Abstract
While the mechanical properties of a substrate or engineered scaffold can govern numerous aspects of cell behavior, cells quickly start to assemble their own matrix and will ultimately respond to their self-made extracellular matrix (ECM) microenvironments. Using fluorescence resonance energy transfer (FRET), we detected major changes in the conformation of a constituent ECM protein, fibronectin (Fn), as cells fabricated a thick three-dimensional (3D) matrix over the course of three days. These data provide the first evidence that matrix maturation occurs and that aging is associated with increased stretching of fibronectin fibrils, which leads to at least partial unfolding of the secondary structure of individual protein modules. A comparison of the conformations of Fn in these 3D matrices with those constructed by cells on rigid and flexible polyacrylamide surfaces suggests that cells in maturing matrices experience a microenviroment of gradually increasing rigidity. In addition, further matrix stiffening is caused by active Fn fiber alignment parallel to the contractile axis of the elongated fibroblasts, a cell-driven effect previously described for other fibrillar matrices. The fibroblasts, therefore, not only cause matrix unfolding, but reciprocally respond to the altered Fn matrix properties by up-regulating their own rigidity response. Consequently, our data demonstrate for the first time that a matured and aged matrix has distinctly different physical and biochemical properties compared to a newly assembled matrix. This might allow cells to specifically recognise the age of a matrix.
Introduction
The mechanical properties of a cell's environment can govern numerous aspects of cell behavior, including cell migration,1–3 gene expression,4,5 cell signaling and differentiation.6–9 Different cell types can be impacted in fundamentally different ways by mechanical factors, which can trigger highly specific functional changes in a manner similar to those induced by soluble biochemical factors.10,11 For example, the optimal substrate stiffness that promotes normal physiological cell function is cell specific, ranging from stiff for bone cells to soft for nerve cells.12–14 The differentiation of mesenchymal stem cells is also reported to be regulated by matrix rigidity. These stem cells differentiate preferentially into a neurogenic linage on soft and an osteogenic linage on rigid substrates.9 However, if cells are cultured over several days, do they still respond to the rigidity of their initial substrate, or do other factors have to be considered? When cells are seeded on tissue culture plates or within engineered 3D scaffolds, they will quickly start to assemble their own matrix and will ultimately respond to their self-made ECM microenvironment. In most mature three-dimensional cell cultures, tissue-engineered scaffolds, and in vivo tissue environments, it is therefore ultimately the ECM which provides the cell with its mechanical and biochemical cues. How do the macroscopically measured materials characteristics of the substrate relate to the microscopic characteristics of cell's self-made matrix? Since cells constantly assemble and remodel their ECM, it is of great importance to establish how ECM is assembled and how the properties of 3D matrices change over time.
While the ECM has been traditionally conceived of as a simple scaffold functioning mainly as a mechanical support for cells, a far more complex picture is emerging in which the ECM is a dynamic entity with physiological functions well beyond simple cell anchorage.9,15–17 For example, fibronectin (Fn) matrices can act as a storage depot for a variety of growth factors, thus regulating cell growth and morphogenesis according to their timed release.18–20 Degradation products are also known to trigger numerous physiological responses, such as antibacterial activity21 and inflammation.22 Furthermore, cell interactions with the ECM can trigger a variety of signaling events via integrins, receptors that provide a molecular bridge between the external environment of the cell and its internal actin cytoskeleton.23–27
Fibronectin (Fn) is a major component of the ECM, and its expression is up-regulated in embryogenesis, at wound sites and during angiogenesis.28 It is a large (450 kD), multimodular protein composed of homologous repeating structural motifs called type I, II, and III modules, which have binding sites for other ECM proteins and for cells as seen in Fig. 1. Fibronectin also has a number of cryptic sites, which remain hidden when the protein is in its globular, soluble form with its two dimeric arms tightly crossed over each other. These cryptic sites become exposed as Fn undergoes conformational changes. Major structural changes have been shown to occur as Fn is polymerized by cells into matrix fibers.29–34 Moreover, the exposure of cryptic sites in Fn modules is an important event regulating integrin binding, cell signaling and the ability of cells to construct new matrix fibrils.30,35–37 Conversely, changes in cell contractility have been shown to alter access to these same sites within matrix fibrils. Inhibition of Rho A, an agonist of cell contractility, prevents the exposure of a cryptic site in FnIII1, and disruption of the actin cytoskeleton with cytochalasin D causes refolding of Fn type III modules in fibrils assembled between cells and an underlying glass substrate.31,32,38 This suggests that alterations to Fn's structure might play an important role in the feedback between cells and the ECM.
Fig. 1.
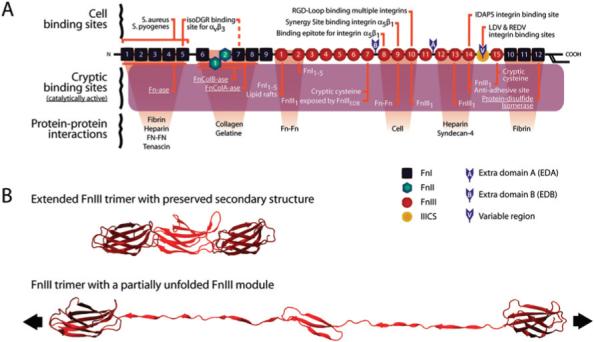
Fibronectin's major binding sites and an example of module unfolding under tensile stress. (A) Fibronectins are dimeric molecules composed of over 50 repeats of three different β-sheet modules (FnI, FnII, and FnIII). One monomer of the type of fibronectin found in human blood plasma is shown in (A); fibronectin produced by cells may contain additional alternatively spliced modules, as indicated. Fibronectins contain a large number of molecular recognition and cryptic sites, including the cell binding site RGD, which is recognized by multiple integrins;77 the synergy site PHSRN, which is recognized by α5β1 and αIIbβ3 integrins;78,79 the sequence IDAPS at the FnIII13−14 junction in the Heparin II region of fibronectin, which supports α4β1-dependent cell adhesion;80,81 and the NGR motif in FnI5, which is non-enzymatically converted to isoDGR and can then bind the αvβ3 integrin.82,83 A similar, highly conserved, NGR motif occurs in FnI7, but has not been extensively studied.83 The cryptic sites include various Fn self-assembly sites whose exposure is needed to induce fibronectin fibrillo-genesis,51 a cryptic fragment from FnIII1 that localizes to lipid rafts and stimulates cell growth and contractility,84 and a binding site for tenascin.85 One cryptic site with enzymatic activity is the FnCol-ase, which is a metalloprotease in the collagen binding domain of plasma fibronectin capable of digesting gelatin, helical type II and type IV collagen, α- and β-casein, and insulin β-chain.86 Other enzymatically active cryptic sites include Fn-ase, a proteinase specific to Fn, actin, and myosin;87 and a disulfide isomerase.88 Finally, there are two cryptic, non-disulfide-bonded cysteines on each monomer, in modules FnIII7 and FnIII15 which are utilized in this study to site-specifically attach the acceptor fluorophores. (B) Tensile stress applied to Fn fibers causes changes in the quaternary, tertiary, and secondary structure of Fn molecules.33 Figure B shows three FnIII modules with intact secondary structure (top) and with the partial unfolding of one module due to increased tensile stress (bottom). (Unfolding of the center module was simulated with the molecular dynamics program NAMD. The ribbon diagrams were assembled using Maya (Autodesk) software.).
We hypothesized that interactions between cells and their environment could alter the rigidity of the ECM, and this would be reflected in the molecular structure of constituent proteins like Fn. In this study, we therefore investigated the dynamic conformational changes Fn undergoes in gradually maturing ECM by using fluorescence resonance energy transfer (FRET) from fluorescently labeled Fn as a probe of structural changes (Fig. 1). In our previous work, we have used FRET to discriminate between compact, extended and partially unfolded conformations of Fn, both in cell culture32,33 and upon adsorption to surfaces.39–41 Here, we first examined how FRET changed as the FRET-labeled Fn probe, together with unlabeled Fn, was harvested from solution and assembled into the ECM by fibroblasts seeded on soft and stiff polyacrylamide surfaces. This established that Fn conformation in matrix fibrils was sensitive to the mechanical characteristics of the underlying substrate. We then asked how the Fn unfolding progressed as cells created their own 3D ECM. Furthermore, Fn extension and unfolding was correlated with known responses of cells to environmental stiffness, such as the length of β1 fibrillar adhesions and actin polymerization. We found that cells actively change the conformation of Fn within the evolving matrix in a manner that is reminiscent of an increasing rigidity response over time. Our data thus suggest that mechanical forces play a significant role in dynamically altering not only the physical properties of ECM fibres, but also the display of molecular and cellular recognition sites through protein stretching and unfolding.
Materials and methods
Fibronectin labeling
Human plasma Fn (>95% purity, Chemicon) was labeled for FRET using a two-step process, similar to previous experiments.32 In the first step, four free cysteine residues located on the Fn modules FnIII7 and FnIII15 within each Fn monomer (Fig. 1) were site-specifically labeled with acceptors by denaturing Fn in 4 M guanidine hydrochloride (GdnHCl) and adding a 10-fold excess of the acceptor, Alexa 546 maleimide (Molecular Probes, Eugene, OR). After incubation for 1 h at room temperature the mixture was dialyzed for 4 h (Slide-a-Lyzer dialysis cassette, 10 000 MW cutoff; Pierce Biotechnology, Rockford, IL) against a PBS buffered solution of 0.1 M sodium bicarbonate, pH 8.6, which served as the labeling buffer for the second labeling step: conjugating unspecified lysine residues with the donor, Alexa 488 succinimidyl ester (Molecular Probes, Eugene, OR). A 60-fold molar excess of Alexa 488 was used to label the protein according to the standard amine-labeling protocol supplied by the manufacturer (Molecular Probes). The resulting doubly labeled protein Fn-D/A was purified by size-exclusion chromatography using a PD-10 gel filtration column (Amersham Pharmacia). The labeling ratio of donors to acceptors per Fn dimer was determined by measuring the absorbance of the labeled protein at 280, 496 and 556 nm and using published extinction coefficients for the dyes and Fn. We have found the optimal labeling for matrix assembly experiments to be 7 to 9 amines conjugated with the donor and all four cysteines conjugated with the acceptor.
