Abstract
The phosphotransferases of the haloalkanoate dehalogenase superfamily
(HADSF) act upon a wide range of metabolites in all eukaryotes and prokaryotes
and thus constitute a significant force in cell function. The challenge posed
for biochemical function assignment of HADSF members is the identification of
the structural determinants that target a specific metabolite. The
“8KDOP” subfamily of the HADSF is defined by the known structure
and catalytic activity of
2-keto-3-deoxy-8-phospho-d-manno-octulosonic
acid (KDO-8-P) phosphatase. Homologues of this enzyme have been uniformly
annotated as KDO-8-P phosphatase. One such gene, BT1713, from the
Bacteroides thetaiotaomicron genome was recently found to encode the
enzyme
2-keto-3-deoxy-d-glycero-d-galacto-9-phosphonononic
acid (KDN-9-P) phosphatase in the biosynthetic pathway of the 9-carbon
α-keto acid,
2-keto-3-deoxy-d-glycero-d-galactonononic
acid (KDN). To find the structural elements that provide substrate-specific
interactions and to allow identification of genomic sequence markers, the
x-ray crystal structures of BT1713 liganded to the cofactor Mg2+and
complexed with tungstate or  /Neu5Ac
were determined to 1.1, 1.85, and 1.63 Å resolution, respectively. The
structures define the active site to be at the subunit interface and, as
confirmed by steady-state kinetics and site-directed mutagenesis, reveal
Arg-64*, Lys-67*, and Glu-56 to be the key residues
involved in sugar binding that are essential for BT1713 catalytic function.
Bioinformatic analyses of the differentially conserved residues between BT1713
and KDO-8-P phosphatase homologues guided by the knowledge of the
structure-based specificity determinants define Glu-56 and Lys-67*
to be the key residues that can be used in future annotations.
/Neu5Ac
were determined to 1.1, 1.85, and 1.63 Å resolution, respectively. The
structures define the active site to be at the subunit interface and, as
confirmed by steady-state kinetics and site-directed mutagenesis, reveal
Arg-64*, Lys-67*, and Glu-56 to be the key residues
involved in sugar binding that are essential for BT1713 catalytic function.
Bioinformatic analyses of the differentially conserved residues between BT1713
and KDO-8-P phosphatase homologues guided by the knowledge of the
structure-based specificity determinants define Glu-56 and Lys-67*
to be the key residues that can be used in future annotations.
Polyhydroxylated α-keto acids are incorporated into glycoproteins and glycolipids for the purpose of cell-surface display in prokaryotic and eukaryotic organisms (1). The two most common polyhydroxylated α-keto acids are 9-carbon N-acetylneuraminate (Neu5Ac)3 and 8-carbon 2-keto-3-deoxy-d-manno-octulosonic acid (KDO) (Fig. 1). KDO is synthesized in Gram-negative bacteria and in plants from phosphoenolpyruvate (PEP) and arabinose 5-phosphate through the consecutive actions of KDO-8-phosphate (KDO-8-P) synthase and KDO-8-P phosphatase (Fig. 1) (2). Neu5Ac, on the other hand, is synthesized in certain animals and specialized bacteria (viz. animal pathogens) by two distinct pathways (3). In animals, Neu5Ac is derived from PEP and N-acetylmannose-2-amine-6-phosphate through the consecutive actions of N-acetylneuraminate-9-phosphate (Neu5Ac-9-P) synthase and Neu5Ac-9-P phosphatase (Fig. 1), whereas in bacteria, Neu5Ac is formed directly from N-acetylmannose-2-amine and PEP through the action of Neu5Ac synthase, obviating the need for a phosphatase.
FIGURE 1.
Schematic of the pathways for biosynthesis and charging of the sialic acid derivatives KDO-8-P in bacteria (left), Neu5Ac-9-P in eukaryotes (center), and KDN-9-P in bacteria (right).
The bacterial KDO-8-P phosphatase has been characterized in terms of both unliganded structure and catalytic activity (4, 5).4 The enzyme falls within and indeed has been used to define a specific clade (8KDOP, also known as the Yrb subfamily) of the C0 subfamily of the haloacid dehalogenase superfamily (HADSF) (6). Thus, within the genomic sequence libraries, homologues of this α-keto acid biosynthetic enzyme have been uniformly identified as KDO-8-P phosphatase. Recently, our laboratories discovered that one such gene, BT1713, within the capsular polysaccharide biosynthetic locus 7 of the Bacteroides thetaiotaomicron genome encodes the enzyme 2-keto-3-deoxy-d-glycero-d-galacto-9-phosphonononic acid (KDN-9-P) phosphatase (7). This 9-carbon α-keto acid, 2-keto-3-deoxy-d-glycero-d-galacto-nononic acid (KDN), structurally related to KDO-8-P, is also formed in Neu5Ac-synthesizing animals (8). The presence of a second putative KDO-8-P phosphatase BT1677 (Expasy accession Q8A748; 24.8% sequence identity with BT1713; and 29.8 and 25.1% sequence identity with KDO-8-P phosphatase from Escherichia coli and Haemophilus influenzae, respectively) in B. thetaiotaomicron alerted us to the fact that BT1713 most likely had a related but not identical function to KDO-8-P phosphatase. Subsequent kinetic analysis of the gene products of the locus demonstrated that in this human gut symbiont KDN is biosynthesized from PEP and mannose 6-phosphate via the consecutive actions of KDN-9-P synthase (BT1714) and the KDN-9-P phosphatase (BT1713) (Fig. 1) (7).
These findings highlight the need for distinctive sequence markers for the assignment of biochemical function. Despite the relatively high sequence identity (28.2%) with the E. coli KDO-8-P phosphatase, the divergence of function in the B. thetaiotaomicron KDN-9-P phosphatase is evidenced by a 50-fold greater efficiency in hydrolysis of KDN-9-P versus KDO-8-P (7). Thus, significant alterations in the ancestral KDO-8-P phosphatase catalytic scaffold have occurred that are not obvious from sequence gazing. One approach to this problem is to utilize the three-dimensional structure of KDN-9-P phosphatase complexed with ligands to find the structural elements that provide substrate-specific interactions. Those structurally identified residues that are conserved differentially between KDO-8-P phosphatases and KDN-9-P phosphatases in the genome sequences can then be considered markers. The markers can be connected definitively to function by observed spatial juxtaposition of the phosphatase genes with genes encoding the corresponding synthase homologue.
Here, we provide the results of this line of attack for KDN-9-P phosphatase. Three x-ray crystallographic structures of KDN-9-P phosphatase bound to Mg2+ and acetate, the product analogue tungstate, and the intermediate analogue vanadate plus N-acetylneuraminate (Neu5Ac) are used to delineate and analyze the structural basis for substrate recognition and catalysis in KDN-9-P phosphatase. The analysis provides sequence markers that may be useful in the future annotation of KDN-9-P phosphatase sequences.
EXPERIMENTAL PROCEDURES
Except where indicated, all chemicals and enzymes were obtained from Sigma. The Malachite Green phosphate assay kit was purchased from Biomol. Recombinant wild-type BT1713 was isolated from transformed and induced E. coli cells and purified to homogeneity (as judged by SDS-PAGE analysis) as described previously (7).
BT1713 Site-directed Mutant—All BT1713 mutant genes were prepared by a PCR-based strategy with commercial primers and the WT-BT1713 plasmid described previously (7) serving as template. The gene sequences were confirmed by DNA sequencing carried out by the Center for Genetics in Medicine at the University of New Mexico. All BTI713 mutants were isolated and purified to homogeneity (as judged by SDS-PAGE analysis) from transformed and induced E. coli cells using the same protocol used for wild-type BT1713.
Native Molecular Weight Determination—The molecular weight of native BT1713 was estimated by fast protein liquid chromatography gel filtration column chromatography against protein standards (25-232 kDa from Amersham Biosciences) using a 2.5 × 120-cm Sephacryl S-200 column (Amersham Biosciences) eluted at 25 °C with 50 mm HEPES, 100 mm NaCl (pH 7.5). The mass was calculated from a plot of log molecular weight versus elution volume from the column.
