Over the past 2 decades, the molecular mechanisms by which cells acquire, distribute, and utilize copper have been under intense investigation. Significant progress has been made in the identification of genes encoding copper homeostasis proteins and in fundamental aspects of their structure, function, and mechanisms of action in copper balance. A number of more comprehensive reviews of the field with respect to the genetics, structure, function, and physiology of copper metabolism have recently appeared elsewhere (1–4). Here, we review general mechanisms for eukaryotic copper metabolism at the cellular level in the context of recent discoveries in the field, identifying potential new functions for copper and copper metabolism proteins in cell signaling, gene expression, tumor cell metastasis, and resistance to anti-neoplastic drugs.
Copper as a Catalytic and Structural Cofactor in Biology
Copper is a redox-active metal that is predominantly used by organisms living in oxygen-rich environments and that fluctuates between the oxidized (Cu2+) and reduced (Cu+) states (5). With these changes in redox state, copper can coordinate to a range of ligands that include carboxylate oxygen, imidazole nitrogen, cysteine thiolate, and methionine thioether groups and engages in cation-π interactions (6–8). Cu+ plays a well established structural role in proteins such as the fungal copper-activated metalloregulatory transcription factors (9). For example, Ace1 in Saccharomyces cerevisiae acquires profound conformational changes upon the cooperative binding of a tetra-Cu+ cluster, activating sequence-specific DNA binding (10).
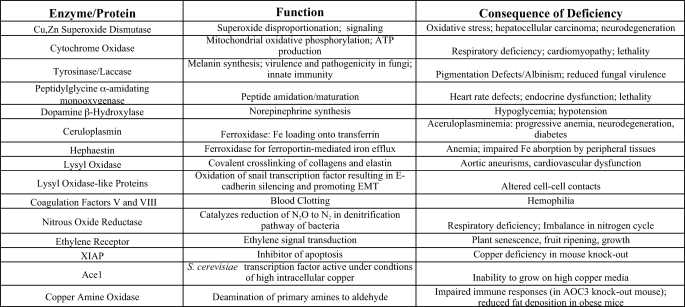
Many enzymes harness the changes in bound copper oxidation state, in the presence of oxygen, to catalyze redox chemistry for a wide range of chemical transformations that are central to biology (Table 1). Significant phenotypes result from a diminution in these enzymatic activities (Table 1). Although copper-binding proteins and enzymes have been characterized for over a century, it is likely that a number of copper-dependent proteins and their functions have yet to be identified. The diversity of potential copper ligands along a polypeptide primary structure, in addition to the proper juxtaposition of bona fide ligands in the correct three-dimensional orientation in proteins, makes prediction of the entire copper proteome a challenging area for future studies (5).
TABLE 1.
Examples of copper-binding proteins
Copper Homeostasis in Eukaryotic Cells
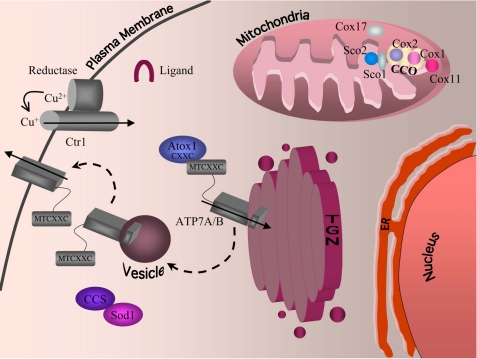
Over the past few decades, critical progress has been made in the identification of genes encoding proteins that function in copper uptake, intracellular distribution, and efflux and in the regulation of the copper homeostasis machinery (1–4). Many of the structural, functional, and regulatory details have been strikingly conserved from microbes to humans. Fig. 1 shows a model for copper homeostasis in a typical mammalian cell. Ctr1, an integral membrane protein, functions as a major copper importer at the plasma membrane. Genetic, biochemical, and structural studies support a model in which Ctr1 homotrimerizes to form a central region of low electron density through which copper may traverse the plasma membrane (11–13). The genetic requirement for a metalloreductase in yeast for Ctr1-mediated high affinity copper uptake, in addition to other data (1, 14), supports the transport of Cu+ rather than Cu2+. Conserved methionine residues are present in the Ctr1 extracellular domain as Met-X-Met or Met-X2-Met and, although not essential for activity, are needed for high affinity Cu+ import. Additionally, a conserved Met motif, Met-X3-Met, present in the Ctr1 second transmembrane domain, is essential for Cu+ import (13). These and other experimental results are consistent with thioether-Cu+ coordination to Ctr1 at one or more steps in the import process.
