Abstract
Radiosensitive T–B– severe combined immunodeficiency (RS-SCID) is caused by defects in the nonhomologous end-joining (NHEJ) DNA repair pathway, which results in failure of functional V(D)J recombination. Here we have identified the first human RS-SCID patient to our knowledge with a DNA-PKcs missense mutation (L3062R). The causative mutation did not affect the kinase activity or DNA end-binding capacity of DNA-PKcs itself; rather, the presence of long P-nucleotide stretches in the immunoglobulin coding joints indicated that it caused insufficient Artemis activation, something that is dependent on Artemis interaction with autophosphorylated DNA-PKcs. Moreover, overall end-joining activity was hampered, suggesting that Artemis-independent DNA-PKcs functions were also inhibited. This study demonstrates that the presence of DNA-PKcs kinase activity is not sufficient to rule out a defect in this gene during diagnosis and treatment of RS-SCID patients. Further, the data suggest that residual DNA-PKcs activity is indispensable in humans.
Introduction
SCID is an inherited primary immunodeficiency. SCID patients present in the first year of life with severe opportunistic infections, chronic diarrhea, and failure to thrive. The total group of SCID patients can be divided in 2 main categories: those with T–B+ SCID, who have a T cell signaling defect (70%), and those with T–B– SCID, who have a defect in V(D)J recombination (30%). V(D)J recombination assembles variable (V), diversity (D), and joining (J) gene segments of the Ig and TCR genes during B and T cell differentiation in order to generate a broad repertoire of antigen-specific receptors. V(D)J recombination starts with introduction of DNA breaks at the border of the gene segments and the flanking recombination signal sequences (RSSs) by the RAG1 and RAG2 proteins (1). The resulting blunt signal ends are ligated directly, forming a signal joint. The hairpin sealed coding ends require further processing before coding joint formation can occur. Recognition and repair of the DNA ends occur via the general nonhomologous end-joining (NHEJ) pathway of DNA double-strand break (DSB) repair (2, 3).
DSBs induce ATM kinase activity, which phosphorylates histone H2AX (4), followed by binding of 53BP1, MDC1, and a complex of MRE11, RAD50, and NBS1 (MRN complex) (5, 6). These proteins form a microenvironment that holds together the DNA ends over a relatively large distance but still allows some degree of freedom for movement of the DNA ends and access of NHEJ proteins (7). DNA ends are first recognized by the NHEJ factor DNA-dependent protein kinase (DNA-PK), which is composed of the DNA-PK catalytic subunit (DNA-PKcs) and the Ku70/Ku80 heterodimer (8). After initial loading of the Ku70/Ku80 heterodimer onto DNA ends, DNA-PKcs is recruited to form a DNA end synapsis, ensuring protection from exonuclease activities and juxtaposition of DNA ends (9). The presence of Ku70/Ku80 and DNA-PKcs at DNA ends is not rigid but constitutes a dynamic equilibrium of DNA-bound and DNA-free protein (10, 11). Trans autophosphorylation of the ABCDE cluster of DNA-PKcs (7 phosphorylation sites between residues 2,609 and 2,647) causes a conformational change that facilitates Artemis nuclease activity (12–14).
During V(D)J recombination, Artemis nuclease activity is required for opening of the hairpin sealed coding ends (15). During processing of coding ends, nucleotides can be lost due to exonuclease activity, and nontemplated nucleotides (N-nucleotides) can be added by terminal deoxynucleotidyl transferase (TdT) (16, 17). This contributes tremendously to the diversity of the antigen receptor repertoire. Trans autophosphorylation of the PQR cluster (6 phosphorylation sites between residues 2,023 and 2,056) functions to limit further end processing and to specifically promote end-joining. Activation (T3950) loop autophosphorylation is also required for efficient end-joining (18). Finally, the DNA ends are ligated by the XRCC4–ligase IV complex, which is promoted by XLF (Cernunnos) (19, 20) The requirement of XRCC4-like factor (XLF) can differ between various species and/or various cell types (21).
In approximately 70% of T–B– SCID patients, mutations are found in the RAG1 and RAG2 genes (22, 23). The majority of the remaining patients show hypersensitivity to ionizing radiation, due to mutations in Artemis or LIG4 (24–29); mutations in the XLF/Cernunnos gene have been found in radiosensitive (RS) patients with growth retardation, microcephaly, and immunodeficiency due to profound T and B cell lymphocytopenia (19, 20). We here present what we believe to be a new type of RS-SCID with a defect in DNA-PKcs.
Results
Flow cytometric analysis of peripheral blood and bone marrow before and after stem cell transplantation.
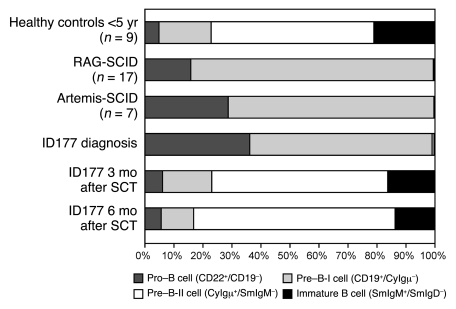
A girl (patient ID177) from consanguineous parents of Turkish origin (first-degree relatives) was clinically diagnosed with SCID when she was 5 months old. B and T cells were virtually absent from peripheral blood, whereas NK cells were within the normal range (Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI37141DS1). The precursor B cell compartment in bone marrow showed a complete block in B cell differentiation before the pre–B-II cell stage that was comparable to the phenotype in Artemis-deficient SCID (25) and RAG-deficient SCID (30), which suggests that the patient has a defect in V(D)J recombination (Figure 1). Patient ID177 underwent hematopoietic stem cell transplantation (SCT) with cells from her HLA-identical cousin, which resulted in full reconstitution of the precursor B cell compartment and production of all lymphocyte subsets after 3 and 6 months.
Figure 1. Flow cytometric analysis of the precursor B cell compartment.
At diagnosis, patient ID177 had a complete block in precursor B cell differentiation before the pre–B-II cell stage, comparable to RAG- and Artemis-deficient SCID patients (25, 30). As early as 3 months after SCT, the precursor B cell compartment showed a normal composition.
DNA repair defect in patient fibroblasts.