Kinetic Constant Determination—The purified recombinant enzyme was concentrated with an Amicon Ultrafiltration apparatus (PM10) or Centricon-10 (Millipore) and dialyzed against 50 mM K+HEPES, 5 mM MgCl2 and 0.5 mM DTT before use in kinetic studies. The steady-state kinetic parameters (Km and kcat) of phosphorylated substrates were determined from initial reaction velocities measured at varying substrate concentrations for reactions containing 5 mm MgCl2 in 50 mm K+HEPES buffer (pH 7.0) at 37 °C. The assay methods used for the various substrates are described below. Protein concentrations were determined by the Bradford assay (9), and absorbance measurements were performed on a PerkinElmer Life Sciences λ25 UV-visible spectrophotometer or a Beckman DU800 spectrophotometer. Data were fit to Equation 1 with the KinetAsyst I program,
 |
(Eq.1) |
where V0 = initial velocity, Vmax = maximum velocity, [S] = substrate concentration, and Km = Michaelis-Menten constant for substrate. The kcat value was calculated from Vmax and [E] according to the equation kcat = Vmax/[E], where [E] is the free enzyme concentration.
Inhibition constants for acetate, N-acetylneuraminate, tungstate, and vanadate were determined from initial velocity data measured in the presence and absence of inhibitors and fitted to Equation 2 (for competitive inhibition),
 |
(Eq.2) |
where [I] is the inhibitor concentration and Ki is the inhibition constant. The Km for Mg2+ activation was measured using reaction solutions initially containing 1.5, 2, 5, 10, or 20 μm MgCl2, 0.1 μm BT1713, 300 μm KDN 9-P, 5 units of Neu5Ac aldolase, 10 units of lactate dehydrogenase, and 0.2 mm NADH in 50 mm K+HEPES (pH 7.0, 25 °C). Reactions were monitored at 340 nm (Δε = 6200 m-1 cm-1), and the initial velocity data were fitted to Equation 1.
Phosphatase Discontinuous Assays—Phosphate ester hydrolysis for all substrates was monitored using the Biomol green kit to detect total phosphate release. The 1-ml assay mixture, containing 50 mm K+HEPES buffer (pH 7.0), 5 mm MgCl2, 1 mm substrate, and 1.9 μm BT1713, was incubated at 37 °C for 10 min. In parallel, the background level of inorganic phosphate was measured using a reaction that excluded BT1713. For analysis, 100 μl of the mixture was added to 1 ml of Biomol green reagent. After 30 min of incubation at room temperature, the absorbance of the solution at 620 nm was measured. Steady-state kinetic constant determinations were carried out using reaction solutions containing BT1713 (0.1-0.4 μm) and varying concentrations of phosphate ester (Km 0.5-5).
Phosphatase Continuous Assays—The rate of p-nitrophenyl phosphate (pNPP) hydrolysis was determined by monitoring the increase in absorbance at 410 nm (Δε = 18.4 mm-1 cm-1) at 25 or 37 °C. The 0.5-ml assay mixtures contained 50 mm HEPES buffer (pH 7.0), 5 mm MgCl2, various concentrations (Km 0.5-5) of pNPP, and 1.9 μm BT1713.
The rate of β-d-glucose 6-phosphate hydrolysis was determined by monitoring the rate of NADPH formation at 340 nm (Δε = 6.22 mm-1 cm-1) at 37 °C in a 0.5-ml coupled assay solution containing 50 mm K+HEPES buffer (pH 7.0), 5 mm MgCl2, 0.2 mm NADP+, and 0.5 unit of glucose dehydrogenase (EC 1.1.1.119) and various concentrations (Km 0.5-5) of β-d-glucose 6-phosphate. The kinetic constants determined using this assay agreed with those determined using the discontinuous phosphate assay described above.
The rate of PEP (phosphoenolpyruvate) hydrolysis was determined by monitoring the rate of NADH (340 nm; Δε = 6.22 mm-1 cm-1) decrease at 37 °C in a 0.5-ml coupled assay solution initially containing 50 mm K+HEPES buffer (pH 7.0), 5 mm MgCl2, 0.2 mm NADH, and 2 units of l-lactate dehydrogenase (EC 1.1.1.27) and various concentrations (Km 0.5-5) of PEP. The kinetic constants determined with this assay agreed with those determined using the discontinuous phosphate assay described above.
Crystallization and X-ray Diffraction Data Collection—BT1713 crystals were obtained at room temperature using the vapor diffusion method with hanging-drop geometry. The protein solution (35 mg/ml protein in 1 mm HEPES buffer (pH 7.0)) was screened for crystallization by sparse-matrix screening using Crystal Screen Kits I and II (Hampton Research). Large crystals grew in 1 day at room temperature (25 °C) with overall dimensions >0.3 × 0.3 × 0.2 mm under several conditions. Crystals from the following two conditions were found to yield the best diffraction: 1) 0.2 m calcium chloride, 0.1 m sodium acetate (pH 4.6), and 20% v/v isopropyl alcohol; and 2) 30% w/v polyethylene glycol (PEG) 4000, 0.1 m Tris HCl (pH 8.5), and 0.2 m sodium acetate.
Crystals from the calcium chloride/sodium acetate/isopropyl alcohol condition were frozen for data collection by passing through 100% Paratone-N (Hampton Research) and freezing directly in a stream of N2 gas at 100 K. Diffraction data were collected to 2.13-Å resolution using CuKα radiation from a Rigaku RUH3 generator equipped with the R axis IV++ image plate located at Boston University School of Medicine. Diffraction data were collected at 100 K and indexed and scaled using DENZO and SCALEPACK (10). The crystals are orthorhombic, belonging to space group C2221 with unit cell dimensions a = 112.01 Å, b = 118.46 Å, and c = 116.35 Å. The unit cell volume is consistent with the presence of a tetramer in the asymmetric unit assuming a Matthew's coefficient of 2.3.
Crystals grown from PEG were refined by using a protein solution containing 40 mg/ml BT1713, 1 mm HEPES (pH 7.0), and 10 mm MgCl2. Crystals were frozen for data collection as for the calcium chloride condition. X-ray diffraction data were collected at the National Synchrotron Light Source, Beamline X12C equipped with an ADSC CCD Quantum 210 detector at Brookhaven National Laboratory. Diffraction data were collected at 100 K at 1.1-Å resolution and indexed and scaled using DENZO and SCALEPACK (10). The crystals are orthorhombic, belonging to space group P21212 with unit cell dimensions a = 81.24 Å, b = 107.48 Å, and c = 75.09 Å. Data collection statistics are summarized in Table 1.
TABLE 1.
