Abstract
Group A Streptococcus (GAS) is a leading human bacterial pathogen capable of producing invasive infections even in previously healthy individuals. As frontline components of host innate defense, macrophages play a key role in control and clearance of GAS infections. We find GAS induces rapid, dose-dependent apoptosis of primary and cultured macrophages and neutrophils. The cell death pathway involves apoptotic caspases, is partly dependent on caspase-1, and requires GAS internalization by the phagocyte. Analysis of GAS virulence factor mutants, heterologous expression, and purified toxin studies identified the pore-forming cytolysin streptolysin O (SLO) as necessary and sufficient for the apoptosis-inducing phenotype. SLO-deficient GAS mutants induced less macrophage apoptosis in vitro and in vivo, allowed macrophage cytokine secretion, and were less virulent in a murine systemic infection model. Ultrastructural evidence of mitochondrial membrane remodeling, coupled with loss of mitochondrial depolarization and cytochrome c release, suggests a direct attack of the toxin initiates the intrinsic apoptosis pathway. A general caspase inhibitor blocked SLO-induced apoptosis and enhanced macrophage killing of GAS. We conclude that accelerated, caspase-dependent macrophage apoptosis induced by the pore-forming cytolysin SLO contributes to GAS immune evasion and virulence.
Group A Streptococcus (GAS)4 is a leading human pathogen that annually infects hundreds of millions of people worldwide (1). The last 3 decades have witnessed a marked increase in severe, invasive forms of GAS infection, many attributable to a single globally disseminated clone of the M1T1 serotype (2). Invasive GAS infection defines a capacity of the pathogen to resist host innate defense mechanisms designed to prevent microbial spread beyond epithelial surfaces.
Macrophages are critical host defense cells involved directly in bacterial clearance and also in alerting other immune system components to invading pathogens. Macrophage microbicidal activity is accomplished by phagocytic uptake coupled with the action of reactive oxygen species, enzymatic proteolysis, and cationic antimicrobial peptides; their role in amplification of the innate and adaptive immune responses is achieved through release of soluble factors such as cytokines and nitric oxide. Mice depleted of macrophages or treated with inhibitors of macrophage phagocytosis cannot clear GAS infections even at relatively low challenge doses (3), demonstrating the essential first line defense function of these immune cells against the pathogen.
We sought to explore the interaction of the highly virulent GAS M1T1 clone with macrophages to better understand its propensity to produce invasive human infection. A prominent regulatory feature of macrophage biology in the context of infectious disease and inflammation is the process of apoptosis, mediated by caspase family proteases. Although a number of highly adapted intracellular bacterial pathogens, including Mycobacterium tuberculosis, Legionella pneumophila, and Brucella spp., have evolved mechanisms to block macrophage apoptosis and use the host cell as a vehicle for in vivo dissemination (4–6), a recent study of GAS M1T1 interactions with another host phagocytic cell type suggested a different outcome. In contrast to other prominent Gram-positive pathogens, including Staphylococcus aureus and Listeria monocytogenes, GAS induced an accelerated program of apoptosis in human neutrophils (7), although the specific virulence factor(s) involved, effects on caspase activation, and contribution to disease outcome were not studied.
Here we report that GAS rapidly induces macrophage apoptosis through caspase-dependent pathways, promoted by release of cytochrome c and permeabilization of mitochondrial outer membranes. GAS-induced macrophage apoptosis is mediated by the cytolysin streptolysin O (SLO), which is both necessary and sufficient for the phenotype. SLO-mediated macrophage apoptosis leads to enhanced GAS survival, dampened cytokine responses, and increased virulence during systemic infection.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Cell Culture Conditions—Serotype M1T1 GAS strain 5448 is an isolate from a patient with necrotizing fasciitis and toxic shock syndrome (8). Lactococcus lactis strain NZ9000 was derived from MG1363 and lacks the nis operon (9). WT GAS, isogenic gene deletion mutants, and L. lactis were grown in Todd-Hewitt broth (THB) (Difco); when required, retention of an slo or nga expression plasmid or empty vector control was ensured by addition of 2 μg/ml (GAS) or 5 μg/ml (L. lactis) erythromycin to the media. For all in vitro and in vivo assays, overnight cultures were diluted 1:10, grown to early logarithmic phase (∼2 h), pelleted, and resuspended to A600 nm = 0.4 (∼2 × 108 cfu/ml GAS, 108 cfu/ml L. lactis) and diluted to desired concentrations. The J774 murine macrophage cell line and the THP1 human monocyte cell line were grown in RPMI supplemented with 10% FBS (Invitrogen). Primary bone marrow-derived macrophages were prepared as described (10) with slight modification. Bone marrow cells were collected from mice and cultured in Dulbecco's modified Eagle's medium (high glucose) supplemented with 20% L-929 cell conditioned medium for 7 days. The adherent cells (bone marrow-derived macrophages) were then collected and cultured in Dulbecco's modified Eagle's medium (high glucose) with 10 ng/ml macrophage colony-stimulating factor (Pepro-Tech) overnight before bacterial infection. Human peripheral blood mononuclear cells (PBMCs) and neutrophils were isolated from fresh whole blood (Ficoll-Paque and Polymorph Prep) and cultured in RPMI containing 10% FBS or 2% autologous serum, respectively.
Generation of GAS SLO and NADase Allelic Exchange Mutants—PCR was used to amplify upstream and downstream DNA fragments from the M1 GAS 5448 chromosome immediately flanking the genes of interest. Primers pairs used were as follows: (a) slo-upF (5′-gtatcactagaagagactct-3′) + slo-upR (5′-gtccttcatacctttttatc-3′ with 30-bp 5′ extension matching 5′ end of the cat gene); (b) slo-downF (5′-gactggttcaagagataagc-3′ with 30-bp 5′ extension matching 3′ end of the cat gene) + slo-downR (5′-gacagttggggtcaaatcag-3′); (c) nga-upF (5′-gggctggttcgtcctccaaac-3′) + nga-upR (5′-taaaccaccttatattatt-3′ with 30-bp 5′ extension matching 5′ end of the cat gene); (d) nga-downF (5′-caatatgtataaggtgccaaaggg-3′ with 30-bp 5′ extension matching 3′ end of the cat gene) + nga-downR (5′-cagtgtgatcttcttcgctc-3′). PCR was performed using the respective upstream and downstream fragments and an amplicon of the cat gene to yield a fusion product in which cat replaced the gene of interest precisely in GAS chromosomal context. This fusion product was T-A-cloned into pCR2.1-Topo (Invitrogen) and then subcloned into pHY304, a temperature-sensitive vector with an erythromycin (Em) resistance marker. These knockout plasmids were transformed into GAS M1 WT and single recombination events identified at 37 °C under Em + chloramphenicol selection. Selection was relaxed by serial passage at 30 °C without antibiotics, and double-crossover events were identified by screening for colonies with chloramphenicol-resistant but EmS phenotype. Precise, in-frame allelic exchange of slo and nga with cat in the GAS chromosome was confirmed by PCR analysis.
