Abstract
Mutations in SHP-2 phosphatase (PTPN11) that cause hyperactivation of its catalytic activity have been identified in Noonan syndrome and various childhood leukemias. Recent studies suggest that the gain-of-function (GOF) mutations of SHP-2 play a causal role in the pathogenesis of these diseases. However, the molecular mechanisms by which GOF mutations of SHP-2 induce these phenotypes are not fully understood. Here, we show that GOF mutations in SHP-2, such as E76K and D61G, drastically increase spreading and migration of various cell types, including hematopoietic cells, endothelial cells, and fibroblasts. More importantly, in vivo angiogenesis in SHP-2 D61G knock-in mice is also enhanced. Mechanistic studies suggest that the increased cell migration is attributed to the enhanced β1 integrin outside-in signaling. In response to β1 integrin cross-linking or fibronectin stimulation, activation of ERK and Akt kinases is greatly increased by SHP-2 GOF mutations. Also, integrin-induced activation of RhoA and Rac1 GTPases is elevated. Interestingly, mutant cells with the SHP-2 GOF mutation (D61G) are more sensitive than wild-type cells to the suppression of cell motility by inhibition of these pathways. Collectively, these studies reaffirm the positive role of SHP-2 phosphatase in cell motility and suggest a new mechanism by which SHP-2 GOF mutations contribute to diseases.
SHP-2, a multifunctional SH2 domain-containing protein-tyrosine phosphatase implicated in diverse cell signaling processes (1–3), plays a critical role in cellular function. Homozygous deletion of Exon 2 (4) or Exon 3 (5) of the SHP-2 gene (PTPN11) in mice leads to early embryonic lethality prior to and at midgestation, respectively. SHP-2 null mutant mice die much earlier, at peri-implantation (4). Exon 3 deletion mutation of SHP-2 blocks hematopoietic potential of embryonic stem cells both in vitro and in vivo (6–8), whereas SHP-2 null mutation causes inner cell mass death and diminished trophoblast stem cell survival (4). Recent studies on SHP-2 conditional knock-out or tissue-specific knock-out mice have further revealed an array of important functions of this phosphatase in various physiological processes (9–12). The phenotypes demonstrated by loss of SHP-2 function are apparently attributed to the role of SHP-2 in the cell signaling pathways induced by growth factors/cytokines. SHP-2 generally promotes signal transmission in growth factor/cytokine signaling in both catalytic-dependent and -independent fashion (1–3). The positive role of SHP-2 in the intracellular signaling processes, in particular, the ERK3 and PI3K/Akt kinase pathways, has been well established, although the underlying mechanism remains elusive, in particular, the signaling function of the catalytic activity of SHP-2 in these pathways is poorly understood.
In addition to the role of SHP-2 in cell proliferation and differentiation, the phenotypes induced by loss of SHP-2 function may be associated with its role in cell migration. Indeed, dominant negative SHP-2 disrupts Xenopus gastrulation, causing tail truncations (13, 14). Targeted Exon 3 deletion mutation in SHP-2 results in decreased cell spreading, migration (15, 16), and impaired limb development in the chimeric mice (7). The role of SHP-2 in cell adhesion and migration has also been demonstrated by catalytically inactive mutant SHP-2-overexpressing cells (17–20). The molecular mechanisms by which SHP-2 regulates these cellular processes, however, have not been well defined. For example, the role of SHP-2 in the activation of the Rho family small GTPases that is critical for cell motility is still controversial. Both positive (19, 21, 22) and negative roles (18, 23) for SHP-2 in this context have been reported. Part of the reason for this discrepancy might be due to the difference in the cell models used. Catalytically inactive mutant SHP-2 was often used to determine the role of SHP-2 in cell signaling. In the catalytically inactive mutant SHP-2-overexpressing cells, the catalytic activity of endogenous SHP-2 is inhibited. However, as SHP-2 also functions independent of its catalytic activity, overexpression of catalytically deficient SHP-2 may also increase its scaffolding function, generating complex effects.
The critical role of SHP-2 in cellular function is further underscored by the identification of SHP-2 mutations in human diseases. Genetic lesions in PTPN11 that cause hyperactivation of SHP-2 catalytic activity have been identified in the developmental disorder Noonan syndrome (24) and various childhood leukemias, including juvenile myelomonocytic leukemia (JMML), B cell acute lymphoblastic leukemia, and acute myeloid leukemia (25, 26). In addition, activating mutations in SHP-2 have been identified in sporadic solid tumors (27). The SHP-2 mutations appear to play a causal role in the development of these diseases as SHP-2 mutations and other JMML-associated Ras or Neurofibromatosis 1 mutations are mutually exclusive in the patients (24–27). Moreover, single SHP-2 gain-of-function (GOF) mutations are sufficient to induce Noonan syndrome, cytokine hypersensitivity in hematopoietic progenitor cells, and JMML-like myeloproliferative disease in mice (28–32). Gain-of-function cell models derived from the newly available SHP-2 GOF mutation (D61G) knock-in mice (28) now provide us with a good opportunity to clarify the role of SHP-2 in cell motility. Unlike the dominant negative approach in which overexpression of mutant forms of SHP-2 generates complex effects, the SHP-2 D61G knock-in model eliminates this possibility as the mutant SHP-2 is expressed at the physiological level (28). Additionally, defining signaling functions of GOF mutant SHP-2 in cell movement can also help elucidate the molecular mechanisms by which SHP-2 mutations contribute to the relevant diseases.