FIGURE 1.
Mammalian intracellular copper homeostasis. Copper is reduced by an as yet unidentified reductase and imported into cells by the high affinity copper transporter Ctr1. Once inside the cell, copper is escorted to destinations via the action of copper chaperones. CCS incorporates copper into the cytosolic protein Cu,Zn-SOD. Atox1 delivers copper to the secretory compartment; the protein possesses an N-terminal copper-binding motif (CXXC) that directly interacts with the N-terminal copper-binding domain of ATP7A and ATP7B (MTCXXC), facilitating copper delivery to ATP7A and ATP7B. ATP7A and ATP7B are P-type copper-transporting proteins that transport copper into the lumen of the Golgi, where the metal can then be incorporated into copper-dependent proteins such as the secreted form of LOX. The P-type ATPases also export copper out of the cell by translocating to the plasma membrane when intracellular copper levels are high. Copper is escorted to the mitochondria by an as yet uncharacterized ligand. Once inside the intermembrane space, copper is handed off to Cox17 and then passed onto either Sco1, which transfers copper to the Cox2 subunit of cytochrome oxidase, or Cox11, which transfers copper to the Cox1 subunit of cytochrome oxidase (CCO). TGN, trans-Golgi network; ER, endoplasmic reticulum.
Copper that is imported by Ctr1, or as yet unidentified, low affinity, Ctr1-independent activities (15), is delivered to intracellular proteins and compartments by the action of copper chaperones (1, 2, 16). CCS (copper chaperone for Cu,Zn-superoxide dismutase) delivers copper to Cu,Zn-SOD2 via direct interactions and a complex mechanism that has been recently reviewed (17). Atox1 (Atx1 in bakers' yeast, S. cerevisiae) is a small soluble protein that binds a single Cu+ atom in a specific but kinetically labile manner. As shown in Fig. 1, Atox1 directly interacts with repeated cytosolic metal-binding domains of the Cu+-transporting ATPases ATP7A and ATP7B to facilitate its delivery across the secretory compartment or, under conditions of high copper, across the basolateral membrane of cells.
In addition to delivery of copper to cytosolic proteins and to the secretory compartment, copper must be targeted to mitochondria, where cytochrome oxidase uses copper for oxidative phosphorylation. Genetic studies in microbes and the mapping of mutated genes that are responsible for defects in cytochrome oxidase assembly have identified many proteins involved in cytochrome oxidase copper metallation (2). These include Cox17 in the mitochondrial intermembrane space; Cox11 in the mitochondrial inner membrane; and Sco1 and Sco2, structurally similar proteins that play a more proximal role in copper delivery to cytochrome oxidase. Defects in Sco1 and Sco2 cause catastrophic defects in cytochrome oxidase assembly and result in severe human disease (18).
A New Role for Ctr1 in Cisplatin Accumulation
Although Cu+ is the only known physiological substrate for Ctr1, recent studies strongly suggest that Ctr1 in yeast and in mammals facilitates uptake of the metal-based anti-neoplastic drug cisplatin (19, 20). Cisplatin is an effective chemotherapeutic agent due to its ability to intercalate in DNA and thereby preferentially inhibit the growth of rapidly proliferating cells (21). Transposon mutagenesis studies in yeast identified Ctr1 as a loss-of-function mutation rendering these mutants highly resistant to cisplatin toxicity (19). Furthermore, Ctr1+/+, Ctr1+/–, or Ctr1–/– mouse embryonic fibroblasts exhibit Ctr1 gene dose-dependent acquisition of cisplatin, with Ctr1–/– cells exhibiting elevated resistance to cisplatin compared with wild-type cells (19). Moreover, cell lines overexpressing human Ctr1 accumulate elevated cisplatin compared with controls cells (20). How does a dedicated Cu+ transporter facilitate the uptake of the structurally unrelated metal complex cisplatin? Recent mutagenesis experiments and FRET studies between yeast Ctr1 subunits suggest distinct mechanisms for Cu+ import compared with cisplatin uptake (22). Lee and co-workers (22) demonstrated that copper enhances the FRET signal of functional Ctr1-enhanced cyan fluorescent protein and Ctr1-enhanced yellow fluorescent protein monomers coexpressed in yeast cells in a manner that is dependent on copper transport-competent Ctr1, suggesting that copper-induced conformational changes occur in concert with Cu+ movement through the plasma membrane. In contrast, although the Ctr1 FRET pairs were capable of stimulating cisplatin uptake, cisplatin neither induced FRET nor abrogated the copper-induced FRET signal. Moreover, a Ctr1 mutant lacking the Met-rich ectodomain was competent for Cu+ uptake but completely defective in cisplatin accumulation, further underscoring differences between Cu+ uptake and cisplatin accumulation. Similar mutations in human Ctr1 extracellular Met-X-Met motifs abrogated cisplatin-stimulated Ctr1 multimerization (23), suggesting that cisplatin may interact with these methionine thioether groups. Taken together, these studies suggest that Ctr1 may function similarly to a transporter for Cu+ acquisition but may enhance cisplatin uptake in a methionine-rich, ectodomain-dependent, receptor-mediated endocytic mechanism. Although there are conflicting reports on whether, like copper, cisplatin enhances Ctr1 endocytosis (19, 22–24), this question must be resolved to understand how Ctr1 facilitates cisplatin accumulation in a manner distinct from that of copper.
