Abstract
Rab proteins influence vesicle trafficking pathways through the assembly of regulatory protein complexes. Previous investigations have documented that Rab11a and Rab8a can interact with the tail region of myosin Vb and regulate distinct trafficking pathways. We have now determined that a related Rab protein, Rab10, can interact with myosin Va, myosin Vb, and myosin Vc. Rab10 localized to a system of tubules and vesicles that have partially overlapping localization with Rab8a. Both Rab8a and Rab10 were mislocalized by the expression of dominant-negative myosin V tails. Interaction with Rab10 was dependent on the presence of the alternatively spliced exon D in myosin Va and myosin Vb and the homologous region in myosin Vc. Yeast two-hybrid assays and fluorescence resonance energy transfer studies confirmed that Rab10 binding to myosin V tails in vivo required the alternatively spliced exon D. In contrast to our previous work, we found that Rab11a can interact with both myosin Va and myosin Vb tails independent of their splice isoform. These results indicate that Rab GTPases regulate diverse endocytic trafficking pathways through recruitment of multiple myosin V isoforms.
Eukaryotic cells are comprised of networks of highly organized membranous structures that require the efficient and timely movement of diverse intracellular proteins for proper function. Molecular motors provide the physical force needed to move these materials along microtubules and actin microfilaments. Unconventional myosin motors, such as those belonging to classes V, VI, and VII, have roles in the trafficking and recycling of membrane-bound structures in eukaryotic cells (1) and are recruited to discrete vesicle populations. Myosin VI is involved in clathrin-mediated endocytosis (2), whereas myosin VIIa participates in the proper development of stereocilia of inner ear hair cells and the transport of pigment granules in retinal pigmented epithelial cells (3, 4). Similarly, the three members of vertebrate class V myosins, myosin Va, myosin Vb, and myosin Vc, are required for the proper transport of a wide array of membrane cargoes, such as the melanosomes of pigment cells, synaptic vesicles in neurons, apical recycling endosomes in polarized epithelial cells, and bulk recycling vesicles in non-polarized cells (5).
Members of the Rab family of small GTPases regulate many cellular systems, including membrane trafficking (6, 7). Certain Rab proteins associate with and regulate the function of class V myosins. Rab27a, in a complex with the adaptor protein melanophilin/Slac2-a, is required to localize myosin Va to the surface of melanin-filled pigment granules in vertebrates (8-10), whereas Rab27a and Slac2-c/MyRIP associate with both Myosin Va and myosin VIIa (3, 11). Rab11a, in a complex with its adaptor protein Rab11-FIP2, associates with myosin Vb on recycling endosomes (12-14) where the tripartite complex regulates the recycling of a variety of cargoes (15-19). In addition, Rab8a associates with both myosin Vb (20) and myosin Vc (21) as part of the non-clathrin-mediated tubular recycling system (20). Recently, Rab11a has also been shown to associate with myosin Va in the transport of AMPA receptors in dendritic spines (22), contributing to the model of myosin V regulation by multiple Rab proteins.
Previous investigations have documented alternative splicing of myosin Va in a tissue-specific manner (23-28). Alternate splicing occurs in a region lying between the coiled-coil region of the neck of the motor and the globular tail region. Three exons in particular are subject to alternative splicing: exons B, D, and F (23-25). Exon F is critical for association with melanophilin/Slac2 and Rab27a (8, 9, 29, 30). Additionally, exon B is required for the interaction of myosin Va with dynein light chain 2 (DLC2) (27, 28). Currently no function for the alternatively spliced exon D has been reported. Similar to myosin Va, myosin Vb contains exons A, B, C, D, and E, whereas no exon F has yet been identified in myosin Vb (Fig. 1A). In addition, exon B in myosin Vb does not resemble the dynein light chain 2 (DLC2) binding region in myosin Va (27, 28), and therefore, it likely does not interact with DLC2. On the other hand, exon D is highly conserved among Myosin Va, myosin Vb, and myosin Vc, suggesting a common function in these molecular motors.
FIGURE 1.
Tissue distribution of human myosin Va and myosin Vb splice isoforms. A, schematic of the alternative exon organization in the tails of myosin Va and myosin Vb. It is known that exons B, D, and F are subject to alternative splicing in myosin Va, whereas there is only evidence that exon D is alternatively spliced in myosin Vb, which does not contain exon F. B, alignment of exon D sequences from mouse and human myosin V's. myosin Va and myosin Vb both contain exon D (amino acids 1320-1346 of myosin Va and 1315-1340 of myosin Vb), whereas myosin Vc contains an exon D-like region (amino acids 1124-1147 of human myosin Vc) that is not known to be alternatively spliced. Alignment of the exon D regions from all three motors reveals a high degree of homology, especially in the center of the exon. Asterisks indicate amino acid identities. C, PCR-based analysis of human tissue panels reveals the alternative splicing pattern of exon D in myosin Va and myosin Vb. Primers flanking the region encoding exon D for both motors were used to amplify cDNA from human MTC™ panels (Clontech). cDNA amplified from HeLa cell RNA as well as myosin Va and myosin Vb tail constructs were used as positive controls. Variants expressing exon D (upper bands) and lacking exon D (lower bands) were visible. Per., peripheral; Pos., positive.
Here we report that Rab10, a protein related to Rab8a and thought to have similar function (31-35), localizes to a system of tubules and vesicles overlapping in distribution with Rab8a in HeLa cells. Utilizing dominant-negative myosin V tail constructs, we show that Rab8a and Rab10 can interact with Myosin Va, myosin Vb, and myosin Vc in vivo. In addition, we have determined that the alternatively spliced exon D in both myosin Va and myosin Vb is required for interaction with Rab10. In contrast to our previous findings, we demonstrate that Rab11a is able to interact with both myosin Va and myosin Vb tails in an exon independent-manner. These results reveal that multiple Rab proteins potentially regulate all three class V myosin motors.
