Abstract
Current treatment for chronic hepatitis C is expensive, is often accompanied by burdensome side effects, and, sadly, fails in almost half of cases. The ability to predict such failures prior to treatment could save a great deal of pain and expense for the patient with HCV. In this issue of the JCI, Aurora and colleagues describe the development of genetic markers predictive of treatment response based on a study of viral sequence variation (see the related article beginning on page 225). Genome-wide covariation analyses of pretreatment virus sequences from 94 patients showed distinct patterns of mutations strongly associated with the ultimate success or failure of treatment. Such analyses suggest markers predictive of response to therapy and may lead to new insights into the underlying biology of hepatitis C.
An estimated 130 million people worldwide (1) and nearly 4 million in the United States are chronically infected with HCV, leading to liver damage and increased risk of hepatocellular carcinoma. In the United States, 10,000 deaths each year are attributed to chronic HCV infection (2). The current treatment regime, pegylated IFN-α and ribavirin, is long and difficult, requiring months of weekly injections, with serious side effects ranging from flu-like symptoms to depression and autoimmune disorders. Success of treatment is far from guaranteed: in HCV genotype 1 infections, which account for the majority of cases in the US, only about half of patients display the long-term suppression of virus indicative of cure.
Numerous studies in recent years have proposed markers for predicting HCV patient response to therapy. Markers may be based on viral factors, such as viral sequence variation (3); host factors, such as gene expression profiles (4) or polymorphisms in specific host genes (5); or combinations thereof (6, 7). Interestingly, very different types of biomarkers can give similar results, indicative of the intimate interactions between the manifold host and viral players in virus replication and disease progression.
In this issue of the JCI, Aurora et al. define a set of biomarkers predictive of the response to HCV therapy (8). These markers are purely viral factors, composed of sets of varying residues in the HCV amino acid sequence identified by covariation analysis.
Covariation analysis reveals functional relationships
A statistical measure, covariance quantifies the degree of linkage between 2 variables; variables that are completely independent have a low covariance, whereas variables that vary synchronously have a high covariance.
Covariance between residues in a protein or set of proteins can be estimated from the variation observed in a population. An alignment of multiple HCV sequences shows both conserved and varying residues. The varying positions are compared in pairwise fashion; for each pair of positions, the linkage between the 2 residues will affect the pattern of variation observed. For a pair of positions with a 10% mutation frequency at each site, both mutations would be shared by 1% of sequences if they are perfectly independent and 10% if they are perfectly covariant.
Because covariation implies a relation between 2 residues in a sequence, it has been used to infer information about direct interactions in the 3-dimensional structure of a protein (9) and to identify protein-protein interactions (10). However, covariance arises from all functional interactions between residues, both direct and indirect, as well as from phylogenetic relationships (Figure 1). Distinguishing between the many sources of covariance is a continuing challenge for anyone wishing to use this technique (11, 12).
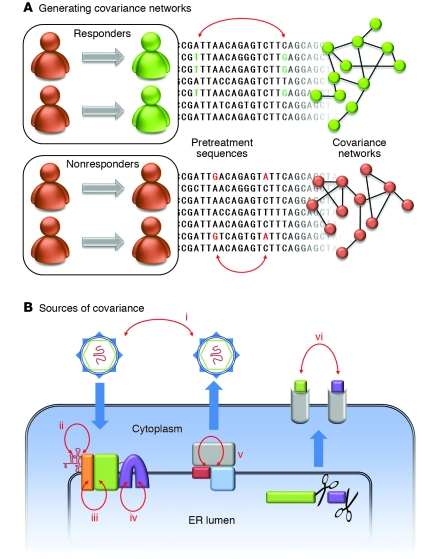
Figure 1. Covariation in HCV.
(A) In the study reported in this issue of the JCI by Aurora et al. (8), patients were grouped according to their treatment response. The sequences of the complete HCV open reading frame obtained from each group of patients prior to treatment were aligned and analyzed for covariance. An example covariant pair is shown in each alignment (red arrows). The set of all covariant pairs forms a network in which each node is an amino acid position and each connecting line represents covariance between 2 positions. The networks differ by treatment response class and may be used to generate markers indicative of HCV treatment outcome. (B) Various causes of covariance in HCV (red arrows). (i) Phylogenetic covariance is an artifact of a shared ancestry, but does not reflect any functional relationship. (ii) RNA secondary structure gives rise to nucleotide-level covariance. (iii) Protein-protein interaction residues covary. (iv) Intraprotein covariance may indicate direct residue contact or indirect interaction through the protein. (v) Variation in a shared interaction partner (host or viral) may result in coordinated variation in a pair of residues. (vi) MHC epitopes will covary across hosts with different HLA alleles.
