Abstract
eIF2α is part of a multimeric complex that regulates cap-dependent translation. Phosphorylation of eIF2α (phospho-eIF2α) is induced by various forms of cell stress, resulting in changes to the proteome of the cell with two diametrically opposed consequences, adaptation to stress or initiation of programmed cell death. In contrast to the robust eIF2α phosphorylation seen in response to acute insults, less is known about the functional role of basal levels of eIF2α phosphorylation. Here we show that mouse embryonic fibroblasts expressing a nonphosphorylatable eIF2α have enhanced sensitivity to diverse toxic insults, including amyloid β-(1–42) peptide (Aβ), a key factor in the pathogenesis of Alzheimer disease. This correlates with impaired glutathione metabolism because of down-regulation of the light chain, xCT, of the cystine/glutamate antiporter system X-c. The mechanistic link between the absence of phospho-eIF2α and xCT expression is nuclear factor ATF4. Consistent with these findings, long term activation of the phospho-eIF2α/ATF4/xCT signaling module by the specific eIF2α phosphatase inhibitor, salubrinal, induces resistance against oxidative glutamate toxicity in the hippocampal cell line HT22 and primary cortical neurons. Furthermore, in PC12 cells selected for resistance against Aβ, increased activity of the phospho-eIF2α/ATF4/xCT module contributes to the resistant phenotype. In wild-type PC12 cells, activation of this module by salubrinal ameliorates the response to Aβ. Furthermore, in human brains, ATF4 and phospho-eIF2α levels are tightly correlated and up-regulated in Alzheimer disease, most probably representing an adaptive response against disease-related cellular stress rather than a correlate of neurodegeneration.
eIF2α is part of the multimeric eIF2 complex that is involved in the initiation of cap-dependent protein translation (for reviews see Refs. 1, 2). The eIF2 complex brings the 40 S ribosomal subunit together with the initiating tRNAMet when eIF2 is bound to GTP. Upon hydrolysis of GTP to GDP, the complex is no longer active, and protein synthesis is not initiated. GDP/GTP exchange requires the activity of the guanine nucleotide exchange factor eIF2B. However, when eIF2α is phosphorylated on Ser-51 (phospho-eIF2α), the affinity of eIF2α for eIF2B increases, and thus it can sequester eIF2B, thereby inhibiting GDP/GTP exchange. As cells have considerably higher amounts of eIF2α compared with eIF2B, even modest increases in phospho-eIF2α can modulate eIF2B reactivation. Although these changes slow down cap-dependent initiation, they favor cap-independent translation. Proteins up-regulated by this mechanism include transcription factors such as activating transcription factor-4 (ATF4). Therefore, eIF2α phosphorylation orchestrates significant changes in the proteome of the cell.
There are four known eIF2α kinases as follows: protein kinase R, heme-regulated eIF2α kinase, protein kinase R-like kinase (PERK), and GCN2 (general control nonderepressible-2) (for reviews see Refs. 1–3), all of which are activated by distinct forms of stress. In addition, two different phosphatase complexes have been described that can mediate eIF2α dephosphorylation (4). In general, the changes to the proteome induced by changes in eIF2α phosphorylation lead to adaptation of the cell to stress with two possible, diametrically opposed consequences, survival or initiation of programmed cell death. The outcome seems to be determined by the duration of the insults, the interplay of different branches of the stress response, and their time courses (5). In addition to a response to cellular stress, basal levels of phospho-eIF2α are present in vitro (6) and in vivo (7, 8), and eIF2α phosphorylation was shown to be involved in biochemical processes as diverse as cell cycle regulation (9), glucose homeostasis (10), and synaptic plasticity (11).
There is good evidence that eIF2α phosphorylation can modulate the resistance of nerve cells to oxidative stress. For example, in early work from our laboratory (12), infection of the HT22 nerve cell line with a construct expressing the S51D mutant of eIF2α, which acts as a constitutively phosphorylated form of the protein, was shown to bring about an increase in the resistance of the cells to oxidative stress, which correlated with an ability to maintain a high GSH concentration in the presence of oxidative stress. Further studies from the David Ron laboratory using a different approach to generate constitutively phosphorylated eIF2α in the HT22 cells confirmed these results (13).
