Abstract
Huntington's disease (HD) is a fatal inherited neurodegenerative disorder that gradually robs affected individuals of memory, cognitive skills, and normal movements. While research has identified a single faulty gene, the huntingtin gene, as the cause of the disease, a cure remains elusive. Strong evidence indicates that mitochondrial impairment plays a key role in HD pathogenesis. Here, we highlight how mtHtt might cause mitochondrial dysfunction by either perturbing transcription of nuclear-encoded mitochondrial proteins or by direct interaction with the organelle and modulation of respiration, membrane potential and Ca2+ buffering. In addition, we propose that mtHtt might convey its neurotoxicity by evoking defects in mitochondrial dynamics, organelle trafficking and fission and fusion, which in turn might result in bioenergetic failure and HD-linked neuronal dysfunction and cell death. Finally, we speculate how mitochondria may dictate selective vulnerability of long projection neurons, such as medium spiny neurons, which are particularly affected in HD.
INTRODUCTION
The discovery of the gene that causes Huntington's disease (HD) in 1993 sparked tremendous hope that a cure was just around the corner.1, 2 However, this initial enthusiasm was soon diminished and a cure remains elusive as current treatments reduce the symptoms, but do not prevent neuronal loss. Unexpectedly, the faulty gene product, mtHtt, is an extremely large protein of 350 kiloDalton and might act as a scaffold protein regulating vesicle and organelle trafficking and signaling pathways. Although the protein is mostly cytoplasmic, it is also present in smaller amounts in multiple subcellular compartments including the plasma membrane, nucleus, endoplasmic reticulum, endocytic vesicles, the Golgi and mitochondria, and interferes with multiple cellular pathways.3-8 Thus, these properties have made it difficult to pin down Htt's normal and neurotoxic function(s). Now, new evidence has emerged that implicates malfunctioning mitochondria in HD pathogenesis. This article focuses on these new observations because they might bring us closer to effective treatments that can either slow or stall the relentless neuronal loss in HD. Specifically, we will focus on the potential pathways by which mtHtt could cause mitochondrial dysfunction.
HD is an autosomal dominant neurodegenerative disorder that affects approximately 30,000 patients in the United States alone. Clinical symptoms of HD include progressive chorea (involuntary dance-like movements), rigidity, weight loss, dementia, seizures and psychiatric disturbances such as depression, withdrawal and irritability. Symptoms result from the selective loss of long projection neurons, known as medium spiny neurons, which are GABA-releasing neurons in the striatal brain regions that control movement, memory and emotions. In addition, early neuronal loss and dysfunction occurs not only in the striatum, but also in the cortex in HD patients.9 HD is slowly progressive and patients succumb to the disease within 15 to 20 years of symptom onset. Thus, HD takes a tremendous toll on patients and their families.
HD is one of nine neurodegenerative disorders caused by an abnormal expansion of CAG repeats that encode a poly-glutamine stretch. A poly-glutamine expansion of less than 35 in the amino-terminus of Htt is normal and does not cause disease. However, Htt with an abnormal stretch of 36 or more glutamines causes the neurodegenerative condition. Clinical symptoms start in most cases between the ages of 30 and 40, but the disease can strike, in some rare cases, young patients whose Htt molecule has 60 or more glutamine residues.10, 11 Thus, there is an inverse relationship between the length of the poly-glutamine stretch and age of disease onset, with longer glutamine stretches resulting in earlier disease onset and increased severity.
A puzzle that has long occupied neurologists is why disease onset is so late despite the presence of the mutation at birth. In addition to the inherited CAG expansion, new observations indicate that somatic CAG expansions occur during aging in the striatum, the brain region which is most affected in HD, modulating the disease onset and progression.12 The underlying mechanism responsible for the somatic expansion of CAG is activation of the error-prone base pair repair enzyme, 7,8-dihydro-8-oxoguanine-DNA glycolylase 1 (OGG1) that is induced by oxidative stress.12 As oxidative damage accumulates with age, a somatic expansion of the CAG repeat might trigger the delayed onset of the disorder.
Mechanism of mtHtt action: gain of function or loss of function?
How mtHtt elicits neurodegeneration remains a mystery, but wild-type Htt exhibits anti-apoptotic properties.13, 14 Htt is a remarkably large molecule with ubiquitous expression. Its domain model does not suggest any particular functions aside from three HEAT domains that may mediate protein interactions.15, 16 Indeed, numerous reports indicate that Htt interacts with over 200 proteins and abnormal protein interactions might in part mediate its toxicity.17, 18 For example, Htt interacts with nuclear transcription factors in the nucleus.19 Specifically, Htt interacts with cAMP response element binding-protein (CREB)-binding protein and specificity protein 1 (Sp1).17, 18 Furthermore, mtHtt reduces the binding of Sp1 to DNA.20, 21 In addition, Htt interacts with the tumor suppressor and transcription factor, p53.22 Finally, a new study found that many Htt-interacting proteins are genetic modifiers of HD neurodegeneration.23
Because HD inheritance is autosomal dominant, caused by a single copy of the mutant gene, the prevailing view is that mtHtt-mediated neuronal dysfunction results from a toxic gain of function. There is ample evidence supporting this view. For example, a common characteristic of HD pathology in both mice and humans is formation of protein aggregates and inclusion bodies.24-26 While the mechanism of toxicity (if any) of these aggregates is unclear, there presence hints at a toxic gain of function in HD. Transgenic mouse models expressing full-length or exon 1 of mtHtt, produce neuropathology resembling the human disorder and provide additional support for the toxic gain-of-function hypothesis.27
The proposed mechanisms of mtHtt gain-of-function neurotoxicity include transcriptional modulation,28-30 protein aggregation,31 and excitotoxicity.32, 33 NMDAR currents and Ca2+ are increased in medium spiny neurons of mtHtt transgenic mice.34-36 In addition, caspase 6 cleavage of mtHtt is a required event for mtHtt-mediated toxicity, with the fragmented mtHtt confering greater neurotoxicity.37
Although the majority of the findings are consistent with a toxic gain of function for mtHtt, there is also evidence that mtHtt conveys neurodegeneration by a loss-of-function mechanism. For example, expression and transport of brain-derived neurotrophic factor (BDNF) requires Htt and mtHtt impairs BDNF gene expression and BDNF vesicle trafficking.30, 38 Thus gain-of-function and loss-of-function mechanisms may co-exist and are not mutually exclusive, thereby adding to the complexity of the potential molecular pathways underlying HD.
