissue was taken from matched cancer-related or unmatched cancer-unrelated specimens. Given that no contaminating tumor cells had been present in their normal breast tissues this might be due to different locations of the recruited tissues (i.e. distance to the tumor margin), and may address a phenomenon that in cancer research is currently being discussed as "field defect" [57,58]. Evidence of such field defect in breast cancer was brought up by Yan et al. [59] showing that RASSF1A promoter methylation in breast carcinoma may progressively diffuse outwards to surrounding normal tissue, establishing a sphere of methylation gradient around the primary tumor. Recently, such gradient was also detected for RUNX3 methylation [60], which together with RASSF1A methylation is among the earliest carcinogenetic events in breast tumor transformation. SFRP2 methylation may be implicated in such field defect in breast cancer, yet dense methylation of the SFRP2 promoter was restricted to carcinoma in our study, and thus it may display important clinical specificity. In bladder cancer, SFRP2 methylation was shown to represent an independent predictor of malignancy, although in multivariate logistic regression analysis it was not a reliable biomarker because of a limited sensitivity/specificity due to some extent of methylation in normal bladder mucosa [22]. In contrast, in faecal DNA SFRP2 methylation was proven to be a highly promising screening marker for colorectal cancer [34], even potent to detect early lesions like adenoma, aberrant crypt foci [35] and colorectal polyps [36] due to the absence of SFRP2 methylation in normal colonic mucosa. In breast cancer, the accurate specificity and sensitivity of SFRP2 methylation remains to be determined by quantitative methods in a future study, for instance by qMSP (MethyLight) [61] or the Pyrosequencing technique [62], integrating receiver-operating characteristic (ROC) curve analyses. This may potentially lead to a valuable early tumor detection marker that will ideally be assessable in patients' body fluids like blood serum, plasma or nipple aspirate.
Contrasting the view that SFRP2 acts as a tumor suppressor gene, Lee and co-workers [42,63] suggested that SFRP2 exhibits rather an oncogenic property in breast tissue since this group detected strong upregulation of SFRP2 protein in canine mammary tumors relative to normal canine breast tissues. In addition, SFRP2 overexpression in a human breast cancer cell line (MCF7) inhibited apoptosis following UV light exposure, while increasing cell-substrate adhesion capacity [64]. It is worthy to note that these experiments were carried out with a canine homologue of SFRP2 cDNA. However, five lines of evidence propose a tumor suppressive role of SFRP2 in human breast carcinogenesis: (1.) Our and another independent study [37] demonstrate that SFRP2 is very frequently targeted by promoter methylation in human breast carcinomas as compared to normal human breast tissues, disposing breast cancer to the large number of human tumor entities for which SFRP2 methylation has already been described. (2.) We found a strong correlation and functional association of SFRP2 methylation with loss of SFRP2 mRNA expression in breast cell lines. (3.) Our study reveals a common SFRP2 protein loss in human breast carcinomas with comparable frequency to promoter methylation, notably by applying the identical SFRP2-antibody that was used for the study of canine mammary tumors. (4.) We detected a weak trend towards adverse clinical patient outcome in case of SFRP2 protein expression loss. (5.) Functional analyses in human breast [37], gastric [23,24] and colorectal cancer cell lines [19] revealed a pro-apoptotic and anti-proliferative capacity of (human) SFRP2 associated with the ability to inhibit activated Wnt signaling, altogether supporting a tumor suppressive rather than an oncogenic function of this gene. These discrepancies to canine mammary tumors may reflect subtle distinctions in the function of structurally related molecules, or alternative activities of molecules when expressed in different contexts and organisms. Furthermore, it emphasizes that study results of SFRP2 from canine breast cancer models may not be generally transferable to human breast carcinogenesis. In conclusion, SFRP2 may represent a candidate class II tumor suppressor gene whose altered expression is caused by epigenetic changes (class II) rather than by mutation (class I) [65]. Class II tumor suppressor genes are particularly interesting drug targets since reversing the block of their gene expression, e.g. by DNA methyltransferase (DNMT) inhibitors or histone deacetylase (HDAC) inhibitors could lead to tumor regression. Furthermore such a treatment could be appropriate to eliminate minimal residual cancer disease after surgical resection of the tumor.