Fabrication of polyacrylamide surfaces
Rigid and flexible polyacrylamide surfaces were engineered by modifying previously published protocols.1,7,42 The surfaces were made on glass-bottomed petri dishes (35 mm diameter petri dish, 14 mm diameter glass, MatTek, Ashland, MA) by first pipetting 150 μl of 0.1 N NaOH on to the glass, air drying, then covering the surface with aminopropyltriethoxysilane and incubating for 4 minutes. The glass was then washed with distilled water and incubated with a 0.5% solution of glutaraldehyde for 30 minutes, taking care to avoid the plastic edges of the chamber. The surfaces were washed with distilled water and air dried. 10 μl of a 10% polyacrylamide solution with either 0.03% bisacrylamide (flexible surface or soft surface) or 0.26% bisacrylamide (rigid surface or stiff surface) was pipetted on to the activated glass and a round 12 mm diameter unactivated coverslip was placed on the droplet. To ensure that the weight of the unactivated coverslip did not cause the acrylamide solution to collect at the periphery, the entire culture dish was inverted. Polymerization occurred in about 1 h, after which the unactivated coverslip was easily removed, since the acrylamide did not attach to it. The surfaces were rinsed and stored in PBS at 4 °C. The finished surfaces were about 90 μm thick. The elasticity of the acrylamide gel has been shown to be independent of film thickness for both the crosslinking densities used in this study.43
The Young's modulus of the surfaces was measured macroscopically on sheets of acrylamide gel (∼50 × 17 × 2 mm) by keeping one end of the substrate fixed and attaching a known weight to the other end. The Young's modulus can be calculated as E = (F/A)/(Δl/l), where F is the applied force, A is the cross-sectional area of the substrate, Δl is the change in length of the substrate, and l is the original length of the substrate. This method was applied to more than three different substrates, and yielded a Young's modulus of ∼7500 Pa for the flexible surface and ∼33 000 Pa for the rigid surface. The Young's modulus was also calculated using atomic force microscopy (AFM) with a rounded SiO2 tip (radius 5 μm, measured spring constant 85 pN nm−1) and a modified Hertz model as previously described.44 The AFM measurements were in good agreement with the macroscopic results, yielding a Young's modulus of ∼7 200 Pa for the flexible surface and ∼33 000 Pa for the rigid surface.
Cell culture on activated polyacrylamide surfaces and on glass surfaces
Since cells are unable to adhere to bare polyacrylamide surfaces, primarily due to low protein adsorption, unlabeled Fn was crosslinked to the polyacrylamide surfaces to allow for cell attachment. First, the polyacrylamide surfaces were activated with a 1 mM solution of sulfosuccinimidyl-6(4′-azido-2′-nitrophenylamino)hexanoate (sulfo-SANPAH, Pierce) as described previously.1,45 Sulfo-SANPAH is a heterobi-functional crosslinker that can bind to polyacrylamide with one reactive group upon photoactivation and then interact with amines on proteins with its second reactive group. UV photoactivation was achieved in a tissue culture hood by exposing the solution to UV for 15 minutes, rinsing with PBS and repeating the procedure. After the final rinse, a 25 μg ml−1 solution of Fn in PBS was incubated with the polyacrylamide surfaces at 4 °C overnight.
NIH 3 T3 cells in growth medium (Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum; Invitrogen, Carlsbad, CA) containing penicillin (100 U ml−1), streptomycin (100 μg ml−1), and fungizone (250 ng ml−1)) were plated on the prepared MatTek petri dishes (1.5 × 105 cells per well). The cells were allowed to adhere for 30−45 minutes, after which the medium, along with any unbound cells was removed by gentle pipetting and replaced with growth medium supplemented with a mixture of Fn-D/A and a 20-fold excess of unlabeled Fn (uFn) yielding a final added Fn concentration of 100 μg ml−1. Excess unlabeled Fn was used to prevent energy transfer between adjacent Fn proteins in matrix fibrils.32 Cells were allowed to grow and assemble matrix for 4 h.
For 3D matrix experiments, sterile glass-bottomed LabTek 4-well chambers (VWR International, West Chester, PA) were coated with unlabeled Fn by adsorption from a 25 μg ml−1 Fn solution in PBS for 60 min at 20 °C. NIH3 T3 fibroblasts were plated at a density of 8.3 × 104 cells cm−2 (1.5 × 105 cells per well) and allowed to grow as described above. Samples were incubated between 24 to 96 h (4 days). Mixtures of labeled and unlabeled Fn were supplied to the cell culture at various times depending on the type of experiment.
Fluorescence microscopy and spectroscopy
All FRET spectra and images were taken from living NIH 3 T3 fibroblasts. Samples were removed from the growth medium and rinsed 3 times with warm (37 °C) PBS by gentle pipetting. Care was taken to avoid pipetting fluids directly over the cells. After the last rinse, PBS supplemented with 1.5 mM of the antioxidant Trolox (Sigma) was added to wells to reduce photobleaching. Fluorescence images and spectra were collected within 30 minutes of removing the cells from the incubator to reduce artifacts that may be caused by the cells dying. Imaging and spectroscopy of Fn-D/A in cell matrices was performed using an inverted epifluorescence microscope (TE 2000E, Nikon; 100× oil PlanFluor objective, 1.3 NA, Nikon) with an attached spectrometer (Acton 150, Roper Scientific, Acton, MA) and camera (TEK512, Princeton Instruments, Trenton, NJ). The experimental system has been described in detail elsewhere.32 Spectra were acquired using Metamorph software (Universal Imaging, Media, PA). Spectra and fluorescence and phase contrast images were taken from between 15 and 20 different regions of each sample so that statistically representative populations of cells and their matrices were acquired. Light exposure was minimized and spectra were collected only once from each sample spot to prevent artifacts due to differential photobleaching of donors and acceptors. Images were processed using Adobe Photoshop 6.0 software and spectra were analyzed using custom software created in IGOR Pro (WaveMetrics, Lake Oswego, OR). The program imported a 512 × 512 16-bit image created in Metamorph and assigned a calibrated wavelength to the x-axis, vertical position to the y-axis, and intensity to the z-axis. Spectra were produced by taking a line scan across the x-axis for a given pixel on the y-axis. The program then calculated the intensities of the donor and acceptor peaks. To exclude out of focus light, only data above a minimum donor peak intensity value were used (between 100 and 500 counts, depending on the sample and the region within the sample). The IA/ID ratios taken from the randomly chosen sample spots were complied into box and whisker plots. Each box and whisker plot represents data taken from ∼200 to 500 different fibrils, depending on the density of the matrix.
Estimation of area covered by Fn matrix and area of fibrils underneath cells
In order to compare the matrix assembled by cells on rigid and flexible surfaces, we estimated the total area covered by fluorescent Fn fibrils on the two surfaces. Fluorescence images were first put through a high-pass filter, and then thresholded on the basis of intensity (Adobe Photoshop). This procedure generated a binary image that showed either the absence of presence of fibrils in the region that was imaged. The total number of pixels that were covered by the fibrils was then measured and converted to square microns (Igor Pro). Division of this total area by the number of cells generating the matrix gave the approximate area of matrix assembled per cell. Since the diameters of the fibers may be below the diffraction limit, this total can only be understood as an upper estimate for comparison between the two surfaces rather than an absolute measure.
We also determined the percentage of the total matrix that was present underneath cells. Fibrils were identified and total area covered by the matrix was calculated as described above. The position of cells was determined by interactively drawing their outlines from the phase contrast image corresponding to the fluorescence image of the Fn fibrils. The outlines of the cells defined a region of interest (ROI), which was then used by the software to calculate the area of fibrils only within that ROI (Igor Pro).
Staining for actin and β1 integrins
Samples were rinsed with warm (37 °C) PBS, then incubated for 30 min with cold (4 °C) 2% paraformaldehyde (Sigma). Cells were permeabilized with 0.2 % Triton X-100 in PBS for 3 minutes. For actin staining, TRITC-phalloidin (Molecular Probes) at a concentration of 1 μg ml−1 in PBS at pH 7.4 was added for 45 minutes, then thoroughly rinsed and viewed in a 1.5 mM solution of Trolox in PBS. For integrin staining, cells were permeabilized for 15 minutes with a 0.2% Triton-X100 (Sigma) and 10% goat serum (Jackson ImmunoResearch Laboratories, West Grove, PA) solution in PBS. Incubation with rat anti-mouse β1 (Pharmingen, San Jose, CA) antibody occurred over 3 h at a 1 : 1000 dilution in a solution of 0.1% Triton-X100 and 5% goat serum. Following primary antibody incubation, cells were rinsed thoroughly with a solution of 0.01% Triton-X100 and 1% goat serum in PBS and incubated with appropriate secondary antibodies (Jackson ImmunoResearch Laboratories Inc) at a 1 : 250 dilution in 0.1% Triton-X100 and 1% goat serum in PBS. After rinsing again with PBS, samples were mounted for imaging using Fluoromount-G (Southern Biotechnology Associates, Birmingham, AL), an anti-photobleaching agent.
Cell extraction
Fibroblasts were extracted from 24 h and 96 h cultures as described by Yamada and co-workers.15 Briefly, each sample was rinsed twice with warm PBS (pH 7.4), then incubated for 3 min. in 300 μl warm extraction buffer (0.5% Triton X-100, 20 mM NH4OH, in PBS). Samples were then rinsed three times with PBS (pH 7.4). A final aliquot of PBS supplemented with 1.5 mM Trolox was added to each well for viewing. Spectroscopy and microscopy of samples were performed immediately following extraction.