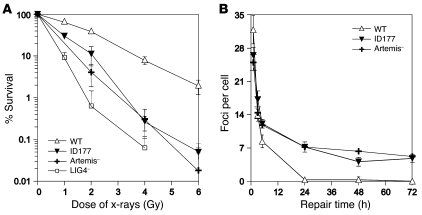
Mutations in RAG1 and RAG2 were excluded by sequencing analysis. Subsequently, clonogenic survival assays showed that ID177 fibroblasts were x-ray sensitive (Figure 2A), indicating that patient ID177 has RS-SCID due to a defect in NHEJ. However, known candidate genes for RS-SCID (Artemis, LIG4, XLF-Cernunnos) were not mutated. Therefore, we analyzed the DSB repair defect in more detail by counting foci of the DSB marker γ-H2AX at various time points after irradiation. The kinetics of induction and disappearance of γ-H2AX foci at early time points (<6 hours) did not differ significantly in patient, control (VH10), and Artemis-deficient cells (Figure 2B). However, γ-H2AX foci disappeared with delayed kinetics in Artemis-deficient and patient fibroblasts, and 15% of residual foci were still present after 72 hours, suggesting that a comparable level of unrepairable DSBs existed in patient ID177 and in Artemis-deficient cells.
Figure 2. ID177 harbors a DSB repair defect.
(A) Clonogenic survival assay using wild-type (FN1) fibroblasts, patient fibroblasts (ID177), and fibroblasts from an Artemis-deficient patient (Artemis-1; ref. 25) and an LIG4-deficient patient (SC2; ref. 29). Fibroblasts were irradiated with increasing x-ray doses. ID177 fibroblasts were RS. Error bars represent the SD from 3 independent experiments. (B) Numbers of γ-H2AX foci per nucleus were determined at indicated time points after irradiation (average number of foci per nucleus in 40 cells) in wild-type (VH10), ID177, and Artemis-deficient fibroblasts (Artemis-6; ref. 31). Error bars represent the SD from 2–4 independent experiments.
Composition of Dh-Jh coding joints from bone marrow mononuclear cells.
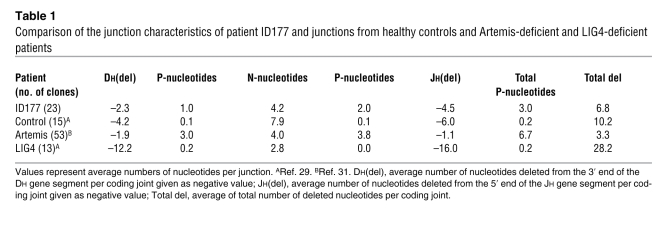
Additional information on the repair defect was obtained by analyzing incomplete IGH gene rearrangements (i.e., Dh-Jh) derived from bone marrow mononuclear cells. These junctions showed an average total number of 3.0 palindromic nucleotides (P-nucleotides), which is significantly higher than the observed 0.2 P-nucleotides in healthy controls (P = 0.025), although not as high as in Artemis-deficient patients (6.7 nucleotides) (31) (Table 1 and Supplemental Table 2). Artemis-deficient patients have a defect in the DNA hairpin opening of the coding end (9), which results in exceptionally high numbers of P-nucleotides, with a high frequency of microhomology in the P-nucleotide tracts (31, 32). The shorter P-tracts and the absence of P-tract microhomologies in ID177 junctions suggest that the hairpin opening defect is somewhat different from Artemis deficiencies.
Table 1 .
Comparison of the junction characteristics of patient ID177 and junctions from healthy controls and Artemis-deficient and LIG4-deficient patients
Analysis of NHEJ components Ku70, Ku80, XRCC4, and DNA-PKcs.
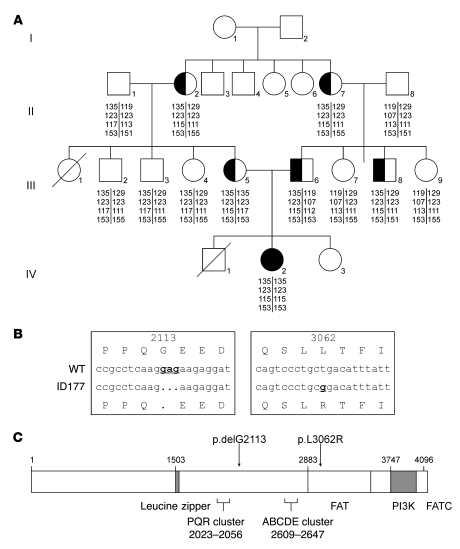
Since Artemis activity depends on DNA-PKcs autophosphorylation, we analyzed the DNA-PKcs locus. Before sequencing this large gene, short tandem repeat (STR) analysis was performed in the patient and 12 family members using 4 polymorphic markers surrounding the DNA-PKcs gene (Figure 3A). Patient ID177 was homozygous for all 4 markers, and the allele defined by those 4 markers was present heterozygously in both parents, in the 2 related grandmothers (sisters), and in 1 uncle. No family members were homozygous for this genotype. Therefore, we sequenced the 12.3 kb DNA-PKcs cDNA and found 2 homozygous mutations (Figure 3B): a 3-nucleotide deletion (c.6338_6340delGAG) resulting in the deletion of a glycine (p.delG2113) and a missense mutation, resulting in the replacement of 1 amino acid (c.9185T→G; p.L3062R) in the DNA-PKcs FAT (FRAP, ATM, and TRRAP) domain (Figure 3C). The mutations were confirmed to be present heterozygously in the parents, the 2 grandmothers, and the uncle and were not present in the other family members. To exclude that these mutations are recurrent polymorphisms, 100 Turkish controls were analyzed, and the mutations were not detected. Mutations in the other NHEJ components (KU70, KU80, and XRCC4) were excluded by sequencing.
Figure 3. DNA-PKcs mutation analysis.
(A) Family tree of patient ID177 and 12 family members. STR analysis was performed using the polymorphic markers D8S1460, D8S359, D8S531, and AF075268 surrounding the DNA-PKcs gene. Patient ID177 (IV-2) was homozygous for all 4 markers. This allele, defined by the 4 markers, was present heterozygously in both parents (III-5 and -6), the 2 grandmothers who are sisters (II-2 and -7), and 1 uncle (III-8). None of the family members were homozygous for this genotype. (B) Detection of 2 homozygous mutations in the DNA-PKcs gene in patient ID177: a deletion of 3 nucleotides (c.6338_6340delGAG) resulting in deletion of glycine 2113 and a missense mutation (c.9185T→G) resulting in replacement of leucine by arginine at position 3,062. (C) Schematic representation of the DNA-PKcs protein and the 2 identified mutations, delG2113 and L3062R. FATC, FAT C-terminal domain.