Complementation and Heterologous Expression Studies—The M1 5448 slo gene was amplified using primers sloF (5′-gcttgatatggtcgaaagaac-3′) and sloR (5′-ggagtgggcacaaggcctca-3′), and the nga+ifs genes using primers ngaF (5′-gtttctcatgtaaaccacc-3′) and ifsR (5′-gaacaataaaaacattttag-3′). Each amplicon was T-A-cloned into pCR2.1-TOPO (Invitrogen), then subcloned into streptococcal expression vector pDCerm (11), before being introduced by electroporation into GAS mutants or L. lactis, and transformants were identified by Em selection.
Apoptosis Measurement by TUNEL Assay—Macrophages were plated at specific concentrations 1 day prior to infection. J774 macrophages and THP1 monocytes were plated at 1.5 × 106 cells/well in 12-well plates, and human PBMCs were plated at 2.5 × 106 cells/well and murine BMD macrophages at 2 × 106 cells/well in 6-well plates. Neutrophils were plated at 4 × 106 cells per well in 6-well plates on the day of the assay. Bacteria were grown to log phase and diluted in cell culture media to allow 2 × 107 cfu (4 × 107 for neutrophil assays) to be added to each well. Plates were centrifuged at 2,000 rpm for 5 min to ensure bacterial contact with cells and then incubated at 37 °C, 5% CO2. One hour after infection, penicillin (5 μg/ml) and gentamicin (100 μg/ml) were added to the media to kill residual extracellular bacteria. At 4 h after infection, cells were collected, fixed, and permeabilized according to the apo-bromodeoxyuridine TUNEL assay protocol (BD Biosciences). Cells were kept in 70% ethanol at –20 °C until they were stained for DNA fragmentation following the TUNEL protocol (BD Biosciences). Apoptosis was quantified by flow cytometry (FACSCalibur, BD Biosciences) with analysis using FlowJo software. In apoptosis inhibition studies, all inhibitors were added to cells for 1 h prior to infection and left for the duration of the assay. Equal volumes of DMSO alone had no effect on macrophage apoptosis. Concentrations used were as follows: 5 μg/ml cytochalasin D (Sigma), 200 μm quinolyl-valyl-O-methylaspartyl[-2,6-difluorophenoxy]-methyl ketone (MP Biomedicals), 20 μg/ml YVAD-CHO (Calbiochem), and 1 μg/ml actinomycin D (Sigma).
Mitochondrial Membrane Depolarization—J774 cells were plated as for the TUNEL assay. Thirty minutes prior to infection with GAS, cells were preloaded with tetramethylrhodamine ethyl ester (TMRE) (Invitrogen) at 100 nm. Cells were washed once with PBS and then infected as usual. Antibiotics were added at 1 h, and at 4 h cells were collected and washed with PBS before assessing fluorescence by flow cytometry (FACSCalibur, BD Biosciences). The mitochondrial membrane uncoupler, carbonylcyanide p-(trifluoromethoxy)phenylhydrazone (Invitrogen), was added at 5 μm for 2 has a positive control.
Caspase Activity Assays—J774 cells were plated and infected as for the TUNEL assay. At 4 h, cells were collected, washed once with PBS, and resuspended in 100 μl of modified RIPA buffer. After 10 min of incubation on ice, samples were centrifuged at full speed, and lysates were collected and kept on ice. Lysates were diluted in water and mixed with the caspase substrate Ac-DEVD-AFC (synthesized in-house) at a final concentration of 100 μm in caspase buffer (12). AFC kinetics were measured immediately using the fmax fluorescent plate reader (Molecular Devices) at 37 °C with 405 nm excitation and 510 nm emission. Recombinant caspase-3 was used as a positive control.
Cytokine ELISA—Supernatants were collected at 4 h from the apoptosis assays using murine BMD macrophages. ELISA was done on cell supernatants for TNFα and IL-1β (R & D Systems). TNFα levels in mouse serum were quantified by ELISA (BD Biosciences). Standards were also diluted in serum as a control for mouse experiments.
Quantitative RT-PCR—Primary mouse peritoneal macrophages at 4 × 106 cells/well incubated in RPMI + 0.5% FBS overnight were infected with WT GAS or the ΔSLO mutant at m.o.i. = 10, and antibiotics were added at 1 h to kill extracellular bacteria. Cells were collected by scraping at 4 h and then pelleting. RNA was collected using the Qiagen RNeasy mini kit, and cDNA was made using SuperScript III First-Strand Synthesis SuperMix (Invitrogen), followed by quantitative RT-PCR (SYBR GreenER qPCR SuperMix, Invitrogen) using established TNFα, IL-1β, and glyceraldehyde-3-phosphate dehydrogenase (control) primer pairs.
Expression and Purification of Recombinant SLO—The SLO gene was cloned into vector pET15b and transformed into BL21 DE3 Escherichia coli. Bacteria expressing SLO were cultured in 3 liters of Difco 2× YT medium (BD Biosciences) and incubated at 37 °C with shaking. Expression was induced in cultures at 0.8 A600 with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside (Bio-Vectra) and maintained at 30 °C for 4 h. Bacterial pellets were disrupted by sonication, and soluble 6× histidine-tagged SLO was purified using nickel-nitrilotriacetic acid-agarose (Invitrogen). Further purification was achieved using Q-Sepharose ion exchange chromatography (GE Healthcare). Proteins were eluted with a salt gradient and monitored by SDS-PAGE. Fractions corresponding to the full-length SLO were pooled, quantitated by A280, and frozen in aliquots at –80 °C. Assays were performed in the presence of 10 mm dithiothreitol (reducing conditions).
Murine Infection Model—Eight-week-old female C57B/6 mice were injected intraperitoneally with 3.3 × 107 cfu suspended in 100 μl of PBS of GAS WT or the isogenic ΔSLO mutant. Blood was collected at 24 h and plated to quantify bacterial load. Survival was monitored for 5 days. For intravenous infections, C57B/6 mice were injected intravenously with 1 × 108 cfu, and blood was collected at 6 h. Blood was plated for quantification of bacterial load, and centrifuged at 500 × g to separate serum that was frozen for ELISA. For in vivo TUNEL and macrophage depletion, mice were injected intraperitoneally with 3 ml of 3% thioglycollate (Difco). Three days later, 109 cfu of either WT GAS or the ΔSLO mutant were injected intraperitoneally, and peritoneal cells were collected 4 h later. Cells were quantitated by flow cytometry and fixed for TUNEL staining.
Macrophage Killing Assays—J774 cells were seeded at 7.5 × 105 cells per well in 24-well plates the day prior to the assay. Bacteria were added at an m.o.i. of 10 in 350 μl of assay media (RPMI 2% FBS), and plates were centrifuged at 2000 rpm to ensure contact of the bacteria to the macrophages. Plates were incubated for 30 min at 37 °C, 5% CO2, and then penicillin (5 μg/ml) and gentamicin (100 μg/ml) were added to each well to kill extracellular bacteria, and the plates were incubated for an additional 1 h. Each well was then rinsed once with PBS to remove extracellular dead bacteria, and 350 μl of fresh assay media was added to each well. This was the 1.5-h time point. At various time points, cells were pipetted off wells and transferred with supernatants to 1.5-ml Eppendorf tubes. Cells and bacteria were pelleted at 7,000 rpm for 5 min and supernatants removed. Pellets were lysed using 0.025% Triton X-100, and dilutions were plated to quantify bacteria.