Summarized crystallographic data collection and refinement statistics
| Data collection statistics | Native | Native-Mg2+ | Mg2+-WO42− | Mg2+-VO3−-Neu5Ac |
|---|---|---|---|---|
| Resolution (last shell) (Å) | 50.00-2.13 (2.21-2.13) | 50.00-1.10 (1.14-1.10) | 50.00-1.85 (1.92-1.85) | 50.00-1.63 (1.69-1.63) |
| X-ray source | CuKα | Synchrotron | CuKα | CuKα |
| Wavelength (Å) | 1.5418 | 0.9000 | 1.5418 | 1.5418 |
| Space group | C2221 | P21212 | P21212 | P21212 |
| Cell dimension (Å) | a = 112.01, b = 118.46, c = 116.35 | a = 81.24, b = 107.48, c = 75.09 | a = 82.13, b = 107.32, c = 75.36 | a = 81.96, b = 106.66, c = 74.90 |
| Reflections observed (unique) | 223,065 (43,289) | 7,042,808 (258,326) | 297,997 (57,453) | 974,651 (78,700) |
| Completeness (%) | 99.8 (100) | 97.4 (84.8) | 99.6 (99.8) | 98.4 (91.2) |
| Rmergea (%) | 6.8 (49.4) | 4.7 (29.0) | 6.4 (50.6) | 4.6 (50.8) |
| I/σ (I) | 13.5 (1.9) | 43.5 (3.8) | 15.7 (2.2) | 35.4 (2.4) |
| Redundancy | 5.2 (4.7) | 13.4 (4.2) | 5.2 (4.7) | 12.2 (6.0) |
| Refinement statistics | ||||
| No. of protein/water atoms per asymmetric unit | 5180/1086 | 5180/512 | 5180/482 | |
| No. of other ligands | Acetate, 6; Mg2+, 4; Cl−, 5; PEG, 5; | WO42−, 4; Mg2+, 4; | VO3−, 4; Mg2+, 4; Neu5Ac, 4; | |
| PG4, 1; EDO, 11 | PEG, 4; EDO, 1 | PEG, 6; EDO, 6 | ||
| No. of reflections (work/free) | 489,603/24,703 | 108,134/5502 | 155,655/7786 | |
| Rwork/Rfree (%) | 12.8/14.1 | 17.6/21.8 | 17.4/20.7 | |
| Resolution (Å) | 28.8-1.10 | 21.9-1.85 | 49.0-1.63 | |
| Average B-factor (Å2) | ||||
| Protein | 14.8 | 25.6 | 30.8 | |
| Ligands | Acetate/Mg2+/Cl−, 15.3/10.4/12.5 | WO42−/Mg2+, 32.8/23.7 | VO3−,/Mg2+/Neu5Ac 28.3/29.8/40.3 | |
| Water | 29.2 | 41.4 | 41.5 | |
| Root mean square deviation | ||||
| Bond length (Å) | 0.008 | 0.011 | 0.007 | |
| Bond angle (°) | 1.1 | 1.2 | 1.1 |
Rmerge = ∑hkl ∑i|Ihkl,i - <Ihkl> I|/∑hkl ∑i|Ihkl,i|, where <Ihkl> is the mean intensity of the multiple Ihkl,i observations for symmetry-related reflections.
Co-crystallization of BT1713 with various ligands failed initially. To
obtain the BT1713 ligand complex, crystals grown from PEG were soaked to
remove bound acetate in either 35% PEG 4000, 0.06 m TAPS (pH 8.5),
50 mm sodium tungstate plus 10 mm MgCl2 or in
35% PEG 4000, 0.06 m TAPS (pH 8.5), 20 mm activated
NaVO3, 100 mm Neu5Ac plus 10 mm
MgCl2 for 15 min at room temperature. Soaked crystals were frozen
for data collection as for the calcium chloride condition. Diffraction data
were collected to 1.85-Å resolution (for the tungstate complex) or
1.63-Å resolution (for the
 -Neu5Ac complex) using
CuKα radiation from a Rigaku RUH3 generator equipped with the
R-Axis IV++ image plate located at Boston University School of
Medicine. Data were indexed and scaled using DENZO and SCALEPACK
(10). Both crystals are
orthorhombic, similar to the native PEG crystals, belonging to space group
P21212 with unit cell dimensions a = 82.13
Å, b = 107.32 Å, c = 75.36 Å (for the
tungstate complex) and a = 81.96 Å, b = 106.66 Å, and
c = 74.90 Å (for the
-Neu5Ac complex) using
CuKα radiation from a Rigaku RUH3 generator equipped with the
R-Axis IV++ image plate located at Boston University School of
Medicine. Data were indexed and scaled using DENZO and SCALEPACK
(10). Both crystals are
orthorhombic, similar to the native PEG crystals, belonging to space group
P21212 with unit cell dimensions a = 82.13
Å, b = 107.32 Å, c = 75.36 Å (for the
tungstate complex) and a = 81.96 Å, b = 106.66 Å, and
c = 74.90 Å (for the
 /Neu5Ac complex). Data collection
statistics are summarized in Table
1.
/Neu5Ac complex). Data collection
statistics are summarized in Table
1.
Phase Determination and Model Refinement—The phase problem was solved via molecular replacement using the structure of KDO-8-P phosphatase (Protein Data Bank accession code 1K1E (4)) as the search model (30% sequence identity to BT1713). Because the unit cell volume of BT1713 is consistent with the presence of a tetramer in the asymmetric unit and because KDO-8-P phosphatase exists as a tetramer in solution (5), the tetramer of KDO-8-P phosphatase was used as the search model. The program MOLREP (11) in the CCP4 program suite was used to solve the rotation and translation functions, yielding a solution with a correlation coefficient of 38.8% and an R-factor of 60.5% at 2.85-Å resolution. Although the R-factor was high, the difference in correlation coefficient between this solution and the next best solution was large, and the resulting model gave no overlap between symmetry mates. In addition the R-factor dropped significantly to 40% during the initial stages of rigid body refinement and model building. Successive rounds of manual rebuilding were performed using the molecular graphics program O (12) alternated with minimization and simulated annealing in CNS (13).
The model obtained for crystals grown from the calcium chloride/sodium acetate/isopropyl alcohol solution was used as the search model for the BT1713 data sets collected on the crystals grown from PEG. Successive rounds of manual rebuilding were performed using the molecular graphics program COOT (14) alternated with minimization and simulated annealing in PHENIX (15). To avoid model bias, ligand molecules were added when Rfree (16) was <30%; waters were also added at this stage. Analysis of the Ramachandran plot as defined by PROCHECK (17) showed that 97.2-97.5% of residues fall in the most favored regions with 2.8 to 2.5% in the additionally allowed regions and with no residues falling in the generously allowed or disallowed regions. Refinement statistics are summarized in Table 1.
Bioinformatics—The “8KDO” clade sequences containing the Glu-56 sequence marker were identified as follows. The sequences annotated as KDO-8-P phosphatase contained in the KEGG gene data bank were aligned, and the sequences containing Glu in place of the H. influenza KDO-8-P phosphatase Arg-60 were selected. Next, the BT1713 sequence was used as query in a Phyre search (18), and the sequences identified in the multisequence alignment to contain Glu-56 were selected. A nonredundant list of sequences was constructed from which a few sequences were selected and used as query sequences in Expasy/SIB BLAST searches. The final set of 25 sequences (from a pool of ∼400 8KDO clade sequences) were aligned using ClustalW (19).
RESULTS AND DISCUSSION
BT1713(Mg2+) Crystal Structure—The three-dimensional structure of KDN-9-P phosphatase complexed with ligands was determined to find the structural elements that provide substrate-specific interactions. As for all HADSF phosphotransferases, the cofactor Mg2+ is required for BT1713 catalysis; the Km value for Mg2+ activation of BT1713-catalyzed hydrolysis of 300 μm KDN-9P measured at pH 7 is 3.3 ± 0.1 μm. The x-ray structure of BT1713 complexed with Mg2+ was determined at 1.1-Å resolution (Table 1; Fig. 2). The final model includes four BT1713 subunits (all 164 residues from each subunit), four Mg2+, five Cl-, and six acetate molecules. The N-terminal Met residue and C-terminal Gln residue are visible but have a higher B-factor as compared with the average B-factor for the entire protein. The tetramer exhibits pseudo P4 symmetry; the four monomers (independently refined with no NCS averaging) are nearly identical with root mean square deviation of 0.17-0.26 Å2 for all residues. The molecular mass of native BT1713 was determined to be 72 kDa by gel filtration, and from the subunit mass of 18.4 kDa it follows that BT1713 is a tetramer in solution, consistent with the quaternary structure observed in the crystal.
FIGURE 2.
X-ray crystal structure of wild-type KDN-9-P phosphatase liganded to the cofactor Mg2+ depicted as a ribbon diagram. A, single protomer is depicted highlighting the conserved phosphoryl transfer residues in the active site loops as follows: loop 1, Asp-10 (red) and Asp-12 (silver); loop 2, Thr-54 (green); loop 3, Lys-80 (yellow); and loop 4, Asp-103 (cyan). The β2 and β3 insert domain forms the barrel domain that allows oligomerization. B, biological tetramer with each subunit differentially colored. The active site is indicated by the position of Mg2+ (magenta sphere). C, close-up view of the hydrophobic packing interactions in the barrel domain. All protein images were rendered using MOLSCRIPT (30) and POVRAY.