Measurement of angles between fibrils of different ages
Fluorescently labeled Fn matrices were prepared as described above. For determining the angle between matrix at different timepoints, a single dye, Alexa 488, was used to label full-length Fn. Alexa 488-labeled Fn was added to cultures during either of two time periods: 0−24 h or 24−48 h. Unlabeled Fn was substituted in all other time periods. During the last hour of assembly, a 70 kDa N-terminal fragment of Fn labeled with Alexa 546 was added to cultures. The binding of the Fn N-terminus to assembly sites on the cell surface is known to be an essential step in Fn fibrillo-genesis.46 Thus, we used the 70 kDa fragment as a marker of new matrix fibrils and compared their arrangement with fibrils constructed during earlier time periods. Fluorescence images were taken of Alexa 488 labeled fibrils and the matrix stained with the 70 kDa fragment. These were processed and overlaid in Adobe Illustrator. Angles between the different fibrils were measured using Igor Pro 4.05A (Wavemetrics Inc.) and displayed as histograms. Each set of data includes observations from two separate experiments and different regions within each sample.
Results
Identifying FRET ratios in solution that correspond to partial unfolding of Fn and loss of secondary structure
In our previous work, we demonstrated that FRET from doubly labeled Fn is sensitive to unfolding of Fn type III modules.32,33,41 Prior to adding Fn to cell cultures, we correlated the FRET from doubly labeled Fn (Fn-D/A) with the conformation of Fn as it was progressively denatured in solution. FRET was defined as the ratio of acceptor to donor peak intensities, or IA/ID.
The structural changes that Fn undergoes upon chemical denaturing have been well documented. Fn has a compact conformation in solution, where its two dimeric arms are crossed over each other (Fig. 1). Under mild denaturing conditions (below 1 M GdnHCl), the electrostatic interactions that stabilize the folding of Fn's dimeric arms upon each other are broken, but the structure of individual Fn modules remained intact.33 Thus, the initial reduction in FRET from 0 to 1 M GdnHCl is due to the loss of intramolecular protein contacts between the two overlapping arms of the dimer, as confirmed by circular dichroism spectroscopy.41,47,48 Circular dichroism spectroscopy also shows that Fn starts losing secondary structure at concentrations of GdnHCl greater than 1 M, and this is accompanied by a further decrease in FRET due to the unfolding of FnIII modules.41,48
For each batch of labeled Fn, we measured the IA/ID ratio at GdnHCl concentrations between 0 and 4 M, and used this curve to interpret the FRET measured in Fn matrices. This calibration is not exact, since chemical denaturants and tensile stress likely unfold Fn via different pathways. However recent experiments from our laboratory support this mode of calibration by demonstrating that Fn fibrils that are stretched to 2−3× their fully relaxed length have an IA/ID ratio similar to Fn in 1 M GdnHCl. At these levels of tension, Fn modules experience significant unfolding, such that the buried, free cysteine residues on FnIII7 and FnIII15 are exposed.49 The IA/ID ratio at 1 M GdnHCl was chosen as a benchmark for the beginning of Fn unfolding in regions within ∼10 nm of FnIII7 and FnIII15 where acceptor fluorophores are located (see Materials and Methods). Fn is thus referred to as “extended” (open dimer arms) if it exhibits IA/ID values observed in solution for 0.5−1 M GdnHCl, and as “partially unfolded” (loss of secondary structure) if IA/ID values are for Fn are > 1 M GdnHCl. The IA/ID levels corresponding to Fn structure in GdnHCl are indicated on all subsequent figures that show FRET in fibrils. In all experiments involving FRET, Fn-D/A was added into the cell culture medium with a 20-fold excess of unlabeled Fn in order to prevent energy transfer between Fn-D/A molecules (see Materials and Methods).
Fibronectin exhibited greater unfolding on rigid polyacrylamide surfaces compared to flexible polyacrylamide surfaces
Many cells, when seeded on surfaces, will start to assemble their own matrix. Consequently, they will replace the cell–surface contact by cell–matrix contacts. We thus asked whether the ECM offers cells an environment that is more compliant to mechanical forces as compared to the glass or plastic substrates typically used for cell culture. To determine whether we could detect structural differences in Fn assembled on soft versus rigid substrates, the cells were cultured on chemically identical polyacrylamide surfaces having two different values for their elastic modulus. Rigid surfaces were created with an elastic modulus of 33 kPa, while the flexible surfaces had an elastic modulus of 7.5 kPa. These values were chosen based on previous experiments which showed differences in cell behavior at these two substrate rigidities,1,11 and based on substrate compliances at which cells were able to assemble sufficient matrix for FRET measurements. These values for elastic modulus are also within the range of those measured for different types of ECM in vivo.9,50
We first confirmed that cells on both surfaces were able to fabricate an Fn matrix after 4 h. The 4 h time point was chosen because it was sufficient time for cells to make enough matrix to obtain FRET data, but not so long that cells were no longer in direct contact with the substrate. Cells on both rigid and flexible acrylamide surfaces were able to assemble a matrix, albeit with measurable differences (Fig. 2). The matrix was more abundant on rigid surfaces compared to flexible surfaces. On flexible surfaces, the area covered by Fn fibrils was only 78 μm2 per cell, compared to 425 μm2 per cell on rigid surfaces. Furthermore, the matrix on flexible surfaces tended to be concentrated underneath the cell body, with about 65% of total fibrillar area found under cells, compared to about 40% under cells on rigid surfaces. This suggests that matrix assembly is enhanced by increased substrate rigidity. Morphologically, both the matrix and the cells on rigid surfaces looked very similar to those on glass surfaces (data not shown).
Fig. 2.
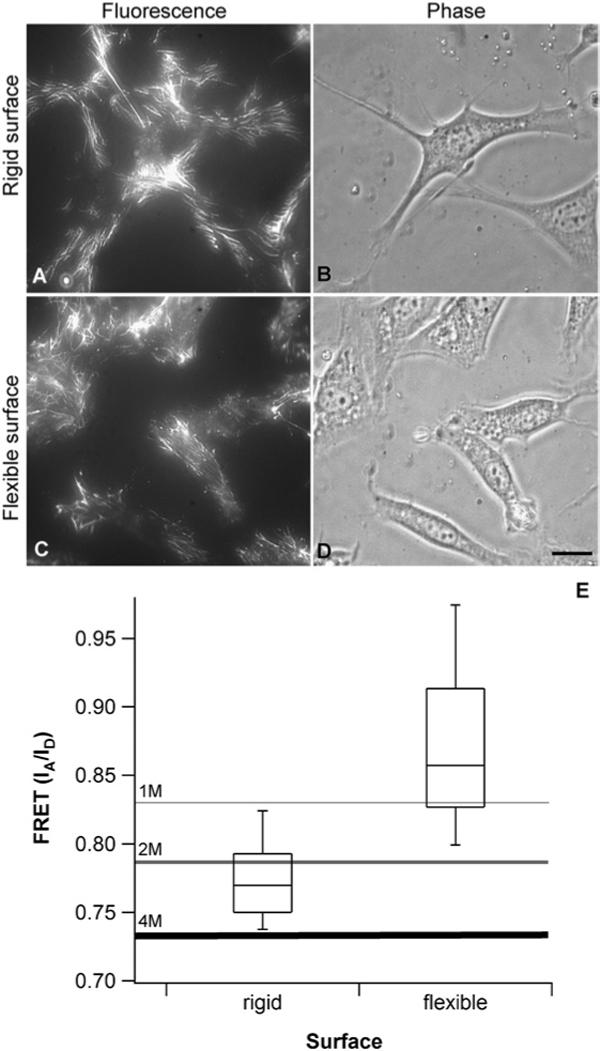
Fn matrix assembly and unfolding on rigid and flexible polyacrylamide surfaces. (A–D) Fluorescence and phase contrast images of Fn matrices and fibroblasts on rigid and flexible polyacrylamide surfaces (4 h culture). Fn matrix assembly occurs on both rigid and flexible polyacrylamide surfaces with differences in matrix organization and cell shape. (A) On rigid surfaces, matrix is abundant, and fibrils appear linear and are formed both around the cell periphery and underneath cells. (B) Cells on rigid surfaces are well spread, with many protrusions extending from the main cell body. (C) On flexible surfaces, the matrix is more sparse and concentrated mostly underneath the cell body. (D) Cells on flexible surfaces are not well spread and cover a smaller surface area than cells on rigid surfaces. Scale bar 15 mm. (E) FRET distributions from matrix fibrils on rigid and flexible surfaces after 4 h of culture (approximately 200−400 fibrils). The boxes represent the 25th to the 75th percentile and the ‘whiskers’ show the 2nd and 98th percentiles. FRET from fibrils on the rigid surface falls below the 1 M calibration point, indicating that Fn is partially unfolded on rigid surfaces. FRET values on the flexible surface shows that Fn is primarily extended. Differences are statistically significant (ANOVA test, p ⪡ 0.001).
The molecular structure of Fn in matrix fibrils on rigid surfaces was markedly more stretched than that on flexible surfaces, as measured by FRET. Fig. 2E shows the IA/ID values from Fn-fibrils assembled by cells on rigid and flexible surfaces. For the data shown on the rigid and flexible surfaces, Fn in matrix fibrils is extended if it exhibits an IA/ID above 0.83, and partially unfolded if IA/ID is below 0.83. Virtually all the Fn in fibrils on the rigid surfaces are partially unfolded. In contrast to rigid surfaces, IA/ID from Fn in fibrils assembled by cells on flexible surfaces mostly lie around the point where Fn is extended in solution. The distributions are significantly different (ANOVA test, p ⪡ 0.001). The data show that Fn-D/A in ECM fibrils is sensitive to substrate rigidity. The data furthermore suggest that cells in direct contact with rigid surfaces are able to partially unfold Fn while cells on flexible surfaces are less likely to do so. The unfolding of Fn also may explain why cells on rigid surfaces make more matrix than cells on flexible surfaces. It is known that cells need to unfold Fn in order to generate matrix fibers,31,51,52 thus it is possible that on flexible surfaces, the inability of cells to unfold Fn leads to reduced matrix assembly. These findings have been reproduced using three different batches of Fn.
As the 3D matrix matures, the newly assembled matrix is increasingly unfolded
Cells in a mature 3D ECM should experience a much softer environment than those grown in direct contact with the glass or plastic surfaces normally used for tissue culture. Because only minimal Fn unfolding was detected on artificial, soft polyacrylamide surfaces, we next tested whether Fn in newly assembled fibrils created by cells in an existing matrix exhibited similarly little unfolding. We cultured cells for 24, 48, and 72 h on glass to generate matrices of varying thickness. In order to measure FRET in Fn in only the fibers produced in the last hour of the culture, the cells were cultured first with unlabeled fibronectin and then given trace amounts of Fn-DA for the final hour (Fig. 3A). This allowed us to measure the unfolding of Fn in newly-fabricated fibrils as cells experienced an increasingly mature matrix environment.