Covariation patterns are highly correlated with treatment outcome
The Viral Resistance to Antiviral Therapy of Chronic Hepatitis C (Virahep-C) clinical study (13) evaluated the efficacy of treatment in HCV genotype 1a and 1b patients. The complete HCV coding sequence was determined for pretreatment isolates from each of 94 patients, who were followed during and after treatment to determine the final outcome of therapy.
In the present study, Aurora et al. analyzed the 94 HCV sequences obtained during the Virahep-C study for amino acid covariance in each of the genotype 1 subtypes as well as stratified within each subtype by treatment response (8). From this analysis they made an important, and perhaps surprising, observation: the sets of covariant pairs were markedly different between the responsive and nonresponsive patient groups. In the HCV genotype 1a sequences, about 2,000 covariant residue pairs were identified; three-quarters of the covariant pairs found in the responsive genomes did not appear in the nonresponsive sequence set, and vice versa. The results of the HCV genotype 1b sequence analysis was even more striking: 90% of the residue pairs identified as being covariant in one response group were independent in the other group.
The strong correlation between covariance sets and therapeutic outcome immediately suggests the possibility of finding a reliable predictor for response to therapy in the pretreatment HCV sequence. However, there is a still an additional step that must be made; a patient coming in for treatment generally harbors a range of closely related viral sequences. Covariance, on the other hand, is an aggregate property determined from a sequence alignment of an entire group of responders or nonresponders. The covariance sets reported by Aurora et al. showed a clear difference between groups of sequences depending on response to therapy (8), but a biomarker must be able to place a single sequence of unknown response into the correct group. In order to bridge this gap, the authors looked to the interconnected nature of the covariance sets they had generated.
Covariance networks
Each covariation analysis performed by Aurora et al. identified on the order of 2,000 pairs of correlated residues (8). However, this set of 2,000 pairs is composed of only about 200 unique residues. Clearly a residue may appear multiple times; in fact, each residue in the set was connected to anywhere between 1 and 100 other residues. The resulting networks are shown in detail in ref. 8.
Because covariant pairs by definition vary, any one pair will appear in only a fraction of sequences. Similarly, a combination of residues correlated with one outcome can appear in a sequence of the opposite outcome, not because the residues are functionally linked, but simply by chance. For this reason, the authors searched for small collections of interconnected pairs, or subnetworks, which were correlated with outcome. By means of exhaustive search, they identified several hundred such subnetworks, which appeared in greater than 95% of sequences of one therapeutic outcome and never appeared in sequences of the opposite outcome (8).
The attentive reader will note — and the authors are quick to point out — that the sequences for which the markers are evaluated are the same sequences used to generate the markers. This is attributed to the unavailability of other sequence sets for which the treatment outcome is known. Nevertheless, the authors provide evidence that the differences observed in the covariance networks are real and will translate into markers that will hold up outside the initial data set. First, the difference in the covariance sets between the 2 possible outcomes is quite large, as much as 90%. Second, the subnetwork analysis yielded not a handful of potential markers, but hundreds of subnetworks with 100% correlation to treatment outcome. Finally, and most interestingly, the chemical makeup of the covariant pairs is significantly different; the nonresponsive sequences contain 3 times as many hydrophobic covariant amino acid pairs as the responsive sequences. This unexpected result implies that the differences in the covariance networks are directly reflective of an underlying physical phenomenon. The authors suggest that the higher fraction of correlated hydrophobic residues is evidence for more stable protein-protein complexes in the nonresponsive strains. This could be envisioned to result in viral replication complexes that are more resistant to antiviral effectors, or even to alter interactions of immunomodulatory HCV proteins with their target host factors. Analysis of covariance networks may therefore not only reveal biomarkers for therapeutic outcome, but also shed light on the mechanistic bases for resistance to treatment and even identify novel targets for antiviral drugs.
Conclusions
Although it still remains for these markers to be validated, the early results presented in this study are promising (8). It is interesting to speculate on the relationship between these markers and other markers, particularly those based on host characteristics. The circulating virus is not an independent entity, but is continually shaped by host selective pressures even as it in turn modulates its host environment. Viral sequences observed prior to treatment may very well represent the success or failure of the host in selecting against the most treatment-resistant variants. Covariance networks may serve as an exciting new tool in further studies along this avenue; networks generated from viral sequences obtained during acute viral infection should be particularly informative.
