Abstract
Lignin is incorporated into plant cell walls to maintain plant architecture and to ensure long-distance water transport. Lignin composition affects the industrial value of plant material for forage, wood and paper production, and biofuel technologies. Industrial demands have resulted in an increase in the use of genetic engineering to modify lignified plant cell wall composition. However, the interaction of the resulting plants with the environment must be analyzed carefully to ensure that there are no undesirable side effects of lignin modification. We show here that Arabidopsis thaliana mutants with impaired 5-hydroxyguaiacyl O-methyltransferase (known as caffeate O-methyltransferase; COMT) function were more susceptible to various bacterial and fungal pathogens. Unexpectedly, asexual sporulation of the downy mildew pathogen, Hyaloperonospora arabidopsidis, was impaired on these mutants. Enhanced resistance to downy mildew was not correlated with increased plant defense responses in comt1 mutants but coincided with a higher frequency of oomycete sexual reproduction within mutant tissues. Comt1 mutants but not wild-type Arabidopsis accumulated soluble 2-O-5-hydroxyferuloyl-l-malate. The compound weakened mycelium vigor and promoted sexual oomycete reproduction when applied to a homothallic oomycete in vitro. These findings suggested that the accumulation of 2-O-5-hydroxyferuloyl-l-malate accounted for the observed comt1 mutant phenotypes during the interaction with H. arabidopsidis. Taken together, our study shows that an artificial downregulation of COMT can drastically alter the interaction of a plant with the biotic environment.
Author Summary
Lignin is an essential component of wood and the second most abundant terrestrial biopolymer. Plants synthesize lignin to strengthen cell walls and to resist pathogen attack. Because the technological value of plants is determined by the amount and composition of lignin, genetic engineering approaches frequently aim at altering these parameters. However, data showing how plants with modified lignin content interact with the biotic environment are still scarce. This study is the first of its kind to evaluate how a model plant, which was mutated for a key enzyme in lignin biosynthesis, interacts with pathogens from different kingdoms. We found that the mutants were generally more susceptible to bacteria and fungi. Additionally, the mutation altered the plant physiology in a manner that elevated sexual reproduction of an oomycete pathogen. Oospores resulting from this mode of reproduction are the most persisting propagules of this pathogen. Their elevated formation correlated with the accumulation of a soluble phenylpropanoid derivative in the mutants, which we synthesized and found to be able to stimulate sexual oomycete reproduction in vitro. Our study indicates that interfering with lignin composition may drastically alter the outcome of plant–pathogen interactions.
Introduction
Lignin accounts for about 30% of the organic carbon in the biosphere and is the second most abundant terrestrial biopolymer after cellulose [1]. Lignin fortifies plant cell walls, is the essential composite of wood, and allows terrestrial plants to gain size and volume. This polymer renders the walls of water-conducting cells impermeable, making it possible for the xylem vessels to transport water [2]. Plants with high lignin content are less accessible to microbes and insect herbivores, and the incorporation of this polymer into cell walls is an important mechanism of plant defense against pathogen attack [3]. Lignin is a complex aromatic heteropolymer composed of the three phenylalanine-derived monolignols, p-coumaryl-, coniferyl-, and sinapyl alcohol [1],[4] (Figure 1). Polymers composed of these monolignols are referred to as p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) lignin, respectively. Dicotyledonous plants contain mostly G and S lignin, whereas monocotyledonous plants contain H, G, and S lignin [5]. The amount of lignin and the proportions of the H, G, and S subunits within the lignocellulosic biomass determine the technological value of the raw plant material for forage, biofuel distillation, wood, or paper production. Attempts to modify lignin composition have been a major focus of research for decades, with the first mutant described as early as 1935 [6]. The brown midrib-3 (bm3) maize mutant has a lower S-lignin content than normal maize [7], due to the downregulation of caffeate O-methyltransferase (COMT) activity [8] (Figure 1). COMT has since been targeted repeatedly in efforts to improve the technological quality of plant material by transgenic approaches. The downregulation of COMT activity leads to a decrease in the proportion of S-lignin, with no major change in overall lignin content [9]. As previously reported for bm3 maize [10], an increase in the G/S lignin ratio generally seems to improve the digestibility of transgenic forage [11]. Poplar trees in which COMT expression has been silenced have been shown to generate higher pulp yields, although the lignins they contain are less amenable to industrial degradation [12]. In field trials, the transgenic trees grew normally and displayed no obvious physiological alterations [13]. However, studies investigating the side effects of changes in COMT gene expression in mutant and transgenic plants remain scarce, although it was shown 30 years ago that the bm3 mutation decreases vascular integrity and stalk robustness in maize [14]. Even less is known about the outcome of biotic interactions involving plants with genetically modified COMT expression and S-lignin content. Arabidopsis thaliana appears to be an excellent tool for addressing this issue. This plant has a typical dicotyledonous lignification pattern [15], the COMT gene has been clearly identified [16], tools for functional analyses have been generated, and A. thaliana has been established as a host for a large number of microorganisms. In this study, we used wild-type A. thaliana and mutant A. thaliana lines with inactivated COMT1 for analyzing their phenotypes of interaction with the bacterial pathogens Xanthomonas campestris pv. campestris and Pseudomonas syringae pv. tomato, the fungal pathogens Alternaria brassicicola, Blumeria graminis, and Botrytis cinerea, the oomycete Hyaloperonospora arabidopsidis (formerly H. parasitica), and the root-knot nematode Meloidogyne incognita. The comt1a mutant was found to have much lower levels of S-lignin than the wild type, although its overall lignin content was similar [15]. Knockout of comt1 led to a strong decrease in soluble 2-O-sinapoyl-L-malate (sinapoyl malate) levels and the accumulation of 2-O-5-hydroxyferuloyl-L-malate (hydroxyferuloyl malate), which is not detectable in wild-type plants ([4],[15] and this study). We paid particular attention to interactions with the oomycete H. arabidopsidis, because comt1 mutants displayed an unexpected phenotype when inoculated with this pathogen. Oomycetes have devastating effects on crops, forests, and natural ecosystems, and there are currently no efficient methods for their control [17]. These fungus-like pathogens have unique physiological characteristics and can undergo vegetative or sexual reproduction. Asexual reproduction leads to the creation of clonal populations well adapted to a given host and environment. Sexual reproduction can occur in a single strain, when the species is homothallic. In heterothallic species, two strains of opposite mating types are required for fertilization. In both homothallic and heterothallic species, fertilization results in thick-walled zygotes called oospores, which are produced in infected plant tissue, and released into the soil as the plant tissue degrades. Oospores are highly resistant against environmental influences and can persist in the soil for several years [18]. Sexual reproduction is frequently initiated in response to selective pressure from the environment [19]. The findings presented here indicate that mutations in COMT1 alter the plant physiology in a way that stimulates the oomycete to reproduce sexually. Risk assessments for plants with modified lignin biosynthesis should, therefore, go for quantitative studies of the interaction with the biotic environment, and beyond.
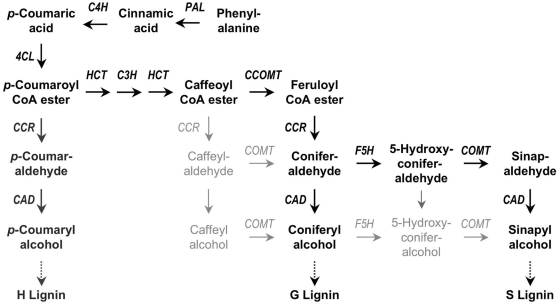
Figure 1. Simplified scheme for monolignol synthesis.
The main pathway in dicotyledonous plants is highlighted in black, involving phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H), 4-coumarate CoA ligase (4CL), p-hydroxycinnamoyl-CoA: quinate shikimate p-hydroxycinnamoyltransferase (HCT), p-coumarate 3-hydroxylase (C3H), caffeoyl-CoA O-methyltransferase (CCOMT), hydroxycinnamyl-CoA reductase (CCR), ferulate 5-hydroxylase (F5H), caffeate O-methyltransferase (COMT), and cinnamyl alcohol dehydrogenase (CAD). Alternate pathways are in light grey. H subunits are only minor lignin components in dicots. Adapted from [1],[4].