Fig. 3.
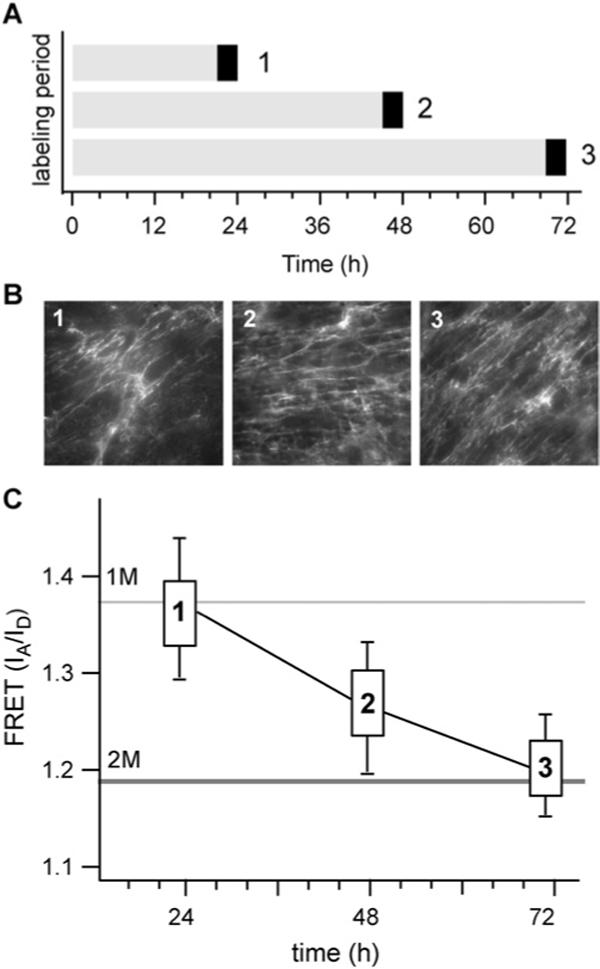
Matrix assembled by cells during the last 1 h of assembly. (A) Labeled Fn (black bars) was added to 24, 48 or 72 h matrices during the last hour of assembly. (B) The morphology of newly added fibrils appears similar at each time point. (C) FRET measurements show major differences in unfolding in Fn in newly assembled matrix. At 72 h, Fn is unfolded to a much greater extent than in either 48 or 24 h cultures.
As the matrix grew from 2D to 3D during the first 24 h, the resident cells increasingly interacted with their matrix and less with the glass substrate (Fig. 3B). It has been shown that cells in such thicker matrices attain a different phenotype than cells grown on the underlying 2D substrate.15,53,54 We therefore hypothesized that as the matrix thickness increased, the cells would become de-coupled from the stiff substrate and interact more with the soft ECM and that this would be reflected in a decrease in Fn unfolding as seen on the polyacrylamide gels. Initially, our hypothesis appeared to hold true: the Fn in a large proportion of newly made fibrils on a 24 h matrix showed little unfolding and was similar in structure to the Fn made by cells on flexible surfaces (elastic modulus = 7.5 kPa) (Fig. 3C). However, at later time points this trend no longer persisted. Surprisingly, the new fibrils produced in the thicker 48 h matrix actually exhibited more Fn unfolding than in the relatively thinner 24 h matrix. On the thickest, 72 h matrix, the degree of Fn unfolding resembled that seen on rigid matrices (elastic modulus = 33 kPa). These results demonstrated that a thick, 3D matrix environment was not necessarily more compliant than that of a thinner, 2D matrix.
Markers of cellular rigidity response: cells on rigid surfaces and in mature matrices have longer β1 integrin adhesions and more actin stress fibers
The FRET data suggest that, at least structurally, the Fn in new fibrils made on thick, mature 3D matrices bear more resemblance to Fn in fibrils assembled on rigid surfaces than on flexible surfaces. We therefore asked whether cellular markers of environmental rigidity were also present in cells in older matrices. Since Fn matrix assembly is initiated by the formation of fibrillar adhesions containing α5β1,29,55–57 we compared the formation of these adhesions on rigid and flexible polyacrylamide surfaces with β1 integrin-containing adhesions in 3D matrices at various time points. On polyacrylamide, after 4 h of culture, fibrillar adhesions containing β1 were visible on both surfaces. However, the β1 staining appeared brighter and more elongated on rigid surfaces (Fig. 4). To quantify these observations, we measured the length of the fibrillar adhesions 4 h after cells were seeded on the two surfaces. The mean length on rigid surfaces was 3.4 μm (n = 242), compared to 2.6 μm (n = 309) on flexible surfaces. The 98th percentile value for rigid surfaces was 8.3 μm compared to 6.4 mm for flexible surfaces. The differences were statistically significant, with p < 0.001 (ANOVA test).
Fig. 4.
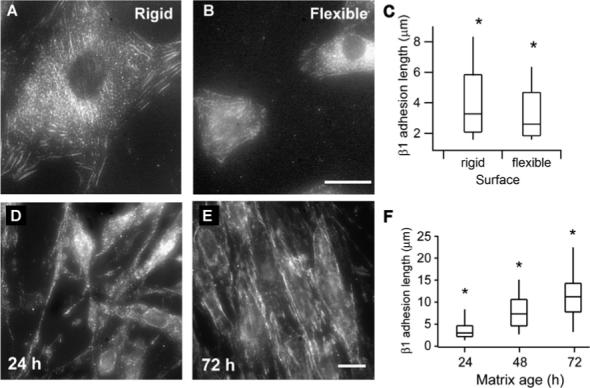
Staining for β1 integrin on rigid and flexible polyacrylamide surfaces and in 3D matrices. Top: Cells on polyacrylamide gels. (A) Vigorous recruitment of β1 into fibrillar adhesions on rigid surfaces. (B) Reduced integrin β1 staining is visible on flexible surfaces in structures reminiscent of fibrillar adhesions. Scale bar is 15 μm. (C) Box and whisker plot of adhesion lengths on rigid (n = 242) and flexible surfaces (n = 309). The top and bottom vertical lines denote the 98th and 2nd percentile values. Distributions are significantly different, the * denotes p < 0.001. Bottom: Cells on glass. (D–F) Typical adhesion structures containing β1 integrins shown over time. (D) At 24 h very few adhesion-like structures were present and those tended to be very short. (E) Over time the β1-containing adhesions became increasingly elon-gated, seeming to reach a kind of steady state by 72 h. Scale bar is 10 mm. (F) Box and whisker plots of integrin lengths at different timepoints. The * denotes a significance of p < 0.001; N = 195, 200, 160, 160, and 143 for each time point, respectively.
The length of β1-containing adhesions was also measured in cell-derived matrices as they matured from 24 h to 96 h (Fig. 4). At 24 h, both the number and length of β1-containing structures was quite low. By 48 h, the length of β1-containing adhesions began to increase again until seeming to reach a steady state by 72 h. By this time, highly elongated β1 integrin-containing adhesions were found on both the basal and dorsal surfaces of cells, ranging up to 20 mm in length. The wide distribution in the length of β1 adhesions may reflect the diversity of adhesion types and maturities which have been observed in 2D 58, or alternatively, might be due to spatial heterogeneity of tensile forces acting within individual cells. No concomitant change in the width of β1-containing adhesions was observed. The adhesions found in mature matrices were even longer than those found on rigid surfaces, and may reflect not only the stiffness of the substrate, but also the enhanced protein recruitment to those adhesion sites over time, and the three-dimensional nature of the ECM.15 Despite the differences in the actual magnitudes of adhesion lengths on the polyacrylamide gels and the ECMs, the pattern of adhesion formation as the matrix matured over 72 h was consistent with cells experiencing an increasingly rigid environment.
Actin stress fiber formation was also measured in cells on both rigid and flexible polyacrylamide surfaces after 4 h of culture. These actin stress fibers were then compared to stress fibers in cells residing in 24 and 96 h matrices. Virtually all cells on rigid surfaces showed well-developed stress fibers (data not shown). By contrast, on flexible surfaces no robust stress fibers were detected, although some cells did show signs of smaller, thinner actin filaments. Actin staining in cells resident in 24 h matrices and 96 h matrices confirmed the trend that the older matrices resembled stiffer environments (data not shown). At 24 h, staining for actin revealed that cells had almost no stress fibers. However, at 96 h, cells stained strongly for actin stress fibers in the direction of their long axis. These results again suggest that cells at 96 h resemble cells on stiff surfaces, and cells at 24 h resemble cells on a soft surface.
Fn unfolding averaged over the entire matrix progressively increased as the ECM matured
In order to further investigate whether the trends seen in the previous sections were symptomatic only for newly assembled matrix or held true even when averaging all fibrils in a culture, we looked at Fn unfolding in the entire matrix over the course of one to four days for fibroblasts cultured on glass (Fig. 5 and 6). For these experiments, labeled Fn was added to the culture medium for either 24, 48, 72 or 96 h. We found that Fn was gradually more stretched and that unfolding increased as the matrix developed over time. At each time point, FRET varied from region to region and from fibril to fibril within a given population which is reflected in the distribution of FRET values and is in agreement with previous observations.38 In the 24 h matrix, the average FRET (IA/ID) ratio was 1.3 (varying between 1.1 and 1.45, Fig. 6). Most of these FRET ratios correspond to Fn conformations that are extended, when compared to the calibration of Fn unfolding in GdnHCl.
Fig. 5.
