SUMMARY
Organ growth is influenced by organ patterning, but the molecular mechanisms that link patterning to growth have remained unclear. We show that the Dpp morphogen gradient in the Drosophila wing influences growth by modulating the activity of the Fat signaling pathway. Dpp signaling regulates the expression and localization of Fat pathway components, and Fat signaling through Dachs is required for the effect of the Dpp gradient on cell proliferation. Juxtaposition of cells that express different levels of the Fat pathway regulators four-jointed and dachsous stimulates expression of Fat/Hippo pathway target genes and cell proliferation, consistent with the hypothesis that the graded expression of these genes contributes to wing growth. Moreover, uniform expression of four-jointed and dachsous in the wing inhibits cell proliferation. These observations identify Fat as a signaling pathway that links the morphogen-mediated establishment of gradients of positional values across developing organs to the regulation of organ growth.
INTRODUCTION
One of the remarkable features of animal development is the achievement of consistent proportions in organ size among individuals of a species. Developmental biologists have long hypothesized that the achievement of consistent sizes and proportions requires links between the regulation of organ patterning and the regulation of organ growth. Studies of appendage regeneration first led to a class of models which posit that growth can be regulated by the steepness of a gradient of positional values (Bohn, 1974; Day and Lawrence, 2000; French et al., 1976; Garcia-Bellido and Garcia-Bellido, 1998; Lawrence, 1970). Recently, it was demonstrated that in the developing wing of Drosophila, the steepness of the gradient of Decapentaplegic (Dpp) pathway activity influences cell proliferation (Rogulja and Irvine, 2005). Dpp is normally distributed in a gradient along the anterior-posterior (A–P) axis of the developing wing, and acts as a long-range morphogen to pattern the wing (Lecuit et al., 1996; Nellen et al., 1996). Juxtaposition of cells that perceive different levels of Dpp signaling stimulates cell proliferation, whereas flattening the normal gradient of Dpp pathway activity inhibits cell proliferation (Rogulja and Irvine, 2005). These observations implied that a gradient of positional values, established by Dpp signaling, influences wing growth, but the molecular mechanism by which this is achieved remained unknown.
A pathway or process that links a gradient of positional values to growth should fulfill three criteria. First, it would be expected to involve cell surface molecules that could be used by cells to compare their relative positional values. Second, the expression or activity of these cell surface molecules should be regulated downstream of the morphogen gradients that establish positional values. Third, this pathway should regulate growth. In considering how wing growth might be stimulated by the juxtaposition of cells that perceive different levels of Dpp signaling, we considered the Fat signaling pathway because of its potential to fulfill these three criteria.
fat encodes an atypical cadherin molecule that functions as a transmembrane receptor for an intercellular signaling pathway (Bennett and Harvey, 2006; Cho et al., 2006; Cho and Irvine, 2004; Fanto et al., 2003; Feng and Irvine, 2007; Mao et al., 2006; Matakatsu and Blair, 2006; Silva et al., 2006; Willecke et al., 2006). Two genes that influence Fat activity have been identified, four-jointed (fj) and dachsous (ds). fj and ds act genetically upstream of fat in the regulation of tissue polarity (Yang et al., 2002), act non-autonomously to influence the expression of Fat target genes (Cho et al., 2006; Cho and Irvine, 2004), and modulate the subcellular localization of Fat (Ma et al., 2003; Mao et al., 2006; Strutt and Strutt, 2002). ds encodes an atypical cadherin (Ds) that appears to associate with Fat (Matakatsu and Blair, 2006) and might act as a Fat ligand. fj encodes a Golgi protein (Fj) (Strutt et al., 2004) and so might modify Fat and/or Ds to modulate their interactions. Both fj and ds are expressed in gradients in developing imaginal discs (Brodsky and Steller, 1996; Clark et al., 1995; Villano and Katz, 1995), suggesting that their expression is regulated downstream of the morphogen gradients responsible for disc patterning.
fat is a Drosophila tumor suppressor, and thus normally functions to limit growth. Fat has recently been linked to several other Drosophila tumor suppressors, including components of the Hippo pathway, and together they form a Fat/Hippo signaling network that regulates a common set of downstream target genes (reviewed in Edgar, 2006; Pan, 2007). Upstream components of this signaling network all impinge on the Warts tumor suppressor. Both Warts protein levels and Warts activity are regulated by Fat/Hippo signaling (Edgar, 2006; Pan, 2007), and fat mutants can be partially rescued by Warts over-expression (Feng and Irvine, 2007). Warts is a kinase, and activated Warts phosphorylates and thereby inactivates a transcriptional co-activator protein, Yorkie (Yki) (Huang et al., 2005). yki is genetically required for the influence of fat on growth (Bennett and Harvey, 2006; Silva et al., 2006; Willecke et al., 2006), and the subcellular localization of Yki is influenced by upstream pathway components, including fat and warts (Dong et al., 2007; Oh and Irvine, 2008). The basic outlines of Fat/Hippo signaling have been worked out in Drosophila, but homologous genes have been identified in mammals, and at least for the Hippo branch of this signaling network, an analogous mammalian tumor suppressor pathway exists and influences growth (Dong et al., 2007; Zhao et al., 2007).
The influence of Fat on downstream gene expression and growth is absolutely dependent upon the unconventional myosin Dachs, as dachs mutation completely suppresses the fat tumor suppressor phenotype (Cho and Irvine, 2004; Feng and Irvine, 2007; Mao et al., 2006). Dachs protein can localize to the plasma membrane, but this membrane localization is inhibited by Fat (Mao et al., 2006). In addition to their influence on growth, fat, fj, and ds also affect planar cell polarity (PCP) (Casal et al., 2002; Strutt and Strutt, 2002; Yang et al., 2002). Interestingly, the localization of Dachs on the membrane is normally polarized, such that Dachs preferentially localizes to the distal sides of cells. This polarized localization is influenced by fat, fj, and ds (Mao et al., 2006), and currently constitutes the most immediate known response to Fat activity.
We have assessed the contribution of Fat signaling to Dpp-regulated growth by examining the influence of Dpp signaling on both regulators and readouts of Fat signaling, including fj and ds expression, Dachs and Yki localization, and transcriptional targets of the Fat/Hippo signaling network. We have also used dachs mutants to examine genetically the contribution of Fat signaling to the influence of Dpp signaling on wing growth, cell proliferation, and gene expression. And we have examined the influence of fj and ds expression patterns on cell proliferation and Fat signaling in the wing. Our results establish that morphogen gradients influence growth in part via the Fat signaling pathway, and emphasize that Fat signaling is modulated by juxtaposition of cells that express different levels of Fat pathway regulators. Finally, we propose a model to explain how the graded expression of Fj and Ds could influence Fat/Hippo signaling.
RESULTS
Dpp signaling influences the expression and localization of Fat pathway components
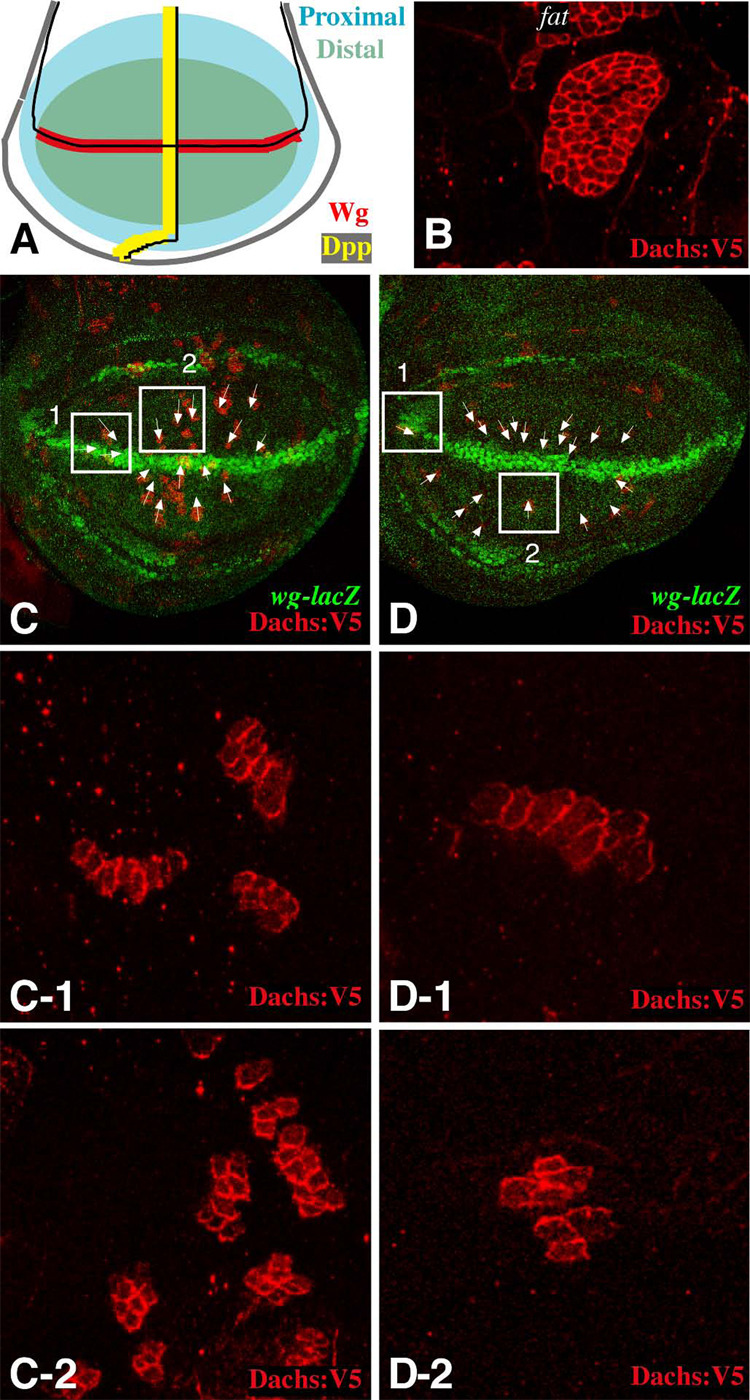
To characterize the potential relationship between Dpp signaling and Fat signaling, we examined the consequences of manipulations of Dpp pathway activity on the expression and localization of Fat pathway components. The most immediate known response to Fat signaling is the localization of Dachs at the membrane, which can be visualized using an epitope-tagged protein, Dachs:V5 (Mao et al., 2006). When expressed in clones of cells, a polarized localization of Dachs within cells is evidenced by the preferential accumulation of Dachs:V5 at the membrane on one side of a clone and not the other. Genetic experiments confirm that Dachs polarization is completely dependent upon fat (Fig. 1B). We have previously noted that, in the wing disc, Dachs preferentially accumulates on the distal sides of cells (Mao et al., 2006). We have since extended these observations by examining a much larger number of small clones throughout the developing wing, to create a Dachs polarization map. (Fig. 1C,D and data not shown). While confirming the general proximal-distal asymmetry in Dachs localization, this study also revealed finer details of Dachs polarization. For example, near the A–P compartment boundary and away from the dorsal-ventral (D–V) compartment boundary, Dachs is polarized along the D–V axis (Fig. 1C-2, D-2), whereas far from the A–P compartment boundary and near the D–V compartment boundary, it is polarized along the A–P axis (Fig. 1C-1, D-1). This pattern suggests that Dachs polarization is influenced downstream of cues emanating from both the A–P and D–V compartment boundaries.