Results
Characteristics of comt1 mutant and transgenic plants
Two allelic mutant lines for COMT1 in the Wassilewskija (WS) background were identified from the Versailles Arabidopsis insertion mutant collection [20]. The comt1a and comt1b mutants were homozygous for unique T-DNA insertions in the first exon and the first intron of the At5g54160 locus, respectively (Figure S1A). Comt1a has been shown to be a positive promoter-trap line expressing a functional ß-glucuronidase (GUS) gene under the control of the COMT1 promoter [15]. Comt1b harbors the GUS gene as an in-frame insertion in the first intron (Figure S1A), providing GUS activity upon transcriptional activation of COMT1. For mutant phenotype complementation, a poplar PtOMT1 cDNA was placed under the control of a constitutive promoter and transferred into the comt1a mutant. PtOMT1 was used to avoid the silencing effects observed in Arabidopsis when the COMT1 gene is overexpressed [15]. The complementation line, CpOMT14, had almost normal levels of COMT activity and S-lignin [15]. The exon-tagged comt1a mutant lacked the COMT1 mRNA signal found in wild-type and CpOMT14 plants on reverse transcription-PCR analysis (RT-PCR, Figure S1B). The T-DNA insertion in comt1b did not lead to a full gene knock-out, but did decrease levels of COMT1 mRNA (Figure S1B), which was correctly spliced and had a sequence identical to that of the wild-type COMT1 mRNA (data not shown).
Transcriptional activation of COMT1
The expression profile of COMT1 was analyzed by GUS staining plant tissues from the promoter-trap lines comt1a and comt1b. Identical profiles of GUS activity were detected in tissues from both lines (data not shown). Under normal growth conditions, GUS activity was restricted to tissues undergoing lignification, such as the vascular systems within aerial organs (Figure 2A–D) and the root central cylinder (Figure 2J). Additional constitutive expression was observed within the distal focus [21] of cotyledons (Figure 2C) and developing young leaves, within leaf primordia (Figure 2D) and root cap columella cells (Figure 2I), suggesting that COMT1 expression responds to venation patterning and auxin signaling [22]. Upon infection with the biotrophic oomycete leaf pathogen, H. arabidopsidis, COMT1 expression extended to mesophyll parenchyma in leaves (Figure 2E and 2F), cotyledons (Figure 2G), and the hypocotyl (Figure 2H). Expression was restricted to cells close to invading hyphae (Figure 2F). In roots, the biotrophic root-knot nematode, M. incognita, invades cortex cells, migrates to the root tip, enters the central cylinder, and moves upward before settling and inducing the formation of a feeding site [23]. This nematode activated COMT1 expression early in cells of the swelling gall (Figure 2K), in giant cells and surrounding dividing cells. COMT1 expression was maintained in the mature root gall until 21 days after inoculation (Figure 2L), in both the giant cells and their neighboring cells. Thus, parenchyma cells from different plant organs did not express COMT1 under normal growth conditions. However, the same cells responded to pathogen infection by local transcriptional activation of this gene.
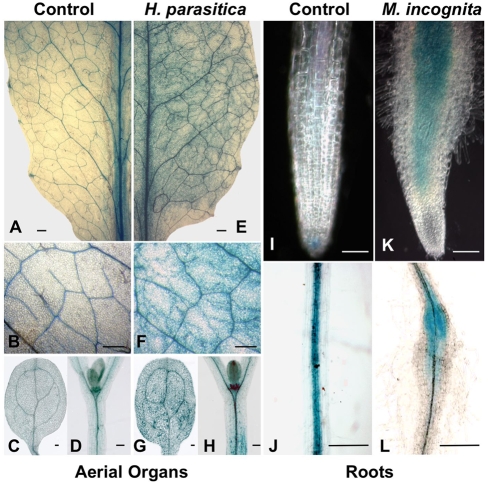
Figure 2. GUS histochemical analyses reporting pathogen-inducible transcriptional activation of COMT1 in A. thaliana.
(A–H) GUS activity in aerial organs, before (A–D), and 4 days post inoculation (dpi) with H. arabidopsidis (E–H). GUS staining occurred in the vascular system (A) and in veins (B) of adult leaves and cotyledons (C), as well as in leaf primordia (D), in non-infected plantlets. Upon infection, GUS staining extended to cells close to oomycete hyphae in the leaf lamina (E,F), in cotyledons (G), and in the hypocotyl (H). (I–L) COMT1 expression in roots, before (I,J), and after inoculation with M. incognita (K,L). Constitutive GUS activity was found in columella root cap cells (I), and in the central cylinder (J). Upon inoculation, GUS staining extended to all cells of the swelling gall, 7 dpi (K). After 21 days, GUS activity was found in nematode-induced giant cells and neighboring parenchyma cells of the feeding site (L). Scale bars represent 1 mm in (A,B,E,F), and 100 µm in (C,D,G–L).
Susceptibility of a comt1 mutant to microbial and nematode pathogens
We compared the susceptibility of wild-type plants and of the comt1a mutant to diverse pathogenic microorganisms, including fungi, bacteria, and nematodes (for details, see Protocol S1). We found the mutant significantly more susceptible to a strain of the necrotrophic fungus, Botrytis cinerea [24] (Figure S2A). Increased susceptibility was also observed for two other analyzed fungal pathogens, the necrotrophic Alternaria brassicicola [25] and the biotrophic Blumeria graminis f. sp. hordei (Bgh) [26] (Figure S2B and S2C). In addition, the comt1 mutant was found to be significantly more susceptible to the xylem-colonizing systemic bacterial pathogen Xanthomonas campestris pathovar campestris (Xcc) [27], and the bacterial speck agent, Pseudomonas syringae pv. tomato (Pst) (Figure S3A and S3B). The conclusion from these experiments was that a mutation of COMT1 generally weakens plant resistance to microbial pathogens. An exception from this observation was only found for the root-knot nematode M. incognita. Although the M. incognita infection activated COMT1 (Figure 2), a gene knockout did not influence the mean number of galls established three weeks after inoculation. It did neither affect nematode ability to complete its life cycle, which was characterized by similar amounts of egg masses produced by M. incognita two months after inoculation in mutant and wild-type plants (Figure S2D).
Susceptibility of comt1 mutants to H. arabidopsidis
Transcriptional activation of COMT1 occurred as a consequence of oomycete infection (Figure 2). We therefore analyzed whether an inactivation of COMT1 influences the interaction between H. arabidopsidis and A. thaliana under laboratory conditions. The virulent isolate Emwa1 completed its infection cycle on wild-type and comt1 mutant plants, resulting in asexual reproduction and the formation of conidia. However, sporulation levels were 40 to 50% lower on comt1 mutants than on wild-type plants (Figure 3A). This enhanced resistance phenotype of seedlings was partly complemented by expressing PtOMT1 in the comt1a mutant background (Figure 3A), and was confirmed on adult plants, with true leaves from different rosette stages displaying significantly lower levels of H. arabidopsidis sporulation in the absence of COMT1 (Figure 3B).
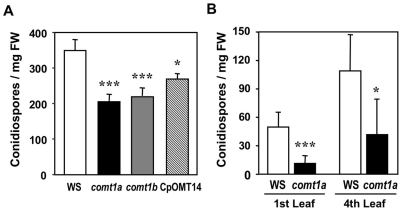
Figure 3. The mutation of COMT1 affects asexual sporulation of H. arabidopsidis.
(A) Sporulation of H. arabidopsidis isolate Emwa1 on cotyledons of Arabidopsis wild-type plantlets (WS), mutants (comt1a and comt1b), and complemented mutants (CpOMT14), 6 dpi. Conidiospores were harvested from 8 pooled plants and counted. The bars represent mean values±SD for 20 repetitions. (B) Sporulation of Emwa1 on the 1st and 4th fully developed leaf of 4 week-old Arabidopsis, 6 dpi. The bars represent mean values±SD from 10 individual leaves. All experiments were repeated three times and gave similar results. Statistically significant differences for values compared with the wild type were determined by Student's t-test (* P<0.01, ** P<0.001, *** P<0.0001).
