Abstract
Mycobacterium avium subsp. paratuberculosis (MAP) is the etiologic agent of Johne’s disease in cattle and other farm ruminants, and is also a suspected pathogen of Crohn’s disease in humans. Development of diagnostic methods for MAP infection has been a challenge over the last few decades. The objective of this study was to investigate the relationship between different methods for detection of MAP in milk and fecal samples. A total of 134 milk samples and 110 feces samples were collected from 146 individual cows in 14 MAP-infected herds in southwestern Ontario. Culture, IS900 polymerase chain reaction (PCR) and nested PCR methods were used for detecting MAP in milk; results were compared with those of fecal culture. A significant relationship was found between milk culture, direct PCR, and nested PCR (P < 0.05). The fecal culture results were not related to any of the 3 assay methods used for the milk samples (P > 0.10). Although fecal culture showed a higher sensitivity than the milk culture method, the difference was not significant (P = 0.2473). The number of MAP colony-forming units (CFU) isolated by culture from fecal samples was, on average, higher than that isolated from milk samples (P = 0.0083). There was no significant correlation between the number of CFU cultured from milk and from feces (Pearson correlation coefficient = 0.1957, N = 63, P = 0.1243). The animals with high numbers of CFU in milk culture may not be detected by fecal culture at all, and vise versa. A significant proportion (29% to 41%) of the positive animals would be missed if only 1 culture method, instead of both milk and feces, were to be used for diagnosis. This suggests that the shedding of MAP in feces and milk is not synchronized. Most of the infected cows were low-level shedders. The proportion of low-level shedders may even be underestimated because MAP is killed during decontamination, thus reducing the chance of detection. Therefore, to identify suspected Johne’s-infected animals using the tests in this study, both milk and feces samples should be collected in duplicate to enhance the diagnostic rate. The high MAP kill rate identified in the culture methods during decontamination may be compensated for by using the nested PCR method, which had a higher sensitivity than the IS900 PCR method used.
Résumé
Mycobacterium avium subsp. paratuberculosis (MAP) est l’agent étiologique de la maladie de Johne chez les bovins et autres ruminants, et est également suspecté d’être l’agent pathogène de la maladie de Crohn chez l’humain. Le développement de méthodes diagnostiques pour les infections par MAP a posé un défi au cours des dernières décennies. L’objectif de la présente étude était de valider la relation entre différentes méthodes de détection de MAP dans le lait et les fèces. Au total, 134 échantillons de lait et de 110 échantillons de fèces ont été prélevés de 146 vaches réparties dans 14 troupeaux infectés par MAP dans le sud-ouest ontarien. Pour la détection de MAP dans le lait on utilisa la culture ainsi que les méthodes PCR pour IS900 et le PCR niché, et les résultats ont été comparés à ceux obtenus pour la culture fécale. Une relation significative a été notée entre les résultats de la culture du lait, le PCR direct et le PCR niché (P < 0,05). Les résultats de culture fécale n’étaient reliés d’aucune façon aux trois méthodes d’analyse utilisées pour les échantillons de lait (P > 0,10). Bien que la culture de fèces avaient une plus grande sensibilité que la méthode de culture du lait, la différence n’était pas significative (P = 0,2473). Le nombre d’unité formatrice de colonie (CFU) de MAP isolée par culture d’échantillons de fèces était en moyenne supérieur à celui dans le lait (P = 0,0083). Il n’y avait aucune corrélation significative entre le nombre de CFU cultivé du lait et celui cultivé des fèces (coefficient de corrélation de Pearson = 0,1957, N = 63, P = 0,1243). Les animaux avec des dénombrements élevés de CFU dans le lait peuvent ne pas être détectés par culture de fèces, et vice versa. Une proportion significative (29–41 %) de ces animaux positifs serait manquée si une seule méthode de culture était utilisée, au lieu de la culture simultanée du lait et des fèces. Ce résultat suggère que l’excrétion de MAP dans les fèces et le lait n’est pas synchronisée. La plupart des vaches infectées excrétaient MAP à de faibles niveaux. La proportion d’excréteurs de faibles niveaux pourrait même être sous-estimée étant donné la destruction de MAP durant l’étape de décontamination et les plus faibles chances d’être détecté. Ainsi, afin d’identifier les animaux individuels suspects d’être infectés par MAP au moyen des analyses utilisées dans la présente étude, du lait et des fèces devraient être prélevés, et l’échantillonnage répété afin d’augmenter le taux de succès du diagnostic. Le haut taux de mortalité identifié durant l’étape de décontamination lors de la culture pourrait être compensé par l’utilisation de PCR niché, qui a démontré une meilleure sensibilité parmi les méthodes de PCR utilisées.
