Abstract
The pathogen Staphylococcus aureus uses iron-regulated surface determinant (Isd) proteins to scavenge the essential nutrient iron from host hemoproteins. The IsdH protein (also known as HarA) is a receptor for hemoglobin (Hb), haptoglobin (Hp), and the Hb-Hp complex. It contains three NEAT (NEAr Transporter) domains: IsdHN1, IsdHN2, and IsdHN3. Here we show that they have different functions; IsdHN1 binds Hb and Hp, whereas IsdHN3 captures heme that is released from Hb. The staphylococcal IsdB protein also functions as an Hb receptor. Primary sequence homology to IsdH indicates that it will also employ functionally distinct NEAT domains to bind heme and Hb. We have used site-directed mutagenesis and surface plasmon resonance methods to localize the Hp and Hb binding surface on IsdHN1. High affinity binding to these structurally unrelated proteins requires residues located within a conserved aromatic motif that is positioned at the end of the β-barrel structure. Interestingly, this site is quite malleable, as other NEAT domains use it to bind heme. We also demonstrate that the IsdC NEAT domain can capture heme directly from Hb, suggesting that there are multiple pathways for heme transfer across the cell wall.
Staphylococcus aureus causes a wide range of life threatening diseases such as pneumonia, septicemia, osteomyelitis, toxic shock syndrome, and cardiomyelitis (1). The rise of antibiotic resistance has created a pressing need for new drugs to treat infections caused by this microbe as methicillin-resistant S. aureus (MRSA) now accounts for up to ∼65% of all clinical isolates, and new multidrug resistant strains have emerged that are resistant to vancomycin, the preferred antibiotic of last resort to treat MRSA infections (2). Bacterial growth is limited by the availability of iron, which is an essential cofactor for enzymes involved in microbial metabolism. Although the human body contains large quantities of iron, little is directly available to S. aureus because it is sequestered intracellularly or bound to the carrier glycoproteins transferrin and lactoferrin. To overcome this bacteriostatic limitation, S. aureus has evolved a variety of mechanisms to assimilate iron (3, 4). Of special interest are recently discovered iron-regulated surface determinant (Isd)5 proteins, which extract heme bound iron from host hemoproteins (5-8). The Isd system likely plays a significant role in virulence as heme contains ∼75% of the body's total iron, and it is the S. aureus preferred source of iron (9). As sequence homologs of Isd proteins are also found in a number of other important pathogens (e.g. Listeria monocytogenes, Bacillus anthracis, and Streptococcus pyogenes), a greater understanding of the Isd system could lead to the development of new therapeutically useful anti-infective agents (10-12).
Hemoglobin (Hb) in red blood cells contains large quantities of heme iron to which S. aureus gains access by secreting cytolytic toxins (leukocidin and α- and δ-hemolysin) (13). Release of Hb from lysed erythrocytes significantly dilutes the protein, causing it to dissociate and oxidize into methemoglobin (MetHb), a dimeric form of Hb that contains single α and β globin chains as a heterodimer, each bound to a ferric heme (14, 15). Because heme readily dissociates from MetHb and it can cause severe oxidative damage to host tissues or serve as a microbial nutrient, several host mechanisms exist to remove it from circulation (16). First, heme-laden MetHb present in the blood is tightly bound by the serum glycoprotein haptoglobin (Hp), and the Hp-MetHb complex is cleared from circulation by receptor-mediated endocytosis in the liver parenchymal cells (17, 18). Second, free heme in the blood is eliminated when it binds to hemopexin and serum albumin proteins (15). Recent studies have begun to elucidate how S. aureus circumvents these clearance systems to gain access to heme iron. Free heme present in the blood can be scavenged by the heme transport system, a membrane protein complex that pumps heme into the cytoplasm (9). Heme is also aggressively acquired by capturing it from circulating MetHb and the Hp-MetHb complex using the Isd system (6, 19). Although both the heme transport and Isd systems import heme and are important for virulence, Staphylococcal strains with disrupted Isd components cause less persistent infections in mice than wild-type strains and are unable to utilize Hb as an iron source in cell cultures (20, 21).
The Isd pathway consists of nine proteins that work in concert to acquire heme (5-8). Three receptor proteins are covalently anchored to the cell wall by the SrtA sortase and are displayed on or near the cell surface (IsdA, IsdB, and IsdH (also known as HarA (haptoglobin receptor A))) (6, 19, 22). In addition, a fourth receptor, IsdC, is attached to the cell wall by the SrtB sortase enzyme (21). MetHb is captured by IsdH and IsdB (6, 19, 20). Although these proteins share a high degree of sequence homology with one another, only IsdH is capable of binding Hp (19, 23). In addition, full-length IsdB binds to heme (6), whereas it is unknown if IsdH can also bind to this molecule. The IsdA protein binds heme and may play a general role in bacterial adhesion, as the isolated protein interacts with an array of extracellular matrix proteins (22). After capture by the surface receptors, heme is thought to be transferred to IsdC, which is presumably embedded within the cell wall because it resists proteolytic degradation in whole cell digestion studies (6). The IsdC protein then passes the heme molecule to the membrane transporter IsdDEF complex where it is imported into the cytoplasm. Finally, iron is released when the tetrapyr-role ring is cleaved by the monooxygenase IsdG or its paralog IsdI (24). Recent studies also suggest that heme imported by the Isd system can be directly incorporated in microbial proteins (8).
The Isd receptors IsdA, IsdB, IsdC, and IsdH bind to proteins and heme using NEAT (NEAR iron transporter) domains. These conserved binding modules are ∼125 residues in length and are named for their prevalence in bacterial genes whose genomic positioning is proximal to putative Fe3+ siderophore transporter genes (10). NEAT domains are also found in putative surface proteins in a number of other Gram-positive pathogens where they presumably function in iron import. Studies of isolated NEAT domains from S. aureus have revealed that different domains bind to distinct ligands. For example, some NEAT domains only bind heme (e.g. IsdC) or only to other proteins (e.g. the first and second domains of IsdH), whereas other NEAT domains can bind both of these ligands (e.g. IsdA) (19, 22, 23, 25). NMR and crystallography studies have revealed that NEAT domains adopt a β-sandwich fold that shares structural homology with immunoglobin-like proteins (23, 25-28). In addition, the structures of the IsdC- and IsdA-heme complexes have revealed the mechanism of heme binding (26-28). However, it remains unknown how NEAT domains interact with other proteins.
The IsdH protein captures MetHb and Hp on the cell surface (19). Sequence analysis indicates that it contains three NEAT domains: IsdHN1, IsdHN2, and IsdHN3 (Fig. 1A). Although the IsdHN1 and IsdHN2 domains bind to MetHb and Hp, the function of the C-terminal IsdHN3 domain is unknown (19). In this study we systematically investigated the functions of NEAT domains within IsdH. We show using alanine scanning mutagenesis and surface plasmon resonance (SPR) that Hp and MetHb bind at the same site on IsdHN1. Interestingly, this site is also used by the IsdA and IsdC NEAT domains to bind heme, suggesting that that this surface is particularly well suited for recognizing a range of distinct ligands (26-28). We also show that the previously uncharacterized IsdHN3 domain binds heme and that it can capture this molecule from MetHb. A model of heme transfer is proposed in which domains IsdHN1 and IsdHN2 first bind MetHb or the Hp-MetHb complex on the cell surface. Heme is then captured by the adjacent IsdHN3 domain before subsequent transfer through the cell wall.
