Abstract
The assembly and activity of cytochrome c oxidase is dependent on the availability of heme A, one of its essential cofactors. In eukaryotes, two inner mitochondrial membrane proteins, heme O synthase (Cox10) and heme A synthase (Cox15), are required for heme A biosynthesis. In this report, we demonstrate that in Saccharomyces cerevisiae the transcription of COX15 is regulated by Hap1, a transcription factor whose activity is positively controlled by intracellular heme concentration. Conversely, COX10, the physiological partner of COX15, does not share the same regulatory mechanism with COX15. Interestingly, protein quantification identified an 8:1 protein ratio between Cox15 and Cox10. Together, these results suggest that heme A synthase and/or heme O synthase might play a new, unidentified role in addition to heme A biosynthesis.
The biosynthesis of heme A, an obligatory cofactor of cytochrome c oxidase (CcO),2 requires two nuclear-encoded proteins, heme O synthase (HOS) and heme A synthase (HAS) (1–5). Like CcO, both HOS and HAS are integral membrane proteins localized to the inner mitochondrial membrane. First, heme O synthase catalyzes the conversion of heme B to heme O via the farnesylation of a vinyl group at position C-2 (5). Second, heme A synthase converts heme O to heme A by oxidizing the C-8 methyl substituent on heme O to an aldehyde group (1, 2). Heme A has no known physiological function other than as a cofactor in the terminal heme-copper oxidases.
COX10 and COX15, encoding eukaryotic HOS and HAS, respectively, are functionally conserved from yeast to humans (6, 7). In yeast, heme A deficiency leads to respiratory incompetence and the degradation of CcO (8). Mutations in either COX10 or COX15 in humans lead to metabolic diseases associated with isolated CcO deficiency, including Leigh syndrome and hypertrophic cardiomyopathy (9, 10). Recently, a deficiency in heme A biosynthesis was suggested to be involved in age-related decline of CcO in patients with Alzheimer disease (11).
Assembly of the multisubunit CcO complex is a complicated and sequential process (12) that requires more than two dozen nuclear-encoded accessory proteins (13). The crystal structure of bovine heart CcO reveals that the two heme A molecules are deeply buried in the hydrophobic pocket of CcO (14), implying that the insertion of heme A probably occurs early during the CcO assembly process. The identification of a heme a-Cox1 intermediate and the lack of Cox1-Cox4-Cox5 subassembly complex in COX10-deficient human cells further support the notion that heme A insertion occurs at an early step during CcO assembly (12, 15, 16). In Rhodobacter sphaeroides, a deletion of Surf1 results in a 50% loss of heme a3, indicating that Surf1 might be involved in heme a3 maturation (17).
Because the availability of heme A is a key determinant in the assembly of CcO, the reactions catalyzed by HOS and HAS are likely targets for regulation. Because of its high reduction potential (18), free heme A is potentially even more toxic than free heme B. Therefore, cells presumably must have a controlled mechanism to regulate the synthesis, transport, and insertion of heme A to ensure that there is sufficient but not excess heme A available in cells. Although the presence of heme A is strongly connected to copper availability under most circumstances, neither the transcription, translation, stability, nor the activity of either HOS or HAS is affected by intracellular copper levels (19).
Our previous data support a model whereby bacterial HOS and HAS interact to form a complex in which heme O is channeled directly from HOS to HAS (20). Because either the conversion of heme O to heme A or the release of the heme A product appears to be rate-limiting, we hypothesize that in bacteria this HAS-HOS complex might partially regulate the flux of hemes through the heme A biosynthetic pathway. Our hypothesis is consistent with the studies of Barros and Tzagoloff in Saccharomyces cerevisiae suggesting that the activity of HOS is down-regulated either by HAS or the product of HAS (21). They also hypothesized that the activity of HAS is positively regulated by a downstream CcO assembly intermediate or subunit of CcO, because most cox mutants have significantly decreased levels of heme A (21).
Despite these recent advances, the detailed regulatory mechanism of heme A biosynthesis is still far from clear. Large scale genetic, molecular, and physiological approaches seem to suggest that O2 concentrations, fermentable versus non-fermentable carbon sources, rich versus minimal medium, and respiratory competence have no dramatic effect on the transcription of either HOS or HAS (22–25). Although these results might initially seem surprising because of the key role that heme A plays in respiration, the fact that no significant signals were detected in the large scale screening is perhaps to be expected given the low mRNA levels of COX10 and COX15. Therefore, individual analysis is required to determine the regulation of these two genes.
In this study, we examine the regulatory mechanism of Cox15 (HAS) and Cox10 (HOS) in S. cerevisiae. We demonstrate that the two physiological partners do not share the same regulatory mechanism, perhaps suggesting an additional function for HAS and/or HOS. Interestingly, the stoichiometry between Cox15 and Cox10 is ∼8:1, not 1:1 as it has generally been assumed.
EXPERIMENTAL PROCEDURES
Materials—The following yeast strains and plasmids were obtained from other laboratories: MHY100 and MHY200 (Li Zhang, University of Texas at Dallas); mss51Δ (Antoni Barrientos, University of Miami); YEp354 (Dennis R. Winge, University of Utah); plasmid pBS1539 (Min-Hao Kuo, Michigan State University). All other strains and plasmids were generated in this study, and their preparation is described below. The characteristics of all strains and plasmids used in this study are listed in Table 1. Chemicals were purchased from Sigma unless otherwise noted.
TABLE 1.
Strains and plasmids utilized in this study
| Strain or plasmid | Genotype or characteristics | Source |
|---|---|---|
| Strains | ||
| DY5113 | MATa ade2 can1 his3 leu2 ura3 trp1Δ | W303a WT strain |
| cox15-Myc | DY5113, cox15::Myc-HIS3 | This study |
| cox10-Myc | DY5113, cox10::Myc-HIS3 | This study |
| cox15Δ cox10-Myc | DY5113, cox10::Myc-HIS3, cox15::URA3 | This study |
| hem1Δ cox15-Myc | DY5113, hem1::URA3; cox15::cox15-Myc-HIS3 | This study |
| hem1Δ cox10-Myc | DY5113, hem1::URA3; cox10::cox10-Myc-HIS3 | This study |
| coq9Δ | W303a, coq9::KANMX6 | —a |
| qcr7Δ | W303a, qcr7::TRP1 | (53) |
| mss51Δ | W303a, mss51::HIS3 | (54) |
| mss51Δ cox15-Myc | W303a, mss51::HIS3; cox15::cox15-Myc-HIS3 | This study |
| mss51Δ cox10-Myc | W303a, mss51::HIS3; cox10::cox10-Myc-HIS3 | This study |
| BWG1-7a | MATa, ura3-52, leu2-3, 112, his4-519, ade1-100 | WT strain |
| MHY100 | BWG1-7a, hem1-Δ100 | (55) |
| MHY200 | MHY100, hap1::LEU2 | (52) |
| Plasmids | ||
| YEp354 | A reporter plasmid containing the E. coli lacZ gene without a promoter | (28) |
| pCOX15/lacZ | The 1000 bp COX15 promoter fused in-frame to the lacZ gene in YEp354 | This study |
| pCOX10/lacZ | The 1000 bp COX10 promoter fused in-frame to the lacZ gene in YEp354 | This study |
| pMCOX15/lacZ | Derived from pCOX15/lacZ by mutating the potential Hap1 binding site | This study |
| pBS1539 | Contains the TAP tag cassette followed by a URA3+ selection marker |
J. Cricco, D. R. Winge, and E. L. Hegg, unpublished results.
Strains and Growth Conditions—The Myc-HIS3 cassette was amplified from plasmid pFA6a-13Myc-HIS3MX6 (26) via PCR, and homologous recombination was used to attach the 13Myc epitope tag to the C terminus of genomic Cox15 and Cox10. The primers used for Cox15 were described previously (19). The primers for Cox10 were 5′-GATTGGTATTATCCTGGTGAAGCAAAGCGACCACAGGAACGATTTCGGATCCCCGGGTTAATTAA-3′ and 5′-GACTGCCCTTTAAGCGTTGTCTCTTTATCTCATTGTAACTAATGGAATTCGAGCTCGTTTAAAC-3′.