EXPERIMENTAL PROCEDURES
Mice, Cell Lines, and Reagents—SHP-2D61G/+ mice (28) originally imported from Beth Israel Deaconess Medical Center were used to backcross with pure C57BL/6J mice at the Animal Resources Center, Case Western Reserve University. F4 mice were used in this study. All animal procedures complied with the NIH Guideline for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee. Ba/F3, an IL-3-dependent murine pro-B lymphoma cell line, was maintained in RPMI 1640 medium with 10% fetal bovine serum (FBS) and 10% conditioned medium produced by murine IL-3 cDNA-transfected X630 hematopoietic cells. Anti-SHP-2, anti-ERK, and anti-phospho-ERK antibodies (Abs) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-Gab2 and anti-phosphotyrosine (Tyr(P)) Abs were provided by Upstate Biotechnology Inc. (Lake Placid, NY). Anti-phospho-Akt and anti-Akt Abs were obtained from Cell Signaling Technology (Beverly, MA). Human fibronectin was supplied by Sigma. The MTS cell proliferation assay (a colorimetric method for determining numbers of viable cells) kit was obtained from Promega Life Science (Madison, WI). The small GTPase assay kit was purchased from Cytoskeleton Inc. (Denver, CO). Matrigel, basic fibroblast growth factor, and vascular endothelial growth factor were provided by BD Biosciences (Bedford, MA). Rabbit anti-Von Willebrand factor (vWF) antibody was from ABR-Affinity Bio-Reagents (Golden, CO).
Generation of Bone Marrow-derived Mast Cells and Macrophages—Bone marrow cells freshly isolated from 10- to 12-week-old SHP-2D61G/+ mice and wild-type (WT) littermates were cultured in RPMI 1640 medium supplemented with 10% FBS and mouse recombinant IL-3 (10 ng/ml) for 4 weeks. Mast cell phenotype was confirmed by flow cytometry analysis with Abs specific for c-kit and FcγRII/RIII. At the time of use, greater than 98% of the cultured cells were mast cells. To generate bone marrow-derived macrophages, bone marrow cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS and 20% L-cell conditioned medium (as a source of mouse colony-stimulating factor 1). After 24 and 48 h, non-adherent cells were collected and seeded into new tissue culture plates. Following 5 to 7 days of culture, cells were confirmed as macrophages as more than 90% of semi-adherent cells were positive for Mac-1 and F4/80.
Adhesion/Spreading and Migration Assay—For the adhesion/spreading assay, 24-well plates were coated with 10 μg/ml fibronectin at 4 °C overnight and washed twice with phosphate-buffered saline (PBS). Plates were then incubated with 1% bovine serum albumin (BSA) in PBS at 37 °C for 1 h to block nonspecific binding. Wells coated with 1% BSA/PBS alone were used as controls. Test cells (5 × 105) suspended in 200 μl of RPMI 1640 medium with 2% BSA were seeded into each well and centrifuged at 600 × g for 1 min to allow attachment of cells to the bottom of the wells. After 30 min of incubation at 37 °C in a5%CO2 incubator, unattached cells were removed by washing twice with pre-warmed RPMI 1640 medium containing 2% BSA. Adherent cells were quantified using the MTS assay. Migration assays were performed using transwells (8 μm pore size, 6.5 mm diameter; Corning Costa, Cambridge, MA) pre-coated on both sides of the transwell membranes with fibronectin (10 μg/ml) at 4 °C overnight. The lower chambers contained 600 μl of RPMI 1640 or DMEM with 2% FBS. Test cells (2 × 105 cells suspended in 100 μl of RPMI 1640 or DMEM with 2% BSA) were loaded into the upper chambers. They were allowed to migrate into lower chambers for 7 h in a 37 °C, 5% CO2 incubator. Cells randomly migrating to the lower chambers were collected and counted on a hemacytometer. In the meantime, cells on the upper surface of the transwell membranes were mechanically removed. Migrated cells adhering to the lower side of the membranes were fixed in methanol, stained with Giemsa, and enumerated under a microscope at ×200 magnification. In some experiments, transwell membranes were stained with 0.4% crystal violet in 20% methanol. Migrated cells adhering to the lower side of the membranes were quantified by measuring OD595 of the dye eluted with 10% acetic acid (50 μl).
Immunoprecipitation and Immunoblotting—To induce integrin signaling, exponentially growing Ba/F3 or mast cells were starved in RPMI 1640 medium with 2% BSA for 5 h. Cells were collected and suspended in PBS (1 × 107 cells/ml) and incubated on ice for 15 min with monoclonal Ab against β1 integrin (anti-CD29, Ha/5, Pharmingen) or hamster IgM as a control. Cells were washed once with PBS and then stimulated by cross-linking using anti-hamster IgM monoclonal Ab at 37 °C for 5 and 15 min as reported (33). Cells were lysed in RIPA buffer (50 mm Tris-HCl, pH 7.4, 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 mm NaCl, 1 mm EDTA, 1 mm NaF, 2 mm Na3VO4, 10 μg/ml leupeptin, 10 μg/ml aprotin, and 1 mm phenylmethylsulfonyl fluoride). Whole cell lysates (500 μg) were immunoprecipitated with 1–2 μg of purified Abs. Immunoprecipitates were washed three times with HNTG buffer (20 mm HEPES, pH 7.5, 150 mm NaCl, 1% glycerol, 0.1% Triton X-100, and 1 mm Na3VO4) and resolved by SDS-PAGE followed by immunoblotting with the indicated Abs.