Like other HADSF C0 subfamily members, the BT1713 monomer consisted of a
single α/β domain (modified Rossmann fold) consisting of a
six-stranded parallel β-sheet (β1 and β4-β8) surrounded by
six helices (α1-α3 and α5-α7)
(Fig. 2A). A
β-loop-β motif (residues 18-34) is inserted between β1 and
α1, the same insertion point as for the cap domain in HADSF C1 subfamily
members (6). The BT1713
tetramer is built by packing the β-loop-β motif of each monomer
together to form a single β-barrel that serves as the tetramer interface
(Fig. 2B). The
β-barrel cavity (∼10 × 10 × 10 Å in dimension) is
packed with four Phe side chains and four Trp side chains
(Fig. 2C). The overall
structure (i.e. oligomerization and monomer fold) observed for the
BT1713(Mg2+) complex is retained in the structures of the
BT1713(Mg2+)(tungstate) and
BT1713(Mg2+)( )(Neu5Ac)
complexes (see below). This finding indicates that the BT1713 structure does
not change upon substrate binding.
)(Neu5Ac)
complexes (see below). This finding indicates that the BT1713 structure does
not change upon substrate binding.
The BT1713(Mg2+) structure shows that the active site is located at the subunit-subunit interface (Fig. 3). Thus, each dimer within the tetramer is the catalytic unit, with four active sites per tetramer in total. The subunit that binds the Mg2+ and the KDN-9-P (or Neu5Ac-9-P) substrate will be referred to as the “core subunit” and the subunit that serves to cover a portion of the active site entrance as the “cap subunit.” The catalytic scaffold observed in the core subunit consists of a 4-loop platform on which the core residues (i.e. the residues conserved among all HADSF phosphotransferases for catalysis) are located (Fig. 2A). Loop 1 positions the Asp-10 nucleophile and Asp-12 general acid/base; loop 2 holds Thr-54 that binds the substrate phosphoryl group; loop 3 positions Lys-80 that bridges the Asp-10 carboxylate and the substrate phosphoryl group; and loops 1 and 4 bind the Mg2+ cofactor. The Mg2+-binding site is typical of HADSF phosphohydrolases with octahedral geometry (see supplemental Fig. SI1 for electron density map).
FIGURE 3.
The active site of KDN-9-P phosphatase liganded to the cofactor Mg2+ highlighting the position of the active site at the subunit interface of the Rossmann fold domains. The ribbon diagram with residues depicted in ball-and-stick shows the interaction of the “catalytic domain” (gray) and “cap domain” (brown with residues denoted as *). The active site Mg2+ is depicted as a magenta sphere.
BT1713(Mg2+)(Tungstate) and
BT1713(Mg2+)(VO-3)-(Neu5Ac)
Structures—The residues comprising the entire active site are best
defined by the structures of the BT1713-(Mg2+)(tungstate) and
BT1713(Mg2+)( )(Neu5Ac)
complexes determined at 1.85 and 1.63 Å resolution, respectively
(Fig. 4). Tungstate, an
analogue of the product orthophosphate, is a competitive inhibitor of BT1713
versus pNPP, and Ki = 350 μm
(measured using assay solutions containing 5 mm MgCl2
and 50 mm K+HEPES at pH 7 and 25 °C). Tungstate has
been used previously as a product analogue in structures of hexose phosphate
phosphatase of HADSF members (C2B subfamily; Ki = 65
μm) (20) and
phosphonatase (C1A subfamily; Ki = 50 μm)
(21), the phosphatase MDP-1
(C0 subfamily) (22), and the
nonspecific acid phosphatase
(23) (C0 subfamily;
Ki = 50 μm).
)(Neu5Ac)
complexes determined at 1.85 and 1.63 Å resolution, respectively
(Fig. 4). Tungstate, an
analogue of the product orthophosphate, is a competitive inhibitor of BT1713
versus pNPP, and Ki = 350 μm
(measured using assay solutions containing 5 mm MgCl2
and 50 mm K+HEPES at pH 7 and 25 °C). Tungstate has
been used previously as a product analogue in structures of hexose phosphate
phosphatase of HADSF members (C2B subfamily; Ki = 65
μm) (20) and
phosphonatase (C1A subfamily; Ki = 50 μm)
(21), the phosphatase MDP-1
(C0 subfamily) (22), and the
nonspecific acid phosphatase
(23) (C0 subfamily;
Ki = 50 μm).
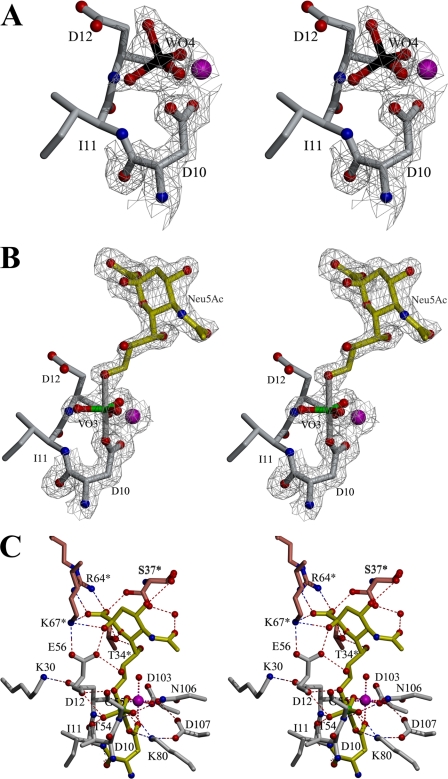
FIGURE 4.
KDN-9-P phosphatase liganded complexes. The 2Fo
- Fc composite omit electron density map contoured at
1.0σ (gray mesh) is shown for all ligands and Asp-10 residue in
A and B. A, close up stereo view of the active site of the
cofactor Mg2+ and product analogue tungstate. The tungstate ion
(W in black) is shown as ball-and-stick and the
cofactor Mg2+ ion as a magenta sphere. B, close up stereo
view of the active site of the complex with
 (green V) and Neu5Ac
(yellow). C, stereo view of the active site of KDN-9-P
phosphatase liganded to
(green V) and Neu5Ac
(yellow). C, stereo view of the active site of KDN-9-P
phosphatase liganded to  /Neu5Ac
(yellow) and consisting of a catalytic domain (gray) from
one subunit and a cap domain (brown and residues labeled as
*) from the adjacent subunit.
/Neu5Ac
(yellow) and consisting of a catalytic domain (gray) from
one subunit and a cap domain (brown and residues labeled as
*) from the adjacent subunit.
Vanadate is a competitive inhibitor of BT1713 versus pNPP; Ki = 350 μm (measured using assay solutions containing 5 mm MgCl2 and 50 mm K+HEPES at pH 7 and 25 °C). Vanadate is a popular structural probe of phosphotransferase mechanism (24), because it readily accepts ligands to form a pentavalent coordination complex with trigonal bipyramidal geometry (25). Structure determination of the HADSF member hexose phosphate phosphatase crystallized from solution containing vanadate and Mg2+ revealed a trigonal bipyramidal complex in which an oxygen atom from the Asp nucleophile (Asp-10) assumes an apical position at a distance of 2.0 Å from the vanadium (20). This finding suggested that vanadate might be used in combination with the sialic acid unit of the BT1713 substrate to form a pentavalent vanadate complex within the BT1713 active site. The sialic acid used for this purpose was Neu5Ac. This was a fortunate choice because the absence of interactions between the BT1713 active site and the Neu5Ac C(5) N-acetyl moiety accounts for the observation that BT1713 does not discriminate between the Neu5Ac-9-P and its physiological substrate KDN-9-P (in which the C(5) substituent is a hydroxyl group).