Electron Microscopy—J774 macrophages were plated at 107 cells in 10-cm tissue culture plates. The following day, plates were treated with media control, GAS (m.o.i. 20), 200 μm Q-VD-OPH and GAS (m.o.i. 20), or 16 μg/ml of purified recombinant SLO. Antibiotics were added at 1 h, and cells were collected and pelleted at 4 h. Pellets were then fixed on ice in 2.5% paraformaldehyde, 2.5% glutaraldehyde, 0.1 m sodium cacodylate buffer, pH 7.4, at least overnight. After washing three times in ice-cold 0.1 m sodium cacodylate buffer containing 2.5% sucrose, fixed cells were incubated with 1% osmium tetroxide in acetate/veronal solution (13) for 1 h on ice in the dark. After washing with acetate/veronal solution three times for 5 min each, fixed cells were stained and stabilized en bloc in 0.5% uranyl acetate in acetate/veronal solution, pH 6, overnight at room temperature in the dark. Cells were then rinsed with double distilled H2O and dehydrated in an ice-cold ethanol series of 70, 90, and 100% successively on ice for 5 min each. The cells were then washed three times for 5 min each in fresh 100% ethanol and three times for 5 min each in fresh 100% acetone at room temperature. The cells were next infiltrated in 67% ethanol, 33% Epon for 3 h with agitation at room temperature followed by 34% ethanol, 66% Epon overnight with agitation at room temperature. The next day, the cells were infiltrated in 100% epoxy Epon with agitation at room temperature for 24 h, after which the samples were placed in an oven and allowed to polymerize in 100% Epon blocks at 60 °C for 48 h. Thin sections (∼70 nm) were stained with uranyl acetate and lead citrate before examination in an FEI Tecnai 12 TEM. Images were recorded on a Teitz 214 digital camera.
Statistics—All statistics were performed using the Student's t test; p values <0.05 were considered significant. Kaplan-Meyer statistics were applied to the mouse survival curve.
RESULTS
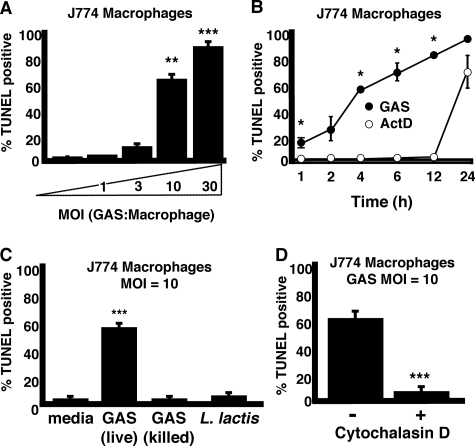
GAS Induces Rapid Dose-dependent Macrophage Apoptosis—To assess the ability of GAS to trigger macrophage apoptosis, murine macrophage cells (J774) were infected with GAS at various multiplicities of infection (m.o.i.) for 1 h before addition of antibiotics to kill extracellular bacteria. At 4 h, macrophages were collected, fixed, permeabilized, and analyzed by terminal dUTP nick-end labeling (TUNEL) staining to quantify DNA fragmentation, a hallmark of apoptosis. GAS induced significant macrophage apoptosis in an inoculum-dependent fashion (Fig. 1A). The kinetics of GAS-induced macrophage apoptosis (60% by 4 h at m.o.i. = 10) were more rapid than control treatments with the RNA synthesis inhibitor actinomycin D (significant macrophage apoptosis seen beginning at 12–24 h) (Fig. 1B). Macrophage apoptosis was not induced by penicillin-killed GAS nor by the nonpathogenic Gram-positive bacterium, L. lactis (Fig. 1C), suggesting an association to a specific factor elaborated by living GAS. Inhibition of phagocytosis by an actin cytoskeleton inhibitor, cytochalasin D, markedly reduced GAS-triggered macrophage apoptosis (Fig. 1D), indicating that GAS must be internalized to exert their proapoptotic effect(s).
FIGURE 1.
Live intracellular GAS induce rapid dose-dependent macrophage apoptosis. A, dose-dependent induction of macrophage apoptosis by GAS (t = 4 h). B, GAS induction of apoptosis proceeds more rapidly than control induction by actinomycin D, multiplicity of infection (MOI) = 10 bacteria/cell. C, GAS must be viable to induce macrophage apoptosis. D, internalization (phagocytosis) of GAS by the macrophage is required for apoptosis induction. * = p < 0.05, ** = p < 0.005, and *** = p < 0.0005. All experiments were performed in triplicate and repeated three times.
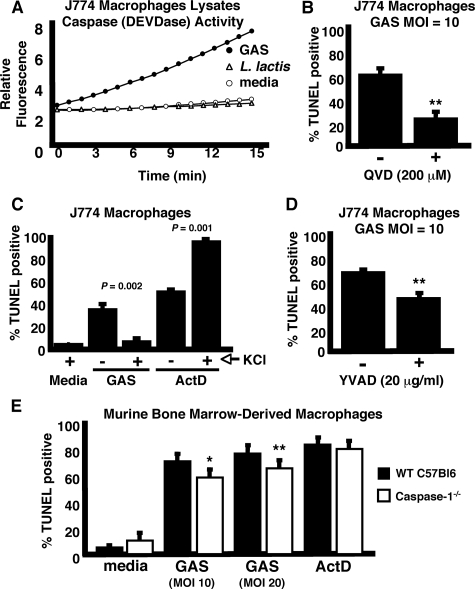
GAS-induced Macrophage Apoptosis Involves Caspase Activation—During apoptosis, stimulation of upstream, or initiator, caspases through multiple pathways ultimately converges upon activation of effector caspases responsible for protein and organelle dismantling, and cell death. Activity on the fluorogenic reporter substrate Ac-DEVD-AFC, selective for apoptotic effector caspases, was increased in lysates from macrophages infected with GAS but not in macrophages infected with L. lactis or media control (Fig. 2A). Pretreatment of J774 cells with the general caspase inhibitor Q-VD-OPH significantly reduced apoptosis induced by GAS infection (Fig. 2B). Because TUNEL activity correlates with apoptotic caspase activation, the macrophage death induced by GAS is verified as apoptotic.
FIGURE 2.
GAS-induced macrophage apoptosis is caspase-dependent. A, activity of apoptotic caspases (DEVDase) was measured with the fluorescent substrate Ac-DEVD-AFC in lysates of cells infected with GAS but not L. lactis control. B, induction of macrophage apoptosis is inhibited by the general caspase inhibitor Q-VD-OPH. C, high extracellular KCl (130 mm) inhibits GAS-induced macrophage apoptosis. ActD, actinomycin D. D, caspase-1-specific inhibitor YVAD-CHO decreases GAS-induced macrophage apoptosis. E, caspase-1–/– mouse macrophages are more resistant to GAS-induced apoptosis. t = 4 h for all experiments. * = p < 0.05 and ** = p < 0.005. All experiments were performed in triplicate and repeated three times. MOI, multiplicity of infection.