Figure 1. Polarization of Dachs localization in the wing.
A) Schematic of a portion the wing imaginal disc. The approximate location of Wg-expressing cells along the D–V boundary (red) and Dpp-expressing cells along the A–P boundary (yellow) are shown. The region illustrated here as distal (green) corresponds to Vestigial-expressing cells, which give rise to the wing blade. B–D) Show portions of wing imaginal discs with clones of cells expressing Dachs:V5 (red). B) In a clone of cells mutant for fat8 and expressing Dachs:V5, Dachs is on the membrane all around the clone circumference. C,D) Two examples of wild-type discs with many small Dachs:V5-expressing clones, the D–V boundary and wing pouch are demarcated by wg-lacZ[ro216] expression (green). Arrows indicate the vectors of Dachs polarization for selected clones. Panels -1 and -2 show close-ups of the boxed regions; box 1 shows clones near the D–V boundary but far from the A–P boundary and box 2 shows clones near the A–P boundary but far from the D–V boundary.
Dpp is the morphogen produced by A–P compartment boundary cells, and becomes distributed in a gradient that influences patterning and growth (reviewed in Affolter and Basler, 2007). The A–P polarization of Dachs localization thus suggests that Fat signaling is influenced downstream of Dpp signaling. This was investigated by examining the influence of an activated form of the Dpp receptor Thickveins, TkvQ-D (Nellen et al., 1996), on Dachs localization. To parallel our earlier study of the effect of TkvQ-D on BrdU labeling (Rogulja and Irvine, 2005), expression of TkvQ-D was temporally controlled in these experiments using either drug-regulated (AyGal4:PR)(Rogulja and Irvine, 2005) or temperature-regulated (Gal4/Gal80ts)(Buttitta et al., 2007) systems; both methods gave similar results. Dachs was lost from the membrane all around the edges of clones expressing TkvQ-D (Fig. 2A). In complementary experiments, we inhibited Dpp signaling by expressing the transcriptional repressor Brinker (Brk). Since most of the influence of Dpp signaling on patterning and growth can be accounted for by its repression of Brk expression (reviewed in Affolter and Basler, 2007), forced expression of Brk is functionally equivalent to loss of Dpp pathway activity. In clones of cells co-expressing Dachs:V5 and Brk, normal Dachs polarization was lost, as Dachs:V5 was observed on both distal and proximal clone edges (Fig. 2B). The results of these pathway activation and inhibition experiments parallel endogenous Dachs localization, because in all cases Dachs concentrates on the membrane of cells with less Dpp pathway activity when they contact cells with higher Dpp pathway activity, and Dachs is excluded from the membrane of cells with higher Dpp pathway activity when they contact cells with lower Dpp pathway activity. Thus, the Dpp morphogen gradient influences Fat signaling, and this influence can be visualized at the level of Dachs localization.
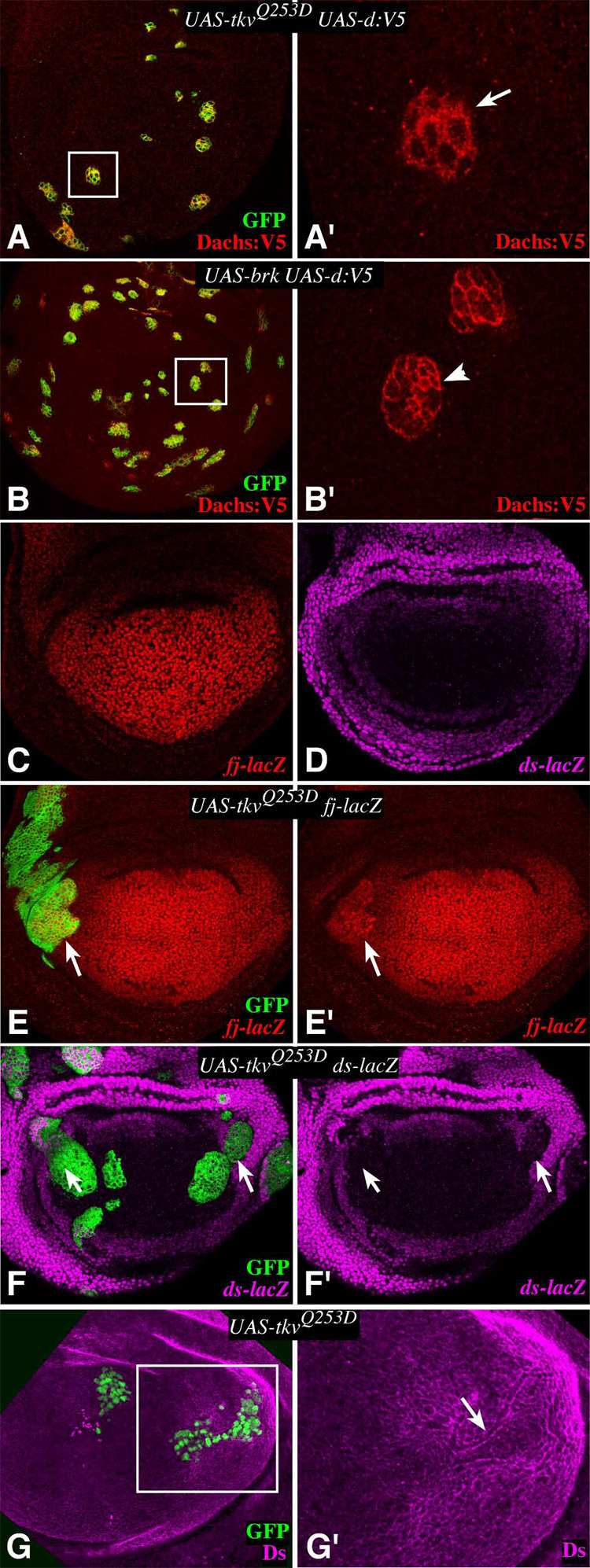
Figure 2. Dpp signaling influences Fat signaling components.
A,B,E,F) Show wing imaginal discs containing tub-Gal4/Gal80ts clones, 24–28h after temperature shift-mediated induction of expression, G) shows a disc with a Gal4:PR-expressing clones at 18 h after RU486-mediated induction of expression; all clones were marked by expression of GFP (green). In this and subsequent figures, panels marked prime show a single channel of the image to the left. A) Clones expressing Dachs:V5 and TKVQ-D. Dachs is not on the membrane on the distal side of the clone (arrow). A’ shows a close-up of the boxed area in A. B) Clones expressing Dachs:V5 and Brinker. B’ shows a close-up of the boxed area in B. Dachs is on the membrane on all sides of the clone, arrowhead points to proximal edge. C) fj expression (fj-lacZ) is highest in distal wing cells, and modestly graded from distal to proximal. D) ds expression (ds-lacZ) is highest in proximal wing cells, and modestly graded from proximal to distal. E) fj-lacZ expression is upregulated within clones expressing TKVQ-D (arrow). F) ds-lacZ expression is repressed in the proximal wing within clones expressing TKVQ-D (arrow). G) Ds protein is relocalized around the edges of clones expressing TkvQ-D and appears diminished within the clone. G’ shows a close-up of the boxed area in G.
To further characterize the relationship between Dpp signaling and Fat signaling, we examined the effect of TkvQ-D on the expression of fj and ds, as they are the only two known Fat regulators. They are expressed in largely complementary patterns in the developing wing, with fj highest in distal cells and ds highest in proximal cells (Fig. 2C,D). Since proximal-distal patterning in the wing is established by the combined action of signals emanating from the A–P and D–V compartment boundaries (Fig. 1A), these expression patterns are suggestive of regulation downstream of Dpp signaling. Indeed, TkvQ-D induced elevated fj expression (Fig. 2E, Supplementary Fig. S1A). This regulation of fj by TkvQ-D presumably contributes to its influence on Dachs localization, as Dachs:V5 is also lost from the edges of clones with elevated Fj (Mao et al., 2006). When Ds expression within TkvQ-D-expressing clones was examined, its levels were reduced (Fig. 2F,G). In addition, there was a relocalization of Ds protein, with a strong ring of Ds detected around the edges of these clones, and a slight halo of decreased Ds staining in immediately adjacent cells (Fig. 2G). Fj affects the localization of Fat and Ds at the membrane (Ma et al., 2003; Mao et al., 2006; Strutt and Strutt, 2002), and thus could contribute to the observed relocalization of Ds. As fat mutants have more severe phenotypes than fj, ds, or fj ds double mutants, there may be additional genes that contribute to Fat regulation. Nonetheless, these observations indicate that the expression of both of the known Fat regulators is regulated by Dpp signaling in a manner consistent with the normal relationship between their expression patterns and the Dpp morphogen gradient.
The D–V compartment boundary is established by Notch activation, and local Notch activation within the wing exerts a non-autonomous influence on wing growth (reviewed in Irvine and Vogt, 1997). Activation of Notch induces a long range morphogen, Wg, but by contrast to the role of Dpp signaling in mediating the influence of the A–P compartment boundary on wing growth, the basis for the influence of Notch on wing growth remains to be elucidated, as it can not be accounted for by Wg (Giraldez and Cohen, 2003; Johnston and Sanders, 2003; Klein and Arias, 1998). Nonetheless, processes downstream of Notch do affect Fat signaling, as expression of fj was induced non-autonomously by clones of cells expressing an activated form of Notch (Nintra)(Supplementary Fig. S1C). Modulation of Fat signaling downstream of Notch was also evidenced by the observation that expression of activated Notch resulted in a loss of membrane localization of Dachs (Fig. S1B), similar to the effects of TkvQ-D (Fig. 2A).