Ctr1 in Growth Factor Signaling
The generation of a mouse Ctr1 knock-out illustrated the essentiality for Ctr1 in metazoan development (25, 26). Ctr1–/– mutant embryos showed growth retardation at embryonic day 7.5, a stage in development when gastrulation begins, with embryonic lethality by embryonic day 12.5. Homozygous mutants histologically demonstrated poorly developed neural ectoderm and mesoderm and a defect in neural tube closure. Although the precise mechanisms underlying the mutant phenotypes have not been elucidated, it is possible that defects in multiple copper-dependent proteins culminate in catastrophic developmental arrest. However, recent work in Xenopus may provide an alternative explanation for the requirement for Ctr1 in normal growth and development (27). Xenopus Ctr1 (Xctr1) was identified as part of a complex consisting of Laloo and SNT-1, proteins required for FGF-mediated mesoderm induction (27). Misexpression of Xctr1 promoted mesodermal and neural ectoderm differentiation, whereas, conversely, loss of Xctr1 through morpholino injection inhibited these processes. Knockdown of Xctr1 also had an effect on pathway activation dynamics as evidenced by partial inhibition of SNT-1 phosphorylation by Laloo and strong inhibition of FGF-dependent ERK phosphorylation. Interestingly, knockdown of Xctr1 at later stages of development only modestly inhibited induction of mesoderm markers, suggesting that Xctr1 is required either before or during gastrulation for mesoderm development, a time window that parallels the developmental arrest of Ctr1–/– mutant embryos.
This study provides a potential explanation for the developmental abnormalities seen in mouse Ctr1–/– mutant embryos and forges a new intersection between copper homeostasis and a cell signaling pathway. However, the role of Xctr1 in mesoderm development is suggested to be independent of its copper-transporting activity, as misexpression of Xctr1 carrying mutations in conserved residues required for copper transport gave the equivalent phenotype as misexpression of wild-type Xctr1 (27). This unanticipated copper transport-independent role for Ctr1 in FGF signaling, morphogenesis, and differentiation must be further explored, as other reports using copper chelators suggest that copper levels modulate hematopoietic stem cell differentiation (28, 29).
Copper-dependent Metabolic Changes in Cancer Cells
Recent exciting work has implicated copper-handling and copper-utilizing proteins in controlling the striking metabolic changes that have long been known to occur in cancer cells. Otto Warburg (30) discovered that respiratory capacity is down-regulated in many cancer cell types with a concomitant dependence on glycolysis for cellular energy generation. Although this phenomenon has been known for over 75 years, the cellular regulatory mechanisms governing this metabolic switch are still not well understood. Matoba et al. (31) reasoned that this metabolic reengineering may result from regulatory changes common to many cancer cell types. Because mutations in the gene encoding the p53 sequence-specific DNA-binding transcription factor are among the most common genetic changes found in a broad range of cancer cells, they explored whether cancer cell lines harboring p53 mutations exhibit changes in their dependence on respiration versus glycolysis. Indeed, human colon cancer cells with p53-inactivating mutations also showed significant reductions in oxygen consumption while generating increased levels of lactate, a by-product of glycolysis, metabolic characteristics reflecting a switch from oxidative phosphorylation to glycolysis typical of the Warburg effect.