GSH and GSH-associated metabolism provide the major line of defense for the protection of cells from oxidative and other forms of stress (14). In addition, the GSSG/GSH redox pair forms the major redox couple in cells and as such plays a critical role in regulating redox-dependent cellular functions. GSH is a tripeptide containing the amino acids cysteine, glutamate, and glycine. Because glutamate and glycine occur at relatively high intracellular concentrations, cysteine is limiting for GSH synthesis in many types of cells. In the extracellular environment, cysteine is readily oxidized to form cystine, so for most cell types cystine transport mechanisms are essential to provide them with the cysteine needed for GSH synthesis.
Cystine uptake in many types of cells is mediated by the Na+-independent cystine/glutamate antiporter, system X-c. System X-c is a member of the disulfide-linked heterodimeric amino acid transporter family and consists of a light chain (xCT) that confers substrate specificity and a heavy chain (4F2hc) that is shared among a number of different amino acid transporters (15). It transports cystine into cells in a 1/1 exchange with glutamate and is thus inhibited by high concentrations of extracellular glutamate (16). The importance of system X-c for the maintenance of GSH levels in cells is demonstrated by the loss of GSH and subsequent cell death seen in neural cells following exposure to millimolar concentrations of extracellular glutamate, a pathway termed oxidative glutamate toxicity or oxytosis (17). System X-c is expressed in the brain (18–21), and mice lacking system X-c function show signs of redox imbalance (22) and brain atrophy (20).
In this study we investigate further the role of eIF2α phosphorylation in regulating GSH metabolism and the response of cells to oxidative stress. Using multiple, distinct cell lines, we show a tight linkage between eIF2α phosphorylation, ATF4, and system X-c expression. We further show that the phospho-eIF2α/ATF4 module is activated in the brains of patients with Alzheimer disease (AD).3 Together, these results suggest that the increase in eIF2α phosphorylation in AD brains that has repeatedly been interpreted as a sign of neuronal degeneration (23–26) should rather be interpreted as an adaptive response to Aβ that promotes neuronal survival.
EXPERIMENTAL PROCEDURES
Cell Culture and Viability Assays—Mouse embryonic fibroblasts derived from embryos genetically engineered to homozygously express the nonphosphorylatable S51A mutation (A/A MEFs) and wild-type controls (S/S MEFs) (10) were a kind gift from Randal J. Kaufman and Donalyn Scheuner (Howard Hughes Medical Institute, University of Michigan, Ann Arbor, MI). MEFs were propagated in high glucose DMEM (Invitrogen) with 10% fetal calf serum (Hyclone, Logan, UT), additionally supplemented with nonessential and essential amino acids (Invitrogen). For passaging, confluent cells were detached with trypsin/EDTA (Invitrogen). Cells were re-plated after trypsinization when confluent for no more than 10 passages. Aβ-resistant PC12 clone r7 was described previously (27–30). Wild-type and Aβ-resistant PC12 cells were grown in high glucose DMEM with 10% horse serum (Hyclone) and 5% fetal calf serum. Cells were split once a week after dislodgement by repeated pipetting. HT22 cells were grown on tissue culture dishes in high glucose DMEM supplemented with 10% fetal calf serum as described (31).
For viability assays, 1 × 104 MEFs, 2.5 × 103 HT22, or 2 × 103 PC12 cells were plated in 96-well plates, in some cases with 30 μm salubrinal (Axxora, San Diego) dissolved in DMSO or DMSO alone. After 24 h of culture, the medium was exchanged with fresh medium and the indicated concentrations of compounds. Sodium glutamate, homocysteic acid, SIN-1, 1,3-bis(2-chloroethyl)-N-nitrosourea (BCNU), ethacrynic acid, and tert-butylhydroperoxide were from Sigma. Amyloid β-(1–42) peptide (Aβ-(1–42)) was obtained from Bachem (Torrance, CA). MEFs and HT22 cells were treated for 24 h and PC12 cells for 48 h. Viability or toxicity of Aβ was measured by the MTT assay as described previously (32).