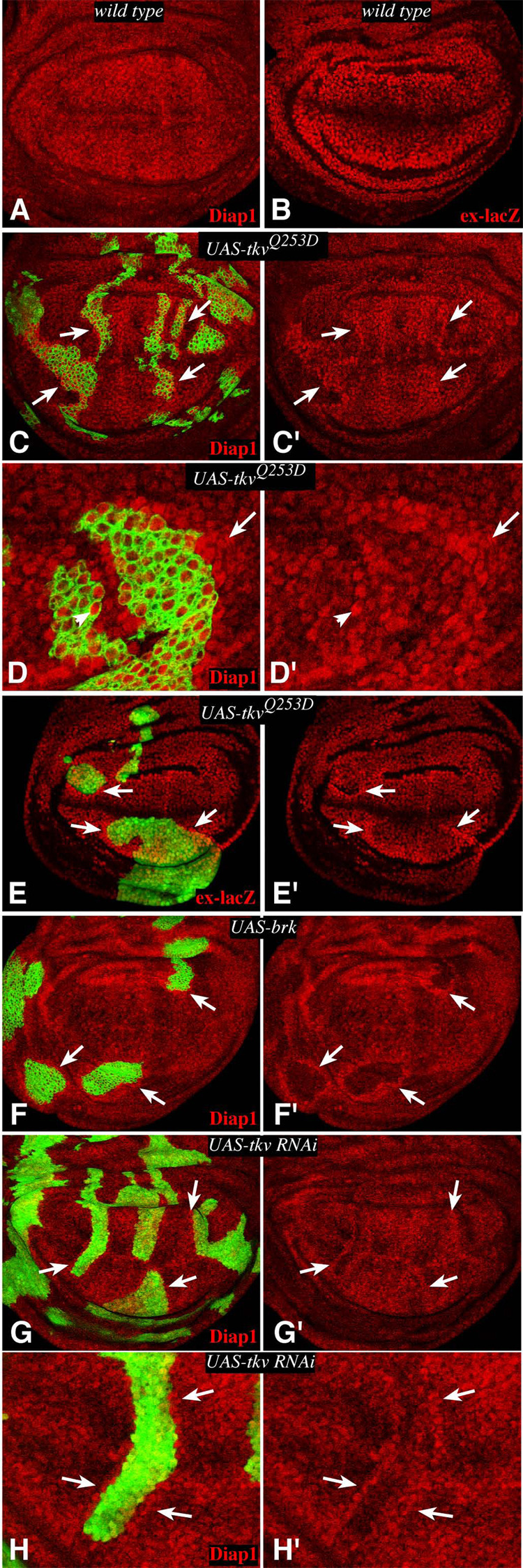
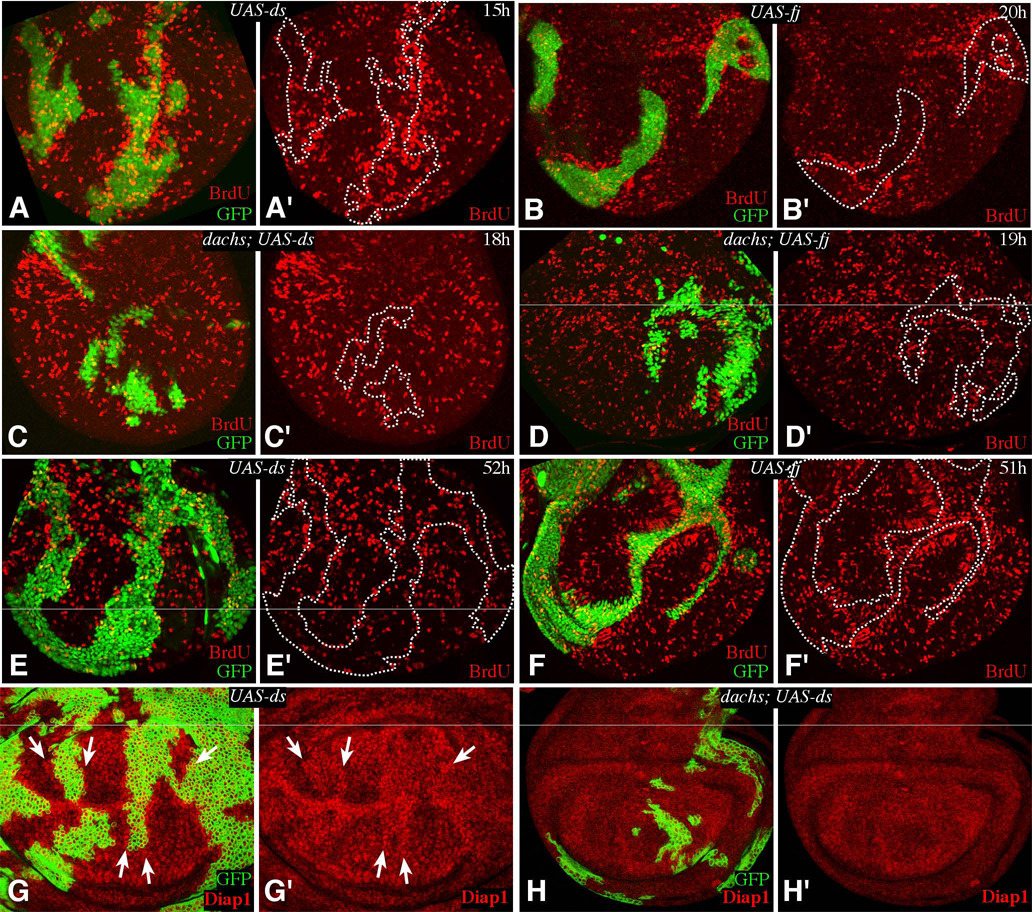
To investigate whether Dpp signaling also impacts transcriptional outputs of the Fat pathway, we examined downstream targets of Fat/Hippo signaling. Expression of the apoptosis inhibitor Diap1 is upregulated within fat mutant clones (Bennett and Harvey, 2006; Cho et al., 2006; Silva et al., 2006; Willecke et al., 2006). Diap1 has been widely used as a cell-autonomous marker of Fat/Hippo signaling (Pan, 2007), and is a direct target of the heterodimeric Yki-Scalloped transcription factor that regulates Fat/Hippo pathway target genes (Wu et al., 2008; Zhang et al., 2008). Diap1 expression was upregulated around the edges of TKVQ-D–expressing clones (Fig. 3C,D). Another downstream target of Fat/Hippo signaling, ex-lacZ (Hamaratoglu et al., 2006), was also upregulated around the edges of TkvQ-D–expressing clones (Fig. 3E). The influence of TKVQ-D–expressing clones on Fat/Hippo signaling was also evidenced by the detection of increased nuclear Yki staining in cells surrounding TkvQ-D-expressing clones (Supplementary Fig. S2), reminiscent of the increased detection of nuclear Yki when fat is downregulated (Oh and Irvine, 2008). The induction of Fat target genes was strongest in cells immediately neighboring TkvQ-D–expressing clones, but could also be observed up to two to three cells away from the clone edge, and in cells just inside of the clone edge (Fig. 3D). The effects of TkvQ-D–expressing clones are strongest in lateral regions, where the endogenous levels of Tkv activity are lowest. Reduction of Dpp-signaling also influenced Diap1 levels, as Brinker-expressing clones were associated with a cell autonomous decrease in Diap1 expression, and a non-autonomous induction of Diap1 (Fig. 3F). Similar effects were observed when endogenous Tkv levels were reduced by RNAi (Fig. 3G,H). Altogether, these results confirm that Dpp signaling modulates transcriptional outputs of Fat signaling, such that Fat pathway activity is inhibited when cells with different levels of Dpp pathway activity are juxtaposed.
Figure 3. Dpp signaling influences Fat/Hippo pathway target genes.
Wing imaginal discs, stained for expression of Diap1 (A, C, D, F–H) or ex-lacZ (B, E) (red). C–H) contain tub-Gal4/Gal80ts clones marked by co-expression of GFP. Arrows point to examples of Fat/Hippo target gene upregulation around clone edges. A,B) Wild-type discs. C) TkvQ-D– expressing clone. Strong Diap1 upregulation is observed in lateral regions, but the effect is subtle in the medial wing, where endogenous Tkv activity is high. D) Close-up of a TkvQ-D-expressing clone, Diap1 upregulation is strongest in cells immediately neighboring the clone, but examples of upregulation two to three cells away (arrow) and inside the clone border (arrowhead) can be observed. E) TkvQ-D–expressing clone, ex-lacZ is upregulated around clone edges, except near the D–V boundary. F) Brinker-expressing clone. Diap1 is downregulated inside the clone, but upregulated just outside, mimicking the effects of Brinker on BrdU labeling (Rogulja and Irvine, 2005). G) Clones in which Tkv levels have been downregulated by RNAi. Diap1 upregulation is observed along clone edges. H) Close-up of a clone in G.