To knock out HEM1 via homologous recombination, the URA3 marker was amplified from plasmid pBS1539. The primers utilized for this deletion were 5′-ATGCAACGCTCCATTTTTGCGAGGTTCGGTAACTCCTCTGCCGCTGTTTCTGAAGCTTGATATCGAAT-3′ and 5′-TTACTGCTTGATACCACTAGAAACCTCTAGTTGTTTAACGATGGGGTCTCTACGACTCACTATAGGGC-3′. Homologous recombination was performed with the method of high lithium transformation as previously described (19).
Yeast strains were cultured at 30 °C either in YP medium (1% yeast extract, 2% peptone) or in synthetic defined (SD) medium with any necessary auxotrophic requirements, both supplemented with either 2% dextrose, 2% galactose, or 2% ethanol-3% glycerol as a carbon source. In this report, 2% dextrose was used as the carbon source unless specifically noted otherwise. When appropriate, media was supplemented with various amounts of δ-aminolevulinic acid (ALA), hemin (chloride salt of ferric heme B), or TEM (0.5% Tween 80, 12 μg/ml ergosterol, and 55.6 μg/ml methionine) (27). Hemin was dissolved in a 0.1% NaOH aqueous solution to obtain a 15 mg/ml 1000× stock solution.
Plasmids—We cloned the upstream 1000-bp promoter region of COX10 and COX15 from genomic DNA, and then subcloned the fragments singly into the yeast plasmid YEp354 (28) containing the Escherichia coli lacZ gene without a promoter. The COX10 and COX15 promoter regions were amplified from genomic yeast DNA using the following primers: 5′-CCCGGGTCGACCCGACGAATACGGCGGAAC-3′ and 5′-CCCGGGAAGCTTCATCTAAAGAGGAAAAAAGGA-3′ (COX10); 5′-CGCGCCCGGGTTATTATTGTCAATAACTGGCC-3′ and 5′-CGCGCCCGGGAGCATGACGTACAGTAGTAGTTAC-3′ (COX15). The lacZ reporter plasmid containing the COX10 promoter sequence is named pCOX10/lacZ, and the lacZ reporter plasmid containing COX15 promoter sequence is named pCOX15/lacZ. Plasmid pMCOX15/lacZ was derived from pCOX15/lacZ by mutating the putative Hap1 binding site on the COX15 promoter region. Mutation of the potential Hap1 binding site was accomplished by site-directed mutagenesis using Expand Long Template PCR system (Roche). The primers were 5′-GATCCGCATTTAAAAAGGCTTACGGCTTTC-3′ and 5′-GAAAGCCGTAAGCCTTTTTAAATGCGGATC-3′. E. coli DH5α was used as the host for plasmid construction and amplification, and unmutated template DNA was cleaved with DpnI (NEB).
β-Galactosidase Activity Assay—Transformants carrying the reporter plasmid (derived from YEp354, which contains a URA3 marker) were selected for URA3 prototrophy. The β-galactosidase activity assay was performed using a modified literature procedure (29). Activity units = (1000 × A420)/(volume of culture assayed × time of reaction × A600).
Total RNA Extraction—The RNeasy Mini Kit from Qiagen was used to purify total RNA from yeast strains grown under various conditions. Usually ∼5 × 107 yeast cells were processed via mechanical disruption to obtain ∼50 μg of RNA, and an on-column DNase digestion was performed to remove any residual DNA contamination. The purity of RNA was analyzed via the A260/A280 ratio.
Reverse Transcriptase (RT) Real-Time PCR—cDNA was synthesized from 5 μg of total RNA by SuperScript™ III reverse transcriptase and Oligo (dT)12–18 primer (Invitrogen) according to the manufacturer's instructions. A 7500 Real Time PCR system and the SYBR GREEN PCR Master Mix from Applied Biosystems were then utilized to quantify the amount of cDNA generated from the previous reverse transcription reaction. ACT1 was used as the endogenous control. Primers were designed by Primer Expression Software (Applied Biosystems). The forward primers were 5′-AGAAGGCCGTAATTCCAAGGA-3′ (COX15), 5′-AAAAATGCCGGATACGTTATGAC-3′ (COX10), and 5′-TCGTTCCAATTTACGCTGGTT-3′ (ACT1), and the reversed primers were 5′-AACGACGCCCATCATAACATG-3′ (COX15), 5′-GACACACGGGCGTTTAGGA-3′ (COX10), and 5′-CGGCCAAATCGATTCTCAA-3′ (ACT1). The data were analyzed using the relative standard curve method, using COX15 from a plasmid to generate the standard curve according to the manufacturer's instructions. After normalization to the endogenous control (ACT1), the relative amounts of COX15 mRNA and COX10 mRNA from different strains were compared. The PCR volume for each well was 25 μl, containing 1–100 ng of cDNA and 400 μm primers. The validation experiment showed that the PCR efficiency of COX10, COX15, and ACT1 are approximately equal. Two independent experiments were performed to quantify the relative amounts of COX15 mRNA and COX10 mRNA in MHY100 and MHY200 strains, and for all experiments, each sample was replicated four times. The S.D. is less than 0.3.
Mitochondria Isolation and Western Analysis—S. cerevisiae cells containing either Cox10 or Cox15 genomically tagged with a C-terminal 13Myc epitope tag were grown for 24 h to exponential phase. Mitochondria were isolated as described previously (19). Mitochondrial proteins were separated via SDS-PAGE using a 12% gel, transferred to a nitrocellulose membrane, and probed with a mouse monoclonal antibody against the Myc epitope tag (Invitrogen). Mitochondrial porin was also probed as the loading control. The secondary antibody was a goat anti-mouse horseradish peroxidase-conjugated antibody (Pierce). The nitrocellulose membrane was then treated with SuperSignal West Pico Chemiluminescent Substrate (Pierce), and exposed to Amersham Hyperfilm ECL (Amersham Biosciences). The intensities of the bands were quantified with ImageJ software (NIH).