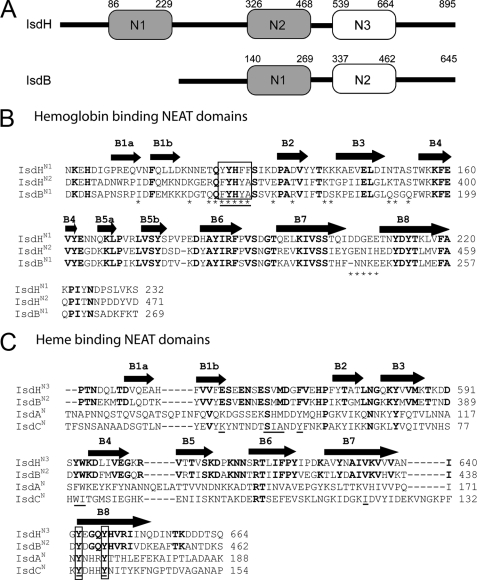
FIGURE 1.
Comparison of the NEAT domains in S. aureus. A, schematic of the IsdB and IsdH proteins showing that they contain two and three NEAT domains, respectively. Domains are shaded based on their relatedness. Domains shaded gray share 46-65% primary sequence identity (IsdHN1, IsdHN2, and IsdBN1). Non-shaded domains share 49% primary sequence identity (IsdHN3 and IsdBN2). Based on studies of the isolated NEAT domains within IsdH, the gray- and non-shaded domains bind Hb and heme, respectively. Both proteins also contain a cell wall sorting signal motif at their C termini that is covalently attached to the cell wall by the SrtA sortase enzyme. B, sequence alignment of related Hb binding NEAT domains within IsdH and IsdB: IsdHN1, IsdHN2, and IsdBN1. These domains are closely related to one another and share 46-65% sequence identity. Residues mutated in this study are indicated with an asterisk, and amino acids are enclosed in a box if their mutation to alanine completely disrupts Hp binding and/or reduces the affinity of IsdHN1 for Hb by at least ×50-fold. The aromatic motif diagnostic of NEAT domains that bind Hb is underlined. The secondary structure of IsdHN1 is shown above the primary sequence. Completely conserved residues are in bold. Only the IsdHN1 and IsdHN2 domains have thus far been shown to bind Hb, whereas the IsdBN1 domain alone has yet to be tested. C, sequence alignment of the IsdHN3 and IsdBN2 NEAT domains, which share 49% sequence identity. The sequences of the distantly related IsdA and IsdC domains are also shown because they bind to heme. Positions within the primary sequence that are within 3.5 Å of the heme in the NMR and crystal structures of the IsdC-heme and IsdA-heme complexes are underlined. The invariant tyrosine residues that coordinate the iron atom of the heme in these structures is enclosed in a box. Residues that are completely conserved in IsdHN3 and IsdBN2 are in bold.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification—The first NEAT domain from IsdH consists of residues Ala86 to Leu229 and is referred to as IsdHN1 (Fig. 1A). Two versions of IsdHN1 were studied. For the heme transfer assay, untagged IsdHN1 was produced from a pet11a vector (Novagen) and purified as previously described (25, 29). For the binding studies using SPR, a histidine-tagged fusion of IsdHN1 was used (his-IsdHN1, residues Ala86 to Leu229 with the sequence Met-Gly-Ser-Ser-His-His-His-His-His-His-Ser-Ser-Gly-Leu-Val-Pro-Arg-Gly-Ser-His-Met at its N terminus). Briefly, the DNA for IsdHN1 was PCR-amplified from S. aureus chromosomal DNA (strain RN4220) using primers that incorporated NdeI and BamHI restriction sites at the 5′ and 3′ ends of the DNA, respectively. This PCR product was then digested with NdeI and BamHI and ligated into a similarly cut pET15b vector. Plasmids encoding single amino acid mutants ofHis-IsdHN1 were constructed from the pET15b vector bearing the IsdHN1 sequence using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's specifications. Escherichia coli BL21(DE3) cells containing the plasmids of either His-tagged wild-type or mutant IsdHN1 were harvested 4 h after the addition of isopropyl β-d-1-thiogalactopyranoside (Sigma) by centrifugation for 15 min at 5000 rpm at 4 °C in a JA-10 rotor, resuspended in lysis buffer (50 mm Tris-HCl, pH 7, 2.5 mm benzamidine (Acros), 1 mm phenylmethylsulfonyl fluoride (Sigma-Aldrich), protease inhibitor mixture II (Roche Applied Science)), and lysed by 3 cycles of sonication. The lysed cells were centrifuged at 4 °C for 1 h at 12,000 rpm in a SS-34 rotor. The supernatant was then applied to Talon nickel affinity beads (Clontech), and the His-tagged fusion proteins were purified according to the manufacturer's instructions. All His-IsdHN1 mutants were checked for proper folding using one-dimensional NMR (data not shown).
Two proteins were used as heme receptors in the heme transfer assay, histidine-tagged IsdC (His-IsdC, residues Ser25 to Gly150), and histidine-tagged C-terminal NEAT domain from IsdH (His-IsdHN3, residues Thr540 to Gln664). Both proteins were expressed from pET15b plasmids (Novagen) and contained the amino acid sequence Met-Gly-Ser-His-His-His-His-His-His-Ser-Ser-Gly-Leu-Val-Pro-Arg-Gly-Ser-His-Met at their N termini. Plasmid construction and protein purification procedures were identical to those used forHis-IsdHN1 (described above). The amount of heme associated with His-IsdC and His-IsdHN3 was determined using a pyridine hemochrome assay (30). After purification, ∼2-3% of His-IsdHN3 and His-IsdC was bound to heme.
Enzyme-linked Immunoabsorbent Assay (ELISA)—An ELISA was used to test His-IsdHN3 binding to a variety of proteins. ELISA plate wells (Nunc microwell plates, Fisher Scientific) were coated with a 150-μl solution containing 20 μg/ml lyophilized bait protein. The bait proteins were dissolved in PBS buffer (10 mm phosphate-buffered saline, 100 mm NaCl, pH 6.0) and included fetuin (Sigma), apotransferrin (Sigma), holo-transferrin (Sigma), Hp (Athens Research), or Hb (Sigma). Additionally, commercially available fibronectin precoated plates (Sigma) were used to test fibronectin interactions. After the bait proteins were incubated at room temperature for 1 h on a Nutator, the wells were washed 3 times with PBS buffer and blocked with 100 μl of a bovine serum albumin solution (10 μg/ml) (Sigma) dissolved in PBS buffer for 1 h with nutation. The wells were then washed 3 times with PBS to remove excess blocking solution, and varying amounts of His-IsdHN3 were added to each well (0 to 30 pmol/well). Interactions between His-IsdHN3 and protein ligands were then detected by adding nickel-labeled horseradish peroxidase according to the manufacturer's directions (Express Detector Nickel-HRP, KPL). After incubating for 1 h at 37 °C, the wells were washed with PBS to remove unbound nickel-labeled horseradish peroxidase, and the binding of His-IsdHN3 was quantified by measuring the conversion of the substrate 3,3′,5,5′-tetramethylbenzidine (Sigma-Aldrich) dissolved in citric phosphate buffer, pH 5, to a colored product using an automated ELISA reader (Molecular Devices, SpectraMax M5). As a positive control similar experiments were performed using His-IsdHN1 protein instead of His-IsdHN3.