dachs is required for the induction of BrdU labeling by TkvQ-D
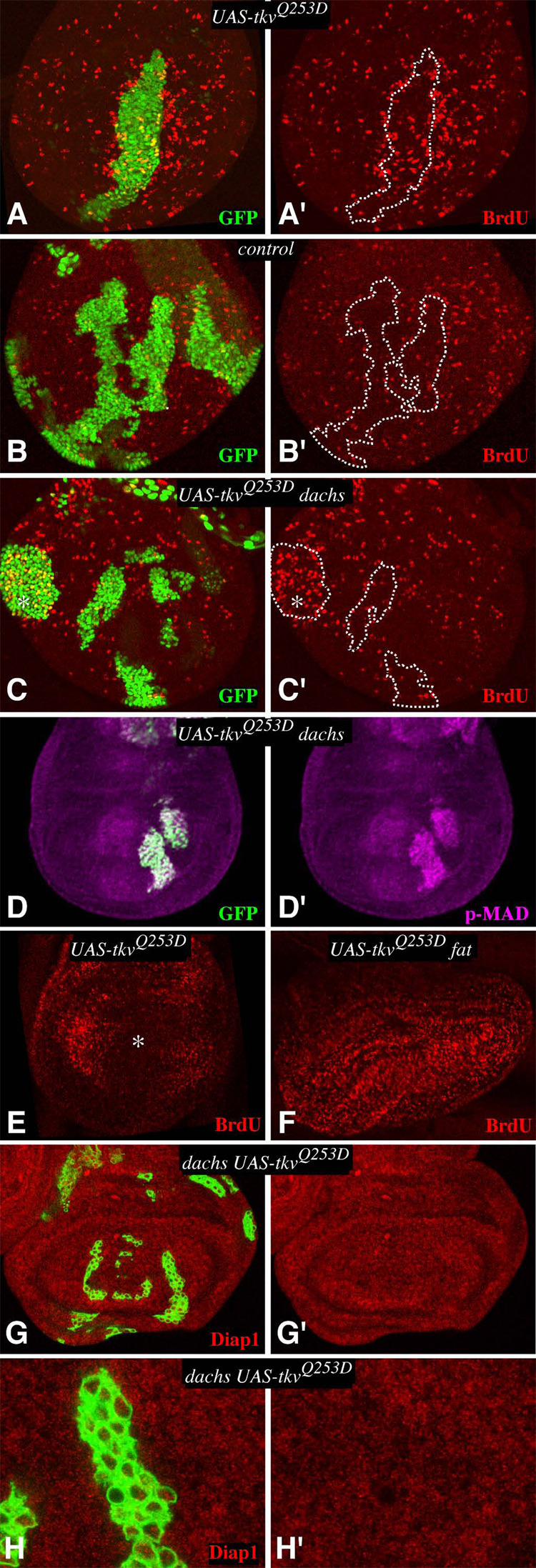
Since loss of Fat/Hippo signaling is associated with overgrowth, the effects of the Dpp pathway on the expression of Fat/Hippo pathway target genes (Fig. 3) correlates with the effects of the Dpp pathway on cell proliferation (Rogulja and Irvine, 2005). This is consistent with the hypothesis that the cell proliferation associated with juxtaposition of cells expressing different levels of TkvQ-D could be accounted for by its influence on Fat signaling. As a critical test of this hypothesis, we investigated the effect of TkvQ-D clones on cell proliferation in dachs mutants, as dachs is genetically required for the influence of Fat on downstream gene expression and growth (Cho and Irvine, 2004; Mao et al., 2006). Thus, if the Dpp gradient acts through its influence on Fat signaling, then the non-autonomous effects of TkvQ-D-expressing clones on cell proliferation and Fat target genes should depend upon dachs. Indeed, upregulation of Diap1 expression around the edges of TkvQ-D expressing clones was lost in dachs mutant animals (Fig. 4G,H).
Figure 4. dachs is required for non-autonomous influences of Tkv on cell proliferation.
A–D Show wing imaginal discs containing Gal4:PR-expressing clones, marked by expression of GFP (green), grown for 15 hours on media containing RU486 and then labeled and stained for BrdU (red), or phospho-Mad (magenta). For ease of comparison, the locations of selected clones are outlined by dashes. Because the nuclei are not all in the same focal plane, we combined staining in different focal planes by maximum projection through confocal sections. A) AyGal4:PR UAS-1tkvQ253D UAS-GFP. BrdU labeling is elevated around the clone. B) AyGal4:PR UAS-GFP. BrdU labeling is normal C) dachsGC13; AyGal4:PR UAS-TkvQ253D UAS-GFP. BrdU labeling is autonomously elevated within a lateral clone (asterisk), but no non-autonomous elevation of labeling is observed. D) dachsGC13; AyGal4:PR UAS-TkvQ253D UAS-GFP, p-MAD staining is elevated in TKVQ-D-expressing clones. E–F Show discs with uniform TkvQ253D expression, induced by actin-Gal4:PR. E) In wild type this represses BrdU labeling in medial cells (asterisk)(Rogulja and Irvine, 2005), but F) in fat no medial repression occurs. G) dachs mutant wing imaginal discs containing tub-Gal4/Gal80ts clones expressing TkvQ253D, stained for expression of Diap1 (red). Diap1 expression is not affected by the clones. H) Close-up of a clone shown in G.
In BrdU labeling experiments, TkvQ-D was expressed under AyGal4:PR control and evaluated at 14–19h after induction of TkvQ-D expression. Because of the intrinsic variability of BrdU labeling, and the incomplete penetrance of BrdU labeling phenotypes associated with TkvQ-D-expressing clones (Rogulja and Irvine, 2005), three types of clones (GFP-expressing, TkvQ-D-expressing in wild type, and TkvQ-D-expressing in dachs) were created by one investigator (DR) and then scored blind by the other (KI). In otherwise wild-type animals, 55% (117/213) of TkvQ-D clones were scored as being associated with non-autonomous induction of BrdU labeling (Fig. 4A), whereas 12% (18/147) of control (GFP-expressing) clones were scored as being associated with elevated BrdU labeling (Fig. 4B). (This compares with 67% of TkvQ-D clones, and 6% of control clones, scored as being associated with non-autonomous elevation of BrdU labeling in a prior series of experiments (Rogulja and Irvine, 2005). conversely, only 11% (34/309) of TkvQ-D clones in dachs mutants were scored as being associated with a non-autonomous elevation of BrdU labeling (Fig. 4C). To quantify these results, we also used an automated image analysis program to count labeled nuclei per unit area, and to compare the frequency of labeled nuclei surrounding clones to the frequency elsewhere in the disc (Supplementary Figure S3). This analysis identified a 2.6-fold increase in labeled nuclei surrounding TkvQ-D –expressing clones in wild-type, whereas in dachs mutant discs the frequency of BrdU labeling was comparable to that in the wild-type control (Fig. S3). The observation that the non-autonomous induction of cell proliferation associated with TkvQ-D-expressing clones is essentially eliminated in dachs mutant animals implies that the Dpp gradient requires the Fat pathway to influence growth. Importantly, Dpp signaling still occurs in dachs mutants, as monitored by the elevated phosphorylation of the Mad transcription factor in TkvQ-D clones (Fig. 4D). Moreover, the ability of TkvQ-D clones to autonomously upregulate BrdU labeling in lateral cells (Martin-Castellanos and Edgar, 2002; Rogulja and Irvine, 2005) is retained in dachs mutants, albeit at a reduced level (Fig. 4C), (38% (35/93) of lateral TkvQ-D-expressing clones were scored as autonomously upregulating BrdU labeling in dachs mutants, compared with 80% (68/85) of TkvQ-D clones and 0% (0/43) control clones in wild-type animals). The persistence of the autonomous mechanism, but not the non-autonomous mechanism, for promotion of cell proliferation by TkvQ-D is consistent with the observation the wing growth is reduced but not eliminated in dachs mutants (Mao et al., 2006).
While juxtaposing cells with different levels of Dpp pathway activity stimulates cell proliferation, uniform activation of Tkv inhibits the proliferation of medial wing cells (Fig. 4E)(Rogulja and Irvine, 2005). To investigate whether this inhibition might require fat, we assayed the influence of uniform TkvQ-D expression on BrdU labeling in fat mutant wing discs. When TkvQ-D was expressed uniformly in fat mutants, strong BrdU labeling was detected throughout the wing (Fig. 4F). Thus, mutation of fat abrogated the inhibition of proliferation normally associated with uniform TkvQ-D expression.
Juxtaposition of cells expressing different levels of Fj or Ds induces elevated BrdU labeling
The observations that TkvQ-D influences Fat signaling, and that dachs is required for the non-autonomous effect of TkvQ-D on BrdU labeling and Fat target gene expression, imply that the Dpp gradient regulates cell proliferation through the Fat pathway. This in turn suggests that the graded expression of the two known Fat regulators, Fj and Ds, could influence wing growth. To investigate this, we examined the consequences of creating sharper than normal juxtapositions between cells expressing different levels of Fj or Ds.
When expression of Fj or Ds was induced in clones of cells, a non-autonomous elevation of BrdU labeling was clearly observed along the edges of these clones by 15 h after induction of expression (Fig. 5). In blind scoring, 57% (106/185) of Fj-expressing clones, and 56% (111/200) of Ds-expressing clones at 15–22 h were identified as causing a non-autonomous elevation of BrdU labeling (Fig. 5A,B), but only 7% (11/148) of GFP-expressing control clones were scored as being associated with elevation of BrdU labeling. Quantitation of this effect by automated image analysis identified a 3.4 fold increase in the frequency of labeled nuclei surrounding Ds-expressing clones, and a 2.6 fold increase in labeled nuclei surrounding Fj-expressing clones (Fig. S3). When we examined Ds or Fj-expressing clones at 51–52h after induction, a robust response to Fj was still detected (62%, or 79/128 clones), although the influence of Ds was reduced (25%, or 23/93 clones) (Fig. 5E,F). To control for the possibility that changes in cell affinity might influence cell proliferation we also included E-cadherin expressing clones at 15–22h in this analysis, but only 8% (7/83) were scored as being associated with a non-autonomous elevation of BrdU labeling in blind scoring (not shown), and no increase in the frequency of BrdU labeled nuclei was detected by automated image analysis (Fig. S3).
Figure 5. Fj- or Ds-expressing clones elevate BrdU incorporation.
A–F show wing imaginal discs containing Gal4:PR-expressing clones, marked by expression of GFP (green), grown for the indicated number of hours on media containing RU486, and labeled and stained for BrdU. For ease of comparison, the locations of selected clones are outlined by dashes. A,E) AyGal4:PR UAS-ds UAS-GFP. Elevated BrdU labeling is evident in A, especially in distal regions, but not in E. B,F) AyGal4:PR UAS-fj UAS-GFP. Elevated BrdU labeling is evident, especially in proximal regions. C) dachsGC13; AyGal4:PR UAS-ds UAS-GFP. BrdU labeling is not affected by the clones. D) dachsGC13; AyGal4:PR UAS-fj UAS-GFP. BrdU labeling is not affected by the clones. G) tub-Gal4/Gal80ts clones expressing ds; Diap1 staining is elevated around the clones (arrows). H) dachsGC13 mutant with tub-Gal4/Gal80ts clones expressing ds, Diap1 staining is not affected by the clones.