Summarizing, our data demonstrate that SFRP2 is a frequent target of epigenetic inactivation in human breast cancer leading to downregulation of SFRP2 expression in mammary tumors. Loss of SFRP2 expression confers a growth advantage to mammary cells, likely due its ability to inhibit oncogenic Wnt signaling. Altogether, our data support the proposed tumor suppressive function of SFRP2 in normal breast tissue. The high incidence and the putative specificity of this epimutation may qualify SFRP2 methylation as potential candidate in a screening marker panel for the early detection of human breast cancer.
Conclusion
Our study on SFRP2 in human breast cancer leads to the following conclusions: SFRP2 expression is very frequently downregulated in breast cancer due to promoter methylation, thus conferring growth advantage to neoplastic mammary cells. Therefore, SFRP2 may be assigned a class II tumor suppressor gene in normal breast tissue, whose block of expression could be reversed by DNA demethylating (DNMT inhibitors) and histone reacetylating (HDAC inhibitors) drugs. In contrast to an adverse prognostic value of SFRP1 or SFRP5 methylation in breast cancer, failure of SFRP2 methylation as a prognostic biomarker may be explained by redundant functions of these closely related SFRP molecules. Alternatively, this failure could be explained by the likely involvement of SFRP2 methylation in the early steps of breast carcinogenesis, rather than being implicated in the development of prognostically adverse tumor subtypes. Nevertheless, SFRP2 methylation may be potentially useful as a molecular tumor biomarker in a DNA methylation biomarker based screening assay, as it may display high clinical sensitivity and specificity in detecting breast cancer cells.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JV participated in the design of the study, carried out the RNA expression and methylation analyses, immunohistochemical studies, in vitro experiments, statistical analysis, and wrote the manuscript. EN performed realtime expression analysis, assisted in Pyrosequencing and data interpretation, and critically revised the manuscript. NB participated in immunohistochemical analysis and data interpretation, and critically revised the manuscript. EJ designed and optimized the SFRP2 Pyrosequencing assay, and critically revised the manuscript. AH participated in collection of clinical data, performed data interpretation, and critically revised the manuscript. RK participated in the design and coordination of the study, and critically revised the manuscript. ED planned and coordinated the study, and critically revised the manuscript. All authors have read and approved the final version of the manuscript.
Supplementary Material
Clinicopathological and immunohistochemical characteristics of primary invasive breast carcinomas (n = 199). The data provided represent the relevant clinicopathological and immunohistochemical patient characteristics used in SFRP2 methylation analysis.
Primer sequences and cycle conditions used in this study. This table provides oligonucleotide primer sequences and PCR cycle conditions that were used throughout this study.
Acknowledgments
Acknowledgements
The expert technical help of Sevim Alkaya, Sonja von Serényi and Inge Losen is greatly appreciated. We thank Dr. Dieter Niederacher (Department of Gynecology and Obstetrics, Heinrich-Heine-University, Düsseldorf, Germany) and Prof. Matthias Dürst (Friedrich-Schiller University, Jena, Germany) for kindly providing patient samples. We are thankful to Dr. Monika Klinkhammer-Schalke and Monika Kerscher from the Tumor Registry Regensburg for continuous help in obtaining clinical follow-up data. The SFRP2/WNT1 vectors were a kind gift from Dr. Hiromu Suzuki (Sapporo Medical University, Sapporo, Japan). This work is a research project within the German Human Genome Project and has been supported by the grant from the Bundesministerium für Bildung und Forschung to Edgar Dahl (01KW0401).
Contributor Information
Jürgen Veeck, Email: juergen.veeck@rwth-aachen.de.