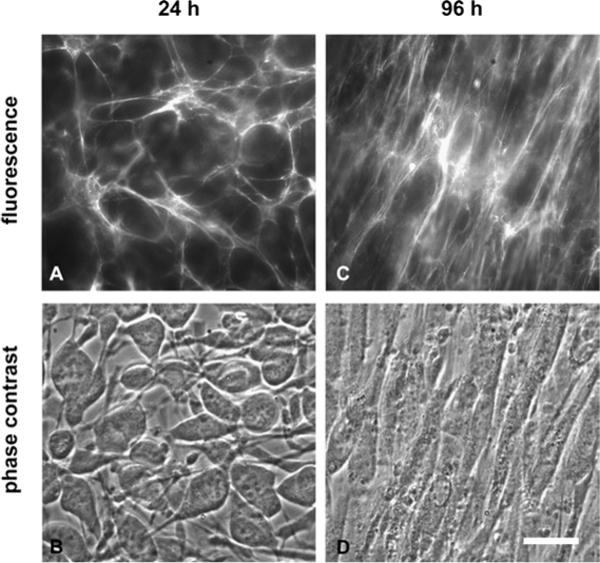
Images of cells and extracellular matrix at 24 and 96 h. (A–B) At 24 h, large numbers of non-polarized cells populate isotropically oriented matrix fibrils. (C–D) At 4 days, the matrix is very dense and aligned with the long axis of cells; cells have acquired a highly elongated in vivo-like spindle shapes. Scale bar is 20 μm.
Fig. 6.
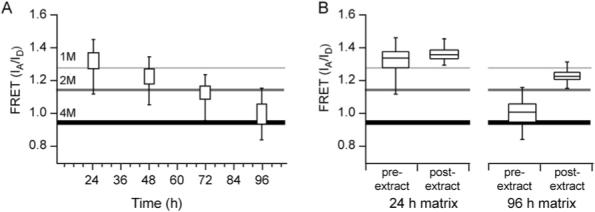
FRET distributions at different time points in Fn matrix assembly. (A) FRET distributions as a function of time shown as percentiles. The vertical lines show the 2nd and the 98th percentile values; the boxes show the 25th and 75th percentile values. (B) FRET from 24 h and 96 h matrices before and after cells were extracted. In 24 h matrices (left) extraction of cells produced a modest increase in FRET mostly due to the loss of the lower FRET values (N = 345). In 96 h matrices (right) a large increase in FRET occurred after removal of cells, although not to the same level as observed in denuded 24 h matrices (N = 455).
Cultured over several days (24−96 h), the cells grew into the third dimension to form at least a cell bilayer and assembled a dense 3D matrix approximately 15 μm deep. The matrix morphology was undergoing another significant transformation: rather than the isotropically arrayed fibrils observed at 24 h, matrix fibrils were aligned with the long axis of cells (Fig. 5). Matrix fibrils also became thicker than at 24 h and lost the fine mesh structure seen at those earlier time points (Fig 5). Corresponding FRET decreased as time progressed (Fig. 6A), with IA/ID = 1.22 at 48 h, IA/ID =1.15 at 72 h, and IA/ID = 0.96 at 96 h, showing that Fn in fibrils became progressively more unfolded. By 96 h, almost all the FRET values were below IA/ID = 1.25, which corresponds to partially unfolded Fn as measured at the 1 M GdnHCl point in the calibration curve (Fig. 6A).
Fn unfolding was controlled by cell contractility
In order to verify that the changes in intensity ratio were not due to artifacts such as partial protein degradation or fluorophore photodamage, the cells were extracted from 24 h and 96 h matrices, leaving behind the intact detergent-insoluble matrix. In agreement with our previous results that showed the unfolding of Fn in 24 h cultures was controlled by contraction of the actin cytoskeleton,33,38 we found that removal of cells at 24 and 96 h caused matrix unfolding, i.e. an increase in FRET (Fig. 6B), although the change within 24 h matrices came primarily from loss of the lower FRET values. Interestingly, FRET in the extracted matrices did not increase to the same absolute level: 24 h extracted matrix exhibited a peak at IA/ID = 1.35 representing a small increase of 0.02 (p < 0.0001, n = 300), while the 96 h extracted matrix had a peak IA/ID = 1.22 with a much larger increase of 0.27 (p < 0.0001, n = 400). This data is particularly significant in light of recent experiments that use cell-derived 3D Fn matrices as scaffolds for seeding cells.15,54,59 To make such cell-derived matrices, cells are extracted from the deposited ECM. Our data show that, although the appearance and overall architecture of the matrix does not change when cells are removed, the molecular structure of Fn is altered: Fn partially refolds when the cell-generated tensile forces are missing. Altered Fn conformation in the ECM could affect all subsequent interactions of newly seeded cells with the matrix.
Old matrix served as scaffold to anchor newly assembled matrix and was thereby progressively stretched
The FRET data shown so far point to a general trend in increased Fn unfolding over time, and that Fn structure in newly laid fibrils depends on the age of the underlying matrix that the new fibrils are assembled on. However, it does not reveal the state of Fn in the older, underlying matrix. To determine whether the progressive decrease in intensity ratio seen in Fig. 6 was due to the addition of new fibers with Fn in an increasingly unfolded state, or due to older matrix that also was continuously changed by cells, we measured the intensity ratio of Fn in fibers of different ages within a 72 h matrix. For these experiments, we examined matrices that had been assembled over 72 h, but given labeled Fn either between 0 to 24 h, 24 to 48 h or 48 to 72 h (Fig. 7A). The FRET data from these experiments (Fig. 7C) show that the matrix made between 0 and 24 h was the most unfolded, while the matrix assembled by cells during the last 24 h of the experiment (48 to 72 h) was the least unfolded. Thus, Fn unfolding progressively increases with time and the oldest fibrils show the greatest amount of unfolding. This showed that cells do not only lay down new fibrils as time progresses. They also continuously remodel older matrix in a way that increasingly stretches and unfolds Fn in the oldest matrix fibrils. The data imply that cells do not endocytose and intracellularly degrade the old matrix as they make new matrix under the chosen experimental conditions, nor do they appear to simply abandon the old matrix. A digested matrix looks visibly different from an intact one and also exhibits very little energy transfer, as indicated by a low intensity ratio (data not shown). An abandoned matrix may visually appear similar to remodeled matrix, but would not show decreases in FRET levels at 48 and 72 h, since we have shown (Fig. 6B) that FRET actually increases when cells no longer exert tension on the matrix. The most likely explanation for the decreased FRET over time is that the old matrix is used as a scaffold for newer fibrils and cells continue to apply tension to the old matrix and further unfold the Fn as they assemble new fibers.
Fig. 7.
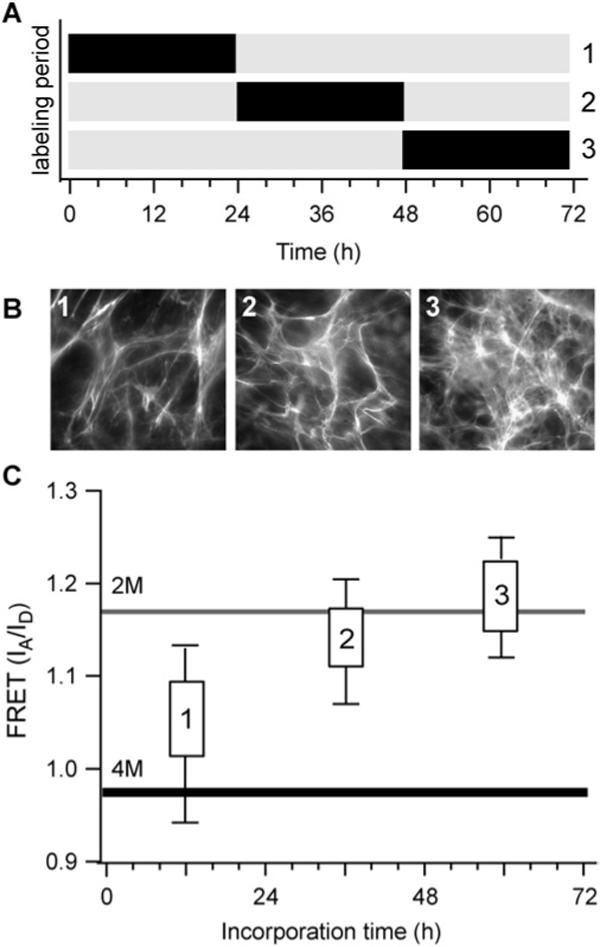
Pulse-chase experiment in a 72 h matrix. (A) Time periods in which samples were given doubly-labeled Fn (black) or unlabled Fn (gray). (B) Representative images of matrix morphology for fibrils made from 0−24 h (1), 24−48 h (2), or 48−72 h (3). (C) FRET (IA/ID) distributions of fibers produced during the three, 24 h time periods. Not all matrix Fn is stretched to the same extent, but rather there is progressive unfolding that results in the older matrix being more unfolded than the recently assembled.
To further determine whether older fibrils in a matrix served as a scaffold for the assembly of new matrix, we measured the angle at which the newly formed fibrils bisected older ones (Fig. 8). We compared the angles formed between new fibrils and the three different groups of fibers studied in Fig. 7: the last 24 h of a 72 h matrix (newest fibrils) to the fibrils made during the first 24 h (oldest fibrils) and the subsequent 24 h (intermediate fibrils). Newly formed fibers were identified by staining with the 70 kD Fn fragment as described in the Materials and Methods section. Although a significant fraction of the newer fibrils appear to align quite well with the older fibrils (angles between 0 and 10 degrees), there is a fairly broad distribution of angles between the two sets of fibrils, with many of the newest fibrils perpendicularly bisecting the oldest ones. In contrast, virtually none of the newest fibrils bisect the intermediate fibrils at high angles. Rather, the newest fibrils appear to be fabricated tangential to the intermediate ones, with most angles falling between 0 and 10 degrees. The data shows that there is only partial overlap between the oldest fibrils in a culture and the newest ones, while there is more overlap between the newest fibrils and those assembled in the time period immediately preceding them (intermediate fibrils). The results support previous experiments which have shown that there is only partial overlap between newly formed Fn fibrils and a pre-formed 3D matrix.59 The data also suggest that a physical connection still exists between older matrix fibrils and cells and that it is mediated by newly assembled fibrils.
Fig. 8.