We investigated whether the observed phenotype of comt1 mutants resulted from a gain-of-function in defense signaling, by assessing constitutive and inducible responses before and after inoculation with H. arabidopsidis. The markers used for the activation of salicylic acid (SA)- and jasmonic acid (JA)-dependent responses were the transcriptional activation of PR-1a and PDF1.2b, respectively. In quantitative RT-PCR experiments, PR-1a mRNA was undetectable in untreated wild-type and mutant plants. PDF1.2b transcripts were present at similar, low levels in both lines tested in the absence of inoculation. The accumulation of transcripts from both genes was induced by the pathogen, to similar levels in wild-type and mutant plants (Figure S4). The enhanced resistance phenotype of comt mutants thus appeared not to be dependent on SA and JA-dependent defense signaling pathways in Arabidopsis.
Development of the oomycete pathogen within comt mutants
Following the inoculation of wild-type A. thaliana, H. arabidopsidis spores germinate on leaf surfaces and form appressoria, enabling the penetration pegs to overcome the cuticle. Once inside the leaf, the hyphae grow intercellular, branch, and establish a filamentous network spanning the entire infected leaf three to four days after the onset of infection (Figure 4A). The growing hyphae locally digest the cell walls of almost all the plant cells they come into contact with, leading to invagination of the host plasma membrane and the formation of intracellular bulbous structures called haustoria (Figure 4D). Haustoria are feeding sites required for the biotrophic lifestyle of the oomycete. Four to six days after inoculation under laboratory conditions, the hyphae use stomatal openings to form conidiophores on the leaf surface and initiate asexual reproduction [28]. In comt1 mutant lines, the initial infection process was identical to that in wild-type plants. H. arabidopsidis penetrated, formed a filamentous network of branched hyphae (Figure 4B), and developed haustoria within host cells. We quantified the development of the oomycete in wild-type and mutant Arabidopsis, by performing RT-PCR with gene-specific primers to generate an amplicon within the intergenic transcribed spacer (ITS2) of H. arabidopsidis ribosomal RNA (Figure 4C). These experiments showed that the oomycete expanded similarly in wild-type and mutant plants (Figure 4C), indicating that hyphal growth and branching were not affected in the mutants. However, microscopic analyses of haustoria indicated that the structure of the feeding sites was less stable in comt1 mutant plant cells than in wild-type cells. Disintegration of the haustorium was observed in mutant cells (Figure 4E) but never in the wild-type background. Even more strikingly, the frequency of sexual reproduction was found to be higher in comt1 mutants, with a significantly larger number of oospores within mutant tissues than within wild-type tissues (Figure 4F). This phenotype was almost complemented in the CpOMT14 line (Figure 4F). Thus, the lower level of conidiospore formation in comt1 mutants (Figure 3) coincided with a higher frequency of sexual reproduction.
Figure 4. The mutation of COMT1 impairs haustorium stability, and favors sexual reproduction.
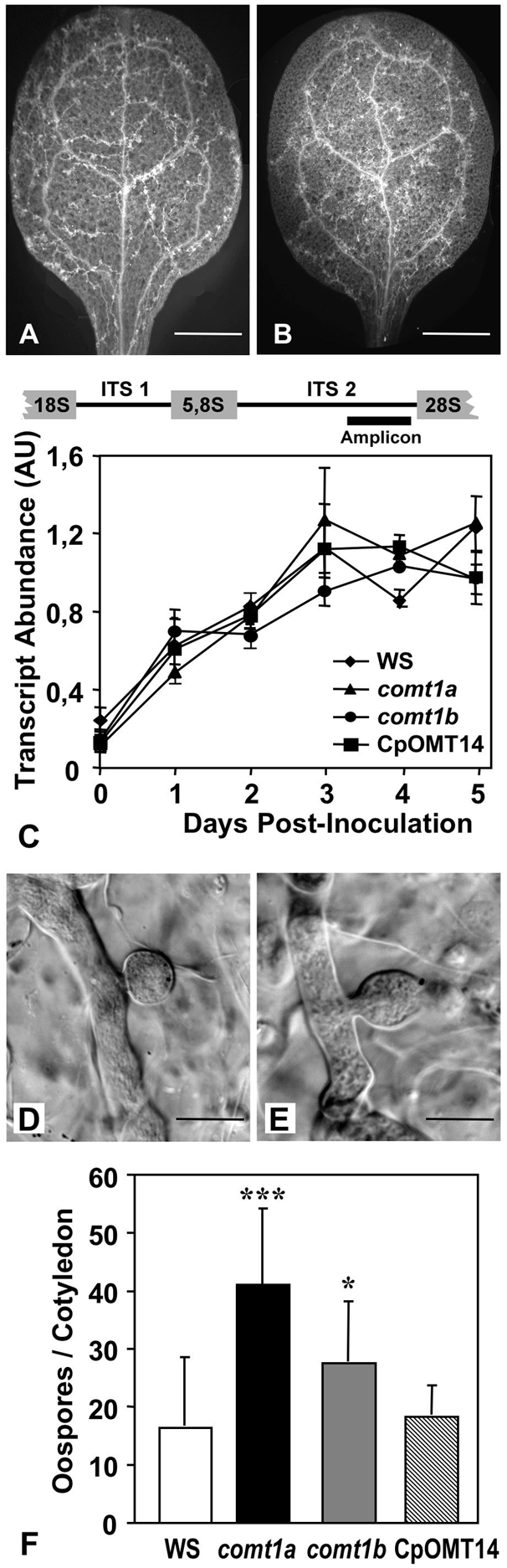
(A,B) General view of the calcofluor-stained H. arabidopsidis hyphal network in cotyledons of wild-type Arabidopsis (A) and the comt1a mutant (B), 3 dpi. (C) Quantification of the H. arabidopsidis ITS2 amplicon as a marker for oomycete biomass. RT-PCR was performed using total RNA, which was extracted at different time points post inoculation from infected cotyledons of Arabidopsis wild-type plants (WS), mutants (comt1a and comt1b), and complemented mutants (CpOMT14). RNA abundance is expressed as the signal intensity ratio between ITS2 and OXA1 amplicons. Displayed are means±SD from 2 repetitions of the experiment. (D,E) Shape of haustoria in host cells from a wild-type (D) and a comt1a mutant (E) plant. Shown are differential interference contrast micrographs of calcofluor-stained hyphae and haustoria within infected tissues, 3 dpi. Scale bars represent 5 µm. (F) Frequency of sexual reproduction of H. arabidopsidis within infected tissues of Arabidopsis wild-type plants, mutants, and the complemented mutant, expressed as the number of oospores formed per infected cotyledon. The bars represent mean values±SD from 20 individual cotyledons. The experiment was repeated three times and gave similar results. Statistically significant differences for values compared with the wild type were determined by Student's t-test (* P<0.01, *** P<0.0001).
Metabolic disequilibrium in comt1 mutants, and its impact on oomycetes
Plant cells store excess monolignol precursors not incorporated into lignin as soluble esters. In the leaves and cotyledons of A. thaliana, the major accumulating soluble hydroxycinnamate ester is sinapoyl malate (SM) [29]. In the comt1a mutant, SM levels are much lower, with these mutants instead accumulating hydroxyferuloyl malate (OH-FM), which is a derivative of the COMT1 substrate, 5-hydroxyconiferaldehyde [4],[15].
We quantified the accumulation of hydroxycinnamoyl malate esters in the various lines used in this study and evaluated the biological activities of these compounds, by synthesizing SM, OH-FM, and 2-O-feruloyl-L-malate (feruloyl malate; FM) (see Protocol S2, and Figure S5). On reverse-phase high-performance liquid chromatography (HPLC) of methanolic extracts from four-week-old plantlets, FM was not detected in any of the Arabidopsis lines. Wild-type plants accumulated soluble SM to a concentration of about 280 nmol/g fresh weight and OH-FM was not detectable in these plants (Figure 5A). By contrast, the mutant lines comt1a and comt1b accumulated OH-FM to concentrations of 200 nmol and 150 nmol/g fresh weight, respectively, whereas SM levels were about 70% lower than wild-type levels in comt1a, and 55% lower in comt1b (Figure 5A and Figure S3). SM was again the main compound detected in seedlings from the complemented mutant line CpOMT14 (Figure 5A). The COMT1 mutation modified the SM/OH-FM ratio, but did not modify total hydroxycinnamoyl malate ester concentrations, which appeared to be similar in plants from all lines.