Erik Noetzel, Email: enoetzel@ukaachen.de.
Nuran Bektas, Email: nbektas@ukaachen.de.
Edgar Jost, Email: ejost@ukaachen.de.
Arndt Hartmann, Email: arndt.hartmann@uk-erlangen.de.
Ruth Knüchel, Email: rknuechel-clarke@ukaachen.de.
Edgar Dahl, Email: edahl@ukaachen.de.
References
- Herman JG, Baylin S. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- Esteller M. Aberrant DNA methylation as a cancer-inducing mechanism. Annu Rev Pharmacol Toxicol. 2005;45:629–656. doi: 10.1146/annurev.pharmtox.45.120403.095832. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Murphy RF, Agrawal DK. DNA methylation in breast and colorectal cancers. Mod Pathol. 2007;20:711–721. doi: 10.1038/modpathol.3800822. [DOI] [PubMed] [Google Scholar]
- Herman JG, Merlo A, Mao L, Lapidus RG, Issa JP, Davidson NE, Sidransky D, Baylin SB. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- Ottaviano YL, Issa JP, Parl FF, Smith HS, Baylin SB, Davidson NE. Methylation of the estrogen receptor gene CpG island marks loss of estrogen receptor expression in human breast cancer cells. Cancer Res. 1994;54:2552–2555. [PubMed] [Google Scholar]
- Graff JR, Herman JG, Lapidus RG, Chopra H, Xu R, Jarrard DF, Isaacs WB, Pitha PM, Davidson NE, Baylin SB. E-cadherin expression is silenced by DNA hypermethylation in human breast and prostate carcinomas. Cancer Res. 1995;55:5195–5199. [PubMed] [Google Scholar]
- Lehmann U, Celikkaya G, Hasemeier B, Länger F, Kreipe H. Promoter hypermethylation of the death-associated protein kinase gene in breast cancer is associated with the invasive lobular subtype. Cancer Res. 2002;62:6634–6638. [PubMed] [Google Scholar]
- Veeck J, Chorovicer M, Naami A, Breuer E, Zafrakas M, Bektas N, Dürst M, Kristiansen G, Wild PJ, Hartmann A, Knuechel R, Dahl E. The extracellular matrix protein ITIH5 is a novel prognostic marker in invasive node-negative breast cancer and its aberrant expression is caused by promoter hypermethylation. Oncogene. 2008;27:865–875. doi: 10.1038/sj.onc.1210669. [DOI] [PubMed] [Google Scholar]
- Umbricht CB, Evron E, Gabrielson E, Ferguson A, Marks J, Sukumar S. Hypermethylation of 14-3-3 sigma (stratifin) is an early event in breast cancer. Oncogene. 2001;20:3348–3353. doi: 10.1038/sj.onc.1204438. [DOI] [PubMed] [Google Scholar]
- Belinsky SA, Liechty KC, Gentry FD, Wolf HJ, Rogers J, Vu K, Haney J, Kennedy TC, Hirsch FR, Miller Y, Franklin WA, Herman JG, Baylin SB, Bunn PA, Byers T. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. Cancer Res. 2006;66:3338–3344. doi: 10.1158/0008-5472.CAN-05-3408. [DOI] [PubMed] [Google Scholar]
- Jones SE, Jomary C. Secreted Frizzled-related proteins: searching for relationships and patterns. Bioessays. 2002;24:811–820. doi: 10.1002/bies.10136. [DOI] [PubMed] [Google Scholar]
- Uren A, Reichsman F, Anest V, Taylor WG, Muraiso K, Bottaro DP, Cumberledge S, Rubin JS. Secreted frizzled-related protein-1 binds directly to Wingless and is a biphasic modulator of Wnt signaling. J Biol Chem. 2000;275:4374–4382. doi: 10.1074/jbc.275.6.4374. [DOI] [PubMed] [Google Scholar]
- Bafico A, Liu G, Yaniv A, Gazit A, Aaronson SA. Novel mechanism of Wnt signalling inhibition mediated by Dickkopf-1 interaction with LRP6/Arrow. Nat Cell Biol. 2001;3:683–686. doi: 10.1038/35083081. [DOI] [PubMed] [Google Scholar]
- Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- Bukholm IK, Nesland JM, Børresen-Dale AL. Re-expression of E-cadherin, alpha-catenin and beta-catenin, but not of gamma-catenin, in metastatic tissue from breast cancer patients. J Pathol. 2000;190:15–19. doi: 10.1002/(SICI)1096-9896(200001)190:1<15::AID-PATH489>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Ryo A, Nakamura M, Wulf G, Liou YC, Lu KP. Pin1 regulates turnover and subcellular localization of beta-catenin by inhibiting its interaction with APC. Nat Cell Biol. 2001;3:793–801. doi: 10.1038/ncb0901-793. [DOI] [PubMed] [Google Scholar]
- Lin SY, Xia W, Wang JC, Kwong KY, Spohn B, Wen Y, Pestell RG, Hung MC. Beta-catenin, a novel prognostic marker for breast cancer: its roles in cyclin D1 expression and cancer progression. Proc Natl Acad Sci USA. 2000;97:4262–4266. doi: 10.1073/pnas.060025397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung GG, Zerkowski MP, Ocal IT, Dolled-Filhart M, Kang JY, Psyrri A, Camp RL, Rimm DL. beta-Catenin and p53 analyses of a breast carcinoma tissue microarray. Cancer. 2004;100:2084–2092. doi: 10.1002/cncr.20232. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, van Engeland M, Toyota M, Tokino T, Hinoda Y, Imai K, Herman JG, Baylin SB. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- Lee AY, He B, You L, Dadfarmay S, Xu Z, Mazieres J, Mikami I, McCormick F, Jablons DM. Expression of the secreted frizzled-related protein gene family is downregulated in human mesothelioma. Oncogene. 2004;23:6672–6676. doi: 10.1038/sj.onc.1207881. [DOI] [PubMed] [Google Scholar]
- Urakami S, Shiina H, Enokida H, Hirata H, Kawamoto K, Kawakami T, Kikuno N, Tanaka Y, Majid S, Nakagawa M, Igawa M, Dahiya R. Wnt antagonist family genes as biomarkers for diagnosis, staging, and prognosis of renal cell carcinoma using tumor and serum DNA. Clin Cancer Res. 2006;12:6989–6997. doi: 10.1158/1078-0432.CCR-06-1194. [DOI] [PubMed] [Google Scholar]
- Urakami S, Shiina H, Enokida H, Kawakami T, Kawamoto K, Hirata H, Tanaka Y, Kikuno N, Nakagawa M, Igawa M, Dahiya R. Combination analysis of hypermethylated Wnt-antagonist family genes as a novel epigenetic biomarker panel for bladder cancer detection. Clin Cancer Res. 2006;12:2109–2116. doi: 10.1158/1078-0432.CCR-05-2468. [DOI] [PubMed] [Google Scholar]
- Cheng YY, Yu J, Wong YP, Man EP, To KF, Jin VX, Li J, Tao Q, Sung JJ, Chan FK, Leung WK. Frequent epigenetic inactivation of secreted frizzled-related protein 2 (SFRP2) by promoter methylation in human gastric cancer. Br J Cancer. 2007;97:895–901. doi: 10.1038/sj.bjc.6603968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima M, Suzuki H, Toyota M, Watanabe Y, Maruyama R, Sasaki S, Sasaki Y, Mita H, Nishikawa N, Yamaguchi K, Hirata K, Itoh F, Tokino T, Mori M, Imai K, Shinomura Y. Frequent epigenetic inactivation of SFRP genes and constitutive activation of Wnt signaling in gastric cancer. Oncogene. 2007;26:4699–4713. doi: 10.1038/sj.onc.1210259. [DOI] [PubMed] [Google Scholar]