Mitochondrial dysfunction
A critical function of mitochondria is the conversion of food into energy or heat by a process known as oxidative phosphorylation. Neurons have intense energy demands that mitochondria must largely fill. Mitochondria in neurons support ATP-dependent processes such as plasma membrane potential maintenance, synaptic neurotransmitter release and re-uptake, and build-up of a reserve pool of vesicles for prolonged or high frequency firing and axonal and dendritic transport of cargo and organelles.39-41 Mitochondria also serve as important Ca2+ buffers, controlling cytoplasmic Ca2+ concentrations after neurotransmission.42 Studies have implicated persistent high cytoplasmic Ca2+ levels in neuronal cell death.43, 44 In sum, mitochondria are crucial for normal neuronal function.45
Despite the critical role of mitochondria to neuronal health, some aspects of mitochondrial function can damage neurons. For example, mitochondria are the main source of free radicals, by-products of respiration that research suggests cause aging and neurodegeneration.42 In addition, mitochondria also play an active role in the execution of cell death by the spill of apoptogenic factors from mitochondria.46-53
Strong evidence indicates that mitochondrial malfunction plays a critical role in HD pathogenesis. First, HD patients exhibit profound weight loss despite a normal diet.54, 55 Furthermore, there is a marked decrease in glucose metabolism and a corresponding increase in lactate in affected brain regions of HD patients, suggesting a bioenergetic defect.56 Indeed, postmortem brain samples of HD patients exhibit decreased activity of mitochondrial respiratory complexes II, III and IV.57 Now, there is new evidence that indicates mitochondrial impairment occurs even in asymptomatic HD carriers; thus, mitochondrial defects may initiate disease onset.58 Moreover, mitochondria isolated from lymphocytes of HD patients have lowered Ca2+ buffering capacity and their mitochondrial membrane potential depolarizes earlier at lower Ca2+ concentrations, possibly due to a direct interaction between the poly-glutamine stretch in mtHtt and the mitochondria.59 Striatal cells from mtHtt mice exhibit impairment of mitochondrial respiration and ATP synthesis.60 Interestingly, expression of complex II subunits in striatal neurons expressing mtHtt exon 1 restores complex II respiratory activity and protects against cell death.56 In addition, recent studies have reported ultrastructral changes in mitochondria in HD.61 Finally, mtDNA deletions occur in HD patients during the progression of the disease.62, 63
Additional support for the notion that mitochondrial respiratory chain inhibition plays a key role in HD pathogenesis stems from experiments with 3-nitropropionic acid (3-NP), a selective inhibitor of succinate dehydrogenase and complex II. 3-NP recapitulates the loss of medium spiny neurons in the substantia nigra and HD-like symptoms in several vertebrate models.27, 64 In addition, humans exposed to 3-NP exhibit motor dysfunction similar to HD patients.27, 65, 66 Finally, cultured cells expressing mtHtt exhibit increased susceptibility to 3-NP.67
In sum, although there is clear evidence that profound mitochondrial impairment is an early and important event in HD pathogenesis, the molecular basis of this deficit remains unclear.
Link between transcriptional deregulation and mitochondrial dysfunction
Aberrant transcriptional regulation is one possible mechanism of mtHtt toxicity because cleaved mtHtt binds to several transcriptional regulators such as p53, Sp1, TAFII130 and CREB-binding protein and interferes with their function .17, 23, 68, 69 For instance, the tumor suppressor p53 regulates genes involved in mitochondrial function. mtHtt binds p53 and increases p53 levels, which in turn results in increased Bax and Puma expression and mitochondrial damage.22 Inhibition of p53 prevents mitochondrial damage.22 Similarly, p53 loss prevents mtHtt-mediated neurodegeneration in Drosophila.22 Thus, altered mtHtt-induced p53 transcriptional activity results in mitochondrial dysfunction and neuronal loss.
Studies with PGC-1α further support a mtHtt-mediated transcriptional block resulting in HD-linked mitochondrial deficits.70-72 PGC-1α (peroxisome proliferator-activated receptor-γ coactivator 1α) is a key transcriptional co-regulator and induces the transcription of cellular programs regulating mitochondrial respiration, oxidative stress defense and adaptive thermogenesis.73 To regulate these programs, PGC-1α binds to transcription factors such as nuclear respiration factors −1 and −2 (NRF-1, 2) and nuclear hormone peroxisome proliferator-activated receptor (PPAR).74 NRF-1 and NRF-2 regulate nuclear-encoded mitochondrial genes such as cytochrome c, mitochondrial transcription factor A (Tfam) and components of the respiratory chain complexes I-V. PGC-1α knockout mice exhibit nervous system and striatal lesions, defective thermoregulation, hyperactivity and limb clasping, a behavioral indicator of neurodegeneration.75, 76 Three reports suggest that PGC-1α is a potent suppressor of free radical production and becomes impaired during HD pathogenesis.70-72
St. Pierre and colleagues found that oxidative stress evoked by hydrogen peroxide triggers increased expression of PGC-1α and PGC-1β, a related gene.71 Concomitant with this increase in PGC-1α is an increase in ROS scavenging enzymes such as Zn/Cu SOD1, SOD2, catalase and glutathione peroxidase. Conversely, PGC-1α knockdown cells exhibit lower SOD1, SOD2 and glutathione peroxidase expression in response to hydrogen peroxide. Furthermore, PGC-1α knockout mice have lower SOD1 and SOD2 levels and are more sensitive to MPTP-, kainic acid- and glutamate receptor agonist-induced neurotoxicity. In sum, these findings suggest that PGC-1α loss may sensitize neurons to neurodegeneration in response to several neurotoxins.
Cui and colleagues tested the role of PGC-1α in HD pathogenesis.70 They observed that PGC-1α was markedly decreased in medium spiny neurons, but not in interneurons, of HD transgenic mice, providing a potential explanation for the selective neuronal loss characteristic of HD. PGC-1α expression reduced neuronal loss in R6/2 HD mice. In contrast, PGC-1α reduction worsened the behavioral and neuropathological abnormalities of R6/2 mice. On the molecular level, mtHtt interferes with the PGC-1α promoter and the formation of CREB/TAF4 complexes, resulting in reduced PGC-1α transcription.