EXPERIMENTAL PROCEDURES
Plasmids and DNA—The following DNA constructs have been previously described: pBD-Rab8a, pBD-Rab11a (12); pBD-Rab11-FIP2 (13); pEGFP-myosin Vc-tail (21); myc-EHD3 (36); mCherry (a gift from Dr. Roger Tsien, University of California San Diego) (37), pmCerulean-C1 (38), pmVenus-C1 (39). The species origin and amino acid boundaries of the myosin V tails used are as follows: mouse myosin Va tail-melanocyte splice form (carboxyl-terminal 637 amino acids including exons A, C, D, E, and F) and brain splice isoform (carboxyl-terminal 588 amino acids including exons A, B, D, and E) were gifts from Dr. John Mercer, McLaughlin Research Institute, Great Falls, MT (24); human myosin Vb tail (GenBank™ accession number NM 001080467, amino acids 1242-1829) was a gift from Dr. Takahiro Nagase, Kazusa DNA Research Institute, Chiba, Japan; human myosin Vc tail is (GenBank™ accession number AL133643, amino acids 902-1741) was a gift from Dr. Richard Cheney, University of North Carolina at Chapel Hill. Myosin Va-tail constructs were subcloned to match the size of the myosin Vb tail constructs using the following primers: 5′-GACGACGAATTCGGTGCGCCTGCTTACCGAGTCC-3′ and 5′-GACGACGTCGACTCAGACCCGTGCGATGAAGCCCAGG-3′. The resulting PCR products were subcloned into pEGFP-C1 (Clontech Laboratories, Mountain View, CA), pAD-GAL4 (Stratagene, La Jolla, Ca), and mCerulean-C1 using the EcoRI and SalI restriction sites. Human myosin Va tail splice isoforms ABCDE and ABCE were cloned from a HeLa cell line, whereas the ACE splice isoform was (GenBank™ accession number EU921839) cloned from a human gastric cancer cell line, MKN7. Myosin Vb tail constructs were created using the following primers: 5′-CCGGGGGTCGACGGCTCCCCAGATAGCTACAGCCTCC-′ and 5′-AAAAAAGGATCCTCAGACTTCATTGAGGAATTCCAG-3′. The PCR product was subcloned into the same plasmids using the SalI and BamHI restriction sites. Myosin Vb-tail lacking exon D (ΔD) was created by nested PCR using the following primers: 5′-CCTATCACGGGGTCTGCCAGACAAACAGGCTGCTGGAGGCTCAGCTGC-3′ and 5′-GCAGCTGAGCCTCCAGCAGCCTGTTTGTCTGGCAGACCCCGTGATAGG-3′.
Tissue Distribution—Alternatively spliced isoforms of myosin Va and myosin Vb were identified in human tissues using cDNA from human MTC™ Panel I and human MTC™ Panel II from Clontech Laboratories Inc. PCR was performed using the following primers for myosin Va (5′-GCCAGAAAGAGGCCATCCAACCCAAGG-3′ and 5′-GGTCAGCCGGGTGATCTCGTGCTGCAGGC-3′) and for myosin Vb (5′-CTCGCCGGCAGGAACGCGGAGCCG-3′ and 5′-CTCGAGCTGAGCCTTGAGATGCTCCACC-3′).
Yeast Two-hybrid Assays—Yeast two-hybrid assays were performed as described previously (13). Briefly, the yeast strain Y190 was co-transformed with various Rab baits in the pBD-GAL4 plasmid and the myosin Va, Vb, and Vc tails (with and without exon D), myosin Vb exons A-B-C-D-E alone, or myosin Vb exons A-B-C-E alone in the pAD-GAL4 prey plasmid. Transformed yeast were plated on dual-deficient synthetic dropout media (SD/-Leu/-Trp) and allowed to grow at 30 °C for 72 h. Surviving colonies were transferred onto filter paper discs (VWR, Westchester, PA), lysed by freezing twice in liquid N2, and incubated in 300 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) for up to 4 h to test for lacZ expression.
Immunocytochemistry—HeLa cells grown on glass coverslips were transiently transfected with plasmids encoding EGFP-,2 mCerulean-, or mVenus-tagged constructs using Effectene transfection reagent (Qiagen, Chatsworth, CA) according to the manufacturer's recommendations. Cells were incubated at 37 °C in 5% CO2 for 18 h after transfection. Coverslips were rinsed once with phosphate-buffered saline and then incubated in 4% paraformaldehyde for 10 min at room temperature to fix the cells. Permeabilization and blocking was performed by incubation of the coverslips in blocking buffer (phosphate-buffered saline with 1% bovine serum albumin and 0.1% Triton-X-100) for 1 h. Blocking buffer was used for all subsequent steps. Cells were incubated with primary antibodies for 1 h, washed 3 times for 10 min, incubated with secondary antibodies for 1 h, washed 3 times for 10 min, and then mounted onto glass microscope slides with Prolong Antifade reagent with DAPI (Invitrogen). Antibodies used in this study have been previously described: rabbit polyclonal anti-Rab8a (40); VU57, rabbit polyclonal anti-Rab11a (41); 8H10, murine monoclonal anti-Rab11a (42); 9E10, murine monoclonal anti-myc (Covance, Berkeley, CA); murine monoclonal anti-CD71 (Invitrogen). The rabbit polyclonal antibody against Rab10 (VU132/134) was raised against a specific peptide sequence at the carboxylterminal variable domain, CKT-PVKEPNSENVDIS. KLH was covalently attached to the amino-terminal cysteine for immunization of the rabbits (Covance, Denver, PA). The antibody was affinity-purified against the immunizing peptide using a SulfoLink Immobilization kit (Thermo Scientific, Waltham, MA) according to the manufacturer's instructions. The purified antibody displayed a single 23-kDa band upon Western blotting of HeLa cell lysates that was sensitive to blocking by the Rab10 peptide (supplemental Fig. 1). Confocal images were captured on an LSM510 confocal microscope (Zeiss, San Jose, CA) using a 60× objective lens. Quantification of the percentage of co-localization was performed using Metamorph software.
Fluorescence Resonance Energy Transfer (FRET)—FRET microscopy was performed as previously described (20, 43). HeLa cells grown on glass coverslips were co-transfected with mCerulean-tagged myosin Va or myosin Vb tails and mVenus-tagged Rab8a, Rab10, or Rab11a as described above. For FRET measurements, coverslips were inverted onto a silicone membrane sealed to a glass slide. Digital images were captured on an LSM510 (Zeiss) equipped with a 40 milliwatt argon laser anda40× objective lens. Three pre-bleach images were taken of both mCerulean (excited at 458 nm) and mVenus (excited at 514 nm) fluorescence. Acceptor photo-bleaching was achieved by exciting with 100 bursts of 514-nm wave-length light at 100% transmission. This was followed by the collection of three post-bleach images. The fluorescence intensity of the photobleached region of interest was measured using the Zeiss LSM software, and the average of the three pre-bleach images was compared with the average of the three post-bleach images after background subtraction. The FRET efficiency was calculated as E = 100(mCeruleanpost - mCeruleanpre)/mCeruleanpost, where mCeruleanpre is the average fluorescence intensity before photobleaching, and mCeruleanpost is the average after photobleaching. FRET data were collected for 10 cells per experimental condition, and each experiment was repeated three times. The results of thirty FRET data sets were averaged for each condition. As a control, the fluorescence intensity of mCerulean-myosin V tails expressed alone was measured both before and after photobleaching.
RESULTS
Distribution of Myosin Va and Myosin Vb Alternative Splice Variants in Human Tissues—Three exons of myosin Va are known to be alternatively spliced in a tissue-specific manner: exons B, D, and F (23-28). Previous studies have established roles for exon F and exon B, yet no function for the alternatively spliced exon D has been reported. Exon D is highly conserved between myosin Va, myosin Vb, and myosin Vc, which contains a region homologous to exon D that is not known to be alternatively spliced (Fig. 1B), suggesting a common function in all three motors.