Anaerobic Growths—Yeast cells were grown in SD medium at room temperature (22–25 °C) for 24 h with spinning in an anaerobic Coy chamber ([O2] = 1 ppm). Media was supplemented with either TEM (27) or hemin (0.5 μg/ml or 15 μg/ml) to support growth under the rigorously anaerobic conditions. Analogous experiments were performed in the air for comparison. In all cases, the media contained 3% glucose to ensure glucose repression of oxidative metabolism, thereby allowing direct comparison of COX15 expression under both aerobic and anaerobic conditions. Quantification of COX15 transcription was monitored via β-galactosidase activity, and Cox15 protein levels were analyzed by Western blotting as described above.
RESULTS
COX15 Is a Heme B-regulated Gene—To define the regulatory mechanism of COX15 transcription, we initially utilized an in vivo transcription assay in which the COX15 promoter was fused to a lacZ reporter gene, thus allowing us to screen a number of different growth conditions. Quantification of COX15/lacZ transcription was monitored via the activity of β-galactosidase, which is encoded by the lacZ gene. The plasmid pCOX15/lacZ was transformed into wild-type strain W303a and several yeast mutant strains (hem1Δ, coq9Δ, qcr7Δ, mss51Δ, and cox10Δ). The strains carrying pCOX15/lacZ were grown in SD medium supplemented with either glucose, galactose, or ethanol/glycerol as the carbon source. HEM1 encodes 5-aminolevulinate synthase (ALA synthase), which catalyzes the first step in the heme B biosynthetic pathway (30). Coq9 is involved in the biosynthesis of coenzyme Q, which competes with heme A biosynthesis for the farnesyl moiety (31, 32). Qcr7 encodes subunit 7 of the ubiquinol cytochrome c reductase complex, one component of the respiratory chain (33). Mss51 is a translational activator of the mitochondrial genome-encoded Cox1 and is also required for the elongation of Cox1 (34). Both mss51Δ and cox10Δ are respiratory incompetent due to the deficiency of CcO, while coq9Δ and qcr7Δ are respiratory incompetent due to the deficiency of the ubiquinol cytochrome c reductase complex. The results show that neither carbon source, nor the presence/absence of Cox10, Mss51, Qcr7, or Coq9 has any effect on the expression of the COX15/lacZ gene fusion (data not shown); conversely, the data indicate that Hem1 might play some role in COX15 regulation.
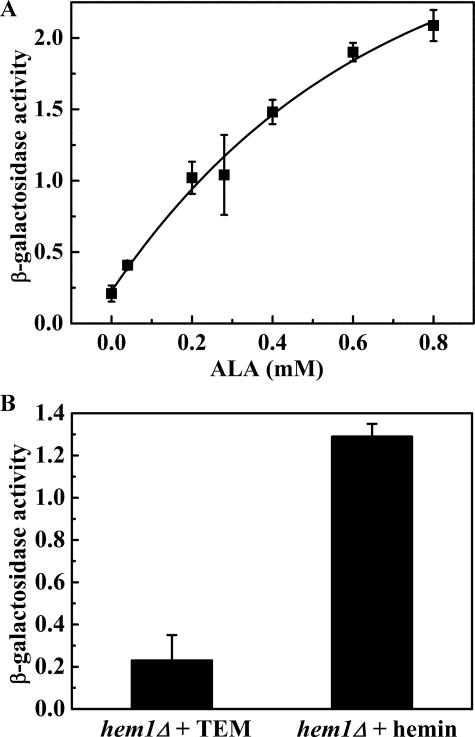
To probe more fully the role of Hem1 on COX15 regulation, hem1Δ cells (DY5113, hem1::URA3) carrying pCOX15/lacZ were grown in SD medium containing various concentrations of ALA (0, 0.04, 0.2, 0.28, 0.4, 0.6, 0.8 mm). In the absence of HEM1, cells can't synthesize heme B and require exogenous heme B, δ-aminolevulinic acid (ALA), or a source of unsaturated fatty acids, sterols, and methionine (TEM) (27). The intracellular heme B concentration can be manipulated by supplementing the growth medium with different amounts of ALA. It has been demonstrated that the addition of 0.5 μg/ml ALA (4 μm) allows for minimal growth, and that the addition of 50 μg/ml ALA (0.4 mm) leads to normal growth (35). In the heme B-depleted condition (neither ALA nor hemin was added to the medium), we supplemented the medium with TEM to obtain viable cells (27). The effect of ALA concentration on β-galactosidase activity is shown in Fig. 1A. The β-galactosidase activity increased as the ALA concentration increased, demonstrating that the transcription of the COX15/lacZ gene fusion is positively regulated either by heme B or a heme precursor.
FIGURE 1.
β-Galactosidase activity assay of the hem1Δ (W303a) cells carrying pCOX15/lacZ in which the lacZ gene is fused to the COX15 promoter region. Activity units of β-galactosidase activity is defined as (1000 × A420)/(time of reaction × A600). A, the cells were grown to mid-exponential phase in SD medium supplemented with various concentration of ALA (0, 0.04, 0.2, 0.28, 0.4, 0.6, 0.8 mm). In the absence of ALA, TEM was added to make the cells viable. B, the cells were grown to mid-exponential phase in SD medium supplemented with either TEM or 15 μg/ml hemin (ferric heme B). In both A and B, the diagrams represent the results of four independent experiments, and the error bars indicate ± S.D.
To clarify whether heme B or a heme precursor regulates the COX15 gene, hem1Δ cells carrying pCOX15/lacZ were grown in SD medium supplemented with either 15 μg/ml hemin (ferric heme B chloride) or TEM, and the β-galactosidase activity of the cells grown under the two different growth conditions was compared. (Adding hemin instead of ALA to the medium bypasses the entire heme B synthetic pathway.) Fig. 1B illustrates the dramatic difference in β-galactosidase activity between cells grown in the absence of heme B (TEM supplemented) and cells grown in the presence of heme B, which clearly demonstrates that it is intracellular heme B, not a heme precursor, which positively regulates the transcription of the COX15 gene.
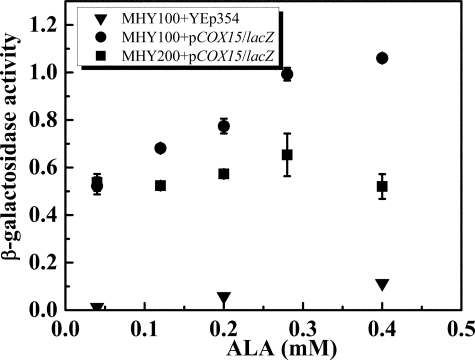
To identify whether this phenomenon is strain-dependent or more general to other yeast strains, the same experiment was performed on another hem1Δ strain, MHY100 (BWG1-7a background). Consistent with previous data shown in Fig. 1A, the β-galactosidase activity increased when the intracellular heme B concentration increased (Fig. 2), indicating that heme B-dependent transcription of HAS is probably a general phenomenon in S. cerevisiae. As expected, the β-galactosidase activity from cells carrying YEp354 alone was close to zero because the lacZ gene lacks a promoter in the plasmid (Fig. 2).
FIGURE 2.