Further support for the idea that PGC-1αfunction is impaired in HD stems from findings made by Weydt and colleagues.72 They noticed that HD transgenic mice develop hypothermia. Uncoupling proteins (UCP) promote mitochondrial heat production by stimulating the production of heat energy at the expense of chemical energy. Recent reports suggest a neuroprotective function of UCPs.77, 78 Weydt et al. found that mtHtt suppresses the UCP1 promoter.72 Furthermore, HD mice had reduced food intake but increased energy expenditure, which may account for their low weight. Importantly, HD patients and mice exhibited reduced expression of PGC-1α target genes. Finally, striatal neuronal cells expressing PGC-1α were less vulnerable to 3-NP, a complex II inhibitor. Collectively, these data suggest an inhibition of PGC-1α function by mtHtt and establish a link between transcriptional inhibition and mitochondrial dysfunction in HD.
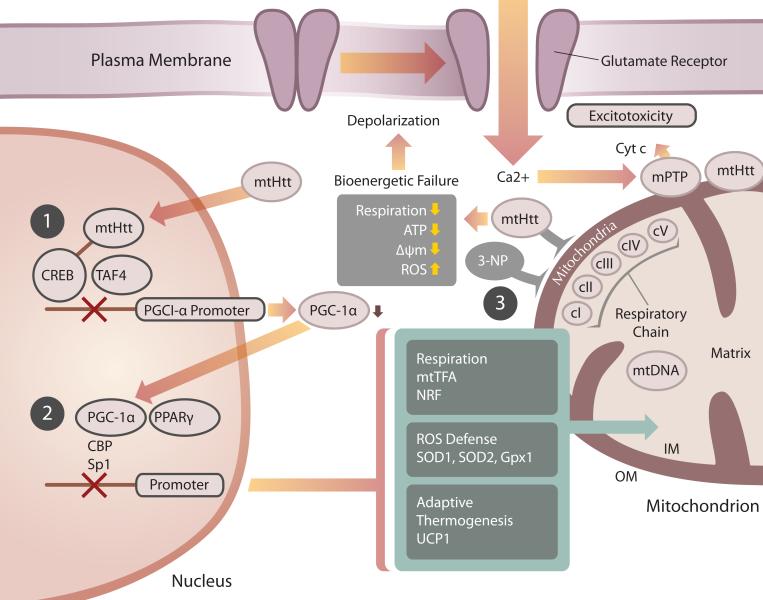
Although the findings of transcriptional dysregulation by mtHtt are of high significance, they cannot entirely explain all the mitochondrial defects observed in HD. For example, mtHtt binds directly to the mitochondrial outer membrane, forming large clusters.79, 80 Furthermore, mtHtt exon 1 directly impairs mitochondrial function, Ca2+ homeostasis and mitochondrial membrane potential in isolated mitochondria 79, 81, 82 and mitochondria in HD have altered ultrastructure and exhibit mtDNA deletions.62, 63 Thus, transcriptional inhibition of nuclear encoded mitochondrial proteins and direct inhibition of mitochondrial function by mtHtt might act in parallel to initiate metabolic defects and a neurodegenerative cascade in HD (Fig. 1).
Figure 1.
mtHtt impairs mitochondrial function by transcriptional dysregulation of nuclear encoded mitochondrial proteins and by direct effects on the organelle. (1) mtHtt blocks the PGC-1α promoter via inhibition of the CREB transcriptional activator, resulting in decreased PGC-1α expression. (2) Lowered PGC-1α will decrease PPARγ-mediated expression of nuclear encoded mitochondrial proteins that are necessary for respiration and oxidative damage defense. (3) mtHtt can have direct effects on mitochondria, blocking the respiratory complex II, just as 3-NP does. Respiratory inhibition, in turn, leads to decreased energy production and ΔΨm as well as increased ROS generation. The bioenergetic decline caused by transcriptional deregulation and direct effects of mtHtt on mitochondria can cause increased vulnerability to excitotoxic stimuli amplification of the mitochondrial damage by Ca2+-mediated mitochondrial permeability transition pore (mPTP) opening.
Mitochondrial trafficking deficits in HD
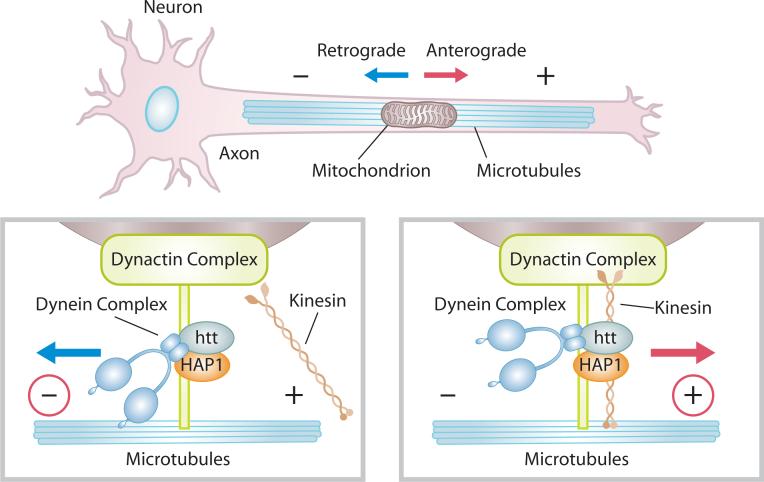
While the bioenergetic deficits in HD are well known, recent studies suggest that impaired mitochondrial trafficking along axons and dendrites may also play a role in disease pathology. Mitochondria in healthy neurons form filaments of variable length (∼2 to 25 μm), which move anterograde, away from the cell body, or retrograde, toward the cell body, along microtubule tracks (Fig. 2). ATP-dependent motor proteins regulate mitochondrial movement. Kinesins mediate anterograde transport and dynein/dynactin regulate retrograde transport (Fig.2).83
Figure 2.
ATP-dependent motor proteins, dynein and kinesin, regulate anterograde and retrograde transport of mitochondria in neurons. Htt binds to HAP1, regulating dynein and kinesin function.
Defects in proper positioning of mitochondria may profoundly affect normal neuronal function. While mitochondrial distribution throughout the long neuronal processes (which extend up to one meter in motor neuron axons) is important, sites of high energy demands, such as synapses and Nodes of Ranvier, possess densely packed mitochondrial populations. Medium spiny neurons, the primary cell type affected by mtHtt, have long projections. Thus, one can speculate that polarized neurons with long axons might be more vulnerable than other cell types to trafficking defects.