Mouse cortical neurons were prepared as described previously (33) and seeded at a density of 1 × 105 cells per well onto poly-l-lysine- and laminin-coated 96-well plates using high glucose DMEM supplemented with 10% fetal calf serum with 30 μm salubrinal dissolved in DMSO or DMSO alone. After 24 h, glutamate was added at the indicated concentrations. Survival was measured by the MTT assay after another 24 h.
Transfection and Luciferase Reporter Assays—For transfection, 5 × 104 S/S and 2 × 106 A/A MEFs were plated in 60-mm dishes and grown for 48 h. Because the two cell lines proliferate at different rates, these conditions yield similar cell densities at the end of the experiment. Transfection was performed with 2 μg of p-SV-β-galactosidase control vector (Promega, Madison, WI), 2 μg of xCT promoter luciferase constructs (generous gifts from Dr. Hideyo Sato, Yamagata University, Tsuruoka, Japan), and 5 μl of Lipofectamine 2000 (Invitrogen) in 2.5 ml of fresh DMEM with 10% fetal calf serum for 6 h. For some experiments, the transfection additionally included 40 pmol of ATF4 siRNA (catalog number sc-35113) or control siRNA (catalog number sc-37007), both from Santa Cruz Biotechnology (Santa Cruz, CA). For overexpression of ATF4, some experiments additionally included 2 μg of empty pRK7 vector (a generous gift form Dr. Robert C. Cumming, University of Western Ontario, London, Ontario, Canada) or a pRK7 vector expressing ATF4. This construct was obtained by excising the ATF4 open reading frame from a pcDNA3-hCD2 plasmid containing ATF4 (a generous gift from Dr. David Ron, Skirball Institute, New York University School of Medicine, New York, NY) and re-ligating into pRK7. To measure the translational activity of the ATF4 5′UTR, luciferase reporter promoter constructs were replaced by a vector containing the ATF4 5′-untranslated region (5′UTR) and AUG fused to luciferase by the TK promoter in a pGL3 backbone (a generous gift from David Ron). 24 h after transfection, the cells were lysed in 1× reporter lysis buffer (Promega), and enzyme activities were measured by luminescence using the Beta Glo and Luciferase Assay Systems (Promega) on a luminometer from Molecular Devices (Madison, WI). The luciferase/β-galactosidase ratio for each condition and cell line was normalized to the ratio obtained by transfection of pGL3 and pSV-β-Gal for the same condition and cell line.
HT22 cells were plated at a density of 4.4 × 105 cells/100-mm dish and grown for 24 h. The medium was exchanged with 5 ml of fresh growth medium, and the cells were transfected with 4 μg of pRK7 or pRK7-ATF4 vector using 5 μl of Lipofectamine 2000 for 6 h. 24 h later, the cells were re-plated for the different assays performed in parallel as follows: 8.8 × 105 in 100-mm dishes for protein preparation; 1.65 × 105 in 60-mm dishes for GSH measurement; 3 × 104/well in 24-well plates for [35S]cystine uptake; and 2.5 × 103/well in 96-well plates for oxidative glutamate toxicity, respectively. Cells were harvested or glutamate was added 24 h after re-plating.
Enzymatic Measurement of Total GSH—For measurement of GSH, 5 × 105 A/A and S/S MEFs, 1.65 × 105 HT22 cells or 4.5 × 105 PC12 cells were plated in 60-mm dishes. MEFs and HT22 cells with or without salubrinal were grown for 24 h and PC12 cells for 48 h. Cells were scraped into ice-cold phosphate-buffered saline, and 10% sulfosalicylic acid was added to a final concentration of 3.3%. GSH was assayed as described (34) and normalized to protein recovered from the acid-precipitated pellet by treatment with 0.2 n NaOH at 37 °C overnight and measured by the bicinchoninic acid assay (Pierce).