Figure 5. 2-O-5-hydoxyferuloyl-L-malate accumulates in comt1 mutants, and promotes oomycete sexual reproduction in vitro.
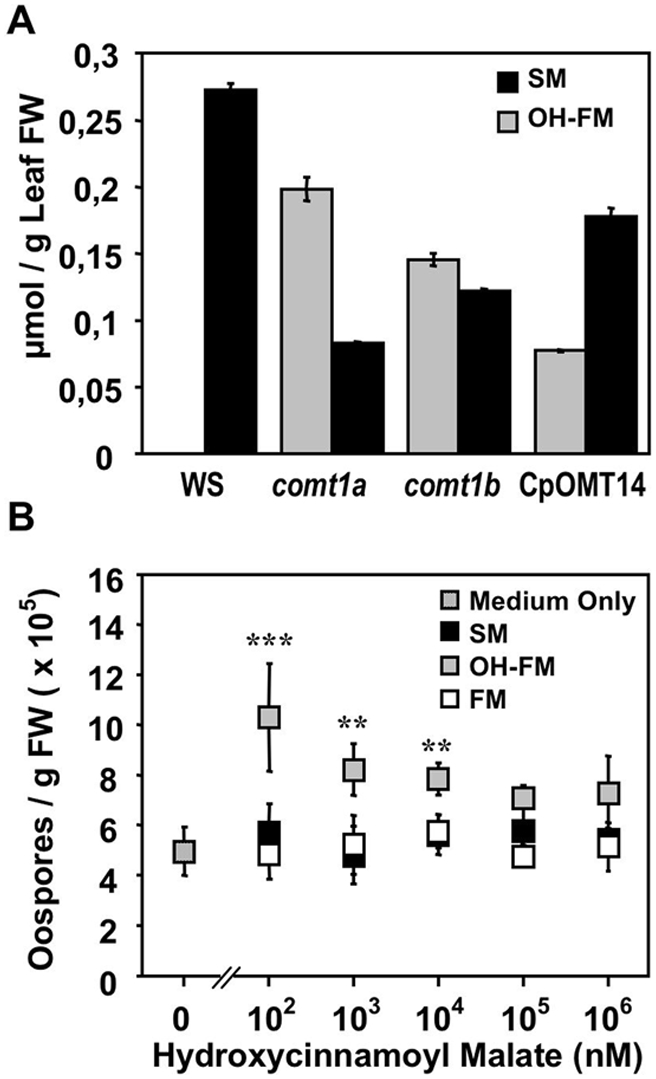
(A) Quantification of sinapoyl malate (SM) and 5-hydroxyferuloyl malate (OH-FM) in methanolic extracts of 4 week-old plantlets from the different A. thaliana lines. The compounds were quantified by HPLC referring to synthetic standards. The bars represent mean values±SD from 3 experiments. (B) Effect of different concentrations of SM, OH-FM, and feruloyl malate (FM) on sexual reproduction of Phytophthora cactorum isolate 723 in vitro. Displayed are means±SD from 4 repetitions. Statistically significant differences for OH-FM values compared with values in the absence of the compound were determined by Student's t-test (** P<0.001, *** P<0.0001). FW = fresh weight.
The correlation between the accumulation of soluble OH-FM (Figure 5A) and the frequency of sexual reproduction (Figure 4F) in the comt1 mutant lines led us to analyze whether this compound was able to stimulate oospore formation. We thus developed an in vitro assay for the sexual reproduction of another homothallic oomycete plant pathogen, Phytophthora cactorum. Using P. cactorum zoopores, we synchronized the age and density of hyphae in titer plate wells before adding OH-FM, SM, or FM at various concentrations. In the absence of these compounds, P. cactorum produced about 5,000 oospores (per g fresh weight of mycelium). The addition of SM or FM did not alter the frequency of sexual reproduction of the oomycete (Figure 5B). In contrast, supplementation of the medium with OH-FM significantly stimulated sexual reproduction, and the number of oospores doubled when OH-FM was added to concentrations of about 0.1 µM (Figure 5B). Beyond 0.1 mM, the effect of OH-FM on sexual reproduction was less obvious. At these concentrations, OH-FM appeared to influence the integrity of the hyphal network, and the mycelium developing in the presence of the compound became frail. These findings strongly suggest that the accumulation of OH-FM can account for the greater sexual activity of H. arabidopsidis in comt1 mutants. However, OH-FM did not stimulate the sexual reproduction of individual mating types of the heterothallic oomycete, P. parasitica (data not shown). OH-FM therefore cannot replace an oomycete mating hormone.
Discussion
In recent decades, lignin has become a privileged target for genetic engineering approaches aiming to increase the industrial processing efficiency of plant biomass. However, the pros and cons of the use of such transgenic plants in open-field situations have only recently begun to be explored [30]–[32]. Systems biology approaches have been used to investigate the extent to which a single engineered gene encoding a protein involved in the lignin biosynthesis pathway interferes with other metabolic pathways, and the extent to which it modifies interactions between the modified plant and its environment [33]. The downregulation of CAD and CCR (Figure 1) in tobacco has been shown to affect not only lignin biosynthesis, but also primary metabolism, stress metabolism, and photorespiration [34]. In poplar, the downregulation of CCR affects the overall metabolism and structure of cell wall polymers [35]. This enzyme also directly regulates pathogen defense signaling in rice [3], indicating that lignin biosynthesis plays a more subtle role in plant defense responses against herbivores [36] and microbes [37] than simply constituting a physical barrier.
This report provides the first large-scale analysis of the role of COMT in interactions between the plant and its biotic environment. We found that the transcriptional activation of COMT1 in parenchyma tissues from roots and aerial organs of Arabidopsis was stimulated by pathogen infection. COMT1 expression was shown before to be enhanced upon Arabidopsis leaf infiltration with either the nonhost bacterium P. syringae pv. phaseolicola [38], bacterial flagellin (flg22) [38],[39], or harpin (HrpZ) [38],[40], or with the necrosis-inducing Phytophthora protein 1 (NPP1) [38],[41]. The transcription of COMT1 appears thus to correlate with the onset of the plant's pathogen-associated molecular pattern (PAMP)-triggered immune (PTI) response [42]. In the Arabidopsis leaf mesophyll, we found an accumulation of toluidine blue-stainable, lignin-like deposits in host cells that surround H. arabidopsidis hyphae (data not shown). Taken together, we suppose that the observed COMT1 promoter activation in response to infection reflects an onset of PTI within host cells and an attempt to reinforce wall rigidity through lignification. Consequently, we found that COMT1 downregulation increased susceptibility to at least three fungal and two bacterial pathogens. Mutants were significantly more susceptible to a moderately aggressive strain of the necrotrophic fungus, B. cinerea, but not to a highly virulent isolate. These findings indicate that COMT1 contributes to limiting the spread of manageable strains of the fungus. The comt1a mutant was also more susceptible to another necrotrophic fungal pathogen, Alternaria brassicicola. This fungus causes black spot disease on almost all cultivated Brassica species including broccoli, cabbage, canola and mustard. It is of worldwide economic importance, because it reduces crop yields and the quality of canola oil [43]. From a human health perspective, Alternaria brassicicola belongs to a genus of fungi considered one of the most potent sources of mold-derived allergens [44]. The resistance of A. thaliana to this fungus requires the phytoalexin camalexin and the signaling molecule JA, but is independent of SA [25]. The downregulation of COMT1 weakened the JA-dependent defenses of Arabidopsis against this pathogen. Unlike B. cinerea and Alternaria brassicicola, Bgh is a biotrophic fungal pathogen causing powdery mildew on barley. Arabidopsis is a nonhost for this pathogen, and SA-dependent defense responses are activated rapidly when the pathogen attempts to penetrate [26]. The higher proportion of successful infections in the mutant indicated that the mutation of COMT1 weakens this penetration resistance of A. thaliana.
Mutants were also more susceptible to the bacterial pathogens Pst carrying avrPphB, and Xcc. AvrPphB is recognized by the corresponding resistance gene product, RPS5, which is present in the WS background, but absent in the Ler background [45]. Plants of these two ecotypes are thus resistant and susceptible, respectively, to Pst avrPphB. The observation that COMT1 downregulation renders WS as susceptible as Ler indicates that the gene plays also an important role in avrPphB-mediated resistance to the bacterial pathogen.