Normal Htt protein regulates anterograde and retrograde transport of endocytic vesicles by interacting with several trafficking mediators: adaptin, Hip1, Hip14, HAP1, HAP 40, SH3GL3 (endophilin 3), clathrin and dynamin.17, 23, 84, 85 Wild-type Htt stimulates trafficking by binding to HAP1, which in turn leads to interaction with the molecular motor dynein/dynactin and kinesin and regulates microtubule-mediated BDNF vesicle transport and mitochondrial transport(Fig. 2).86, 87 Phosphorylation of Htt serves as a molecular decision maker for anterograde versus retrograde direction of vesicle transport.88 While wild-type Htt promotes axonal BDNF vesicle trafficking, mtHtt interacts more tightly with HAP1 and dynactin, thereby leading to de-railing of the molecular motors from microtubule tracks and cessation of transport.86 Additionally, either loss of Htt or over-expression of mtHtt in Drosophila causes vesicle and mitochondrial movement defects.89
While abnormal protein interaction of mtHtt with motor proteins plays a role in trafficking defects in HD, mtHtt aggregates may also act as physical roadblocks. Reynolds and colleagues reported that mtHtt induces a change in mitochondrial morphology from elongated to round phenotype which correlates with a blockage in mitochondrial movement.90, 91 Thus, aggregates in the narrow neuronal projections seem to prevent passage and fragmented mitochondria accumulate around mtHtt aggregates. Besides blocking mitochondrial movement, it is also possible that mtHtt elicits an imbalance in mitochondrial fission and fusion, which may cause the mitochondrial defects underlying HD pathogenesis.
Defects in mitochondrial fission and fusion balance in neurodegeneration
In addition to migration and movement, mitochondria undergo cycles of fusion, combination of two mitochondria, and fission, splitting of a mitochondrial filament into two or more new organelles. An evolutionarily conserved engine consisting of large GTPases of the dynamin family of proteins regulates these processes.40, 92, 93 The key mitochondrial fission regulator is dynamin-related protein 1 (DRP1), which is localized in the cytoplasm and cycles on and off mitochondria to mediate fission events. DRP1 forms large clusters on mitochondria at fission sites. Recent findings indicate that motor proteins such as dynein regulate mitochondrial fission by recruitment of DRP1 to mitochondria.94 Disruption of dynein/dynactin results in loss of DRP1 from mitochondria.94 Thus, there seems to be a link between mitochondrial motors and fission regulators. Recombinant DRP1 protein oligomerizes into large ring structures in vitro, 95, 96 and its yeast ortholog Dnm1 oligomerizes into spirals which have the diameter of mitochondria.97 Thus, it has been proposed that DRP1 may function similar to dynamin by acting as a mechano-enzyme to constrict and divide mitochondria. Fission is important for mitochondrial renewal, redistribution and proliferation into synapses.
Mitofusins (MFN1 and MFN2) located on the mitochondrial outer membrane and optic atrophy 1 (OPA1) located on the inner membrane mediate mitochondrial fusion .98-100 Fusion is important for mitochondrial interaction and communication, which facilitate mitochondrial movement and distribution across long distances.
An emerging concept in studies of neurodegeneration is that an imbalance of mitochondrial fission and fusion can initiate a neurodegenerative cascade.40 The strongest support for this notion originates from studies of hereditary neurodegenerative disease in humans. Mutations in MFN2 cause Charcot-Marie-Tooth syndrome, a progressive peripheral neuropathy linked with axonal sensory and motor neuron degeneration.101-103 In addition, hereditary mutations in OPA1 cause autosomal dominant optic atrophy (ADOA) characterized by retinal ganglion and optic nerve degeneration.101, 104, 105 While most mutations lead to vision loss, some mutations in OPA1 cause an ‘ADOA-plus’ phenotype, which is characterized by additional deafness, sensory-motor neuropathy, and muscle weakness. Because of the similarity of the phenotypes caused by MFN2 and OPA1 mutations, these two gene products might be functionally linked. Mutations in MFN2 and OPA1 have an impact on mitochondrial fusion, trafficking and bioenergetic functionality.106, 107
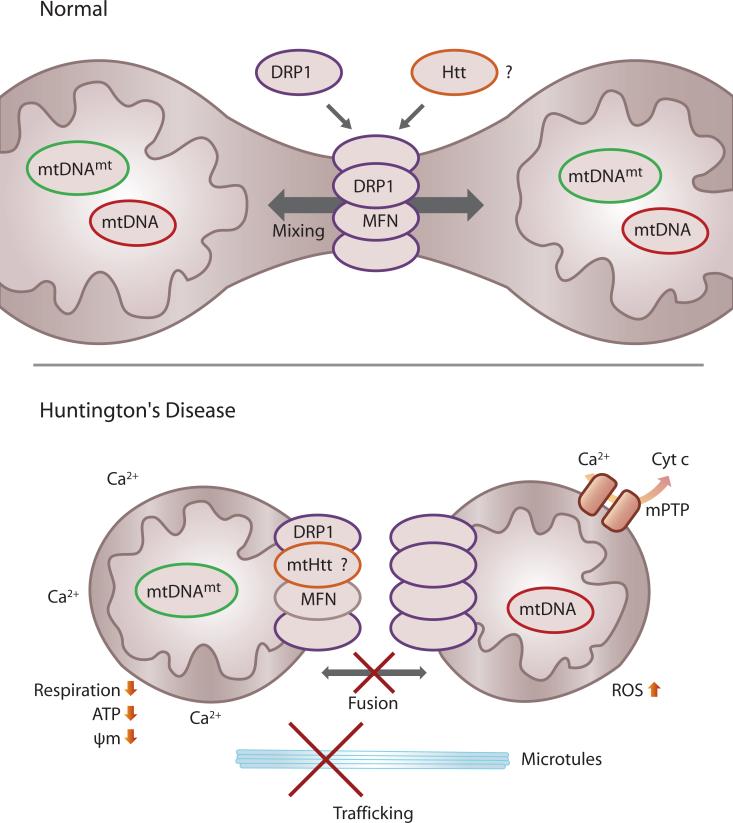
Expression of mtHtt causes abnormal mitochondrial ultrastructure,61 impaired Ca2+ buffering,59 bioenergetic defects,108 and mtDNA deletions.63 In principle, all of these effects are consistent with a defect in maintaining mitochondrial fission and fusion balance, although this hypothesis has not been adequately explored.40 Thus, we hypothesize malfunctions of the mitochondrial fission and fusion machinery might cause the HD-linked mitochondrial defects. The following observations support this hypothesis. First, mtHtt in transgenic mice forms large aggregates on the tips or opposing sides of mitochondria in immuno EM.59 This distribution pattern has striking similarities to DRP1.109 Thus, mtHtt might be part of a DRP1 complex. Second, Htt interacts with dynamin.23 It is therefore conceivable that DRP1, in analogy to dynamin, may form a complex with Htt. Last, Htt interacts with endophilins.110 Endophilins play a role in endocytosis and membrane scission.111, 112 Bax, a pro-apoptotic member of the Bcl-2 family, co-localizes at sites of mitochondrial division during apoptosis that also contain DRP1 and MFN2.109 Bax also binds Endophilin B1 and might regulate apoptotic fission.109, 113-115 Thus, one can speculate that Htt might regulate fission events by interaction with Endophilin family members. In sum, normal Htt may regulate mitochondrial fission and fusion complexes and mtHtt may alter the assembly and function of these complexes. New investigations are needed to test this idea.