(Traduit par Docteur Serge Messier)
Introduction
Mycobacterium avium subsp. paratuberculosis (MAP), the recognized pathogen of Johne’s disease (JD), causes chronic granulomatous enteritis in cattle, sheep, and other ruminants (1,2), and results in significant economical loss to the dairy industry (3). It is also a suspected pathogen of Crohn’s disease in humans (4). This pathogen has been cultured from cows with clinical or subclinical JD in both their milk and feces (5,6).
Unfortunately, the culture methods require 8 to 16 wk to confirm that a sample is negative for MAP (7); therefore, a rapid and sensitive protocol for detection of MAP is important for development of a JD control program. Efforts have been made in the last few decades to develop protocols for the detection of MAP in feces, milk, tissue, food, and environmental samples using various methods. Serology and fecal culture, however, are the most commonly used tests in the field (8,9). Polymerase chain reaction (PCR) is an ideal method for rapid turnaround time, but its test sensitivity is lower than desired for use in a control program (10). Progress has been made recently to improve the sensitivity of PCR-based tests for MAP in milk (11,12,13), and milk from asymptomatic cows infected with MAP has been estimated to contain only 2 to 8 colony-forming units (CFU) per 50 mL of sample (6). Apart from the low number of organisms in the sample, it is also difficult to lyse the MAP present by boiling; therefore, a mechanical force must be used to break up the bacterial cell wall to access the DNA for increased sensitivity in PCR detection (14). While most of the laboratories used only the pellet fraction for isolation of MAP from milk, we found that unlike other bacteria, most of the MAP in raw milk dispersed to the cream fraction instead of the pellet after centrifugation because of the lipid-rich cell walls in cream. Therefore, both pellet and cream should be processed for recovery of MAP in milk. These findings have significantly increased the sensitivity of our protocols for isolation and PCR detection of MAP in our laboratory (13–15). Although a number of reports are available for comparison of protocols for the detection of MAP in milk and feces (8,16), more information is still needed to understand the correlation and agreement between the feces and milk samples, and between the assay methods used. Such information is important for decision-making in protocol selection for screening of MAP in a population and for individual diagnoses. The assessment of a prevalence study on the samples and detection methods used may also be affected. The objective of this study was to investigate the correlation and agreement between milk culture and PCR using updated and optimized protocols, and compare the results with those found using fecal culture.
Materials and methods
Milk and fecal samples
A total of 134 milk samples and 110 feces samples were collected from 146 individual cows in 14 MAP-infected herds in southwestern Ontario. Both milk and fecal samples were available for 99 individuals. All cows enrolled had tested positive on fecal culture, or milk or serum enzyme-linked immunosorbent assay (ELISA) in previous herd screening. The teats of cows were disinfected with a chlorhexidine spray and dried with individual paper towels. A 60-mL composite milk sample was collected using a technique described by the National Mastitis Council (17). Fecal samples (35 mL) were collected per rectum using a new sterile rectal sleeve for each individual cow (18). Collected samples were transported to the laboratory at 4°C and stored at −80°C until being processed.