Metalloprotoporphyrin Affinity Measurements—UV and fluorescence spectroscopy were used to determine the affinity of IsdHN3 for metalloprotoporphyrins. In the UV assay the soret band of His-IsdHN3 at 402 nm was monitored as a function of heme (iron protoporphyrin XI) added. The concentration of His-IsdHN3 was held constant at 1 μm in buffer A (50 mm Tris HCl, pH 7.5, 100 mm NaCl), and hemin was added in small aliquots from a 1 mm stock solution (hemin dissolved in 0.1 m NaOH). After pH adjustment, the UV absorbance was measured and plotted as a function of heme concentration to yield a binding curve. All UV measurements were performed using a PharmaSpec UV-1700 spectrophotometer (Shimadzu). The affinity of IsdHN3 for the heme analog zinc protoporphyrin IX (ZnPPIX) was determined using fluorescence spectroscopy. In this assay the concentration of ZnPPIX was held constant, and its fluorescence was monitored as a function of protein added. Binding reactions contained 150 μl of 7.5 μm ZnPPIX (Sigma-Aldrich) dissolved in binding buffer (50 mm potassium phosphate; 100 mm NaCl, pH 7.5). Aliquots of His-IsdHN3 from a 125 μm stock solution were added to a final concentration of 50 μm. Fluorescence was measured using a Quanta Master Model QM5 spectrofluorimeter (Photon Technology International) at room temperature. The excitation and emission wavelengths were 365 and 585 nm, respectively. Binding data were fit to the equation A = Af + (Aa - Af)(Ka[L]/1+ Ka[L]), where A is the fluorescence emission, Af is the fluorescence emission of free ZnPPIX, Ab is the fluorescence of bound ZnPPIX, L is the concentration of ligand (heme) added, and Ka is the association constant (31). Data were fit using the program SigmaPlot 2000.
SPR Affinity Measurements—Binding of IsdHN1 to immobilized MetHb was measured by SPR. MetHb was immobilized on a CM5 chip using the following procedure. First, a 0.1 mg/ml stock solution of MetHb was prepared by dissolving commercially available MetHb (Sigma) in 50 mm sodium phosphate, pH 6.2, 100 mm NaCl. The stock solution was then diluted to 1 μg/ml in Biacore binding buffer (10 mm sodium acetate, pH 5.5) and immobilized on a CM5 chip using N-ethyl-N-(dimethylaminopropyl)carboiimide/N-hydroxysuccinimide according to the manufacturers instructions. After MetHb immobilization, the final measured response units (RU) of the CM5 chip was ∼1000. Measurement of IsdHN1 binding was performed at 10 °C. Wild-type and mutant IsdHN1 was flowed over the MetHb-conjugated surface at a rate of 85 μl/min with a contact time of 150 s and a dissociation time of 300 s. SPR measurements were taken at concentrations of 0, 50, 100, 200, 300, 400, 500, 1000, 2000, and 4000 nm IsdHN1 using buffer A (50 mm sodium phosphate, pH 6.2, 100 mm NaCl) as the flow buffer. A similar procedure was used to measure IsdHN1 binding to Hp. Haptoglobin from pooled human plasma (Athens research) was immobilized on a CM5 chip using the aforementioned methods. This protein consists of two alleles of haptoglobin, α-1 and α-2. These proteins differ in their oligomerization state as a result of differences in their amino acid sequence. A 100 μg/ml solution of Hp dissolved in Biacore binding buffer was immobilized so as to yield a haptoglobin conjugated chip with a RU of ∼1000. SPR experiments were performed on a Biacore T100 instrument, and the data were analyzed using the program BIAevaluation 4.01 (Amersham Biosciences).
Heme Transfer Assay—A total of 0.3 μmol of either His-IsdHN3 or His-IsdC was incubated with 750 μl of Talon beads (Clontech) dissolved in buffer A (50 mm sodium phosphate, pH 6.0, 100 mm NaCl) for 1 h. The beads were then centrifuged, washed, and equilibrated in buffer B (50 mm Tris HCl, pH 7.2, 300 mm NaCl, 0.8% Triton X-100 (Sigma-Aldrich)). UV measurements and SDS-PAGE analysis of the wash and beads confirmed that the protein was bound. The beads were then incubated on a rotary with a 5 μm solution of MetHb dissolved in 10 ml of buffer B. The incubation time was varied from 0 to 300 min. The beads were then centrifuged, and the absorbance of the supernatant at 411 nm was measured. For transfer experiments that included IsdHN1, MetHb was first incubated with 3 mol eq of IsdHN1 for 90 min. The program SigmaPlot 2000 was used to fit the transfer data to the equation A = (Af - A0)(1 - etkobs) + A0, where A0 is the initial absorbance of the MetHb solution prior to incubating with IsdHN3, and Af is the final absorbance of MetHb in the eluate.
Under the conditions we have used kobs is approximately equal to the rate of heme release from the β subunit. This is because at the end of the assay ∼50% of the MetHb dimers contain a single heme molecule, and previous studies have shown that the β chain within MetHb binds heme less tightly that the α chain (32). Under these conditions the heme transfer reaction is, therefore, described by the following equilibrium equation,
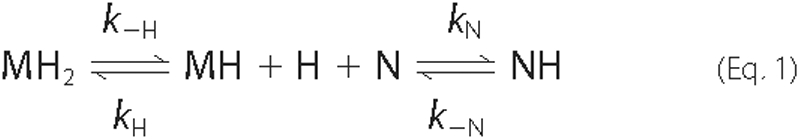
where MH2 is MetHb containing two heme molecules, MH is MetHb with a single heme molecule bound to its α chain, N is the IsdHN3 NEAT domain, H is free heme, and NH is IsdHN3 bound to a single heme molecule. As described by Hargrove et al. (32), a simple expression for the rate of change of MH2 can be derived by assuming that d[H]/dt ∼ 0 and that the concentration of free heme is much smaller than the concentration of heme bound to protein (i.e. [H] [tlt] [MH2] + [MH] + [NH] ∼ [H]total). When these conditions are met the observed rate constant shows the following dependence.
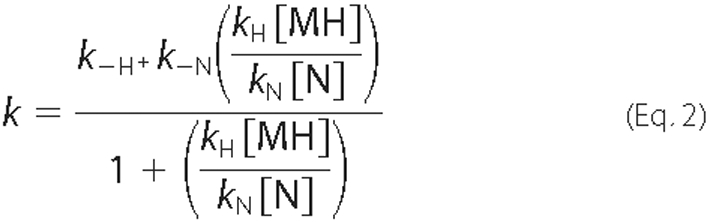
This equation indicates that the initial kinetics of transfer are described by a single exponential where the observed rate constant is equal to the rate at which heme is released from the β subunit of MetHb (kobs ∼ k-H) because [MH]/[N] ∼ 0. In principle, more complex binding behavior will be observed in which the data is biphasic. For example, as the ratio of [MH]/[N] increased, the reaction could accelerate if k-N > k-H, decelerate if k-N < k-H, or remain the same if k-N = k-H. However, these effects are expected to be small and difficult to detect experimentally.