Normal DNA-PKcs expression and activity.
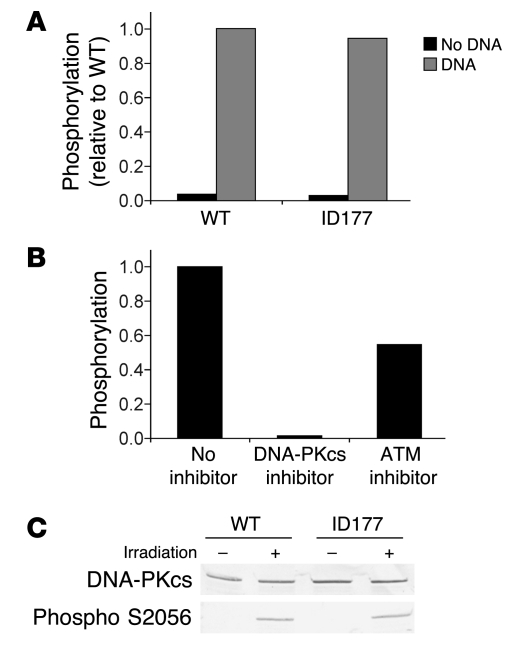
First, we studied the effect of the DNA-PKcs mutations on kinase activity and protein expression. In vitro DNA-PK kinase activity appeared to be similar in extracts from patient and control cell cultures (Figure 4A). The observed phosphorylation could specifically be inhibited by the DNA-PK inhibitor NU7441 (Figure 4B) at a final concentration of 0.5 μM (33). NU7441 is specific for DNA-PK kinase inhibition (IC50, 0.014 μM) and inhibits ATM only at high concentrations (IC50, >100 μM) (33). ATM inhibitor KU-55933 inhibited the phosphorylation activity by 50% (Figure 4B), which can be attributed the fact that this inhibitor is less specific (34). KU-55933 has an IC50 of 0.013 μM for ATM but an IC50 of 2.5 μM for inhibition of DNA-PK. These data show that ID177 nuclear extracts possesses wild-type levels of DNA-PK activity.
Figure 4. Measurement of DNA-PKcs activity.
(A) DNA-PKcs kinase activity of cellular extracts of wild-type (MRC5) and ID177 fibroblasts was measured by quantification of phosphorylation of a p53 peptide in the presence and absence of DNA. (B) DNA-PKcs kinase activity in ID177 cellular extracts was measured by quantification of phosphorylation of a p53 peptide in the presence of DNA without inhibitor or with DNA-PKcs–specific inhibitor NU7441 (33) or ATM-specific inhibitor KU-55933 (34) in a final concentration of 0.5 μM. Phosphorylation is expressed relative to ID177 without inhibitor. (C) Western blot analysis of DNA-PKcs and phosphorylated DNA-PKcs in cellular extracts from untreated (–) and irradiated (2 Gy) fibroblasts (+) of ID177 and wild-type (C5RO) fibroblasts with the DNA-PKcs antibodies 2,208 and 2,129 (1:1) and the phosphospecific DNA-PKcs antibody S2056.
Subsequently, we studied autophosphorylation using a phosphospecific antibody for one of the autophosphorylation sites (S2056) (Figure 4C). The S2056 site is specifically known to be an exclusive DNA-PK target site in vivo (35). DNA-PKcs expression was detected in all fractions, while phosphorylated DNA-PKcs was only found in irradiated control and ID177 fibroblasts. These experiments showed that mutated DNA-PKcs was expressed and retained kinase and autophosphorylation capacity.
Confirmation of the disease-causing DNA-PKcs mutation.
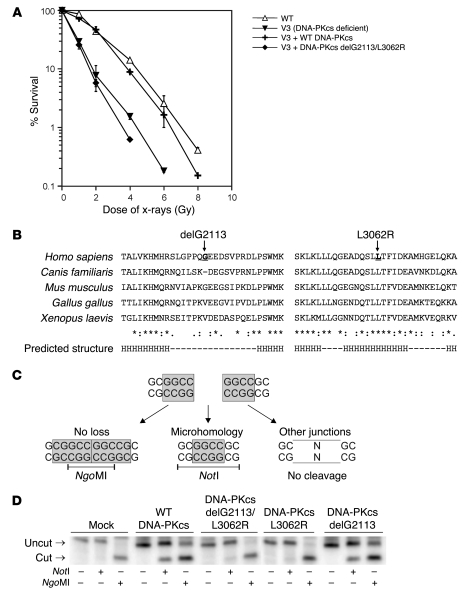
As we did not observe a defect in DNA-PKcs activity, we sought a different way to prove that the DNA-PKcs mutations have a disease-causing effect. Therefore, DNA-PKcs–deficient V3 cells expressing the mutated DNA-PKcs protein were studied in a clonogenic survival assay. Indeed, DNA-PKcs carrying both mutations did not complement the radiosensitivity of V3 cells, whereas wild-type DNA-PKcs did, proving that one or both DNA-PKcs mutations in ID177 are disease causing (Figure 5A).
Figure 5. Confirmation of disease-causing effect of DNA-PKcs mutation.
(A) Clonogenic survival assay of DNA-PKcs–deficient V3 cells (57) complemented with wild-type DNA-PKcs or mutated DNA-PKcs (delG2113/L3062R) compared with wild-type (AA8) cells. In contrast to wild-type DNA-PKcs, mutated DNA-PKcs was not able to complement the radiosensitivity of V3 cells. Error bars represent the SD from 3 independent experiments. (B) Sequence alignment of DNA-PKcs protein sequences of different species. G2113 is located in a region without a predicted secondary structure (indicated as –); L3062 is located in a predicted helix (H). PredictProtein analysis software was used. (C) Coding end structure of recombination substrate (pDVG93) used in the V(D)J recombination assay (31). Depending on the manner of hairpin opening and coding joint formation, 3 different types of coding joints can be generated and discriminated by restriction enzyme digestion. Coding joints with long stretches of homologous P-nucleotides without nucleotide loss results in an NgoMI restriction site. A 4 bp microhomology of the coding ends creates a NotI restriction site. Junctions that have been processed differently will not contain either of the 2 restriction sites. (D) PCR and restriction enzyme digestion of coding joints recovered from DNA-PKcs–deficient V3 cells and V3 cells expressing wild-type DNA-PKcs or mutated DNA-PKcs after cotransfection of pDVG93 and RAG1 and RAG2 expression constructs. Wild-type DNA-PKcs and DNA-PKcs delG2113 were able to restore the shifted junction pattern as observed in DNA-PKcs–deficient V3 cells.