Selective neuronal loss
Why do long projection neurons selectively degenerate in HD when mtHtt is present in all cells? Several cell-type specific characteristics might be responsible. For example, the loss of PGC-1α in medium spiny neurons and not other neurons (e.g., interneurons) might explain their selective loss.70 Another possibility is that impaired mitochondrial Ca2+ buffering affects medium spiny neurons more severely because of the lack of compensatory mechanisms.116 Alternatively, there may be increased expression of cell death mediators in medium spiny neurons as compared to other cell types. Moreover, these neurons might be more sensitive to oxidative stress. Furthermore, because medium spiny neurons have long projections, it may render them more vulnerable to trafficking and mitochondrial fission/fusion defects. Finally, there is increasing evidence that the mitochondrial proteome differs based on cell type.117, 118
Conclusion
Treatment regimes for HD relieve merely the symptoms, but can neither restore neuronal function nor stop the insidious loss of neurons. Clearly, although a single gene mutation causes HD, the outcome is highly complex and it is unlikely that a single drug will undo the damage. Current treatments are nonspecific and include anti-depressants, anti-psychotics and levodopa to ameliorate some of the movement defects. Because HD is an autosomal dominant hereditary disorder with late onset, physicians can start potential treatment regimes prior to the symptomatic stage and follow treatment efficacy over time. New insights towards the cellular and molecular mechanisms underlying HD in the past years have provided important clues for therapeutic intervention. Potential targets include restoring metabolism and mitochondrial function, blocking excitotoxicity, modulating transcription, diminishing aggregate formation and now, increasing axonal organelle dynamics. Because of the complexity of the disease and its long duration, combinatorial treatments might hold the greatest promise.
Figure 3.
Hypothesis that mtHtt impinges on the mitochondrial fission/fusion engine by recruitment into the DRP1/MFN complexes. Mitochondrial fusion allows mixing of metabolites and mtDNA molecules. This process may prevent manifestation of mtDNA mutations by complementation with wild-type mtDNA. mtHtt may abnormally interact with the mitochondrial fission and fusion GTPases, DRP1 or MFN, thereby preventing further mitochondrial fusion or continuous fission. This may have a profound impact on mitochondrial mobility, Ca2+ handling, respiration, ATP production, free radical production and mitochondrial membrane potential. It may also lead to the manifestation of mitochondrial DNA mutations and may sensitize neurons to Ca2+-mediated opening of the mitochondrial permeability transition pore (mPTP) and release of apoptogenic factors like cytochrome c.
ACKNOWLEDGMENTS
We wish to acknowledge the support of NIH grants RO1 EY016164, RO1 NS047456, and RO1 NS055193, and the Hereditary Disease Foundation. We thank Adam Wilson for help with illustrations.
References
- 1.Goldberg YP, et al. Identification of an Alu retrotransposition event in close proximity to a strong candidate gene for Huntington's disease. Nature. 1993;362:370–373. doi: 10.1038/362370a0. [DOI] [PubMed] [Google Scholar]
- 2.A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 3.Kegel KB, et al. Huntingtin is present in the nucleus, interacts with the transcriptional corepressor C-terminal binding protein, and represses transcription. J Biol Chem. 2002;277:7466–7476. doi: 10.1074/jbc.M103946200. [DOI] [PubMed] [Google Scholar]
- 4.Kegel KB, et al. Huntingtin associates with acidic phospholipids at the plasma membrane. J Biol Chem. 2005;280:36464–36473. doi: 10.1074/jbc.M503672200. [DOI] [PubMed] [Google Scholar]
- 5.Rockabrand E, et al. The first 17 amino acids of Huntingtin modulate its sub-cellular localization, aggregation and effects on calcium homeostasis. Hum Mol Genet. 2007;16:61–77. doi: 10.1093/hmg/ddl440. [DOI] [PubMed] [Google Scholar]
- 6.Strehlow AN, et al. Wild-type huntingtin participates in protein trafficking between the Golgi and the extracellular space. Hum Mol Genet. 2007;16:391–409. doi: 10.1093/hmg/ddl467. [DOI] [PubMed] [Google Scholar]
- 7.Atwal RS, et al. Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum Mol Genet. 2007;16:2600–2615. doi: 10.1093/hmg/ddm217. [DOI] [PubMed] [Google Scholar]
- 8.Caviston JP, et al. Huntingtin facilitates dynein/dynactin-mediated vesicle transport. Proc Natl Acad Sci U S A. 2007;104:10045–10050. doi: 10.1073/pnas.0610628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosas HD, et al. Regional and progressive thinning of the cortical ribbon in Huntington's disease. Neurology. 2002;58:695–701. doi: 10.1212/wnl.58.5.695. [DOI] [PubMed] [Google Scholar]
- 10.Nance MA, Myers RH. Juvenile onset Huntington's disease--clinical and research perspectives. Ment Retard Dev Disabil Res Rev. 2001;7:153–157. doi: 10.1002/mrdd.1022. [DOI] [PubMed] [Google Scholar]
- 11.Ribai P, et al. Psychiatric and cognitive difficulties as indicators of juvenile huntington disease onset in 29 patients. Arch Neurol. 2007;64:813–819. doi: 10.1001/archneur.64.6.813. [DOI] [PubMed] [Google Scholar]
- 12.Kovtun IV, et al. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–452. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leavitt BR, et al. Wild-type huntingtin reduces the cellular toxicity of mutant huntingtin in vivo. Am J Hum Genet. 2001;68:313–324. doi: 10.1086/318207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rigamonti D, et al. Wild-type huntingtin protects from apoptosis upstream of caspase-3. J Neurosci. 2000;20:3705–3713. doi: 10.1523/JNEUROSCI.20-10-03705.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takano H, Gusella JF. The predominantly HEAT-like motif structure of huntingtin and its association and coincident nuclear entry with dorsal, an NF-kB/Rel/dorsal family transcription factor. BMC Neurosci. 2002;3:15. doi: 10.1186/1471-2202-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andrade MA, Bork P. HEAT repeats in the Huntington's disease protein. Nat Genet. 1995;11:115–116. doi: 10.1038/ng1095-115. [DOI] [PubMed] [Google Scholar]