Culture of milk sample
Isolation and culture of MAP from the milk samples followed a previously developed protocol (15). Fifty milliliters of the milk sample were transferred to a sterile 50-mL centrifuge tube and centrifuged at 3100 × g (Beckman, GH-3.8 rotor at 3700 rpm) for 30 min at room temperature. The whey was removed and 20 mL of 0.75% hexadecylpyridium chloride (HPC) was added to the combined pellet and cream; the contents were then mixed by vortexing. Samples were incubated at room temperature for 2 h with shaking at intervals of 15 min then centrifuged at 1600 × g for 10 min. The pellet was resuspended in 0.5 mL of antibiotic brew (per liter: amphotericin B 50 mg, vancomycin 100 mg, nalidixic acid 100 mg, BHI 18.5 g). A portion of 125 μL was plated out on each Herrold’s egg yolk medium (HEYM, with mycobactin J 2 μg/mL) slant containing antibiotics (per liter: amphotericin B 50 mg, vancomycin 100 mg, nalidixic acid 100 mg). Two slants were used for each sample. The remaining suspension was stored at −20°C for PCR assay, which was conducted about 2 to 3 mo after. Inoculated slants were incubated at 37°C at slanted position with loose cap for about a week until the excessive liquid had evaporated. Caps were closed tightly and the tubes incubated for up to 24 wk. Slants were examined every 2 to 4 wk for growth and colonies suspected to be MAP were confirmed by IS900 PCR.
PCR assay of milk samples
To prepare template DNA, the samples (~ 300 μL), which were frozen at −20°C after 250 μL had been used for culture, were subjected to bead beating, and DNA was recovered as previously described (14). Twenty microliters of the 50 μL final template DNA solution were used in PCR detection. The PCR reaction mix (50 μL) contained 0.2 mM each of the 4 dNTPs, 0.2 μM each of the 2 primers P90+ and P91+ (19), 1.5 mM MgCl2 and 1 U of Taq polymerase (HotStar Taq; Qiagen, Missisauga, Ontario). The thermocycler was programmed for one cycle at 94°C for 5 min, and 40 cycles of 93°C × 2 min, 58°C × 1 min, and 72°C × 3 min, with a final extension at 72°C for 10 min. A product of 413 base pairs (bp) was obtained from this first round PCR. In the nested PCR reaction, the same PCR conditions were used with following exceptions: 1) 1 μL of the first PCR product served as template in a 15 μL reaction; 2) primers P25 and P26 (19), generating an internal 219 bp product, were used; and 3) programmed for 30 cycles. The PCR products were separated using a 1.8% agarose gel at 5 V/cm for 30 min, stained with ethidium bromide, and visualized using the Gel Doc 1000 system (Bio-Rad, Hercules, California, USA).
Culture of fecal samples
Culture from fecal samples followed the procedures previously described (7,20) with modification. A 6 to 8-mL fecal sample was added to a 50-mL conical tube containing 35 mL of sterile 7H9 broth without supplements. Samples were gently shaken horizontally at room temperature for 30 min. Tubes were then placed upright for 10 min to allow solids to settle. A 2.0-mL aliquot was taken from the top of the sample and transferred to a 15-mL conical tube containing 10 mL of 0.9% HPC. Tube contents were mixed thoroughly by inversion and then incubated overnight at 37°C. After centrifugation at 3000 × g for 20 min, supernatants were discarded, and the pellets were suspended in 1.0 mL of 7H9 broth supplemented with ADC also containing 0.5% glycerol, mycobactin J (2 mg/L), van-comycin (100 mg/L), nalidixic acid (100 mg/L), amphotericin B (50 mg/L) and cycloheximide (500 mg/L), and incubated for 36 to 48 h at 37°C. A 250-μL aliquot from each sample was inoculated onto 2 HEYM slants and incubated in slanted position with caps loosened to dry the surface. Caps were then tightened and the tubes incubated at 37°C in upright position until colonies were visible (approximately 5 to 6 wk), or for 16 wk when no MAP colonies were noticed.
Confirmation of MAP colony on slants
A visible tiny culture was picked from suspected single colony with a needle and suspended in 50 μL of water; 1 μL of the suspension served as the template. The PCR condition was the same as previously mentioned for the first round PCR, except that the PCR reaction volume was reduced to 10 μL and 30 cycles.
Determination of positive and negative culture on slant
In milk culture, the sample was scored positive if at least 1 of the suspected colonies was confirmed to be MAP, and negative if there was no growth or none of the colonies was confirmed to be MAP. In 14 milk samples, 1 of their duplicate slants was overwhelmed by fast growing colonies, and the result was scored based on only 1 slant. Data for culture were lost in 1 sample due to heavy contamination on both slants. Although most of the fast-growing bacteria were inhibited after decontamination there were still many kinds of slow-growing bacteria on the slants, and most of them were not MAP. Some MAP colonies were even buried in other fast-growing colonies or mold. For this reason, slants should be checked at intervals every 2 to 4 wk, especially during such long incubation periods. Milk culture, direct PCR, nested PCR, and fecal culture results were obtained for 99 cows.