The Mg2+ ligands observed in the BT1713(Mg2+)(tungstate) complex structure are the same as those observed in the structure of the BT1713(Mg2+) complex except that one water ligand is replaced by the tungstate (O-Mg 1.9 Å) (Fig. 4A). The tetrahedral BT1713 tungstate ligand is aligned with the nucleophilic Asp-10 carboxylate group (which is the leaving group in the aspartyl 10-phosphate hydrolysis partial reaction), and thus this complex is a good mimic of the BT1713(Mg2+)(phosphate) “product complex.” The tungstate engages in hydrogen bond formation (2.6 Å) with the (loop 3) Lys-80 ammonium group, the (loop 4) Asn-106 side chain NH2 (2.8 Å), and the backbone amide NHs of the (loop 1) Ile-11 (2.9 Å), and Asp-12 (2.9 Å) as well as the backbone amide NH of (loop 2) Gly-55 (2.7 Å). The analogous electrostatic interactions are observed in the structures of tungstate-bound HADSF members phosphonatase and MDP-1. In addition, the distances (3.2 Å) between the Asp-12 carboxylic acid “OH” and two tungstate oxygen atoms (with Asp-12 OH positioned between the two tungstate oxygen atoms) suggest hydrogen bond formation with either oxygen. The geometry of the three does not favor any one bond over the other.
The structure of the
BT1713(Mg2+)( )(Neu5Ac)
complex (Fig. 4, B and
C) reflects the enzyme ligand interactions present in a
pentavalent trigonal bipyramidal intermediate/transition state formed during
catalytic turnover of Neu5Ac-9-P. The Asp-10 nucleophile and the Neu5Ac C(9)O
occupy the apical positions of the vanadium. The distance between the V and O
atoms was 2.1 Å and thus evidence of V-O bonding. One of the three
equatorial oxygen atoms bonds with the Mg2+ (2.0 Å),
analogous to the coordination bond between Mg2+ and the tungstate
oxygen atom in the BT1713(Mg2+)(tungstate) complex (1.9 Å).
The Asp-12 carboxyl group is positioned to form a hydrogen bond with both the
apical Neu5Ac C(9)O (3.2 Å) and the equatorial oxygen (3.0 Å). The
other two equatorial oxygen atoms of the vanadium complex engage in hydrogen
bonds with the (loop 3) Lys-80 ammonium (2.8 Å), the (loop 4) Asn-106
side chain NH2 (3.0 Å), and the backbone amide NHs of the
(loop 1) Ile-11 (3.2 Å) and Asp-12 (2.9 Å), and (loop 2) Gly-55
(2.9 Å).
)(Neu5Ac)
complex (Fig. 4, B and
C) reflects the enzyme ligand interactions present in a
pentavalent trigonal bipyramidal intermediate/transition state formed during
catalytic turnover of Neu5Ac-9-P. The Asp-10 nucleophile and the Neu5Ac C(9)O
occupy the apical positions of the vanadium. The distance between the V and O
atoms was 2.1 Å and thus evidence of V-O bonding. One of the three
equatorial oxygen atoms bonds with the Mg2+ (2.0 Å),
analogous to the coordination bond between Mg2+ and the tungstate
oxygen atom in the BT1713(Mg2+)(tungstate) complex (1.9 Å).
The Asp-12 carboxyl group is positioned to form a hydrogen bond with both the
apical Neu5Ac C(9)O (3.2 Å) and the equatorial oxygen (3.0 Å). The
other two equatorial oxygen atoms of the vanadium complex engage in hydrogen
bonds with the (loop 3) Lys-80 ammonium (2.8 Å), the (loop 4) Asn-106
side chain NH2 (3.0 Å), and the backbone amide NHs of the
(loop 1) Ile-11 (3.2 Å) and Asp-12 (2.9 Å), and (loop 2) Gly-55
(2.9 Å).
Because phosphoryl transfer is the common chemistry catalyzed by the HADSF scaffold, specificity is contributed by the interactions that occur with the Neu5Ac leaving group. Those between the Neu5Ac moiety and residues of the core subunit include hydrogen bonds between C(2)OH (2.6 Å) and C(8)OH (2.8 Å) and the Glu-56 carboxylate group and hydrogen bonds between C(9)OH and the Asp-12 carboxylic acid group (3.1 Å). The interactions that occur with residues of the cap subunit include hydrogen bonds between the ammonium group of cap subunit Lys-67* and the Neu5Ac C(1)OO- (2.8 Å) and C(2)OH (3.0 Å) and a strong salt bridge between the cap subunit Arg-64* guanidinium group and Neu5Ac C(1)OO- (2.9 Å) (in the Mg2+ bound unliganded BT1713 structure, an acetate molecule occupies the same position as the Neu5Ac C(1)OO-; see above). There are additional hydrogen bonds between the cap subunit and the Neu5Ac leaving group formed by Thr-34* and the Neu5Ac C(1)OO- (3.0 Å), C(6)O (ring O) (3.3 Å), and by Ser-37* (rotomer 1; the other rotomer 2 observed in the crystal structure is not within hydrogen-bonding distance) and the Neu5Ac C(1)OO- (2.5 Å). There are also water molecules mediated hydrogen bond interactions from the side chain of Ser-37* (rotomer 2) to the carbonyl oxygen atom of the C(5) NAc group (2.5 Å) and the backbone carbonyl O of Ser-37* to C(4)OH (2.8 Å) (Fig. 4C).
To evaluate the contributions that the Neu5Ac (and by analogy KDN)-binding residues make to BT1713 catalysis, the residues were separately replaced with Ala by site-directed mutagenesis, and the kinetic constants for catalyzed hydrolysis of KDN-9-P was measured (Table 2). Ala replacement of Arg-64* removes detectable catalytic activity, and the replacement of Glu-56* with Ala reduces the kcat/Km for catalyzed hydrolysis of KDN-9-P by 170-fold. Repeated attempts to prepare the K67A mutant failed. It is presumed that the K67A mutant does not fold to a stable native structure, and thus Lys-67 plays an important structural function in addition to the suggested role in catalysis. The kcat/Km values of the S37A and T34A mutants are 2- and 20-fold smaller than those of wild-type BT1713. Thus Arg-64*, Lys-67*, and Glu-56 appear to be the key “non-core” residues (i.e. residues not directly involved in phosphoryl group transfer that are essential for BT1713 catalytic function).
TABLE 2.
Steady-state kinetic constants for wild-type and mutant BT1713-catalyzed hydrolysis of KDN-9 phosphate in 50 mm K+ HEPES containing 2 mm MgCl2 (pH 7.0, 25 °C)
| BT1713 | kcat | Km | kcat/Km |
|---|---|---|---|
| s−1 | mm | m−1s−1 | |
| Wild type | 1.2 ± 0.1 | 0.10 ± 0.01 | 1 × 104 |
| R64A | <10−4 | ||
| E56A | 0.026 ± 0.002 | 0.41 ± 0.01 | 6 × 101 |
| S37A | 1.2 ± 0.1 | 0.20 ± 0.01 | 6 × 103 |
| T34A | 0.09 ± 0.01 | 0.18 ± 0.01 | 5 × 102 |
BT1713 Substrate Specificity—The previously reported
(7) steady-state kinetic
constants for BT1713-catalyzed hydrolysis of KDN-9-P, Neu5Ac-9-P, and KDO-8-P
are listed in Table 3 and
considered here within the context of inhibition data and the BT1713
structure. First, the kcat/Km value
measured for KDN-9-P hydrolysis is only 2-fold greater than that measured for
Neu5Ac-9-P hydrolysis. Analogously, the Ki values measured
for KDN (16 mm) and Neu5Ac (37 mm) as competitive
inhibitors (versus pNPP) suggest only a 2-fold tighter binding of
KDN. These findings are consistent with the
BT1713-(Mg2+)( )(Neu5Ac)
structure and with the corresponding
BT1713(Mg2+)(
)(Neu5Ac)
structure and with the corresponding
BT1713(Mg2+)( )(KDN)
model. Specifically, there is no apparent interaction between the active site
and the C(5)N-acetyl group of the Neu5Ac ligand or between the active
site and the C(5)OH group of the KDN ligand.