Notably, when Fj was expressed, the detection of elevated BrdU labeling was strongest in proximal regions of the wing (62%, 100/161 clones), where endogenous FJ levels are lowest, and weaker in distal regions (43%, 39/90 clones), where endogenous Fj levels are higher (Fig. 5B). Conversely, when Ds was expressed, the detection of elevated BrdU labeling was strongest in distal regions of the wing (69%, 79/115 clones), where endogenous Ds levels are lowest, and weaker in proximal regions (46%, 73/160 clones), where endogenous Ds levels are higher (Fig. 5A). These observations suggest that the ability of Fj or Ds-expressing clones to induce cell proliferation depends on the degree of difference in expression levels between neighboring cells. BrdU labeling was most obviously elevated along the outside edge of clones, but also sometimes appeared elevated along the inside edge of clones.
A variety of observations have indicated that Fj or Ds expression are associated with inhibition of Fat signaling along clone edges. fat mutant clones have been associated with upregulation of wg, Ser, fj, and Diap1 (Cho et al., 2006; Cho and Irvine, 2004; Mao et al., 2006; Yang et al., 2002), and these same genes can be upregulated around the edges of clones of cells expressing Fj or Ds (Fig. 5G, Supplementary Fig. S1D)(Buckles et al., 2001; Cho et al., 2006; Cho and Irvine, 2004; Zeidler et al., 1999). To confirm that the elevated BrdU labeling induced by juxtaposing cells expressing different levels of Fj or Ds is also mediated through the Fat pathway, we made Fj-or Ds-expressing clones in dachs mutants. Indeed, the ability of Fj or Ds-expressing clones to induce BrdU labeling was suppressed in dachs mutants (Fig. 5C,D; in blind scoring, 13% (15/112) of DS-expressing clones and 5% (2/37) of FJ-expressing clones were scored as being associated with elevated BrdU labeling in dachs mutant wing discs). The induction of Diap1 expression around the edges of Ds-expressing clones (Fig. 5G) was also lost in dachs mutants (Fig. 5H).
Uniform expression of Fj and Ds inhibits cell proliferation and growth
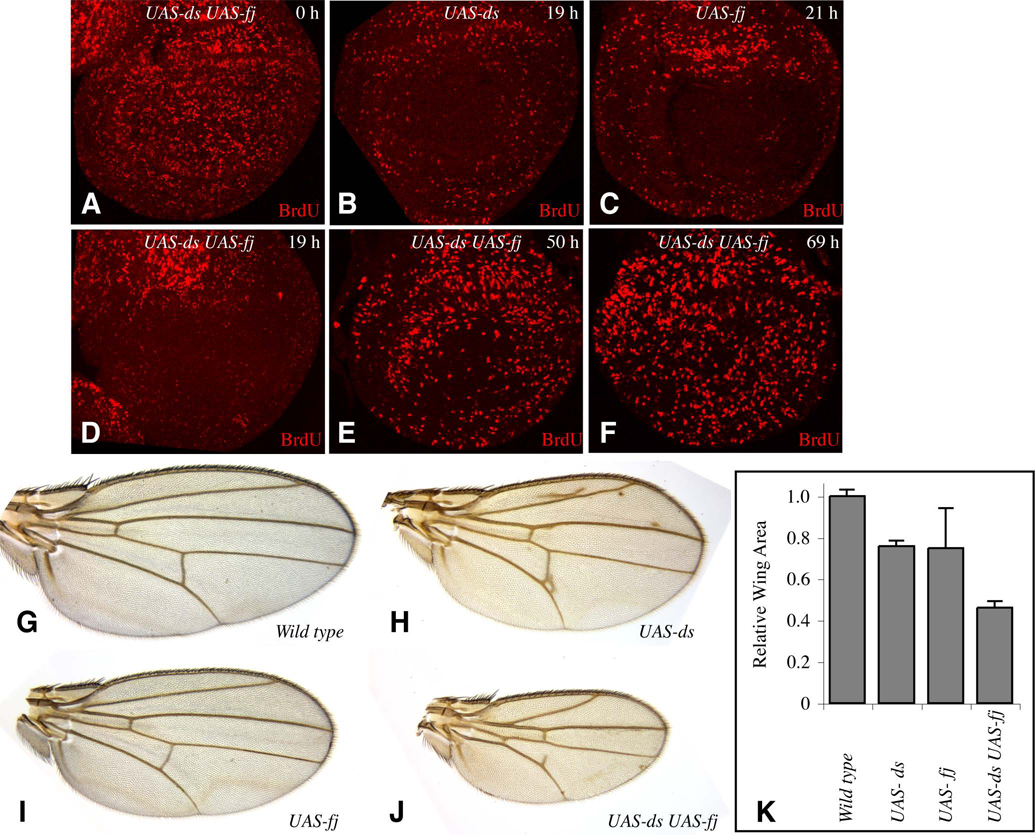
If wing growth is normally influenced by the gradients of fj and ds expression, then flattening these gradients by driving uniform expression of fj or ds should inhibit wing growth. To evaluate this possibility, we expressed fj and ds both alone and together under the control of the actin promoter, using a derivative of AyGal4:PR from which the flip-out cassette has been permanently excised, such that it constitutively expresses a drug-inducible form of Gal4 (act>Gal4:PR) (Rogulja and Irvine, 2005). Uniform expression of either fj or ds alone resulted in inhibition of BrdU labeling by 19 h after the induction of Gal4:PR mediated expression (Fig. 6B,C). Although Fj and Ds have distinct molecular roles within the Fat pathway, studies of tissue polarity suggest that the information provided by their graded expression is partially redundant (Matakatsu and Blair, 2004; Simon, 2004), and co-expression of fj and ds resulted in a stronger decrease in BrdU labeling (Fig. 6D). In all cases, BrdU labeling was strongly decreased in the wing region of the disc, but the notal region was less affected (Fig. 6B–D). Since the same transgene insertions and experimental conditions were used for uniform expression experiments and clonal expression experiments, the levels of Fj or Ds expression induced are expected to be similar. Thus, the observation that uniform induction of Fj or Ds expression inhibits BrdU labeling (Fig. 6), whereas patchy induction of Fj or Ds expression stimulates it (Fig. 5), indicates that the relative levels of Fj or Ds between neighboring cells, and not simply the absolute level of expression, is a critical determinant of whether or not wing cells proliferate. The shut down of BrdU labeling in the wing associated with uniform Fj and Ds expression was transient, as by 50 h, BrdU labeling began to recover, and by 69 h was again detected throughout the disc (Fig. 4E,F and data not shown). The transience of this response suggests that there are alternative processes that can promote cell proliferation in addition to the Fj and Ds gradients.
Figure 6. Uniform Fj and Ds expression inhibits BrdU incorporation & wing growth.
Panels A–F show discs grown for the indicated number of hours on media containing RU486 and then labeled and stained for BrdU (red). A) UAS-ds UAS-fj actin>Gal4:PR UAS-GFP. B) UAS-ds actin>Gal4:PR UAS-GFP. C) UAS-fj actin>Gal4:PR UAS-GFP. D–F) UAS-ds UAS-fj actin>Gal4:PR UAS-GFP. G–J show adult wings, all at the same magnification, from animals with a tub-Gal4 transgene and G) No UAS transgene H) UAS-ds, I) UAS-fj, J) UAS-ds UAS-fj. K) Histogram of the average areas of ten female wings of the genotypes in G–J, normalized to the average area in wild-type. Error bars indicate one standard deviation.
The consequences of uniform co-expression of Fj and Ds on tissue polarity have been examined in recent studies. While their influence on growth was not a focus of those experiments, it does appear that smaller wings can be generated as a consequence of uniform Fj and Ds expression (Matakatsu and Blair, 2004; Simon, 2004). To directly characterize the overall influence of Fj or Ds expression on wing growth, we first created adult wings in which Fj and Ds were expressed either alone or in combination under tub-gal4 control, and measured the area of adult wing blades in comparison to control wings. tub-gal4 UAS-fj UAS-ds wings were ~45% of control size (Fig. 6J,K). This reduction is similar to that observed in dachs null mutants (Mao et al., 2006), suggesting that Fat signaling in animals co-expressing Fj and Ds is comparable to that in the absence of dachs. Uniform expression of either Fj or Ds alone resulted in wing sizes intermediate between wild-type and tub-gal4 UAS-fj UAS-ds wings (Fig. 6H,I,K). The observation that wing size is decreased in animals uniformly expressing Fj or Ds implies that the loss of BrdU labeling detected in the time course experiments described above is reflective of an influence of these manipulations on wing growth.
To establish that the inhibition of growth associated with uniform expression of Fj or Ds is a local, rather than a systemic, response, we examined the consequences of expressing these genes only in the posterior compartment, using an en-Gal4 driver. Expression of fj or ds, or of both genes together, reduced the relative size of the posterior compartment, and decreased expression of Diap1, resulting in a phenotype similar to, though weaker than, downregulation of dachs by en-Gal4 driven RNAi (Supplementary Figure S4). Conversely, downregulation of ds by en-Gal4 driven RNAi increased the relative size of the posterior compartment, and upregulated expression of Diap1 (Supplementary Figure S4).
DISCUSSION
The Dpp morphogen gradient influences Fat signaling
Studies of regeneration first led to models which proposed that growth could be influenced by gradients of positional values, with steep gradients promoting growth and shallow gradients suppressing growth (Bohn, 1974; Day and Lawrence, 2000; French et al., 1976; Garcia-Bellido and Garcia-Bellido, 1998; Lawrence, 1970). Experimental manipulations of Dpp pathway activity in the Drosophila wing supported this concept (Rogulja and Irvine, 2005), but left unanswered the question of how differences in the levels of Dpp pathway activity perceived by neighboring cells are actually linked to growth. Here, we have established that the Fat signaling pathway provides this link. Dpp signaling influences the Fat pathway, as the expression of upstream Fat pathway regulators, the sub-cellular localization of Fat pathway components, and downstream transcriptional outputs of Fat signaling, are all affected by Dpp signaling (Fig 1–Fig 3, Fig S1, S2). The effects that Tkv and Brk expression have on expression of Fat target genes parallels their effects on BrdU labeling (Rogulja and Irvine, 2005), and depend genetically on Fat signaling (Fig. 4).
