Abstract
MicroRNAs (miRNAs) are small, noncoding RNAs that regulate gene expression by targeting complementary sequences, referred to as miRNA recognition elements (MREs), typically located in the 3′ untranslated region of mRNAs. miR-181a is highly expressed in developing thymocytes and markedly downregulated in post-thymic T cells. We investigated whether endogenous miR-181a can be harnessed to segregate expression of chimeric antigen receptors (CARs) and TCRs between developing and mature T cells. Lentiviral-encoded antigen receptors were tagged with a miR-181a–specific MRE and transduced into mouse BM cells that were used to generate hematopoietic chimeras. Expression of a CAR specific for human CD19 (hCD19) was selectively suppressed in late double-negative and double-positive thymocytes, coinciding with the peak in endogenous miR-181a expression. Receptor expression was fully restored in post-thymic resting and activated T cells, affording protection against a subsequent challenge with hCD19+ tumors. Hematopoietic mouse chimeras engrafted with a conalbumin-specific TCR prone to thymic clonal deletion acquired peptide-specific T cell responsiveness only when the vector-encoded TCR transcript was similarly engineered to be subject to regulation by miR-181a. These results demonstrate the potential of miRNA-regulated transgene expression in stem cell–based therapies, including cancer immunotherapy.
Introduction
Stem cell engineering for research or therapeutic applications increasingly requires tight restriction of transgene expression to selected lineages at chosen differentiation stages (1–3). Current strategies rely on RNA polymerase II–dependent (pol II–dependent) transcriptional regulation, which may be in some instances limited by promoter leakiness or the unavailability of promoter/enhancer elements that afford complex expression patterns. Thus, a major limitation of T cell transgenesis is the difficulty in teasing apart the postdevelopmental functions of genes from effects on T cell development, when transgenes are placed under the control of T lineage enhancer/promoters, such as those derived from the Lck, Cd4, or CD2 genes (4–6). Segregating thymic versus post-thymic effects of transgene expression would also be useful to target gene products such as tumor antigen–specific TCRs to post-thymic T cells without perturbing thymocyte development and selection, which could result from the expression of autoreactive, high-affinity TCRs (7, 8).
Posttranscriptional gene repression mediated by microRNAs (miRNAs) has recently emerged as a fundamental physiological mechanism for the modulation of gene expression. miRNAs constitute a phylogenetically conserved class of small (20–25 nucleotides) noncoding RNAs that derive from endogenous hairpin-structured precursor transcripts (9–12). miRNAs function as guide molecules through base pairing with target sequences, referred to as miRNA recognition elements (MREs), typically residing in the 3′ untranslated region (3′ UTR) of native mRNAs (13, 14). This interaction recruits effector complexes mediating mRNA cleavage or translational repression (13, 15–19). The degree of complementarity between the miRNA and the coding mRNA is believed to be a major determinant of the outcome, resulting in mRNA degradation when targeted sequences are near-perfectly complementary (13, 20).
The vast majority of miRNA genes are transcribed by pol II (21, 22). Their expression patterns are therefore amenable to elaborate spatiotemporal control. Indeed, tissue- and/or developmental stage–specific expression has been documented for some miRNA species studied (23–26). Tagging MREs to reporter genes, thus creating miRNA “sensors,” has proven helpful to track miRNA expression patterns in Drosophila melanogaster (23) and mouse embryos (27). The same strategy has been implemented to restrict expression of vector-encoded transgenes in hepatic APCs following systemic administration of a viral vector as well as in hematopoietic and embryonic stem cells, demonstrating the broad applicability of this mechanism of regulation (28, 29). Furthermore, regulation afforded by miR-223, a miRNA with a myeloid-specific expression pattern (30, 31), was shown to downregulate expression of a GFP transgene in myeloid cells in hematopoietic chimeras (29).
Here, we sought to investigate whether miRNA-mediated gene regulation can be exploited to regulate transgenes during T cell development. We specifically investigated miR-181a, which is highly expressed in lymphoid tissues, particularly in the thymus, where it modulates T cell sensitivity to peptide antigens (30, 32, 33). Using a chimeric antigen receptor specific for hCD19 (34), we demonstrated that expression could be selectively silenced in stages of thymocyte development where negative selection occurs and restored in post-thymic T cells, thus providing a level of developmental control that, to our knowledge, could not previously be attained through transcriptional regulation. We show that this strategy enables T lymphocytes expressing a self-reactive TCR to evade thymic selection, which would be very useful in cancer immunotherapy.
Results
Transgene regulation by hematopoietic miRNAs enables lineage-restricted expression in murine BM chimeras.
We initially wanted to assess the potential of miRNA-mediated gene regulation to impart lineage and developmental stage specificity upon transgene expression in hematopoietic mouse chimeras. We generated lentiviral vectors encoding GFP under the transcriptional control of the constitutive human elongation factor 1a (hEF1a) promoter, tagged with MREs specific for miR-223 and miR-181a (Figure 1A). These vectors were initially transduced in a panel of murine and human cell lines, which express miR-223 (K562), miR-181a (Jurkat), both (HL60), or neither (MEL and NIH3T3), as shown in Supplemental Figure 1A (supplemental material available online with this article; doi: 10.1172/JCI37216DS1). Normalized GFP expression was the same for all vectors in MEL and NIH3T3 cells, which do not express any of the 2 miRNAs, whereas it was specifically repressed in cells expressing the MRE-complementary miRNA. GFP expression from the vector encoding 4 tandem miR-223–specific MREs (NG4×223) was reduced 6.3- ± 0.9-fold (n = 4 independent transduction experiments) and 13.1- ± 1.2-fold (n = 3) in K562 and HL60 cells, respectively, whereas expression from the vector encoding the miR-181a–specific MRE (NG4×181a) decreased 18.1- ± 2.3- (n = 7) and 10.3- ± 0.9-fold (n = 5) in Jurkat and HL60 cells, respectively (Supplemental Figure 1B). Similar results were obtained with reporter genes other than GFP (data not shown). Vector titers were not affected by inclusion of 4 copies of the miR-223–specific MRE, but were slightly decreased (2- to 2.5-fold) in the case of NG4×181a (data not shown). The MRE repeats in both vectors were faithfully maintained in most transcripts in both Jurkat and MEL cells (data not shown). This initial in vitro study showed that hematopoietic lineage-specific miRNAs afford potent and highly specific regulation of transgene expression.