- 17.Li SH, Li XJ. Huntingtin-protein interactions and the pathogenesis of Huntington's disease. Trends Genet. 2004;20:146–154. doi: 10.1016/j.tig.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 18.Sugars KL, Rubinsztein DC. Transcriptional abnormalities in Huntington disease. Trends Genet. 2003;19:233–238. doi: 10.1016/S0168-9525(03)00074-X. [DOI] [PubMed] [Google Scholar]
- 19.Thomas EA. Striatal specificity of gene expression dysregulation in Huntington's disease. J Neurosci Res. 2006;84:1151–1164. doi: 10.1002/jnr.21046. [DOI] [PubMed] [Google Scholar]
- 20.Dunah AW, et al. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington's disease. Science. 2002;296:2238–2243. doi: 10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- 21.Li SH, et al. Interaction of Huntington disease protein with transcriptional activator Sp1. Mol Cell Biol. 2002;22:1277–1287. doi: 10.1128/mcb.22.5.1277-1287.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bae BI, et al. p53 mediates cellular dysfunction and behavioral abnormalities in Huntington's disease. Neuron. 2005;47:29–41. doi: 10.1016/j.neuron.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Kaltenbach LS, et al. Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet. 2007;3:e82. doi: 10.1371/journal.pgen.0030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davies SW, et al. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 25.DiFiglia M, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 26.Sapp E, et al. Axonal transport of N-terminal huntingtin suggests early pathology of corticostriatal projections in Huntington disease. J Neuropathol Exp Neurol. 1999;58:165–173. doi: 10.1097/00005072-199902000-00006. [DOI] [PubMed] [Google Scholar]
- 27.Rubinsztein DC. Lessons from animal models of Huntington's disease. Trends Genet. 2002;18:202–209. doi: 10.1016/s0168-9525(01)02625-7. [DOI] [PubMed] [Google Scholar]
- 28.Cha JH. Transcriptional dysregulation in Huntington's disease. Trends Neurosci. 2000;23:387–392. doi: 10.1016/s0166-2236(00)01609-x. [DOI] [PubMed] [Google Scholar]
- 29.Luthi-Carter R, et al. Decreased expression of striatal signaling genes in a mouse model of Huntington's disease. Hum Mol Genet. 2000;9:1259–1271. doi: 10.1093/hmg/9.9.1259. [DOI] [PubMed] [Google Scholar]
- 30.Zuccato C, et al. Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science. 2001;293:493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- 31.Ross CA. Huntington's disease: new paths to pathogenesis. Cell. 2004;118:4–7. doi: 10.1016/j.cell.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 32.DiFiglia M. Excitotoxic injury of the neostriatum: a model for Huntington's disease. Trends Neurosci. 1990;13:286–289. doi: 10.1016/0166-2236(90)90111-m. [DOI] [PubMed] [Google Scholar]
- 33.Fan MM, Raymond LA. N-methyl-D-aspartate (NMDA) receptor function and excitotoxicity in Huntington's disease. Prog Neurobiol. 2007;81:272–293. doi: 10.1016/j.pneurobio.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 34.Fernandes HB, et al. Mitochondrial sensitivity and altered calcium handling underlie enhanced NMDA-induced apoptosis in YAC128 model of Huntington's disease. J Neurosci. 2007;27:13614–13623. doi: 10.1523/JNEUROSCI.3455-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeron MM, et al. Potentiation of NMDA receptor-mediated excitotoxicity linked with intrinsic apoptotic pathway in YAC transgenic mouse model of Huntington's disease. Mol Cell Neurosci. 2004;25:469–479. doi: 10.1016/j.mcn.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 36.Zeron MM, et al. Increased sensitivity to N-methyl-D-aspartate receptor-mediated excitotoxicity in a mouse model of Huntington's disease. Neuron. 2002;33:849–860. doi: 10.1016/s0896-6273(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 37.Graham RK, et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 38.Zuccato C, et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
- 39.Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 40.Knott AB, et al. Mitochondrial fragmentation in neurodegeneration. Nat Rev Neurosci. 2008;9:505–518. doi: 10.1038/nrn2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Chan DC. New insights into mitochondrial fusion. FEBS Lett. 2007;581:2168–2173. doi: 10.1016/j.febslet.2007.01.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 43.Wojda U, et al. Calcium ions in neuronal degeneration. IUBMB Life. 2008 doi: 10.1002/iub.91. [DOI] [PubMed] [Google Scholar]
- 44.Vesce S, et al. Relationships between superoxide levels and delayed calcium deregulation in cultured cerebellar granule cells exposed continuously to glutamate. J Neurochem. 2004;90:683–693. doi: 10.1111/j.1471-4159.2004.02516.x. [DOI] [PubMed] [Google Scholar]
- 45.Nicholls DG, Budd SL. Mitochondria and neuronal survival. Physiol Rev. 2000;80:315–360. doi: 10.1152/physrev.2000.80.1.315. [DOI] [PubMed] [Google Scholar]
- 46.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 47.Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 48.Bossy-Wetzel E, et al. Mitochondrial cytochrome c release in apoptosis occurs upstream of DEVD-specific caspase activation and independently of mitochondrial transmembrane depolarization. Embo J. 1998;17:37–49. doi: 10.1093/emboj/17.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X. The expanding role of mitochondria in apoptosis. Genes Dev. 2001;15:2922–2933. [PubMed] [Google Scholar]
- 50.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 51.Ghafourifar P, et al. Ceramide induces cytochrome c release from isolated mitochondria. Importance of mitochondrial redox state. J Biol Chem. 1999;274:6080–6084. doi: 10.1074/jbc.274.10.6080. [DOI] [PubMed] [Google Scholar]
- 52.Potts MB, et al. Reduced Apaf-1 levels in cardiomyocytes engage strict regulation of apoptosis by endogenous XIAP. J Cell Biol. 2005;171:925–930. doi: 10.1083/jcb.200504082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bao Q, Shi Y. Apoptosome: a platform for the activation of initiator caspases. Cell Death Differ. 2007;14:56–65. doi: 10.1038/sj.cdd.4402028. [DOI] [PubMed] [Google Scholar]