As the SCO2 gene was previously identified as a potential p53 target, the expression of SCO2 was explored as a function of p53 gene dosage (31, 32). Although SCO2 mRNA and protein levels were considerably reduced in p53+/– and p53–/– mouse livers compared with wild-type livers, SCO1 mRNA levels were unchanged. Furthermore, the p53-dependent stimulation of SCO2 expression was dependent on a canonical p53-binding site in the SCO2 promoter, establishing a direct functional link between p53 and SCO2 expression levels. Taken together, these studies identified a link between the p53 status of cancer cells and metabolic reprogramming toward glycolytic energy generation first observed by Warburg. With the essential role for copper in cytochrome oxidase function, it would be interesting to ascertain if p53-dependent changes in the expression of other factors that function in copper acquisition might also play a role in the Warburg effect.
Copper in Signaling and Tumor Cell Metastasis
Cell migration and tissue invasion from the primary tumor site require a loss of integrity of target tissue cell-cell junctions. The epithelial-mesenchymal transition is a critical step in the initiation of cell invasion, as it results in compromised cell-cell junctions due, at least in part, to the down-regulation of E-cadherin, a protein that functions in cell-cell adhesion (33). Normally, the transcription of E-cadherin is regulated by the Snail repressor, the protein stability of which is tightly regulated through its phosphorylation by glycogen synthase kinase 3β (34). Recent work has identified LOX-like proteins to also play a part in the regulation of Snail protein stability. Although the extracellular copper-dependent LOX is well established for its role in the maturation of collagen and elastin via lysine oxidation, four additional members of this protein family, called lysyl oxidase-like proteins (LOXL1–4), have been identified (34–36). All five members of this family harbor signatures for bound copper and lysyl-tyrosyl quinone cofactors, but LOXL1–4 lack a typical secretory propeptide found in extracellular LOX and instead contain scavenger receptor cysteine-rich domains and are predicted to be intracellular proteins (34). Recent studies have demonstrated that LOXL2 and LOXL3 interact with Snail to stabilize the protein in a manner that is dependent on two Snail lysine residues (37). These and other studies support a model to suggest that lysine oxidation diminishes glycogen synthase kinase 3β-mediated phosphorylation, thereby stabilizing Snail, inhibiting E-cadherin expression, and compromising tight junctions (33). Interestingly, LOXL2 expression is strongly elevated in highly invasive forms of metastatic breast, colon, and esophageal cancers (36, 38), consistent with a more general role in cancer cell invasion.
The observation that a family of copper-dependent amine oxidases functions not only in determining the integrity of connective tissues but also in the regulation of the epithelial-mesenchymal transition has several implications in copper biology. It will be important to decipher whether the LOXL1–4 proteins catalyze oxidative deamination of peptidyl lysines in a manner similar to extracellular LOX. Initial experiments with recombinant isoforms suggest mechanistic similarity to LOX in that they are dependent on both copper and the lysyl-tyrosyl quinone cofactor (39–41). With the discovery of a family of LOLX proteins, the field is now poised to gather information on the spectrum of their specific substrates, as there may be a plethora of pathways and processes that are altered under conditions of copper limitation.
Copper Deficiency: Who Goes First?
Important insights into the hierarchy of copper metallation in vertebrates have recently been gained through a small molecule chemical screen in zebrafish (42). A search was conducted for molecules that interfered with copper metabolism, specifically giving rise to copper-deficient phenotypes as evidenced by lack of pigmentation. Zebrafish lateral line pigmentation is mediated by tyrosinase, a copper-dependent enzyme required for melanin synthesis (43). The most severe copper-deficient phenotype, obtained by treating embryos with the copper chelator neocuproine, resulted in a deformed notochord, impaired neurogenesis, inappropriate cartilage maturation, poor vascular development, and hematopoietic defects. Furthermore, it was found that varying copper chelator concentrations would generate a continuum of phenotypes that could be correlated with the severity of copper deficiency within the embryo, from a lack of pigmentation but with a normal notochord to a lack of pigmentation and abnormal notochord development. Continued exposure of embryos to neocuproine resulted in a lack of pigmentation, notochord defects, and neurogenesis defects. These results suggest that, at early stages of copper deficiency, pigmentation processes mediated by tyrosinase would be among the first to be lost, freeing up limited copper reservoirs for more essential developmental processes. As the developing embryo encounters increasingly stringent access to copper, mechanisms appear to be in place that prioritize copper utilization to meet physiological demands.