Alignment of the DNA-PKcs protein sequences of several species showed that leucine 3,062 is a highly conserved amino acid in a predicted α-helix (H), whereas glycine 2,113 is located in a region with little conservation (Figure 5B). Moreover, Canis familiaris lacks this amino acid as well. Therefore, we hypothesized that L3062R is most likely the disease-causing mutation.
We recently developed a V(D)J recombination assay, which allows detection of hairpin opening proficiency by simple analysis of junctions using restriction enzyme digestion (Figure 5C) (31). This assay is based on cotransfection of a V(D)J recombination substrate containing 2 RSSs in inverse orientation and RAG1 and RAG2. Depending on the manner of hairpin opening and coding joint formation, 3 different types of coding joints can be generated and discriminated from each other by restriction enzyme digestion. Coding joints without nucleotide loss are NgoMI sensitive, whereas a 4-base-pair microhomology of the coding ends creates a NotI restriction site. Junctions that have been processed differently will not contain either of the 2 restriction sites. In a wild-type situation, the 3 different types are used at equal frequencies. Similar to Artemis-deficient cells, DNA-PKcs–deficient V3 cells showed a complete shift toward NgoMI-sensitive junctions (Figure 5D). Expression of wild-type DNA-PKcs in V3 cells restored normal distribution of junctions. The normal junction pattern could not be restored in V3 cells by the DNA-PKcs expression construct containing both mutations or containing only the L3062R mutation. However, the DNA-PKcs delG2113 deletion was able to correct the shifted junction pattern, indicating that the DNA-PKcs L3062R missense mutation is the disease-causing mutation.
Accumulation of mutated DNA-PKcs at DSB sites.
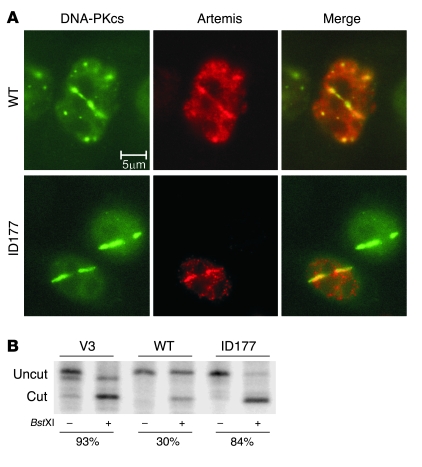
To further pinpoint the effect of the DNA-PKcs mutation, we studied the ability of mutated DNA-PKcs to recruit Artemis to laser-induced DSB sites in living cells (11). Lines of DNA damage were introduced in V3 cells expressing YFP-tagged wild-type DNA-PKcs or mutated DNA-PKcs. In this assay, a Myc-tagged Artemis expression construct was cotransfected. Mutated DNA-PKcs protein was still able to accumulate at DSB sites and to recruit Artemis to these sites (Figure 6A). Colocalization of mutated DNA-PKcs and Artemis suggests that the point mutation did not disrupt interaction with Artemis.
Figure 6. Residual function of DNA-PKcs defect.
(A) Immunostaining of microirradiated wild-type and delG2113/L3062R YFP-DNA-PKcs–expressing V3 cells after cotransfection with a wild-type myc-Artemis expression construct (in red). Cells were microirradiated using a multiphoton laser system. One of the two V3 cells expressing delG2113/L3062R YFP-DNA-PKcs was not transfected with Artemis. This figure shows representative results of 2 independent experiments, each with multiple laser-damaged cells. (B) DNA end-joining assay in DNA-PKcs–deficient V3 cells and V3 cells expressing wild-type or ID177 mutant DNA-PKcs. Linearized pDVG94 can be rejoined via direct or microhomology-directed end-joining. Joining via microhomology results in the generation of a BstXI restriction site (36). Junctions were PCR amplified, and PCR products were digested with BstXI, as indicated.
Impaired end-joining activity of linear substrates.
Finally, the relative level of microhomology use was tested in a plasmid recircularization assay, which is based on transfection of linear DNA. This is a sensitive method to detect subtle NHEJ defects (36). The mutated DNA-PKcs expression construct was not able to correct the increased microhomology use in DNA-PKcs-deficient V3 cells, showing that the overall NHEJ activity was affected by this mutation (Figure 6B).
Discussion
We identified the first human RS-SCID patient to our knowledge with a mutation in the DNA-PKcs gene. Clinically the DNA-PKcs–deficient patient presented with classical SCID with symptoms similar to those of patients with RAG or Artemis mutations, i.e., with a severe immunodeficiency due to lack of B and T cells, but without signs of microcephaly or mental retardation, as found in XLF-deficient patients (20) or in patients with LIG4 syndrome (37). For several years, mutations in this gene have been expected to exist in (SCID) patients, because spontaneous mutations in the DNA-PKcs gene had already been reported in 3 different species — horses of Arabian breed (38), mice (39, 40), and Jack Russell terriers (41) — which indicated that mutations in this gene are not lethal. This notion was further supported by a recent study in which a human adenocarcinoma DNA-PKcs–null cell line was generated (42). This study clearly demonstrated that under cell culture conditions, DNA-PKcs is not essential for human cells.
The disease-causing mutation in the SCID patient described here concerned a hypomorphic L3062R missense mutation in the DNA-PKcs FAT domain. This mutation did not influence in vitro kinase activity, nor did it hamper DNA-PKcs autophosphorylation. Moreover, mutated DNA-PKcs protein was still able to accumulate at DSB sites and to recruit Artemis to these sites in vivo, suggesting that Artemis interaction was not disrupted. However, subtle changes in the interaction might influence correct positioning of the nuclease domain relative to the DNA end. It is interesting to note that a cryo-electron microscopy–based 3-dimensional model of the DNA-PK structure places the FAT domain (which contains the L3062R mutation) on a protrusion that faces the DNA end (43). Therefore, this domain could serve as the Artemis nuclease interaction site. The consequence of the L3062R mutation was insufficient Artemis activation leading to disturbed coding joint formation and concomitantly to strongly reduced numbers of B and T cells in peripheral blood. The coding joints contained long stretches of P-nucleotides, indicating that Artemis nuclease activity was strongly inhibited.