RESULTS
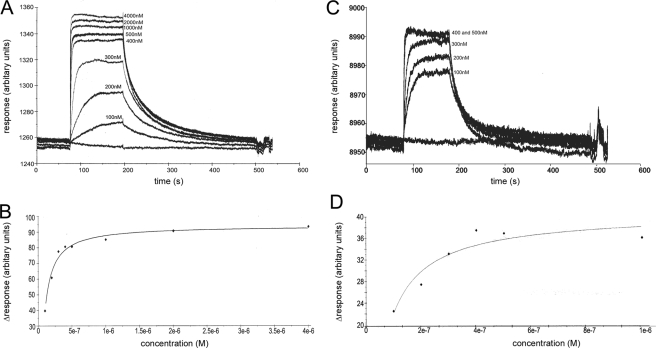
The Same Site on IsdHN1 Binds to MetHb and the Structurally Unrelated Hp Protein—Previous studies have shown that the IsdHN1 and IsdHN2 domains within IsdH bind MetHb and Hp (19, 23, 25). Despite the determination of the three-dimensional structure of IsdHN1, the surface that mediates binding to Hp and MetHb remains unknown because samples of the IsdHN1-Hb and IsdHN1-Hp complexes have thus far proved unsatisfactory for chemical shift mapping by NMR (23, 25). Therefore, we used alanine scanning mutagenesis combined with SPR to probe the surface of IsdHN1 that binds MetHb and Hp. A total of 22 single amino acid mutants were constructed that alter surface-exposed side chains in IsdHN1. The majority of amino acid substitutions are located in the loops that connect strands β1b to β2(β1-β2 loop), β3-β4(β3-β4 loop), and β7 to β8(β7-β8 loop) (see Fig. 5A). The rationale behind altering these sites is based on the solution structure of IsdHN1 and sequence alignments of IsdHN1 with other NEAT domains known to bind MetHb (described under “Discussion”). Each single amino acid mutant contained a histidine tag to facilitate purification and was shown to be properly folded by one-dimensional NMR (data not shown). Fig. 2A shows representative SPR data of wild-type IsdHN1 binding to MetHb adhered to a CM5 chip. These data were interpreted quantitatively by plotting the maximal SPR RU obtained at varying concentrations of IsdHN1 (Fig. 2B). Curve-fitting indicates that wild-type histidine IsdHN1 binds MetHb with a KD of 17 ± 10 nm. Importantly, the presence of the histidine tag does not affect protein binding, as similar affinities for MetHb have been measured for glutathione S-transferase-tagged (23) and untagged variants of IsdHN1 (data not shown). Binding affinities for the mutants were obtained in a similar manner and are listed in Table 1. The most significant changes in affinity occur when residues within the β1-β2 loop are altered, as mutation of 5 of the amino acids in this loop result in a greater than 40-fold reduction in affinity. Alanine substitutions in adjacent loops also cause affinity reductions, albeit to a lesser extent. An interpretation of this data is presented under “Discussion.”
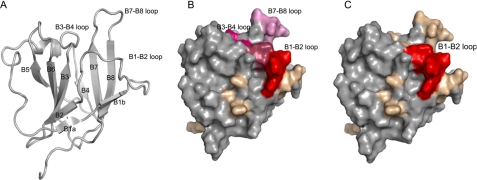
FIGURE 5.
A common surface on the IsdHN1 NEAT domain binds haptoglobin and methemoglobin. A, ribbon drawing of the solution structure of IsdHN1. Strands of β sheet are gray and are indicated by arrows. The three loops involved in protein binding are indicated. B, solvent-exposed surface of IsdHN1 color coded to show the effects of amino acid mutations on MetHb binding. Color coding key: red, > 50× reduction or no detectable binding; dark purple, 10-11× reduction; pink, 2-4× reduction; tan, no significant effect on binding (<2× reduction). C, solvent-exposed surface of IsdHN1 color-coded to show the effects of amino acid mutations on MetHb binding. Color coding is as described in panel B. The view of the structure is identical in all of the panels.
FIGURE 2.
SPR measurements of IsdHN1 binding to MetHb and Hp. A, representative SPR data of wild-type IsdHN1 binding to immobilized MetHb. The panel shows an overlay of several experiments in which the concentration of IsdHN1 was varied (0, 100, 200, 300, 400, 500, 1000, 2000, and 4000 nm). Each curve is a plot of the RU as a function of time. B, representative binding curve of the wild-type IsdHN1 for immobilized MetHb. The curve was generated from the data in panel A by plotting the RU value measured 1 s before dissociation for each concentration of IsdHN1. The solid line shows the best fit of this data to obtain the KD of binding. C, representative SPR data of wild-type IsdHN1 binding to immobilized Hp. The results of six independent experiments are superimposed and differ in the concentration of concentration of IsdHN1 (0, 100,200,300,400,500, and 1000 nm IsdHN1). D, representative binding curve of wild-type IsdHN1 for immobilized Hp by plotting the RU value measured 1 s before dissociation for each concentration of IsdHN1.
TABLE 1.
Hemoglobin and haptoglobin affinities of IsdHN1
|
MetHb binding
|
Hp binding
|
|||
|---|---|---|---|---|
| Protein | Kda | -Fold decreaseb | Kda | -Fold decrease |
| nm | nm | |||
| Wild-type IsdHN1c | 17 ± 10 | 35 ± 10 | ||
| B1-B2 loop | ||||
| N120A | 21 ± 2.4 | 1.2 | 46 ± 4.1 | 1.3 |
| T123A | 80 ± 2.4 | 4 | 45 ± 3.7 | 1.3 |
| Q124A | 62 ± 3.3 | 3 | 24 ± 5.3 | 0.7 |
| Y125A | 2287 ± 1.2 | 134 | 1025 ± 0.4 | 21 |
| Y126A | 2607 ± 6.9 | 153 | No binding | |
| H127A | 1230 ± 0.5 | 72 | No binding | |
| F128A | 720 ± 13 | 42 | No binding | |
| F129A | 698 ± 2.6 | 41 | No binding | |
| B3-B4 loop | ||||
| N151A | 24 ± 1.0 | 1.4 | 33 ± 16 | 0.9 |
| S153A | 219 ± 6.4 | 11 | 46 ± 2.5 | 1.3 |
| T154A | 203 ± 16 | 10 | 29 ± 1.7 | 0.8 |
| B7-B8 loop | ||||
| D205A | 53 ± 3.7 | 3 | 53 ± 10 | 1.5 |
| D206A | 55 ± 4.1 | 3 | 57 ± 12 | 1.6 |
| G207A | 32 ± 7.5 | 2 | 20 ± 7 | 0.6 |
| E208A | 70 ± 7.6 | 4 | 24 ± 12 | 0.7 |
| E209A | 55 ± 8.0 | 3 | 33 ± 15 | 0.9 |
| Surrounding mutants | ||||
| V112A | 20 ± 2.4 | 1.2 | 50 ± 1.0 | 1.4 |
| D133A | 24 ± 7.5 | 1.4 | 52 ± 1.5 | 1.5 |
| D136A | 24 ± 1.0 | 1.4 | 37 ± 5.6 | 1.0 |
| K141A | 20 ± 1.2 | 1.2 | 55 ± 14 | 1.6 |
| K142A | 16 ± 6.9 | 1.0 | 47 ± 16 | 1.3 |
Dissociation constants were measured twice using SPR on a Biacore system. The reported errors are the average fits from the average dissociation constant.
The -fold decrease is the affinity of the mutant that is compared to the wild-type protein affinity for its substrate.
The wild-type protein consists of residues 86-229 of the IsdH protein connected at its N terminus to a 19-residue histidine tag. Each mutant is identical to the wild-type protein but contains a single alanine residue substitution at the indicated position.
The surface mutants of IsdHN1 were also tested for their ability to bind to immobilized Hp using SPR. As shown in Fig. 2C, the SPR data acquired for Hp contains more noise than the MetHb data (∼0.5 and ∼3 RU for Hp and MetHb, respectively). The increase in noise is likely caused by heterogeneity in the commercially acquired Hp protein, which consists of distinct allelic forms that differ in their oligomerization state. However, the binding affinity of IsdHN1 for Hp could still be measured quantitatively from plots of the maximal signal intensity of each sensogram as a function of IsdHN1 concentration (Fig. 2D). The results of this work are reported in Table 1 and reveal that the wild-type protein binds Hp with a KD of ∼35 ± 10 nm. Alanine substitution of Tyr-125, His-127, Phe-128, and Phe-129 within the β1-β2 loop completely abrogates binding to Hp, whereas a Y125A mutation causes a 21-fold reduction in affinity. All other substitutions had a negligible effect on Hp binding. These same five residues within the β1-β2 loop are also important for MetHb binding, indicating that Hp and MetHb bind to the same site on IsdHN1.