Although myosin Va is known to undergo alternative splicing in human tissues, little is known about alternative splicing of myosin Vb. Previous work has suggested that myosin Vb may indeed be subject to alternative splicing (44). To determine the existence and distribution of exon D splice variants of myosin Vb in human tissues, we screened two cDNA tissue panels from Clontech Laboratories) by PCR using primers corresponding to exon C and exon E (Fig. 1A). For comparison, we also screened splicing in myosin Va transcripts. Fig. 1C demonstrates that, as with myosin Va, myosin Vb is indeed alternatively spliced in a tissue-specific manner but with a unique pattern of distribution. Specifically, although myosin Va lacks exon D in the brain, two isoforms of myosin Vb with and without exon D are apparent. Conversely, only the exon D-lacking isoform of myosin Vb is found in tissues such as kidney, pancreas, and testis, whereas both isoforms of myosin Va are present.
Yeast Two-hybrid Assays Reveal a Role for Exon D—Previous work done in our laboratory has utilized a rabbit clone of Myosin Vb, which lacks exon D, and the corresponding chicken brain isoform of myosin Va (which also lacks exon D). To determine if the presence or absence of exon D in both of these motors could affect their ability to interact with various Rab GTPases, we performed yeast two-hybrid assays on splice variants of both motors. These include mouse myosin Va carboxyl-terminal tail isoforms from melanocytes (containing exons ABCDEF or ACDEF), human myosin Va containing exons ABCDE and ABCE cloned from HeLa cells, or a fully spliced isoform lacking exons B, D, and F (containing exons ACE) cloned from a human gastric cancer cell line, similar to a variant identified in human melanocytes (45) and human myosin Vb carboxyl-terminal tail isoforms with (exons ABCDE) (46) and without (ABCE) exon D as well as human myosin Vc carboxyl-terminal tail (21). All were screened against Rab8a, Rab10, Rab11a, and Rab11-FIP2 (Table 1).
TABLE 1.
Exon D is required to interact with Rab10. The table illustrates that only those myosin V tails that contain exon D were able to react with the Rab10 bait in a blue/white yeast two-hybrid assay. In addition, all of the myosin Va and myosin Vb tails tested were able to interact with Rab11a and Rab11-FIP2 regardless of alternate exon usage. Rab8a interacted with myosin Va tails with exon D (ABCDEF, ACDEF, and ABCDE), with myosin Vb tail without exon D (ABCE), and with myosin Vc tail (which expresses exon D) but did not interact with myosin Va tails lacking exon D (ABCE and ACE) and myosin Vb tail with exon D (ABCDE). Similar to the results for the full tail segments, GAL4 fusion constructs expressing only exons A, B, C, D, and E of myosin Vb (lacking the globular tail domain) interacted with Rab10, but not Rab8a, whereas exons A, B, C, and E interacted with Rab8a but not Rab10. Neither set of exons interacted with Rab11a or Rab11-FIP2.
| pAD- | pBD-Rab8a | pBD-Rab10 | pBD-Rab11a | pBD-Rab11-FIP2 |
|---|---|---|---|---|
| Myosin Va tail (ABCDEF) | + | + | + | + |
| Myosin Va tail (ACDEF) | + | + | + | + |
| Myosin Va tail (ABCDE) | + | + | + | + |
| Myosin Va tail (ABCE) | − | − | + | + |
| Myosin Va tail (ACE) | − | − | + | + |
| Myosin Vb tail (ABCDE) | − | + | + | + |
| Myosin Vb tail (ABCE) | − | − | + | + |
| Myosin Vc tail | + | + | − | − |
| Myosin Vb Exons ABCDE | − | +/− | − | − |
| Myosin Vb Exons ABCE | + | − | − | − |
The results in Table 1 demonstrate that Rab10 was only able to interact with those myosin V tail constructs expressing exon D, suggesting that exon D is specifically required for interaction with Rab10. The binding pattern for Rab8a was not as straight-forward. Rab8a is able to interact with those isoforms of myosin Va that contain exon D but not those that lack exon D. In contrast, Rab8a only interacted with the isoform of myosin Vb tail that lacks exon D. Myosin Vc tail interacted with both Rab8a and Rab10, whereas it did not interact with Rab11a. Rab8a also interacted with a construct expressing only the amino-terminal 180 amino acids of human myosin Vb tail lacking exon D (MVb-ABCE) but not a matching construct that included exon D (MVb-ABCDE). These constructs encode sequences from the beginning of the tail in exon A to the beginning of the globular portion of the tail after exon E (corresponding to amino acid 1440 of human full myosin Vb tail). These results indicate that for myosin Vb, exons A, B, C, and E represent a minimal binding domain for Rab8a, and the presence of exon D destabilizes this binding domain.
Surprisingly, both myosin Va and myosin Vb interacted with Rab11a and Rab11-FIP2 regardless of splice isoform. We have previously reported that the chicken myosin Va tail was unable to interact with Rab11a by yeast two-hybrid (12, 13, 20). Scrutiny of the DNA sequences used in those previous experiments revealed an unrecognized mutation in the clone of chicken myosin Va tail which resulted in a frameshift near the carboxyl terminus of the protein. These results are supported by recent evidence that myosin Va and Rab11a form a complex in hippocampal neurons (22) and that yeast class V myosin, Myo2p, interacts with a yeast isoform of Rab11, Ypt32p (47, 48).
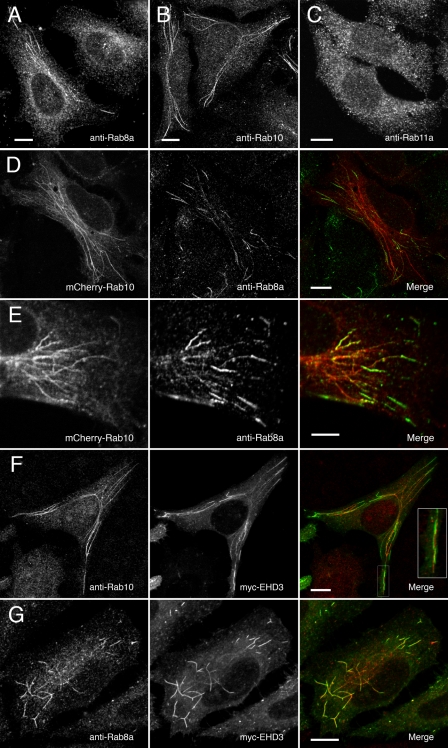
Endogenous Rab10 Shows a Tubular Distribution Similar to Rab8a in HeLa Cells—Rab10 is a mammalian homologue of the yeast Sec4p GTPase and is highly homologous to Rab8a, Rab8b, and Rab13 (49). Like Rab8a, Rab10 is involved with the regulation of membrane trafficking in both polarized and non-polarized cells (31-35). We probed HeLa cells with a specific rabbit polyclonal antibody raised against a peptide sequence unique to human Rab10 to determine endogenous Rab10 localization. As seen in Fig. 2B, Rab10 localized to a network of long tubules in some cells and small vesicles in others, strikingly similar to the pattern of Rab8a (Fig. 2A) localization in HeLa cells (20, 40). This tubular pattern was absent when the antibody was blocked with the specific Rab10 antigen peptide (supplemental Fig. 1). These same tubules were observed in cells expressing a monomeric fluorescent protein (mCherry)-tagged human Rab10 (Fig. 2D). Endogenous Rab8a partially co-localized with these mCherry-Rab10-labeled tubules (Fig. 2, D and E). Interestingly, Rab8a localized to the peripheral ends of the Rab10-labeled tubules. Nevertheless, although some Rab8a-positive tubules were double positive for Rab10, some were single positive for either Rab8a or Rab10. Rab8a colocalizes with EHD3, a marker of the tubular recycling network (20). Similar to Rab8a, Rab10 partially localized to the network of EHD3-positive tubules (Fig. 2F), although not as completely as Rab8a co-localized with EHD3 (Fig. 2G). Thus, these two Rab proteins may associate with overlapping, yet distinct trafficking networks.