Inflammatory caspase-1 has been implicated in programmed cell death during infections (14–17); sometimes the net effect of such activation is proinflammatory, leading to coinage of the term pyroptosis (18). Caspase-1 activation occurs via inflammasomes, complexes of intracellular proteins formed upon detection of danger signals such as microbial factors or low intracellular potassium (19). When assays were repeated in the presence of high extracellular KCl (130 mm), which prevents inflammasome activation (20), we observed a reduction in macrophage apoptosis induced by GAS, but not that induced by actinomycin D (Fig. 2C). Pretreatment of macrophages with the inhibitor Ac-YVAD-CHO, which targets inflammatory caspases, also led to a partial but significant decrease in apoptosis upon GAS infection (Fig. 2D). BMD macrophages from caspase-1–/– mice were slightly more resistant to GAS-induced apoptosis compared with macrophages from wild type (WT) control mice (Fig. 2E). In sum these data demonstrate a caspase-dependent pathway of apoptosis is induced by GAS, and suggest a contributory role of inflammatory caspases to the overall level of GAS-induced macrophage apoptosis. However, (a) the activation of apoptotic caspases, (b) the more effective inhibition of death by the general caspase inhibitor versus the inflammatory caspase inhibitor, and (c) the minimal, although significant, suppression of TUNEL in caspase-1-deficient macrophages all distinguish GAS-induced macrophage apoptosis from pyroptosis. More evidence for this distinction is provided below in studies of inflammatory cytokine production.
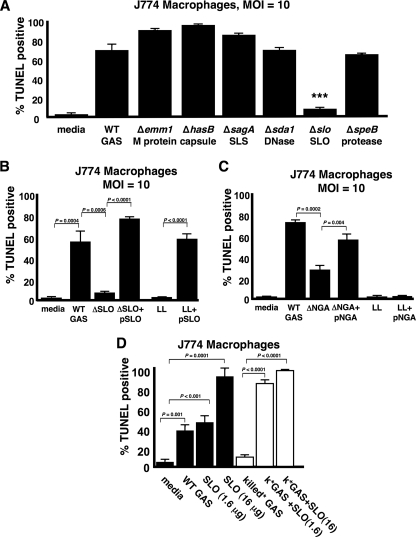
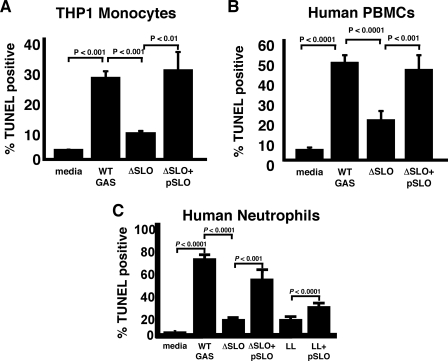
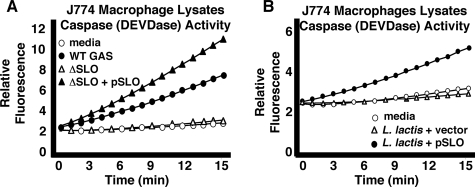
Streptolysin O Is Necessary and Sufficient for GAS-induced Macrophage and Neutrophil Apoptosis—Studies of GAS interactions with epithelial cells have implicated various bacterial factors, potential agents of apoptosis, including the protease (SpeB), pore-forming cytolysin SLO, and nicotinamide dehydrogenase (NADase) (21–24). To ascertain the contributory role of specific virulence factor(s), we compared the relative abilities of a panel of isogenic, single-gene deletion mutants to the WT parent M1T1 GAS strain in the macrophage apoptosis assay (Fig. 3A). Elimination of the cell wall-anchored antiphagocytic M1 protein, the surface-expressed hyaluronic acid capsule, potent β-hemolysin and cytolysin streptolysin S, the phageencoded DNase Sda1, or the cysteine protease SpeB did not significantly diminish GAS-induced macrophage apoptosis. In contrast, an isogenic mutant lacking cytolysin SLO (ΔSLO) was dramatically reduced in its ability to produce apoptosis (Fig. 3A). Single gene complementation of the isogenic ΔSLO knock-out mutant with the slo gene on a multi-copy plasmid restored the ability to induce macrophage apoptosis even beyond WT levels (Fig. 3B). Heterologous expression of the GAS slo gene in the nonpathogenic L. lactis conferred a strong pro-apoptotic phenotype, indicating that the toxin is not only necessary but also sufficient, at least in the context of an intact bacterium, to trigger macrophage programmed cell death (Fig. 3B). The phenotype of SLO-mediated apoptosis was confirmed in the human monocytic cell line THP1 (Fig. 4A), as well as in PBMCs freshly isolated from human blood (Fig. 4B). Parallel studies using purified human neutrophils (Fig. 4C) indicate that SLO is the GAS virulence factor responsible for the striking and rapid neutrophil apoptosis induced by GAS infection (7). A direct link of SLO expression to apoptotic caspases was confirmed in fluorescent substrate assays comparing lysates from macrophages infected with WT GAS versus the ΔSLO mutant (Fig. 5A) or L. lactis expressing the GAS slo gene versus empty vector control (Fig. 5B).
FIGURE 3.
Streptolysin O is necessary and sufficient for GAS-induced macrophage apoptosis. A, screen of isogenic GAS virulence factor mutants reveals SLO is necessary for induction of macrophage apoptosis. B, single gene complementation of SLO rescues the mutant, and SLO confers to L. lactis (LL) the ability to induce macrophage apoptosis. C, streptococcal NADase (nga) plays a contributory, but not sufficient, role in induction of macrophage apoptosis. D, purified recombinant SLO induces dose-dependent macrophage apoptosis, which can be enhanced by the addition of bacterial components. t = 4 h for all experiments. *** = p < 0.0005. All experiments were performed in triplicate and repeated three times. MOI, multiplicity of infection. SLS, streptolysin S.
FIGURE 4.
Streptolysin O mediates apoptosis of human phagocytic cells. SLO expression in GAS is responsible for the induction of apoptosis in the human monocytic cell line THP1 (A) and in freshly isolated PBMCs from human blood (B). C, SLO is also necessary (in GAS) and sufficient (in L. lactis) for induction of neutrophil apoptosis. LL, L. lactis.
FIGURE 5.
SLO mediates activation of effector caspases-3 and -7. A, apoptotic caspase activity (DEVDase) in cell lysates of macrophages infected with WT GAS strains, but not the ΔSLO mutant as assessed by cleavage of fluorescent substrate Ac-DEVD-AFC. B, SLO expression in L. lactis is sufficient to induce apoptotic caspase activity in macrophage lysates.
SLO has been implicated in the generation of membrane pores that allow specific transport of GAS NADase into host cells (25). An isogenic knock-out of the nga gene encoding GAS NADase (ΔNGA) was reduced in its ability to induce macrophage apoptosis, but not as markedly as the GAS ΔSLO mutant (Fig. 3C). In contrast to SLO, heterologous expression of NADase in L. lactis was insufficient to confer a proapoptotic phenotype in the macrophage infection assay (Fig. 3C).