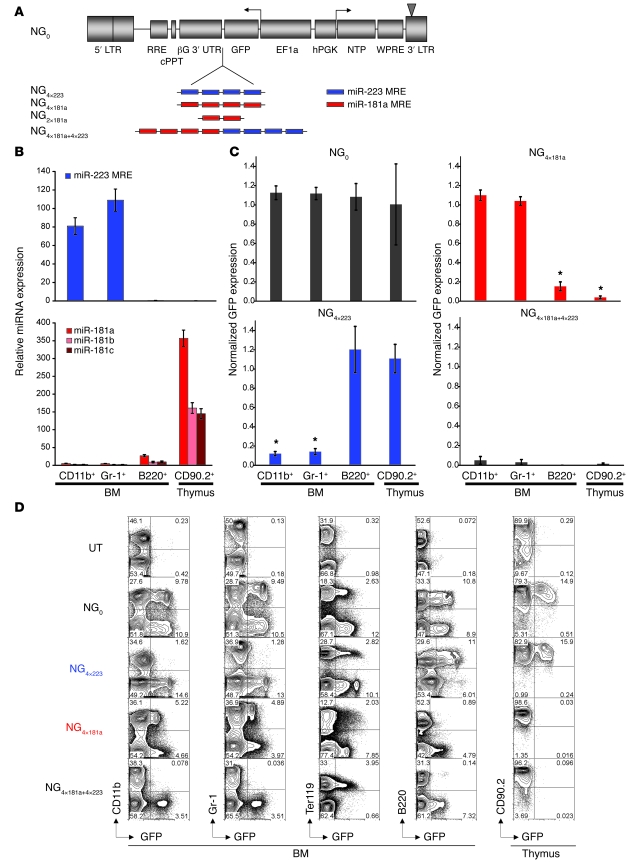
Figure 1. miRNA-mediated regulation effectively restricts transgene expression in lymphoid and myeloid lineages in vivo.
(A) Lentiviral vectors used in this study encode 2 reporters, GFP and NTP, bidirectionally driven by the ubiquitous EF1a and hPGK promoters, respectively. NG4×223 through NG4×181a+4×223 harbor repeats of MREs in the 3′ UTR of GFP, so that GFP functions as the sensor of miRNA regulation. Each MRE consists of a nucleotide sequence with perfect complementarity to miRNA miR-223 or miR-181a. Vectors NG2×181a and NG4×181a contain, respectively, 2 and 4 tandem MREs complementary to mature miR-181a, and NG4×181a+4×223 contains a combination of 4 copies of both MREs. LTR, long-terminal repeat; RRE, Rev responsive element; cPPT, central polypurine tract; βG 3′ UTR, human β-globin 3′ UTR; WPRE, Woodchuck hepatitis virus posttranscriptional regulatory element. (B) Expression of miR-223, miR-181a, miR-181b, and miR-181c in highly purified BM CD11b+, Gr-1+, and B220+ cells and thymocytes derived from hematopoietic mouse chimeras 4–6 weeks after transplantation. Expression levels are normalized to those measured in Ter119+ BM cells of the same mouse. Error bars denote SD. (C) GFP expression in different cell lineages in BM and thymus, normalized to the expression in Ter119+ BM cells. Data are averaged from 9 mice for NG0, 9 for NG4×223, 6 for NG4×181a, and 5 for NG4×181a+4×223. Error bars denote SEM. *P < 0.001 versus respective NG0 value. (D) Representative FACS analysis of BM cells and thymocytes from mouse chimeras reconstituted with BM cells transduced with vectors NG0, NG4×223, NG4×181a, or NG4×181a+4×223. Numbers within plots denote percentage of cells in the respective quadrants. UT, untransduced.
To test the potential of this strategy in vivo, we generated murine BM chimeras harboring the NG4×223 and NG4×181a vectors. At 4–6 weeks after transplantation, we quantified the expression of miR-223 and miR-181 family miRNAs, as well as the expression of the vector-encoded GFP, in different hematopoietic lineages. Endogenous miRNA expression as well as vector-encoded GFP expression in different lineages of the BM and thymus was normalized to the respective expression in Ter119+ BM cells, which express miR-223 and miR-181a at very low levels (data not shown) and were thus used as a reference population. The levels of miRNAs miR-181b and miR-181c, which are highly homologous to miR-181a, were also quantified, because these are likely to additionally target a miR-181a–specific MRE. miR-223 was exclusively expressed in myeloid cells (CD11b+ and Gr-1+), whereas miR-181a, miR-181b, and miR-181c were selectively expressed in B220+ B lymphocytes in BM and, most noticeably, in thymocytes (Figure 1B), corroborating previous reports (30–33). NG4×223-encoded GFP expression was markedly restricted from myeloid cells, as evidenced by a decrease in GFP expression of 88% ± 7% and 86% ± 9% relative to NG0 vector in CD11b+ and Gr-1+ cells, respectively, whereas it remained unaltered in erythroid (Ter119+) and lymphoid progenitors and thymocytes (Figure 1, C and D). Reciprocally, NG4×181a-encoded expression was robust in myeloid and erythroid lineages, but greatly diminished in B220+ cells, and it decreased to almost undetectable levels in thymocytes (Figure 1, C and D). In contrast, mice reconstituted with cells transduced with the control vector NG0 exhibited similar levels of GFP expression in all hematopoietic lineages. The presence of the vector in all lineages was confirmed by vector copy number quantification in sorted CD11b+, Gr-1+, B220+, Ter119+, lin–, and CD90.2+ cells from BM and spleen and in total thymocytes from 8 mice (data not shown). These data demonstrate the robustness of miRNA regulation to direct lineage-specific transgene regulation in vivo in hematopoietic chimeras.
To assess whether regulation by more than one miRNA could be harnessed, we constructed the NG4×181a+4×223 vector, which encodes a composite MRE composed of both miRNA target sequences (4× miR-223–targeted MRE in tandem with 4× miR-223–targeted MRE; Figure 1A). In K562 and Jurkat cells, which respectively express miR-223 and miR-181a, NG4×181a+4×223-encoded GFP was knocked down to the same degree as in the NG4×223 or NG4×181a vectors, respectively (Supplemental Figure 2). In HL60 cells, which express high levels of both miRNAs, the effect was additive, with NG4×223-encoded GFP being downregulated 13.1- ± 1.2-fold, NG4×181a-encoded GFP 10.3- ± 0.9-fold, and NG4×181a+4×223-encoded GFP 24.6- ± 1.2-fold (Supplemental Figure 2). In vivo, expression of NG4×181a+4×223-encoded GFP was profoundly repressed in both the myeloid and lymphoid lineages (decreased by 95% ± 9% in CD11b+ cells, 97% ± 6% in Gr-1+ cells, 99% ± 0.5% in B220+ cells, and 99% ± 2% in CD90.2+ thymocytes) but preserved in the erythroid lineage (Figure 1, C and D). These data indicate that collinear MREs function additively, thus offering the potential to further refine developmental regulation or tissue restriction if needed.
miR-181a–mediated regulation restricts antigen receptor expression in developing thymocytes.