)(KDN)
model. Specifically, there is no apparent interaction between the active site
and the C(5)N-acetyl group of the Neu5Ac ligand or between the active
site and the C(5)OH group of the KDN ligand.
TABLE 3.
Steady-state kinetic constants for BT1713-catalyzed hydrolysis of KDN-9-P, Neu5Ac-9-P, KDO-8-P, and simple organophosphates
Compounds also tested that did not show significant activity are as follows: d-fructose-1,6-bis-P, d-glucitol-6-P, d-fructose-6-P, d-glucose-2-amine-6-P, d-mannose-6-P, trehalose-6-P, sucrose-6-P, d-ribose-5-P, d-arabinose-5-P, d-2-deoxyarabinose-5-P, ATP, ADP, AMP, pyrophosphate, pyridoxal-5-P, dl-glycerol-3-P, dl-glyceraldehyde-3-P, dl-serine phosphate, ethanolamine phosphate, α-d-glucose-1-P, and α-d-mannose-1-P.
| Substrate | kcat | Km | kcat/Km |
|---|---|---|---|
| s−1 | mm | m−1s−1 | |
| KDN-9-Pa | 1.2 ± 0.1 | 0.10 ± 0.01 | 1 × 104 |
| KDN-9-Pb | 2.2 ± 0.1 | 0.087 ± 0.005 | 3 × 104 |
| Neu5Ac-9-Pa | 7.3 (±0.01) × 10−1 | 0.12 ± 0.03 | 6 × 103 |
| KDO-8-Pa | 6.3 (±0.03) × 10−2 | 0.31 ± 0.03 | 2 × 102 |
| pNPPb | 1.5 (±0.1) × 10−1 | 1.1 ± 0.1 | 1 × 101 |
| Glucose-6-Pb | 1.6 (±0.1) × 10−2 | 11.4 ± 0.7 | 1 |
| PEPb | 2.4 (±0.2) × 10−1 | 7 ± 1 | 4 × 101 |
| Tyrosine phosphateb | 3.2 (±0.2) × 10−1 | 5.5 ± 0.8 | 6 × 101 |
| Gluconate-6-Pb | 4.2 (±0.1) × 10−2 | 1.9 ± 0.2 | 2 × 101 |
Steady-state kinetic constants for BT1713-catalyzed hydrolysis of KDN 9-phosphate, Neu5Ac 9-phosphate, and KDO 8-phosphate in 50 mm K+HEPES buffer (pH 7.0), and 2 mm MgCl2 at 25 °C are shown (7).
Steady-state kinetic constants for BT1713-catalyzed hydrolysis of nonspecific phosphate esters in 50 mm Na+HEPES buffer (pH 7.0) and 5 mm MgCl2 at 37 °C.
The kcat/Km value for
BT1713-catalyzed KDO-8-P hydrolysis is 53-fold smaller than that for KDN-9-P
hydrolysis (Table 3). KDN-9-P
and KDO-8-P differ not only in the length of the carbon linker between the
phosphoryl group and the ring C(6) (3 versus 2 carbons) but also in
the stereochemistry at C(5). When KDO-8-P is modeled in place of Neu5Ac in the
BT1713(Mg2+)( )(Neu5Ac)
structure (by overlaying the two carboxylate groups at C(2) and the phosphoryl
group of KDO-8-P with the vanadyl group of the Neu5Ac complex), the ring of
KDO-8-P must be flipped over because of the difference in stereochemistry at
C(2). This places the C(4)OH, C(5)OH, and C(7)OH in the opposite orientation
for KDO-8-P versus Neu5Ac (and by analogy KDN). Comparing these
complexes, only those interactions with the phosphoryl and carboxylate groups
are held in common (although this is a significant number of interactions,
four to the carboxylate group and nine to the phosphoryl group), which
accounts for the lower substrate activity observed for KDO-8-P.
)(Neu5Ac)
structure (by overlaying the two carboxylate groups at C(2) and the phosphoryl
group of KDO-8-P with the vanadyl group of the Neu5Ac complex), the ring of
KDO-8-P must be flipped over because of the difference in stereochemistry at
C(2). This places the C(4)OH, C(5)OH, and C(7)OH in the opposite orientation
for KDO-8-P versus Neu5Ac (and by analogy KDN). Comparing these
complexes, only those interactions with the phosphoryl and carboxylate groups
are held in common (although this is a significant number of interactions,
four to the carboxylate group and nine to the phosphoryl group), which
accounts for the lower substrate activity observed for KDO-8-P.
HADSF phosphatases are typically promiscuous with regard to metabolites that are recognized as “substrate” and hydrolyzed (26, 27). To define the substrate range of BT1713, a broad substrate screen was carried out using a library of common phosphate esters and anhydrides (Table 3 legend). The compounds that displayed “significant” substrate activity at pH 7 and 37 °C were subjected to initial velocity studies (Table 3); the substrate activity profile can be summarized as follows: KDN-9-P, Neu5Ac-9-P > and KDO-8-P > PEP, gluconate 6-phosphate, tyrosine phosphate ester > glucose-6-P. Nucleotides and the other phosphate ester metabolites tested are not substrates. The activities observed for KDN-9-P and Neu5Ac-9-P are physiologically relevant, whereas those observed for the other compounds fall well below those values of kcat/Km expected for a physiological substrate (<60 m-1 s-1).
Structure-based Analysis of Functional Divergence—The biochemical function of BT1713 is KDN-9-P hydrolysis, and not surprisingly KDN-9-P is the most active substrate. The GenBank™ annotation of BT1713 as KDO-8-P phosphatase is therefore clearly incorrect. BT1713 and B. thetaiotamicron KDO-8-P phosphatase BT1677 (with kcat/Km = 5 × 104 m-1 s-1 for KDO-8-P versus kcat/Km = 1 × 101 m-1 s-1 for KDN-9-P)5 share 24.8% sequence identity. BLAST searches carried out with BT1713 or BT1677 as the query do not define a clear boundary between the closest BT1713 and BT1677 sequence homologs. It is likely that the 8KDO clade includes other KDN-9-P phosphatases mis-annotated as KDO-8-P phosphatase.
The search to identify sequence markers that could be used to distinguish
KDN-9-P phosphatase sequences from KDO-8-P phosphatase sequences was started
with the non-core residues unique to the KDN-9-P phosphatase activity and of
demonstrated importance to BT1713 catalytic site function and the non-core
residues unique to a bona fide KDO-8-P phosphatase with demonstrated
importance to its catalytic function. A comparison of the previously reported
unliganded structure of the H. influenzae KDO-8-P phosphatase
(Co2+)
complex4
(4) and the
BT1713(Mg2+)( )(Neu5Ac)
highlighted residues that are positioned to interact with the KDN or KDO
moieties and thus serve as substrate specificity residues
(Fig. 5). Based on the active
site structure of the linear gluconate-6-P modeled with H. influenzae
KDO-8-P phosphatase and residue conservation, Herzberg and co-workers
(4) hypothesized that Arg-60
from the catalytic unit and Arg-68* from the cap subunit engage in
ion pair formation with the C(1)OO- of KDO-8-P. The corresponding
residues in BT1713 are Glu-56 and Arg-64*, respectively. The BT1713
cap subunit conserved Arg-64* interacts with the Nue5Ac
COO- and thus assumes the analogous role of Arg-60 in KDO-8-P
phosphatase. As described, replacement of Arg-64 with Ala destroys catalytic
activity (Table 2). The BT1713
cap subunit Glu-56 engages in an intricate hydrogen bond network between the
Neu5Ac moiety (C(1)OO-, C(2)OH, and C(8)OH) and the cap subunit
Lys-67*. Replacement of Lys-67* or Glu-56 with Ala also
has a devastating effect on BT1713 catalytic activity
(Table 2). Although the BT1713
cap subunit Ser-37* and Thr-34* are positioned to
contribute to binding the sugar unit, replacement of these residues with Ala
had a comparatively small effect on catalysis.