Measurement of System X -c Activity and of Other Amino Acid Uptake Systems—6 × 105 A/A and S/S MEF in parallel, 3 × 105 HT22, or PC12 cells with or without 30 μm salubrinal were plated in 24-well dishes and grown for 24, 24, and 48 h, respectively. For PC12 cells, the plates were coated with poly-l-lysine. Cells were washed three times with sodium-free HBSS. Uptake was performed in triplicate with 25 μm [35S]cystine (PerkinElmer Life Sciences) for 20 min at 37 °C. To specifically measure system X-c activity, 10 mm glutamate was added to parallel wells in triplicate. Cells were washed three times with ice-cold, sodium-free HBSS and lysed in 0.2 n NaOH. Radioactivity was measured by liquid scintillation counting and normalized to protein measured by the Bradford method (Pierce). Because [35S]cystine became unavailable during the course of the experiments, system X-c activity in HT22 cells treated with salubrinal was measured as sodium-insensitive, HCA-inhibitable uptake of [3H]glutamate (PerkinElmer Life Sciences). Cells in triplicate were incubated in 10 μm glutamate ([3H]glutamate/cold glutamate 1/1000) with or without 1 mm HCA adjusted to pH 7.4 in sodium-free HBSS. To measure cysteine uptake, [35S]cystine was preincubated with 10 μm cysteamine, and uptake was performed similarly as for system X-c but with sodium-containing HBSS. This results in glutamate-insensitive, cysteine-sensitive uptake of radioactivity reflecting the function of system ASC and others (34). To measure system L activity, 1 μm [3H]leucine was used in sodium-containing HBSS.
Protein Preparation—For Western blotting, confluent A/A or S/S MEFs in 100-mm dishes, 3.3 × 105 HT22 cells per 60-mm dish grown for 24 h, or 1.2 × 106 wild-type PC12 cells and 6 × 105 PC12r7 cells per 100-mm dish grown for 48 h were used. For whole cell extracts, cells were rinsed twice with ice-cold phosphate-buffered saline and scraped into lysis buffer consisting of 50 mm HEPES, pH 7.5, 50 mm NaCl, 10 mm NaF, 10 mm sodium pyrophosphate, 5 mm EDTA, 1% Triton X-100, 1 mm sodium orthovanadate, 1× protease inhibitor, and phosphatase inhibitor mixtures (Sigma). Cells were sonicated on ice and centrifuged. Cystosolic, membrane, and nuclear fractions were prepared as described previously (35). Protein in the different fractions was quantified by the bicinchoninic acid method (Pierce) and adjusted to equal concentrations. 5× Western blot sample buffer (74 mm Tris-HCl, pH 8.0, 6.25% SDS, 10% β-mercaptoethanol, 20% glycerol) was added to a final concentration of 2.5×, and samples were boiled for 5 min.
Samples of midfrontal cortex from individuals with histologically confirmed Alzheimer disease and age-matched control brains were obtained from Dr. Carol Miller at the Alzheimer Disease Research Center, Los Angeles, CA, and were described in detail previously (29, 30). Frozen tissue samples were partially thawed, minced in 5× weight/volume extraction buffer (50 mm Tris-HCl, pH 7.5, 2% SDS, and protein inhibitor mixture), sonicated on ice, and centrifuged. Protein concentrations were determined using the Lowry assay (Pierce). Samples were boiled in 1× Western blot sample buffer.
Western Blotting—For SDS-PAGE, similar amounts of cellular protein, typically 40 μg per lane were used. In experiments using human brain extracts, 40 μg per lane was loaded for detection of phospho-eIF2α and ATF4 and 20 μg/lane for detection of pan-eIF2α and actin. All samples were separated using 10% Criterion XT Precast BisTris gels (Bio-Rad).