Small GTPase Assay—RhoA/Cdc42/Rac1 GTPase activation was determined by the GST-Rhotekin-RBD (for RhoA) or GST-PAK-CRIB (for Rac1 and Cdc42) pull-down assays. Cells were lysed on ice for 5 min in RIPA buffer containing 0.5% sodium deoxycholate. Supernatants were collected quickly after centrifugation. An equal volume of whole cell lysates (500 μg) were incubated with 60 μg of GST-Rhotekin-RBD or GST-PKA-CRIB glutathione-agarose beads at 4 °C for 3 h. Proteins bound to glutathione-agarose beads were then resolved by SDS-PAGE and immunoblotted with anti-pan-RhoA, anti-pan-Rac1, or anti-pan-Cdc42 Abs. Total cellular RhoA, Rac1, and Cdc42 levels in the whole cell lysates were also determined by immunoblotting.
In Vivo Angiogenesis Assay—In vivo assessment of angiogenesis was performed as previously described (34–36). Matrigel on ice was mixed with heparin (50 μg/ml), basic fibroblast growth factor (800 ng/ml), and vascular endothelial growth factor (300 ng/ml). The Matrigel mixture of 0.5 ml was injected subcutaneously into flank areas (left and right sides) near the bottom and midline of the abdomen of 8–10-week-old SHP-2D61G/+ mice or WT littermates. Animals were sacrificed 9 days after Matrigel injection. The mouse skin was detached along the abdominal midline, and the Matrigel plugs were retrieved. The samples were then fixed in 10% buffered formalin for histology or dispensed in PBS buffer for analysis of hemoglobin levels. For measurement of hemoglobin, each Matrigel plug was dispersed in 1 ml of PBS and homogenized. The resulting lysates were clarified by centrifugation at 13,000 × g for 10 min at 4 °C. The supernatants (50 μl) were mixed (1:5) with Drabkin's solution (Sigma) and incubated at 37 °C for 15 min. The absorbance at 540 nm was measured and the hemoglobin levels were calculated based on the standard curve generated using purified human hemoglobin (Sigma). The results were normalized based on the hemoglobin levels of the mice, as SHP-2D61G/+ mice show slightly decreased hemoglobin in peripheral blood. In addition, Matrigel plugs were processed for cryosections followed by immunostaining with anti-vWF rabbit antiserum at 8 μg/ml. Normal rabbit IgG at the same concentration was used as the negative control. vWF positive endothelial cells/capillary vessels were quantified under a microscope.
RESULTS AND DISCUSSION
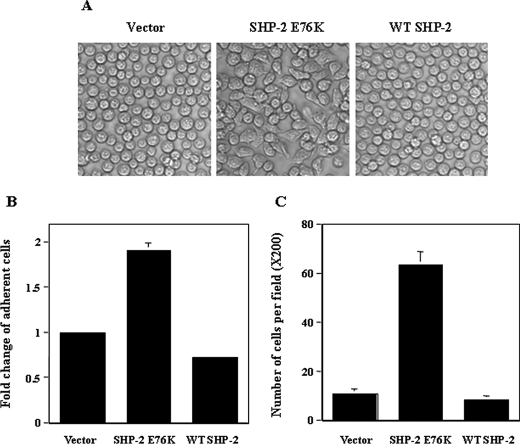
SHP-2 GOF mutations have been identified in Noonan syndrome and childhood leukemias. Recent studies suggest that the SHP-2 GOF mutations play a causal role in the pathogenesis of these diseases. However, biochemical functions of GOF mutant SHP-2 are not fully understood. During our previous studies characterizing the mechanisms by which the SHP-2 GOF mutation E76K impacts IL-3 signaling, we noticed that Ba/F3 cells transduced with SHP-2 E76K displayed enhanced adhesion/spreading to tissue culture dishes (Fig. 1A). By contrast, the adhesion capability of WT SHP-2 overexpressing cells was not altered (Fig. 1, A and B). In addition, the migrating capability of SHP-2 E76K cells was greatly increased compared with the cells overexpressing WT SHP-2 or the cells transduced with the control vector (Fig. 1C). These results indicate that the GOF mutation E76K in SHP-2 impacts cell spreading and movement.
FIGURE 1.
Transduction of SHP-2 E76K but not WT SHP-2 increases Ba/F3 cell adhesion and migration. A, SHP-2 E76K, WT SHP-2, and control vector-transduced Ba/F3 cells were generated in our previous studies (32, 44). Cells cultured in RPMI 1640 medium with 10% FBS and 10% IL-3-conditioned medium were photographed under a microscope. B, cells were assayed in cytokine and serum-free medium for their capabilities to adhere to fibronectin-coated 24-well plates as described under “Experimental Procedures.” The number of adherent cells was determined by the MTS assay. C, cells were assayed for migration using fibronectin-coated transwells as described under “Experimental Procedures.” Seven hours later, cells on the upper surface were mechanically removed. The transwell membranes were fixed in methanol and stained with Giemsa. Migrated cells adhering to the lower side of the membranes were enumerated under a microscope. Two to three independent experiments were performed and similar results were obtained in each. Results shown are the mean ± S.D. of triplicates from one experiment.