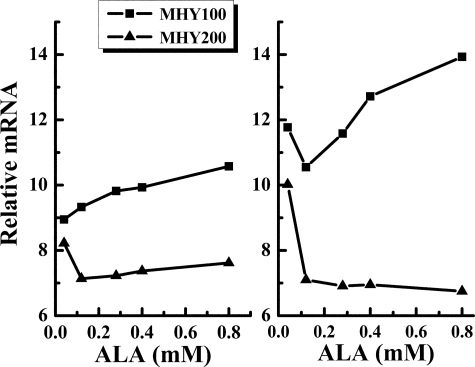
β-Galactosidase activity assay of MHY100 (hem1-Δ100) and MHY200 (MHY100, hap1::LEU2) cells. The cells were grown to mid-exponential phase in SD medium supplemented with various concentrations of ALA (0.04, 0.12, 0.20, 0.28, 0.4 mm). Triangles represent MHY100 carrying YEp354, in which the lacZ gene lacks a promoter. Circles represent MHY100 carrying pCOX15/lacZ, in which lacZ gene is fused to the COX15 promoter region. Squares represent MHY200 carrying pCOX15/lacZ. The diagram represents the results of four independent experiments, and the error bars indicate ± S.D.
COX15 Is a Hap1-regulated Gene—To understand how heme B positively regulates the transcription of COX15, we examined the role of Hap1 in COX15 regulation. In the yeast S. cerevisiae, there are two identified heme-regulated transcription factors, Hap1 and the Hap2/3/4/5 complex (36, 37). Hap1 activation is strictly and precisely regulated by heme concentration (36). However, the connection between the Hap2/3/4/5 complex and heme is not clear. The Hap2/3/4/5 complex activates the genes required for respiratory metabolism when yeast cells are switching from fermentation to respiration (37). It has been identified that this complex is repressed by glucose, and is de-repressed by galactose and non-fermentable carbon sources. Considering the fact that the transcription of COX15 is regulated by intercellular heme, we hypothesized that the COX15 gene might be transcriptionally regulated by Hap1.
To determine if Hap1 plays a role in the regulation of COX15 transcription, we transformed a strain carrying a deletion in both hem1 and hap1, MHY200 (MHY100, hap1::LEU2), with pCOX15/lacZ. The transformed cells were incubated with various concentrations of ALA (0.04, 0.12, 0.20, 0.28, 0.4 mm) in the growth medium. Because MHY200 does not grow well in the absence of heme, 0.00 mm ALA with TEM media was not utilized in this experiment. As shown in Fig. 2, the β-galactosidase activity was constant in the MHY200 (MHY100, hap1::LEU2) strain regardless of the level of ALA in the growth medium, suggesting that transcription is now independent of intracellular heme levels and that the heme B-dependent transcription observed previously must be related to Hap1. Furthermore, compared with MHY100 (hem1Δ), MHY200 cells (hap1Δ and hem1Δ double mutants) have lower β-galactosidase activity in the presence of ALA. Together, these data indicate that COX15/lacZ is positively regulated by Hap1.
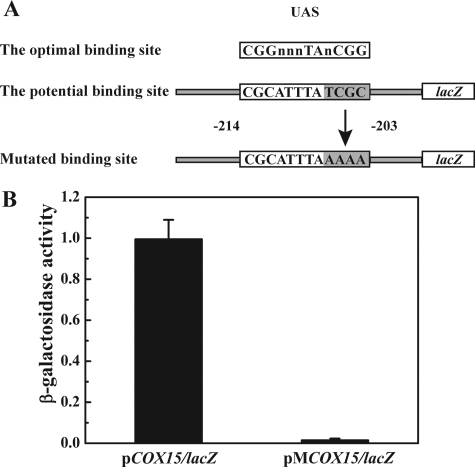
The Consensus Recognition Sequence for Hap1 in the COX15 Promoter Region—While the previous results demonstrated that Hap1 is required for the maximal expression of COX15, they did not ascertain whether Hap1 affects COX15 transcription directly or indirectly through another factor. Further evidence is required to support that Hap1 is the transcription activator of COX15. Previous studies revealed that Hap1 recognizes at least two related DNA sequences, CGGNNNTANCGG and CGCNNNTANCGC (38–40). Inspection of the COX15 promoter region identified one potential Hap1-binding sequence CGCATTTATCGC located –214 to –203 (relative to the A at the +1 of the ATG start condon) (Fig. 3A), which is highly homologous to the UAS of the CYC7 promoter (CGCTATTATCGC), another well-defined Hap1-regulated gene.
FIGURE 3.
Site-directed mutagenesis on the UAS of the COX15 promoter region. A, the top sequence shows the optimal Hap1 binding site. The middle sequence shows the potential Hap1 binding site on COX15/lacZ, which is located –214 to –203 relative to the A at the +1 of the ATG start codon. The bottom sequence shows the mutated binding site on the pMCOX15/lacZ plasmid. B, the β-galactosidase activity assay of the BWG1-7a cells carrying either pCOX15/lacZ or pMCOX15/lacZ containing the mutated binding site. The cells were grown to mid-exponential phase in SD medium. The diagram represents the results of four independent experiments, and the error bars indicate ± S.D.
Because not all potential Hap1 binding sites that are homologous to the consensus sites are functional (39), further analysis was required to verify that this is a true Hap1 binding site. To accomplish this, we mutated the potential Hap1 binding site on plasmid pCOX15/lacZ and inferred the relative expression level by quantifying the corresponding β-galactosidase activity. Previous studies showed that the two CGC triplets are critical for Hap1 binding (38). We mutated the potential binding site CGCATTTATCGC to CGCATTTAAAAA (Fig. 3A) via site-directed mutagenesis. The plasmid containing the mutated promoter was named pMCOX15/lacZ. Both pMCOX15/lacZ and pCOX15/lacZ were transformed into BWG1-7a (wild-type strain). The β-galactosidase activity assay revealed that the mutation of the second CGC triplet completely abolished the transcription of lacZ in the MCOX15/lacZ fusion (Fig. 3B). Together, these results unequivocally establish that the Hap1 binding site is required for the transcription of COX15.
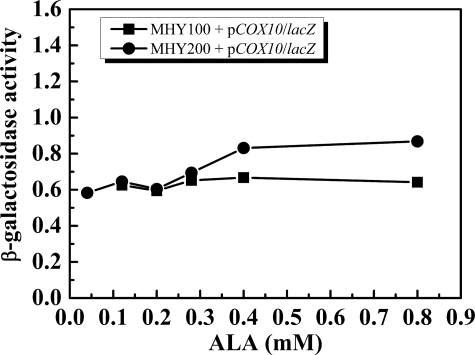
COX10 Is Not a Heme B-regulated Gene—Because Cox10 and Cox15 work in concert to synthesize heme A, we hypothesized that the expression of Cox10 and Cox15 might be coordinated and regulated by a similar mechanism to form a 1:1 stoichiometry. To probe the regulation of COX10, we transformed pCOX10/lacZ into MHY100 and MHY200 strains grown under various ALA concentrations. The β-galactosidase activity assay (Fig. 4) demonstrated that the expression of COX10/lacZ is independent of intracellular heme B levels and Hap1, indicating that COX10 is not utilizing the same regulatory mechanism as COX15. We also searched the promoter region of COX10, and no potential Hap1 binding site was identified. Together, our data indicate that COX10 is regulated neither by intracellular heme B levels nor by Hap1.
FIGURE 4.