Data analysis and statistics
All the data were managed and the charts were created using a computer program (Microsoft Office Excel; Microsoft Corp., Redmond, Washington, USA). Statistical analyses were conducted using a statistics program (SAS; SAS Institute, Cary, North Carolina, USA). The chi-squared test and Kappa coefficient were employed to study the independence and agreement between methods. Pearson’s correlation coefficient was used to analyze the correlation between the numbers of CFU isolated from milk and fecal samples. Paired t-test (2-tailed) was used to compare the numbers of MAP CFU isolated from milk and fecal samples. The Z-test was used to compare the difference between the proportions of positive samples detected in milk and fecal samples. The relative sensitivity was calculated based on the gold standard that the cow was regarded as JD positive if live MAP was isolated from either the milk or fecal sample by culture method.
Results
Identification of positive samples
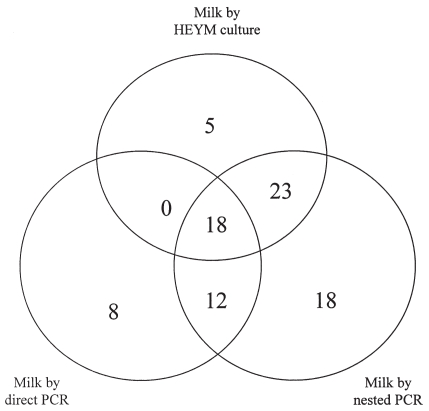
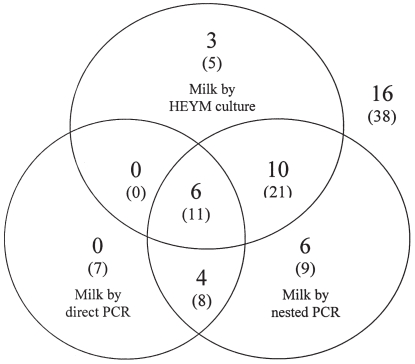
The positive samples detected by culture, direct PCR, and nested PCR in milk samples are presented in Figure 1. The positive samples by fecal culture and their association with those as detected by the 3 methods for milk samples are presented in Figure 2. All positive samples are summarized in Table I.
Figure 1.
Identification of positive milk samples by 3 detection methods (N = 133).
Figure 2.
Identification of 45 positive samples (without parenthesis) by fecal culture and their conjunction with samples as identified by other 3 methods (in parentheses) used for milk samples (N = 99). For example, at the center of the chart 11 (in parentheses) represents samples identified as positive by all the 3 methods employed for milk, and among them, 6 were also identified positive by fecal culture. The upper right corner shows 38 (in parentheses) samples that were negative by all the 3 methods for milk, while 16 of them were identified as positive by fecal culture.
Table I.