Dpp signaling impinges on Fat signaling upstream of Fat, as the expression of both of its known regulators, Fj, and Ds, is regulated by Dpp signaling (Fig. 2). Although the Fat signaling pathway was only recently discovered, and our understanding of Fat signaling and its regulation remains incomplete, the inference that Fat signaling is normally influenced by the Dpp morphogen gradient is supported by the polarized localization of Dachs in wild-type wing discs (Fig. 1). Near the D–V compartment boundary, the vector of Dachs polarization parallels the vector of the Dpp morphogen gradient, and the consequences of altered Dpp pathway activity confirm that the correlation between them is reflective of a functional link. The expression of Fj and Ds, and the localization of Dachs, are also polarized along the D–V axis. The implication that signaling downstream of the D–V compartment boundary thus also impinges on Fat signaling, and indeed may also influence growth through this pathway, is consistent with the observation that normal wing growth requires both A–P and D–V compartment boundary signals, and is further supported here by the observation that Notch activation affects both fj expression and Dachs localization (Supplementary Fig. S1).
Modeling Growth regulation by Fj and Ds gradients
Our results argue that Fat signaling is influenced by the graded expression of its regulators: uniform expression of Fj and Ds can activate Fat signaling and thereby inhibit growth, whereas juxtaposition of cells expressing different levels of Fj or Ds can inhibit Fat signaling and thereby promote growth. Here we propose a model to explain how Fat signaling can be modulated by Fj and Ds gradients. While aspects of our model remain speculative, it provides an explanation for a number of observations that would otherwise appear puzzling, and serves as a useful framework for future studies.
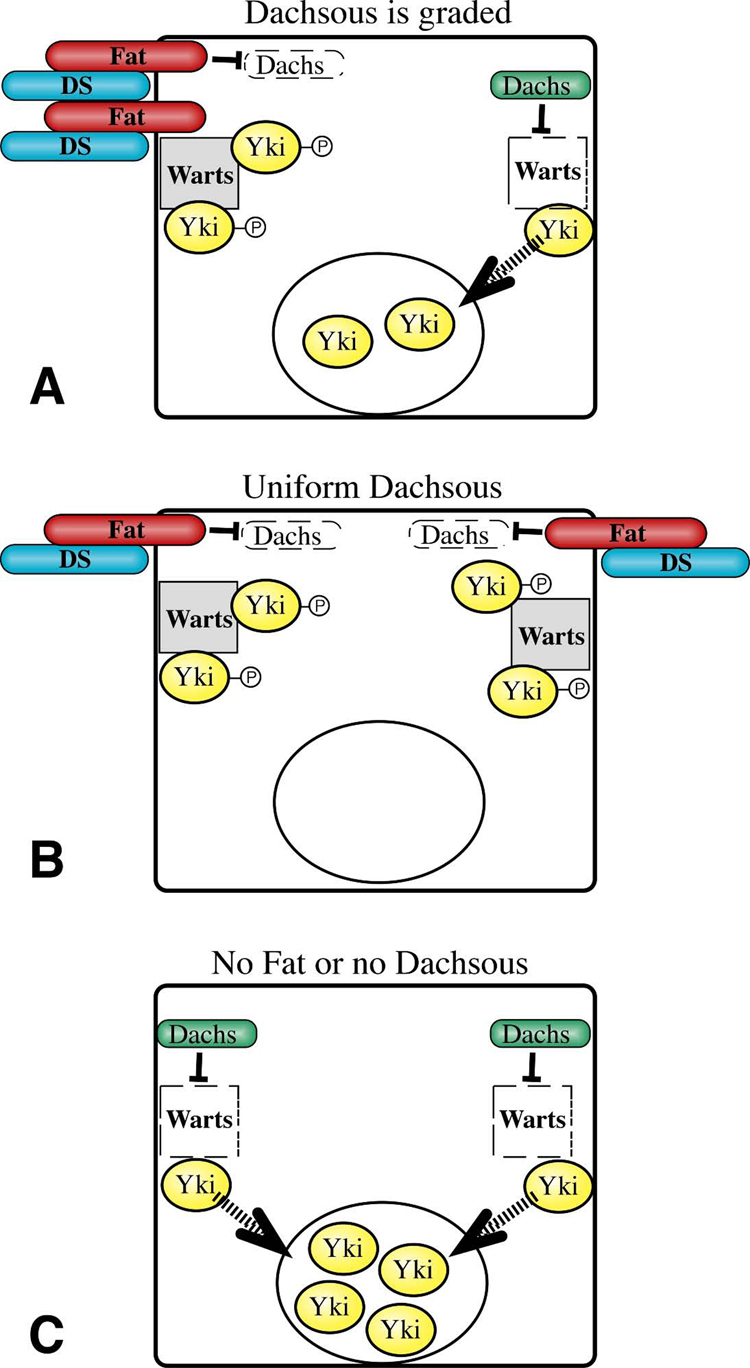
Central to our model (Fig. 7) is the inference that interaction between Ds and Fat activates Fat. This inference is well supported by the observations that mutation or downregulation of ds results in overgrowth and upregulation of Diap1, whereas uniform over-expression of Ds inhibits growth and Diap1 expression (Matakatsu and Blair, 2006)(Fig 6, Fig S3). A second key aspect of our model is that once activated by Ds, Fat locally transmits a signal to a complex at the membrane. An important corollary to this is that if Fat and Ds are not engaged around the entire circumference of a cell, then there could be a region where Fat is locally inactive (Fig. 7A). This is hypothetical, but the Fat-dependent polarization of Dachs implies that there can be regional differences in Fat activity within a cell. Local Fat signaling is then proposed to locally promote Warts stability and activity, and thereby locally antagonize Yki activity. Conversely, a local absence of Fat signaling could result in a local failure to phosphorylate Yki, which could then transit to the nucleus where it would promote the expression of downstream target genes (Dong et al., 2007; Oh and Irvine, 2008). Formally, this model treats Fat signaling like a contact inhibition pathway: if Fat is engaged by Ds around the entire circumference of a cell then Fat is active everywhere and downstream gene expression is off, but if Fat is not active on even one side of a cell, then Yki-dependent gene expression can be turned on and growth promoted.
Figure 7. Model for regulation of Fat signaling by a Dachsous gradient.
A) Schematic of a cell within a Ds gradient. Characterization of Fat staining in discs with clones of cells mutant for or over-expressing Ds indicates that localization of Fat at the membrane can be influenced by the levels of Ds in neighboring cells. Since every cell in a Ds gradient sees more Ds on one side than it does on the other, we suggest that Fat protein could be asymmetrically localized (as indicated). Alternatively, Fat might be uniformly localized but asymmetrically activated. If this asymmetric localization or activity influenced Fat and Ds in neighboring cells, then the asymmetry could be propagated through local cell-cell interactions. The asymmetric localization and/or activity of Fat within a cell results in asymmetric localization of Dachs to the membrane (Fig. 1)(Mao et al., 2006). We suggest that where Dachs can accumulate at the membrane, it locally promotes the degradation and inactivation of Warts (Cho et al., 2006; Feng and Irvine, 2007). We further suggest that Warts locally inhibits the activity of its substrate, Yki (Huang et al., 2005), but where Warts is absent active Yki can be produced, which would then enter the nucleus (arrow) and regulate gene expression to promote growth (Dong et al., 2007; Oh and Irvine, 2008). A transcriptional signal in this context is thus generated from a side of cell opposite to where Fat and Ds are engaged. B) When Ds is uniformly expressed, active Fat would be localized to the membrane around the entire circumference of the cell, where it would antagonize the localization of Dachs to the membrane (Mao et al., 2006). This in turn would allow accumulation of active Warts, and consequently increase inhibition of Yki. C) In the absence of Ds or Fat, Dachs would accumulate at the membrane around the entire circumference of the cell, resulting in uniformly low levels of active Warts, and thereby allowing more Yki to enter the nucleus. By modulating Fat-Ds interactions, a gradient of Four-jointed (not shown) could establish a gradient of Ds-Fat binding activity even under conditions where Ds expression is relatively uniform.
In this model, graded expression of Fat regulators, like Fj and Ds, could modulate Fat signaling by polarizing Fat activity within a cell. In theoretical models of planar cell polarity, even shallow gradients of polarizing activity can be converted to strong polarity responses through positive feedback mechanisms (Klein and Mlodzik, 2005). How this might be achieved in Fat signaling is not yet clear, but the polarized localization of Dachs implies that at some level Fat activity is normally polarized in wild-type animals, even where the Fj and Ds expression gradients appear relatively shallow. Importantly, this polarization hypothesis provides a solution to the puzzle of how Ds could act as a ligand to activate Fat, yet inhibit Fat along the edges of Ds-expressing clones. In our model, Ds over-expression in clones polarizes Fat activity, possibly through its ability to re-localize Fat (Cho and Irvine, 2004; Ma et al., 2003; Mao et al., 2006). This would allow a strong derepression of Yki on the side of the cell opposite to where Ds and Fat are actually bound (Fig. 7A), resulting in the induction of Yki:Scalloped target gene expression and promotion of cell proliferation. Propagation of this polarization, e.g. through the influence of Fat-Ds binding on Fat and Ds localization, might explain the spread of effects beyond immediately neighboring cells. Conversely, uniform expression of Ds would generate cells presenting a ligand that activates Fat and dampens the relative difference in expression levels between neighboring cells. Yki would thus remain sequestered around the entire cell circumference (Fig. 7B), consistent with the reduced growth and Diap1 expression observed. A dampening of gradients could also explain why the induction of Fat/Hippo target gene expression or BrdU labeling associated with clones expressing Ds, Fj, or TkvQ-D is biased towards cells outside of clones.
The hypothesis of Fat polarization and local signal transduction also suggests a solution to another puzzle. In terms of their effects on tissue polarity and Dachs localization, Fj and Ds always behave as though they have opposite effects on Fat (Casal et al., 2002; Mao et al., 2006; Strutt and Strutt, 2002; Yang et al., 2002). Conversely, in terms of their effects on cell proliferation and downstream gene expression, Fj and Ds behave as though they have identical effects on Fat (Fig 5, Fig S1)(Buckles et al., 2001; Cho et al., 2006; Cho and Irvine, 2004). To explain this, we propose that Fj acts oppositely to Ds, by, for example, antagonizing Ds-Fat binding. The influence of Ds and Fj on polarity would be a function of the direction in which they polarize Fat activity, which, based on their effects on Dachs:V5 is opposite (Mao et al., 2006). In contrast, their influence on downstream gene expression and growth would be a function of the degree to which they polarize Fat activity, which could be the same. In other words, their influence on polarity would be a function of the vector of their expression gradients, and their influence on growth would be a function of the slope. However, since Dachs:V5 generally appears to be strongly polarized (Fig. 1), the actual interpretation of Fj and Ds gradients may involve feedback amplification and threshold responses rather than providing a continuous response proportional to the gradient slope.