Copper Chelation and Cancer Chemotherapy
Copper has been proposed to be essential for angiogenesis, the formation of new blood vessels that provide a delivery route for nutrients, growth factors, and other signaling agents important for tumor growth and survival. Although a number of copper-dependent roles in angiogenesis have been proposed (44), the entire range of functions important for efficient angiogenesis that require copper has not been elucidated. This is an important challenge, as TTM, a potent Cu+ chelator, has been reported to be of therapeutic value in the treatment of several types of cancers as an anti-angiogenesis and anti-cancer molecule. Recently, ATN-224, an orally available choline salt derivative of TTM, has been suggested to preferentially target Cu,Zn-SOD in tumor and endothelial cells, with the implication that SOD1 plays a stimulatory role in growth factor signaling (45, 46). SOD1 protects cells from oxidative stress and catalyzes the disproportionation of superoxide to hydrogen peroxide, molecules that have been established to play signaling roles in biology (47).
In an initial report, TTM was shown to potently inhibit Cu,Zn-SOD activity, thereby increasing superoxide anion accumulation and inducing programmed cell death in multiple myeloma cells (45). In parallel with these features, the phosphorylation of ERK, which has critical functions downstream of Ras in cell proliferation, was also inhibited (45). Recently, ATN-224 was reported to inhibit growth factor-stimulated phosphorylation of the EGF and insulin-like growth factor receptors in parallel with an increase in superoxide and a decrease in hydrogen peroxide accumulation (46). This decrease in hydrogen peroxide was proposed to reduce the inactivation of the protein-tyrosine phosphatase PTP1B, thereby inhibiting growth factor-stimulated receptor phosphorylation and attenuating downstream activation of proliferation pathways involving ERK. This is a potentially important mechanism for the action of copper chelators, but many questions remain. It is unclear why ATN-224 is effective for inhibiting SOD1 activity in cell culture in the nanomolar range but exhibits potent anti-proliferative effects at micromolar concentrations. Moreover, RNA interference-mediated knockdown of SOD1 did not exhibit anti-proliferative effects, and Sod1 knock-out mice did not show any obvious growth defects (48). Although Cu,Zn-SOD may be an important target for the anti-angiogenesis and anti-cancer activity of clinically relevant copper chelators, it is not currently clear whether other copper-dependent activities might play a role. A thorough understanding of the anti-proliferative mechanism of copper chelators will require a comprehensive identification of the copper proteome and a deeper understanding of the mechanisms of copper homeostasis and their regulation.
Future Directions
To date, genetic studies in model systems and biochemical, cell biology, and structural studies have identified many proteins involved in eukaryotic copper homeostasis and provided insights into their structures and mechanisms of action. It is likely that there are other functions and mechanisms to be discovered for known copper homeostasis proteins, and new components of this machinery will be discovered and integrated into our current understanding of cellular copper balance. Many questions have yet to be tackled in the field of copper metabolism, from new, exciting, and perhaps serendipitous perspectives. How does copper make its way from the site of import at the plasma membrane, or efflux from intracellular storage vesicles, to the copper chaperones and other intracellular ligands? What is the entire constellation of copper-binding proteins in a mammalian cell? How do the components of the copper homeostasis machinery communicate, and what other signals, including chemical, hormonal, and environmental, trigger changes in copper homeostasis? How do multicellular organisms sense copper deficiency in peripheral tissues and transmit this signal to mobilize copper stores? The field of copper homeostasis is currently similar to a high school graduate going off to college, armed with some basic information; we will see how things develop with additional outside influence.
Supplementary Material
Acknowledgments
We gratefully acknowledge Tracy Nevitt and Byung-Eun Kim for critical comments on this review.
This work was supported, in whole or in part, by National Institutes of Health Grant ES010356. This is the second article of three in the Thematic Minireview Series on Metals in Biology. This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.
Footnotes
The abbreviations used are: SOD, superoxide dismutase; FRET, fluorescence resonance energy transfer; FGF, fibroblast growth factor; ERK, extracellular signal-regulated kinase; LOX, lysyl oxidase; TTM, tetrathiomolybdate.