Summary of positive samples as diagnosed by 4 methods for detection of Mycobacterium avium subsp. paratuberculosis in milk and feces samples collected in dairy herds infected with Johne’s disease in southwestern Ontario
| Method | Milk culture | Milk direct PCR | Milk nested PCR | Fecal culture |
|---|---|---|---|---|
| Positive/Total (percentage) | 46/133 (34.6%) | 38/134 (28.4%) | 72/134 (53.7%) | 46/110 (41.8%) |
Independency, agreement, and correlation between methods
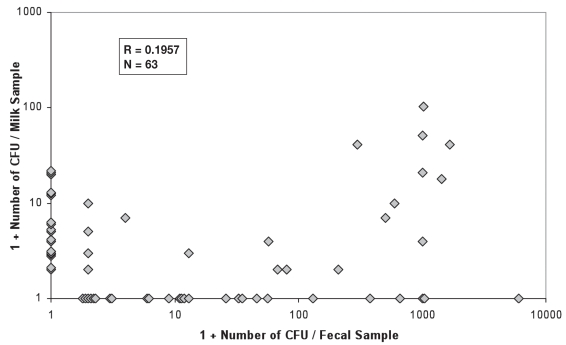
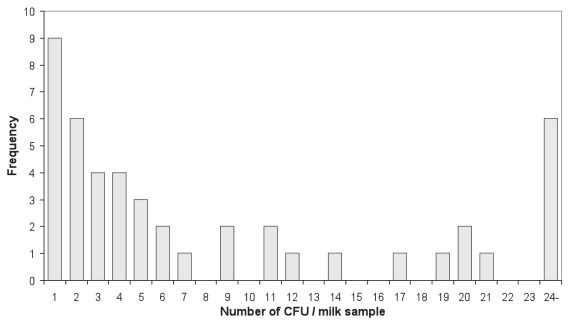
The results for chi-squared tests between the 4 detection methods and Kappa coefficients are presented in Table II. The correlation of the numbers of CFU as isolated by culture method between milk and feces is not significant (P = 0.1243, Figure 3). The average number of MAP CFU isolated by culture method from fecal samples is higher than that from the milk (P = 0.0083). Histograms for the number of CFU isolated from milk and fecal samples are presented in Figures 4 and 5, respectively based on all samples available.
Table II.
Results of chi-squared test and Kappa coefficient among 4 detection methods for detection of Mycobacterium avium subsp. paratuberculosis in milk and fecal samples collected from dairy herds infected with Johne’s disease in southwestern Ontario
| Fecal culture | Milk nested PCR | Milk direct PCR | |
|---|---|---|---|
| Milk culture | |||
| chi-squared | 0.8286 | 36.1096 | 3.8418 |
| P-value | 0.3627 | < 0.0001 | 0.04996 |
| N | 99 | 133 | 133 |
| Kappa | 0.0902 | 0.4844a | 0.1683 |
| Milk direct PCR | |||
| chi-squared | 0.6955 | 13.5661 | |
| P-value | 0.4043 | 0.0002 | |
| N | 99 | 134 | |
| Kappa | −0.0768 | 0.2771 | |
| Milk nested PCR | |||
| chi-squared | 2.2642 | ||
| P-value | 0.1324 | ||
| N | 99 | ||
| Kappa | 0.1507 | ||
The best agreement between milk culture and milk nested PCR.
Figure 3.
Correlation of the number of CFU of MAP isolated per animal between milk and fecal samples, based on the samples that were positive in at least milk or fecal samples (Cf. Figure 2). Note: One is added to the real number of CFU to allow plotting on a logarithmic scale, that is 1 on the axes represents for 0 CFU and 10 for 9 CFU, and so on. Some dots were shifted technically from where they should be to avoid overlap.
Figure 4.
Frequency of the number of CFU isolated per milk sample based on 46 positive samples out of 133 assayed.
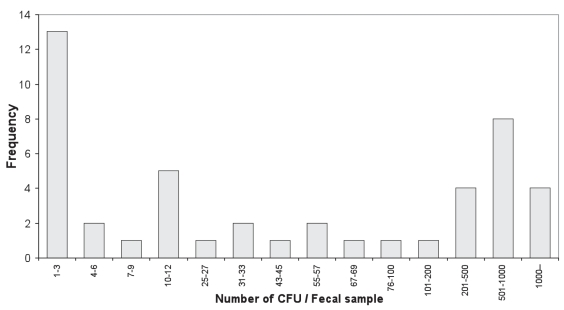
Figure 5.
Frequency of the number of CFU isolated per fecal sample based on 46 positive samples out of 110 assayed. Note: The class ranges are not equal, nor continuous.
Discussion
General relationships among diagnostic methods
A significant relationship was recognized between milk culture and nested PCR, and between milk direct and nested PCR as well (P < 0.001). Milk culture and direct PCR had a significant association (P = 0.04996). The fecal culture was not related to any of the 3 assay methods used for milk samples (P > 0.10, Table II). Results from chi-squared tests are consistent with the Kappa coefficient findings. Generally, the agreement is high between milk tests, and is low between the milk tests and fecal culture. The highest agreement was recognized between milk culture and milk nested PCR, while the lowest agreement was between fecal culture and milk direct PCR. Similar results indicating that the relationship between fecal culture and milk nested PCR was not significant (P = 0.0589) were reported based on samples from 52 herds (8). The results herein differ from a recent report (21), in which milk culture was claimed to be the most sensitive, followed by fecal culture, m-ELISA, and fecal PCR base on 26 lactating dairy cows that had Johne’s disease. Publications on the relationships between milk culture and fecal culture for JD-infected cows are scarce.