Two exceptions to the trend of comt1a mutants being generally more susceptible to pathogens were observed after inoculation with the root-knot nematode, M. incognita, and the biotrophic oomycete pathogen, H. arabidopsidis. M. incognita triggers the transcriptional activation of COMT1, but knocking out this gene had no effect on disease establishment (gall formation), or nematode development (egg masses). However, all the Arabidopsis ecotypes analyzed to date are susceptible to M. incognita. Variety-specific differences in defense and resistance influencing nematode development, such as those within the Solanaceae [46], are not known for A. thaliana. We therefore cannot exclude the possibility that COMT1 may affect resistance phenotypes in other plant species. In this context, it has been reported that a downregulation of the COMT1 ortholog in tobacco [47], leads to increased M. incognita reproduction [48].
The obligate oomycete pathogen H. parasitica causes downy mildew disease on agronomically important Brassicaceae, such as rapeseed and cabbage. Its relative, H. arabidopsidis only infects A. thaliana in clear gene-for-gene relationships and is nonpathogenic on other crucifers tested [49]–[51]. WS harbors genes from the RPP1 group (RPP1-WsA, RPP1-WsB, and RPP1-WsC), which confer resistance to the H. arabidopsidis isolate Noco2, but not to Emwa1 [50]. These two isolates thus give rise to genetically incompatible and compatible interactions, respectively, with WS. In Arabidopsis seedlings inoculated with Noco2, comt1 mutants displayed the same resistance phenotype as WS, with no hyphal development detectable in any of the lines tested (data not shown). COMT1 is thus not a key enzyme for the genetic resistance of Arabidopsis to oomycetes. However, the virulent H. arabidopsidis isolate Emwa1 produced significantly fewer conidiospores on COMT1 mutants than on wild-type plants. According to the generally accepted criterion for analyzing plant susceptibility to this oomycete [52], comt1 mutants were thus more resistant. To date, several Arabidopsis genes have been identified, which are required for full susceptibility to biotrophic fungal and oomycete pathogens, and which confer, when inactivated, increased resistance phenotypes to mutants. Vogel and coworkers identified 26 recessive powdery mildew resistance (pmr) mutants in a genetic screen [53]. PMR6 codes for a pectate lyase [54], and PMR2 is the Arabidopsis ortholog of the barley mlo gene, Atmlo2 [55]. The MLO protein is required for successful entry of Bgh into the host cell [56]. Both pmr6 and Atmlo2 mutants were not affected in susceptibility to H. arabidopsidis [54],[55], indicating that the downy mildew does not require the host cell pectate lyase and MLO for successful infection. However, another pmr mutant, pmr4 (or gsl5) is more resistant to both powdery- and downy mildews [53]. PMR4 codes for a callose synthase, which seems to be involved in yet unknown functions required for functional haustoria formation [57], and which negatively regulates SA defense signaling in the plant [58]. Since, six additional Arabidopsis loci were identified that confer, when mutated, resistance to H. arabidopsidis [59]. For three of the downy mildew resistant (dmr) mutants, enhanced resistance was correlated with a constitutive expression of the SA-dependent PR-1a gene [59]. Of the remaining DMR genes, DMR6 was identified to code for a 2-oxoglutarate (2OG)-Fe(II) oxygenase of unknown function [60]. The role of DMR6 for disease susceptibility is not yet known, but it also appears to be a negative regulator of defense gene activation [60]. Here, we showed that the increased resistance of comt1 mutants was not correlated with a change in PR-1a and PDF1.2 gene expression, when compared to wild-type plants. The inactivation of COMT1 appears, therefore, not to interfere with SA- and JA-dependent defense signaling pathways.
The absence of COMT1 did not impair intercellular hyphal growth and branching, and did not interfere with overall oomycete biomass development in mutant plant tissues. However, H. arabidopsidis interacting with mutant plants displayed haustorial instability and enhanced sexual reproduction as two additional phenotypes to reduced asexual sporulation. We found that the occurrence of these phenotypes correlated with a metabolic difference between comt1 mutants and wild-type plants, i.e. lower levels of SM within mutants and accumulation of OH-FM instead. Synthetic OH-FM promoted the sexual reproduction of P. cactorum in vitro, whereas the closely related derivative SM, which was present in large amounts in wild-type Arabidopsis, and synthetic FM did not. Moreover, high OH-FM concentrations decreased the mechanical resistance of the P. cactorum hyphal network in vitro, indicating that the compound is toxic for oomycetes and likely responsible for the observed haustorial instability phenotype of H. arabidopsidis in comt1 mutants. These findings make OH-FM accumulation a potential candidate cause for the phenotypes we observed during the interaction between comt1 mutants and H. arabidopsidis. In this context, it might be possible that the observed transcriptional activation of COMT1 in host cells close to downy mildew hyphae has another reason than the above discussed stimulated lignification defense. In a hypothetical scenario, an H. arabidopsidis virulence function might promote host COMT1 expression, in order to prevent plant cells harboring oomycete haustoria from any accumulation of detrimental OH-FM. However, OH-FM was not detectable in wild-type Arabidopsis by the means we used for the present study. To prove the hypothetical scenario, more sensitive detection methods for OH-FM need to be developed to compare the accumulation of microquantities of the compound in living cells harboring or not haustoria.
In conclusion, this study demonstrates that manipulating the expression of single genes within the monolignol biosynthetic pathway may affect the interaction of engineered plants with the biotic environment. COMT has been a key target for such manipulation in the past [13]. The downregulation of COMT in agronomically important plants may affect susceptibility to fungal and bacterial attack in a similar manner, as shown here for A. thaliana. The metabolic disequilibrium generated by COMT1 knockout in Arabidopsis probably also creates a selective pressure in other crops that forces oomycete pathogens to undergo sexual reproduction. Sexual reproduction is a source of genetic variation even in homothallic oomycetes [61],[62]. Furthermore, oospores persist in the soil [63],[64] and are insensitive to fungicides [65], and thus make disease control difficult. The stimulation of oomycete sexual reproduction in genetically engineered COMT plants may, therefore, lead to the evolution of novel infection traits.
Materials and Methods
Plant material and growth conditions
All Arabidopsis lines used for the experiments were from the Wassilewskija (WS) genetic background. The comt1a mutant, previously named Atomt1 [15] and comt1 [4], and the complemented line CpOMT14 have been described before [15]. The comt1b mutant from the Versailles T-DNA insertion collection [18] was obtained from Dr. Laurent Nussaume (CEA, Cadarache, France). Analysis of the T-DNA flanking region within comt1b was performed by sequencing the amplicon obtained with primer pair 3 and 4 on genomic DNA as the template (Figure S1). Plants were grown in sand supplemented with MS medium in growth chambers at 20°C with a 12 h photoperiod.
Pathogen assays
H. arabidopsidis isolates Emwa1 and Noco2 were obtained from Dr. Jane Parker (MPIZ, Cologne, Germany), and transferred weekly onto the genetically susceptible Arabidopsis accessions WS and Columbia (Col), respectively, as described [66]. For infection, 10-day-old plants were spray-inoculated to saturation with a spore suspension of 40,000 spores/ml. Plants were kept in a growth cabinet at 16°C for 3 d with a 16 h photoperiod. Sporulation was then induced by spraying plants with water, and keeping them for 48 hours under high humidity. To evaluate conidiospore production, pools of 8 plants were harvested in 1 ml of water. After vortexing, the amount of liberated spores was determined with a hemocytometer. To evaluate oospore production, inoculated cotyledons were destained in 80% ethanol, mounted on glass slides, and analyzed by microscopy.
For nematode infection, A. thaliana were grown in vitro on MS medium containing 1% sucrose and 0.7% plant cell culture-tested agar (Sigma-Aldrich). One hundred surface-sterilized freshly hatched M. incognita J2 larvae were added to each 2-week-old seedling, as described [67]. The plates were kept at 20°C with a 16 h photoperiod.
Pathogenicity assays with B. cinerea, A. brassicicola, B. graminis, P. syringae, and X. campestris were performed as described in Protocol S1.