However, the effect of the L3062R mutation was not limited to the Artemis activation deficit. Although we did not observe a significant difference in DSB repair kinetics in DNA-PKcs–deficient (ID177) and Artemis-deficient cells, joining of transfected linear DNA in ID177 was shifted toward microhomology use. Such a shift in joining products has been observed before in V3 cells that completely lack DNA-PKcs activity (36) but not in Artemis-deficient cells (25). As DNA-PKcs–null mutations have not been found in patients, we assume that residual DNA-PK activity is indispensable in humans. It is possible that DNA-PKcs–dependent NHEJ is partially functional in ID177. However, the essential function in humans could also be unrelated to NHEJ, possibly a DNA-PK–dependent phosphorylation event.
The L3062R mutation differs substantially from the spontaneous mutations described in SCID horses, mice, and dogs, which all concerned DNA-PKcs mutations that resulted in truncated proteins. In SCID foals, a 5 bp deletion resulted in a frameshift and premature stop codon that prevented the translation of the 967 C-terminal amino acids, which resulted in a nonstable protein (44, 45). C.B-17 BALB/c SCID mice have a nonsense mutation in a highly conserved region with a stop codon at a position corresponding to amino acid 4,045, thereby deleting the 83 C-terminal amino acids (46). Jack Russell terriers also have a nonsense mutation that deletes the 517 C-terminal amino acids (41, 47). The human DNA-PKcs–null cell line lacked the kinase catalytic domain (encoded by exons 81, 82, and 83) and did not express detectable levels of DNA-PKcs protein (42). In the 3 mentioned species as well as in the human cell line, the DNA-PKcs defects resulted in absence of kinase activity (41, 42, 44, 48). This is in strong contrast to the human L3062R mutation, which retained kinase activity. It is tempting to speculate that DNA-PKcs kinase activity is essential for human development. The human carcinoma cell line was viable, although these cells had a severe proliferation and genomic stability deficit, suggesting that DNA-PK activity is more important in humans than in most other mammals. It is possible, that DNA-PK has an essential function, e.g., mediating some kinase signaling event that is carried out by ATM in other mammals. However, we cannot exclude that the presence of DNA-PKcs activity is specific for the (hypomorphic) L3062R mutation and that other DNA-PKcs mutations lacking this kinase activity can still be found in SCID patients. This study shows that the presence of DNA-PKcs protein and DNA-PKcs activity is not sufficient to rule out a defect in this gene. This might explain the delay in identification of the first DNA-PKcs–deficient patient and probably of DNA-PKcs mutations in other RS-SCID patients.
The question remains whether mutations in the other NHEJ genes (KU70, KU80, and XRCC4) can also be expected in patients. Ku70- and Ku80-knockout mice are viable and have an RS-SCID phenotype (49–52), but no spontaneous animal models have been reported. In contrast, human cell lines with inactivated KU70 or KU80 are not viable, which suggests that KU70 and KU80 are essential for human cell viability (53, 54). Xrcc4-deficient mice are embryonic lethal due to apoptosis of postmitotic neurons (55), and if this can be extrapolated to humans, functional null mutations in XRCC4 are also unlikely. However, hypomorphic mutations in KU70, KU80, or XRCC4 cannot be excluded, and therefore testing for defects in these genes remains valid in RS-SCID or in patients with immunodeficiency, growth retardation, and/or developmental defects. As shown by this study, the analysis of coding joints from the bone marrow precursor B cells is helpful for identification of the type of V(D)J recombination or NHEJ defect and can therefore guide the molecular testing of candidate genes.
In conclusion, DNA-PKcs is a new candidate gene for human RS-SCID. This study illustrates that the presence of DNA-PKcs kinase activity is not sufficient to rule out a defect in this gene and that coding joint analysis is a powerful tool in the diagnostic process. In addition, our results suggest that residual DNA-PKcs activity is indispensable in humans.
Methods
Case report.
A Turkish girl (patient ID177) presented at the age of 5 months with recurrent oral candidiasis and lower respiratory tract infections since the third month of life, with progressive respiratory distress leading to hypoxia. At the age of 4 month, she had a large oral aphthous lesion. She is the third child of consanguineous parents (first-degree relatives). A male sibling died due to a congenital heart defect at the age of 3 years. She has a healthy 3-year-old sister. She had no adverse reaction to vaccination with bacille Calmette-Guérin. She had minimal tonsillary tissue. She underwent SCT with cells from her HLA-identical cousin (a healthy male) without a pretransplant conditioning regimen.
Cell lines and tissue culture.
Primary fibroblasts were cultured from a skin biopsy sample from patient ID177 and were used in all assays. Informed consent was received from patients under a protocol approved by the medical ethics committees of Hacettepe University Children’s Hospital and Erasmus Medical Center. Wild-type human skin fibroblasts FN1, VH10, C5RO, or MCR5 were used as positive controls. In addition, fibroblasts derived from Artemis-deficient patients (Artemis-1 [ref. 25], Artemis-6 and Artemis-7 [ref. 31]) and a LIG4-deficient patient (SC2 [ref. 29]) were used in several assays. DNA samples of 12 family members and 100 healthy Turkish individuals were used for STR marker analysis and sequence analysis. Other cell cultures used in this study were the hamster cell lines AA8 (56) (wild-type), DNA-PKcs–deficient V3 cells (40, 57), and V3 cells that were stably transfected with the YFP-DNA-PKcs expression construct (11). All cell lines were cultured in a 1:1 mixture of Ham’s F10 medium and DMEM (BioWhittaker), supplemented with 10% FCS, penicillin (100 U/ml), and streptomycin (100 μg/ml).
Flow cytometric analysis of peripheral blood and bone marrow.
At diagnosis and 3 and 6 months after SCT, flow cytometric analysis of peripheral blood was performed, aiming at the analysis of the peripheral lymphocyte subset populations and at assessing the composition of the precursor B cell compartment as previously described (29).
Clonogenic survival assay.
Clonogenic survival assays with primary skin fibroblasts or hamster cells were performed as described previously (25, 29). In short, primary skin fibroblasts in exponential growth or hamster cells were trypsinized, and 500–2,000 cells (5,000–80,000 cells for the highest doses) were seeded into 10 cm plastic dishes (2 dishes per dose, 3 for nonirradiated control) and irradiated at room temperature at a dose of approximately 2.7 Gy/min (200 kV, 4.0 mA). After 12–14 days, the cells were rinsed with 0.9% NaCl and stained with 0.25% methylene blue for survival assessment. Three independent survival experiments were performed. In complementation experiments, only cell lines with homogeneous human YFP-DNA-PKcs expression were analyzed.