- 54.Djousse L, et al. Weight loss in early stage of Huntington's disease. Neurology. 2002;59:1325–1330. doi: 10.1212/01.wnl.0000031791.10922.cf. [DOI] [PubMed] [Google Scholar]
- 55.Morales LM, et al. Nutritional evaluation of Huntington disease patients. Am J Clin Nutr. 1989;50:145–150. doi: 10.1093/ajcn/50.1.145. [DOI] [PubMed] [Google Scholar]
- 56.Jenkins BG, et al. Effects of CAG repeat length, HTT protein length and protein context on cerebral metabolism measured using magnetic resonance spectroscopy in transgenic mouse models of Huntington's disease. J Neurochem. 2005;95:553–562. doi: 10.1111/j.1471-4159.2005.03411.x. [DOI] [PubMed] [Google Scholar]
- 57.Gu M, et al. Mitochondrial defect in Huntington's disease caudate nucleus. Ann Neurol. 1996;39:385–389. doi: 10.1002/ana.410390317. [DOI] [PubMed] [Google Scholar]
- 58.Saft C, et al. Mitochondrial impairment in patients and asymptomatic mutation carriers of Huntington's disease. Mov Disord. 2005;20:674–679. doi: 10.1002/mds.20373. [DOI] [PubMed] [Google Scholar]
- 59.Panov AV, et al. Early mitochondrial calcium defects in Huntington's disease are a direct effect of polyglutamines. Nat Neurosci. 2002;5:731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- 60.Milakovic T, Johnson GV. Mitochondrial respiration and ATP production are significantly impaired in striatal cells expressing mutant huntingtin. J Biol Chem. 2005;280:30773–30782. doi: 10.1074/jbc.M504749200. [DOI] [PubMed] [Google Scholar]
- 61.Squitieri F, et al. Severe ultrastructural mitochondrial changes in lymphoblasts homozygous for Huntington disease mutation. Mech Ageing Dev. 2006;127:217–220. doi: 10.1016/j.mad.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 62.Horton TM, et al. Marked increase in mitochondrial DNA deletion levels in the cerebral cortex of Huntington's disease patients. Neurology. 1995;45:1879–1883. doi: 10.1212/wnl.45.10.1879. [DOI] [PubMed] [Google Scholar]
- 63.Banoei MM, et al. Huntington's disease and mitochondrial DNA deletions: event or regular mechanism for mutant huntingtin protein and CAG repeats expansion?! Cell Mol Neurobiol. 2007;27:867–875. doi: 10.1007/s10571-007-9206-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brouillet E, Hantraye P. Effects of chronic MPTP and 3-nitropropionic acid in nonhuman primates. Curr Opin Neurol. 1995;8:469–473. doi: 10.1097/00019052-199512000-00014. [DOI] [PubMed] [Google Scholar]
- 65.Cattaneo E, et al. Loss of normal huntingtin function: new developments in Huntington's disease research. Trends Neurosci. 2001;24:182–188. doi: 10.1016/s0166-2236(00)01721-5. [DOI] [PubMed] [Google Scholar]
- 66.Sipione S, Cattaneo E. Modeling Huntington's disease in cells, flies, and mice. Mol Neurobiol. 2001;23:21–51. doi: 10.1385/MN:23:1:21. [DOI] [PubMed] [Google Scholar]
- 67.Gines S, et al. Specific progressive cAMP reduction implicates energy deficit in presymptomatic Huntington's disease knock-in mice. Hum Mol Genet. 2003;12:497–508. doi: 10.1093/hmg/ddg046. [DOI] [PubMed] [Google Scholar]
- 68.Harjes P, Wanker EE. The hunt for huntingtin function: interaction partners tell many different stories. Trends Biochem Sci. 2003;28:425–433. doi: 10.1016/S0968-0004(03)00168-3. [DOI] [PubMed] [Google Scholar]
- 69.Shimohata T, et al. Expanded polyglutamine stretches interact with TAFII130, interfering with CREB-dependent transcription. Nat Genet. 2000;26:29–36. doi: 10.1038/79139. [DOI] [PubMed] [Google Scholar]
- 70.Cui L, et al. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 71.St-Pierre J, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 72.Weydt P, et al. Thermoregulatory and metabolic defects in Huntington's disease transgenic mice implicate PGC-1alpha in Huntington's disease neurodegeneration. Cell Metab. 2006;4:349–362. doi: 10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 73.Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocr Rev. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- 74.Lin J, et al. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 75.Leone TC, et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lin J, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 77.Conti B, et al. Uncoupling protein 2 protects dopaminergic neurons from acute 1,2,3,6-methyl-phenyl-tetrahydropyridine toxicity. J Neurochem. 2005;93:493–501. doi: 10.1111/j.1471-4159.2005.03052.x. [DOI] [PubMed] [Google Scholar]
- 78.Li H, et al. Bcl-xL induces Drp1-dependent synapse formation in cultured hippocampal neurons. Proc Natl Acad Sci U S A. 2008;105:2169–2174. doi: 10.1073/pnas.0711647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Choo YS, et al. Mutant huntingtin directly increases susceptibility of mitochondria to the calcium-induced permeability transition and cytochrome c release. Hum Mol Genet. 2004;13:1407–1420. doi: 10.1093/hmg/ddh162. [DOI] [PubMed] [Google Scholar]
- 80.Osuga H, et al. Cyclin-dependent kinases as a therapeutic target for stroke. Proc Natl Acad Sci U S A. 2000;97:10254–10259. doi: 10.1073/pnas.170144197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bezprozvanny I, Hayden MR. Deranged neuronal calcium signaling and Huntington disease. Biochem Biophys Res Commun. 2004;322:1310–1317. doi: 10.1016/j.bbrc.2004.08.035. [DOI] [PubMed] [Google Scholar]
- 82.Tang TS, et al. Disturbed Ca2+ signaling and apoptosis of medium spiny neurons in Huntington's disease. Proc Natl Acad Sci U S A. 2005;102:2602–2607. doi: 10.1073/pnas.0409402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hollenbeck PJ, Saxton WM. The axonal transport of mitochondria. J Cell Sci. 2005;118:5411–5419. doi: 10.1242/jcs.02745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Borrell-Pages M, et al. Huntington's disease: from huntingtin function and dysfunction to therapeutic strategies. Cell Mol Life Sci. 2006;63:2642–2660. doi: 10.1007/s00018-006-6242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wanker EE. Hip1 and Hippi participate in a novel cell death-signaling pathway. Dev Cell. 2002;2:126–128. doi: 10.1016/s1534-5807(02)00121-1. [DOI] [PubMed] [Google Scholar]