Addition of purified SLO protein to macrophages was sufficient to induce dose-dependent macrophage apoptosis (Fig. 3D). Penicillin-killed GAS, unable to induce apoptosis on its own (Fig. 1C), significantly enhanced the apoptosis induced by lower concentrations of purified SLO (Fig. 3D), indicating that other bacterial components can synergize with SLO to induce apoptosis.
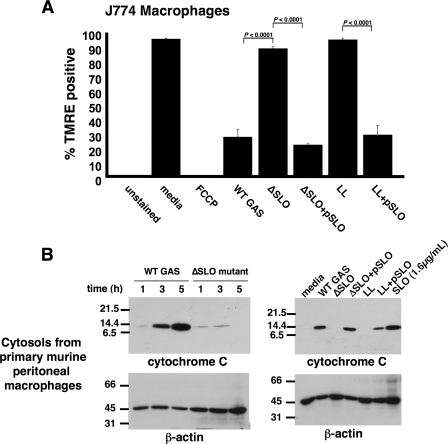
SLO Reduces Mitochondrial Membrane Potential and Triggers Cytochrome c Release—A hallmark of apoptosis mediated through the intrinsic pathway of caspase activation is the release of cytochrome c from the mitochondria into the cytosol (26) where it programs Apaf-1 to activate the initiator caspase-9 (27). Release of cytochrome c precedes a decrease in mitochondrial membrane potential (Δψm) reflecting the opening of mitochondrial membrane transition pores. Using the fluorescent stain TMRE that labels healthy polarized mitochondria, we found that macrophages exposed to WT GAS but not the isogenic ΔSLO mutant showed a rapid loss of TMRE staining indicating reduced Δψm (Fig. 6A). The action of SLO to diminish Δψm was corroborated by complementation of the ΔSLO mutant with the slo gene and heterologous expression of the GAS slo gene in L. lactis (Fig. 6A).
FIGURE 6.
SLO is responsible for disruption of the mitochondrial membrane and release of cytochrome c into the macrophage cytosol. A, SLO-dependent loss of TMRE staining demonstrates perturbation of mitochondrial membrane potential (t = 4 h). Experiments were performed in triplicate and repeated three times. LL, L. lactis; FCCP, carbonylcyanide p-(trifluoromethoxy)phenylhydrazone. B, Western blot of cytosols of macrophages infected with GAS ± SLO, L. lactis ± SLO, or purified SLO illustrates SLO-mediated release of cytochrome c from mitochondria. Actin serves as loading control. Experiments were performed in duplicate.
Next, cell lysates of mouse primary peritoneal macrophages infected with WT GAS or the isogenic ΔSLO mutant were collected at various time points, separated by centrifugation into mitochondrial and cytosolic fractions, and analyzed for cytochrome c by Western blot. Increasing amounts of cytochrome c were detected in the cytosol of WT GAS-infected macrophages over time, but not those infected with the ΔSLO mutant (Fig. 6B, left). Complementation of the ΔSLO mutant, heterologous expression of slo in L. lactis, and addition of purified SLO protein confirmed that the cytolysin was the factor responsible for induction of cytochrome c release into the macrophage cytoplasm (Fig. 6B, right).
Electron Microscopy Reveals Changes in Mitochondrial Ultrastructure Accompanying Apoptosis—Changes in mitochondrial structure occur during apoptosis induced by the intrinsic pathway. These changes include fragmentation of the mitochondria network and remodeling of the inner mitochondrial membrane, the significance of which is debated (28–30). Scorrano et al. (28) observed a dramatic inner membrane remodeling in purified rat liver mitochondria treated with the proapoptotic protein, Bid, to induce cytochrome c release, and proposed that the remodeling was required for efficient and complete release of cytochrome c from the intracristal compartments (29). Subsequently, a correlated three-dimensional light and electron microscopy study of etoposide-induced apoptosis in HeLa cells revealed a transformation of the normal mitochondrial inner membrane into a structure fragmented into numerous individual vesicular matrix compartments. Changes in mitochondrial inner membrane structure have been attributed to changes in the amount and processing of the dynamin-related protein, OPA1 (Mgm1p in yeast), required for mitochondrial inner membrane fusion (28, 31, 32). Sun et al. (30) found that inhibition of caspases efficiently inhibited all changes in the inner membrane conformation without inhibiting release of cytochrome c.
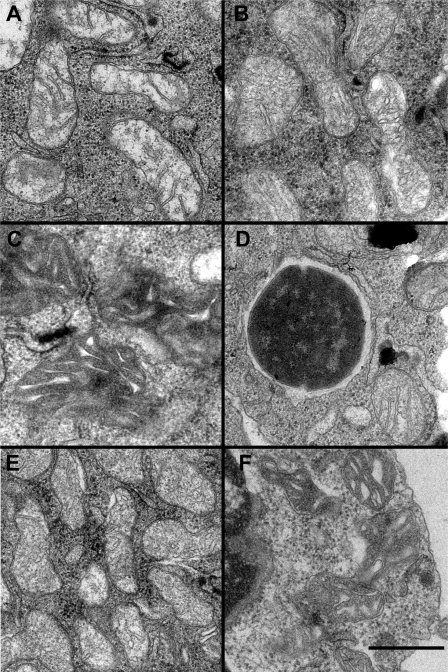
To investigate ultrastructurally the apoptosis induced by GAS infection of macrophages, we examined four samples by electron microscopy of thin sections. Mitochondria in macrophages treated with media contained, as expected, only mitochondria with normal morphology (Fig. 7A), which represents over 95% of 357 mitochondrial images observed in 21 cells. In macrophages infected with GAS, mitochondrial structure was altered; fewer than a quarter of the 300 mitochondrial images observed in 23 cells appeared normal, although approximately half displayed the condensed matrix appearance of the mitochondrion in Fig. 7C, and the remainder displayed dilated cristae similar to those seen in Fig. 7B. Inhibition of caspases with Q-VD-OPH efficiently inhibited these structural changes, and nearly 90% of over 300 mitochondria observed in 26 cells had normal inner membrane morphology as shown in Fig. 7D immediately adjacent to a phagocytosed GAS cells. On the other hand, treatment of macrophages with SLO produced the same morphological changes as infection with GAS with even fewer normal mitochondria and somewhat more with condensed matrix compartments or dilated cristae (Fig. 7, E and F). Samples infected with GAS and those treated with SLO contained a significant number of lysed cells not observed in the media control; treatment with Q-VD-OPH reduced the cell lysis produced by GAS exposure.
FIGURE 7.
Electron microscopy reveals cristae remodeling within mitochondria of GAS-infected cells. A, mitochondria in control cells treated with media display typical ultrastructure with the inner membrane projecting into the matrix at crista junctions to form lamellar cristae. B, mitochondria in GAS-infected cells show altered cristae structure consistent with swollen crista compartments. C, some mitochondria in GAS-treated cells have a different appearance with irregularly shaped cristae cross-sections within a very dense matrix compartment. D, Q-VD-OPH effectively prevents the alterations of mitochondrial structure found in GAS-infected macrophages. Several mitochondria, including the one at lower right, maintain normal crista structure despite being immediately adjacent to a phagocytosed GAS cell slightly left of center in this image. E, cells treated with purified SLO showing mitochondria with dilated cristae similar to those treated with GAS in B. F, mitochondria in SLO-treated cells with condensed matrix compartments similar to those following GAS treatment in C. Scale bar indicates 200 nm.