References
- 1.Kim, B.-E., Nevitt, T., and Thiele, D. J. (2008) Nat. Chem. Biol. 4 176–185 [DOI] [PubMed] [Google Scholar]
- 2.Cobine, P. A., Pierrel, F., and Winge, D. R. (2006) Biochim. Biophys. Acta 1763 759–772 [DOI] [PubMed] [Google Scholar]
- 3.Lutsenko, S., Barnes, N. L., Bartee, M. Y., and Dmitriev, O. Y. (2007) Physiol. Rev. 87 1011–1046 [DOI] [PubMed] [Google Scholar]
- 4.Prohaska, J. R., and Gybina, A. A. (2004) J. Nutr. 134 1003–1006 [DOI] [PubMed] [Google Scholar]
- 5.Ridge, P. G., Zhang, Y., and Gladyshev, V. N. (2008) PLoS ONE 3 e1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertini, I. (2007) Biological Inorganic Chemistry: Structure and Reactivity, University Science Books, Sausalito, CA
- 7.Lippard, S. J., and Berg, J. M. (1994) Principles of Bioinorganic Chemistry, University Science Books, Mill Valley, CA
- 8.Franz, K. J. (2008) Nat. Chem. Biol. 4 85–86 [DOI] [PubMed] [Google Scholar]
- 9.Rutherford, J. C., and Bird, A. J. (2004) Eukaryot. Cell 3 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dameron, C. T., Winge, D. R., George, G. N., Sansone, M., Hu, S., and Hamer, D. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 6127–6131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aller, S. G., and Unger, V. M. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 3627–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dancis, A., Haile, D., Yuan, D. S., and Klausner, R. D. (1994) J. Biol. Chem. 269 25660–25667 [PubMed] [Google Scholar]
- 13.Puig, S., Lee, J., Lau, M., and Thiele, D. J. (2002) J. Biol. Chem. 277 26021–26030 [DOI] [PubMed] [Google Scholar]
- 14.Lee, J., Pena, M. M., Nose, Y., and Thiele, D. J. (2002) J. Biol. Chem. 277 4380–4387 [DOI] [PubMed] [Google Scholar]
- 15.Lee, J., Petris, M. J., and Thiele, D. J. (2002) J. Biol. Chem. 277 40253–40259 [DOI] [PubMed] [Google Scholar]
- 16.Puig, S., and Thiele, D. J. (2002) Curr. Opin. Chem. Biol. 6 171–180 [DOI] [PubMed] [Google Scholar]
- 17.Culotta, V. C., Yang, M., and O'Halloran, T. V. (2006) Biochim. Biophys. Acta 1763 747–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leary, S. C., Kaufman, B. A., Pellecchia, G., Guercin, G. H., Mattman, A., Jaksch, M., and Shoubridge, E. A. (2004) Hum. Mol. Genet. 13 1839–1848 [DOI] [PubMed] [Google Scholar]
- 19.Ishida, S., Lee, J., Thiele, D. J., and Herskowitz, I. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 14298–14302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, X., Okuda, T., Holzer, A., and Howell, S. B. (2002) Mol. Pharmacol. 62 1154–1159 [DOI] [PubMed] [Google Scholar]
- 21.Andrews, P. A., and Howell, S. B. (1990) Cancer Cells 2 35–43 [PubMed] [Google Scholar]
- 22.Sinani, D., Adle, D. J., Kim, H., and Lee, J. (2007) J. Biol. Chem. 282 26775–26785 [DOI] [PubMed] [Google Scholar]
- 23.Guo, Y., Smith, K., and Petris, M. J. (2004) J. Biol. Chem. 279 46393–46399 [DOI] [PubMed] [Google Scholar]
- 24.Holzer, A. K., and Howell, S. B. (2006) Cancer Res. 66 10944–10952 [DOI] [PubMed] [Google Scholar]
- 25.Kuo, Y. M., Zhou, B., Cosco, D., and Gitschier, J. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 6836–6841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, J., Prohaska, J. R., and Thiele, D. J. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 6842–6847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haremaki, T., Fraser, S. T., Kuo, Y. M., Baron, M. H., and Weinstein, D. C. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 12029–12034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peled, T., Glukhman, E., Hasson, N., Adi, S., Assor, H., Yudin, D., Landor, C., Mandel, J., Landau, E., Prus, E., Nagler, A., and Fibach, E. (2005) Exp. Hematol. 33 1092–1100 [DOI] [PubMed] [Google Scholar]