- 86.Gauthier LR, et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118:127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 87.McGuire JR, et al. Interaction of Huntingtin-associated protein-1 with kinesin light chain: implications in intracellular trafficking in neurons. J Biol Chem. 2006;281:3552–3559. doi: 10.1074/jbc.M509806200. [DOI] [PubMed] [Google Scholar]
- 88.Colin E, et al. Huntingtin phosphorylation acts as a molecular switch for anterograde/retrograde transport in neurons. Embo J. 2008 doi: 10.1038/emboj.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gunawardena S, et al. Disruption of axonal transport by loss of huntingtin or expression of pathogenic polyQ proteins in Drosophila. Neuron. 2003;40:25–40. doi: 10.1016/s0896-6273(03)00594-4. [DOI] [PubMed] [Google Scholar]
- 90.Chang DT, et al. Mutant huntingtin aggregates impair mitochondrial movement and trafficking in cortical neurons. Neurobiol Dis. 2006;22:388–400. doi: 10.1016/j.nbd.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 91.Trushina E, et al. Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol Cell Biol. 2004;24:8195–8209. doi: 10.1128/MCB.24.18.8195-8209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chan DC. Mitochondrial fusion and fission in mammals. Annu Rev Cell Dev Biol. 2006;22:79–99. doi: 10.1146/annurev.cellbio.22.010305.104638. [DOI] [PubMed] [Google Scholar]
- 93.Hoppins S, et al. The machines that divide and fuse mitochondria. Annu Rev Biochem. 2007;76:751–780. doi: 10.1146/annurev.biochem.76.071905.090048. [DOI] [PubMed] [Google Scholar]
- 94.Varadi A, et al. Cytoplasmic dynein regulates the subcellular distribution of mitochondria by controlling the recruitment of the fission factor dynamin-related protein-1. J Cell Sci. 2004;117:4389–4400. doi: 10.1242/jcs.01299. [DOI] [PubMed] [Google Scholar]
- 95.Smirnova E, et al. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yoon Y, et al. Mammalian dynamin-like protein DLP1 tubulates membranes. Mol Biol Cell. 2001;12:2894–2905. doi: 10.1091/mbc.12.9.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ingerman E, et al. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J Cell Biol. 2005;170:1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen H, et al. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ishihara N, et al. Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J Cell Sci. 2004;117:6535–6546. doi: 10.1242/jcs.01565. [DOI] [PubMed] [Google Scholar]
- 100.Olichon A, et al. The human dynamin-related protein OPA1 is anchored to the mitochondrial inner membrane facing the inter-membrane space. FEBS Lett. 2002;523:171–176. doi: 10.1016/s0014-5793(02)02985-x. [DOI] [PubMed] [Google Scholar]
- 101.Carelli V, et al. Mitochondrial dysfunction as a cause of optic neuropathies. Prog Retin Eye Res. 2004;23:53–89. doi: 10.1016/j.preteyeres.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 102.Kijima K, et al. Mitochondrial GTPase mitofusin 2 mutation in Charcot-Marie-Tooth neuropathy type 2A. Hum Genet. 2005;116:23–27. doi: 10.1007/s00439-004-1199-2. [DOI] [PubMed] [Google Scholar]
- 103.Zuchner S, et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
- 104.Alexander C, et al. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat Genet. 2000;26:211–215. doi: 10.1038/79944. [DOI] [PubMed] [Google Scholar]
- 105.Delettre C, et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet. 2000;26:207–210. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- 106.Baloh RH, et al. Altered axonal mitochondrial transport in the pathogenesis of Charcot-Marie-Tooth disease from mitofusin 2 mutations. J Neurosci. 2007;27:422–430. doi: 10.1523/JNEUROSCI.4798-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Detmer SA, Chan DC. Functions and dysfunctions of mitochondrial dynamics. Nat Rev Mol Cell Biol. 2007;8:870–879. doi: 10.1038/nrm2275. [DOI] [PubMed] [Google Scholar]
- 108.Benchoua A, et al. Involvement of mitochondrial complex II defects in neuronal death produced by N-terminus fragment of mutated huntingtin. Mol Biol Cell. 2006;17:1652–1663. doi: 10.1091/mbc.E05-07-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Karbowski M, et al. Spatial and temporal association of Bax with mitochondrial fission sites, Drp1, and Mfn2 during apoptosis. J Cell Biol. 2002;159:931–938. doi: 10.1083/jcb.200209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ralser M, et al. Ataxin-2 and huntingtin interact with endophilin-A complexes to function in plastin-associated pathways. Hum Mol Genet. 2005;14:2893–2909. doi: 10.1093/hmg/ddi321. [DOI] [PubMed] [Google Scholar]
- 111.Schuske KR, et al. Endophilin is required for synaptic vesicle endocytosis by localizing synaptojanin. Neuron. 2003;40:749–762. doi: 10.1016/s0896-6273(03)00667-6. [DOI] [PubMed] [Google Scholar]
- 112.Verstreken P, et al. Synaptojanin is recruited by endophilin to promote synaptic vesicle uncoating. Neuron. 2003;40:733–748. doi: 10.1016/s0896-6273(03)00644-5. [DOI] [PubMed] [Google Scholar]
- 113.Cuddeback SM, et al. Molecular cloning and characterization of Bif-1. A novel Src homology 3 domain-containing protein that associates with Bax. J Biol Chem. 2001;276:20559–20565. doi: 10.1074/jbc.M101527200. [DOI] [PubMed] [Google Scholar]
- 114.Pierrat B, et al. SH3GLB, a new endophilin-related protein family featuring an SH3 domain. Genomics. 2001;71:222–234. doi: 10.1006/geno.2000.6378. [DOI] [PubMed] [Google Scholar]
- 115.Karbowski M, et al. Role of Bax and Bak in mitochondrial morphogenesis. Nature. 2006;443:658–662. doi: 10.1038/nature05111. [DOI] [PubMed] [Google Scholar]
- 116.Brustovetsky N, et al. Increased susceptibility of striatal mitochondria to calcium-induced permeability transition. J Neurosci. 2003;23:4858–4867. doi: 10.1523/JNEUROSCI.23-12-04858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Johnson DT, et al. Functional consequences of mitochondrial proteome heterogeneity. Am J Physiol Cell Physiol. 2007;292:C698–707. doi: 10.1152/ajpcell.00109.2006. [DOI] [PubMed] [Google Scholar]
- 118.Johnson DT, et al. Tissue heterogeneity of the mammalian mitochondrial proteome. Am J Physiol Cell Physiol. 2007;292:C689–697. doi: 10.1152/ajpcell.00108.2006. [DOI] [PubMed] [Google Scholar]