Proteins were transferred to nitrocellulose membranes, and the quality of protein measurement, electrophoresis, and transfer was checked by staining with Ponceau S. Membranes were blocked with 5% skim milk in TBS-T (20 mm Tris buffer, pH 7.5, 0.5 m NaCl, 0.1% Tween 20) for 2 h at room temperature and incubated overnight at 4 °C in the primary antibody diluted in 5% bovine serum albumin in TBS, 0.05% Tween 20. The primary antibodies used were as follows: rabbit anti-phospho-Ser-51-eIF2α (catalog number 9721, 1/1000) and rabbit anti-eIF2α (catalog number 9722, 1/500) from Cell Signaling (Beverly, MA); rabbit anti-ATF4 (catalog number sc-200, 1/500) and rabbit anti-Nrf2, (catalog number sc-13032, 1/500) from Santa Cruz Biotechnology; mouse anti-actin (catalog number A5441, 1/200,000) from Sigma; and rabbit anti-xCT (1/1000, a generous gift from Dr. Sylvia Smith, Medical College of Georgia, Augusta, GA). Subsequently, blots were washed in TBS, 0.05% Tween 20 and incubated for 1 h at room temperature in horse-radish peroxidase-goat anti-rabbit or goat anti-mouse (Bio-Rad) diluted 1/5000 in 5% skim milk in TBS, 0.1% Tween 20. After additional washing, protein bands were detected by chemiluminescence using the Super Signal West Pico substrate (Pierce). For relative eIF2α phosphorylation, parallel blots were performed. For all other antibodies, the same membrane was re-probed for actin. Ponceau S-stained membranes and autoradiographs were scanned using a Bio-Rad GS800 scanner. Band density was measured using the manufacturer's software. Relative eIF2α phosphorylation was calculated as the ratio of band densities obtained by the phospho- and the pan-eIF2α antibody. Relative expression of the other proteins was normalized to actin band density, with the exception of the comparison of wild-type and resistant PC12 cells that express highly different amounts of nonmembrane-associated actin (30) that sediments with the nuclear fraction using our technique. Here relative expression was normalized to histone band density obtained by scanning the Ponceau S-stained membranes. Each Western blot was repeated at least three times with independent protein samples.
ATF4 Promoter Analysis—Genomic sequence comparison was done using the NCBI genome data base and the reference sequence and the BLASTn algorithm (blast.ncbi.nlm.nih.gov). The reference sequences for the different genomic sequences are NT 039229.7/Mm3 39269 37 for murine chromosome 3, NW 047625.2/Rn2 WGA21474 for rat chromosome 2, NT 016354.18/Hs4 16510 for human chromosome 4, and NW 001493490.2/Bt17 WGA16084 for bovine chromosome 17.
Statistical Analysis—Data from at least three independent experiments were normalized, pooled, and analyzed using Graph Pad Prism 4 software followed by appropriate statistical tests. For exclusion of outliers, the established definition of an extreme outlier was used as follows: values lower or higher than the 25% percentile (Q1) or 75% percentile (Q3) minus or plus three times the interquartile range (Q3–Q1), respectively.