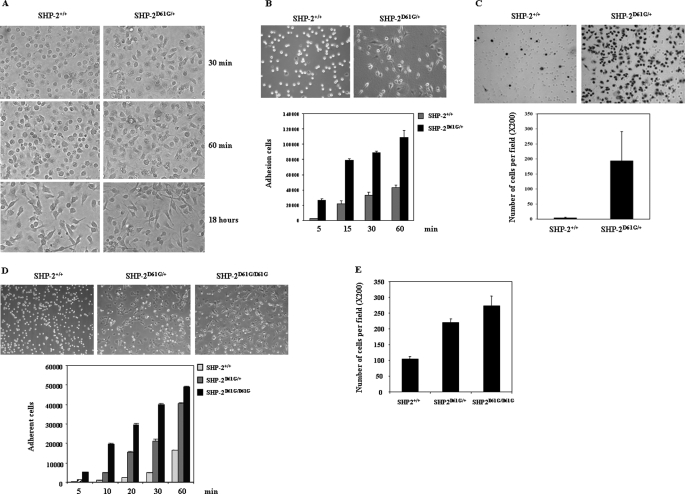
The results described above were obtained from SHP-2 E76K overexpressing cells. To exclude the possibility that the elevated SHP-2 protein level might affect the analyses and to further test whether other GOF mutations in SHP-2 can also enhance cell spreading and migration, we examined hematopoietic cells from SHP-2 D61G knock-in mice in which expression of mutant SHP-2 is at the physiological level (28). Macrophages derived from SHP-2D61G/+ mice (SHP-2D61G/D61G mice are lethal at around embryonic day 13.5) adhered and spread much faster than WT macrophages when replated into non-tissue culture plates (Fig. 2A). Likewise, mast cells derived from SHP-2D61G/+ mice showed markedly increased adhesion/spreading in tissue culture plates (Fig. 2B). We also assessed the migrating capability of SHP-2D61G/+ cells. Compared with WT mast cells, SHP-2D61G/+ cells demonstrated drastically enhanced migration across transwell membranes. The numbers of cells migrating and attaching to the lower side of the membranes (Fig. 2C) as well as in the medium in the lower chambers (data not shown) were substantially increased compared with those of WT cells. To determine whether the effect of SHP-2 D61G mutation on cell motility is cell type-specific, we generated WT, SHP-2D61G/+, and SHP-2D61G/D61G embryonic fibroblasts (MEFs) from embryonic day 13.5 embryos, immortalized the cells by the 3T3 protocol, and evaluated cell behaviors. Spreading of SHP-2 mutant MEFs was also greatly increased compared with WT cells (Fig. 2D). Moreover, homozygous (SHP-2D61G/D61G) mutant cells spread much faster than heterozygous (SHP-2D61G/+) cells, especially at the early time points. Consistent with this result, migration of both homozygous and heterozygous cells was also enhanced (Fig. 2E). We also tested WT and mutant MEFs immortalized with SV40 large T antigen. Similar results were obtained (data not shown). These observations suggest that heterozygous GOF mutations of SHP-2 as identified in human diseases are sufficient to enhance cell motility and that the mutant SHP-2 functions in a dominant manner in this cellular process.
FIGURE 2.
Adhesion and migration of SHP-2 D61G knock-in cells are dramatically enhanced. A, WT and SHP-2D61G/+ bone marrow-derived macrophages were replated into non-tissue culture plates in cytokine-free medium and photographed 30 min, 60 min, and 18 h later. B, upper panel, WT and SHP-2D61G/+ bone marrow-derived mast cells were plated into cytokine-free medium and photographed 30 min later. Lower panel, WT and SHP-2D61G/+ mast cells were allowed to adhere to fibronectin-coated wells in cytokine and serum-free medium for the indicated periods of time. The number of adherent cells was determined by the MTS assay. C, cells were assayed for migration as described in the legend to Fig. 1C. Migrated cells adhering to the lower side of the membranes were photographed and enumerated under a microscope. D, immortalized WT, SHP-2D61G/+, and SHP-2D61G/D61G embryonic fibroblasts were suspended in DMEM with 2% BSA, allowed to adhere to fibronectin-coated plates for 6 h, and then photographed. Adhesion (D) and migration (E) capabilities of the cells were assessed as described under “Experimental Procedures.” Three independent experiments were performed and similar results were obtained in each. Results shown are the mean ± S.D. of triplicates from one experiment.
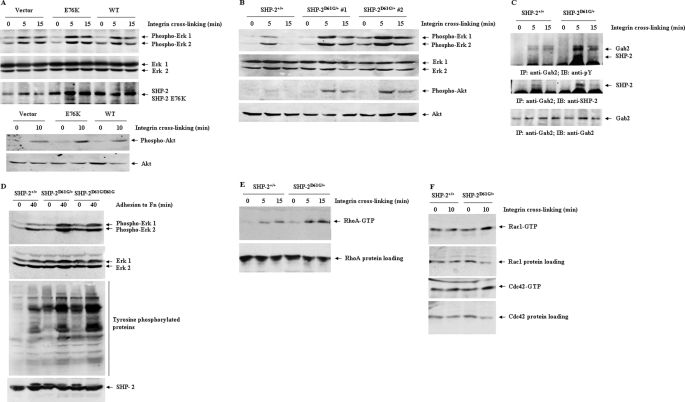
The mechanisms by which GOF mutations in SHP-2 enhance cell spreading and migration were then investigated. We examined the impact of SHP-2 mutations on the signaling pathways activated by β1 integrin that plays a critical role in these cellular processes. In Ba/F3 cells, transduction of SHP-2 E76K, but not WT SHP-2 increased β1 integrin cross-linking-induced activation of ERK and Akt kinases (Fig. 3A). The increased activation of ERK and Akt pathways does not seem to be attributed to the high expression level of the mutant SHP-2 because β1 integrin-induced ERK and PI3K/Akt pathways were also drastically enhanced in SHP-2 D61G knock-in (SHP-2D61G/+) mast cells (Fig. 3B) in which the mutant SHP-2 is expressed at the physiological level. The enhanced activation of ERK and Akt correlated with the increased interaction between mutant SHP-2 (SHP-2 D61G) and the scaffolding protein Gab2 (Fig. 3C) that plays an important role in the ERK and PI3K/Akt pathways (37). The increased interaction between mutant SHP-2 and Gab2 is likely due to the SHP-2 conformational change induced by D61G mutation. D61G mutation disrupts intramolecular binding between the N-terminal SH2 domain (N-SH2) and the protein-tyrosine phosphatase (PTP) domain, making SHP-2 bind more effectively to tyrosine-phosphorylated Gab2 because there is no longer any free-energy cost to disrupting the N-SH2/PTP interface and opening the N-SH2 phosphotyrosine binding pocket. In agreement with the results obtained from hematopoietic cells, MEFs harboring heterozygous and homozygous D61G mutations also showed greatly increased activation of ERK kinases following fibronectin stimulation (Fig. 3D), consistent with a recent study (38). Tyrosine phosphorylation of cellular proteins in the heterozygous or homozygous cells at either the basal levels or in response to fibronectin stimulation was increased, rather than decreased, although SHP-2 D61G expressed in these cells possesses enhanced catalytic activity (25, 39). This result supports the general positive role of SHP-2 catalytic activity in integrin signaling. Yet, the underlying mechanisms remain to be further determined.