Analysis of γ-H2AX foci after ionizing radiation.
X-ray-induced H2AX foci were analyzed in confluent, serum-starved primary human fibroblasts after irradiation at a dose of 1 Gy by staining with monoclonal anti–phospho-H2AX (Ser139) (Upstate) (10 ng/ml) and subsequently with Alexa Fluor 488–conjugated goat anti-mouse IgG (Invitrogen). Cell nuclei were counterstained with DAPI (Sigma-Aldrich). Foci were examined by fluorescence microscopy, and 40 cells per time point were analyzed (29).
Sequence analysis of genes involved in V(D)J recombination and NHEJ.
Sequence analysis of genes involved in V(D)J recombination and NHEJ was performed by PCR analysis (for RAG1 [NCBI M29474], RAG2 [NCBI M94633], Artemis/DCLRE1C [NCBI M94633], XLF/NHEJ1 [NCBI AJ972687], and LIG4 [NCBI X83441]) or RT-PCR analysis (for KU70/XRCC6 [NCBI NM_001469], KU80/XRCC5 [NCBI NM_021141], XRCC4 [NCBI NM_003401], and DNA-PKcs/PRKDC [NCBI NM_006904]) of the coding regions with the TaqGold amplification system (Applied Biosystems) followed by direct sequencing. For primer sequences, see Supplemental Table 2. PredictProtein software (http://www.predictprotein.org/) was used for sequence alignment of DNA-PKcs protein sequences of different species.
Analysis of Dh-Jh junctions from bone marrow.
DNA was isolated from bone marrow mononuclear cells, and Dh-Jh coding joints were amplified by PCR with family-specific Dh primers and a consensus Jh primer (58, 59) as previously described (31), followed by cloning of the PCR products into pGEM-T Easy (Promega). Individual clones were sequenced, and the composition of the junctional region was determined using information from the international ImMunoGeneTics (IMGT) information system (http://imgt.cines.fr/).
Western blot analysis for DNA-PKcs.
Cellular extracts were prepared for Western blot analysis of fibroblasts from patient ID177 and control fibroblasts (C5RO) 30 minutes after irradiation (2 Gy). Nonirradiated cells were used as control. DNA-PKcs protein expression was detected with a mixture of the 2 rabbit polyclonal antibodies produced against amino acids 3,486–3,677 and 356–570, respectively (antibodies 2,208 and 2,129) (29) to detect full-length DNA-PKcs and the phosphospecific antibody against P-S2056-DNA-PKcs (11).
DNA-PK kinase activity assay.
DNA-PK activity was determined using the SignaTECT DNA-Dependent Protein Kinase Assay System (Promega). Cell extracts were prepared from fibroblasts and assayed for DNA-dependent kinase activity according to the supplier’s protocol. DNA-PKcs–specific kinase inhibitor NU7441 (33) and ATM inhibitor KU-55933 (34) were provided by Graeme C.M. Smith of KuDOS Pharmaceuticals and used in a final concentration of 0.5 μM. The inhibitors were dissolved in DMSO, and as control (no inhibitor) DMSO was added to the same concentration.
STR marker analysis.
Four STR markers in the DNA-PKcs gene region were selected (D8S1460, D8S359, D8S531, and AF075268). PCR analysis with one FAM-labeled primer was performed on DNA samples of the patient and 12 family members. The number of STRs was determined using fragment analysis on the ABI PRISM 3100 Genetic Analyzer (Applied Biosystems).
V(D)J recombination assay.
V3 cells with or without DNA-PKcs expression were transfected with a recombination substrate (pDVG93) containing 2 RSSs in inverse orientation and RAG1 and RAG2 expression constructs. Where indicated, a FLAG-tagged DNA-PKcs expression construct was also included. All expression constructs were of human origin. After 48 hours, the fibroblasts were harvested, and extrachromosomal DNA was isolated to amplify the coding joints. PCR products were digested with NotI or NgoMI. After electrophoresis, the gel was dried, and PCR products were visualized by phosphorimaging (31).
DNA-PKcs expression constructs.
Mutated full-length cDNA constructs (delG2113/L3062R, delG2113, and L3062R) with FLAG or EYFP tags were generated in pBluescript II with CMV promoter and stably transfected in V3 cells (11).
End-joining assay.
EcoRV/Eco47III-linearized pDVG94 (with homologous ends [ATCAGC sequence]) was transfected into V3 cells and V3 cells expressing either the wild-type or the mutant human DNA-PKcs construct as described previously (36). Newly formed junctions were PCR amplified, and the relative microhomology use was determined by BstXI digestion.
DNA damage experiments.
DSBs were introduced in a subnuclear volume using a pulsed 800 nm (multiphoton) laser system as described previously (10). Cells were transfected with human Myc-tagged Artemis (pDVG190) (31), and proteins were detected using the EYFP signal on DNA-PKcs and anti-Myc tag antisera (Santa Cruz Biotechnology Inc.). Immunofluorescence was carried out as described (10).
Statistics.
Differences in numbers of P-nucleotides were analyzed using the nonparametric Mann-Whitney U test (1-tailed; P < 0.05 was considered significant) in the GraphPad Prism program (GraphPad Software).
Supplementary Material
Acknowledgments
The authors thank W.M. Comans-Bitter for making the figures and M.C. van Zelm for critical reading of the manuscript. This work was supported by grants from the Dutch Organization for Scientific Research (NWO/ZonMw veni grant 916.56.107 to M. van der Burg), the Dutch Cancer Society (KWF; grant EMCR 2002-2734 to D.C. van Gent, M.Z. Zdzienicka, and J.J.M. van Dongen), the European Community (RISC-RAD: grant FI6R-CT-2003-508842; and DNA repair: grant LSHG-CT-2005-512113; to P.O. Mari and D.C. van Gent), and the NIH (grants 5-R37-CA050519-16 and PO1-CA92584 to D.J. Chen).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: DNA-PKcs, DNA-PK catalytic subunit; DSB, double-strand break; FAT, FRAP, ATM, and TRRAP; NHEJ, nonhomologous end-joining; N-nucleotide, nontemplated nucleotide; P-nucleotide, palindromic nucleotide; RS, radiosensitive; RSS, recombination signal sequence; SCT, stem cell transplantation; STR, short tandem repeat; XLF, XRCC4-like factor.