- 29.Peled, T., Landau, E., Prus, E., Treves, A. J., Nagler, A., and Fibach, E. (2002) Br. J. Haematol. 116 655–661 [DOI] [PubMed] [Google Scholar]
- 30.Warburg, O. (1930) The Metabolism of Tumors, Constable and Co., Ltd., London
- 31.Matoba, S., Kang, J. G., Patino, W. D., Wragg, A., Boehm, M., Gavrilova, O., Hurley, P. J., Bunz, F., and Hwang, P. M. (2006) Science 312 1650–1653 [DOI] [PubMed] [Google Scholar]
- 32.Hwang, P. M., Bunz, F., Yu, J., Rago, C., Chan, T. A., Murphy, M. P., Kelso, G. F., Smith, R. A., Kinzler, K. W., and Vogelstein, B. (2001) Nat. Med. 7 1111–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peinado, H., Portillo, F., and Cano, A. (2005) Cell Cycle 4 1749–1752 [DOI] [PubMed] [Google Scholar]
- 34.Lucero, H. A., and Kagan, H. M. (2006) CMLS Cell. Mol. Life Sci. 63 2304–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molnar, J., Fong, K. S., He, Q. P., Hayashi, K., Kim, Y., Fong, S. F., Fogelgren, B., Szauter, K. M., Mink, M., and Csiszar, K. (2003) Biochim. Biophys. Acta 1647 220–224 [DOI] [PubMed] [Google Scholar]
- 36.Payne, S. L., Hendrix, M. J., and Kirschmann, D. A. (2007) J. Cell. Biochem. 101 1338–1354 [DOI] [PubMed] [Google Scholar]
- 37.Peinado, H., Del Carmen Iglesias-de la Cruz, M., Olmeda, D., Csiszar, K., Fong, K. S., Vega, S., Nieto, M. A., Cano, A., and Portillo, F. (2005) EMBO J. 24 3446–3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fong, S. F., Dietzsch, E., Fong, K. S., Hollosi, P., Asuncion, L., He, Q., Parker, M. I., and Csiszar, K. (2007) Genes Chromosomes Cancer 46 644–655 [DOI] [PubMed] [Google Scholar]
- 39.Pouyssegur, J., Dayan, F., and Mazure, N. M. (2006) Nature 441 437–443 [DOI] [PubMed] [Google Scholar]
- 40.Kim, M. S., Kim, S. S., Jung, S. T., Park, J. Y., Yoo, H. W., Ko, J., Csiszar, K., Choi, S. Y., and Kim, Y. (2003) J. Biol. Chem. 278 52071–52074 [DOI] [PubMed] [Google Scholar]
- 41.Jung, S. T., Kim, M. S., Seo, J. Y., Kim, H. C., and Kim, Y. (2003) Protein Expression Purif. 31 240–246 [DOI] [PubMed] [Google Scholar]
- 42.Mendelsohn, B. A., Yin, C., Johnson, S. L., Wilm, T. P., Solnica-Krezel, L., and Gitlin, J. D. (2006) Cell Metab. 4 155–162 [DOI] [PubMed] [Google Scholar]
- 43.Rawls, J. F., Mellgren, E. M., and Johnson, S. L. (2001) Dev. Biol. 240 301–314 [DOI] [PubMed] [Google Scholar]
- 44.Finney, L., Vogt, S., Fukai, T., and Glesne, D. (2008) Clin. Exp. Pharmacol. Physiol., in press [DOI] [PMC free article] [PubMed]
- 45.Juarez, J. C., Betancourt, O., Jr., Pirie-Shepherd, S. R., Guan, X., Price, M. L., Shaw, D. E., Mazar, A. P., and Donate, F. (2006) Clin. Cancer Res. 12 4974–4982 [DOI] [PubMed] [Google Scholar]
- 46.Juarez, J. C., Manuia, M., Burnett, M. E., Betancourt, O., Boivin, B., Shaw, D. E., Tonks, N. K., Mazar, A. P., and Donate, F. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 7147–7152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rhee, S. G. (2006) Science 312 1882–1883 [DOI] [PubMed] [Google Scholar]
- 48.Reaume, A. G., Elliott, J. L., Hoffman, E. K., Kowall, N. W., Ferrante, R. J., Siwek, D. F., Wilcox, H. M., Flood, D. G., Beal, M. F., Brown, R. H., Jr., Scott, R. W., and Snider, W. D. (1996) Nat. Genet. 13 43–47 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.