β-Galactosidase activity assay of the MHY100 (hem1-Δ100) and MHY200 (MHY100, hap1::LEU2) cells. The cells were grown to mid-exponential phase in SD media supplemented with various concentrations of ALA (0.04, 0.12, 0.20, 0.28, 0.4 mm). Squares represent MHY100 carrying pCOX10/lacZ in which lacZ gene is fused to the COX10 promoter, and circles represent MHY200 carrying pCOX10/lacZ. The diagram represents the average results of two independent experiments.
Quantification of the COX15 and COX10 mRNA Levels—To confirm the previous results obtained from the β-galactosidase activity assay, the endogenous mRNA levels of COX15 and COX10 in MHY100 and MHY200 were quantified by RT real-time PCR. MHY100 and MHY200 cells were grown in medium supplemented with various concentrations of ALA (0.04–0.8 mm), and the total RNA was extracted from cells harvested at mid-exponential phase. The housekeeping gene ACT1 was utilized as an endogenous control. The relative amounts of ACT1 mRNA, COX15 mRNA, and COX10 mRNA were quantified using the standard curve obtained from a plasmid containing the COX15 gene.
The RT real-time PCR data reveal that neither intracellular heme levels nor the absence of Hap1 has any effect on the COX10 mRNA levels (data not shown), which is consistent with the data acquired from our previous β-galactosidase activity assays (Fig. 4). Therefore, we subsequently used COX10 as the endogenous control to calculate the relative mRNA levels of COX15 by dividing the calculated mRNA level of COX15 by the calculated mRNA level of COX10. The results (Fig. 5) demonstrate that while COX15 mRNA levels increase with increasing intracellular heme levels in the presence of Hap1, the mRNA levels of COX15 are depressed and independent of intracellular heme levels in the absence of Hap1. Both of these results are consistent with the result obtained from the β-galactosidase activity assay. It is not currently understood why the lowest ALA concentration (0.04 mm) gave the highest amount of COX15 mRNA in MHY200 cells.
FIGURE 5.
The mRNA level of COX15 in MHY100 (hem1-Δ100) and MHY200 (MHY100, hap1::LEU2) cells. The cells were grown to mid-exponential phase in SD media supplemented with various concentrations of ALA (0.04, 0.12, 0.28, 0.4, 0.8 mm). Squares represent MHY100 and triangles represent MHY200. Each point is the average of four replicates. Each experiment was repeated twice and a similar trend was observed. The particular diagram shown here represents the results of the two experiments.
The RT real-time PCR data also reveal an interesting aspect concerning the ratio of COX15 mRNA to COX10 mRNA. The mRNA ratio between COX15 and COX10 in both wild-type (BWG1-7a) and MHY100 cells (grown in the presence of 0.8 mm ALA) is close to 10:1. Although RT real-time PCR is generally applied to compare the expression level of a single gene in different samples, we hypothesize that the ∼10:1 ratio might shed some light on the relative stoichiometry of Cox15 and Cox10 protein levels, which is generally believed to be close to 1:1.
The Protein Levels of Cox15 and Cox10 Were Quantified—To confirm that the changes in the mRNA levels of COX15 are reflected in the protein levels, we inserted a 13Myc epitope tag to the C terminus of genomic Cox15. The same epitope-tagged Cox15 in a yeast vector was found to complement the cox15Δ mutant phenotype,3 suggesting that the addition of the 13Myc epitope tag has no effect on either the activity or the stability of Cox15. We then quantified the Cox15 levels by immunoblotting. Because Cox15 is located in the inner mitochondrial membrane, mitochondria were isolated from hem1Δ (W303a, hem1::URA3; cox15::cox15-Myc-HIS3) cells before Cox15 protein levels were quantified via Western blot analysis. Porin, an outer mitochondrial membrane protein, was utilized as the loading control. The intensity of each band was quantified with ImageJ.
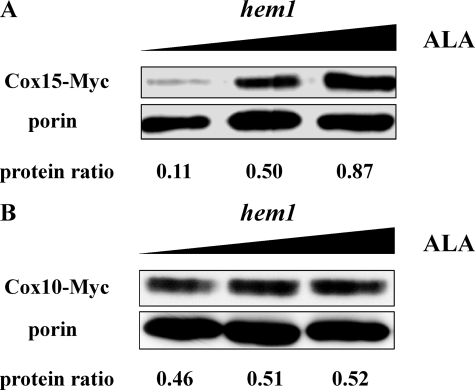
Cox15 protein levels from hem1Δ cells grown with low (0.04 mm), medium (0.2 mm), and high (2 mm) concentrations of ALA are compared in Fig. 6A. Cox15 protein levels increase nearly 8-fold in heme-replete cells relative to heme-depleted cells. Interestingly, the RT real-time PCR data reveal that the mRNA level of Cox15 increases only 10% when ALA concentration is increased from 0.05 mm to 0.80 mm. Cox15 is believed to utilize heme B as the cofactor (41), and in addition to activating Hap1, this heme cofactor may also be important in stabilizing Cox15.
FIGURE 6.
Western blot analysis of the relative protein levels of Cox15 and Cox10 in wild-type and hem1Δ stains. The C terminus of both Cox15 and Cox10 were genomically linked to a 13Myc epitope tag. Porin (bottom band), an outer mitochondrial membrane protein, was utilized as the loading control. The relative intensity (protein ratio) of Cox15 or Cox10 to porin was quantified with ImageJ. The protein ratios were calculated by dividing the intensity of upper band by the intensity of porin band. A, the relative protein levels of Cox15 (upper band) from hem1Δ cells (W303a, hem1::URA3; cox15::cox15-Myc-HIS3) grown under 0.04 mm, 0.2 mm and 2.0 mm ALA. B, relative protein levels of Cox10 (upper band) from hem1Δ cells (W303a, hem1::URA3; cox10::cox10-Myc-HIS3) were analyzed via Western blot. The protein ratios are the average results of two experiments.
We performed the same experiment to quantify the Cox10 levels in hem1Δ cells grown with low (0.04 mm), medium (0.2 mm), and high (2 mm) concentrations of ALA. The Western blot (Fig. 6B) demonstrates that the protein level of Cox10 is heme B-independent, consistent with the COX10/lacZ and COX10 mRNA data. Significantly, together with Fig. 6A, these data also indicate that Cox10 is not affected by the presence/absence of its physiological partner, Cox15.
Protein Ratio between Cox15 and Cox10—As previously mentioned, the RT real-time PCR data suggested that COX15 mRNA levels are about 10 times higher than COX10 mRNA levels in wild-type cells. However, because of the controversial evidence regarding the relationship between mRNA and protein levels (42, 43), whether the mRNA ratio is also reflected in the protein ratio needed to be determined. Western blot analysis was utilized to quantify protein abundance. We compared the protein levels of Cox15 and Cox10 that were genomically fused to a C-terminal Myc epitope tag. Because the molecular weights of Cox15 and Cox10 are similar, we genomically tagged them separately in two wild-type cells, DY5113, COX15::Myc-HIS3 and DY5113, COX10::Myc-HIS3. The two strains were grown under identical conditions to mid-exponential phase, and the purified mitochondria were analyzed via Western blot analysis. The results (Fig. 7) revealed a protein ratio of 8:1 between Cox15 and Cox 10, indicating a good correlation between the mRNA abundance and the protein abundance.
FIGURE 7.