- Nomoto S, Kinoshita T, Kato K, Otani S, Kasuya H, Takeda S, Kanazumi N, Sugimoto H, Nakao A. Hypermethylation of multiple genes as clonal markers in multicentric hepatocellular carcinoma. Br J Cancer. 2007;97:1260–1265. doi: 10.1038/sj.bjc.6604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl E, Wiesmann F, Woenckhaus M, Stoehr R, Wild PJ, Veeck J, Knüchel R, Klopocki E, Sauter G, Simon R, Wieland WF, Walter B, Denzinger S, Hartmann A, Hammerschmied CG. Frequent loss of SFRP1 expression in multiple human solid tumours: association with aberrant promoter methylation in renal cell carcinoma. Oncogene. 2007;26:5680–5691. doi: 10.1038/sj.onc.1210345. [DOI] [PubMed] [Google Scholar]
- Veeck J, Niederacher D, An H, Klopocki E, Wiesmann F, Betz B, Galm O, Camara O, Dürst M, Kristiansen G, Huszka C, Knüchel R, Dahl E. Aberrant methylation of the Wnt antagonist SFRP1 in breast cancer is associated with unfavourable prognosis. Oncogene. 2006;25:3479–3488. doi: 10.1038/sj.onc.1209386. [DOI] [PubMed] [Google Scholar]
- Veeck J, Geisler C, Noetzel E, Alkaya S, Hartmann A, Knüchel R, Dahl E. Epigenetic inactivation of the Secreted frizzled-related protein-5 (SFRP5) gene in human breast cancer is associated with unfavorable prognosis. Carcinogenesis. 2008;29:991–998. doi: 10.1093/carcin/bgn076. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Gabrielson E, Chen W, Anbazhagan R, van Engeland M, Weijenberg MP, Herman JG, Baylin SB. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet. 2002;31:141–149. doi: 10.1038/ng892. [DOI] [PubMed] [Google Scholar]
- Zou H, Molina JR, Harrington JJ, Osborn NK, Klatt KK, Romero Y, Burgart LJ, Ahlquist DA. Aberrant methylation of secreted frizzled-related protein genes in esophageal adenocarcinoma and Barrett's esophagus. Int J Cancer. 2005;116:584–591. doi: 10.1002/ijc.21045. [DOI] [PubMed] [Google Scholar]
- Marsit CJ, Karagas MR, Andrew A, Liu M, Danaee H, Schned AR, Nelson HH, Kelsey KT. Epigenetic inactivation of SFRP genes and TP53 alteration act jointly as markers of invasive bladder cancer. Cancer Res. 2005;65:7081–7085. doi: 10.1158/0008-5472.CAN-05-0267. [DOI] [PubMed] [Google Scholar]
- Fukui T, Kondo M, Ito G, Maeda O, Sato N, Yoshioka H, Yokoi K, Ueda Y, Shimokata K, Sekido Y. Transcriptional silencing of secreted frizzled related protein 1 (SFRP1) by promoter hypermethylation in non-small-cell lung cancer. Oncogene. 2005;24:6323–6327. doi: 10.1038/sj.onc.1208777. [DOI] [PubMed] [Google Scholar]
- Qi J, Zhu YQ, Luo J, Tao WH. Hypermethylation and expression regulation of secreted frizzled-related protein genes in colorectal tumor. World J Gastroenterol. 2006;12:7113–7117. doi: 10.3748/wjg.v12.i44.7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller HM, Oberwalder M, Fiegl H, Morandell M, Goebel G, Zitt M, Mühlthaler M, Ofner D, Margreiter R, Widschwendter M. Methylation changes in faecal DNA: a marker for colorectal cancer screening? Lancet. 2004;363:1283–1285. doi: 10.1016/S0140-6736(04)16002-9. [DOI] [PubMed] [Google Scholar]