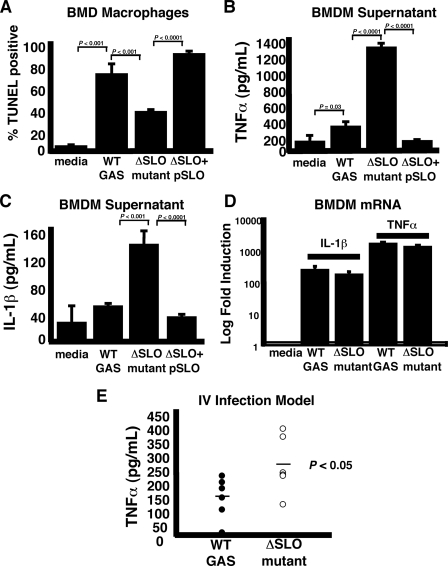
SLO Blunts Macrophage Immune Response and Contributes to GAS Virulence—The rapid nature of GAS SLO-induced apoptosis suggested the process could adversely impact macrophage function in innate immune response activation. When BMD macrophages from C57B/6 mice were infected by WT M1T1 GAS, the isogenic ΔSLO mutant, or the complemented mutant, we found a pattern of rapid SLO-dependent apoptosis resembling that seen in the J774 cell studies (Fig. 8A). TNFα and interleukin-1β (IL-1β) are key inflammatory cytokines that contribute to broader immune system activation in response to pathogens. When TNFα and IL-1β release from infected macrophages was measured by ELISA, the levels detected were more than 3-fold higher in macrophages exposed to the ΔSLO mutant than to the WT strain or complemented mutant (Fig. 8, B and C). To determine whether this difference was a result of increased cytokine gene expression in macrophages infected with the ΔSLO mutant, we performed quantitative real time RT-PCR of primary macrophage cytokine transcripts. TNFα and IL-1β mRNA increased over 200- and 1,000-fold, respectively, in macrophages infected with WT GAS, and these levels were not increased, but rather slightly decreased, in macrophages infected with the ΔSLO mutant (Fig. 8D). SLO was also seen to blunt the early cytokine response to M1T1 GAS infection in vivo; in C57B/6 mice challenged by intravenous injection, serum TNFα levels 6 h post-infection were significantly lower in animals challenged with the WT strain than those challenged with the isogenic ΔSLO mutant (Fig. 8E), although bacterial levels in the blood at this time point were not significantly different (data not shown).
FIGURE 8.
SLO induction of macrophage apoptosis blunts immune activation. A, role for SLO in apoptosis induction observed upon GAS infection of primary BMD macrophages. B, reduced TNFα levels in supernatants of primary mouse macrophages infected with SLO-expressing strains. C, accelerated apoptosis induced by WT GAS reduces levels of inflammatory cytokine IL-1β from macrophages compared with infection with the ΔSLO mutant. D, TNFα and IL-1β gene transcripts are significantly up-regulated by infection with both WT GAS and the ΔSLO mutant. Experiments performed in triplicate and repeated three times. E, decreased TNFα detected in the blood of mice 6 h after intravenous infection with WT GAS versus the ΔSLO mutant. IV, intravenous.
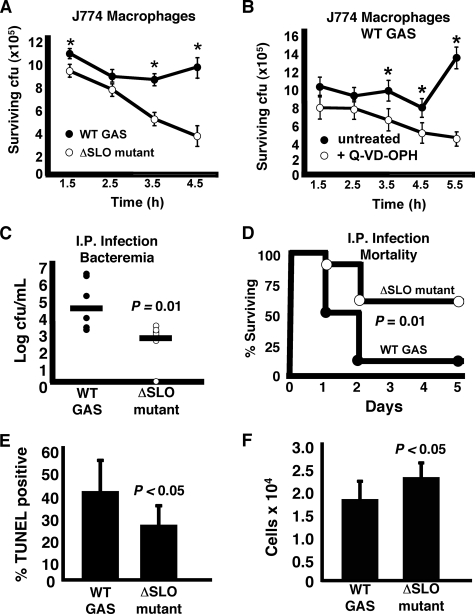
Furthermore, we found that macrophage apoptosis mediated by SLO contributed directly to GAS evasion of macrophage killing. Upon coincubation with J774 macrophages, the WT GAS strain survived better than the isogenic ΔSLO mutant (Fig. 9A). Corroborating a role for accelerated apoptosis in this survival phenotype, addition of the pan-caspase inhibitor Q-VD-OPH to retard apoptosis enhanced the ability of the macrophages to kill the WT GAS strain (Fig. 9B). Ultimately, impairment of host macrophage innate immune response correlated with increased virulence of the WT GAS M1T1 strain in the mouse intraperitoneal infection model. Compared with the ΔSLO mutant, the WT parent strain reached significantly higher bacterial loads at 24 h (Fig. 9C) and produced greater overall mortality (90% versus 40% by day 5) (Fig. 9D). To determine whether SLO was mediating apoptosis in vivo, we elicited peritoneal macrophage accumulation in mice with thioglycollate, and then injected those mice intraperitoneally with either the WT GAS strain or the ΔSLO mutant. Fig. 9E demonstrates a significant increase in apoptosis of peritoneal cells in vivo in mice infected with the WT GAS compared with the ΔSLO mutant. Significantly fewer cells were collected from the peritoneal cavity of WT GAS-infected mice compared with those infected with the ΔSLO mutant (Fig. 9F), demonstrating an overall depletion of immune cells by the WT SLO-expressing strain. Thus SLO, one of the most strongly up-regulated genes in vivo during M1T1 GAS infection (33), is an important virulence factor in the pathogenesis of systemic infection, acting to impair macrophage bactericidal function and cytokine responses.
FIGURE 9.
SLO induction of macrophage apoptosis enhances bacterial survival and promotes virulence in vivo. A, higher survival of WT GAS versus ΔSLO mutant in the presence of macrophages. Experiment was performed in triplicate and repeated three times. B, caspase inhibitor Q-VD-OPH enhances the ability of macrophages to kill WT GAS. Experiment was performed in triplicate and repeated three times. * = p < 0.05. C, higher numbers of WT GAS than ΔSLO mutant recovered from blood of mice injected intraperitoneally (I.P.). t = 24 h. D, ΔSLO mutant is significantly attenuated (60% survival) versus the WT GAS (10% survival) in a mouse systemic infection model. E, higher level of apoptosis was detected in the peritoneal cells of mice infected with GAS WT compared with the ΔSLO mutant. F, fewer peritoneal cells were collected from mice infected with WT GAS, demonstrating SLO-mediated immune cell depletion.