With the sustained and rapid growth of both computational power and sequencing capabilities, we expect covariation analyses to become increasingly common as a tool to study different aspects of HCV biology (14). The high mutation rate of RNA viruses and the intense competition within the quasispecies makes them particularly amenable to this technique. We look forward to seeing further application of covariance networks to questions ranging from protein structure and protein-protein interactions to drug resistance, host selection pressures, and viral evolution.
Acknowledgments
The authors acknowledge Catherine Murray for her assistance in the preparation of this manuscript. Work in the laboratory of C.M. Rice is supported by the Greenberg Medical Research Institute and the Starr Foundation.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: Virahep-C, Viral Resistance to Antiviral Therapy of Chronic Hepatitis C [study].
Citation for this article: J. Clin. Invest. 119:5–7 (2009).doi:10.1172/JCI38069.
See the related article beginning on page 225.
References
- 1.Wasley A., Alter M.J. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin. Liver Dis. 2000;20:1–16. doi: 10.1055/s-2000-9506. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong G.L., et al. The prevalence of hepatitis C virus infection in the United States, 1999 through 2002. Ann. Intern. Med. 2006;144:705–714. doi: 10.7326/0003-4819-144-10-200605160-00004. [DOI] [PubMed] [Google Scholar]
- 3.Moreau I., Levis J., Crosbie O., Kenny-Walsh E., Fanning L.J. Correlation between pre-treatment quasispecies complexity and treatment outcome in chronic HCV genotype 3a. Virol. J. 2008;5:78. doi: 10.1186/1743-422X-5-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Torres-Puente M., et al. Genetic variability in hepatitis C virus and its role in antiviral treatment response. J. Viral Hepat. 2008;15:188–199. doi: 10.1111/j.1365-2893.2007.00929.x. [DOI] [PubMed] [Google Scholar]
- 5.Persico M., et al. Elevated expression and polymorphisms of SOCS3 influence patient response to antiviral therapy in chronic hepatitis C. . Gut. 2008;57:507–515. doi: 10.1136/gut.2007.129478. [DOI] [PubMed] [Google Scholar]
- 6.
- 7.El-Shamy A., et al. Prediction of efficient virological response to pegylated interferon/ribavirin combination therapy by NS5A sequences of hepatitis C virus and anti-NS5A antibodies in pre-treatment sera. Microbiol. Immunol. 2007;51:471–482. doi: 10.1111/j.1348-0421.2007.tb03922.x. [DOI] [PubMed] [Google Scholar]
- 8.Aurora R., Donlin M.J., Cannon N.A., Tavis J.E., for the Virahep-C Study Group. Genome-wide hepatitis C virus amino acid covariance networks can predict response to antiviral therapy in humans. J. Clin. Invest. 2009;119:225–236. doi: 10.1172/JCI37085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gobel U., Sander C., Schneider R., Valencia A. Correlated mutations and residue contacts in proteins. Proteins. 1994;18:309–317. doi: 10.1002/prot.340180402. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y.E., DeLisi C. Inferring protein-protein interactions in viral proteins by co-evolution of conserved side chains. Genome Inform. 2006;17:23–35. [PubMed] [Google Scholar]
- 11.Eyal E., Frenkel-Morgenstern M., Sobolev V., Pietrokovski S. A pair-to-pair amino acids substitution matrix and its applications for protein structure prediction. Proteins. 2007;67:142–153. doi: 10.1002/prot.21223. [DOI] [PubMed] [Google Scholar]
- 12.Dunn S.D., Wahl L.M., Gloor G.B. Mutual information without the influence of phylogeny or entropy dramatically improves residue contact prediction. Bioinformatics. 2008;24:333–340. doi: 10.1093/bioinformatics/btm604. [DOI] [PubMed] [Google Scholar]
- 13.Donlin M.J., et al. Pretreatment sequence diversity differences in the full-length Hepatitis C Virus open reading frame correlate with early response to therapy. J. Virol. 2007;81:8211–8224. doi: 10.1128/JVI.00487-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campo D.S., Dimitrova Z., Mitchell R.J., Lara J., Khudyakov Y. Coordinated evolution of the hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 2008;105:9685–9690. doi: 10.1073/pnas.0801774105. [DOI] [PMC free article] [PubMed] [Google Scholar]