IsdH Contains a Third NEAT Domain That Selectively Binds Heme—Inspection of the IsdH amino acid sequence suggests that it contains a third NEAT domain at its C terminus (IsdHN3, residues Thr-540 to Gln-664) (Fig. 1A). This putative domain has not been characterized previously and shares only ∼15% sequence identity with the IsdHN1 and IsdHN2 domains. Moreover, it shares limited sequence homology with all other NEAT domains of known function (23 and 15% sequence identity with IsdA and IsdC, respectively, Fig. 1C). To investigate its function, a histidine-tagged variant of IsdHN3 was overexpressed and purified (His-IsdHN3, residues Thr-540 to Gln-664 containing a 21-residue histidine tag at its N terminus). Centrifuged cell pellets overexpressing His-IsdHN3 were colored red, a phenomena characteristic of many hemoproteins (data not shown). Heme binding was further substantiated by the UV-visible spectrum of purified His-IsdHN3, which exhibits a Soret band at 402 nm along with additional Q-band peaks at 511, 548, and 625 nm (Fig. 3A). The amount of heme bound to His-IsdHN3 was quantified using a pyridine hemochrome assay and revealed that after purification from E. coli, ∼2-3% of the protein contains heme (assuming 1:1 binding of heme to protein). NMR spectra of 15N-labeled IsdHN3 indicate that it is folded and that it binds heme but is of insufficient quality for detailed structural studies using this technique.
FIGURE 3.
The IsdHN3 NEAT domain binds heme. A, UV spectrum of purified IsdHN3 that show characteristics of a heme-binding protein. These include a Soret band at 402 nm along with additional Q-band peaks at 511, 548, and 625 nm. Abs, absorbance. B, quantitative measurement of protoporphyrin binding using a fluorescence assay. The panel shows a plot of ZnPPIX fluorescence emission at 585 nm as a function of IsdHN3 added. The solid line shows a best fit of this data which yields a KD of ZnPPIX binding to IsdHN3 of 2.8 ± 0.1 μm. C, an ELISA assay showing that the IsdHN1 NEAT domain binds MetHb, whereas the IsdHN3 does not. IsdHN3 binding to a range of other proteins was also tested, but no interaction could be detected.
The affinity of His-IsdHN3 for metalloprotoporphyrins was measured using two techniques. Initially, binding of His-IsdHN3 to heme (iron-substituted protoporphyrin IX in its ferric form) was measured by monitoring changes in the Soret band of the protein-heme complex after the addition of varying amounts of hemin (data not shown). Analysis of this data yielded an approximate dissociation constant (KD) of ∼1-20 μm; however, a more quantitative analysis could not be performed because heme aggregates at higher concentrations needed to saturate this protein. Therefore, a fluorescence-based assay was used to monitor the binding of His-IsdHN3 to a close analog of hemin that replaces the iron atom with zinc (ZnPPIX). In this assay the concentration of ZnPPIX is held constant at 7.5 μm, and changes in its fluorescence spectrum are monitored after adding varying amounts of His-IsdHN3. As shown in Fig. 3B, the addition of protein caused a concentration-dependent increase in the fluorescence of ZnPPIX that saturates when 10 μm His-IsdHN3 is present. A plot of the fluorescence change as a function of the protein:ligand ratio revealed a binding stoichiometry of 1:1 (data not shown), whereas curvefitting of the data in Fig. 3B yielded a KD of 2.8 ± 0.1 μm. Similar experiments performed using an untagged variant of IsdHN3 yielded a KD of 2.3 ± 0.4 μm, indicating that the presence of the histidine tag does not affect metalloprotoporphyrin binding.
We also performed an ELISA to determine whether IsdHN3 was capable of binding to proteins previously demonstrated to interact with other NEAT domains. These included holo-transferrin, apotransferrin, fetuin, fibronectin, fibrinogen, Hp, and MetHb (19, 22). When probed by ELISA, IsdHN3 did not interact with MetHb (Fig. 3C, closed squares) nor did it bind to any of the other proteins tested (data not shown). This is in marked contrast to IsdHN1, which as expected binds MetHb when tested by ELISA (Fig. 3C, closed diamonds). Thus far we have only shown that IsdHN3 binds heme.
IsdHN3 and IsdC Capture Heme from MetHb—Because the IsdHN1 and IsdHN2 NEAT domains within IsdH binds to MetHb, we wondered whether the IsdHN3 domain could capture heme from MetHb. To measure heme transfer, His-IsdHN3 was adhered to a cobalt resin (Talon beads, Clontech) and then incubated with MetHb (6:1 molar ratio of His-IsdHN3 to MetHb). The two proteins were then separated by washing the resin to elute MetHb. Maximum absorbance of the eluate at 411 nm is diagnostic for the presence of MetHb bound to heme, as free heme does not absorb maximally at this wavelength, and IsdHN3 and the IsdHN3-heme complex should remain bound to the beads. When a solution of heme-saturated MetHb is incubated with only the cobalt resin, ∼96% of the heme bound MetHb is recovered in the eluate (Fig. 4A). In contrast, incubation of MetHb with apoIsdHN3 for 60 min significantly reduces the amount of MetHb-bound heme that is obtained in the eluate when the resin is washed (Fig. 4A). Two control experiments verify that heme loss from MetHb is a result of transfer to IsdHN3. First, SDS-PAGE analysis of the eluate indicates that similar levels of MetHb are recovered in either the presence or absence of IsdHN3 on the cobalt resin (data not shown). This indicates that the proteins do not interact with each another, consistent with the ELISA data (Fig. 3C). Second, when His-IsdHN3 is eluted from the resin with imidazole after incubation with MetHb, the UV spectrum of the eluate indicates that IsdHN3 has acquired heme (data not shown). Taken together these results indicate that the IsdHN3 domain is capable of acquiring heme from MetHb and that heme transfer to IsdHN3 does not require stable protein-protein interactions.
FIGURE 4.
Transfer of heme from MetHb to IsdHN3 and IsdC. A, bar graph showing the percentage of heme recovered after MetHb is incubation with talon beads for 120 min. The negative control indicates that the talon beads were naked. IsdHN3 and IsdC indicate that the talon beads used in this experiment were preincubated with the His-tagged versions of these proteins. B, quantitative measurement of the rate of heme transfer from MetHb to IsdHN3. The open circles represent heme transfer from MetHb to IsdHN3 as a function of time. The closed circles indicate transfer from MetHb when it is bound to IsdHN1 in the IsdHN1-MetHb complex. The solid line is the best fit of these data and was used to extract the rate of transfer (described under Heme Transfer Assay (see “Experimental Procedures”)). C, quantitative measurement of heme transfer from MetHb to IsdC. Results are as described in panel B, except that His-IsdC protein was used as the receptor for heme. The open and closed squares represent heme transfer from MetHb and the IsdHN1-MetHb complex, respectively.
The rate of heme transfer from MetHb to IsdHN3 was determined by varying the amount of time the proteins were incubated with one another before separation (Fig. 4B, open circles). The transfer data can be adequately fit to the single exponential equation A = (Af - A0)(1 - etkobs) + A0, where A0 is the initial absorbance of the MetHb solution before incubating with IsdHN3, and Af is the final absorbance of MetHb in the eluate. This yields a rate constant (kobs), which under the experimental conditions used describes the rate at which a single heme molecule is released from the β subunit of the MetHb heterodimer (detailed under “Experimental Procedures”). The value of kobs is 4.4 ± 0.33 h-1, which is similar to the previously reported rate of spontaneous heme release from the β chain of MetHb (7.8 ± 2.0 h-1 at pH 7.0, 37 °C) (32). We next investigated whether IsdHN1 binding to MetHb accelerates the rate at which heme is transferred to IsdHN3. The heme transfer assay was repeated where MetHb was first preincubated with a 3-fold molar excess of IsdHN1. At this ratio we have previously shown that a IsdHN1-MetHb complex forms that has a stoichiometry of 2:1 (25). Thus, in this experiment the rate of transfer from the IsdHN1-MetHb complex to His-IsdHN3 is measured. As shown in Fig. 4B, closed circles), IsdHN1 binding to MetHb increases the rate of hemin transfer to His-IsdHN3 by ∼2-fold (kobs = 10.9 ± 0.65 h-1).