Western blot analysis of protein ratio between Cox15 and Cox10. A 13Myc epitope tag was fused to the C terminus of both genomic COX10 and genomic COX15. Porin (bottom band), an outer mitochondrial membrane protein, was utilized as the loading control. The relative intensity (protein ratio) of Cox15 or Cox10 to porin was quantified with ImageJ. The relative protein levels of Cox15 (upper, right band) and Cox10 (upper, left band) from two strains, W303a, cox15::cox15-Myc-HIS3 and W303a, cox10::cox10-Myc-HIS3, were quantified. The protein ratios were calculated by dividing the intensity of the upper band by the intensity of the porin band, and the results presented here are the average of two independent experiments.
The Stability of Cox15 or Cox10 Is Not Decreased in the Absence of Cytochrome c Oxidase—It has been reported previously that heme A levels decreased dramatically in cells deficient in CcO (21). It is therefore reasonable to hypothesize that the stability of Cox15 might be down-regulated by a key subassembly complex of CcO, especially the one containing Cox1, the final heme A acceptor. To ascertain if Cox15 stability is decreased in the absence of CcO, we genomically inserted a 13Myc epitope tag to the C terminus of Cox15 in an mss51Δ strain (W303a, mss51::HIS3; cox15::cox15-Myc-HIS3). MSS51 is a nuclear gene encoding for a translational activator of Cox1, subunit I of CcO (44). In the absence of Mss51, the translation of Cox1 is completely blocked, resulting in total loss of CcO activity. We utilized quantitative Western blot analysis to compare the Cox15 levels in the wild-type strain versus the mss51Δ strain (Fig. 8). The data demonstrated that there is not a decrease in Cox15 levels in an mss51Δ strain, suggesting that the stability of Cox15 is not affected by Mss51, Cox1, or a Cox1-associated subassembly complex of CcO.
FIGURE 8.
Western blot analysis of the steady state level of Cox15 in the absence of Mss51. A 13Myc epitope tag was fused to the C terminus of genomic COX15. Porin (bottom band), an outer mitochondrial membrane protein, was utilized as the loading control. The relative intensity (protein ratio) of Cox15-13Myc to porin was quantified with ImageJ. The protein level of Cox15-13Myc from mss51Δ(W303a, mss51::HIS3; cox15::cox15-Myc-HIS3) (upper, left band) and W303a, cox15::cox15-Myc-HIS3 (upper, right band) were compared. The protein ratios are calculated by dividing the intensity of the upper band by the intensity of the porin band, and the results presented here are the average of two experiments.
To identify whether the stability of Cox10 is altered by the presence of its physiological partner Cox15 or by CcO, we determined the Cox10 protein level in cox15 Δ and mss51Δ strains. The comparison between mss51Δ cells and wild-type cells demonstrates that the protein level of Cox10 is affected neither by Mss51 nor by the presence/absence of CcO. Similarly, the stability of Cox10 is not affected by the absence of its physiological partner Cox15 (data not shown). Thus, HOS appears to be regulated completely independently of HAS.
Regulation of COX15 under Anaerobic Conditions—Because of the central role that heme A plays in the assembly and function of CcO, it is reasonable to ask whether COX15 might also be regulated by O2. Clearly O2 will indirectly affect COX15 transcription because intracellular heme levels in yeast are dependent on O2 concentrations, and heme obviously regulates COX15 transcription via Hap1. However, might O2 also play a more direct role in regulating COX15?
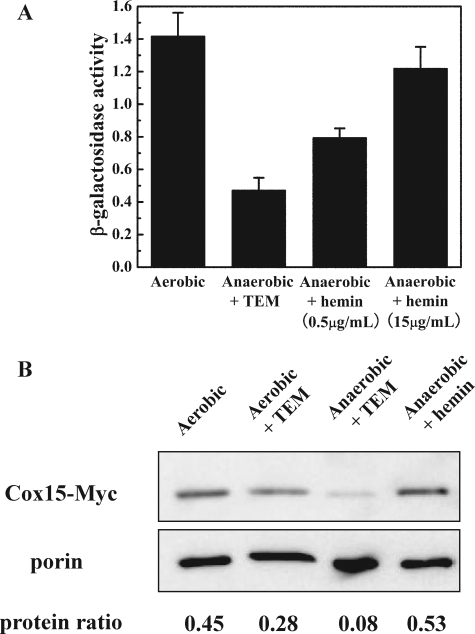
To address this question, pCOX15/lacZ was transformed into W303a cells, and the cells were incubated under either aerobic or rigorously anaerobic conditions. The β-galactosidase activity (Fig. 9a) revealed that the transcription of lacZ is significantly lower in cells grown under anaerobic conditions (+TEM). To ascertain if the decrease in β-galactosidase activity is due solely to a decrease in intracellular heme levels or whether O2 concentrations play a more direct role, cells were grown anaerobically in the presence of hemin. The addition of 15 μg/ml of hemin to the growth medium restored β-galactosidase activity to aerobic levels. Analysis of the Cox15 protein levels (genomically fused to a C-13Myc epitope tag) revealed that Cox15 protein levels were also significantly depressed under anaerobic conditions, but restored upon the addition of hemin to the growth medium, consistent with the results obtained using the pCOX15/lacZ reporter plasmid. Together these results demonstrate that while O2 does play a role in COX15 regulation, it does so indirectly through intracellular heme levels.
FIGURE 9.
Dependence of COX15 on O2. A, β-Galactosidase activity assay of W303a cells carrying the reporter plasmid pCOX15/lacZ. Cells were grown in SD medium containing 3% glucose either aerobically, anaerobically in the presence of TEM, or anaerobically with varying concentrations of hemin added to the growth medium. The graph represents the average results of four independent experiments, and the error bars indicate ± S.D. B, Western blot analysis of Cox15 genomically fused to a C-terminal 13Myc epitope tag. Cox15 proteins levels were determined for cells grown under both aerobic and anaerobic conditions. Porin (bottom band), an outer mitochondrial membrane protein, was utilized as the loading control. The relative intensity (protein ratio) of Cox15-13Myc to porin was quantified with ImageJ. The protein ratios are calculated by dividing the intensity of the upper band by the intensity of the porin band.
DISCUSSION
In the yeast S. cerevisiae, CcO is composed of 11 subunits, which are mainly regulated by ATP concentration, oxygen availability, intracellular heme levels, carbon sources, and retrograde regulation (45, 46). In addition to the structural subunits, more than two dozen accessory proteins are required for CcO assembly and function, and their regulatory mechanism remains largely unknown (47). Among these accessory proteins are HOS and HAS, which are required for the biosynthesis of the heme A cofactor.
In this study, we demonstrated that the transcription of COX15 (HAS) is positively regulated by heme B via Hap1, a transcription factor which widely regulates many genes involved in respiration and oxygen damage control. Interestingly, the transcription of COX15/lacZ decreased to zero when the HAP1-binding site on the promoter region of COX15 was mutated, while the transcription of COX15/lacZ only decreased to ∼50% of wild-type levels when HAP1 was deleted. These two seemingly contradictory results suggest that there may be another unidentified transcription factor which is competing for the same binding site with Hap1, and that they work together to strictly regulate COX15. This scenario would explain why we did not observe as dramatic a decrease in the mRNA levels of COX15 in the absence of Hap1.