FIGURE 3.
SHP-2 D61G mutation greatly enhancesβ1 integrin-induced outside-in signaling. Ba/F3 cells transduced with WT SHP-2, SHP-2 E76K, or control vector (A) and mast cells derived from WT and SHP-2D61G/+ bone marrow cells (B and C) were starved in serum and cytokine-free medium for 5 h and then subjected to β1 integrin cross-linking for the indicated periods of time as described under “Experimental Procedures.” D, WT, SHP-2D61G/+, and SHP-2D61G/D61G fibroblasts were trypsinized and resuspended in DMEM with 2% BSA for 1 h. The cells were then stimulated by adhering to fibronectin-coated plates in serum-free medium for the indicated periods of time. Whole cell lysates were prepared and examined for Akt and ERK activities by immunoblotting with anti-phospho-ERK, anti-phospho-Akt, and anti-Tyr(P) Abs. Blots were stripped and reprobed with anti-pan-ERK and anti-pan-Akt Abs to check protein loadings. The cell lysates were also immunoprecipitated with anti-Gab2 Ab followed by anti-Tyr(P) immunoblotting. Blots were stripped and re-probed with anti-SHP-2 and then anti-Gab2 Abs. E and F, WT and SHP-2D61G/+ mast cells stimulated by β1 integrin cross-linking were assayed for activation of small GTPases. Activated GTP-bound RhoA (E), Rac1, or Cdc42 (F) in the cell lysates were determined as described in the text. The lysates were also examined for protein loading by immunoblotting with anti-pan-RhoA, -Rac1, or Cdc42 Abs. Results shown are representative from two to three independent experiments.
Furthermore, we assessed the impact of SHP-2 activating mutations on activation of the Rho family small GTPases that have been shown to be critical in regulating cell motility. The role of SHP-2 phosphatase in activation of the Rho family small GTPases has been controversial. Both positive and negative roles for SHP-2 in RhoA activation have been reported (18, 19, 21–23). These results were obtained from calpeptin (a reported tyrosine phosphatase inhibitor)-treated cells, SHP-2 loss-of-function mutant cells, or dominant negative mutant SHP-2-overexpressing cells. The SHP-2 GOF mutation knock-in cells now provide an alternative model to clarify the role of SHP-2 in small GTPase activation. Accordingly, we determined small GTPase activities in SHP-2D61G/+ mast cells. As shown in Fig. 3, E and F, integrin cross-linking-induced activation of RhoA and Rac1 was greatly enhanced in SHP-2 D61G mutant cells, whereas activation of Cdc42 was slightly elevated. We also assessed RhoA GTPase activation in the SHP-2 E76K-transduced Ba/F3 cells in response to β1 integrin cross-linking. Similar results were obtained (data not shown). These data reaffirm that SHP-2 plays a positive role in activation of this family of small GTPases.
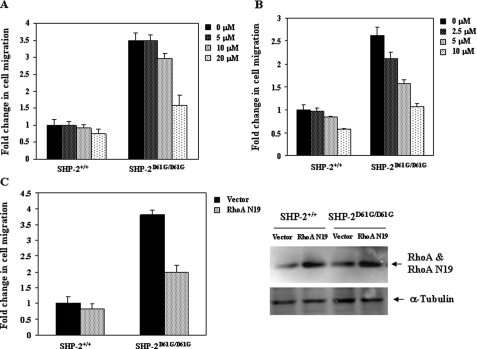
To determine whether increased activation of RhoA, ERK, and PI3K/Akt pathways is responsible for the enhanced spreading and migration of SHP-2 D61G mutant cells, we treated WT and mutant MEFs with MEK1 inhibitor PD98059, PI3K inhibitor LY294002, or transduced the cells with a dominant negative RhoA mutant (RhoA N19) (40) and then assessed cell migration. The results showed that inhibition of these pathways decreased motility of both WT and mutant cells. Interestingly, SHP-2 D61G mutant cells were more sensitive than WT cells to the inhibition of these pathways (Fig. 4). These results suggest that enhanced activation of ERK, Akt, and RhoA pathways contributes to the increased motility of SHP-2 mutant cells.
FIGURE 4.