FIGURE 2.
Endogenous Rab10 localizes to a system of tubules and vesicles and partially co-localizes with Rab8a- and myc-EHD3-labeled tubules. A-C, HeLa cells stained with anti-Rab8a, anti-Rab10, or anti-Rab11a polyclonal primary and Cy3-labeled secondary antibodies were imaged by confocal microscopy. In a typical HeLa culture ∼17% (±1.2 S.E.) of cells showed a tubular Rab8a or Rab10 pattern, whereas the rest showed a vesicular distribution. D and E, HeLa cells transfected with mCherry-Rab10 (left) and stained with anti-Rab8a polyclonal primary and Alexa488-labled secondary antibodies (center). The merged image (right) shows that Rab8a localized with Rab10 on the peripheral ends of some but not all tubules. F and G, HeLa cells transfected with Myc-tagged EHD3 and stained with Alexa-488-labeled secondary antibodies (center) were co-stained with anti-Rab10 or anti-Rab8a polyclonal primary antibodies and Alexa-568-labeled secondary antibodies (left). Merged images (right) demonstrate that Rab10 only partially co-localizes with EHD3 (see inset). Scale bars represent 10 μmin panels A-D, F, and G and 5 μm in panel E.
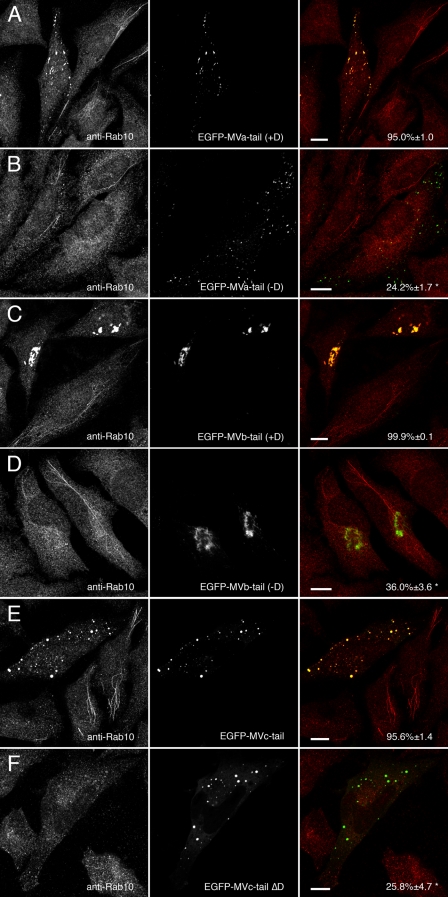
Myosins Va, Vb, and Vc Require Exon D to Colocalize with Rab10—To determine how the alternate usage of exon D in myosin Va, Vb, and Vc affected association with Rab8a, Rab10, and Rab11a in vivo, HeLa cells were transfected with EGFP-tagged myosin Va, Vb, and Vc tails with or without exon D. Transfected cells were probed for endogenous Rab10 using a specific polyclonal antibody. Fig. 3A shows that endogenous Rab10 localization was altered by the expression of dominant-negative myosin Va tail containing exon D (+D). In cells expressing myosin Va tail +D, Rab10 did not localize to tubules but, rather, co-localized with the large, labeled foci throughout the cytoplasm. In contrast, myosin Va tail lacking exon D (-D) when expressed in HeLa cells did not alter Rab10 localization (Fig. 3B). Similarly, myosin Vb tail with exon D (Fig. 3C) but not myosin Vb tail lacking exon D (Fig. 3D) altered Rab10 localization. EGFP-myosin Vb tail +D caused endogenous Rab10 to collapse into EGFP-positive perinuclear clusters. EGFP-myosin Vb tail -D still formed these clusters but was unable to recruit Rab10. In agreement with our observations for myosins Va and Vb, wild-type myosin Vc tail expressing the exon D-like region altered Rab10 localization (Fig. 3E), whereas a synthetic construct of EGFP-myosin Vc tail lacking exon D (ΔD) did not affect Rab10 localization (Fig. 3F). These data confirmed our yeast two-hybrid findings that exon D was critical for Rab10 association with the tails of class V myosin motors. It was notable, however, that the distributions of the myosin V tails were strikingly different. Supplemental Videos 1-3 demonstrate that the peripheral myosin Va tail puncta and pericentrosomal myosin Vb tail clusters were dynamic structures. Myosin Vc tail localized to more static structures.
FIGURE 3.
Only myosin V tails expressing exon D alter endogenous Rab10 distribution. A, HeLa cells transfected with EGFP-myosin Va tail splice isoform containing exon D (EGFP-MVa-tail +D) and stained for Rab10. EFGP-myosin Va tail +D caused endogenous Rab10 to mislocalize to EGFP-labeled puncta. B, EGFP-myosin Va tail-brain splice isoform, which lacks exon D (EGFP-MVa-tail -D), also localized to disperse puncta but was unable to recruit endogenous Rab10. C and D, HeLa cells transfected with EGFP-myosin Vb tail expressing exon D (EGFP-MVb-tail +D) or lacking exon D (EGFP-MVb-tail -D) and stained for Rab10. Both splice variants of EGFP-myosin Vb tail localized to a perinuclear cisternum, but only myosin Vb tail expressing exon D caused Rab10 to mislocalize to the same cisternum. E and F, HeLa cells transfected with wild-type EGFP-myosin Vc tail, which contains an exon D-like domain (EGFP-MVc-tail) or a synthetic construct lacking exon D (EGFP-MVc-tail ΔD) and stained for Rab10. Similar to myosin Va and myosin Vb, myosin Vc tail required exon D to recruit Rab10. Scale bars in all panels represent 10 μm. Percent co-localization (±S.E.) are listed in the merged images on the right of each panel (n ≥ 10). *, statistically significant difference comparing +D and -D constructs (p < 0.001).