RESULTS
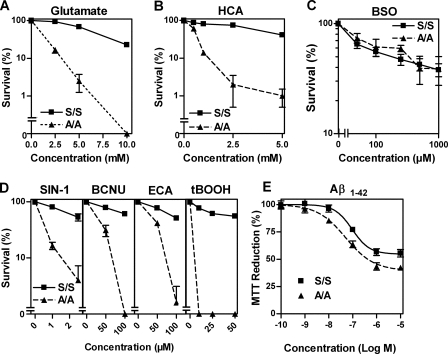
In our earlier study on eIF2α and GSH metabolism (12), we showed that nerve cells engineered to express decreased levels of eIF2α were much less sensitive than wild-type cells to oxidative glutamate toxicity. This decrease in sensitivity to glutamate correlated with an increase in the levels of GSH that the cells were able to maintain in the presence of glutamate. Similar results were obtained following overexpression of a constitutively phosphorylated form of eIF2α (13). To explore further the role of eIF2α phosphorylation in modulating the response of cells to toxic stress, we utilized MEFs derived from mice homozygously expressing a nonphosphorylatable form of eIF2α (A/A MEFs) and compared them with wild-type fibroblasts (S/S MEFs) (10). When S/S and A/A MEFs were subjected to oxidative glutamate toxicity by exposure to increasing concentrations of glutamate and cell survival assessed after 24 h by the MTT assay, we observed a striking difference in sensitivity between the two cell lines (Fig. 1A). Whereas S/S MEFs were relatively resistant to glutamate treatment with an EC50 of ∼7.5 mm for cell death, A/A MEFs showed a significant enhancement of sensitivity with an EC50 of ∼1 mm (Fig. 1A). Similar results were obtained following treatment with HCA, another system X-c inhibitor (Fig. 1B). Of note, the sensitivity of both cell lines against inhibition of GSH synthesis by the glutamate cysteine ligase inhibitor, buthionine sulfoximine (36), was similar (Fig. 1C). On the other hand, A/A MEFs were significantly more sensitive to a variety of other insults that decrease GSH levels in cells, including the peroxynitrite generator SIN-1 (37), BCNU (38), ethacrynic acid (38), and the peroxidizing agent tert-butylperoxide (39) (Fig. 1D). Interestingly, MTT reduction in the A/A MEFs also showed increased sensitivity to Aβ-(1–42) (Fig. 1E), a peptide implicated in the pathophysiology of AD. Although AD is a central nervous system disease, numerous other tissues have been reported to be affected, including fibroblasts (40). These data not only indicate that the fibroblast lines could provide a useful starting point for understanding the mechanisms underlying the regulation of GSH metabolism by eIF2α phosphorylation, but they also suggest that the regulation of eIF2α phosphorylation could provide an important line of defense for cells against both exogenous and endogenous forms of toxic stress.
FIGURE 1.
Defective eIF2α phosphorylation increases the sensitivity of mouse embryonic fibroblasts to system X-c inhibition and oxidative stress. Embryonic fibroblasts derived from homozygous eIF2α-S51A mice (A/A) and wild-type embryonic fibroblasts (S/S) were plated at a density of 1 × 104 cells per well in 96-well plates. After 24 h of culture, cells were treated with the system X-c inhibitors glutamate (A) and homocysteic acid (HCA) (B); glutamate cysteine ligase (GCL) inhibitor buthionine sulfoximide (BSO)(C); SIN-1, BCNU, ethacrynic acid (ECA), and tert-butylhydroperoxide (tBOOH)(D) at the indicated concentrations. Survival was measured after 24 h using the MTT assay. E, 2.5 × 105 A/A and S/S cells per well were grown for 24 h in 96-well plates and treated with Aβ-(1–42) peptide (Aβ1–42) for 48 h. The effect of Aβ-(1–42) was measured by the MTT assay. All single experiments were performed in quadruplicate, and the MTT value without treatment was normalized to 100%. Each data point represents the means of at least three experiments ± S.E. Statistical analysis was performed by two-way ANOVA. Column factor for A/A versus S/S was highly significant (p < 0.0001) for all insults tested except buthionine sulfoximine, which showed no statistical difference.
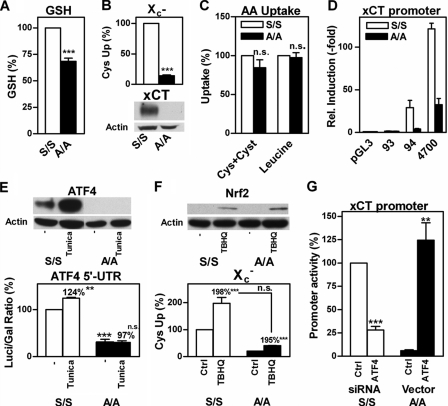
Consistent with our previous results (12), A/A MEFs have lower average levels of GSH than S/S MEFs (Fig. 2A), and this is likely to contribute to their enhanced sensitivity to multiple forms of stress. To determine which step(s) in GSH metabolism were affected by eIF2α phosphorylation, we began by looking at the first step in GSH metabolism which, in most cells, is cystine uptake via system X -c. Differences in expression of the specific subunit of system X-c have been shown to modulate the sensitivity of cells to oxidative glutamate toxicity (41). Indeed, we found a significant impairment of system X-c function measured as glutamate-sensitive, sodium-independent 35S-labeled cystine uptake associated with a striking reduction of xCT protein expression in the A/A compared with S/S MEFs (Fig. 2B). In contrast, the uptake of 35S-labeled cystine in combination with cysteamine, which converts cystine to cysteine, which can then be taken up by a variety of other transporters (34), and the uptake of [3H]leucine were not different in A/A and S/S MEFs (Fig. 2C). Thus, the impairment of system X-c shows clear specificity in A/A MEFs, is due to reduced expression of xCT, and most possibly results in their increased sensitivity to stress.