- Huang ZH, Li LH, Yang F, Wang JF. Detection of aberrant methylation in fecal DNA as a molecular screening tool for colorectal cancer and precancerous lesions. World J Gastroenterol. 2007;13:950–954. doi: 10.3748/wjg.v13.i6.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberwalder M, Zitt M, Wöntner C, Fiegl H, Goebel G, Zitt M, Köhle O, Mühlmann G, Ofner D, Margreiter R, Müller HM. SFRP2 methylation in fecal DNA-a marker for colorectal polyps. Int J Colorectal Dis. 2008;23:15–19. doi: 10.1007/s00384-007-0355-2. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Toyota M, Caraway H, Gabrielson E, Ohmura T, Fujikane T, Nishikawa N, Sogabe Y, Nojima M, Sonoda T, Mori M, Hirata K, Imai K, Shinomura Y, Baylin SB, Tokino T. Frequent epigenetic inactivation of Wnt antagonist genes in breast cancer. Br J Cancer. 2008;98:1147–1156. doi: 10.1038/sj.bjc.6604259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobin LH, Wittekind C, eds . UICC: TNM classification of malignant tumors. 5. New York: Wiley-Liss; 1997. [Google Scholar]
- Elston EW, Ellis IO. Method for grading breast cancer. J Clin Pathol. 1993;46:189–190. doi: 10.1136/jcp.46.2.189-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmele W, Stegner HE. Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Pathologe. 1987;8:138–40. [PubMed] [Google Scholar]
- Dahl E, Kristiansen G, Gottlob K, Klaman I, Ebner E, Hinzmann B, Hermann K, Pilarsky C, Dürst M, Klinkhammer-Schalke M, Blaszyk H, Knuechel R, Hartmann A, Rosenthal A, Wild PJ. Molecular profiling of laser-microdissected matched tumor and normal breast tissue identifies karyopherin alpha2 as a potential novel prognostic marker in breast cancer. Clin Cancer Res. 2006;12:3950–3960. doi: 10.1158/1078-0432.CCR-05-2090. [DOI] [PubMed] [Google Scholar]
- Lee JL, Chang CJ, Wu SY, Sargan DR, Lin CT. Secreted frizzled-related protein 2 (SFRP2) is highly expressed in canine mammary gland tumors but not in normal mammary glands. Breast Cancer Res Treat. 2004;84:139–149. doi: 10.1023/B:BREA.0000018412.83348.ff. [DOI] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galm O, Herman JG. Methylation-specific polymerase chain reaction. Methods Mol Med. 2005;113:279–291. doi: 10.1385/1-59259-916-8:279. [DOI] [PubMed] [Google Scholar]
- Liu TH, Raval A, Chen SS, Matkovic JJ, Byrd JC, Plass C. CpG island methylation and expression of the secreted frizzled-related protein gene family in chronic lymphocytic leukemia. Cancer Res. 2006;66:653–658. doi: 10.1158/0008-5472.CAN-05-3712. [DOI] [PubMed] [Google Scholar]
- CpG Island Searcher http://cpgislands.usc.edu/
- Ensembl Genome Browser http://www.ensembl.org/index.html
- Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci USA. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Lukas J, Müller H, Lützhøft D, Strauss M, Bartek J. Cyclin D1 protein expression and function in human breast cancer. Int J Cancer. 1994;57:353–361. doi: 10.1002/ijc.2910570311. [DOI] [PubMed] [Google Scholar]
- Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- Issa JP, Garcia-Manero G, Giles FJ, Mannari R, Thomas D, Faderl S, Bayar E, Lyons J, Rosenfeld CS, Cortes J, Kantarjian HM. Phase 1 study of low-dose prolonged exposure schedules of the hypomethylating agent 5-aza-2'-deoxycytidine (decitabine) in hematopoietic malignancies. Blood. 2004;103:1635–1640. doi: 10.1182/blood-2003-03-0687. [DOI] [PubMed] [Google Scholar]
- Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- Osborne CK, Hobbs K, Trent JM. Biological differences among MCF-7 human breast cancer cell lines from different laboratories. Breast Cancer Res Treat. 1987;9:111–121. doi: 10.1007/BF01807363. [DOI] [PubMed] [Google Scholar]