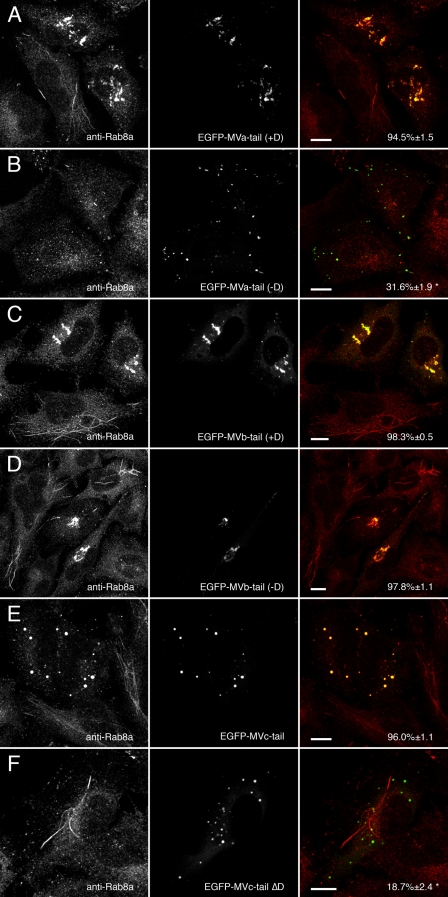
Rab8a Association with Myosin Va and Myosin Vb Is Not Strictly Dependent on Exon D—Similar to the results observed in yeast two-hybrid assays, Rab8a association with myosin Va tail and myosin Vb tail was not simply determined by the presence or absence of exon D. Expression of EGFP-myosin Va tail with exon D (Fig. 4A) caused the redistribution of endogenous Rab8a, whereas EGFP-myosin Va tail lacking exon D (Fig. 4B) did not, in agreement with our previous data (20). In contrast, expression of EGFP-myosin Vb tail caused the redistribution of Rab8a regardless of whether exon D was present or not (Fig. 4, C and D), whereas expression of myosin Vc tail lacking exon D (ΔD) abolished co-localization with Rab8a (Fig. 4F). With the exception of the effects of the loss of exon D on Rab8a association, these findings generally agreed with the yeast two-hybrid results (Table 1). The discrepancy in the case of Rab8a binding to myosin Vb with exon D may reflect a change in affinity between Rab8a and the myosin V tail or a global change in myosin Vb conformation rather than a simple disruption of a Rab8a binding domain.
FIGURE 4.
Myosin Va, Vb, and Vc tails alter endogenous Rab8a distribution. A and B, HeLa cells transfected with EGFP-myosin Va tail +D or EGFP-myosin Va tail -D and stained for Rab8a. As with Rab10, EGFP-myosin Va tail +D caused endogenous Rab8a to mislocalize to EGFP-labeled puncta, whereas EGFP-myosin Va tail -D did not co-localize with Rab8a. C and D, expression of either splice variant of EGFP-myosin Vb tail (+Dor -D) caused Rab8a to redistribute to the EGFP-labeled perinuclear cisternum. E and F, only wild-type EGFP-myosin Vc tail, which contains an exon D-like domain, was able to recruit Rab8a, whereas MVc-tail-ΔD did not. Scale bars in all panels represent 10 μm. Percent co-localization (±S.E.) are listed in the merged images on the right of each panel (n ≥ 10). *, statistically significant difference comparing +D and -D constructs (p < 0.001).
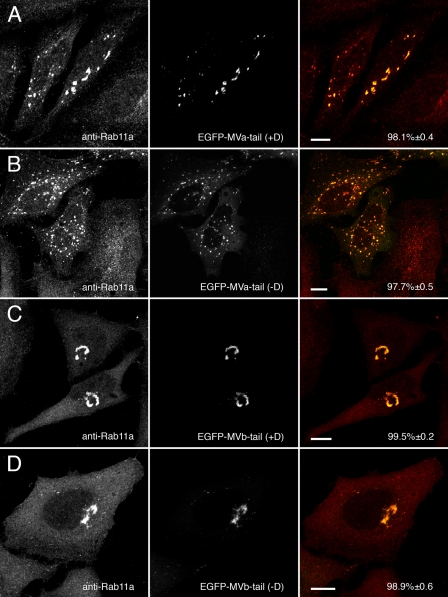
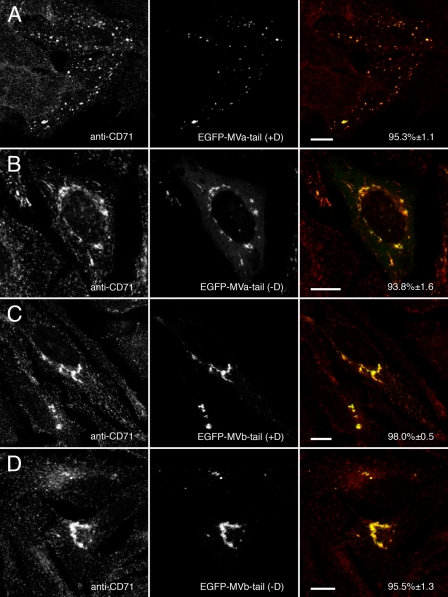
Rab11a Co-localizes with Myosin Va and Myosin Vb in an Exon-independent Manner—Expression of EGFP-tagged Myosin Va and myosin Vb tails revealed that Rab11a associates in an exon-independent manner. In contrast to our previous reports using chicken myosin Va (12, 13), Rab11a co-localized with both the exon D-containing (Fig. 5A) and exon D-lacking (Fig. 5B) splice isoforms of myosin Va tail. Similarly, Rab11a co-localized with both isoforms of EGFP-myosin Vb tail (Fig. 5, C and D). Although both myosin Vb tails containing or lacking exon D accumulated Rab11a in a pericentrosomal collapsed cisternum, both myosin Va tail constructs accumulated Rab11a in smaller puncta located more peripherally throughout the cells, similar to the distributions noted for the myosin V tails in previous figures. In addition, both EGFP-myosin Va and myosin Vb tails caused the accumulation of transferrin receptor (CD71) in transfected cells independent of splice isoform (Fig. 6). Similarly, major histocompatibility complex class I recycling was altered by the expression of any of the myosin Va and myosin Vb tails (supplemental Fig. 2). As observed with the Rab proteins, both myosin Vb tail isoforms accumulated transferrin and major histocompatibility complex class I receptors in a perinuclear tubular cisternum, whereas the myosin Va tails concentrated both receptors in puncta dispersed in the cell periphery. Thus, both myosin Va and myosin Vb can be utilized for Rab11a-mediated recycling.
FIGURE 5.
Myosin Va and myosin Vb tails alter endogenous Rab11a distribution in an exon-independent manner. A and B, HeLa cells transfected with EGFP-myosin Va tail +D or EGFP-myosin Va tail -D and stained for endogenous Rab11a. Unlike Rab8a and Rab10, Rab11a co-localized with both splice isoforms of myosin Va tail in scattered puncta. C and D, similarly, expression of either splice isoform of EGFP-myosin Vb tail (+Dor -D) caused endogenous Rab11a to localize to the EGFP-labeled perinuclear cisterna. Scale bars in all panels represent 10 μm. Percent co-localization (±S.E.) are listed in the merged images on the right of each panel (n ≥ 10).
FIGURE 6.