Conversely, the transcription of COX10 (HOS) is not dependent on the intracellular heme levels or Hap1. Interestingly, it was recently reported that the mRNA level of COX10 increases 2-fold when Hap4 is overexpressed (48). (Hap4 is the subunit of the transcription factor complex Hap2/3/4/5 that specifically binds to the CCAAT consensus sequence.) However, the physiological consequence of this finding and the specific relationship between Hap4 and COX10 has not yet been fully explored.
Our RT real-time PCR data revealed that the mRNA level of COX10 is approximately ten times less than that of COX15, and protein quantification confirmed that the ratio of the mRNA is reflected in the ratio of the two proteins. Previously it was generally believed that the transcription and translation of HOS and HAS are coordinated to obtain a 1:1 stoichiometry, because these two enzymes work cooperatively to synthesize heme A, and a HAS-HOS complex in bacteria is supported by our previous studies (20). There are three possible explanations concerning the 8:1 stoichiometry between HAS and HOS protein levels. 1) The first possibility is that the turnover number of HAS is much lower than that of HOS, and therefore more HAS is required to synthesize enough heme A for CcO. 2) The second possibility is that HAS is in excess, and that HOS is limiting in the heme A biosynthetic pathway. This possibility is perhaps supported by the fact that co-overexpression of Mss51 and Cox10 increases both Cox1 and heme A levels in shy1Δ cells (49). (If Cox10 is limiting, however, why does the majority of the regulation appear to be at Cox15?) Finally, 3) Cox15 and/or Cox10 may have dual, non-overlapping functions involving both heme A biosynthesis and some other unknown function. Recently, Shy1 and Cox14, two other CcO accessory proteins, were determined to promote the assembly of CcO with cytochrome bc1 to form a respiratory chain supercomplex (50). In addition, human Sco1 and Sco2 (51) and Cox11 (15) were reported to have multiple functions in addition to aiding in the maturation of the CuA and CuB sites of CcO, respectively. Further studies are underway to ascertain whether HAS and/or HOS have a secondary function.
Quantifying the protein levels of Cox15 and Cox10 in the mss51Δ strain and the wild-type strain demonstrates that the steady-state levels of Cox15 and Cox10 are not down-regulated by the absence of Cox1, a Cox1-associated subassembly complex, or respiratory deficiency. This is contrary to the structural subunits of CcO, which are generally degraded by mitochondrial proteolysis if the respiratory chain is not competent. Our observation, coupled to the results reported by Barros and Tzagoloff (21), suggest that the decreased amount of heme A observed in the absence of subunit I could be partially caused by a decrease in the turnover number of the heme A biosynthetic pathway. One logical possibility is that heme A release from HAS is a target for regulation, and that this release is facilitated either by Cox1 or by some subassembly intermediate of CcO. The downstream regulation mechanism prevents uncontrolled release of heme A and blocks the accumulation of unassociated heme A in the mitochondrion, thus avoiding potential deleterious damage to DNA and membranes. A second possibility is that either HAS or a mitochondrial ferredoxin, a potential electron donor for HAS in yeast (1), can sense the mitochondrial redox potential (which is dependent on the efficiency of the respiratory chain), and that electron flow through HAS (and therefore the activity of HAS) is inhibited by respiratory deficiency. Currently we cannot distinguish between these two different possibilities.
In summary, our results conclusively demonstrate that COX15 is positively regulated by intracellular heme levels via Hap1. In addition, our data suggest that there is an additional, as of yet unidentified, factor that also controls the transcription of COX15, sharing with Hap1 an overlapping recognition sequence in the COX15 promoter region. Conversely, the transcription of COX10 is completely independent of both Cox15 and Hap1, and Cox10 is naturally present at much lower levels than Cox15, highlighting the divergent regulation of these two partner enzymes.
This work was supported, in whole or in part, by National Institutes of Health Grant GM66236 (to E. L. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: CcO, cytochrome c oxidase; ALA, δ-aminolevulinic acid; HAS, heme A synthase; HOS, heme O synthase; IM, inner membrane; IMS, intermembrane space; RT, reverse transcriptase.
J. Cricco and E. L. Hegg, unpublished data.
References
- 1.Barros, M. H., Carlson, C. G., Glerum, D. M., and Tzagoloff, A. (2001) FEBS Lett. 492 133–138 [DOI] [PubMed] [Google Scholar]
- 2.Brown, K. R., Allan, B. A., Do, P., and Hegg, E. L. (2002) Biochemistry 41 10906–10913 [DOI] [PubMed] [Google Scholar]
- 3.Mogi, T., Saiki, K., and Anraku, Y. (1994) Mol. Microbiol. 14 391–398 [DOI] [PubMed] [Google Scholar]
- 4.Svensson, B., Lubben, M., and Hederstedt, L. (1993) Mol. Microbiol. 10 193–201 [DOI] [PubMed] [Google Scholar]
- 5.Puustinen, A., and Wikstrom, M. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 6122–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murakami, T., Reiter, L. T., and Lupski, J. R. (1997) Genomics 42 161–164 [DOI] [PubMed] [Google Scholar]
- 7.Valnot, I., von Kleist-Retzow, J. C., Barrientos, A., Gorbatyuk, M., Taanman, J. W., Mehaye, B., Rustin, P., Tzagoloff, A., Munnich, A., and Rotig, A. (2000) Hum. Mol. Genet. 9 1245–1249 [DOI] [PubMed] [Google Scholar]
- 8.Glerum, D. M., Muroff, I., Jin, C., and Tzagoloff, A. (1997) J. Biol. Chem. 272 19088–19094 [DOI] [PubMed] [Google Scholar]
- 9.Antonicka, H., Mattman, A., Carlson, C. G., Glerum, D. M., Hoffbuhr, K. C., Leary, S. C., Kennaway, N. G., and Shoubridge, E. A. (2003) Am. J. Hum. Genet. 72 101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antonicka, H., Leary, S. C., Agar, J. N., Horvath, R., Kennaway, N. G., Harding, C. O., Jaksch, M., and Shoubridge, E. A. (2003) Hum. Mol. Genet. 12 2693–2702 [DOI] [PubMed] [Google Scholar]
- 11.Atamna, H. (2004) Ageing Res. Rev. 3 303–318 [DOI] [PubMed] [Google Scholar]