Multiple mechanisms contribute to wing growth
Our results have provided a molecular understanding of a how a gradient of positional values, established by the morphogen Dpp and reflected at least in part in the graded expression of Fj and Ds, can influence growth. However, it is clear that other mechanisms must also contribute to the regulation of wing growth. The relative contribution of Fat gradients to wing growth can be estimated by considering the size of the wing in dachs mutants, or when Fj and Ds are expressed ubiquitously, as in either case we would expect the de-repression of Yki associated with normal Fat signaling gradients to be abolished. In both cases, the wing is less than half its normal size (Fig. 6)(Mao et al., 2006). Fat signaling could thus be considered a major, but by no means the sole, mechanism regulating wing growth. The determination that not all wing growth depends on the regulation of Fat activity fits with the observation that Dpp signaling promotes growth in at least two distinct ways, one dependent upon its gradient, and the other dependent upon its levels (Rogulja and Irvine, 2005). Other models for wing growth, including a Vestigial-dependent recruitment of new cells into the wing (Zecca and Struhl, 2007), and an inhibition of Dpp-promoted wing growth by mechanical strain (Aegerter-Wilmsen et al., 2007; Hufnagel et al., 2007), have also been proposed. We emphasize that these models are not incompatible with the conclusion that a Fat gradient influences growth. Rather, it is plausible, and even likely, that multiple mechanisms contribute to the appropriate regulation of wing growth. Indeed, we expect that a critical challenge for the future will be to define not only the respective contributions of these or other mechanisms to growth control, but also to understand feed-back and cross-talk processes that influence how these different mechanisms interact with each other.
EXPERIMENTAL PROCEDURES
Clone generation and transgene induction
For generation of Flp-out clones, flies of the following genotypes: y w hs-Flp[122];GS-ds, y w hs-Flp[122];UAS-ds(III), y w hs-Flp[122];UAS-fj (II), y w hs-Flp[122];UAS-fj (III), UAS-ds UAS-fj/TM6b, y w hs-Flp[122];UAS-brk, y w hs-Flp[122];UAS-TkvQ253D, or y w hs-Flp[122] were crossed to UAS-GFP; AyGal4:PR[3]/TM6b, AyGal4 UAS-GFP, tub>CD2>Gal4 UAS-CD8:GFP; tub-Gal80ts/TM6b, y w hs-Flp; IF/CyO; AyGal4:PR[3] UAS-GFP or y w hs-Flp; AyGal4 UAS-GFP/CyO flies.
tkv RNAi clones were generated by crossing y w hs-Flp; AyGal4 UAS-GFP/CyO; UAS-dcr2/TM6b flies to UAS-hairpin RNAi line 862 (tkv) from the VDRC.
For the generation of Flp-out clones in a dachs mutant background using the AyGal4:PR method, flies of the following genotypes:
y w hs-Flp[122];dGC13; UAS-tkvQ253D / L14, y w hs-Flp[122];dGC13; UAS-fj / L14, y w hs-Flp[122];dGC13; UAS-ds / L14, y w hs-Flp[122];dGC13 were crossed to: dGC13; AyGal4:PR[3] UAS-GFP / L14 flies. For the generation of Flp-out clones in a dachs mutant background using the tub-Gal4/Gal80ts method, flies of the following genotypes: y w hs-Flp[122];dGC13; UAS-tkvQ253D / L14, y w hs-Flp[122];dGC13/CyO-GFP; UAS-ds/TM6b, y w hs-Flp[122];dGC13 were crossed to: dGC13 tub>CD2>Gal4 UAS-CD8:GFP; tub-Gal80ts/+ flies.
To assess the influence of TKVQ253D Flp-out clones on fj-lacZ, ds-lacZ or ex-lacZ expression, y w hs-Flp[122];ds-lacZ/Cyo; UAS-tkvQ253D/TM6b, y w hs-Flp[122];ex-lacZ/Cyo; UAS-tkvQ253D/TM6b or y w hs-Flp[122];fj-lacZ/Cyo; UAS-tkvQ253D/TM6b flies were crossed to UAS-GFP; AyGal4:PR[3]/TM6b or tub>CD2>Gal4 UAS-CD8:GFP; tub-Gal80ts/TM6b flies.
To assess the influence of Dpp signaling on Dachs localization, y w hs-Flp[122];UAS-D:V5/Cyo; UAS-tkvQ253D/TM6b, y w hs-Flp[122];UAS-D:V5/Cyo; UAS-brk/TM6b or y w hs-Flp[122];UAS-D:V5/CyO flies were crossed to UAS-GFP; AyGal4:PR[3]/TM6b or tub>CD2>Gal4 UAS-CD8:GFP; tub-Gal80ts/TM6b flies. Wild-type Dachs localization was examined in crosses of y w hs-Flp; wg-LacZ[rO216]; UAS-D:V5 to y w; act>y+>Gal4[25]. Dachs localization in fat8 mutant clones was assessed using MARCM clones.
The influence of activated Notch was assessed using y w hs-Flp[122]; act>y+>Gal4 UAS-GFP to UAS-N:Δ[34a] ap-lacZ[rK568]; UAS-D:V5 [9F] or UAS-N:Δ[34a] fj-lacZ[P1].
Regulated induction of transgene expression in clones was achieved using the AyGal4:PR method (Rogulja and Irvine, 2005), or the tub-Gal4/Gal80ts method (Buttitta et al., 2007). Flies of the appropriate genotype were crossed to tub>CD2>Gal4 UAS-CD8:GFP/ CyO; tub-Gal80ts/ TM6b at 18.5°C. After 5–6 days larvae were heat-shocked at 37C for 8 min. to induce clones. After 4 days at 18.5°C, larvae were transferred to 29.5°C for 28 h, and then dissected. Alternatively flies of the appropriate genotype were crossed to y w hs-FLP; AyGal4:PR UASGFP/ TM6b flies at 25°C. Gal4:PR was activated by transfer of larvae to instant food (Instant Drosophila Medium, Connecticut Valley Biological) containing RU486 (Mifepristone, Sigma). Two grams instant food was mixed with 7.5 mL RU486 in water, resulting in a final medium volume of approximately 8.5 mL. The RU486 solution was 24 µg/mL resulting in final effective concentrations of 20 µg/mL RU486. After 2 days larvae were heat-shocked at 36°C for 7–10min. After 2 days at 25C larvae were transferred to media containing 12ug/ml RU486, for the indicated time intervals, and then dissected.
For temporally controlled ubiquitous transgene expression, y w hs-Flp[122];UAS-ds, y w hs-Flp[122];UAS-fj (III), UAS-ds UAS-fj/TM6b, UAS-GFP, UAS-tkvQ253D/TM6b, or fat8/Cyo; UAS-tkvQ253D/TM6b flies were crossed to: y w hs-Flp[122]; UAS-GFP; actin>Gal4:PR[3]/TM6b or fatG-rv/CyO; actin>Gal4:PR[3]/TM6b flies. For permanent expression of transgenes in broad domains, these same UAS lines were crossed to en-Gal4 UAS-GFP or tub-Gal4 drivers. RNAi was conducted using en-Gal4 UAS-GFP UAS-dcr2 flies, and UAS-hairpin RNAi lines 12555 (dachs), or 36219 (ds) from the VDRC.
Wing areas were measured in pixels by tracing wing outlines in ImageJ, and normalized to the average wing area in wild-type.
Tissue staining and BrdU labeling
For BrdU labeling, larvae were dissected in Ringers solution, and then incubated in M3 complete medium containing 0.1mg/ml BrdU (BD Pharmingen) for 30 min. at room temperature. After three rinses with cold PTW (PBS, 0.1% Tween-20), larvae were fixed for 20 min. in 4% paraformaldehyde plus 0.1% Tween-20. Larvae were then washed four times for 20 min. in PTW, and then treated with 20 units DNAse I (Promega) in 400 µL DNase buffer + PBS for 1.5 h at 37°C. After three washes in PTW, larvae were incubated with anti-BrdU.
Primary antibodies used were mouse anti-BrdU (BD Pharmingen), goat anti-αGal (Biogenesis), rat anti-Ds (gift of M. Simon), Mouse anti-Diap1 (gift of B. Hay), mouse anti-V5 (Invitrogen), Rabbit anti-Yki (Oh and Irvine, 2008) and guinea pig anti-Phospho-Mad (gift of E. Laufer). Secondary antibodies were from Jackson ImmunoResearch.