Sensitivity of the detection methods
Nested PCR resulted in the highest number of positive milk samples (Tables I and III), which is consistent with the finding that the milk nested PCR yielded more positive samples than did fecal culture (8). The availability of both milk culture and fecal culture for the same animals allowed us to evaluate the relative sensitivity of these 2 culture methods. Although higher sensitivity was found for fecal culture than the milk culture method (Tables I and III, Figure 2), the difference is not significant (P = 0.2473). A close look of Figure 2 reveals that among the 63 (37 positive in milk and 45 positive in feces with an overlap of 19) culture positive samples, 18 (28.6%) of the positive animals would remain undetected if only the fecal culture method were used, or 26 (41.3%) missed if only the milk culture were used for diagnosis. The correlation between the number of CFU from milk and fecal samples was not significant, and the animals with a high number of CFU in milk culture might not be detected by fecal culture at all, and vise versa. All the spots plotted on the axes represent the animals, which were detected by only milk or fecal sample, and missed by the other (Figure 3). This suggests that the shedding of MAP organism in feces is not synchronized with the shedding in milk. According to our experience, if a sample was scored as positive based on only 1 or a few CFU, the result had low reproducibility. A similar situation was found in another study with the semi-nested PCR for detection of MAP in fecal samples (22). Therefore, it is possible that some positive animals were missed due to low MAP-shedding levels and competitive growth of background flora. Of the 46 positive milk samples, 26 were determined to be positive based on 1–5 CFU (Table I, Figure 4); a similar situation was found in the fecal culture (Figure 5). This may be an important finding in the context of developing a control program for JD. The results suggest that both milk and feces might be useful for identifying suspected individuals. In addition, there is added value in using duplicate samples, to enhance the detection rate of the low-level shedders.
Table III.
Relative sensitivity based on 99 matched feces and milk samples (Golden standard: A sample is regarded as positive when MAP was isolated from either milk or fecal sample and confirmed to be MAP by IS900 PCR)
| Method | Fecal culture | Milk culture | Milk direct PCR | Milk nested PCR |
|---|---|---|---|---|
| Relative sensitivity | 71.4% | 58.7% | 41.3% | 77.8% |
Detection of MAP by PCR
The protocol for PCR detection was an optimized procedure developed in our laboratory (13,14). The limit of detection, as evaluated by IS900 PCR, was as low as ≤ 1 CFU/mL in an initial 50-mL sample. Although 50-mL milk samples were used, about half of the final suspension, 250 μL, was used for MAP culture. Therefore, the limit of detection in direct PCR was about ≤ 2 CFU/mL. Most of the 38 positive milk samples in direct PCR had weak to very weak bands (Table I). Among them, 8 were negative in both milk culture and nested PCR (Figure 1), suggesting that the DNA bands amplified in direct PCR were not from the IS900 sequence. We also had difficulties in reliably scoring results for another 13 of the milk samples due to a low amplified DNA signal. Of the 13 samples, 4 were positive in fecal culture, 6 positive in milk culture, 7 positive by nested PCR, while 5 were positive by both milk culture and nested PCR. These 13 samples are not included in the 38 positive milk samples by direct PCR in Table I, because the amplified DNA bands were too weak to be reliable. Our result is consistent with the estimation by Sweeney et al (6) that the concentration of organisms in milk samples was low (2 to 8 CFU/50 mL). Therefore, we suggest that nested PCR should be applied for detection of MAP in milk samples for clinical diagnosis or prevalence studies, and both the cream and pellet from a large volume be processed for organism isolation or template DNA preparation (15), otherwise, the organism could be missed easily. Nested PCR produced a strong band in gel electrophoresis, and the highest sensitivity among all the detection methods (Tables I and III, Figure 1). As the most sensitive method, nested PCR detected 18 milk samples, which were not detected by culture and direct PCR. Consistent with our results from milk, more fecal samples were scored positive by semi-nested real-time PCR than by culture, and the nested PCR was considered a suitable alternative method to culture for detection of MAP in a surveillance program (22). These nested PCR positive, culture negative samples may represent the cows shedding MAP at levels around or below the detection limit of culture methods.