- Wistuba II, Behrens C, Milchgrub S, Syed S, Ahmadian M, Virmani AK, Kurvari V, Cunningham TH, Ashfaq R, Minna JD, Gazdar AF. Comparison of features of human breast cancer cell lines and their corresponding tumors. Clin Cancer Res. 1998;4:2931–2938. [PubMed] [Google Scholar]
- Shen L, Kondo Y, Rosner GL, Xiao L, Hernandez NS, Vilaythong J, Houlihan PS, Krouse RS, Prasad AR, Einspahr JG, Buckmeier J, Alberts DS, Hamilton SR, Issa JP. MGMT promoter methylation and field defect in sporadic colorectal cancer. J Natl Cancer Inst. 2005;97:1330–1338. doi: 10.1093/jnci/dji275. [DOI] [PubMed] [Google Scholar]
- Maekita T, Nakazawa K, Mihara M, Nakajima T, Yanaoka K, Iguchi M, Arii K, Kaneda A, Tsukamoto T, Tatematsu M, Tamura G, Saito D, Sugimura T, Ichinose M, Ushijima T. High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res. 2006;12:989–995. doi: 10.1158/1078-0432.CCR-05-2096. [DOI] [PubMed] [Google Scholar]
- Yan PS, Venkataramu C, Ibrahim A, Liu JC, Shen RZ, Diaz NM, Centeno B, Weber F, Leu YW, Shapiro CL, Eng C, Yeatman TJ, Huang TH. Mapping geographic zones of cancer risk with epigenetic biomarkers in normal breast tissue. Clin Cancer Res. 2006;12:6626–6636. doi: 10.1158/1078-0432.CCR-06-0467. [DOI] [PubMed] [Google Scholar]
- Cheng AS, Culhane AC, Chan MW, Venkataramu CR, Ehrich M, Nasir A, Rodriguez BA, Liu J, Yan PS, Quackenbush J, Nephew KP, Yeatman TJ, Huang TH. Epithelial progeny of estrogen-exposed breast progenitor cells display a cancer-like methylome. Cancer Res. 2008;68:1786–1796. doi: 10.1158/0008-5472.CAN-07-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh BN, Long TI, Laird PW. DNA methylation analysis by MethyLight technology. Methods. 2001;25:456–462. doi: 10.1006/meth.2001.1268. [DOI] [PubMed] [Google Scholar]
- Colella S, Shen L, Baggerly KA, Issa PJ, Krahe R. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35:146–150. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- Lee JL, Chang CJ, Chueh LL, Lin CT. Secreted Frizzled Related Protein 2 (sFRP2) Decreases Susceptibility to UV-Induced Apoptosis in Primary Culture of Canine Mammary Gland Tumors by NF-kappaB Activation or JNK Suppression. Breast Cancer Res Treat. 2006;100:49–58. doi: 10.1007/s10549-006-9233-9. [DOI] [PubMed] [Google Scholar]
- Lee JL, Lin CT, Chueh LL, Chang CJ. Autocrine/paracrine secreted Frizzled-related protein 2 induces cellular resistance to apoptosis: a possible mechanism of mammary tumorigenesis. J Biol Chem. 2004;279:14602–14609. doi: 10.1074/jbc.M309008200. [DOI] [PubMed] [Google Scholar]
- Sager R. Tumor suppressor genes: the puzzle and the promise. Science. 1989;246:1406–1412. doi: 10.1126/science.2574499. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinicopathological and immunohistochemical characteristics of primary invasive breast carcinomas (n = 199). The data provided represent the relevant clinicopathological and immunohistochemical patient characteristics used in SFRP2 methylation analysis.
Primer sequences and cycle conditions used in this study. This table provides oligonucleotide primer sequences and PCR cycle conditions that were used throughout this study.