Citation for this article: J. Clin. Invest. 119:91–98 (2009). doi:10.1172/JCI37141
References
- 1.Jung D., Giallourakis C., Mostoslavsky R., Alt F.W. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu. Rev. Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- 2.Lees-Miller S.P., Meek K. Repair of DNA double strand breaks by non-homologous end joining. Biochimie. 2003;85:1161–1173. doi: 10.1016/j.biochi.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 3.Weterings E., van Gent D.C. The mechanism of non-homologous end-joining: a synopsis of synapsis. DNA Repair. (Amst.). 2004;3:1425–1435. doi: 10.1016/j.dnarep.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 4.McManus K.J., Hendzel M.J. ATM-dependent DNA damage-independent mitotic phosphorylation of H2AX in normally growing mammalian cells. Mol. Biol. Cell. 2005;16:5013–5025. doi: 10.1091/mbc.E05-01-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ward I.M., Minn K., Jorda K.G., Chen J. Accumulation of checkpoint protein 53BP1 at DNA breaks involves its binding to phosphorylated histone H2AX. J. Biol. Chem. 2003;278:19579–19582. doi: 10.1074/jbc.C300117200. [DOI] [PubMed] [Google Scholar]
- 6.Lee A.C., Fernandez-Capetillo O., Pisupati V., Jackson S.P., Nussenzweig A. Specific association of mouse MDC1/NFBD1 with NBS1 at sites of DNA-damage. Cell Cycle. 2005;4:177–182. doi: 10.4161/cc.4.1.1354. [DOI] [PubMed] [Google Scholar]
- 7.Moreno-Herrero F., et al. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature. 2005;437:440–443. doi: 10.1038/nature03927. [DOI] [PubMed] [Google Scholar]
- 8.Smith G.C., Jackson S.P. The DNA-dependent protein kinase. Genes Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- 9.van Gent D.C., van der Burg M. Non-homologous end-joining, a sticky affair. Oncogene. 2007;26:7731–7740. doi: 10.1038/sj.onc.1210871. [DOI] [PubMed] [Google Scholar]
- 10.Mari P.O., et al. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc. Natl. Acad. Sci. U. S. A. 2006;103:18597–18602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uematsu N., et al. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J. Cell Biol. 2007;177:219–229. doi: 10.1083/jcb.200608077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodarzi A.A., et al. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. EMBO J. 2006;25:3880–3889. doi: 10.1038/sj.emboj.7601255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui X., et al. Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Mol. Cell. Biol. 2005;25:10842–10852. doi: 10.1128/MCB.25.24.10842-10852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meek K., Douglas P., Cui X., Ding Q., Lees-Miller S.P. trans Autophosphorylation at DNA-dependent protein kinase’s two major autophosphorylation site clusters facilitates end processing but not end joining. Mol. Cell. Biol. 2007;27:3881–3890. doi: 10.1128/MCB.02366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma Y., Pannicke U., Schwarz K., Lieber M.R. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/S0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 16.Grawunder U., Harfst E. How to make ends meet in V(D)J recombination. Curr. Opin. Immunol. 2001;13:186–194. doi: 10.1016/S0952-7915(00)00203-X. [DOI] [PubMed] [Google Scholar]
- 17.Benedict C.L., Gilfillan S., Thai T.H., Kearney J.F. Terminal deoxynucleotidyl transferase and repertoire development. Immunol. Rev. 2000;175:150–157. doi: 10.1111/j.1600-065X.2000.imr017518.x. [DOI] [PubMed] [Google Scholar]
- 18.Douglas P., et al. The DNA-dependent protein kinase catalytic subunit is phosphorylated in vivo on threonine 3950, a highly conserved amino acid in the protein kinase domain. Mol. Cell. Biol. 2007;27:1581–1591. doi: 10.1128/MCB.01962-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahnesorg P., Smith P., Jackson S.P. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 20.Buck D., et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 21.Li G., et al. Lymphocyte-specific compensation for XLF/cernunnos end-joining functions in V(D)J recombination. Mol. Cell. 2008;31:631–640. doi: 10.1016/j.molcel.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwarz K., et al. RAG mutations in human B cell-negative SCID. Science. 1996;274:97–99. doi: 10.1126/science.274.5284.97. [DOI] [PubMed] [Google Scholar]
- 23.Schatz D.G. V(D)J recombination. Immunol. Rev. 2004;200:5–11. doi: 10.1111/j.0105-2896.2004.00173.x. [DOI] [PubMed] [Google Scholar]
- 24.Moshous D., et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/S0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 25.Noordzij J.G., et al. Radiosensitive SCID patients with Artemis gene mutations show a complete B-cell differentiation arrest at the pre-B-cell receptor checkpoint in bone marrow. Blood. 2003;101:1446–1452. doi: 10.1182/blood-2002-01-0187. [DOI] [PubMed] [Google Scholar]
- 26.Li L., et al. A founder mutation in Artemis, an SNM1-like protein, causes SCID in Athabascan-speaking Native Americans. J. Immunol. 2002;168:6323–6329. doi: 10.4049/jimmunol.168.12.6323. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi N., et al. Novel Artemis gene mutations of radiosensitive severe combined immunodeficiency in Japanese families. Hum. Genet. 2003;112:348–352. doi: 10.1007/s00439-002-0897-x. [DOI] [PubMed] [Google Scholar]
- 28.O’Driscoll M., Gennery A.R., Seidel J., Concannon P., Jeggo P.A. An overview of three new disorders associated with genetic instability: LIG4 syndrome, RS-SCID and ATR-Seckel syndrome. DNA Repair. (Amst.). 2004;3:1227–1235. doi: 10.1016/j.dnarep.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 29.van der Burg M., et al. A new type of radiosensitive T-B-NK+ severe combined immunodeficiency caused by a LIG4 mutation. J. Clin. Invest. 2006;116:137–145. doi: 10.1172/JCI26121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noordzij J.G., et al. The immunophenotypic and immunogenotypic B-cell differentiation arrest in bone marrow of RAG deficient SCID patients corresponds to residual recombination activities of mutated RAG proteins. Blood. 2002;100:2145–2152. [PubMed] [Google Scholar]