Assay limitations
The culture method used in our laboratory recovered 16% of MAP, on average, in raw milk; however, most of the organisms were killed during decontamination (at 21°C for 2–4 h), or lost during processing. The recovery rate decreased when the sample was decontaminated at a higher temperature (37°C), or for longer a incubation time, or both (15). The commonly used fecal culture protocol should kill more MAP at decontamination due to the use of a higher temperature (37°C) and longer incubation time (overnight). The sensitivity of a fecal culture would be greatly improved if a mild-to-MAP decontamination protocol or more efficient recovery protocol such as immunomagnetic separation were available. Due to limitation of each assay tested, some infected animals, especially the low-shedders, would be missed, and disease prevalence be underestimated if screening relies on a single method, or screens either milk or feces but not both. Since most of the infected cows were light shedders, development of a more sensitive method is still needed. The high rate of MAP killed though decontamination and the low rate of recovery from samples must be overcome or compensated by using a more sensitive protocol for detection of MAP, such as the nested PCR in milk and fecal samples at this stage.
Acknowledgments
The authors acknowledge the financial support received from the Special Research Fund Program of Ontario Ministry of Agriculture and Food, Health Canada, the Dairy Farmers of Ontario and from Elanco Animal Health.
References
- 1.Beard PM, Daniels MJ, Henderson D, et al. Paratuberculosis infection of nonruminant wildlife in Scotland. J Clin Microbiol. 2001;39:1517–1521. doi: 10.1128/JCM.39.4.1517-1521.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiodini RJ, van Kruiningen HJ. Characterization of Mycobacterium paratuberculosis of bovine, caprine, and ovine origin by gas-liquid chromatographic analysis of fatty acids in whole-cell extracts. Am J Vet Res. 1985;46:1980–1989. [PubMed] [Google Scholar]
- 3.Ott SL, Wells SJ, Wagner BA. Herd-level economic losses associated with Johne’s disease on US dairy operations. Prev Vet Med. 1999;40:179–192. doi: 10.1016/s0167-5877(99)00037-9. [DOI] [PubMed] [Google Scholar]
- 4.Hermon-Taylor J, Bull TJ, Sheridan JM, et al. Causation of Crohn’s disease by Mycobacterium avium subspecies paratuberculosis. Can J Gastroenteriol. 2000;14:521–539. doi: 10.1155/2000/798305. [DOI] [PubMed] [Google Scholar]
- 5.Streeter RN, Hoffsis GF, Bech-Nielsen S, et al. Isolation of Mycobacterium paratuberculosis from colostrum and milk of subclinically infected cows. Am J Vet Res. 2995;56:1322–1324. [PubMed] [Google Scholar]
- 6.Sweeney RW, Whitlock RH, Rosenberger AE. Mycobacterium paratuberculosis cultured from milk and supramammary lymph nodes of infected asymptomatic cows. J Clin Microbiol. 1992;30:166–171. doi: 10.1128/jcm.30.1.166-171.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whipple D, Callihan D, Jarnagin J. Cultivation of Mycobacterium paratuberculosis from bovine faecal specimens and a suggested standardized procedure. J Vet Diag Invest. 1991;3:368–373. doi: 10.1177/104063879100300424. [DOI] [PubMed] [Google Scholar]
- 8.Stabel JR, Wells SJ, Wagner BA. Relationships between fecal culture, ELISA, and bulk tank milk test results for Johne’s disease in US dairy herds. J Dairy Sci. 2002;85:525–531. doi: 10.3168/jds.S0022-0302(02)74104-0. [DOI] [PubMed] [Google Scholar]
- 9.Whittington RJ, Sergeant ESG. Progress towards understanding the spread, detection and control of Mycobacterium avium subsp. paratuberculosis in animal populations. Aust Vet J. 2001;4:267–278. doi: 10.1111/j.1751-0813.2001.tb11980.x. [DOI] [PubMed] [Google Scholar]