Histochemical analyses
GUS activity in comt1a mutants was analyzed histochemically [68]. For the observation of oomycete development in infected tissues, cotyledons were fixed in 0.1 M glutaraldehyde and 1.5 M formaldehyde in phosphate buffer, bleached in a series of increasing ethanol concentrations, and stained with 0.3% Fluorescent Brightener 28 [69]. Calcofluor-stained hyphae were observed by fluorescence binocular microscopy, or a Zeiss Axioplan 2 fluorescence microscope configured for brightfield, darkfield, and differential interference contrast (excitation 480 nm, barrier filter 510 nm).
RNA methods
Total RNA was extracted from A. thaliana seedlings using TRIZOL Reagent (Invitrogen) following the instructions of the manufacturer. One µg RNA was reverse-transcribed using the iScript cDNA Synthesis Kit (Biorad). For H. arabidopsidis ITS2 RNA quantification within infected tissues, PCR (25 cycles) was performed with 8 ng cDNA as template using the ITS2 forward primer 5′-TGTGGTAGACGAATGGGTGA-3′, and the ITS2 reverse primer 5′-AAGTGCAGCCGAAGCTTTAC-3′. PCR with the primer pairs OXA1-a and OXA1-b (Figure S1) was used as a quantitative control. Aliquots of individual PCR products were resolved by agarose gel electrophoresis, visualized with ethidium bromide and quantified using a Fujifilm FLA-3000 Phospho/Fluoroimager. The expression of PR-1a and PDF1.2b was analyzed by real-time quantitative PCR using the forward primer 5′-GGAGCTACGCAGAACAACTAAGA-3′ and reverse primer 5′-CCCACGAGGATCATAGTTGCAACTGA-3′ for PR-1a, and the forward primer 5′-TCATGGCTAAGTTTGCTTCC-3′, and reverse primer 5′-AATACACACCACGATTTAGCACC-3′ for PDF1.2b. Amplification and detection were performed in the Chromo4 detection system (Biorad). Reactions were done in a final volume of 15 µl containing 10 µl qPCR MasterMix Plus For SYBRGreen I No Rox (Eurogentec), 0.5 mM of each primer, and 8 ng of cDNA template. PCR conditions were as follows: 95°C for 15 min, followed by 40 cycles of 95°C for 15 s, 56°C for 30 s and 72°C for 30 s. At the end of the program a melting curve (from 60°C to 95°C, read every 0.5°C) was determined to ensure that only single products were formed. Ubiquitin-specific protease 22 (UBP22) expression was used to normalize the transcript level in each sample with the primer pairs 5′-GCCAAAGCTGTGGAGAAAAG-3′ and 5′-TGTTTAGGCGGAACGGATAC-3′. Data analysis was performed using the MJ OpticonMonitor Analysis software (version 3.1; Biorad).
Extraction and analysis of hydroxycinnamoyl malates
Extraction of soluble phenolics from fresh plants was carried out in 100% methanol (0.4 ml/100 mg of fresh weight). After centrifugation, the cleared supernatant was adjusted to 80% aequeous methanol, and passed through a 200 mg C18 (Nucleodur 100-30, Macherey-Nagel, Düren, Germany), in order to stop chorophyll and lipidic compounds. The unretained fraction was directly analyzed by HPLC. The liquid chromatograph (System Controller 680 with two 510 pumps; Waters Millipore, Milford, MA) was equipped with an Inertsil 5ODS3 C18 column (5 µm, 250×4,6 mm i.d.; Interchim, Montluçon, France). Samples were chromatographied with the following gradient (flow rate l ml/min): 1 min isocratic 15% solvent A (methanol) and 85% solvent B (water, 0.5% H3PO4), then within 29 min to 75% solvent A, then within 3 additional min to 100% solvent A, followed by a 3 min isocratic step in 100% solvent A. Compounds were detected by a Waters TM 996 Photodiode Array Detector (200 to 500 nm). Peaks were identified and quantified using the Empower software (Waters), after external standardization with synthetic compounds.
Analysis of oomycete reproduction in vitro
The P. cactorum isolate 723 was from the Sophia Antipolis Phytophthora collection, and was sampled in 2005 from Fragaria in France. Propagation and zoospore production were performed as described [70], and 5,000 zoospores were pipetted into 0.5 ml of V8 medium [70] in 24-well titer plates. After 24 h at 24°C, another 0.5 ml of V8 medium containing or not the hydroxycinnamoyl malates were added. After an incubation for further 48 h at 24°C, mycelium was picked out of the wells, rinsed, dried on filter paper, weighed, placed into 1 ml of water, and macerated in a potter. Liberated oospores were enumerated with a hemocytometer and the number of oospores was expressed per g mycelium fresh weight.
Gene accession numbers
Sequences used in this article were derived from gene IDs 835504 (COMT1), 169452 (PtOMT1), 1815949 (PR1-a), 817143 (PDF1.2b), 836325 (OXA1), 830946 (UBP22).
Supporting Information
Molecular analyses of the mutants and transgenic lines used in this study.
(0.57 MB PDF)
Susceptibility of COMT1 knock-outs to fungal pathogens and a nematode.
(0.82 MB PDF)
Susceptibility of COMT1 knock-outs to bacterial pathogens.
(0.11 MB PDF)
Comt1a mutants are not altered in SA- and JA-dependent defense responses.
(0.10 MB PDF)
Reversed-phase HPLC of synthetic hydroxycinnamoyl malate esters and of methanolic plant extracts.
(0.30 MB PDF)
Characteristics of fungal and bacterial pathogens, and inoculation procedures.
(0.03 MB DOC)
Synthesis and spectral analyses of hydroxycinnamoyl malates.
(0.03 MB DOC)
Acknowledgments
The authors thank Marie-Line Kuhn and Catherine Mura for maintenance of oomycetes in vitro, Nicolas Marfaing for care of Arabidopsis, Maurice Tronchet for bacterial tests, Pascal Le Pècheur for analyzing Botrytis resistance, and Benoit Industri for help with the HPLC analyses. We are grateful to Dr. Laurent Nussaume (CEA, Cadarache, France) for A. thaliana line comt1b, to Dr Jane Parker (MPIZ Cologne, Germany) for H. arabidopsidis isolates Noco2 and Emwa1, to Dr. Franck Panabières for discussions concerning oomycete lifestyles, and to Philippe Lecomte for critical reading of the manuscript.
Footnotes
The authors have declared that no competing interests exist.
This work was supported with funding by a joined French-German plant genomics program (Génoplante-GABI 2003-14), by the German Federal Ministry of Education and Research (0313096), and by the French National Research Agency (ANR-GNP05024G).