Increased activation of ERK, Akt, and small GTPases contributes to the enhanced motility of SHP-2 D61G mutant cells. WT and SHP-2D61G/D61G MEFs were preincubated with the MEK1 inhibitor PD98059 (A) or the PI3K inhibitor LY294002 (B) in serum-free medium at the indicated concentrations for 1 h prior to the migration assay. C, WT and SHP-2D61G/D61G MEFs grown in 60-mm dishes were transfected with 5 μg of RhoA N19 expression plasmid or the control vector by Lipofectamine 2000. Transfected cells were assayed for migration. Migrated cells adhering to the lower side of the membranes were quantified as described under “Experimental Procedures.” Two-three independent experiments were performed and similar results were obtained in each. Results shown are the mean ± S.D. of triplicates from one experiment.
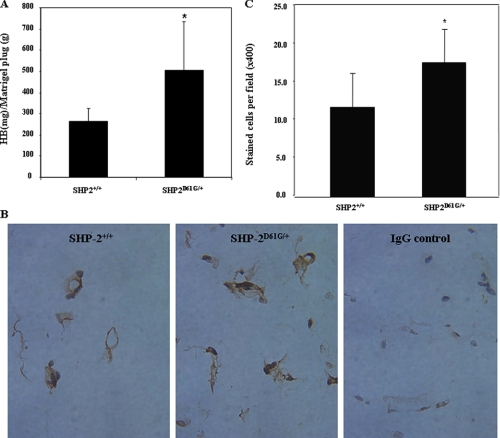
Finally, to confirm the effect of GOF mutations in SHP-2 on cell motility, we assessed angiogenesis of SHP-2 D61G mutant mice using a well characterized in vivo assay of angiogenesis (35, 36). Matrigel containing basic fibroblast growth factor and vascular endothelial growth factor was injected subcutaneously into SHP-2D61G/+ mice or WT littermates. The Matrigel plugs were retrieved 9 days later. Blood vessel formation in the Matrigel plugs was determined based on their hemoglobin content. As shown in Fig. 5A, angiogenesis in the Matrigel plugs in SHP-2D61G/+ mice was significantly increased. To further verify this result, cryosections prepared from the Matrigel plugs were immunostained for vWF, a marker protein of endothelial cells (41, 42). As demonstrated in Fig. 5, B and C, vWF positive endothelial cells/capillary vessels are significantly increased in the Matrigel plugs dissected from SHP-2D61G/+ mice. These results suggest that endothelial cells in the mutant mice have increased migration, although the increased angiogenesis in SHP-2D61G/+ mice may be due to enhanced integrin signaling and growth factor signaling.
FIGURE 5.
In vivo angiogenesis in SHP-2 D61G knock-in mice is enhanced. SHP-2D61G/+ mice (n = 10) or WT (n = 12) littermates were tested using an in vivo angiogenesis assay as described under “Experimental Procedures.” Nine days following the injection of Matrigel, animals were sacrificed. A, the Matrigel plugs were retrieved and vascularization of the plugs was determined by measuring their hemoglobin content. The results were normalized based on the hemoglobin levels of SHP-2D61G/+ mice. B, cryosections prepared from the Matrigel plugs were immunostained with anti-vWF Ab and photographed at ×400 magnification. Normal rabbit IgG was used as the negative control. C, vWF positive endothelial cells/capillary vessels were quantified at ×400 magnification under a microscope. Results shown are the mean ± S.D. from two independent experiments. The asterisk indicates significance between WT and SHP-2D61G/+ mice (A, p = 0.0178; C, p = 0.0032).
In summary, in this report we have shown that Noonan syndrome/leukemia mutations in SHP-2 phosphatase dramatically enhance cell motility. Our findings clarify the role of SHP-2 in adhesion-dependent activation of the RhoA family small GTPases. Moreover, they provide new insights into the pathogenesis of SHP-2 GOF mutation-associated diseases. Many developmental processes such as heart morphogenesis and vasculogenesis require the precise regulation of cell motility. Conceivably, certain phenotypes of SHP-2-associated Noonan syndrome or JMML might arise from aberrant cell motility induced by SHP-2 mutations. For example, lymphedema, one of the prominent phenotypes of Noonan syndrome, the pathogenesis of which has been unclear, might be induced by the increased motility of endothelial cells and thereby aberrant development of capillary vessels of the lymphatic system. Also, hepatomegaly caused by massive leukocyte infiltration into the liver in JMML is likely attributable to the increased motility of leukocytes. Additionally, as in vivo angiogenesis plays an important role in cancer formation and metastasis, our data indicate that GOF mutations in SHP-2 may contribute to tumorigenesis by enhancing tumor angiogenesis.
The enhanced cell motility by GOF mutations in SHP-2 is apparently associated with the increased integrin outside-in signaling. Integrin-induced ERK, PI3K/Akt, as well as small GTPase pathways are hyper-activated by SHP-2 GOF mutations. Moreover, SHP-2 mutant cells are more sensitive than WT cells to the inhibition of these pathways in terms of the suppression of cell motility. However, detailed signaling mechanisms by which GOF mutations in SHP-2 enhance integrin signaling remain to be further determined. It seems that GOF mutations in SHP-2 impact adhesion-induced signaling by either promoting activation of upstream kinases or decreasing its efficiency of dephosphorylation of its downstream substrates. Physical interaction between SHP-2 D61G and Gab2 is significantly prolonged. Moreover, tyrosine phosphorylation of Gab2 is greatly increased, even though Gab2 is a target of the catalytic activity of SHP-2 and the catalytic activity of SHP-2 D61G is increased (25, 39). Thus, altered physical and functional interactions between mutant SHP-2 and its target proteins may play an important role. It is conceivable that the substrate specificity of the mutant SHP-2 is changed as the consequence of the mutation in the N-SH2 domain. This notion is also supported by previous observations that interaction between SHP-2 with Gab1 in response to epidermal growth factor stimulation is enhanced by Noonan syndrome/leukemia-associated SHP-2 mutations (28, 39, 43). Moreover, a recent study showed that SHP-2 D61G mutation induced hyper-tyrosyl phosphorylation of the transmembrane glycoproteins, SIRPα and PZR (38), the two prominent targets of SHP-2 involved in cell adhesion and migration. Clearly, further studies are needed to define how GOF mutations in the N-SH2 domain change the accessibility of the catalytic site of SHP-2 to substrate proteins and how these changes diminish its efficiency in dephosphorylating the targets.