- 12.Williams, S. L., Valnot, I., Rustin, P., and Taanman, J. W. (2004) J. Biol. Chem. 279 7462–7469 [DOI] [PubMed] [Google Scholar]
- 13.Barrientos, A., Barros, M. H., Valnot, I., Rotig, A., Rustin, P., and Tzagoloff, A. (2002) Gene 286 53–63 [DOI] [PubMed] [Google Scholar]
- 14.Tsukihara, T., Aoyama, H., Yamashita, E., Tomizaki, T., Yamaguchi, H., Shinzawaitoh, K., Nakashima, R., Yaono, R., and Yoshikawa, S. (1995) Science 269 1069–1074 [DOI] [PubMed] [Google Scholar]
- 15.Khalimonchuk, O., Bird, A., and Winge, D. R. (2007) J. Biol. Chem. 282 17442–17449 [DOI] [PubMed] [Google Scholar]
- 16.Nijtmans, L. G. J., Taanman, J. W., Muijsers, A. O., Speijer, D., and Van den Bogert, C. (1998) Eur. J. Biochem. 254 389–394 [DOI] [PubMed] [Google Scholar]
- 17.Smith, D., Gray, J., Mitchell, L., Antholine, W. E., and Hosler, J. P. (2005) J. Biol. Chem. 280 17652–17656 [DOI] [PubMed] [Google Scholar]
- 18.Zhuang, J. Y., Reddi, A. R., Wang, Z. H., Khodaverdian, B., Hegg, E. L., and Gibney, B. R. (2006) Biochemistry 45 12530–12538 [DOI] [PubMed] [Google Scholar]
- 19.Morrison, M. S., Cricco, J. A., and Hegg, E. L. (2005) Biochemistry 44 12554–12563 [DOI] [PubMed] [Google Scholar]
- 20.Brown, B. M., Wang, Z. H., Brown, K. R., Cricco, J. A., and Hegg, E. L. (2004) Biochemistry 43 13541–13548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barros, M. H., and Tzagoloff, A. (2002) FEBS Lett. 516 119–123 [DOI] [PubMed] [Google Scholar]
- 22.Kwast, K. E., Lai, L. C., Menda, N., James, D. T., 3rd, Aref, S., and Burke, P. V. (2002) J. Bacteriol. 184 250–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeRisi, J. L., Iyer, V. R., and Brown, P. O. (1997) Science 278 680–686 [DOI] [PubMed] [Google Scholar]
- 24.Gasch, A. P., Spellman, P. T., Kao, C. M., Carmel-Harel, O., Eisen, M. B., Storz, G., Botstein, D., and Brown, P. O. (2000) Mol. Biol. Cell 11 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Epstein, C. B., Waddle, J. A., Hale, W. t., Dave, V., Thornton, J., Macatee, T. L., Garner, H. R., and Butow, R. A. (2001) Mol. Biol. Cell 12 297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wach, A., Brachat, A., Alberti-Segui, C., Rebischung, C., and Philippsen, P. (1997) Yeast 13 1065–1075 [DOI] [PubMed] [Google Scholar]
- 27.Crisp, R. J., Pollington, A., Galea, C., Jaron, S., Yamaguchi-Iwai, Y., and Kaplan, J. (2003) J. Biol. Chem. 278 45499–45506 [DOI] [PubMed] [Google Scholar]
- 28.Rutherford, J. C., Jaron, S., and Winge, D. R. (2003) J. Biol. Chem. 278 27636–27643 [DOI] [PubMed] [Google Scholar]
- 29.Silverman, S. J., Rose, M., Botstein, D., and Fink, G. R. (1982) Mol. Cell Biol. 2 1212–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Urban-Grimal, D., Volland, C., Garnier, T., Dehoux, P., and Labbe-Bois, R. (1986) Eur. J. Biochem. 156 511–519 [DOI] [PubMed] [Google Scholar]
- 31.Goldstein, J. L., and Brown, M. S. (1990) Nature 343 425–430 [DOI] [PubMed] [Google Scholar]
- 32.Johnson, A., Gin, P., Marbois, B. N., Hsieh, E. J., Wu, M., Barros, M. H., Clarke, C. F., and Tzagoloff, A. (2005) J. Biol. Chem. 280 31397–31404 [DOI] [PubMed] [Google Scholar]
- 33.Lee, S. Y., Raha, S., Nagar, B., and Robinson, B. H. (2001) Arch. Biochem. Biophys. 393 207–214 [DOI] [PubMed] [Google Scholar]
- 34.Barrientos, A., Zambrano, A., and Tzagoloff, A. (2004) EMBO J. 23 3472–3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kennedy, M. A., Barbuch, R., and Bard, M. (1999) Biochim. Biophys. Acta 1445 110–122 [DOI] [PubMed] [Google Scholar]
- 36.Mense, S. M., and Zhang, L. (2006) Cell Res. 16 681–692 [DOI] [PubMed] [Google Scholar]
- 37.McNabb, D. S., and Pinto, I. (2005) Eukaryotic Cell 4 1829–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hach, A., Hon, T., and Zhang, L. (2000) J. Biol. Chem. 275 248–254 [DOI] [PubMed] [Google Scholar]
- 39.Ter Linde, J. J., and Steensma, H. Y. (2002) Yeast 19 825–840 [DOI] [PubMed] [Google Scholar]
- 40.Pfeifer, K., and Guarente, L. (1987) Cell 49 19–27 [DOI] [PubMed] [Google Scholar]
- 41.Svensson, B., and Hederstedt, L. (1994) J. Bacteriol. 176 6663–6671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Futcher, B., Latter, G. I., Monardo, P., McLaughlin, C. S., and Garrels, J. I. (1999) Mol. Cell Biol. 19 7357–7368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gygi, S. P., Rochon, Y., Franza, B. R., and Aebersold, R. (1999) Mol. Cell Biol. 19 1720–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zambrano, A., Fontanesi, F., Solans, A., de Oliveira, R. L., Fox, T. D., Tzagoloff, A., and Barrientos, A. (2007) Mol. Biol. Cell 18 523–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beauvoit, B., Bunoust, O., Guerin, B., and Rigoulet, M. (1999) Eur. J. Biochem. 263 118–127 [DOI] [PubMed] [Google Scholar]
- 46.Butow, R. A., and Avadhani, N. G. (2004) Mol. Cell 14 1–15 [DOI] [PubMed] [Google Scholar]
- 47.Tzagoloff, A., and Dieckmann, C. L. (1990) Microbiol. Rev. 54 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fontanesi, F., Jin, C., Tzagoloff, A., and Barrientos, A. (2008) Hum. Mol. Genet. 17 775–788 [DOI] [PubMed] [Google Scholar]
- 49.Pierrel, F., Bestwick, M. L., Cobine, P. A., Khalimonchuk, O., Cricco, J. A., and Winge, D. R. (2007) EMBO J. 26 4335–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mick, D. U., Wagner, K., van der Laan, M., Frazier, A. E., Perschil, I., Pawlas, M., Meyer, H. E., Warscheid, B., and Rehling, P. (2007) EMBO J. 26 4347–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leary, S. C., Cobine, P. A., Kaufman, B. A., Guercin, G. H., Mattman, A., Palaty, J., Lockitch, G., Winge, D. R., Rustin, P., Horvath, R., and Shoubridge, E. A. (2007) Cell Metab. 5 9–20 [DOI] [PubMed] [Google Scholar]
- 52.Haldi, M. L., and Guarente, L. (1995) Mol. Gen. Genet. 248 229–235 [DOI] [PubMed] [Google Scholar]
- 53.Snyder, C., and Trumpower, B. L. (1998) Biochim. Biophys. Acta 1365 125–134 [DOI] [PubMed] [Google Scholar]
- 54.Barrientos, A., Korr, D., and Tzagoloff, A. (2002) EMBO J. 21 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hon, T., Dodd, A., Dirmeier, R., Gorman, N., Sinclair, P. R., Zhang, L., and Poyton, R. O. (2003) J. Biol. Chem. 278 50771–50780 [DOI] [PubMed] [Google Scholar]