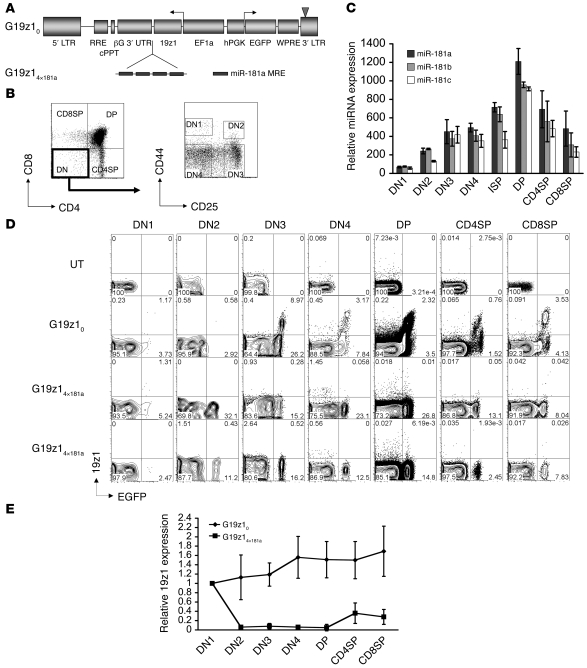
Expression of miRNA miR-181a is dynamically regulated during T cell development (32, 33). To closely decipher the expression pattern afforded by the miR-181a MRE, we quantified expression of miR-181a miRNA in sorted cell subpopulations in all stages of thymocyte development, as well as in post-thymic T lymphocytes. We found that miR-181 miRNAs progressively accumulated in CD4/CD8 double-negative (DN) thymocytes at stages DN1–DN4, peaked at the double-positive (DP) stage, and were subsequently downmodulated in CD4+ single-positive (CD4SP) or CD8SP cells before markedly decreasing in resting post-thymic T cells (Figure 2C and see below). This pattern suggested that miR-181a–specific MREs could be used to specifically restrict transgene expression in developing T cells but yet allow transgene expression in post-thymic, mature T lymphocytes. This possibility was investigated with vectors encoding 19z1, a chimeric antigen receptor (CAR) specific for hCD19, which is currently used in clinical studies to target T cells against B cell malignancies (34). We placed 19z1 under posttranscriptional control of miR-181a using the lentiviral vectors shown in Figure 2A. Expression of the 19z1 receptor was assessed in the thymus and other hematopoietic tissues of murine BM chimeras. In contrast to mice bearing the control G19z10 vector, cell surface expression of 19z1 was diminished to almost undetectable levels in DN and DP cells of mice harboring the G19z14×181a vector (Figure 2, B, D, and E). In 2 of 10 mice harboring the G19z14×181a vector, a small number of 19z1dull cells was detectable, predominantly falling within the CD4SP and CD8SP populations (data not shown). The same profound suppression of expression compared with control vector was observed in 5 mouse chimeras bearing a GFP sensor encoded by vector NG4×181a (data not shown). These data demonstrate that miR-181a–mediated regulation can be effectively harnessed to silence transgene expression in developing thymocytes.
Figure 2. miR-181a MRE restricts tumor antigen–specific receptor expression in developing thymocytes.
(A) Lentiviral vectors bidirectionally expressing EGFP (as the neutral reporter) and the 19z1 CAR under posttranscriptional regulation by miR-181a. (B) Gating example of flow cytometry analysis of thymocyte subpopulations: DN (CD4–CD8–), DN1 (CD4–CD8–CD44+CD25–), DN2 (CD4–CD8–CD44+CD25+), DN3 (CD4–CD8–CD44–CD25+), DN4 (CD4–CD8–CD44–CD25–), DP (CD4+CD8+), CD4SP (CD4+CD8–), and CD8SP (CD4–CD8+). (C) Expression of miR-181 family miRNAs in sorted murine thymocyte subsets relative to expression in Ter119+ BM cells. ISP, intermediate SP. Error bars denote SD. (D) Representative FACS profile of thymocytes from 2 mice harboring the miR-181a–regulated vector G19z14×181a, 1 mouse harboring the control vector G19z10, and 1 nontransplanted mouse. Shown are cells gated on subpopulations exemplified in B. Numbers within plots denote percentage of cells in the respective quadrants. (E) Average 19z1 expression, quantified by FACS analysis, at different stages of thymocyte development relative to expression in DN1 stage in mouse chimeras reconstituted with BM cells transduced with vectors G19z10 and G19z14×181a. Data are mean ± SEM from 20 mice.
miR-181–regulated antigen receptor expression is restored in resting and activated T lymphocytes.
Because miR-181a expression persists in post-thymic T lymphocytes, albeit at greatly reduced levels compared with thymocytes (Figure 2C and see below), we investigated whether miR-181a–regulated expression of an antigen receptor can be restored in resting or activated mature T cells. We further hypothesized that altering the number of MRE repeats in the 3′ UTR of the receptor transcript may be exploited to dose antigen receptor expression. We therefore quantified receptor repression mediated by either 2 or 4 copies of the miR-181a–specific MRE. An initial in vitro study using vectors NG2×181a and NG4×181a (Figure 1A) showed that increasing the number of MRE repeats from 2 to 4 approximately doubled the level of miRNA-dependent downregulation in HL60 (from 7.6- to 13.1-fold) and Jurkat cells (from 7.2- to 18.1-fold; Supplemental Figure 3). In vivo, murine BM chimeras harboring 19z1-encoding vectors 19z12 and 19z14 (Figure 3A), with 2 and 4 MRE copies, respectively, showed similarly potent repression of receptor expression in thymocytes (Figure 3B). In peripheral T cells, however, expression from 19z14 greatly increased compared with thymocyte levels, but did not reach the same levels as did 19z12-encoded expression (Figure 3C).
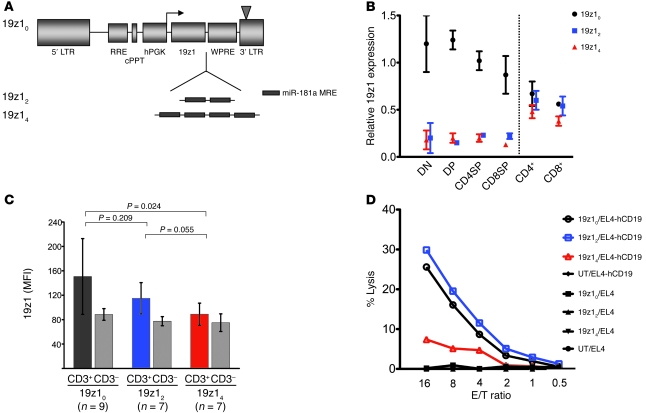
Figure 3. CAR expression and function in peripheral T lymphocytes is fully restored by 2-MRE copy regulation.