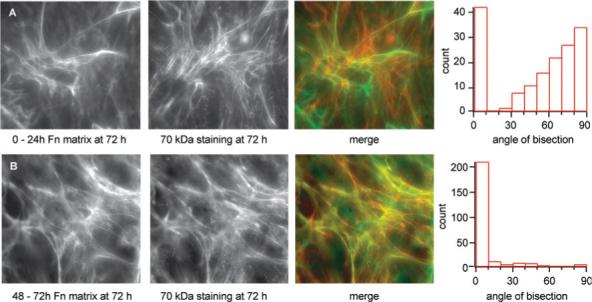
Location and angular arrangement of newly made fibrils compared to the oldest (0−24 h) and intermediate (24−48 h) fibrils in a 72 h matrix. (A) Comparison of matrix fibrils made from 0−24 h and with matrix undergoing active assembly at the 72 h time point based on staining with the 70 kDa Fn fragment (see Materials and Methods). Since the angle of bisection can be measured from either side, in order to increase signal to noise the acute angle was chosen. The angular distribution clustered around 0 and 90 degrees. N = 124. (B) Same comparison as described above, but newly made fibrils are compared to intermediate fibrils (24−72 h). The distribution is sharply centered around 0 degrees, indicating that most fibrils are made tangentially to their immediate predecessors. N = 115. Representative images are also shown.
Discussion
Through the introduction of a conformationally sensitive probe, we showed for the first time that the maturation and remodeling of the matrix is manifested at a molecular level in the progressive unraveling of the secondary structure of one of its major constituent proteins, the multimodular protein Fn. We also showed that fibroblasts respond to the altered Fn matrix properties. The ECM is thus more than a scaffold with a static Fn conformation that provides only structural support to cells. We also found that cells cultured over several days do not solely respond to the rigidity of the initial substrate. Nor do they only sense the increased compliance of their self-made ECM. Rather, the cells increasingly stretched Fn in the matured matrix and showed a gradually up-regulated rigidity response. Our data showed that Fn module unfolding in fibrils is affected by the rigidity of the underlying substrate only at very early time points, when cells had direct contact with the surfaces upon which they are seeded. The Fn matrix was significantly more stretched and unfolded on the rigid (33 kPa) than on the soft (7.5 kPa) substrate (Fig. 2). In this context, it should be briefly noted that the question of cell's ability to unfold Fn in matrix fibers was controversial32,38,60–62 and was recently answered positively,33,63 thereby confirming previous results.32,38,52
A closer look at the time course of the reciprocity between matrix maturation and the cellular response revealed how the cell's response to rigidity was altered when the cell increasingly interacted with self-made matrix rather than the provided substrate. Once a confluent fibroblast monolayer had been formed on glass after 24 h, new fibrils deposited on 3D matrix (Fig. 3) did indeed contain Fn that was similarly stretched to Fn deposited on soft surfaces (Fig. 2). However, the trend did not hold for Fn newly deposited on 48 and 72 h matrices. The overall trend instead pointed to increased unfolding of newly deposited Fn, reminiscent of an increasingly rigid ECM environment. The results show that thicker, mature matrices that decouple cells from their initial substrates can constantly evolve over time, possibly providing environments of decreasing compliance to the resident cells.
Several pieces of evidence support the notion that the ECM environment elicits an increasing cellular rigidity response over time. Fibroblasts display longer β1 integrin adhesions as the matrix ages and show more actin stress fibers (Fig. 4). Moreover, cells do not abandon older matrix with the passage of time. Rather, the older matrix serves as scaffold to anchor a newly-assembled matrix (Fig. 8) and, as tensile forces are applied, Fn in the older matrix becomes continuously more stretched and unfolded (Fig. 7). Matrix unfolding can at least be partially reversed when cell contractility is inhibited,38 or when the cells are removed (Fig. 6B). The presence of this increasingly unfolded Fn could thus account at least partially for the increased cellular rigidity response. The large extensibility range of single fibrils of Fn and of other ECM proteins is thought to involve the unraveling of their secondary structure.33,38,62,64,65 While the stress–strain relationship has not yet been established for Fn fibers, fibrin fibers (which have a comparable extensibility to those of Fn) show a stiffness increase at the presumed onset of module unfolding.66,67 Our data may also offer a molecular level explanation for other experiments that have shown increases in ECM stiffness over time.66,67
An additional contribution to the upregulated rigidity response comes from a second effect often previously described for fibrous matrices, namely matrix stiffening through fiber alignment. In the maturing ECM, the Fn fibers of the older matrix are increasingly aligned parallel to the long axis of the fibroblasts (Fig. 5). Tension-induced stiffening is thus caused by an anisotropic alignment of fibers along the force vector and has also been observed to alter the micro- or macroscopic properties of polymeric materials.68 Tension stiffening also plays an important physiological role, as cell contractility leads to the local compression and stiffening of a variety of biological matrices (for reviews see ref. 69 & 70). Alterations of collagen and elastin fibers, for example, are involved in arterial stiffening that is associated with the aging process and disease states such as hypertension, diabetes, atherosclerosis, and chronic renal failure.71 The process is driven by reciprocal interactions between cells and their surrounding matrices. Even within the cell, cytoskeletal stiffening also occurs, and is caused by alignment of the force-bearing actin stress fibers along the force vector.72–74 In our experiments, we see both an enhanced alignment of the fibronectin fibrils as well as of stress fibers over time, suggesting that the cell is causing tension stiffening of the matrix as it matures.
Three major conclusions can thus be drawn from these findings: one, the rigidity of a cell's substrate can affect the conformation of Fn in the ECM; two, the ECM is constantly changing at the conformational level of its constituent proteins; and three, thick, mature matrices contain more unfolded Fn and up-regulate the rigidity response of cells compared to younger matrices.
What might be the role of increasingly unfolded Fn in matrix fibrils? Several molecular recognition sites as well as catalytically-active sites have been identified that are cryptic in native Fn (Fig. 1). Future research has to address whether they can be exposed by mechanical force and whether they are still catalytically active in strained Fn. Another possibility by which aging might modulate Fn function is illustrated by recent results from our laboratory that the stretching and unfolding of Fn fibers regulates the binding of serum proteins in a protein-specific manner (W.C. Little, M. L. Smith, R. Schwartlander, U. Ebneter, D. Gourdon, V. Vogel, in prepration). Aging of the matrix may thus change the composition of serum proteins that are transiently bound to intact Fn fibers. Enzymatic degradation of the ECM could also be modulated by the force-regulation of Fn unfolding if enzymatic cleavage sites exist that could be exposed in a strain-dependent manner.22 Growth factors sequestered in the ECM can be released via enzymatic degradation20 and Fn degradation products themselves are known to trigger numerous physiological responses, such as antibacterial activity21 and inflammation.22
Our observations of changing Fn conformation during matrix maturation further suggest a new mechanism by which the binding and release of growth factors, peptides and other soluble proteins might be regulated in a matrix age-dependent manner. Moreover, all proteins are known to have finite lifetimes before they are degraded by proteolytic or endocytotic mechanisms. The ECM also is known to be continuously remodeled, with fibronectin degradation occurring intracellularly after endocytosis.75,76 Again, the mechanisms that control the balance between the degradation and the assembly process are incompletely understood. Increasingly unfolded Fn in the matrix could thus act as a marker for aged matrix and subject it to degradation when other conditions are appropriate. Such a marker for degradation could potentially exist by unmasking protease binding sites in stretched Fn that would otherwise be hidden. Future research is needed to determine whether the age-dependent unfolding of proteins in the ECM might impact cell signaling, matrix degradation, and other matrix-regulated pathways.
Acknowledgements
We thank William Little and Manu Forero for performing the AFM measurements of the Young's modulus of the polyacrylamide surfaces, Sheila Luna and John Sager for assistance with figures, and Greg Young for assistance with staining of integrins. This work was supported by grants from the NIH (NIH 8 R01 EB00249-09, 3 P50 HG 02360 03S1), ETH Zurich, the Nanotechnology Center for Mechanics in Regenerative Medicine (NIH grant PN2 EY016586; part of the NIH Nanomedicine Development Center Network), and in part by a grant from A*STAR, Singapore, through the Singapore University of Washington Alliance.