DISCUSSION
Our data demonstrate that live GAS induce rapid and significant dose-dependent macrophage apoptosis upon phagocytosis by macrophages, characterized by the definitive readouts of DNA fragmentation and apoptotic caspase activation. This cell death is induced by the pore-forming protein toxin SLO, which is both necessary and sufficient for this pathogenic phenotype, and is expressed by all GAS strains. We further identify SLO as the specific virulence determinant responsible for marked neutrophil apoptosis observed upon coincubation with GAS compared with other bacteria (7), identifying a mechanism by which the pathogen may overcome both major classes of human phagocytic cells. The rapid proapoptotic potential of SLO appears unique and specific, because induction of macrophage apoptosis by GAS was not reduced upon elimination of the potent membrane active toxin streptolysin S. GAS NADase contributes to the magnitude of macrophage apoptosis, but itself is not sufficient to produce the phenotype, likely because of the observation that NADase is dependent on SLO-induced pores for membrane translocation (25). By depleting intracellular energy stores, the NADase may diminish the ability of the host to combat or repair the membrane toxic effects of SLO (34).
Some previous studies of GAS interactions with epithelial cells assessed apoptosis as an outcome variable. In respiratory epithelial cell lines, internalization of the bacteria was observed to be critical for induction of the apoptotic phenotype (24), and a provocative role was identified for the GAS protease, SpeB (21). In contrast, internalization was said to inhibit GAS-induced death of keratinocytes, where the phenotype was attributed to SLO-dependent influx of extracellular calcium (35). In our GAS macrophage infection models, we find no requirement for SpeB, and we show that cytochalasin D inhibition of bacterial uptake markedly reduces apoptosis. These findings are consistent with the partial contribution of inflammasomes and inflammatory caspases, which rely on intracellular signaling for activation and suggest an intracellular target for SLO membrane toxicity may participate in apoptosis induction.
We established that GAS-induced macrophage apoptosis is triggered by the activation of apoptotic caspases, including a partial contributory role for inflammatory caspase-1. Thus, cell death is partly dependent on caspase-1, which may contribute a supporting role to the main death trigger, which activates the apoptotic caspases. Gurcel et al. (36) demonstrated that another bacterial pore-forming toxin, aerolysin, stimulated the activation of caspase-1, with the net effect of promoting cell survival in the presence of the purified toxin. Conversely, we find that in the context of live infection with GAS expressing SLO, inhibition of caspase-1 results in reduced apoptosis and increased macrophage survival. The opposite results may reflect the small size (1–2 nm) of aerolysin-induced pores (37), compared with the much larger size (25–30 nm) of pores produced by SLO and other cholesterol-dependent pore-forming toxins (38), which could more rapidly effect intracompartmental fluxes of a larger array of host cell and bacterial components and thereby influence differential activation of signal pathways (39).
GAS-induced macrophage apoptosis differs significantly from another form of programmed cell death induced by Salmonella and Shigella, termed pyroptosis (40, 41). Pyroptosis is an inflammatory cell death completely dependent on caspase-1 and resulting in a strong inflammatory cytokine response (18). Our ELISA data, in vitro and in vivo, contradict the possibility that GAS also induces pyroptosis, because we observed very low levels of inflammatory cytokines released from dying macrophages following infection with WT GAS. Our quantitative PCR data demonstrate that macrophages are activated to markedly increase expression of the corresponding cytokines in response to WT (and ΔSLO) GAS infection, so low levels found in the WT infected cells are because of post-transcriptional events, namely accelerated apoptosis. The blunting of the macrophage-elicited systemic immune response to the invading bacterium reveals a novel virulence mechanism for GAS to undermine important host leukocyte functions.
Based on the knowledge that GAS must be internalized to induce apoptosis, and the SLO-dependent release of cytochrome c from the mitochondria, we propose that SLO effects accelerated macrophage apoptosis by forming pores in the membranes of the endocytic vesicle, followed by direct attack of SLO upon the mitochondrial outer membrane, leading to release of proteins, including cytochrome c, into the cytosol, activating the intrinsic pathway of apoptosis. This is supported by electron microscopy of macrophages infected with GAS or treated with SLO that show changes in mitochondrial structure characteristic of apoptosis. The observation that apoptosis induced by purified SLO protein is further enhanced in the presence of bacterial components suggests the cooperation of additional pathways of apoptosis induction. SLO membrane toxicity can allow GAS escape from the phagocytic vacuole (42), or form pores that allow passage of inflammasome-activating cellular components into the cell cytosol. Cytosolic delivery of bacterial cells or cell components could activate additional caspase-dependent pathways that synergize with SLO-mediated mitochondrial membrane damage to produce rapidly accelerated apoptosis.
Through induction of apoptosis, WT GAS were better able to survive in the presence of macrophages than the isogenic SLO-deficient mutant. This difference was corroborated in vivo where the WT GAS blunted host cytokine responses and demonstrated enhanced bloodstream survival and lethality compared with the toxin-negative strain. Addition of a pan-caspase inhibitor protected macrophages from apoptosis and enhanced GAS killing. A deeper understanding of the pathway of GAS-induced macrophage apoptosis could aid in the identification of novel targets for immunomodulatory therapy of invasive GAS infections, estimated to strike >600,000 individuals each year worldwide (1). Neutralization of the GAS SLO toxin production may itself represent a virulence factor-based therapy to support innate immune clearance of the invasive pathogen.
Acknowledgments
We acknowledge the assistance of Steven Barlow with preparation of samples for electron microscopy and Annelies Zinkernagel for intravenous injection of mice.
This work was supported, in whole or in part, by National Institutes of Health grants (to V. N., M. K., T. G. F., and G. S. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: GAS, group A Streptococcus; SLO, streptolysin O; WT, wild type; TNFα, tumor necrosis factor α; IL-1β, interleukin-1β; ELISA, enzyme-linked immunosorbent assay; PBS, phosphate-buffered saline; cfu, colony-forming unit; TUNEL, terminal dUTP nick-end labeling; TMRE, tetramethylrhodamine ethyl ester; PBMC, peripheral blood mononuclear cell; FBS, fetal bovine serum; m.o.i., multiplicity of infection; RT, reverse transcription; BMD, bone marrow-derived; AFC, 7-amino-4-trifluoromethylcoumarin.