(A) Lentiviral vectors used in these experiments express 19z1 CAR driven by hPGK promoter under regulation by either no (19z10), 2 (19z12), or 4 (19z14) copies of miR-181a–specific MRE. (B) Mean fluorescence intensity (MFI) expression of 19z1 in DN, DP, CD4SP, and CD8SP thymocytes as well as CD4+ and CD8+ splenocytes normalized to expression in CD4–CD8– splenocytes for each mouse. Data are averaged from 4 mice harboring each of vectors 19z10, 19z12, and 19z14 analyzed 3–4 months after BM transplantation. Error bars denote SEM. (C) Expression of 19z1 in peripheral blood 19z1+CD3+ T cells in a total of 23 mouse chimeras harboring vectors 19z10, 19z12, and 19z14 analyzed 4 weeks after BM transplantation. Expression levels in CD3– cells indicate comparable levels of gene transfer. Error bars denote SD. Repeated analysis at week 10 after BM transplantation (not shown) corroborated the data presented here. (D) T cells expressing a miR-181–regulated 19z1 receptor specifically lysed a hCD19+ tumor cell line. 51Cr-release assays of T cells isolated from spleens of mouse chimeras harboring vectors 19z10, 19z12, and 19z14 targeting EL4-hCD19+ tumor cells or unmodified EL4 (hCD19–) tumor cells, as well as T cells from a mouse reconstituted with untransduced BM cells. E/T ratio, effector-to-target ratio of CD8+19z1+ T cells to tumor cells.
These disparate expression levels resulted in functional differences: cytotoxic activity of T lymphocytes expressing 19z1 under regulation by 4 MRE repeats was lower than that of T lymphocytes expressing the receptor tagged with a 2-copy MRE (Figure 3D). Regulation by 2 miR-181a–specific MRE copies allowed restoration of both expression and function of the receptor to levels similar to the control (non–miR-181–regulated) vector 19z10 (Figure 3, C and D). In addition, miR-181–regulated receptor expression was not diminished throughout ex vivo activation of T lymphocytes (Figure 4, A and B). Furthermore, secondary BM recipients exhibited similar segregation of miR-181–regulated 19z1 receptor, demonstrating efficient miR-181a–mediated transgene regulation in long-term repopulating hematopoietic stem cells (Supplemental Figure 4).
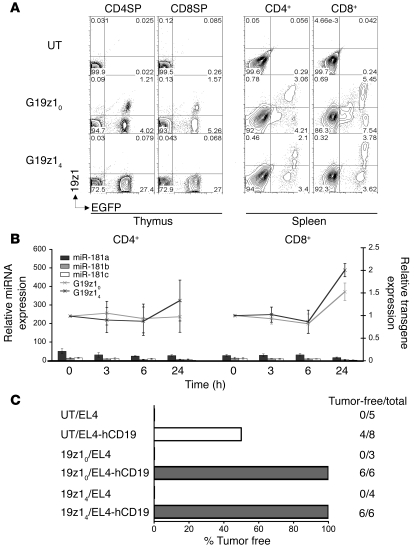
Figure 4. miR-181a–regulated 19z1 expression is maintained in activated T lymphocytes and mediates antitumor response.
(A) FACS profile of CD4SP and CD8SP thymocytes (left) and CD4+ and CD8+ splenocytes polyclonally activated in vitro for 48 hours (right) from 1 mouse harboring vector 19z14 and 1 mouse harboring the control vector 19z10. Numbers within plots denote percentage of cells in the respective quadrants. (B) miR-181–regulated antigen receptor expression was maintained throughout activation of T lymphocytes. Relative expression of miR-181 miRNA family members (bars; left axis) and relative 19z1 expression (lines; right axis) in CD4+ and CD8+ T cells from LNs of murine BM chimeras harboring vectors 19z10 and 19z14 stimulated in vitro for 3, 6, and 24 hours, normalized to unstimulated T cells. Error bars denote SD. (C) miR-181–regulated 19z1 receptor prevents tumor formation in mouse chimeras challenged with hCD19+ EL4 tumor cells. Outcome of systemic tumor challenge of chimeras reconstituted with BM cells harboring either no vector or vectors 19z10 and 19z14 with EL4 or EL4-hCD19+ tumor cells.
miR-181–regulated antigen receptor expression confers protection against a CD19+ tumor.
The function of a miR-181a–regulated 19z1 receptor was tested in a tumor challenge experiment, using syngeneic EL4 tumor cells that express hCD19 (34). Mouse chimeras reconstituted with BM cells harboring either no vector or vectors 19z10 and 19z14 were intravenously injected with EL4 or EL4-hCD19+ tumor cells. The mice that received the unmodified EL4 tumors rapidly developed hind-limb paralysis and died within 2 weeks. The mice that received EL4-hCD19+ tumor cells survived longer, probably owing to the antigenicity of hCD19, and were sacrificed 40 days after tumor challenge. Cell suspensions from the BM, liver, and spleen were analyzed by flow cytometry for the presence of hCD19+ cells. None of the chimeras reconstituted with vectors expressing 19z1 were found to have detectable tumors, whereas half of the mice harboring no vector had established tumors (Figure 4C). The results presented thus far collectively demonstrated that subjecting a lentiviral-encoded antigen receptor to endogenous miR-181a regulation enabled specific receptor downregulation in developing thymocytes as well as restoration of its expression and function in mature and activated T lymphocytes.
miR-181 regulation enables peripheral T cell expression and antigen responsiveness of a self-reactive αβ TCR targeted to hematopoietic stem cells.
In view of the above findings, we hypothesized that miR-181a regulation could allow peripheral T cell expression of an αβ TCR normally deleted during thymic development. We therefore constructed vectors encoding miR-181–regulated α and β chains linked by a 2A peptide (Figure 5A) of a TCR derived from the D10.G4.1 (referred to herein as D10) T cell clone, which responds to a peptide derived from chicken conalbumin (CA-wt) bound to the MHC class II allele I-Ak and I-Ad (35). We used these vectors to generate hematopoietic mouse chimeras of the B10.D2 strain harboring the I-Ad allele, which induces negative selection of thymocytes expressing the D10 TCR (34). As expected, the number of EGFP+ T cells was severely reduced in chimeras repopulated by cells transduced with the control GD100 vector, consistent with clonal deletion of these autoreactive cells (Figure 5B). Cells transduced with the miR-181–regulated vector, however, gave rise to CD4+ and CD8+ T cells in peripheral blood and spleen, which were present in similar fractions in the transduced (EGFP+) and nontransduced (EGFP–) cells present in the same mouse (Figure 5B). T cells expressing the D10 TCR were detected in the peripheral blood and spleen of these chimeras, were detected at low levels in resting T cells, and were markedly upregulated after T cell activation (Figure 5C). Functional responsiveness to the cognate peptide–MHC complex was demonstrated by mixed cultures of T cells isolated from the spleens of these mice with irradiated I-Ak APCs loaded with titrated amounts of CA-wt peptide (Figure 5D). These data demonstrate that cells harboring the miR-181–regulated D10 TCR, but not those harboring the non–miR-181a–regulated vector, evade negative selection and acquire peripheral responsiveness to the antigen-MHC complex.