References
- 1.Pelham RJ, Jr, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl. Acad. Sci. U. S. A. 1997;94(25):13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peyton SR, Putnam AJ. Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion. J. Cell. Physiol. 2005;204(1):198–209. doi: 10.1002/jcp.20274. [DOI] [PubMed] [Google Scholar]
- 3.Lo CM, Wang HB, Dembo M, Wang YL. Cell movement is guided by the rigidity of the substrate. Biophys. J. 2000;79(1):144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang JH, Thampatty BP, Lin JS, Im HJ. Mechanoregulation of gene expression in fibroblasts. Gene. 2007;391(1−2):1–15. doi: 10.1016/j.gene.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacKenna D, Summerour SR, Villarreal FJ. Role of mechanical factors in modulating cardiac fibroblast function and extracellular matrix synthesis. Cardiovasc. Res. 2000;46(2):257–263. doi: 10.1016/s0008-6363(00)00030-4. [DOI] [PubMed] [Google Scholar]
- 6.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8(3):241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 7.Wang HB, Dembo M, Wang YL. Substrate flexibility regulates growth and apoptosis of normal but not transformed cells. Am. J. Physiol.: Cell Physiol. 2000;279(5):C1345–1350. doi: 10.1152/ajpcell.2000.279.5.C1345. [DOI] [PubMed] [Google Scholar]
- 8.Tamada M, Sheetz MP, Sawada Y. Activation of a signaling cascade by cytoskeleton stretch. Dev. Cell. 2004;7(5):709–718. doi: 10.1016/j.devcel.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 9.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 10.Janmey PA, Weitz DA. Dealing with mechanics: mechanisms of force transduction in cells. Trends Biochem. Sci. 2004;29(7):364–70. doi: 10.1016/j.tibs.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, Funaki M, Zahir N, Ming W, Weaver V, Janmey PA. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil. Cytoskeleton. 2005;60(1):24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- 12.Georges PC, Janmey PA. Cell type-specific response to growth on soft materials. J. Appl. Physiol. 2005;98(4):1547–53. doi: 10.1152/japplphysiol.01121.2004. [DOI] [PubMed] [Google Scholar]
- 13.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310(5751):1139–43. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 14.Semler EJ, Lancin PA, Dasgupta A, Moghe PV. Engineering hepatocellular morphogenesis and function via ligand-presenting hydrogels with graded mechanical compliance. Biotechnol. Bioeng. 2005;89(3):296–307. doi: 10.1002/bit.20328. [DOI] [PubMed] [Google Scholar]
- 15.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294(5547):1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 16.Wozniak MA, Desai R, Solski PA, Der CJ, Keely PJ. ROCK-generated contractility regulates breast epithelial cell differentiation in response to the physical properties of a three-dimensional collagen matrix. J. Cell Biol. 2003;163(3):583–95. doi: 10.1083/jcb.200305010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolf K, Wu YI, Liu Y, Geiger J, Tam E, Overall C, Stack MS, Friedl P. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat. Cell Biol. 2007;9(8):893–904. doi: 10.1038/ncb1616. [DOI] [PubMed] [Google Scholar]
- 18.Gregory KE, Ono RN, Charbonneau NL, Kuo CL, Keene DR, Bachinger HP, Sakai LY. The prodomain of BMP-7 targets the BMP-7 complex to the extracellular matrix. J. Biol. Chem. 2005;280(30):27970–80. doi: 10.1074/jbc.M504270200. [DOI] [PubMed] [Google Scholar]
- 19.Dallas SL, Miyazono K, Skerry TM, Mundy GR, Bonewald LF. Dual role for the latent transforming growth factor-beta binding protein in storage of latent TGF-beta in the extracellular matrix and as a structural matrix protein. J. Cell Biol. 1995;131(2):539–49. doi: 10.1083/jcb.131.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taipale J, Keski-Oja J. Growth factors in the extracellular matrix. FASEB J. 1997;11(1):51–9. doi: 10.1096/fasebj.11.1.9034166. [DOI] [PubMed] [Google Scholar]
- 21.Sarikaya A, Record R, Wu CC, Tullius B, Badylak S, Ladisch M. Antimicrobial activity associated with extracellular matrices. Tissue Eng. 2002;8(1):63–71. doi: 10.1089/107632702753503063. [DOI] [PubMed] [Google Scholar]
- 22.Barilla ML, Carsons SE. Fibronectin fragments and their role in inflammatory arthritis. Semin. Arthritis Rheum. 2000;29(4):252–65. doi: 10.1016/s0049-0172(00)80012-8. [DOI] [PubMed] [Google Scholar]
- 23.Bailly M. Connecting cell adhesion to the actin polymerization machinery: vinculin as the missing link? Trends Cell Biol. 2003;13(4):163–5. doi: 10.1016/s0962-8924(03)00030-8. [DOI] [PubMed] [Google Scholar]
- 24.Brakebusch C, Fassler R. The integrin–actin connection, an eternal love affair. EMBO J. 2003;22(10):2324–2333. doi: 10.1093/emboj/cdg245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen CS, Tan J, Tien J. Mechanotransduction at cell–matrix and cell–cell contacts. Annu. Rev. Biomed. Eng. 2004;6:275–302. doi: 10.1146/annurev.bioeng.6.040803.140040. [DOI] [PubMed] [Google Scholar]
- 26.Choquet D, Felsenfeld DP, Sheetz MP. Extracellular matrix rigidity causes strengthening of integrin–cytoskeleton linkages. Cell. 1997;88(1):39–48. doi: 10.1016/s0092-8674(00)81856-5. [DOI] [PubMed] [Google Scholar]
- 27.Geiger B, Bershadsky A, Pankov R, Yamada KM. Transmembrane crosstalk between the extracellular matrix–cytoskeleton crosstalk. Nat. Rev. Mol. Cell Biol. 2001;2(11):793–805. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- 28.Hynes RO. Fibronectins. Springer; New York: 1990. [Google Scholar]
- 29.Zamir E, Katz M, Posen Y, Erez N, Yamada KM, Katz BZ, Lin S, Lin DC, Bershadsky A, Kam Z, Geiger B. Dynamics and segregation of cell–matrix adhesions in cultured fibroblasts. Nat. Cell Biol. 2000;2(4):191–196. doi: 10.1038/35008607. [DOI] [PubMed] [Google Scholar]
- 30.Hocking DC, Sottile J, McKeown-Longo PJ. Fibronectin's III-1 module contains a conformation-dependent binding site for the amino-terminal region of fibronectin. J. Biol. Chem. 1994;269(29):19183–19187. [PubMed] [Google Scholar]
- 31.Zhong C, Chrzanowska-Wodnicka M, Brown J, Shaub A, Belkin AM, Burridge K. Rho-mediated contractility exposes a cryptic site in fibronectin and induces fibronectin matrix assembly. J. Cell Biol. 1998;141(2):539–551. doi: 10.1083/jcb.141.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baneyx G, Baugh L, Vogel V. Coexisting conformations of fibronectin in cell culture imaged using fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. U. S. A. 2001;98(25):14464–14468. doi: 10.1073/pnas.251422998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith ML, Gourdon D, Little WC, Kubow KE, Eguiluz RA, Luna-Morris S, Vogel V. Force-induced unfolding of fibronectin in the extracellular matrix of living cells. PLoS Biol. 2007;5(10):e268. doi: 10.1371/journal.pbio.0050268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mao Y, Schwarzbauer JE. Fibronectin fibrillogenesis, a cell-mediated matrix assembly process. Matrix Biol. 2005;24(6):389–99. doi: 10.1016/j.matbio.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 35.Gao M, Craig D, Lequin O, Campbell ID, Vogel V, Schulten K. Structure and functional significance of mechanically unfolded fibronectin type III1 intermediates. Proc. Natl. Acad. Sci. U. S. A. 2003;100(25):14784–14789. doi: 10.1073/pnas.2334390100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ingham KC, Brew SA, Huff S, Litvinovich SV. Cryptic self-association sites in type III modules of fibronectin. J. Biol. Chem. 1997;272(3):1718–1724. doi: 10.1074/jbc.272.3.1718. [DOI] [PubMed] [Google Scholar]
- 37.Krammer A, Lu H, Isralewitz B, Schulten K, Vogel V. Forced unfolding of the fibronectin type III module reveals a tensile molecular recognition switch. Proc. Natl. Acad. Sci. U. S. A. 1999;96(4):1351–1356. doi: 10.1073/pnas.96.4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baneyx G, Baugh L, Vogel V. Fibronectin extension and unfolding within cell matrix fibrils controlled by cytoskeletal tension. Proc. Natl. Acad. Sci. U. S. A. 2002;99(8):5139–5143. doi: 10.1073/pnas.072650799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Antia M, Islas LD, Boness DA, Baneyx G, Vogel V. Single molecule fluorescence studies of surface-adsorbed fibronectin. Biomaterials. 2006;27(5):679–90. doi: 10.1016/j.biomaterials.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 40.Halter M, Antia M, Vogel V. Fibronectin conformational changes induced by adsorption to liposomes. J. Controlled Release. 2005;101(1−3):209–22. doi: 10.1016/j.jconrel.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 41.Baugh L, Vogel V. Structural changes of fibronectin adsorbed to model surfaces probed by fluorescence resonance energy transfer. J. Biomed. Mater. Res. 2004;69A(3):525–534. doi: 10.1002/jbm.a.30026. [DOI] [PubMed] [Google Scholar]
- 42.Beningo KA, Dembo M, Kaverina I, Small JV, Wang YL. Nascent focal adhesions are responsible for the generation of strong propulsive forces in migrating fibroblasts. J. Cell Biol. 2001;153(4):881–888. doi: 10.1083/jcb.153.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Engler A, Richert L, Wong J, Picart C, Discher D. Surface probe measurements of the elasticity of sectioned tissue, thin gels and polyelectrolyte multilayer films: Correlations between substrate stiffness and cell adhesion. Surf. Sci. 2004;570:142–154. [Google Scholar]
- 44.Mahaffy RE, Shih CK, MacKintosh FC, Kas J. Scanning probe-based frequency-dependent microrheology of polymer gels and biological cells. Phys. Rev. Lett. 2000;85(4):880–883. doi: 10.1103/PhysRevLett.85.880. [DOI] [PubMed] [Google Scholar]
- 45.Beningo KA, Lo CM, Wang YL. Flexible polyacrylamide substrata for the analysis of mechanical interactions at cell-substratum adhesions. Methods Cell Biol. 2002;69:325–339. doi: 10.1016/s0091-679x(02)69021-1. [DOI] [PubMed] [Google Scholar]
- 46.McKeown-Longo PJ, Mosher DF. Interaction of the 70,000-mol-wt amino-terminal fragment of fibronectin with the matrix-assembly receptor of fibroblasts. J. Cell Biol. 1985;100(2):364–74. doi: 10.1083/jcb.100.2.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson KJ, Sage H, Briscoe G, Erickson HP. The compact conformation of fibronectin is determined by intramolecular ionic interactions. J. Biol. Chem. 1999;274(22):15473–15479. doi: 10.1074/jbc.274.22.15473. [DOI] [PubMed] [Google Scholar]