References
- 1.Carapetis, J. R., Steer, A. C., Mulholland, E. K., and Weber, M. (2005) Lancet Infect. Dis. 5 685–694 [DOI] [PubMed] [Google Scholar]
- 2.Cunningham, M. W. (2000) Clin. Microbiol. Rev. 13 470–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldmann, O., Rohde, M., Chhatwal, G. S., and Medina, E. (2004) Infect. Immun. 72 2956–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loeuillet, C., Martinon, F., Perez, C., Munoz, M., Thome, M., and Meylan, P. R. (2006) J. Immunol. 177 6245–6255 [DOI] [PubMed] [Google Scholar]
- 5.Banga, S., Gao, P., Shen, X., Fiscus, V., Zong, W.-X., Chen, L., and Luo, Z.-Q. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 5121–5126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He, Y., Reichow, S., Ramamoorthy, S., Ding, X., Lathigra, R., Craig, J. C., Sobral, B. W. S., Schurig, G. G., Sriranganathan, N., and Boyle, S. M. (2006) Infect. Immun. 74 5035–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi, S. D., Braughton, K. R., Whitney, A. R., Voyich, J. M., Schwan, T. G., Musser, J. M., and DeLeo, F. R. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 10948–10953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kansal, R. G., McGeer, A., Low, D. E., Norrby-Teglund, A., and Kotb, M. (2000) Infect. Immun. 68 6362–6369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiupers, O. P., de Ruyter, P. G., Kleerebezem, M., and de Vos, W. M. (1998) J. Biotechnol. 64 15–21 [Google Scholar]
- 10.Hsu, L. C., Park, J. M., Zhang, K., Luo, J. L., Maeda, S., Kaufman, R. J., Eckmann, L., Guiney, D. G., and Karin, M. (2004) Nature 428 341–345 [DOI] [PubMed] [Google Scholar]
- 11.Jeng, A., Sakota, V., Li, Z., Datta, V., Beall, B., and Nizet, V. (2003) J. Bacteriol. 185 1208–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denault, J. B., and Salvesen, G. S. (2008) Methods Mol. Biol. 414 191–220 [DOI] [PubMed] [Google Scholar]
- 13.Palade, G. E. (1952) Anat. Rec. 114 427–451 [DOI] [PubMed] [Google Scholar]
- 14.Bergsbaken, T., and Cookson, B. T. (2007) PLoS Pathog. 3 e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hilbi, H., Moss, J. E., Hersh, D., Chen, Y., Arondel, J., Banerjee, S., Flavell, R. A., Yuan, J., Sansonetti, P. J., and Zychlinsky, A. (1998) J. Biol. Chem. 273 32895–32900 [DOI] [PubMed] [Google Scholar]
- 16.Monack, D. M., Detweiler, C. S., and Falkow, S. (2001) Cell. Microbiol. 3 825–837 [DOI] [PubMed] [Google Scholar]
- 17.Sarkar, A., Hall, M. W., Exline, M., Hart, J., Knatz, N., Gatson, N. T., and Wewers, M. D. (2006) Am. J. Respir. Crit. Care Med. 174 1003–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fink, S. L., and Cookson, B. T. (2005) Infect. Immun. 73 1907–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Creagh, E. M., and O'Neill, L. A. (2006) Trends Immunol. 27 352–357 [DOI] [PubMed] [Google Scholar]
- 20.Petrilli, V., Papin, S., Dostert, C., Mayor, A., Martinon, F., and Tschopp, J. (2007) Cell Death Differ. 14 1583–1589 [DOI] [PubMed] [Google Scholar]
- 21.Tsai, P. J., Lin, Y. S., Kuo, C. F., Lei, H. Y., and Wu, J. J. (1999) Infect. Immun. 67 4334–4339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bricker, A. L., Cywes, C., Ashbaugh, C. D., and Wessels, M. R. (2002) Mol. Microbiol. 44 257–269 [DOI] [PubMed] [Google Scholar]
- 23.Kuo, C.-F., Wu, J.-J., Tsai, P.-J., Kao, F.-J., Lei, H.-Y., Lin, M. T., and Lin, Y.-S. (1999) Infect. Immun. 67 126–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakagawa, I., Nakata, M., Kawabata, S., and Hamada, S. (2001) Cell. Microbiol. 3 395–405 [DOI] [PubMed] [Google Scholar]
- 25.Madden, J. C., Ruiz, N., and Caparon, M. (2001) Cell 104 143–152 [DOI] [PubMed] [Google Scholar]
- 26.Liu, X., Kim, C. N., Yang, J., Jemmerson, R., and Wang, X. (1996) Cell 86 147–157 [DOI] [PubMed] [Google Scholar]
- 27.Riedl, S. J., and Salvesen, G. S. (2007) Nat. Rev. Mol. Cell. Biol. 8 405–413 [DOI] [PubMed] [Google Scholar]
- 28.Frezza, C., Cipolat, S., Martins de Brito, O., Micaroni, M., Beznoussenko, G. V., Rudka, T., Bartoli, D., Polishuck, R. S., Danial, N. N., De Strooper, B., and Scorrano, L. (2006) Cell 126 177–189 [DOI] [PubMed] [Google Scholar]
- 29.Scorrano, L., Ashiya, M., Buttle, K., Weiler, S., Oakes, S. A., Mannella, C. A., and Korsmeyer, S. J. (2002) Dev. Cell 2 55–67 [DOI] [PubMed] [Google Scholar]
- 30.Sun, M. G., Williams, J., Munoz-Pinedo, C., Perkins, G. A., Brown, J. M., Ellisman, M. H., Green, D. R., and Frey, T. G. (2007) Nat. Cell Biol. 9 1057–1065 [DOI] [PubMed] [Google Scholar]
- 31.Arnoult, D., Rismanchi, N., Grodet, A., Roberts, R. G., Seeburg, D. P., Estaquier, J., Sheng, M., and Blackstone, C. (2005) Curr. Biol. 15 2112–2118 [DOI] [PubMed] [Google Scholar]
- 32.Griparic, L., van der Wel, N. N., Orozco, I. J., Peters, P. J., and van der Bliek, A. M. (2004) J. Biol. Chem. 279 18792–18798 [DOI] [PubMed] [Google Scholar]
- 33.Sumby, P., Whitney, A. R., Graviss, E. A., DeLeo, F. R., and Musser, J. M. (2006) PLoS Pathog. 2 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michos, A., Gryllos, I., Hakansson, A., Srivastava, A., Kokkotou, E., and Wessels, M. R. (2006) J. Biol. Chem. 281 8216–8223 [DOI] [PubMed] [Google Scholar]
- 35.Cywes Bentley, C., Hakansson, A., Christianson, J., and Wessels, M. R. (2005) Cell. Microbiol. 7 945–955 [DOI] [PubMed] [Google Scholar]
- 36.Gurcel, L., Abrami, L., Girardin, S., Tschopp, J., and van der Goot, F. G. (2006) Cell 126 1135–1145 [DOI] [PubMed] [Google Scholar]
- 37.Mota, L. J., Sorg, I., and Cornelis, G. R. (2005) FEMS Microbiol. Lett. 252 1–10 [DOI] [PubMed] [Google Scholar]
- 38.Shatursky, O., Heuck, A. P., Shepard, L. A., Rossjohn, J., Parker, M. W., Johnson, A. E., and Tweten, R. K. (1999) Cell 99 293–299 [DOI] [PubMed] [Google Scholar]
- 39.McCaffrey, R. L., Fawcett, P., O'Riordan, M., Lee, K. D., Havell, E. A., Brown, P. O., and Portnoy, D. A. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 11386–11391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cookson, B. T., and Brennan, M. A. (2001) Trends Microbiol. 9 113–114 [DOI] [PubMed] [Google Scholar]
- 41.Boise, L. H., and Collins, C. M. (2001) Trends Microbiol. 9 64–67 [DOI] [PubMed] [Google Scholar]
- 42.Nakagawa, I., Amano, A., Mizushima, N., Yamamoto, A., Yamaguchi, H., Kamimoto, T., Nara, A., Funao, J., Nakata, M., Tsuda, K., Hamada, S., and Yoshimori, T. (2004) Science 306 1037–1040 [DOI] [PubMed] [Google Scholar]