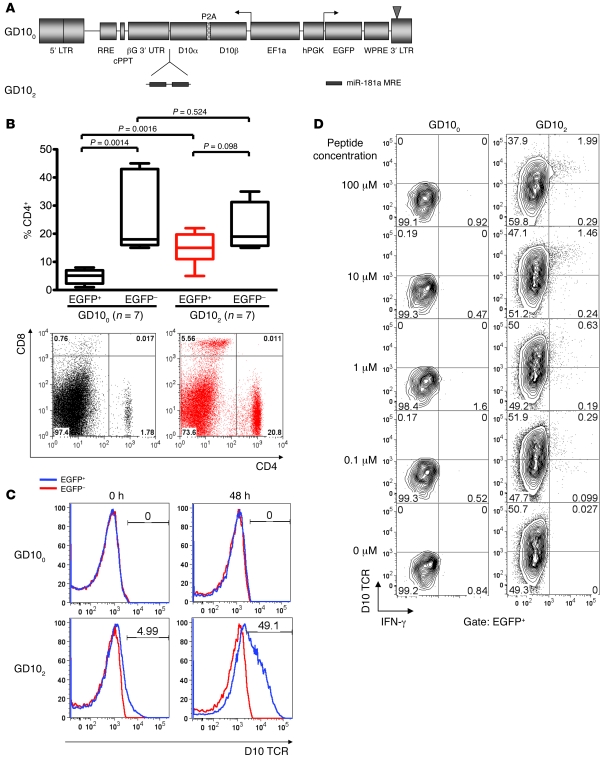
Figure 5. Lymphocytes harboring a miR-181a–regulated self-reactive D10 TCR evade negative selection.
(A) Vectors used in this study encode the α and β chain of the D10 TCR linked by the P2A peptide (49) that allows stoichiometric expression of both genes. In vector GD102, the D10 TCR is subject to regulation by 2 copies of the miR-181a–specific MRE. (B) Top: Percentage of CD4+ cells in the EGFP+ and EGFP– cell subsets of PBMCs of mouse chimeras reconstituted with BM cells transduced with either GD100 or GD102 vectors 5 weeks after BM transplantation. Boxes denote interquartile range; lines within boxes indicate mean; whiskers define the fifth and ninety-fifth percentiles. Bottom: CD4/CD8 T cell distribution in EGFP+ gated PBMCs in 1 mouse harboring vector GD100 (left) and 1 mouse with vector GD102 (right). Numbers within plots denote percentages of cells in the respective quadrants. (C) Expression of D10 TCR in gated EGFP+ or EGFP– CD4+ splenocytes, either resting or activated for 48 hours, from 1 mouse chimera with vector GD100 and 1 mouse with vector GD102. Numbers within histograms denote percentage of cells within the indicated gate. (D) T cells expressing the miR-181–regulated D10 TCR specifically responded to CA-wt peptide presented on MHC I-Ak allele. T cells were isolated from the spleens of mouse chimeras harboring vectors GD100 and GD102 stimulated on APCs expressing the I-Ak allele loaded with the indicated amounts of peptide and analyzed by flow cytometry for IFN-γ production. Numbers within plots denote percentage of cells in the respective quadrants.
pol II–driven MREs borne on lentiviral vectors do not detectably deregulate miRNA target repression.
Although we found that hematopoietic lineage distribution was largely unperturbed in mouse chimeras harboring miRNA-regulated vectors NG4×181a and NG4×223 (Supplemental Figure 5), we sought to investigate the safety of using vectors harboring MREs for hematopoietic stem cell–based therapies. It has recently been reported that exogenously expressed MREs can function as competitive inhibitors of endogenous miRNAs in cultured animal cells (36). In these studies, miRNA inhibition was achieved by transient expression systems that drive very high levels of expression from very high copy numbers of the MRE-bearing transcripts (36). We wished to investigate the potential saturation of miRNA function in a more physiologically relevant setting, whereby endogenously expressed MREs compete with exogenous MREs expressed from a low number of copies of stably integrated vectors for binding of endogenously expressed miRNAs. We set a competitive reporter assay in a system where endogenous miRNAs are physiologically induced during a developmental process, namely terminal erythroid differentiation of the murine erythroleukemia cell line MEL. Following chemically induced MEL cell differentiation, expression of miRNAs miR-144 and miR-451 was upregulated from almost undetectable levels up to 60-fold (ref. 37 and data not shown), mediating repression of a fluorescent red reporter (DsRed) harboring MREs complementary to miR-144 or miR-451 (Figure 6A, left, and data not shown). Superinfection of the cells with high MOI of a vector harboring 4 copies of an MRE complementary to miR-144 driven by hEF1a promoter did not derepress DsRed upon differentiation (Figure 6A, right), although pol III–driven MREs specific for either miR-144 or miR-451 did cause some degree of derepression of DsRed tagged with the respective MRE (Figure 6B). The average vector copy number was between 12 and 39 (data not shown). These data suggest that MREs harbored by lentiviral vectors, even at high copy numbers and driven by a strong constitutive pol II promoter, are not likely to interfere with normal miRNA function and repression of physiological targets. These findings support the further investigation of MRE-regulated vectors to regulate transgene expression in the hematopoietic system.
Figure 6. MREs driven by pol II do not cause derepression of a miRNA target.
(A) hEF1a-driven MREs did not deregulate a heterologous miRNA target. Left: MEL clones harboring DsRed reporters encoded by vectors R144 or R223 were generated. Upon induction of differentiation with hexamethylene bisacetamide (HMBA), miR-144 was upregulated, resulting in specific repression of DsRed tagged with the miR-144–specific MRE (R144), but not with control miR-223–specific MRE (R223). Right: Superinfection at high MOI of the same clones with a lentiviral vector encoding 4 copies of the miR-144–specific MRE tagged to GFP (G144) did not detectably alter DsRed repression (compare bottom overlays). Shown are the results of 3 independent induction experiments (induced 1–induced 3). Numbers in parentheses indicate mean fluorescence intensity of DsRed expression. (B) MREs driven by U6 promoter mediated derepression of the respective miRNA target. MEL clones harboring DsRed sensors regulated by either miR-144 or miR-451 (top left) were superinfected at high MOI with vectors harboring 4 copies of the respective MREs driven by the strong pol III U6 promoter (top right). Transduction with both vectors mediated derepression of DsRed targeted by the respective miRNA, from 2.3-fold to 1.7-fold in the case of the miR-144–specific MRE (left overlays) and from 2-fold to 1.4-fold in the case of the miR-451–specific MRE (right overlays). Numbers in parentheses indicate mean fluorescence intensity of DsRed expression. Fold DsRed repression is expressed as ratio of mean fluorescence intensities before and after induction.