- 10.van der Giessen JWB, Haring RM, Vauclare E, et al. Evaluation of the abilities of three diagnostic tests based on the polymerase chain reaction to detect Mycobacterium paratuberculosis in cattle: Application in a control program. J Clin Microbiol. 1992;30:1216–1219. doi: 10.1128/jcm.30.5.1216-1219.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khare S, Ficht T, Santos R, et al. Rapid and sensitive detection of Mycobacterium avium subsp. paratuberculosis in bovine milk and feces by a combination of immunomagnetic bead separation-conventional PCR and real-time PCR. J Clin Microbiol. 2004;42:1075–1081. doi: 10.1128/JCM.42.3.1075-1081.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Mahony J, Hill C. Rapid real-time PCR assay for detection and quantitation of Mycobacterium avium subsp. paratuberculosis DNA in artificially contaminated milk. Appl Envron Microbiol. 2004;70:4561–4568. doi: 10.1128/AEM.70.8.4561-4568.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao A, Mutharia L, Raymond M, Odumeru J. Improved template DNA preparation procedure for detection of Mycobacterium avium subsp. paratuberculosis in milk by PCR. J Microbiol Methods. 2007;69:417–420. doi: 10.1016/j.mimet.2006.10.019. [DOI] [PubMed] [Google Scholar]
- 14.Odumeru J, Gao A, Chen S, Raymond M, Mutharia L. Use of the bead beater for preparation of Mycobacterium paratuberculosis template DNA in milk. Can J Vet Res. 2001;65:201–205. [PMC free article] [PubMed] [Google Scholar]
- 15.Gao A, Odumeru J, Raymond M, Mutharia L. Development of improved method for isolation of Mycobacterium avium subsp. paratuberculosis from bulk tank milk: Effect of age of milk, centrifugation, and decontamination. Can J Vet Res. 2005;69:81–87. [PMC free article] [PubMed] [Google Scholar]
- 16.Paolicchii F, Zumarraga M, Gioffre A, et al. Application of different methods for the diagnosis of paratuberculosis in a dairy cattle herd in Argentina. J Vet Med B Infect Dis Vet Public Health. 2003;50:20–26. doi: 10.1046/j.1439-0450.2003.00606.x. [DOI] [PubMed] [Google Scholar]
- 17.National Mastitis Council. Microbiological Procedures for the Diagnosis of Bovine Udder Infection. 3rd ed. Verona; Wisconsin, USA: 1990. [Google Scholar]
- 18.Hendrick SH, Kelton DF, Leslie KE, et al. Efficacy of monensin sodium for the reduction of fecal shedding of Mycobacterium avium subsp. paratuberculosis in infected dairy cattle. Prev Vet Med. 2006;75:206–220. doi: 10.1016/j.prevetmed.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Millar D, Ford J, Sanderson J, et al. IS900 PCR to detect Mycobacterium paratuberculosis in retail supplies of whole pasteurized cows’ milk in England and Wales. Appl Environ Microbiol. 1996;62:3446–3452. doi: 10.1128/aem.62.9.3446-3452.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stabel JR. An improved method for cultivation of Mycobacterium paratuberculosis from bovine fecal samples and comparison to three other methods. J Vet Diagn Invest. 1997;9:375–380. doi: 10.1177/104063879700900406. [DOI] [PubMed] [Google Scholar]
- 21.Singh SV, Singh AV, Singh R, et al. Evaluation of highly sensitive indigenous milk ELISA kit with fecal culture, milk culture and fecal-PCR for the diagnosis of bovine Johne’s disease (BJD) in India. Comp Immunol Microbiol Infect Dis. 2007;30:175–186. doi: 10.1016/j.cimid.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 22.Bögli-Stuber K, Kohler C, Seitert G, et al. Detection of Mycobacterium avium subspecies paratuberculosis in Swiss dairy cattle by real-time PCR and culture: A comparison of the two assays. J App Microbiol. 2005;98:587–597. doi: 10.1111/j.1365-2672.2005.02645.x. [DOI] [PubMed] [Google Scholar]