Acknowledgments
We thank Dr. Toshiyuki Araki (Ontario Cancer Institute) for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants HL068212 and HL082670 (to C. K. Q.), HL076810 (to K. R. M.), CA049152 and CA66600 (to B. G. N.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: ERK, extracellular signal-regulated kinase; PI3K, phosphatidylinositol 3-kinase; JMML, juvenile myelomonocytic leukemia; GOF, gain-of-function; IL, interleukin; FBS, fetal bovine serum; Ab, antibody; wWF, von Willebrand factor; WT, wild type; DMEM, Dulbecco's modified Eagle's medium; PBS, phosphate-buffered saline; BSA, bovine serum albumin; MEF, mouse embryonic fibroblasts; PTP, protein-tyrosine phosphatase; SH2, Src homology domain 2; MTS, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium inner salt.
References
- 1.Tonks, N. K. (2006) Nat. Rev. Mol. Cell. Biol. 7 833–846 [DOI] [PubMed] [Google Scholar]
- 2.Neel, B. G., Gu, H., and Pao, L. (2003) Trends Biochem. Sci. 28 284–293 [DOI] [PubMed] [Google Scholar]
- 3.Qu, C. K. (2002) Biochim. Biophys. Acta 1592 297–301 [DOI] [PubMed] [Google Scholar]
- 4.Yang, W., Klaman, L. D., Chen, B., Araki, T., Harada, H., Thomas, S. M., George, E. L., and Neel, B. G. (2006) Dev. Cell 10 317–327 [DOI] [PubMed] [Google Scholar]
- 5.Saxton, T. M., Henkemeyer, M., Gasca, S., Shen, R., Rossi, D. J., Shalaby, F., Feng, G. S., and Pawson, T. (1997) EMBO J. 16 2352–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qu, C. K., Shi, Z. Q., Shen, R., Tsai, F. Y., Orkin, S. H., and Feng, G. S. (1997) Mol. Cell. Biol. 17 5499–5507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qu, C. K., Yu, W. M., Azzarelli, B., Cooper, S., Broxmeyer, H. E., and Feng, G. S. (1998) Mol. Cell. Biol. 18 6075–6082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qu, C. K., Nguyen, S., Chen, J., and Feng, G. S. (2001) Blood 97 911–914 [DOI] [PubMed] [Google Scholar]
- 9.Zhang, E. E., Chapeau, E., Hagihara, K., and Feng, G. S. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 16064–16069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fornaro, M., Burch, P. M., Yang, W., Zhang, L., Hamilton, C. E., Kim, J. H., Neel, B. G., and Bennett, A. M. (2006) J. Cell Biol. 175 87–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ke, Y., Lesperance, J., Zhang, E. E., Bard-Chapeau, E. A., Oshima, R. G., Muller, W. J., and Feng, G. S. (2006) J. Biol. Chem. 281 34374–34380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ke, Y., Zhang, E. E., Hagihara, K., Wu, D., Pang, Y., Klein, R., Curran, T., Ranscht, B., and Feng, G. S. (2007) Mol. Cell. Biol. 27 6706–6717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang, T. L., Freeman, R. M., Jr., O'Reilly, A. M., Neel, B. G., and Sokol, S. Y. (1995) Cell 80 473–483 [DOI] [PubMed] [Google Scholar]
- 14.O'Reilly, A. M., and Neel, B. G. (1998) Mol. Cell. Biol. 18 161–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu, D. H., Qu, C. K., Henegariu, O., Lu, X., and Feng, G. S. (1998) J. Biol. Chem. 273 21125–21131 [DOI] [PubMed] [Google Scholar]
- 16.Oh, E. S., Gu, H., Saxton, T. M., Timms, J. F., Hausdorff, S., Frevert, E. U., Kahn, B. B., Pawson, T., Neel, B. G., and Thomas, S. M. (1999) Mol. Cell. Biol. 19 3205–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manes, S., Mira, E., Gomez-Mouton, C., Zhao, Z. J., Lacalle, R. A., and Martinez, A. C. (1999) Mol. Cell. Biol. 19 3125–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kodama, A., Matozaki, T., Fukuhara, A., Kikyo, M., Ichihashi, M., and Takai, Y. (2000) Mol. Biol. Cell 11 2565–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inagaki, K., Noguchi, T., Matozaki, T., Horikawa, T., Fukunaga, K., Tsuda, M., Ichihashi, M., and Kasuga, M. (2000) Oncogene 19 75–84 [DOI] [PubMed] [Google Scholar]
- 20.Lacalle, R. A., Mira, E., Gomez-Mouton, C., Jimenez-Baranda, S., Martinez, A. C., and Manes, S. (2002) J. Cell Biol. 157 277–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Reilly, A. M., Pluskey, S., Shoelson, S. E., and Neel, B. G. (2000) Mol. Cell. Biol. 20 299–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kontaridis, M. I., Eminaga, S., Fornaro, M., Zito, C. I., Sordella, R., Settleman, J., and Bennett, A. M. (2004) Mol. Cell. Biol. 24 5340–5352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schoenwaelder, S. M., Petch, L. A., Williamson, D., Shen, R., Feng, G. S., and Burridge, K. (2000) Curr. Biol. 10 1523–1526 [DOI] [PubMed] [Google Scholar]