Discussion
The genetic targeting of gene products to T cells, but not their thymic precursors, is a lingering challenge in animal transgenesis and stem cell therapies. We therefore sought to modulate expression from a constitutive promoter by subjecting it to endogenous miR-181a regulation. We show here that lentiviral vector–encoded antigen receptor expression that is regulated by miR-181a enabled robust segregation between developing thymocytes and mature T cells. miR-181a–targeted transcripts were maximally repressed in late DN and DP cells (Figure 2, D and E), as expected from our miR-181a expression studies (Figure 2C) and the previous studies of Neilson et al. (33) and Li et al. (32). Our functional analyses show that antigen receptor expression was fully repressed in DP cells (Figure 2, D and E), a developmental stage in which miR-181a, which downregulates phosphatase expression, renders DP thymocytes exceptionally sensitive to TCR signaling (32). The profound downregulation of the CAR 19z1 demonstrated that miR-181–mediated silencing of a relatively highly expressed transcript driven by the hEF1a promoter was sufficient to functionally prevent expression at the stages of positive and negative selection. Our studies with the D10 TCR further compound that antigen receptor is profoundly suppressed in all stages of thymocyte development in which negative selection occurs (including DN, DP, and the DP-to-SP transition; ref. 38), averting the deletion of D10 TCR+ T cells in B10.D2 recipient mice (Figure 5 and ref. 35).
Transgene expression was readily detectable in peripheral T cells, as found with the 19z1 CAR (Figures 3 and 4) and the D10 TCR (Figure 5). Expression was reinstated in peripheral T cells, consistent with the major decrease of miR-181a levels in post-thymic T cells (Figure 4B). Expression of both antigen receptors was functional, enabling mice harboring the 19z1 receptor to kill CD19+ tumors in vitro (Figure 3D) and resist challenge with CD19+ tumor cells (Figure 4C). Similarly, B10.D2 mice engrafted with the GD102 vector acquired the ability to respond to the cognate peptide, which was precluded in mice harboring the GD100 vector (Figure 5D). These results demonstrate the ability of the miR-181a MRE to developmentally regulate receptor expression within the T cell lineage, preserving receptor functionality.
Our findings also underscore the importance of MRE copy number to achieving desirable levels of transgene expression. Whereas vector-encoded transcripts bearing either 2 or 4 miR-181a MRE copies under the transcriptional control of either a human phosphoglycerate kinase (hPGK; vectors 19z12 and 19z14) or a hEF1a (vector G19z14×181a) promoter were similarly downregulated in thymocytes, the presence of either 2 or 4 copies yielded different outcomes in peripheral T cells (Figure 3). Expression of 19z1 was lower in T cells expressing transcripts encoding 4 MRE copies, resulting in diminished cytolytic activity. This functional difference is consistent with the known importance of antigen receptor levels for T cell responsiveness. Higher expression of TCR chains may also be important for the transgenic TCR to successfully compete with endogenous TCR chains for rate-limiting amounts of CD3 to achieve cell surface expression (39, 40). miR-181a expression did not increase upon T cell activation (Figure 4B), whereas cell surface expression of the 19z1 (Figure 4B) and D10 (Figure 5C) receptors increased within 24 hours of mitogen activation, consistent with promoter activation (41).
MRE-mediated repression may also vary in developing thymocytes. Whereas 19z1 expression was profoundly repressed in SP cells (Figure 2, D and E), the receptor was detected at very low levels in 2 of 10 mice, which also expressed low 19z1 levels in DP-stage cells (data not shown). The latter expressed reduced levels of both CD4 and CD8 (DPdull), a phenotype corresponding to the immediate progeny of positive selection (42), which possess reduced sensitivity to TCR signaling, potentially owing to reduced levels of expression of miR-181a (43). Our in vitro and in vivo findings support the notion of MRE dose dependence of knockdown efficiency, corroborating data from transient reporter assays (44) and stably integrated miRNA target sites in cell lines (29).
MRE-regulated transgene expression is poised to find applications in basic biology as well as in therapeutic stem cell engineering. Posttranscriptional control superimposed onto classic transcriptional regulation is highly valuable to correct undesirable expression patterns or fine-tune developmentally regulated or inducible gene expression. Preventing transgene expression in professional APCs may attenuate the immunogenicity of foreign antigens (28). As we showed here, miR-181–regulated transgene expression was effective in allowing temporal transgene regulation throughout lymphopoiesis, which to our knowledge was previously unattainable through pol II–mediated transcriptional regulation.
A major challenge of cancer immunotherapy is the generation of large numbers of T cells with the desired antitumor specificities that can persist long-term in vivo. Delivery of tumor-specific TCR genes into hematopoietic stem cells constitutes a recent promising strategy in this direction (7, 8). Antigen-specific T cells may also be provided by infusion of lymphoid progenitors that are educated and tolerized according to the recipient’s genetic background (45). A predictable problem of this approach is negative selection of T lymphocytes recognizing self tumor antigens (46) as a consequence of clonal deletion of developing thymocytes expressing autoreactive receptors. miR-181a regulation of transgenic TCRs targeted to either the hematopoietic stem cell or the lymphoid progenitor compartment may thus be very useful to circumvent negative selection of tumor-reactive T lymphocytes and afford durable immune surveillance.
Methods
Plasmid construction and vector production.
Vector NG0 was derived from T9.FIX (47) by placing the GFP cDNA in reverse orientation upstream of the β-globin 3′ UTR, under the transcriptional control of the hEF1a promoter, and adding the hPGK-NTP-WPRE cassette. NTP is an inactive human low-affinity nerve growth factor receptor (48). NG2×181a and the intermediate vector NG2×223 were constructed by annealing oligonucleotides O3, O4 and O1, O2, respectively (Supplemental Table 1), which consist of 2 repeats of a sequence perfectly complementary to the respective mature miRNA separated by 4 nucleotides (49). These adapters were inserted into XmaI and AvrII sites in a linker previously inserted downstream of GFP cDNA. NG4×181a and NG4×223 were derived from NG2×181a and NG2×223 by inserting annealed oligonucleotides O7, O8 and O5, O6, respectively (Supplemental Table 1), and NG4×181a+4×223 was derived from NG4×181a after insertion of annealed oligonucleotides O9 and O10 (Supplemental Table 1). Vectors G19z10 and G19z14×181a were derived from NG0 and NG4×181a, respectively, by replacing NTP with EGFP and GFP with 19z1. For the construction of vectors GD100 and GD102, the D10 TCR α and β chain cDNAs (35) were inserted in G19z10 and G19z14×181a, respectively, in place of the 19z1 cDNA linked by the P2A peptide preceded by a Gly-Ser-Gly linker (50). GD102 was derived from an intermediate GD104 vector after removal of 2 MRE copies as a ClaI fragment. Vectors R144, R451, and R223 were derived from NG0 after replacing GFP with the DsRed monomer cDNA (Clontech) and inserting annealed oligonucleotides O11, O12 (for R144) and O13, O14 (for R451) downstream of DsRed. For R223, a XmaI/AvrII fragment containing 4 copies of miR-223 MRE derived from NG4×223 was inserted in the respective sites in the 3′ end of DsRed cDNA. G144 was derived from NG0 after removing the hPGK-NTP-WPRE cassette and inserting annealed oligos O11, O12 downstream of GFP. For GU6-144 and GU6-451, U6 promoter was amplified from human genomic DNA and inserted in a NheI site in the U3 region of the 3′ long-terminal repeat, followed by insertion of oligonucleotides O11, O12 and O13, O14, respectively, in a downstream ClaI site. All oligonucleotide sequences are provided in Supplemental Table 1. Vector production was performed by triple transfection of the plasmid DNA encoding the vector pCMVΔR8.91 and pUCMD.G into 293T cells, as previously described (51).