References
- 1.Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annu Rev Plant Biol. 2003;54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- 2.Boyce CK, Zwieniecki MA, Cody GD, Jacobsen C, Wirick S, et al. Evolution of xylem lignification and hydrogel transport regulation. Proc Natl Acad Sci U S A. 2004;101:17555–17558. doi: 10.1073/pnas.0408024101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawasaki T, Koita H, Nakatsubo T, Hasegawa K, Wakabayashi K, et al. Cinnamoyl-CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice. Proc Natl Acad Sci U S A. 2006;103:230–235. doi: 10.1073/pnas.0509875103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Do CT, Pollet B, Thévenin J, Sibout R, Denoue D, et al. Both caffeoyl Coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta. 2007;226:1117–1129. doi: 10.1007/s00425-007-0558-3. [DOI] [PubMed] [Google Scholar]
- 5.Lewis NG, Yamamoto E. Lignin: occurrence, biogenesis and biodegradation. Annu Rev Plant Physiol Plant Mol Biol. 1990;41:455–496. doi: 10.1146/annurev.pp.41.060190.002323. [DOI] [PubMed] [Google Scholar]
- 6.Emerson RA, Beadle GW, Fraser AC. A summary of linkage studies in maize. Cornell Univ Agric Exp Stn Memoirs. 1935;180:1–83. [Google Scholar]
- 7.Lapierre C, Tollier MT, Monties B. Mise en évidence d'un nouveau type d'unité constitutive dans les lignines d'un mutant de maïs bm3. C R Acad Sci Paris. 1988;307:723–728. [Google Scholar]
- 8.Grand C, Parmentier P, Boudet A, Boudet AM. Comparison of lignins and of enzymes involved in lignification in normal and brown midrib (bm3) mutant corn seedlings. Physiol Veg. 1985;23:905–911. [Google Scholar]
- 9.Anterola AM, Lewis NG. Trends in lignin modification: a comprehensive analysis of the effects of genetic manipulations/mutations on lignification and vascular integrity. Phytochemistry. 2002;61:221–294. doi: 10.1016/s0031-9422(02)00211-x. [DOI] [PubMed] [Google Scholar]
- 10.Barnes RF, Muller LD, Bauman LF, Colenbrander VF. In vitro dry matter disappearance of brown midrib mutants of maize (Zea mays L.). J Anim Sci. 1971;33:881–884. [Google Scholar]
- 11.Guo D, Chen F, Wheeler J, Winder J, Selman S, et al. Improvement of in-rumen digestibility of alfalfa forage by genetic manipulation of lignin O-methyltransferases. Transgenic Res. 2001;10:457–464. doi: 10.1023/a:1012278106147. [DOI] [PubMed] [Google Scholar]
- 12.Jouanin L, Goujon T, de Nadaï V, Martin MT, Mila I, et al. Lignification in transgenic poplars with extremely reduced caffeic acid O-methyltransferase activity. Plant Physiol. 2000;123:1363–1374. doi: 10.1104/pp.123.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pilate G, Guiney E, Holt K, Petit-Conil M, Lapierre C, et al. Field and pulping performances of transgenic trees with altered lignification. Nat Biotechnol. 2002;20:607–612. doi: 10.1038/nbt0602-607. [DOI] [PubMed] [Google Scholar]
- 14.Zuber MS, Colbert TR, Bauman LF. Effect of brown-midrib-3 mutant in maize (Zea mays L.) on stalk strength. Z Pflanzenzüchtg. 1977;79:310–314. [Google Scholar]
- 15.Goujon T, Sibout R, Pollet B, Maba B, Nussaume L, et al. A new Arabidopsis thaliana mutant deficient in the expression of O-methyltransferase impacts lignins and sinapoyl esters. Plant Mol Biol. 2003;51:973–989. doi: 10.1023/a:1023022825098. [DOI] [PubMed] [Google Scholar]
- 16.Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W. Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 2003;133:1051–1071. doi: 10.1104/pp.103.026484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Attard A, Gourgues M, Galiana E, Panabières F, Ponchet M, et al. Strategies of attack and defense in plant-oomycete interactions, accentuated for Phytophthora parasitica Dastur (syn. P. Nicotianae Breda de Haan). J Plant Physiol. 2008;165:83–94. doi: 10.1016/j.jplph.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 18.McKay R. The longevity of the oospores of onion downy mildew Peronospora destructor (Berk.) Casp. Sci Proc R Dublin Soc Nat Sci. 1957;27:295–307. [Google Scholar]
- 19.Judelson HS, Blanco FA. The spores of Phytophthora: weapons of the plant destroyer. Nat Rev Microbiol. 2005;3:47–58. doi: 10.1038/nrmicro1064. [DOI] [PubMed] [Google Scholar]
- 20.Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris. 1993;316:1194–1199. [Google Scholar]
- 21.Mattsson J, Ckurshumova W, Berleth T. Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol. 2003;131:1327–1339. doi: 10.1104/pp.013623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cnops G, Neyt P, Raes J, Petrarulo M, Nelissen H, et al. The TORNADO1 and TORNADO2 genes function in several patterning processes during early leaf development in Arabidopsis thaliana. Plant Cell. 2006;18:852–866. doi: 10.1105/tpc.105.040568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caillaud MC, Dubreuil G, Quentin M, Perfus-Barbeoch L, Lecomte P, et al. Root-knot nematodes manipulate plant cell functions during a compatible interaction. J Plant Physiol. 2008;165:104–113. doi: 10.1016/j.jplph.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 24.Choquer M, Fournier E, Kunz C, Levis C, Pradier JM, et al. Botrytis cinerea virulence factors: new insights into a necrotrophic and polyphageous pathogen. FEMS Microbiol Lett. 2007;277:1–10. doi: 10.1111/j.1574-6968.2007.00930.x. [DOI] [PubMed] [Google Scholar]
- 25.Van Wees SC, Chang HS, Zhu T, Glazebrook J. Characterization of the early response of Arabidopsis to Alternaria brassicicola infection using expression profiling. Plant Physiol. 2003;132:606–617. doi: 10.1104/pp.103.022186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zimmerli L, Stein M, Lipka V, Schulze-Lefert P, Somerville S. Host and non-host pathogens elicit different jasmonate/ethylene responses in Arabidopsis. Plant J. 2004;40:633–646. doi: 10.1111/j.1365-313X.2004.02236.x. [DOI] [PubMed] [Google Scholar]
- 27.Lummerzheim M, De Oliveira D, Castresana C, Miguens FC, Louzada E, et al. Identification of compatible and incompatible interactions between Arabidopsis thaliana and Xanthomonas campestris pv. campestris and characterization of the hypersensitive response. Mol Plant Microbe Interact. 1993;6:532–544. [Google Scholar]
- 28.Koch E, Slusarenko A. Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell. 1990;2:437–445. doi: 10.1105/tpc.2.5.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nair RB, Bastress KL, Ruegger MO, Denault JW, Chapple C. The Arabidopsis thaliana REDUCED EPIDERMAL FLUORESCENCE1 gene encodes an aldehyde dehydrogenase involved in ferulic acid and sinapic acid biosynthesis. Plant Cell. 2004;16:544–554. doi: 10.1105/tpc.017509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herrera S. Struggling to see the forest through the trees. Nat Biotechnol. 2005;23:165–167. doi: 10.1038/nbt0205-165. [DOI] [PubMed] [Google Scholar]
- 31.Talukder K. Low-lignin wood—a case study. Nat Biotechnol. 2006;24:395–396. doi: 10.1038/nbt0406-395. [DOI] [PubMed] [Google Scholar]
- 32.Hopkins DW, Webster EA, Boerjan W, Pilate G, Halpin C. Genetically modified lignin below ground. Nat Biotechnol. 2007;25:168–169. doi: 10.1038/nbt0207-168. [DOI] [PubMed] [Google Scholar]
- 33.Vanholme R, Morreel K, Ralph J, Boerjan W. Lignin engineering. Curr Opin Plant Biol. 2008;11:278–285. doi: 10.1016/j.pbi.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 34.Dauwe R, Morreel K, Goeminne G, Gielen B, Rohde A, et al. Molecular phenotyping of lignin-modified tobacco reveals associated changes in cell-wall metabolism, primary metabolism, stress metabolism and photorespiration. Plant J. 2007;52:263–285. doi: 10.1111/j.1365-313X.2007.03233.x. [DOI] [PubMed] [Google Scholar]
- 35.Leplé JC, Dauwe R, Morreel K, Storme V, Lapierre C, et al. Downregulation of cinnamoyl-coenzyme A reductase in poplar: multiple-level phenotyping reveals effects on cell wall polymer metabolism and structure. Plant Cell. 2007;19:3669–3691. doi: 10.1105/tpc.107.054148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wainhouse D, Cross DJ, Howell RS. The role of lignin as a defence against the spruce bark beetle Dendroctonus micans: effect on larvae and adults. Oecologia. 1990;85:257–265. doi: 10.1007/BF00319411. [DOI] [PubMed] [Google Scholar]
- 37.Mörschbacher B, Noll U, Gorrichon L, Reisener HJ. Specific inhibition of lignification breaks hypersensitive resistance of wheat to stem rust. Plant Physiol. 1990;93:465–470. doi: 10.1104/pp.93.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brunner F, Nürnberger T. AtGenExpress: Response to bacterial-(LPS, HrpZ, Flg22) and oomycete-(NPP1) derived elicitors. 2004. Available: http://www.arabidopsis.org; ExpressionSet:1008080727. Accessed November 2004.