)(Neu5Ac)
highlighted residues that are positioned to interact with the KDN or KDO
moieties and thus serve as substrate specificity residues
(Fig. 5). Based on the active
site structure of the linear gluconate-6-P modeled with H. influenzae
KDO-8-P phosphatase and residue conservation, Herzberg and co-workers
(4) hypothesized that Arg-60
from the catalytic unit and Arg-68* from the cap subunit engage in
ion pair formation with the C(1)OO- of KDO-8-P. The corresponding
residues in BT1713 are Glu-56 and Arg-64*, respectively. The BT1713
cap subunit conserved Arg-64* interacts with the Nue5Ac
COO- and thus assumes the analogous role of Arg-60 in KDO-8-P
phosphatase. As described, replacement of Arg-64 with Ala destroys catalytic
activity (Table 2). The BT1713
cap subunit Glu-56 engages in an intricate hydrogen bond network between the
Neu5Ac moiety (C(1)OO-, C(2)OH, and C(8)OH) and the cap subunit
Lys-67*. Replacement of Lys-67* or Glu-56 with Ala also
has a devastating effect on BT1713 catalytic activity
(Table 2). Although the BT1713
cap subunit Ser-37* and Thr-34* are positioned to
contribute to binding the sugar unit, replacement of these residues with Ala
had a comparatively small effect on catalysis.
FIGURE 5.
Overlay of the active site of KDN-9-P phosphatase (catalytic domain, gray; cap domain, dark gray) liganded to VO -3/Neu5Ac (yellow backbone) with that of KDO-8-P phosphatase (catalytic domain, cyan; cap domain, blue). The C terminus of KDO-8-P phosphatase is longer than KDN-9-P phosphatase, and partially overlaps the position of the sialic acid ring. The active site Mg2+ is depicted as a magenta sphere.
Based on these results, a search of gene data banks was carried out to identify other KDN-9-P or Neu5Ac-9-P phosphatases (“Experimental Procedures”) by looking for KDO-8-P phosphatase homologs that substitute Arg-60 with a Glu residue. The product of this search is a representative (not necessarily a complete) set of KD8O clade sequences that conserve the “Glu-56” residue (supplemental Fig. SI2). The pairwise sequence dissimilarity among the 25 proteins extends to 70%, and thus the conservation of residues is well defined. The biochemical function as KDN-9-P phosphatase in 13 of the proteins is evidenced by the juxtaposition of the gene with a gene encoding a Neu5Ac-9-P synthase homologue (which based on our previous biochemical characterization (7) probably acts as a KDN-9-P synthetase). The KDN-9P/Neu5Ac-9-P synthase can be easily distinguished from the KDO-8-P synthase, although they are members of the same α/β barrel superfamily (28) because of the presence of an additional “antifreeze” domain in KDN-9P/Neu5Ac-9-P synthase (29).
The core residues responsible for catalysis of the phosphoryl transfer
(Asp-10, Asp-12, Thr-54, Lys-80, Asp-103, Asp-107) are conserved in all 25
proteins consistent with the assumed function as a phosphatase. Some but not
all of the residues observed in the
BT1713(Mg2+)( )(Neu5Ac)
structure to interact with the Neu5Ac moiety are conserved. In addition to
Arg-64* and the sequence marker Glu-56, the Glu-56 salt-bridge
partner Lys-67 is stringently conserved. Thus it can be concluded that the key
amino acid residues that can be used to a distinguish KDN-9-P phosphatase from
KDO-8-P phosphatase are Glu-56 and Lys-67*. Notably, the percent
sequence identity cannot be used to make this distinction because some members
of the KDN-9-P phosphatase group share as much if not more sequence identity
with members of the KDO-8-P phosphatase group than they do with one
another.
)(Neu5Ac)
structure to interact with the Neu5Ac moiety are conserved. In addition to
Arg-64* and the sequence marker Glu-56, the Glu-56 salt-bridge
partner Lys-67 is stringently conserved. Thus it can be concluded that the key
amino acid residues that can be used to a distinguish KDN-9-P phosphatase from
KDO-8-P phosphatase are Glu-56 and Lys-67*. Notably, the percent
sequence identity cannot be used to make this distinction because some members
of the KDN-9-P phosphatase group share as much if not more sequence identity
with members of the KDO-8-P phosphatase group than they do with one
another.
Conclusion—The structure of the
BT1713(Mg2+)( )(Neu5Ac)
complex and the models of
BT1713(Mg2+)(
)(Neu5Ac)
complex and the models of
BT1713(Mg2+)( )(KDN) and
BT1713(Mg2+ (KDO-8-P) provide a structural context to the relative
substrate activities of KDN-9-P, Neu5Ac-9-P, and KDO-8-P (53:26:1). BT1713 has
not evolved to discriminate between KDN-9-P and Neu5Ac-9-P, and this is
evident from the absence of residue binding partners to either the C(5)OH of
KDN-9-P or the C(5)NAc of Neu5Ac-9-P. However, such discrimination is not
necessary in the context of the cell because of the far greater activity of
the synthase enzyme (300:1) in producing KDN-9-P versus Neu5Ac-9-P
(7). In contrast, BT1713 has
evolved to enhance KDN-9-P phosphatase activity and suppress KDO-8-P
phosphatase activity. The comparison between the
BT1713(Mg2+)(KDO-8-P) and
BT1713(Mg2+)(
)(KDN) and
BT1713(Mg2+ (KDO-8-P) provide a structural context to the relative
substrate activities of KDN-9-P, Neu5Ac-9-P, and KDO-8-P (53:26:1). BT1713 has
not evolved to discriminate between KDN-9-P and Neu5Ac-9-P, and this is
evident from the absence of residue binding partners to either the C(5)OH of
KDN-9-P or the C(5)NAc of Neu5Ac-9-P. However, such discrimination is not
necessary in the context of the cell because of the far greater activity of
the synthase enzyme (300:1) in producing KDN-9-P versus Neu5Ac-9-P
(7). In contrast, BT1713 has
evolved to enhance KDN-9-P phosphatase activity and suppress KDO-8-P
phosphatase activity. The comparison between the
BT1713(Mg2+)(KDO-8-P) and
BT1713(Mg2+)( )(KDN)
structures illuminates the conservation of interactions with the substrate
phosphoryl group (core subunit residues) and the carboxylate group (cap
subunit residues) and the loss of interactions of the cap subunit residues
with the KDO-8-P hydroxyl groups. The structures also suggests a key switch of
residues that in BT1713 form a hydrogen bond network extending between the
KDN-9-P carboxylate group and the hexose ring hydroxyl groups. The BT1713
switch residues Glu-56 and Lys-67 allowed the identification of sequences of
the 8KDOP clade that, based on the conservation of these residues, are likely
to function as a KDN-9-P or Neu5Ac-9-P phosphatase, a conclusion supported by
the genome context of the encoding gene.
)(KDN)
structures illuminates the conservation of interactions with the substrate
phosphoryl group (core subunit residues) and the carboxylate group (cap
subunit residues) and the loss of interactions of the cap subunit residues
with the KDO-8-P hydroxyl groups. The structures also suggests a key switch of
residues that in BT1713 form a hydrogen bond network extending between the
KDN-9-P carboxylate group and the hexose ring hydroxyl groups. The BT1713
switch residues Glu-56 and Lys-67 allowed the identification of sequences of
the 8KDOP clade that, based on the conservation of these residues, are likely
to function as a KDN-9-P or Neu5Ac-9-P phosphatase, a conclusion supported by
the genome context of the encoding gene.
The biological range of the “KDN-9-P or Neu5Ac-9-P phosphatase” as evidenced by the 25 putative bacterial hosts (supplemental Fig. SI2) extends beyond the three human symbionts B. thetaiotaomicron, Bacteroides stercoris, and Bacteroides intestinalis to a variety of soil, fresh water, and marine bacteria that also belong to the superphylum Bacteroidetes/Chlorobi and beyond. Examples of more distantly related bacterial hosts include Nitrosospira multiformis and Pelobacter carbinolicus, Desulfuromonas acetoxidans (Proteobacteria β and δ subdivisions), Acidobacteria bacterium (Bacteria Firmicutes), and Methanosscarcina acetivorans (Archaea). The annotated genomes of these evolutionary outliers evidence the juxtaposition of the genes encoding the BT1713 and BT1714 orthologues, suggestive of KDN synthesis in these bacteria. How KDN might serve these bacteria, which live outside the animal host, is presently unclear.