Analysis of miRNA expression.
RNA was isolated from cell lines and primary murine cells with TRIzol reagent (Invitrogen). Quantitative RT-PCR for the detection of mature miRNAs miR-223, miR-181a, miR-181b, and miR-181c was performed with the mirVana qRT-PCR miRNA detection kit (Ambion). Reactions were carried out in triplicate in an ABI 7500 system (Applied Biosystems). Quantification was performed with the relative standard curve method using serial dilutions of RNA isolated from HL60 cells as standards and U6 snRNA as endogenous control.
Cell purification.
Cell suspensions were prepared from BM (extracted from femurs and tibias), thymus, spleen, and LNs and filtered through a 40-μm nylon cell strainer. Positive or negative selection of lin–, Sca1–, Ter119+, CD11b+, Gr-1+, B220+, CD4+, and CD8+ cells was carried out by immunomagnetic separation (Miltenyi Biotec) according to the manufacturer’s instructions. Purity in all cases was between 91% and 99%.
Generation of murine BM chimeras.
C57BL/6 and B10.D2 mice were purchased from The Jackson Laboratory. lin– cells isolated from 8- to 10-week-old Ly5.1 C57BL/6 or B10.D2 mice were prestimulated for 12 hours in X-VIVO 10 medium (BioWhittaker) supplemented with l-glutamine (2 mM), penicillin (100 IU/ml), streptomycin (100 μg/ml), b-mercaptoethanol (0.5 mM; Invitrogen), 100 ng/ml recombinant mouse stem cell factor, and 100 ng/ml recombinant mouse thrombopoietin (rmSCF and rmTPO, respectively; R&D Systems). Transduction with viral supernatants was performed in retronectin-coated (TAKARA) 24-well plates in 400 μl total volume at MOI between 20 and 30 for 8 hours. After the end of transduction, 6 × 104 cells were mixed with 4 × 105 supportive Sca-1–depleted cells per recipient mouse and were injected via the retroorbital route into lethally irradiated Ly5.2 C57BL/6 or B10.D2 mice. All animal work was approved by and in compliance with the Institutional Animal Care and Use Committee guidelines of MSKCC.
Flow cytometry and cell sorting.
A PE-conjugated anti–low-affinity nerve growth factor receptor antibody (anti-LNGFR; BD Biosciences — Pharmingen) was used for FACS analysis of cell lines. Mouse BM cells and thymocytes were blocked in 1% mouse serum, stained with the antibodies anti-Ter119, anti-B220, anti–Gr-1, anti-CD11b, and anti-CD90.2 (all allophycocyanin-conjugated) as well as with 19E3-PE, an anti-idiotypic monoclonal antibody specific for the 19z1 receptor (provided by I. Riviere, MSKCC) and analyzed on a FACSCalibur cytometer (BD Biosciences). Analysis of DN, DP, and SP thymocyte subpopulations, as well as CD4+ and CD8+ lymphocytes, was performed as previously described (52). Briefly, linage-negative cells were detected by staining with anti-Ter119, anti–Gr-1, anti-NK1.1, anti-CD11b, anti-γδTCR, and anti–B220-biotin followed by PE-Cy7–conjugated streptavidin. The following antibody combinations were used: PE-conjugated 19E3 or Ly5.1, PerCP5.5-conjugated anti-CD8, allophycocyanin-conjugated anti-CD4, allophycocyanin-Cy7–conjugated anti-CD25, AF700-conjugated anti-TCRβ, and FITC- or Pacific Blue–conjugated anti-CD44. Dead cells were excluded by DAPI staining. Antibodies were purchased from BD Biosciences. Detection of the D10 TCR was done with a clonotypic antibody (3D3) (35). Detection of intracellular IFN-γ was performed with an allophycocyanin-conjugated antibody after fixation and permeabilization (Cytofix/Cytoperm; BD). FACS analysis was performed in a LSRII cytometer (BD Biosciences) and analyzed using FlowJo software (version 8.5.3; Tree Star). For cell sorting, DN thymocytes were enriched by depletion of DP and CD8SP thymocytes with rat anti-CD8 (TIB210) followed by magnetic depletion with Biomag anti-rat IgG (Polysciences Inc.). Cell sorting was performed on a MoFlo cell sorter (DakoCytomation).
T cell activation and cytotoxicity assay.
In vitro stimulation of T lymphocytes for FACS analysis was performed with cell suspensions prepared from spleens or LNs of mice in the presence of soluble anti-CD3 (2 μg/ml) and soluble anti-CD28 (2 μg/ml). For miRNA expression analysis, CD4+ and CD8+ cells were negatively selected by immunomagnetic separation (Miltenyi Biotec) and cultured in anti-CD3–coated plates with soluble anti-CD28. For peptide-specific stimulation, T cells were immunomagnetically negatively selected from splenocytes and mixed with Dynabeads CD3/CD28 (Invitrogen) in a 1:1 ratio in the presence of 100 U/ml recombinant IL-2. After 5 days, cells were restimulated on APCs prepared from irradiated splenocytes of B10.BR animals (expressing the I-Ak allele) pulsed with titrated amounts of CA-wt peptide. Brefeldin A (BD GolgiPlug) was added 2 hours later, and FACS analysis was performed 6 hours after the beginning of stimulation. The cytotoxic activity of T cells isolated from the spleens of the chimeras was determined by standard 51Cr-release assays (34).
Statistics.
Statistical analysis was done by 2-tailed Student’s t test using Prism software (version 5.0a; GraphPad). A P value less than 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Isabelle Riviere for the 19E3 anti-idiotypic monoclonal antibody, Renier Brentjens for helpful discussion, and Gertrude Gunset for excellent technical assistance. This work was supported in part by NIH grants RO1 HL57612 and PO1 CA59350 and by the Starr Foundation through the Tri-Institutional Stem Cell initiative.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: CAR, chimeric antigen receptor; DN, double-negative; DP, double-positive; EF1a, elongation factor 1a; miRNA, microRNA; MRE, miRNA recognition element; PGK, phosphoglycerate kinase; pol II, RNA polymerase II; SP, single-positive; 3′ UTR, 3′ untranslated region.