- 48.Khan MY, Medow MS, Newman SA. Unfolding transitions of fibronectin and its domains. Stabilization and structural alteration of the N-terminal domain by heparin. Biochem. J. 1990;270(1):33–38. doi: 10.1042/bj2700033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Little WC, Smith ML, Ebneter U, Vogel V. Assay to mechanically tune and optically probe fibrillar fibronectin conformations from fully relaxed to breakage. Matrix Biol. 2008 doi: 10.1016/j.matbio.2008.02.003. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gueta R, Barlam D, Shneck RZ, Rousso I. Measurement of the mechanical properties of isolated tectorial membrane using atomic force microscopy. Proc. Natl. Acad. Sci. U. S. A. 2006;103(40):14790–5. doi: 10.1073/pnas.0603429103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sechler JL, Rao H, Cumiskey AM, Vega-Colon I, Smith MS, Murata T, Schwarzbauer JE. A novel fibronectin binding site required for fibronectin fibril growth during matrix assembly. J. Cell Biol. 2001;154(5):1081–1088. doi: 10.1083/jcb.200102034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barker TH, Baneyx G, Cardo-Vila M, Workman GA, Weaver M, Menon PM, Dedhar S, Rempel SA, Arap W, Pasqualini R, Vogel V, Sage EH. SPARC regulates extracellular matrix organization through its modulation of integrin-linked kinase activity. J. Biol. Chem. 2005;280(43):36483–93. doi: 10.1074/jbc.M504663200. [DOI] [PubMed] [Google Scholar]
- 53.Mao Y, Schwarzbauer JE. Accessibility to the fibronectin synergy site in a 3D matrix regulates engagement of a5b1 versus avb3 integrin receptors. Cell Commun. Adhes. 2006;13(5−6):267–77. doi: 10.1080/15419060601072215. [DOI] [PubMed] [Google Scholar]
- 54.Amatangelo MD, Bassi DE, Klein-Szanto AJ, Cukierman E. Stroma-derived three-dimensional matrices are necessary and sufficient to promote desmoplastic differentiation of normal fibroblasts. Am. J. Pathol. 2005;167(2):475–88. doi: 10.1016/S0002-9440(10)62991-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang JT, Hynes RO. Fibronectin receptor functions in embryonic cells deficient in a 5 b 1 integrin can be replaced by a V integrins. Mol. Biol. Cell. 1996;7(11):1737–1748. doi: 10.1091/mbc.7.11.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu C, Bauer JS, Juliano RL, McDonald JA. The a 5 b 1 integrin fibronectin receptor, but not the a 5 cytoplasmic domain, functions in an early and essential step in fibronectin matrix assembly. J. Biol. Chem. 1993;268(29):21883–21888. [PubMed] [Google Scholar]
- 57.Wennerberg K, Lohikangas L, Gullberg D, Pfaff M, Johansson S, Fassler R. b 1 integrin-dependent and -independent polymerization of fibronectin. J. Cell Biol. 1996;132(1−2):227–238. doi: 10.1083/jcb.132.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zaidel-Bar R, Ballestrem C, Kam Z, Geiger B. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J. Cell Sci. 2003;116(22):4605–4613. doi: 10.1242/jcs.00792. [DOI] [PubMed] [Google Scholar]
- 59.Mao Y, Schwarzbauer JE. Stimulatory effects of a three-dimensional microenvironment on cell-mediated fibronectin fibrillogenesis. J. Cell Sci. 2005;118(Pt 19):4427–36. doi: 10.1242/jcs.02566. [DOI] [PubMed] [Google Scholar]
- 60.Abu-Lail NI, Ohashi T, Clark RL, Erickson HP, Zauscher S. Understanding the elasticity of fibronectin fibrils: Unfolding strengths of FN-III and GFP domains measured by single molecule force spectroscopy. Matrix Biol. 2006;25(3):175–184. doi: 10.1016/j.matbio.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 61.Erickson HP. Reversible unfolding of fibronectin type III and immunoglobulin domains provides the structural basis for stretch and elasticity of titin and fibronectin. Proc. Natl. Acad. Sci. U. S. A. 1994;91(21):10114–10118. doi: 10.1073/pnas.91.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ohashi T, Kiehart DP, Erickson HP. Dynamics and elasticity of the fibronectin matrix in living cell culture visualized by fibronectin–green fluorescent protein. Proc. Natl. Acad. Sci. U. S. A. 1999;96(5):2153–2158. doi: 10.1073/pnas.96.5.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Little WC, Smith ML, Ebneter U, Vogel V. Assay to mechanically tune and optically probe fibrillar fibronectin conformations from fully relaxed to breakage. Matrix Biol. 2008 doi: 10.1016/j.matbio.2008.02.003. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vogel V. Mechanotransduction involving multimodular proteins: converting force into biochemical signals. Annu. Rev. Biophys. Biomol. Struct. 2006;35:459–88. doi: 10.1146/annurev.biophys.35.040405.102013. [DOI] [PubMed] [Google Scholar]
- 65.Liu W, Jawerth LM, Sparks EA, Falvo MR, Hantgan RR, Superfine R, Lord ST, Guthold M. Fibrin fibers have extraordinary extensibility and elasticity. Science. 2006;313(5787):634. doi: 10.1126/science.1127317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leung LY, Tian D, Brangwynne CP, Weitz DA, Tschumperlin DJ. A new microrheometric approach reveals individual and cooperative roles for TGF-b1 and IL-1b in fibroblast-mediated stiffening of collagen gels. FASEB J. 2007 doi: 10.1096/fj.06-7510com. [DOI] [PubMed] [Google Scholar]
- 67.Sivakumar P, Czirok A, Rongish BJ, Divakara VP, Wang YP, Dallas SL. New insights into extracellular matrix assembly and reorganization from dynamic imaging of extracellular matrix proteins in living osteoblasts. J. Cell Sci. 2006;119(Pt 7):1350–60. doi: 10.1242/jcs.02830. [DOI] [PubMed] [Google Scholar]
- 68.Wu X, Dzenis YA. Size effect in polymer nanofibers under tension. J. Appl. Phys. 2007;102(4):044306–044312. [Google Scholar]
- 69.Storm C, Pastore JJ, MacKintosh FC, Lubensky TC, Janmey PA. Nonlinear elasticity in biological gels. Nature. 2005;435(7039):191–4. doi: 10.1038/nature03521. [DOI] [PubMed] [Google Scholar]
- 70.Wang N, Butler JP, Ingber DE. Mechanotransduction across the cell surface and through the cytoskeleton. Science. 1993;260(5111):1124–7. doi: 10.1126/science.7684161. [DOI] [PubMed] [Google Scholar]
- 71.Diez J. Mechanisms of cardiac fibrosis in hypertension. J. Clin. Hypertens. 2007;9(7):546–50. doi: 10.1111/j.1524-6175.2007.06626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Meshel AS, Wei Q, Adelstein RS, Sheetz MP. Basic mechanism of three-dimensional collagen fibre transport by fibroblasts. Nat. Cell Biol. 2005;7(2):157–64. doi: 10.1038/ncb1216. [DOI] [PubMed] [Google Scholar]
- 73.Fernandez P, Pullarkat PA, Ott A. A master relation defines the nonlinear viscoelasticity of single fibroblasts. Biophys. J. 2006;90(10):3796–805. doi: 10.1529/biophysj.105.072215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wakatsuki T, Elson EL. Reciprocal interactions between cells and extracellular matrix during remodeling of tissue constructs. Biophys. Chem. 2003;100(1−3):593–605. doi: 10.1016/s0301-4622(02)00308-3. [DOI] [PubMed] [Google Scholar]
- 75.Streuli C. Extracellular matrix remodelling and cellular differentiation. Curr. Opin. Cell Biol. 1999;11(5):634–40. doi: 10.1016/s0955-0674(99)00026-5. [DOI] [PubMed] [Google Scholar]
- 76.Sottile J, Chandler J. Fibronectin matrix turnover occurs through a caveolin-1-dependent process. Mol. Biol. Cell. 2005;16(2):757–68. doi: 10.1091/mbc.E04-08-0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pierschbacher MD, Ruoslahti E. Variants of the cell recognition site of fibronectin that retain attachment-promoting activity. Proc. Natl. Acad. Sci. U. S. A. 1984;81(19):5985–8. doi: 10.1073/pnas.81.19.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takagi J, Strokovich K, Springer TA, Walz T. Structure of integrin a5b1 in complex with fibronectin. EMBO J. 2003;22(18):4607–15. doi: 10.1093/emboj/cdg445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Redick SD, Settles DL, Briscoe G, Erickson HP. Defining fibronectin's cell adhesion synergy site by site-directed mutagenesis. J. Cell Biol. 2000;149(2):521–7. doi: 10.1083/jcb.149.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sharma A, Askari JA, Humphries MJ, Jones EY, Stuart DI. Crystal structure of a heparin- and integrin-binding segment of human fibronectin. EMBO J. 1999;18(6):1468–79. doi: 10.1093/emboj/18.6.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mould AP, Humphries MJ. Identification of a novel recognition sequence for the integrin a 4 a 1 in the COOH-terminal heparin-binding domain of fibronectin. EMBO J. 1991;10(13):4089–95. doi: 10.1002/j.1460-2075.1991.tb04985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Takahashi S, Leiss M, Moser M, Ohashi T, Kitao T, Heckmann D, Pfeifer A, Kessler H, Takagi J, Erickson HP, Fassler R. The RGD motif in fibronectin is essential for development but dispensable for fibril assembly. J. Cell Biol. 2007;178(1):167–78. doi: 10.1083/jcb.200703021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Curnis F, Longhi R, Crippa L, Cattaneo A, Dondossola E, Bachi A, Corti A. Spontaneous formation of L-isoaspartate and gain of function in fibronectin. J. Biol. Chem. 2006;281(47):36466–76. doi: 10.1074/jbc.M604812200. [DOI] [PubMed] [Google Scholar]
- 84.Hocking DC. Fibronectin matrix deposition and cell contractility: implications for airway remodeling in asthma. Chest. 2002;122(6 Suppl):275S–278S. doi: 10.1378/chest.122.6_suppl.275s. [DOI] [PubMed] [Google Scholar]
- 85.Ingham KC, Brew SA, Erickson HP. Localization of a cryptic binding site for tenascin on fibronectin. J. Biol. Chem. 2004;279(27):28132–5. doi: 10.1074/jbc.M312785200. [DOI] [PubMed] [Google Scholar]
- 86.Schnepel J, Tschesche H. The proteolytic activity of the recombinant cryptic human fibronectin type IV collagenase from E. coli expression. J. Protein Chem. 2000;19(8):685–92. doi: 10.1023/a:1007104420017. [DOI] [PubMed] [Google Scholar]
- 87.Schnepel J, Unger J, Tschesche H. Recombinant cryptic human fibronectinase cleaves actin and myosin: substrate specificity and possible role in muscular dystrophy. Biol. Chem. 2001;382(12):1707–14. doi: 10.1515/BC.2001.206. [DOI] [PubMed] [Google Scholar]
- 88.Langenbach KJ, Sottile J. Identification of protein-disulfide isomerase activity in fibronectin. J. Biol. Chem. 1999;274(11):7032–8. doi: 10.1074/jbc.274.11.7032. [DOI] [PubMed] [Google Scholar]