To determine whether the IsdC NEAT domain also acquires heme from MetHb, the transfer assay was repeated using His-IsdC as the receptor attached to talon beads instead of His-IsdHN3 (Fig. 4C, open squares). Again, the transfer data can be adequately fit to a single exponential equation with a rate constant of transfer equal to 3.8 ± 0.42 h-1. Intriguingly, the rate of heme capture is similar to that observed for IsdHN3 and is compatible with the rate at which heme is released from the β subunit of MetHb (see above). Similar experiments were performed after preincubating MetHb with IsdHN1. The rate of hemin transfer from the MetHb-IsdHN1 complex to His-IsdC was 10.0 ± 0.84 h-1. Similar to the case for capture by IsdHN3, this rate is ∼2-3-fold faster than from MetHb.
DISCUSSION
To cause disease, S. aureus needs to acquire iron from its human host. Heme is the preferred source of iron and is harvested from hemoglobin using Isd proteins (5-7, 9). One of the first steps in this process is the capture of Hb or the Hb-Hp complex on the cell surface by the IsdH and IsdB proteins (6, 19). These bacterial proteins are closely related to one another yet are structurally distinct from microbial Hb and Hp receptors found in Gram-negative bacteria (23, 25). Thus far, only IsdH has been studied in detail in vitro (19, 23, 25). Analysis of its primary sequence reveals that it contains three NEAT domains: IsdHN1, IsdHN2, and IsdHN3 (Fig. 1A). Previously, the structure of IsdHN1 has been determined, and the isolated IsdHN1 and IsdHN2 domains have been shown to bind Hb and Hp (19, 23, 25). However, the mechanism of Hb or Hp binding is not known because the structure of IsdHN1 was solved in isolation.
Because IsdHN1 and IsdHN2 are closely related to one another, we hypothesized that they would use a common set of surface-exposed side chains to mediate Hb and Hp binding. To identify these residues, we compared the primary sequences of NEAT domains whose ligand specificities had been determined experimentally; that is, IsdHN1, IsdHN2, and the NEAT domains from the IsdA and IsdC proteins, which do not bind Hb with high affinity (22, 25). This comparison revealed the presence of an aromatic motif within IsdHN1 and IsdHN2 (Fig. 1B, boxed residues). A related motif was also found in the N-terminal domain of IsdB (IsdBN1). In IsdHN1, the motif consists of residues 125YYHFF129, which in the NMR structure are located within a loop that connects strands β1b to β2(β1-β2 loop) (Fig. 5A). In conjunction with the surface loops that connect strands β3 to β4 (β3-β4 loop) and strands β7 to β8 (β7-β8 loop), it forms a large surface located at the end of the β-barrel structure. Intriguingly, the backbone atoms of many of the residues within this area of the protein are undetectable in the NMR data of IsdHN1, presumably because they undergo slow time scale motional fluctuations that are characteristic of ligand binding sites (23, 25).
Our results indicate that residues within the aromatic motif and the surrounding surface loops are important for binding MetHb (the oxidized dimeric form of Hb that is prevalent in blood serum after erythrocyte cell lysis) (Fig. 5B). The most important binding surface is formed by the exposed side chains of Tyr-125, Tyr-125, and His-127 within the aromatic motif. As compared with the wild-type protein, which binds MetHb with a KD of ∼17 ± 10 nm, mutation of these residues causes more than a 100-fold decrease in binding affinity (Fig. 5B, in red). However, not all residues in the β1-β2 loop that houses the aromatic motif are important, as amino acids immediately before the motif in the primary sequence can be altered without affecting binding (tan surface near the β1-β2 loop in Fig. 5B, N120A, T123A, and Q124A mutations). In the structure, the β3-β4 and β7-β8 loops are in close proximity to the aromatic motif (Fig. 5A). Surface-exposed residues within these sites also contribute to MetHb binding, although to a lesser extent. The β3-β4 loop appears to be more important as mutations in it reduce binding ∼10-fold (Fig. 5B, dark purple) as compared with 3-4-fold reductions when residues within the β7-β8 loop are altered (Fig. 5B, pink). Because mutations that alter surface-exposed side chains elsewhere in the protein have little effect on binding, we conclude that the primary interface for MetHb binding is located at the end of the β-barrel structure. IsdHN1 and MetHb form a complex that has a stoichiometry of 2:1 (25). This suggests that IsdHN1 interacts with both the α and β subunits of MetHb. Residues within the aromatic motif may adaptively recognize the distinct binding surfaces of these subunits, as NMR studies suggest that residues in the motif are mobile in the apo state (26-28).
IsdH is also the sole Hp receptor in S. aureus as isdH- strains are unable to bind Hp (19). Because Hp binds MetHb with a KD ∼ 10-15 m, IsdH binding to Hp may enable S. aureus to coat itself with this high affinity MetHb receptor. It is also possible that Hp coating of the cell surface enables the bacterium to disguise itself immunologically to prevent targeting by the host immune response (23). Our results indicate that IsdHN1 binds Hp with a KD of 35 ± 10 nm. This affinity is higher than previously reported values determined by SPR (23). Presumably this difference is caused by the way we performed the binding assay. In our assay, Hp is conjugated to a CM5 chip, and binding is measured by passing different concentrations of IsdHN1 over it. In contrast, previous studies have used the reverse approach; IsdHN1 was adhered to the chip, and Hp was passed over it (23). With this prior approach a biphasic association curve was observed that contained both a fast and slow association phase. The slower phase describes a weaker binding event with a measured KD of ∼5 μm, whereas the faster phase characterizing a stronger interaction was not quantified. Because our sensograms have single phase behavior, they enable us to determine the dissociation constant for the high affinity interaction and reveal that IsdHN1 is capable of binding Hp and MetHb with similar nanomolar affinities (35 and 17 nm, respectively).
Interestingly, residues in the aromatic motif required for Hb binding also play an essential role in Hp recognition (Table 1 and Fig. 5C). The most important residues are Tyr-125, His-127, Phe-128, and Phe-129, which when mutated completely disrupt binding. Surprisingly, amino acid mutations in the surrounding β3-β4 and β7-β8 loops as well as in other regions of IsdHN1 have no detectable effect on Hp binding (Fig. 5C, tan). This suggests that the interface between IsdHN1 and Hp is largely restricted to residues in the aromatic motif, in contrast to the MetHb binding surface, which also includes residues located in the surrounding loops (compare Figs. 5, B and C). Interestingly, the residues in the aromatic motif of IsdHN2 and IsdBN1 are identical (see Fig. 1B, boxed sequence). However, only IsdHN2 binds Hp, whereas the full-length IsdB protein does not (19, 23, 25). This argues against the β1-β2 loop being the sole binding surface for Hp. It is possible that more drastic non-conservative amino acid substitutions are required to identify residues important for Hp that are located outside of the aromatic motif.