Myosin Va and myosin Vb tails alter endogenous transferrin receptor (CD71) distribution. A and B, HeLa cells transfected with EGFP-myosin Va tail +D or EGFP-myosin Va tail -D were stained for endogenous transferrin receptor (CD71). Similar to Rab11a, CD71 co-localized with both splice isoforms of myosin Va tail in scattered puncta. C and D, likewise, expression of EGFP-myosin Vb tail +D or EGFP-myosin Vb tail -D caused endogenous CD71 to be recruited to the EGFP-labeled perinuclear cisterna. Scale bars in all panels represent 10 μm. Percent co-localization (±S.E.) are listed in the merged images on the right of each panel (n ≥ 10).
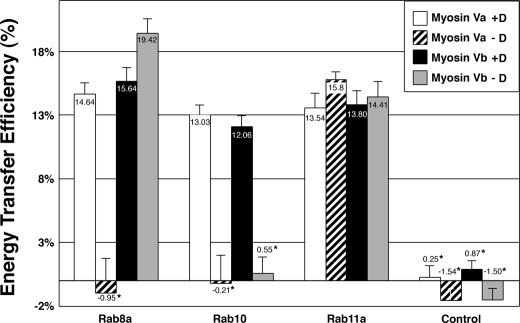
In Situ Association of Myosin V Isoforms with Rab Proteins—To evaluate protein-protein interactions between myosin V tails and Rab proteins in situ, we performed FRET studies between the Rab proteins and myosin V tails using acceptor photobleaching FRET (43). In this method FRET was measured by analyzing the increase in the fluorescent signal of the donor tag before and after the acceptor tag is photobleached. Myosin Va and Vb tails were tagged with the monomeric cyan fluorescent protein derivative, mCerulean (38), whereas Rab8a, Rab10, and Rab11a were tagged with the monomeric yellow fluorescent protein derivative, mVenus (39). HeLa cells were co-transfected with mCerulean-tagged myosin V tails with or without exon D and a monomeric Venus-tagged Rab construct. Fig. 7 illustrates that FRET was observed between the myosin Va or Vb tails expressing exon D and Rab8a, Rab10, and Rab11a. Similarly, FRET was noted between the myosin Vb tail lacking exon D and either Rab8a or Rab11a. The mCerulean-tagged myosin V tails expressed alone were subject to the same photobleaching procedures. These controls showed no FRET, with percent efficiencies of energy transfer similar to those values observed for myosin V tails lacking exon D and Rab10. These results agree with and support the yeast two-hybrid and in vivo data, demonstrating that class V myosin motors are able to associate with multiple Rab proteins.
FIGURE 7.
Myosin V tails FRET with Rab8a, Rab10, and Rab11a in vivo. FRET microscopy was performed on HeLa cells grown on coverslips and cotransfected with mCerulean-tagged myosin Va tail with or without exon D (myosin Va +Dor -D), myosin Vb tail with or without exon D (myosin Vb +D or -D), and mVenus-tagged Rab8a, Rab10, or Rab11a. The fluorescence intensity of a photobleached region of interest was measured before (pre-) and after (post-) photobleaching, and the average of pre-bleach images was compared with the average of the post-bleach images after background subtraction. As a control, FRET was measured on cells expressing the mCerulean-tagged myosin V tails alone. Error bars indicate the S.E. Rab11a produced a positive FRET signal with all four myosin V tail constructs regardless of splicing, whereas Rab10 produced a positive FRET signal only with myosin Va and myosin Vb tails expressing exon D. In contrast, Rab8a produced a positive FRET signal only with myosin Va tail +D but with both myosin Vb tail +Das well as -D. No FRET was observed between myosin Va tail -D and either Rab8a or Rab10 or between myosin Vb tail -D and Rab10. *, statistically significant difference comparing +D and -D constructs (p < 0.001).
DISCUSSION
Unconventional myosin motors, especially class V myosins, play critical roles in the proper trafficking and recycling of a large number of diverse intracellular cargoes. Many investigators have elucidated the roles that members of the Rab family of small GTPases play in the regulation and localization of myosin V in a variety of organisms. Previous studies have revealed that Rab27a, along with its adaptor proteins melanophilin/Slac2-a and Slac2-c/MyRIP, link myosin Va to distinct cargo membranes. Similarly, Rab11a and its adaptor protein Rab11-FIP2 form a complex with myosin Vb that is critical for the recycling of many different receptors. Recent work from our laboratory has demonstrated that myosin Vb is utilized in two distinct intracellular recycling systems through its interactions with either Rab8a or Rab11a (20). Our current studies demonstrate that all three vertebrate class V myosins, myosin Va, myosin Vb, and myosin Vc, can interact with Rab8a and the related protein, Rab10. Rab10 is thought to act in a similar recycling pathway as Rab8a (32, 35), and the two Rabs share a common GTPase-activating protein (GAP), AS160 (34). Rab8a and Rab10 have partially overlapping localizations in HeLa cells, and both proteins are mislocalized by expression of dominant-negative EGFP-myosin Va, Vb, and Vc tail fusion proteins.
Previous investigations have determined that a single exon in myosin Va (designated exon F) determines interaction with Rab27a in conjunction with melanophilin (8, 9, 29, 30). We have now identified the structural basis of the interaction of Rab10 with myosin V tails. The alternatively spliced exon D found in myosin Va and myosin Vb is required for Rab10 binding. In addition, we have determined that Rab8a interacts with myosin Vb in the region of the alternate exons and that exon D perturbs this interaction in yeast two-hybrid assays with myosin Vb, although this perturbation is overcome in vivo. Conversely, exon D is required for Myosin Va interaction with both Rab10 and Rab8a. Likewise, myosin Vc is also able to interact with both Rab8a and Rab10, and deletion of the exon D-like domain of myosin Vc disrupted association with both proteins. These results demonstrate that alternative exon splicing can determine the interaction of particular isoforms of myosin V motors with specific Rab proteins. It remains unclear whether myosin V isoforms containing exon D can bind simultaneously to both Rab8a and Rab10.
In contrast to our previous reports, we were able to demonstrate that myosin Va is indeed able to interact with Rab11a and Rab11-FIP2 in the same manner as myosin Vb. Unlike Rab8a and Rab10, the presence of exon D was dispensable for interaction with Rab11a, which associates with the globular tail regions distal to exon E of myosin Vb (12). Although expression of the different EGFP-myosin Va tail or myosin Vb tail isoforms caused accumulation of endogenous Rab proteins in discrete puncta, it is notable that the patterns of distribution were remarkably different. Although the myosin Vb tail constructs formed large perinuclear tubular cisternae, myosin Va tail constructs elicited the formation of small membrane puncta throughout the periphery of the cells. Nevertheless, expression of any of the myosin Va and myosin Vb tail isoforms could alter transferrin receptor trafficking. These results indicate that the potential functions of myosin Va and myosin Vb may lie in differentiable steps in membrane-trafficking pathways. The different patterns of distribution for the tail constructs may reflect the association of motor complexes with specific regulatory proteins. Therefore, although the present studies suggest that myosin V motors can be utilized in a number of contexts by multiple Rab proteins in the same cells, some specificity of functions must account for the differences in regional distributions.