Supplementary Material
Acknowledgments
We thank Dr. Tracy Arakaki and Dr. Nicholas Silvaggi for their generous assistance in data processing, molecular replacement, and refinement. We also gratefully acknowledge Dr. Ezra Peisach for assistance in figure preparation and Jonathan Vural for NMR assistance. We thank Dr. Karen Anderson for providing 8-phospho-2-keto-3-deoxy-d-manno-octulosonic acid synthase. Some data for this study were measured at Beamline X12C of the National Synchrotron Light Source, Brookhaven National Laboratories. Financial support comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the United States Department of Energy, and from the National Center for Research Resources of the National Institutes of Health.
The atomic coordinates and structure factors (codes 3E8M, 3E84, and 3E81) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health GM61099 (to K. N. A. and D. D.-M.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental SI1 and SI2.
Footnotes
The abbreviations used are: Neu5Ac, N-acetylneuraminate; HADSF, haloalkanoic acid dehalogenase superfamily; KDN, 2-keto-3-deoxy-d-glycero-d-galactonononic acid; KDN-9-P, 2-keto-3-deoxy-d-glycero-d-galacto nononate 9-phosphate; KDN-9-P phosphatase, 2-keto-3-deoxy-d-glycero-d-galactonononate-9-phosphate phosphohydrolase; KDN-9-P synthase, 2-keto-3-de-oxy-d-glycero-d-galactonononate-9-phosphate synthase; Neu5Ac-9-P, N-acetylneuraminate 9-phosphate; Neu5Ac-9-P synthase, N-acetylneuraminate-9-phosphate synthase; Neu5Ac-9-P phosphatase, N-acetylneuraminate-9-phosphate phosphohydrolase; KDO, 2-keto-3-deoxy-d-manno-octulosonic acid; KDO-8-P, 2-keto-3-deoxy-d-manno-octulosonate 8-phosphate; KDO-8-P phosphatase, 2-keto-3-deoxy-d-manno-octulosonate-8-phosphate phosphohydrolase; KDO-8-P synthase, 2-keto-3-deoxy-d-manno-octulosonate-8-phosphate synthase; PEP, phosphoenolpyruvate; P, phosphate; pNPP, p-nitrophenyl phosphate; TAPS, 2-hydroxy-1,1-bis(hydroxymethyl)ethyl)-amino]-1-propanesulfonic acid.
At the time that this structure was published, the enzyme function was unknown. However, its high sequence identity with the known E. coli KDO-8-P phosphatase and the juxtaposition of the encoding gene with the KDO-8-P synthase gene is strongly suggestive of KDO-8-P phosphatase catalytic function. Since that time in vitro activity measurements have shown that this H. influenza KDO-8-P phosphatase enzyme is indeed KDO-8-P phosphatase (O. Herzberg, personal communication).
R. Wu, unpublished results.
References
- 1.Angata, T., and Varki, A. (2002) Chem. Rev. 102 439-469 [DOI] [PubMed] [Google Scholar]
- 2.Raetz, C. R. (1990) Annu. Rev. Biochem. 59 129-170 [DOI] [PubMed] [Google Scholar]
- 3.Tanner, M. E. (2005) Bioorg. Chem. 33 216-228 [DOI] [PubMed] [Google Scholar]
- 4.Parsons, J. F., Lim, K., Tempczyk, A., Krajewski, W., Eisenstein, E., and Herzberg, O. (2002) Proteins 46 393-404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu, J., and Woodard, R. W. (2003) J. Biol. Chem. 278 18117-18123 [DOI] [PubMed] [Google Scholar]
- 6.Burroughs, A. M., Allen, K. N., Dunaway-Mariano, D., and Aravind, L. (2006) J. Mol. Biol. 361 1003-1034 [DOI] [PubMed] [Google Scholar]
- 7.Wang, L., Lu, Z., Allen, K. N., Mariano, P., and Dunaway-Mariano, D. (2008) Chem. Biol. 15 893-897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inoue, S., and Kitajima, K. (2006) Glycoconj. J. 23 277-290 [DOI] [PubMed] [Google Scholar]
- 9.Bradford, M. M. (1976) Anal. Biochem. 72 248-254 [DOI] [PubMed] [Google Scholar]
- 10.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307-326 [DOI] [PubMed] [Google Scholar]
- 11.Vagin, A., and Teplyakov, A. (1997) J. Appl. Crystallogr. 30 1022-1025 [Google Scholar]
- 12.Jones, T. A., Zou, J. Y., Cowan, S. W., and Kjeldgaard, M. (1991) Acta Crystallogr. Sect. A 47 110-119 [DOI] [PubMed] [Google Scholar]
- 13.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta Crystallogr. Sect. D Biol. Crystallogr. 54 905-921 [DOI] [PubMed] [Google Scholar]
- 14.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 15.Adams, P. D., Grosse-Kunstleve, R. W., Hung, L. W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K., and Terwilliger, T. C. (2002) Acta Crystallogr. Sect. D Biol. Crystallogr. 58 1948-1954 [DOI] [PubMed] [Google Scholar]
- 16.Brünger, A. T. (1992) Nature 355 472-474 [DOI] [PubMed] [Google Scholar]
- 17.Laskowski, R. A., MacArthur, M. W., Moss, D. S., and Thornton, J. M. (1993) J. Appl. Crystallogr. 26 283-291 [Google Scholar]
- 18.Bennett-Lovsey, R. M., Herbert, A. D., Sternberg, M. J., and Kelley, L. A. (2008) Proteins 70 611-625 [DOI] [PubMed] [Google Scholar]
- 19.Higgins, D. G. (1994) Methods Mol. Biol. 25 307-318 [DOI] [PubMed] [Google Scholar]
- 20.Lu, Z., Dunaway-Mariano, D., and Allen, K. N. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 5687-5692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morais, M. C., Zhang, W., Baker, A. S., Zhang, G., Dunaway-Mariano, D., and Allen, K. N. (2000) Biochemistry 39 10385-10396 [DOI] [PubMed] [Google Scholar]
- 22.Peisach, E., Selengut, J. D., Dunaway-Mariano, D., and Allen, K. N. (2004) Biochemistry 43 12770-12779 [DOI] [PubMed] [Google Scholar]
- 23.Felts, R. L., Ou, Z., Reilly, T. J., and Tanner, J. J. (2007) Biochemistry 46 11110-11119 [DOI] [PubMed] [Google Scholar]
- 24.Veige, A. S., Slaughter, L. M., Lobkovsky, E. B., Wolczanski, P. T., Matsunaga, N., Decker, S. A., and Cundari, T. R. (2003) Inorg. Chem. 42 6204-6224 [DOI] [PubMed] [Google Scholar]
- 25.Crans, D. C., Smee, J. J., Gaidamauskas, E., and Yang, L. (2004) Chem. Rev. 104 849-902 [DOI] [PubMed] [Google Scholar]
- 26.Lu, Z., Dunaway-Mariano, D., and Allen, K. N. (2005) Biochemistry 44 8684-8696 [DOI] [PubMed] [Google Scholar]
- 27.Tremblay, L. W., Dunaway-Mariano, D., and Allen, K. N. (2006) Biochemistry 45 1183-1193 [DOI] [PubMed] [Google Scholar]
- 28.Radaev, S., Dastidar, P., Patel, M., Woodard, R. W., and Gatti, D. L. (2000) J. Biol. Chem. 275 9476-9484 [DOI] [PubMed] [Google Scholar]
- 29.Reaves, M. L., Lopez, L. C., and Daskalova, S. M. (2008) BMB Rep. 41 72-78 [DOI] [PubMed] [Google Scholar]
- 30.Kraulis, P. J. (1991) J. Appl. Crystallogr. 24 946-950 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.