- 31.van der Burg M., et al. Defective Artemis nuclease is characterized by coding joints with microhomology in long palindromic-nucleotide stretches. Eur. J. Immunol. 2007;37:3522–3528. doi: 10.1002/eji.200737624. [DOI] [PubMed] [Google Scholar]
- 32.Rooney S., et al. Defective DNA repair and increased genomic instability in Artemis-deficient murine cells. J. Exp. Med. 2003;197:553–565. doi: 10.1084/jem.20021891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leahy J.J., et al. Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg. Med. Chem. Lett. 2004;14:6083–6087. doi: 10.1016/j.bmcl.2004.09.060. [DOI] [PubMed] [Google Scholar]
- 34.Hickson I., et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 35.Chen B.P., et al. Ataxia telangiectasia mutated (ATM) is essential for DNA-PKcs phosphorylations at the Thr-2609 cluster upon DNA double strand break. J. Biol. Chem. 2007;282:6582–6587. doi: 10.1074/jbc.M611605200. [DOI] [PubMed] [Google Scholar]
- 36.Verkaik N.S., et al. Different types of V(D)J recombination and end-joining defects in DNA double-strand break repair mutant mammalian cells. Eur. J. Immunol. 2002;32:701–709. doi: 10.1002/1521-4141(200203)32:3<701::AID-IMMU701>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 37.O’Driscoll M., et al. DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency. Mol. Cell. 2001;8:1175–1185. doi: 10.1016/S1097-2765(01)00408-7. [DOI] [PubMed] [Google Scholar]
- 38.McGuire T.C., Poppie M.J. Hypogammaglobulinemia and thymic hypoplasia in horses: a primary combined immunodeficiency disorder. Infect. Immun. 1973;8:272–277. doi: 10.1128/iai.8.2.272-277.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bosma G.C., Custer R.P., Bosma M.J. A severe combined immunodeficiency mutation in the mouse. Nature. 1983;301:527–530. doi: 10.1038/301527a0. [DOI] [PubMed] [Google Scholar]
- 40.Peterson S.R., et al. Loss of the catalytic subunit of the DNA-dependent protein kinase in DNA double-strand-break-repair mutant mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 1995;92:3171–3174. doi: 10.1073/pnas.92.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meek K., et al. SCID in Jack Russell terriers: a new animal model of DNA-PKcs deficiency. . J. Immunol. 2001;167:2142–2150. doi: 10.4049/jimmunol.167.4.2142. [DOI] [PubMed] [Google Scholar]
- 42.Ruis B.L., Fattah K.R., Hendrickson E.A. The catalytic subunit of DNA-dependent protein kinase regulates proliferation, telomere length, and genomic stability in human somatic cells. Mol. Cell. Biol. 2008;28:6182–6195. doi: 10.1128/MCB.00355-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spagnolo L., Rivera-Calzada A., Pearl L.H., Llorca O. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol. Cell. 2006;22:511–519. doi: 10.1016/j.molcel.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 44.Wiler R., et al. Equine severe combined immunodeficiency: a defect in V(D)J recombination and DNA-dependent protein kinase activity. Proc. Natl. Acad. Sci. U. S. A. 1995;92:11485–11489. doi: 10.1073/pnas.92.25.11485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shin E.K., Perryman L.E., Meek K. A kinase-negative mutation of DNA-PK(CS) in equine SCID results in defective coding and signal joint formation. J. Immunol. 1997;158:3565–3569. [PubMed] [Google Scholar]
- 46.Blunt T., et al. Identification of a nonsense mutation in the carboxyl-terminal region of DNA-dependent protein kinase catalytic subunit in the scid mouse. Proc. Natl. Acad. Sci. U. S. A. 1996;93:10285–10290. doi: 10.1073/pnas.93.19.10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bell T.G., et al. Autosomal recessive severe combined immunodeficiency of Jack Russell terriers. J. Vet. Diagn. Invest. 2002;14:194–204. doi: 10.1177/104063870201400302. [DOI] [PubMed] [Google Scholar]
- 48.Blunt T., et al. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 49.Gu Y., et al. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity. 1997;7:653–665. doi: 10.1016/S1074-7613(00)80386-6. [DOI] [PubMed] [Google Scholar]
- 50.Ouyang H., et al. Ku70 is required for DNA repair but not for T cell antigen receptor gene recombination In vivo. J. Exp. Med. 1997;186:921–929. doi: 10.1084/jem.186.6.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nussenzweig A., et al. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature. 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- 52.Zhu C., Bogue M.A., Lim D.S., Hasty P., Roth D.B. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell. 1996;86:379–389. doi: 10.1016/S0092-8674(00)80111-7. [DOI] [PubMed] [Google Scholar]
- 53.Fattah K.R., Ruis B.L., Hendrickson E.A. Mutations to Ku reveal differences in human somatic cell lines. DNA Repair (Amst.). 2008;7:762–774. doi: 10.1016/j.dnarep.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li G., Nelsen C., Hendrickson E.A. Ku86 is essential in human somatic cells. Proc. Natl. Acad. Sci. U. S. A. 2002;99:832–837. doi: 10.1073/pnas.022649699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao Y., et al. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell. 1998;95:891–902. doi: 10.1016/S0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- 56.van Ankeren S.C., Murray D., Meyn R.E. Induction and rejoining of gamma-ray-induced DNA single- and double-strand breaks in Chinese hamster AA8 cells and in two radiosensitive clones. Radiat. Res. 1988;116:511–525. doi: 10.2307/3577393. [DOI] [PubMed] [Google Scholar]
- 57.Whitmore G.F., Varghese A.J., Gulyas S. Cell cycle responses of two X-ray sensitive mutants defective in DNA repair. Int. J. Radiat. Biol. 1989;56:657–665. doi: 10.1080/09553008914551881. [DOI] [PubMed] [Google Scholar]
- 58.Szczepañski T., et al. Precursor-B-ALL with DH-JH gene rearrangements have an immature immunogenotype with a high frequency of oligoclonality and hyperdiploidy of chromosome 14. Leukemia. 2001;15:1415–1423. doi: 10.1038/sj.leu.2402206. [DOI] [PubMed] [Google Scholar]
- 59.Szczepanski T., et al. Cross-lineage T cell receptor gene rearrangements occur in more than ninety percent of childhood precursor-B acute lymphoblastic leukemias: alternative PCR targets for detection of minimal residual disease. Leukemia. 1999;13:196–205. doi: 10.1038/sj.leu.2401277. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.