The present investigations also suggest that Rab proteins determine the majority of trafficking specificity of the Rab/myosin complexes rather than myosin V motors. Thus, Rab8a, Rab10, and Rab11a seem to define specific vesicular, tubulovesicular, or tubular membrane domains. The dynamics of these domains are regulated by the class V myosins, as expression of the motor-deficient tail domains elicit prominent alterations in their distribution. For example, previous studies have suggested that Rab27a is not present in the brain (50-52), although a large amount of myosin Va lacking exon F is expressed in the brain (53, 54). Brown et al. (55) have demonstrated that both Rab8a and Rab11a are critical to the proper trafficking of glutamate receptors in neurons, whereas Correia et al. (22) have shown that myosin Va and Rab11a co-purify with glutamate receptors. Recently, yeast Myo2p has been reported to interact with the Rab11-related protein, Ypt32p (47), and mutations that inhibit Myo2 interaction with Ypt32p map to the globular tail domain (48). These recent investigations and the present results clarify the likely role for an interaction of Rab11a and Rab8a with myosin Va and possibly myosin Vb in neuronal vesicle trafficking and post-Golgi trafficking in yeast.
In summary, the results presented here demonstrate that isoforms of myosin V can interact with Rab10 based on the presence of an alternatively spliced exon sequence. Previous investigations and the present findings also demonstrate that exon choice can influence interactions of myosin V motors with Rab8a, whereas alternate exon usage has no influence on interaction with Rab11a. These results as well as previous studies suggest that myosin V isoforms and splice variants of those isoforms have differential distributions within tissues. Accordingly, cell-specific expression may account for the use of particular pathways in various cells. Nevertheless, it is also clear that multiple forms of these myosin V motors are simultaneously expressed in certain cells. Thus, although cell-specific expression of myosin V species may dictate some of their interactions with particular Rab proteins, it remains possible that the myosin V motors are also involved in the transfer of cargoes between steps along particular trafficking pathways. In this model, interactions of specific myosin V motors with specific Rab proteins may define zones of trafficking pathways as well as their distribution and complexity. Moreover, interactions with adapter proteins or posttranslational modifications could regulate the affinities of myosin V motors for particular Rab proteins. All of this work further supports a model where class V Myosins are multifunctional motors recruited by a variety of Rab proteins to perform multiple tasks in a variety of distinct and overlapping recycling systems.
Supplementary Material
Acknowledgments
We are grateful to Dr. Gustav Leinhard and Dr. Richard Cheney for continuing discussions and cDNA sequences. We are also very appreciative of the gift of Rab8a antibodies from Dr. Johan Peranen. Confocal images were generated through the use of the VUMC Cell Imaging Shared Resource (supported by National Institutes of Health Grants CA68485, DK20593, DK58404, HD15052, DK59637, and EY08126).
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DK070856 (NIDDK) and R01 DK48370 (to J. R. G.) and F32 DK072789 (to J. T. R.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and 2 and Movies 1-3.
Footnotes
The abbreviations used are: EGFP, enhanced green fluorescent protein; FRET, fluorescence resonance energy transfer.
References
- 1.Sellers, J. R. (2000) Biochim. Biophys. Acta 1496 3-22 [DOI] [PubMed] [Google Scholar]
- 2.Sweeney, H. L., and Houdusse, A. (2007) Curr. Opin. Cell Biol. 19 57-66 [DOI] [PubMed] [Google Scholar]
- 3.El-Amraoui, A., and Petit, C. (2005) J. Cell Sci. 118 4593-4603 [DOI] [PubMed] [Google Scholar]
- 4.Futter, C. E. (2006) Pigm. Cell Res. 19 104-111 [DOI] [PubMed] [Google Scholar]
- 5.Desnos, C., Huet, S., and Darchen, F. (2007) Biol. Cell 99 411-423 [DOI] [PubMed] [Google Scholar]
- 6.Markgraf, D. F., Peplowska, K., and Ungermann, C. (2007) FEBS Lett. 581 2125-2130 [DOI] [PubMed] [Google Scholar]
- 7.Grosshans, B. L., Ortiz, D., and Novick, P. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 11821-11827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Provance, D. W., James, T. L., and Mercer, J. A. (2002) Traffic 3 124-132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strom, M., Hume, A. N., Tarafder, A. K., Barkagianni, E., and Seabra, M. C. (2002) J. Biol. Chem. 277 25423-25430 [DOI] [PubMed] [Google Scholar]
- 10.Fukuda, M., Kuroda, T. S., and Mikoshiba, K. (2002) J. Biol. Chem. 277 12432-12436 [DOI] [PubMed] [Google Scholar]
- 11.Fukuda, M., and Kuroda, T. S. (2002) J. Biol. Chem. 277 43096-43103 [DOI] [PubMed] [Google Scholar]
- 12.Lapierre, L. A., Kumar, R., Hales, C. M., Navarre, J., Bhartur, S. G., Burnette, J. O., Provance, D. W., Jr., Mercer, J. A., Bahler, M., and Goldenring, J. R. (2001) Mol. Biol. Cell 12 1843-1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hales, C. M., Vaerman, J. P., and Goldenring, J. R. (2002) J. Biol. Chem. 277 50415-50421 [DOI] [PubMed] [Google Scholar]
- 14.Lindsay, A. J., and McCaffrey, M. W. (2002) J. Biol. Chem. 277 27193-27199 [DOI] [PubMed] [Google Scholar]
- 15.Volpicelli, L. A., Lah, J. J., Fang, G., Goldenring, J. R., and Levey, A. I. (2002) J. Neurosci. 22 9776-9784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hobdy-Henderson, K. C., Hales, C. M., Lapierre, L. A., Cheney, R. E., and Goldenring, J. R. (2003) Traffic 4 681-693 [DOI] [PubMed] [Google Scholar]
- 17.Fan, G. H., Lapierre, L. A., Goldenring, J. R., Sai, J., and Richmond, A. (2004) Mol. Biol. Cell 15 2456-2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore, R. H., Millman, E. E., Alpizar-Foster, E., Dai, W., and Knoll, B. J. (2004) J. Cell Sci. 117 3107-3117 [DOI] [PubMed] [Google Scholar]