Citation for this article: J. Clin. Invest. 119:157–168 (2009). doi:10.1172/JCI37216
References
- 1.von Kalle C., Baum C., Williams D.A. Lenti in red: progress in gene therapy for human hemoglobinopathies. J. Clin. Invest. 2004;114:889–891. doi: 10.1172/JCI23132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang A.H., Sadelain M. The genetic engineering of hematopoietic stem cells: the rise of lentiviral vectors, the conundrum of the LTR, and the promise of lineage-restricted vectors. Mol. Ther. 2007;15:445–456. doi: 10.1038/sj.mt.6300060. [DOI] [PubMed] [Google Scholar]
- 3.Nienhuis A.W. Development of gene therapy for blood disorders. Blood. 2008;111:4431–4444. doi: 10.1182/blood-2007-11-078121. [DOI] [PubMed] [Google Scholar]
- 4.Orban P.C., Chui D., Marth J.D. Tissue- and site-specific DNA recombination in transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 1992;89:6861–6865. doi: 10.1073/pnas.89.15.6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolfer A., et al. Inactivation of Notch 1 in immature thymocytes does not perturb CD4 or CD8T cell development. Nat. Immunol. 2001;2:235–241. doi: 10.1038/85294. [DOI] [PubMed] [Google Scholar]
- 6.de Boer J., et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 2003;33:314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- 7.Stolzer A.L., Sadelain M., Sant’Angelo D.B. Fulminant experimental autoimmune encephalo-myelitis induced by retrovirally mediated TCR gene transfer. Eur. J. Immunol. 2005;35:1822–1830. doi: 10.1002/eji.200526123. [DOI] [PubMed] [Google Scholar]
- 8.Yang L., Baltimore D. Long-term in vivo provision of antigen-specific T cell immunity by programming hematopoietic stem cells. Proc. Natl. Acad. Sci. U. S. A. 2005;102:4518–4523. doi: 10.1073/pnas.0500600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He L., Hannon G.J. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 10.Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 11.Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 12.Lee R.C., Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 13.Yekta S., Shih I.-H., Bartel D.P. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 14.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 15.Rehwinkel J., Behm-Ansmant I., Gatfield D., Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giraldez A.J., et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 17.Bagga S., et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 18.Meister G., et al. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Olsen P.H., Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 20.Hutvagner G., Zamore P.D. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 21.Lee Y., et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai X., Hagedorn C.H., Cullen B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brennecke J., Hipfner D.R., Stark A., Russell R.B., Cohen S.M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/S0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 24.Lagos-Quintana M., et al. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/S0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 25.Aravin A.A., et al. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. doi: 10.1016/S1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 26.Landgraf P., et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mansfield J.H., et al. MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat. Genet. 2004;36:1079–1083. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- 28.Brown B.D., Venneri M.A., Zingale A., Sergi L.S., Naldini L. Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat. Med. 2006;12:585–591. doi: 10.1038/nm1398. [DOI] [PubMed] [Google Scholar]
- 29.Brown B.D., et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat. Biotechnol. 2007;25:1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- 30.Chen C.-Z., Li L., Lodish H.F., Bartel D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 31.Fazi F., et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBP[alpha] regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 32.Li Q.-J., et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 33.Neilson J.R., Zheng G.X.Y., Burge C.B., Sharp P.A. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 2007;21:578–589. doi: 10.1101/gad.1522907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brentjens R.J., et al. Eradication of systemic B-cell tumors by genetically targeted human T lymphocytes co-stimulated by CD80 and interleukin-15. Nat. Med. 2003;9:279–286. doi: 10.1038/nm827. [DOI] [PubMed] [Google Scholar]
- 35.Sant’Angelo D.B., Janeway C.A., Jr. Negative selection of thymocytes expressing the D10 TCR. Proc. Natl. Acad. Sci. U. S. A. 2002;99:6931–6936. doi: 10.1073/pnas.102182499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ebert M.S., Neilson J.R., Sharp P.A. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhan M., Miller C.P., Papayannopoulou T., Stamatoyannopoulos G., Song C.Z. MicroRNA expression dynamics during murine and human erythroid differentiation. Exp. Hematol. 2007;35:1015–1025. doi: 10.1016/j.exphem.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hogquist K.A., Baldwin T.A., Jameson S.C. Central tolerance: learning self-control in the thymus. Nat. Rev. Immunol. 2005;5:772–782. doi: 10.1038/nri1707. [DOI] [PubMed] [Google Scholar]
- 39.Sommermeyer D., et al. Designer T cells by T cell receptor replacement. Eur. J. Immunol. 2006;36:3052–3059. doi: 10.1002/eji.200636539. [DOI] [PubMed] [Google Scholar]
- 40.Abad J.D., et al. T-cell receptor gene therapy of established tumors in a murine melanoma model. J. Immunother. 2008;31:1–6. doi: 10.1097/CJI.0b013e31815c193f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cooper L.J., et al. Enhanced transgene expression in quiescent and activated human CD8+ T cells. Hum. Gene Ther. 2004;15:648–658. doi: 10.1089/1043034041361217. [DOI] [PubMed] [Google Scholar]
- 42.Sant’Angelo D.B., et al. A molecular map of T cell development. Immunity. 1998;9:179–186. doi: 10.1016/S1074-7613(00)80600-7. [DOI] [PubMed] [Google Scholar]
- 43.Wong P., Barton G.M., Forbush K.A., Rudensky A.Y. Dynamic tuning of T cell reactivity by self-peptide-major histocompatibility complex ligands. J. Exp. Med. 2001;193:1179–1188. doi: 10.1084/jem.193.10.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doench J.G., Petersen C.P., Sharp P.A. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zakrzewski J.L., et al. Tumor immunotherapy across MHC barriers using allogeneic T-cell precursors. Nat. Biotechnol. 2008;26:453–461. doi: 10.1038/nbt1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Houghton A.N., Gold J.S., Blachere N.E. Immunity against cancer: lessons learned from melanoma. Curr. Opin. Immunol. 2001;13:134–140. doi: 10.1016/S0952-7915(00)00195-3. [DOI] [PubMed] [Google Scholar]
- 47.Chang A.H., Stephan M.T., Sadelain M. Stem cell-derived erythroid cells mediate long-term systemic protein delivery. Nat. Biotechnol. 2006;24:1017–1021. doi: 10.1038/nbt1227. [DOI] [PubMed] [Google Scholar]
- 48.Gallardo H.F., Tan C., Ory D., Sadelain M. Recombinant retroviruses pseudotyped with the vesicular stomatitis virus G glycoprotein mediate both stable gene transfer and pseudotransduction in human peripheral blood lymphocytes. Blood. 1997;90:952–957. [PubMed] [Google Scholar]
- 49.Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szymczak A.L., et al. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat. Biotechnol. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- 51.May C., et al. Therapeutic haemoglobin synthesis in [beta]-thalassaemic mice expressing lentivirus-encoded human [beta]-globin. Nature. 2000;406:82–86. doi: 10.1038/35017565. [DOI] [PubMed] [Google Scholar]
- 52.Dose M., et al. c-Myc mediates pre-TCR-induced proliferation but not developmental progression. Blood. 2006;108:2669–2677. doi: 10.1182/blood-2006-02-005900. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.