Our finding that the same surface on IsdHN1 contacts MetHb and Hp is surprising considering that these proteins are structurally unrelated to one another. Although their names imply that they both adopt a globin fold, only the α and β subunits within the MetHb heterodimer possess a globin structure. In contrast, the structure of Hp is not known; however, based on primary sequence homology, its β-chain may adopt a fold related to the serine proteases (33). Previous modeling studies of Hp have suggested that it contains two hydrophobic surfaces that may enable it to function as a protein chaperone (34). To further investigate how IsdHN1 might interact with MetHb and Hp, we used the program ClusPro to model the structures of the IsdHN1-MetHb and IsdHN1-Hp complexes (35). The docking calculations made use of the NMR structure of IsdHN1, the crystal structure of carboxyhemoglobin (pdb 1IRD), and a model of the structure of Hp generated using the structure of the human complement C1S protease as a template (pdb 1ELV, which shares 33% sequence identity with residues 91 to 406 of Hp) (36, 37). In both sets of docking calculations, the predominant solution obtained positioned the aromatic motif within IsdHN1 at the protein-protein interface. In addition, this structural element tended to be inserted between the α and β domains of Hb and the protease-like and complement-like modules in Hp (data not shown). However, a molecular level understanding of recognition will require the determination of the three-dimensional structures of the IsdHN1-MetHb and IsdHN1-Hp complexes by either NMR spectroscopy or x-ray crystallography.
Our results indicate that IsdH contains a third, previously uncharacterized heme binding NEAT domain at its C terminus (IsdHN3) (Fig. 3C). Its mode of heme binding can be predicted from the recently determined structures of the metalloprotoporphyrin complexes of the IsdA and IsdC NEAT domains (26-28). Although IsdHN3 shares only 15-23% sequence identity with IsdA and IsdC, residues that form the heme binding pocket in these proteins are generally conserved (residues are underlined in Fig. 1C). Of particular interest is the presence of an invariant tyrosine residue that coordinates the central metal ion of the heme: Tyr166 (IsdA), Tyr132 (IsdC), and Tyr642 (IsdHN3) (26-28). Assuming that IsdHN3 adopts a conventional NEAT domain fold, it seems likely that it will bind heme in a similar manner through an exposed pocket located at the end of a β-barrel structure. The walls of the heme binding pocket would be formed by residues within strands β7 and β8 as well as residues within the loops that connect strands β1b to β2(β1-β2 loop) and strands β3 to β4(β3-β4 loop). Our data indicate that IsdHN3 binds metalloprotoporphyrins with a KD = 2.8 ± 0.1 μm. The IsdC proteins from S. aureus and B. anthracis also bind heme weakly (12, 28). This is compatible with the IsdH and IsdC physiological role in heme transport, because they presumably need to rapidly bind and release heme as it is transferred across the cell wall (12, 28). The weak affinity of these proteins is also consistent with the recently determined structures of IsdA and IsdC, as in them only ∼35% of the solvent-accessible surface area of the protoporphrin ring is contacted by the protein (26-28).
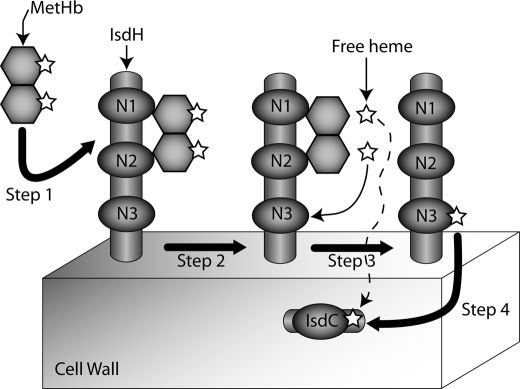
An overview of how the NEAT domains within IsdH might function in heme capture is shown in Fig. 6. Upon erythrocyte lysis large quantities of tetrameric ferrous Hb is released into the blood, which rapidly oxidizes and dissociates into dimeric MetHb (14). The IsdHN1 and IsdHN2 domains bind MetHb with nanomolar affinity, tethering it to the cell surface (step 1). Heme is then released and captured by the adjacent IsdHN3 domain (step 2). This process is passive as the measured rate of transfer from MetHb to IsdHN3 is ∼4.4/h (Fig. 4B), which is similar to the rate at which heme spontaneous dissociates from MetHb (12, 28). Heme bound to IsdHN3 then presumably dissociates where it can be either recaptured by MetHb or transferred to either IsdA or IsdC within the cell wall (step 3). It is also conceivable that heme is directly transferred from IsdHN3 to IsdA and/or IsdC through activated protein-protein complexes, as this mechanism of transfer has been shown to occur between the IsdA and IsdC proteins (38). In addition, our results indicate that IsdC can also directly capture heme that is released from MetHb (Fig. 4C); however, this process would seem to occur less frequently as IsdC is presumably positioned more distal to MetHb than IsdHN3. This model of heme transfer is compatible with recent studies by the Lei and co-workers (39), which have shown that the IsdA and IsdC proteins can acquire heme that is released from MetHb.
FIGURE 6.
Schematic showing how the IsdH protein may capture and transfer heme from hemoglobin. Step 1, MetHb is tethered to IsdH via the N-terminal IsdHN1 and IsdHN2 NEAT domains. MetHb is shown as two hexagons representing the α and β globin chains. Heme molecules are represented by five-pointed stars. Step 2, heme is released from MetHb. Our data indicates that this process is passive when NEAT domains are in isolation. Step 3, heme diffuses and is captured by the C-terminal IsdHN3 NEAT domain. Step 4, heme is released from IsdHN3 and subsequently captured by IsdC located within the cell wall. Our results also indicate that IsdC can directly capture heme that is passively released from MetHb (dashed line). Note that IsdH and IsdC are covalently attached to the cell wall by the SrtA and SrtB sortases, respectively.
The IsdB protein also functions as a MetHb and heme receptor in S. aureus (19, 23, 25). As shown in Fig. 1A, it contains two NEAT domains, IsdBN1 and IsdBN2. The IsdBN1 domain is most closely related to the IsdHN1 and IsdHN2 domains. In contrast, the C-terminal IsdBN2 domain is most closely related to IsdHN3, and it contains conserved residues that presumably enable it to bind heme (Fig. 1B). This suggests IsdB and IsdH capture heme from MetHb through a similar mechanism; MetHb is bound by IsdBN1, and released heme is scavenged by the IsdBN2 domain. Interestingly, Lei and co-workers have shown that heme transfer from MetHb to the full-length IsdB protein occurs at rates that are much faster than the rate of spontaneous heme release from MetHb (38, 39). This suggests that MetHb binding to the full-length IsdB protein stimulates heme release. Our finding that binding of the isolated IsdHN1 domain to MetHb only modestly increases the rate of heme transfer to IsdHN3 may explain why IsdB has been proposed to play a more important role than IsdH in the in vivo capture of heme from hemoglobin (19, 23, 25). Alternatively, it suggests that the NEAT domains within IsdH and IsdB need to be part of the same polypeptide, that is, act in cis to effectively capture heme. Unfortunately, we have not been able to test the degree of synergy between the NEAT domains within IsdH because the full-length protein degrades during purification.
In conclusion, we have shown that IsdH captures heme from MetHb using three NEAT domains that have different ligand binding specificities. The IsdHN1 and IsdHN2 bind MetHb and Hp, whereas a third C-terminal IsdHN3 domain captures released heme. Analogous NEAT domains are present in IsdB based on primary sequence homology, suggesting that it will capture heme from MetHb through a similar mechanism. We have also shown that IsdHN1 employs a common surface located at the end of its β structure to interact with the structurally distinct Hp and MetHb proteins. Interestingly, structural studies of the IsdA and IsdC proteins indicate that they use the same site bind heme. This suggests that this part of the protein scaffold is acquiescent to change that ultimately determines the function of the NEAT domain.