- 19.Lise, M. F., Wong, T. P., Trinh, A., Hines, R. M., Liu, L., Kang, R., Hines, D. J., Lu, J., Goldenring, J. R., Wang, Y. T., and El-Husseini, A. (2006) J. Biol. Chem. 281 3669-3678 [DOI] [PubMed] [Google Scholar]
- 20.Roland, J. T., Kenworthy, A. K., Peranen, J., Caplan, S., and Goldenring, J. R. (2007) Mol. Biol. Cell 18 2828-2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodriguez, O. C., and Cheney, R. E. (2002) J. Cell Sci. 115 991-1004 [DOI] [PubMed] [Google Scholar]
- 22.Correia, S. S., Bassani, S., Brown, T. C., Lise, M. F., Backos, D. S., El-Husseini, A., Passafaro, M., and Esteban, J. A. (2008) Nat. Neurosci. 11 457-466 [DOI] [PubMed] [Google Scholar]
- 23.Mercer, J. A., Seperack, P. K., Strobel, M. C., Copeland, N. G., and Jenkins, N. A. (1991) Nature 349 709-713 [DOI] [PubMed] [Google Scholar]
- 24.Seperack, P. K., Mercer, J. A., Strobel, M. C., Copeland, N. G., and Jenkins, N. A. (1995) EMBO J. 14 2326-2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang, J. D., Mermall, V., Strobel, M. C., Russell, L. B., Mooseker, M. S., Copeland, N. G., and Jenkins, N. A. (1998) Genetics 148 1963-1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambert, J., Naeyaert, J. M., Callens, T., De Paepe, A., and Messiaen, L. (1998) Biochem. Biophys. Res. Commun. 252 329-333 [DOI] [PubMed] [Google Scholar]
- 27.Wagner, W., Fodor, E., Ginsburg, A., and Hammer, J. A., III (2006) Biochemistry 45 11564-11577 [DOI] [PubMed] [Google Scholar]
- 28.Hodi, Z., Nemeth, A. L., Radnai, L., Hetenyi, C., Schlett, K., Bodor, A., Perczel, A., and Nyitray, L. (2006) Biochemistry 45 12582-12595 [DOI] [PubMed] [Google Scholar]
- 29.Wu, X. S., Rao, K., Zhang, H., Wang, F., Sellers, J. R., Matesic, L. E., Copeland, N. G., Jenkins, N. A., and Hammer, J. A., III (2002) Nat. Cell Biol. 4 271-278 [DOI] [PubMed] [Google Scholar]
- 30.Au, J. S., and Huang, J. D. (2002) Cell Motil Cytoskeleton 53 89-102 [DOI] [PubMed] [Google Scholar]
- 31.Chen, Y. T., Holcomb, C., and Moore, H. P. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 6508-6512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Babbey, C. M., Ahktar, N., Wang, E., Chen, C. C., Grant, B. D., and Dunn, K. W. (2006) Mol. Biol. Cell 17 3156-3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishikura, S., Bilan, P. J., and Klip, A. (2007) Biochem. Biophys. Res. Commun. 353 1074-1079 [DOI] [PubMed] [Google Scholar]
- 34.Sano, H., Eguez, L., Teruel, M. N., Fukuda, M., Chuang, T. D., Chavez, J. A., Lienhard, G. E., and McGraw, T. E. (2007) Cell Metab. 5 293-303 [DOI] [PubMed] [Google Scholar]
- 35.Schuck, S., Gerl, M. J., Ang, A., Manninen, A., Keller, P., Mellman, I., and Simons, K. (2007) Traffic 8 47-60 [DOI] [PubMed] [Google Scholar]
- 36.Naslavsky, N., Rahajeng, J., Sharma, M., Jovic, M., and Caplan, S. (2006) Mol. Biol. Cell 17 163-177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shu, X., Shaner, N. C., Yarbrough, C. A., Tsien, R. Y., and Remington, S. J. (2006) Biochemistry 45 9639-9647 [DOI] [PubMed] [Google Scholar]
- 38.Rizzo, M. A., Springer, G. H., Granada, B., and Piston, D. W. (2004) Nat. Biotechnol. 22 445-449 [DOI] [PubMed] [Google Scholar]
- 39.Nagai, T., Ibata, K., Park, E. S., Kubota, M., Mikoshiba, K., and Miyawaki, A. (2002) Nat. Biotechnol. 20 87-90 [DOI] [PubMed] [Google Scholar]
- 40.Hattula, K., Furuhjelm, J., Tikkanen, J., Tanhuanpaa, K., Laakkonen, P., and Peranen, J. (2006) J. Cell Sci. 119 4866-4877 [DOI] [PubMed] [Google Scholar]
- 41.Lapierre, L. A., Avant, K. M., Caldwell, C. M., Ham, A. J., Hill, S., Williams, J. A., Smolka, A. J., and Goldenring, J. R. (2007) Am. J. Physiol. Gastrointest. Liver Physiol. 292 1249-1262 [DOI] [PubMed] [Google Scholar]
- 42.Goldenring, J. R., Smith, J., Vaughan, H. D., Cameron, P., Hawkins, W., and Navarre, J. (1996) Am. J. Physiol. 270 G515-G525 [DOI] [PubMed] [Google Scholar]
- 43.Kenworthy, A. K. (2001) Methods 24 289-296 [DOI] [PubMed] [Google Scholar]
- 44.Swiatecka-Urban, A., Talebian, L., Kanno, E., Moreau-Marquis, S., Coutermarsh, B., Hansen, K., Karlson, K. H., Barnaby, R., Cheney, R. E., Langford, G. M., Fukuda, M., and Stanton, B. A. (2007) J. Biol. Chem. 282 23725-23736 [DOI] [PubMed] [Google Scholar]
- 45.Westbroek, W., Lambert, J., Bahadoran, P., Busca, R., Herteleer, M. C., Smit, N., Mommaas, M., Ballotti, R., and Naeyaert, J. M. (2003) J. Investig. Dermatol. 120 465-475 [DOI] [PubMed] [Google Scholar]
- 46.Bement, W. M., Hasson, T., Wirth, J. A., Cheney, R. E., and Mooseker, M. S. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 6549-6553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Casavola, E. C., Catucci, A., Bielli, P., Di Pentima, A., Porcu, G., Pennestri, M., Cicero, D. O., and Ragnini-Wilson, A. (2008) Mol. Microbiol. 67 1051-1066 [DOI] [PubMed] [Google Scholar]
- 48.Lipatova, Z., Tokarev, A. A., Jin, Y., Mulholland, J., Weisman, L. S., and Segev, N. (2008) Mol. Biol. Cell 19 4177-4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pereira-Leal, J. B., and Seabra, M. C. (2001) J. Mol. Biol. 313 889-901 [DOI] [PubMed] [Google Scholar]
- 50.Nagata, K., Satoh, T., Itoh, H., Kozasa, T., Okano, Y., Doi, T., Kaziro, Y., and Nozawa, Y. (1990) FEBS Lett. 275 29-32 [DOI] [PubMed] [Google Scholar]
- 51.Seabra, M. C., Ho, Y. K., and Anant, J. S. (1995) J. Biol. Chem. 270 24420-24427 [DOI] [PubMed] [Google Scholar]
- 52.Chen, D., Guo, J., Miki, T., Tachibana, M., and Gahl, W. A. (1997) Biochem. Mol. Med. 60 27-37 [DOI] [PubMed] [Google Scholar]
- 53.Espindola, F. S., Espreafico, E. M., Coelho, M. V., Martins, A. R., Costa, F. R., Mooseker, M. S., and Larson, R. E. (1992) J. Cell Biol. 118 359-368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Espreafico, E. M., Cheney, R. E., Matteoli, M., Nascimento, A. A., De Camilli, P. V., Larson, R. E., and Mooseker, M. S. (1992) J. Cell Biol. 119 1541-1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown, T. C., Correia, S. S., Petrok, C. N., and Esteban, J. A. (2007) J. Neurosci. 27 13311-13315 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.