Blind scoring of BrdU labeling was accomplished by D.R. taking confocal micrographs of all clones in a set of experimental and control stains, assigning them random numbers, and then having K.I score clones for effects on staining. Because nuclei are in different focal planes, the images scored were maximum projections through a series of confocal sections. In experiment 1, GFP-expressing clones in wild type, TkvQ-D-expressing clones in wild type, and TkvQ-D-expressing clones in dachs were assigned random numbers, combined, and then scored blind. In experiment 2, fj-expressing, ds-expressing, GFP-expressing, E-cadherin expressing, shaggy-expressing, and activated Arm-expressing clones were all assigned random numbers, combined, and then scored blind. For purposes of the localization of effects to distal versus proximal regions, the wing pouch was defined as distal and clones were scored separately for distal and proximal effects. The automated image analysis is described in the legend to Supplementary Fig. S3.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Sheyum Syed for writing the image analysis program, K. Basler, B. Hay, B. Edgar, M. Simon, S. Blair, M. Crickmore, L. Johnston, the Developmental Studies Hybridoma Bank and the Bloomington and Vienna stock centers for antibodies and Drosophila stocks, and M. Crickmore and G. Struhl comments on the manuscript. This work was supported by the Howard Hughes Medical Institute (KDI), a Benedict Michaels Fellowship (DR), and NIH grant GM078620 (KDI).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aegerter-Wilmsen T, Aegerter CM, Hafen E, Basler K. Model for the regulation of size in the wing imaginal disc of Drosophila. Mech Dev. 2007;124:318–326. doi: 10.1016/j.mod.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Affolter M, Basler K. The Decapentaplegic morphogen gradient: from pattern formation to growth regulation. Nat Rev Genet. 2007;8:663–674. doi: 10.1038/nrg2166. [DOI] [PubMed] [Google Scholar]
- Bennett FC, Harvey KF. Fat Cadherin Modulates Organ Size in Drosophila via the Salvador/Warts/Hippo Signaling Pathway. Curr Biol. 2006;16:2101–2110. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- Bohn H. Extent and properties of the regeneration field in the larval legs of cockroaches (Leucophaea maderae) III. Origin of the tissues and determination of symmetry properties in the regenerates. J Embryol Exp Morphol. 1974;32:81–98. [PubMed] [Google Scholar]
- Brodsky MH, Steller H. Positional information along the dorsal-ventral axis of the Drosophila eye: graded expression of the four-jointed gene. Dev Biol. 1996;173:428–446. doi: 10.1006/dbio.1996.0038. [DOI] [PubMed] [Google Scholar]
- Buckles GR, Rauskolb C, Villano JL, Katz FN. four-jointed interacts with dachs, abelson and enabled and feeds back onto the Notch pathway to affect growth and segmentation in the Drosophila leg. Development. 2001;128:3533–3542. doi: 10.1242/dev.128.18.3533. [DOI] [PubMed] [Google Scholar]
- Buttitta LA, Katzaroff AJ, Perez CL, de la Cruz A, Edgar BA. A double-assurance mechanism controls cell cycle exit upon terminal differentiation in Drosophila. Dev Cell. 2007;12:631–643. doi: 10.1016/j.devcel.2007.02.020. [DOI] [PubMed] [Google Scholar]
- Casal J, Struhl G, Lawrence P. Developmental compartments and planar polarity in Drosophila. Curr Biol. 2002;12:1189–1198. doi: 10.1016/s0960-9822(02)00974-0. [DOI] [PubMed] [Google Scholar]
- Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nat Genet. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- Cho E, Irvine KD. Action of fat, four-jointed, dachsous and dachs in distal-to-proximal wing signaling. Development. 2004;131:4489–4500. doi: 10.1242/dev.01315. [DOI] [PubMed] [Google Scholar]
- Clark HF, Brentrup D, Schneitz K, Bieber A, Goodman C, Noll M. Dachsous encodes a member of the cadherin superfamily that controls imaginal disc morphogenesis in Drosophila. Genes Dev. 1995;9:1530–1542. doi: 10.1101/gad.9.12.1530. [DOI] [PubMed] [Google Scholar]
- Day SJ, Lawrence PA. Measuring dimensions: the regulation of size and shape. Development. 2000;127:2977–2987. doi: 10.1242/dev.127.14.2977. [DOI] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA. From cell structure to transcription: hippo forges a new path. Cell. 2006;124:267–273. doi: 10.1016/j.cell.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Fanto M, Clayton L, Meredith J, Hardiman K, Charroux B, Kerridge S, McNeill H. The tumor-suppressor and cell adhesion molecule Fat controls planar polarity via physical interactions with Atrophin, a transcriptional co-repressor. Development. 2003;130:763–774. doi: 10.1242/dev.00304. [DOI] [PubMed] [Google Scholar]
- Feng Y, Irvine KD. Fat and expanded act in parallel to regulate growth through warts. Proc Natl Acad Sci U S A. 2007;104:20362–20367. doi: 10.1073/pnas.0706722105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French V, Bryant PJ, Bryant SV. Pattern regulation in epimorphic fields. Science (New York, NY. 1976;193:969–981. doi: 10.1126/science.948762. [DOI] [PubMed] [Google Scholar]
- Garcia-Bellido AC, Garcia-Bellido A. Cell proliferation in the attainment of constant sizes and shapes: the Entelechia model. Int J Dev Biol. 1998;42:353–362. [PubMed] [Google Scholar]
- Giraldez AJ, Cohen SM. Wingless and Notch signaling provide cell survival cues and control cell proliferation during wing development. Development. 2003;130:6533–6543. doi: 10.1242/dev.00904. [DOI] [PubMed] [Google Scholar]
- Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Hufnagel L, Teleman AA, Rouault H, Cohen SM, Shraiman BI. On the mechanism of wing size determination in fly development. Proc Natl Acad Sci U S A. 2007;104:3835–3840. doi: 10.1073/pnas.0607134104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine KD, Vogt TF. Dorsal-ventral signaling in limb development. Curr Opin Cell Biol. 1997;9:867–876. doi: 10.1016/s0955-0674(97)80090-7. [DOI] [PubMed] [Google Scholar]
- Johnston LA, Sanders AL. Wingless promotes cell survival but constrains growth during Drosophila wing development. Nat Cell Biol. 2003;5:827–833. doi: 10.1038/ncb1041. [DOI] [PubMed] [Google Scholar]
- Klein T, Arias AM. Different spatial and temporal interactions between Notch, wingless, and vestigial specify proximal and distal pattern elements of the wing in Drosophila. Dev Biol. 1998;194:196–212. doi: 10.1006/dbio.1997.8829. [DOI] [PubMed] [Google Scholar]
- Klein TJ, Mlodzik M. Planar cell polarization: an emerging model points in the right direction. Annu Rev Cell Dev Biol. 2005;21:155–176. doi: 10.1146/annurev.cellbio.21.012704.132806. [DOI] [PubMed] [Google Scholar]
- Lawrence PA. Polarity and patterns in the postembryonic development of insects. Adv Insect Physiol. 1970;7:197–266. [Google Scholar]
- Lecuit T, Brook WJ, Ng M, Calleja M, Sun H, Cohen SM. Two distinct mechanisms for long-range patterning by Decapentaplegic in the Drosophila wing. Nature. 1996;381:387–393. doi: 10.1038/381387a0. [DOI] [PubMed] [Google Scholar]
- Ma D, Yang CH, McNeill H, Simon MA, Axelrod JD. Fidelity in planar cell polarity signalling. Nature. 2003;421:543–547. doi: 10.1038/nature01366. [DOI] [PubMed] [Google Scholar]
- Mao Y, Rauskolb C, Cho E, Hu WL, Hayter H, Minihan G, Katz FN, Irvine KD. Dachs: an unconventional myosin that functions downstream of Fat to regulate growth, affinity and gene expression in Drosophila. Development. 2006;133:2539–2551. doi: 10.1242/dev.02427. [DOI] [PubMed] [Google Scholar]
- Martin-Castellanos C, Edgar BA. A characterization of the effects of Dpp signaling on cell growth and proliferation in the Drosophila wing. Development. 2002;129:1003–1013. doi: 10.1242/dev.129.4.1003. [DOI] [PubMed] [Google Scholar]
- Matakatsu H, Blair SS. Interactions between Fat and Dachsous and the regulation of planar cell polarity in the Drosophila wing. Development. 2004;131:3785–3794. doi: 10.1242/dev.01254. [DOI] [PubMed] [Google Scholar]
- Matakatsu H, Blair SS. Separating the adhesive and signaling functions of the Fat and Dachsous protocadherins. Development. 2006;133:2315–2324. doi: 10.1242/dev.02401. [DOI] [PubMed] [Google Scholar]
- Nellen D, Burke R, Struhl G, Basler K. Direct and long-range action of a DPP morphogen gradient. Cell. 1996;85:357–368. doi: 10.1016/s0092-8674(00)81114-9. [DOI] [PubMed] [Google Scholar]
- Oh H, Irvine KD. In vivo regulation of Yorkie phosphorylation and localization. Development. 2008;135:1081–1088. doi: 10.1242/dev.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- Rogulja D, Irvine KD. Regulation of cell proliferation by a morphogen gradient. Cell. 2005;123:449–461. doi: 10.1016/j.cell.2005.08.030. [DOI] [PubMed] [Google Scholar]
- Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The Tumor-Suppressor Gene fat Controls Tissue Growth Upstream of Expanded in the Hippo Signaling Pathway. Curr Biol. 2006;16:2081–2089. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Simon MA. Planar cell polarity in the Drosophila eye is directed by graded Four-jointed and Dachsous expression. Development. 2004;131:6175–6184. doi: 10.1242/dev.01550. [DOI] [PubMed] [Google Scholar]
- Strutt H, Mundy J, Hofstra K, Strutt D. Cleavage and secretion is not required for Four-jointed function in Drosophila patterning. Development. 2004;131:881–890. doi: 10.1242/dev.00996. [DOI] [PubMed] [Google Scholar]
- Strutt H, Strutt D. Nonautonomous planar polarity patterning in Drosophila: dishevelled-independent functions of frizzled. Dev Cell. 2002;3:851–863. doi: 10.1016/s1534-5807(02)00363-5. [DOI] [PubMed] [Google Scholar]
- Villano JL, Katz FN. four-jointed is required for intermediate growth in the proximal-distal axis in Drosophila. Development. 1995;121:2767–2777. doi: 10.1242/dev.121.9.2767. [DOI] [PubMed] [Google Scholar]
- Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, Zhang X, Halder G. The Fat Cadherin Acts through the Hippo Tumor-Suppressor Pathway to Regulate Tissue Size. Curr Biol. 2006;16:2090–2100. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell. 2008;14:388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Yang C, Axelrod JD, Simon MA. Regulation of Frizzled by Fat-like Cadherins during Planar Polarity Signaling in the Drosophila Compound Eye. Cell. 2002;108:675–688. doi: 10.1016/s0092-8674(02)00658-x. [DOI] [PubMed] [Google Scholar]
- Zecca M, Struhl G. Recruitment of cells into the Drosophila wing primordium by a feed-forward circuit of vestigial autoregulation. Development. 2007;134:3001–3010. doi: 10.1242/dev.006411. [DOI] [PubMed] [Google Scholar]
- Zeidler MP, Perrimon NQ, Strutt DI. The four-jointed gene is required in the Drosophila eye for ommatidial polarity specification. Curr Biol. 1999;9:1363–1372. doi: 10.1016/s0960-9822(00)80081-0. [DOI] [PubMed] [Google Scholar]
- Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell. 2008;14:377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.