- 39.Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, et al. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
- 40.Engelhardt S, Lee J, Gäbler Y, Kemmerling B, Haapalainen M-L, et al. Separable roles of the Pseudomonas syringae pv. phaseolicola accessory protein HrpZ1 in ion-conducting pore formation and activation of plant immunity. Plant J. 2008 doi: 10.1111/j.1365-313X.2008.03723.x. In press. [DOI] [PubMed] [Google Scholar]
- 41.Fellbrich G, Romanski A, Varet A, Blume B, Brunner F, et al. NPP1, a Phytophthora-associated trigger of plant defense in parsley and Arabidopsis. Plant J. 2002;32:375–390. doi: 10.1046/j.1365-313x.2002.01454.x. [DOI] [PubMed] [Google Scholar]
- 42.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 43.Hodgkins T, MacDonald MV. The effect of a phytotoxin from Alternaria brassicicola on Brassica pollen. New Phytol. 1986;104:631–636. doi: 10.1111/j.1469-8137.1986.tb00663.x. [DOI] [PubMed] [Google Scholar]
- 44.Cramer RA, Lawrence CB. Cloning of a gene encoding an Alt a1 isoallergen differentially expressed by the necrotrophic fungus Alternaria brassicicola during Arabidopsis infection. Appl Environ Microbiol. 2003;69:2361–2364. doi: 10.1128/AEM.69.4.2361-2364.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Henk AD, Warren RF, Innes RW. A new Ac-like transposon of Arabidopsis is associated with a deletion of the RPS5 disease resistance gene. Genetics. 1999;151:1581–1589. doi: 10.1093/genetics/151.4.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pegard A, Brizzard G, Fazari A, Soucaze O, Abad P, et al. Histological characterization of resistance to different root-knot nematode species related to phenolics accumulation in Capsicum annuum. Phytopathology. 2005;95:158–165. doi: 10.1094/PHYTO-95-0158. [DOI] [PubMed] [Google Scholar]
- 47.Atanassova R, Favet N, Martz F, Chabbert B, Tollier MT, et al. Altered lignin composition in transgenic tobacco expressing O-methyltransferase sequences in sense and antisense orientation. Plant J. 1995;8:465–477. [Google Scholar]
- 48.Wuyts N, Lognay G, Swennen R, De Waele D. Nematode infection and reproduction in transgenic and mutant Arabidopsis and tobacco with an altered phenylpropanoid metabolism. J Exp Bot. 2006;57:2825–2835. doi: 10.1093/jxb/erl044. [DOI] [PubMed] [Google Scholar]
- 49.Göker M, Riethmüller A, Voglmayr H, Weiss M, Oberwinkler F. Phylogeny of Hyaloperonospora based on nuclear ribosomal internal transcribed spacer sequences. Mycol Prog. 2004;3:83–94. [Google Scholar]
- 50.Slusarenko A, Schlaich N. Downy mildew of Arabidopsis thaliana caused by Hyaloperonospora parasitica (formerly Peronospora parasitica). Mol Plant Pathol. 2003;4:159–170. doi: 10.1046/j.1364-3703.2003.00166.x. [DOI] [PubMed] [Google Scholar]
- 51.Holub EB. Natural history of Arabidopsis thaliana and oomycete symbioses. Eur J Plant Pathol. 2008;122:91–109. [Google Scholar]
- 52.Kwon C, Neu C, Pajonk S, Yun HS, Lipka U, et al. Co-option of a default secretory pathway for plant immune responses. Nature. 2008;451:835–840. doi: 10.1038/nature06545. [DOI] [PubMed] [Google Scholar]
- 53.Vogel J, Somerville S. Isolation and characterization of powdery mildew-resistant Arabidopsis mutants. Proc Natl Acad Sci U S A. 2000;97:1897–1902. doi: 10.1073/pnas.030531997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vogel JP, Raab TK, Schiff C, Somerville SC. PMR6, a pectate lyase-like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell. 2002;14:2095–2106. doi: 10.1105/tpc.003509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Consonni C, Humphry ME, Hartmann HA, Livaja M, Durner J, et al. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat Genet. 2006;38:716–720. doi: 10.1038/ng1806. [DOI] [PubMed] [Google Scholar]
- 56.Panstruga R. Serpentine plant MLO proteins as entry portals for powdery mildew fungi. Biochem Soc Trans. 2005;33:389–392. doi: 10.1042/BST0330389. [DOI] [PubMed] [Google Scholar]
- 57.Jacobs AK, Lipka V, Burton RA, Panstruga R, Strizhov N, et al. An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. Plant Cell. 2003;15:2503–2513. doi: 10.1105/tpc.016097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nishimura MT, Stein M, Hou BH, Vogel JP, Edwards H, et al. Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science. 2003;301:969–972. doi: 10.1126/science.1086716. [DOI] [PubMed] [Google Scholar]
- 59.Van Damme M, Andel A, Huibers RP, Panstruga R, Weisbeek PJ, et al. Identification of Arabidopsis loci required for susceptibility to the downy mildew pathogen Hyaloperonospora parasitica. Mol Plant Microbe Interact. 2005;18:583–592. doi: 10.1094/MPMI-18-0583. [DOI] [PubMed] [Google Scholar]
- 60.Van Damme M, Huibers RP, Elberse J, Van den Ackerveken G. Arabidopsis DMR6 encodes a putative 2OG-Fe(II) oxygenase that is defense-associated but required for susceptibility to downy mildew. Plant J. 2008;54:785–793. doi: 10.1111/j.1365-313X.2008.03427.x. [DOI] [PubMed] [Google Scholar]
- 61.Förster H, Tyler BM, Coffey MD. Phytophthora sojae races may have arisen by clonal evolution and by rare outcrosses. Mol Plant Microbe Interact. 1994;7:780–791. [Google Scholar]
- 62.Francis DM, Gehlen MF, St Clair DA. Genetic variation in homothallic and hyphal swelling isolates of Pythium ultimum var. ultimum and P. utlimum var. sporangiferum. Mol Plant Microbe Interact. 1994;7:766–775. doi: 10.1094/mpmi-7-0766. [DOI] [PubMed] [Google Scholar]
- 63.Drenth A, Janssen EM, Govers F. Formation and survival of oospores of Phytophthora infestans under natural conditions. Plant Pathol. 1995;44:86–94. [Google Scholar]
- 64.Medina MV, Platt HW. Viability of oospores of Phytophthora infestans under field conditions in northeastern North America. Can J Plant Pathol. 1999;21:137–143. [Google Scholar]
- 65.Duncan JM. Effect of fungicides on survival, infectivity and germination of Phytophthora fragariae oospores. Trans Br Mycol Soc. 1985;85:585–593. [Google Scholar]
- 66.Dangl JL, Holub EB, Debener T, Lehnackers H, Ritter C, et al. Genetic definition of loci involved in Arabidopsis-pathogen interactions. In: Koncz C, Chua N-H, Schell J, editors. Methods in Arabidopsis Research. Singapore: World Scientific Publishers; 1992. pp. 393–418. [Google Scholar]
- 67.Caillaud MC, Lecomte P, Jammes F, Quentin M, Pagnotta S, et al. MAP65-3 microtubule-associated protein is essential for nematode-induced giant cell ontogenesis in Arabidopsis. Plant Cell. 2008;20:423–437. doi: 10.1105/tpc.107.057422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rohringer R, Kim WK, Samborski DJ, Howes NK. Calcofluor: an optical brightener for fluorescence microscopy of fungal plant parasites in leaves. Phytopathology. 1977;67:808–810. [Google Scholar]
- 70.Keller H, Blein JP, Bonnet P, Ricci P. Physiological and molecular characteristics of elicitin-induced systemic acquired resistance in tobacco. Plant Physiol. 1996;110:365–376. doi: 10.1104/pp.110.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Molecular analyses of the mutants and transgenic lines used in this study.
(0.57 MB PDF)
Susceptibility of COMT1 knock-outs to fungal pathogens and a nematode.
(0.82 MB PDF)
Susceptibility of COMT1 knock-outs to bacterial pathogens.
(0.11 MB PDF)
Comt1a mutants are not altered in SA- and JA-dependent defense responses.
(0.10 MB PDF)
Reversed-phase HPLC of synthetic hydroxycinnamoyl malate esters and of methanolic plant extracts.
(0.30 MB PDF)
Characteristics of fungal and bacterial pathogens, and inoculation procedures.
(0.03 MB DOC)
Synthesis and spectral analyses of hydroxycinnamoyl malates.
(0.03 MB DOC)