- 24.Tartaglia, M., Mehler, E. L., Goldberg, R., Zampino, G., Brunner, H. G., Kremer, H., van der Burgt, I., Crosby, A. H., Ion, A., Jeffery, S., Kalidas, K., Patton, M. A., Kucherlapati, R. S., and Gelb, B. D. (2001) Nat. Genet. 29 465–468 [DOI] [PubMed] [Google Scholar]
- 25.Tartaglia, M., Niemeyer, C. M., Fragale, A., Song, X., Buechner, J., Jung, A., Hahlen, K., Hasle, H., Licht, J. D., and Gelb, B. D. (2003) Nat. Genet. 34 148–150 [DOI] [PubMed] [Google Scholar]
- 26.Loh, M. L., Vattikuti, S., Schubbert, S., Reynolds, M. G., Carlson, E., Lieuw, K. H., Cheng, J. W., Lee, C. M., Stokoe, D., Bonifas, J. M., Curtiss, N. P., Gotlib, J., Meshinchi, S., Le Beau, M. M., Emanuel, P. D., and Shannon, K. M. (2004) Blood 103 2325–2331 [DOI] [PubMed] [Google Scholar]
- 27.Bentires-Alj, M., Paez, J. G., David, F. S., Keilhack, H., Halmos, B., Naoki, K., Maris, J. M., Richardson, A., Bardelli, A., Sugarbaker, D. J., Richards, W. G., Du, J., Girard, L., Minna, J. D., Loh, M. L., Fisher, D. E., Velculescu, V. E., Vogelstein, B., Meyerson, M., Sellers, W. R., and Neel, B. G. (2004) Cancer Res. 64 8816–8820 [DOI] [PubMed] [Google Scholar]
- 28.Araki, T., Mohi, M. G., Ismat, F. A., Bronson, R. T., Williams, I. R., Kutok, J. L., Yang, W., Pao, L. I., Gilliland, D. G., Epstein, J. A., and Neel, B. G. (2004) Nat. Med. 10 849–857 [DOI] [PubMed] [Google Scholar]
- 29.Mohi, M. G., Williams, I. R., Dearolf, C. R., Chan, G., Kutok, J. L., Cohen, S., Morgan, K., Boulton, C., Shigematsu, H., Keilhack, H., Akashi, K., Gilliland, D. G., and Neel, B. G. (2005) Cancer Cell 7 179–191 [DOI] [PubMed] [Google Scholar]
- 30.Chan, R. J., Leedy, M. B., Munugalavadla, V., Voorhorst, C. S., Li, Y., Yu, M., and Kapur, R. (2005) Blood 105 3737–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schubbert, S., Lieuw, K., Rowe, S. L., Lee, C. M., Li, X., Loh, M. L., Clapp, D. W., and Shannon, K. M. (2005) Blood 106 311–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu, W. M., Daino, H., Chen, J., Bunting, K. D., and Qu, C. K. (2006) J. Biol. Chem. 281 5426–5434 [DOI] [PubMed] [Google Scholar]
- 33.Yu, W. M., Hawley, T. S., Hawley, R. G., and Qu, C. K. (2002) Blood 99 2351–2359 [DOI] [PubMed] [Google Scholar]
- 34.Juarez, J. C., Guan, X., Shipulina, N. V., Plunkett, M. L., Parry, G. C., Shaw, D. E., Zhang, J. C., Rabbani, S. A., McCrae, K. R., Mazar, A. P., Morgan, W. T., and Donate, F. (2002) Cancer Res. 62 5344–5350 [PubMed] [Google Scholar]
- 35.Zhang, J. C., Claffey, K., Sakthivel, R., Darzynkiewicz, Z., Shaw, D. E., Leal, J., Wang, Y. C., Lu, F. M., and McCrae, K. R. (2000) FASEB J. 14 2589–2600 [DOI] [PubMed] [Google Scholar]
- 36.Zhang, J. C., Donate, F., Qi, X., Ziats, N. P., Juarez, J. C., Mazar, A. P., Pang, Y. P., and McCrae, K. R. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 12224–12229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gu, H., Maeda, H., Moon, J. J., Lord, J. D., Yoakim, M., Nelson, B. H., and Neel, B. G. (2000) Mol. Cell. Biol. 20 7109–7120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eminaga, S., and Bennett, A. M. (2008) J. Biol. Chem. 283 15328–15338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keilhack, H., David, F. S., McGregor, M., Cantley, L. C., and Neel, B. G. (2005) J. Biol. Chem. 280 30984–30993 [DOI] [PubMed] [Google Scholar]
- 40.Nobes, C. D., and Hall, A. (1999) J. Cell Biol. 144 1235–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oswald, J., Boxberger, S., Jorgensen, B., Feldmann, S., Ehninger, G., Bornhauser, M., and Werner, C. (2004) Stem Cells 22 377–384 [DOI] [PubMed] [Google Scholar]
- 42.Jin, K., Zhu, Y., Sun, Y., Mao, X. O., Xie, L., and Greenberg, D. A. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 11946–11950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fragale, A., Tartaglia, M., Wu, J., and Gelb, B. D. (2004) Hum. Mutat. 23 267–277 [DOI] [PubMed] [Google Scholar]
- 44.Chen, J., Yu, W. M., Bunting, K. D., and Qu, C. K. (2004) Oncogene 23 3659–3669 [DOI] [PubMed] [Google Scholar]