This work was supported, in whole or in part, by National Institutes of Health Grant AI52217. This work was also supported by Department of Energy Grant DE-FC-03-87ER60615. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: Isd, iron-regulated surface determinant; Hb, hemoglobin; MetHb, methemoglobin; Hp, haptoglobin; NEAT, NEAR iron transporter; SPR, surface plasmon resonance; PBS, phosphate-buffered saline; ZnPPIX, zinc protoporphyrin IX; RU, response units; ELISA, enzyme-linked immunoabsorbent assay.
References
- 1.Lowy, F. D. (1998) N. Engl. J. Med. 339 520-532 [DOI] [PubMed] [Google Scholar]
- 2.Kluytmans-Vandenbergh, M. F., and Kluytmans, J. A. (2006) Clin. Microbiol. Infect. 12 Suppl. 1, 9-15 [DOI] [PubMed] [Google Scholar]
- 3.Andrews, S. C., Robinson, A. K., and Rodriguez-Quinones, F. (2003) FEMS Microbiol. Rev. 27 215-237 [DOI] [PubMed] [Google Scholar]
- 4.Brown, J. S., and Holden, D. W. (2002) Microbes Infect. 4 1149-1156 [DOI] [PubMed] [Google Scholar]
- 5.Maresso, A. W., and Schneewind, O. (2006) Biometals 19 193-203 [DOI] [PubMed] [Google Scholar]
- 6.Mazmanian, S. K., Skaar, E. P., Gaspar, A. H., Humayun, M., Gornicki, P., Jelenska, J., Joachmiak, A., Missiakas, D. M., and Schneewind, O. (2003) Science 299 906-909 [DOI] [PubMed] [Google Scholar]
- 7.Skaar, E. P., and Schneewind, O. (2004) Microbes Infect 6 390-397 [DOI] [PubMed] [Google Scholar]
- 8.Reniere, M. L., Torres, V. J., and Skaar, E. P. (2007) Biometals 20 333-345 [DOI] [PubMed] [Google Scholar]
- 9.Skaar, E. P., Humayun, M., Bae, T., DeBord, K. L., and Schneewind, O. (2004) Science 305 1626-1628 [DOI] [PubMed] [Google Scholar]
- 10.Andrade, M. A., Ciccarelli, F. D., Perez-Iratxeta, C., and Bork, P. (2002) Genome Biology 3(9), RESEARCH0047 [DOI] [PMC free article] [PubMed]
- 11.Skaar, E. P., Gaspar, A. H., and Schneewind, O. (2006) J. Bacteriol. 188 1071-1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maresso, A. W., Chapa, T. J., and Schneewind, O. (2006) J. Bacteriol. 188 8145-8152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiseman, G. M. (1975) Bacteriol. Rev. 39 317-344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Umbreit, J. (2007) Am. J. Hematol. 82 134-144 [DOI] [PubMed] [Google Scholar]
- 15.Ascenzi, P., Bocedi, A., Visca, P., Altruda, F., Tolosano, E., Beringhelli, T., and Fasano, M. (2005) IUBMB Life 57 749-759 [DOI] [PubMed] [Google Scholar]
- 16.Wandersman, C., and Delepelaire, P. (2004) Annu. Rev. Microbiol. 58 611-647 [DOI] [PubMed] [Google Scholar]
- 17.Kristiansen, M., Graversen, J. H., Jacobsen, C., Sonne, O., Hoffman, H. J., Law, S. K., and Moestrup, S. K. (2001) Nature 409 198-201 [DOI] [PubMed] [Google Scholar]
- 18.Okuda, M., Tokunaga, R., and Taketani, S. (1992) Biochim. Biophys. Acta 1136 143-149 [DOI] [PubMed] [Google Scholar]
- 19.Dryla, A., Gelbmann, D., von Gabain, A., and Nagy, E. (2003) Mol. Microbiol. 49 37-53 [DOI] [PubMed] [Google Scholar]
- 20.Torres, V. J., Pishchany, G., Humayun, M., Schneewind, O., and Skaar, E. P. (2006) J. Bacteriol. 188 8421-8429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mazmanian, S. K., Ton-That, H., Su, K., and Schneewind, O. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 2293-2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clarke, S. R., Wiltshire, M. D., and Foster, S. J. (2004) Mol. Microbiol. 51 1509-1519 [DOI] [PubMed] [Google Scholar]
- 23.Dryla, A., Hoffmann, B., Gelbmann, D., Giefing, C., Hanner, M., Meinke, A., Anderson, A. S., Koppensteiner, W., Konrat, R., von Gabain, A., and Nagy, E. (2007) J. Bacteriol. 189 254-264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skaar, E. P., Gaspar, A. H., and Schneewind, O. (2004) J. Biol. Chem. 279 436-443 [DOI] [PubMed] [Google Scholar]
- 25.Pilpa, R. M., Fadeev, E. A., Villareal, V. A., Wong, M. L., Phillips, M., and Clubb, R. T. (2006) J. Mol. Biol. 360 435-447 [DOI] [PubMed] [Google Scholar]
- 26.Grigg, J. C., Vermeiren, C. L., Heinrichs, D. E., and Murphy, M. E. (2007) Mol. Microbiol. 63 139-149 [DOI] [PubMed] [Google Scholar]
- 27.Sharp, K. H., Schneider, S., Cockayne, A., and Paoli, M. (2007) J. Biol. Chem. 282 10625-10631 [DOI] [PubMed] [Google Scholar]
- 28.Villareal, V. A., Pilpa, R. M., Robson, S. A., Fadeev, E. A., and Clubb, R. T. (2008) J. Biol. Chem. 283 31591-31600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pilpa, R. M., and Clubb, R. T. (2005) J. Biomol. NMR 33 137. [DOI] [PubMed] [Google Scholar]
- 30.Berry, E. A., and Trumpower, B. L. (1987) Anal. Biochem. 161 1-15 [DOI] [PubMed] [Google Scholar]
- 31.Lundblad, J. R., Laurance, M., and Goodman, R. H. (1996) Mol. Endocrinol. 10 607-612 [DOI] [PubMed] [Google Scholar]
- 32.Hargrove, M. S., Singleton, E. W., Quillin, M. L., Ortiz, L. A., Phillips, G. N., Jr., Olson, J. S., and Mathews, A. J. (1994) J. Biol. Chem. 269 4207-4214 [DOI] [PubMed] [Google Scholar]
- 33.Kurosky, A., Barnett, D. R., Lee, T. H., Touchstone, B., Hay, R. E., Arnott, M. S., Bowman, B. H., and Fitch, W. M. (1980) Proc. Natl. Acad. Sci. U. S. A. 77 3388-3392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ettrich, R., Brandt, W., Jr., Kopecky, V., Baumruk, V., Hofbauerova, K., and Pavlicek, Z. (2002) Biol. Chem. 383 1667-1676 [DOI] [PubMed] [Google Scholar]
- 35.Comeau, S. R., Gatchell, D. W., Vajda, S., and Camacho, C. J. (2004) Bioinformatics 20 45-50 [DOI] [PubMed] [Google Scholar]
- 36.Gaboriaud, C., Rossi, V., Bally, I., Arlaud, G. J., and Fontecilla-Camps, J. C. (2000) EMBO J. 19 1755-1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arnold, K., Bordoli, L., Kopp, J., and Schwede, T. (2006) Bioinformatics 22 195-201 [DOI] [PubMed] [Google Scholar]
- 38.Liu, M., Tanaka, W. N., Zhu, H., Xie, G., Dooley, D. M., and Lei, B. (2008) J. Biol. Chem. 283 6668-6676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu, H., Xie, G., Liu, M., Olson, J. S., Fabian, M., Dooley, D. M., and Lei, B. (2008) J. Biol. Chem. 283 18450-18460 [DOI] [PMC free article] [PubMed] [Google Scholar]