FIGURE 2.
Glutathione metabolism in A/A cells is compromised by decreased ATF4 translation and subsequent down-regulation of system X -c. A, GSH is decreased in A/A cells. Exponentially dividing A/A and S/S cells in 60-mm dishes were harvested, and GSH was measured enzymatically. For each experiment, the GSH content of S/S cells was normalized to 100%. B, system X-c activity and xCT expression are down-regulated in A/A cells. A/A and S/S cells were plated in 24-well plates, and glutamate-sensitive uptake of [35S]cystine in sodium-free HBSS was measured for 20 min and normalized to total protein (upper panel). Uptake of S/S cells was normalized to 100% for every individual experiment. Similar amounts of purified membranes from A/A and S/S cells were separated by SDS-PAGE, transferred to nitrocellulose, and hybridized with an anti-xCT antibody. Comparable loading was verified by staining of the same membrane with an anti-actin antibody (lower panel). C, amino acid uptake systems distinct from system X-c are not compromised in A/A cells. A/A and S/S cells were plated as in B, upper panel. Uptake of 25 μm [35S]cystine co-incubated with 10 μm cysteamine (Cys+Cyst) or 1 μm [3H]leucine was measured for 20 min. Uptake in S/S cells was normalized to 100% for each experiment. D, proximal xCT promoter activity is decreased in A/A cells. 5 × 104 A/A or 2 × 105 S/S cells were grown in 60-mm dishes for 48 h and then transfected with either control plasmid (pGL3) or pGL3 containing the proximal 93, 94, or 4700 bp of the xCT promoter together with the pSV-β-GAL vector. After 24 h, cells were harvested, and luciferase and β-galactosidase activity were measured. Relative luciferase induction by xCT promoter fragments was calculated by normalizing individual luciferase/β-galactosidase ratios of cells transfected with pGL3 to one. E, ATF4 translation is defective in A/A cells. Nuclear extracts of S/S and A/A cells either treated with vehicle or 5 μg/ml tunicamycin (Tuni) for 2 h were separated by SDS-PAGE, transferred to nitrocellulose, and incubated with anti-ATF4. Membranes were hybridized with anti-actin as a loading control (upper panel). S/S and A/A cells were co-transfected as in D with the ATF4 5′UTR luciferase or empty pGL3 vector and the pSV-β-GAL vector. Cells were treated with vehicle or 5 μg/ml tunicamycin (Tuni) for 2 h. The relative increase in luciferase activity by the ATF4 5′UTR compared with pGL3 in control S/S cells was normalized to 100% for every individual experiment (lower panel). F, Nrf2 is similarly induced in A/A and S/S cells. Nrf2 expression in S/S and A/A cells with or without treatment with 25 μm tBHQ for 4 h was analyzed using nuclear extracts and Western blotting (upper panel). System X-c activity with and without tBHQ treatment was measured using glutamate-sensitive uptake of [35S]cystine (lower panel). G, ATF4 mediates differential xCT promoter activity in S/S and A/A cells. S/S and A/A cells were co-transfected with the 4700-bp xCT promoter-luciferase construct, pSV-β-GAL, and either ATF4 or control siRNA for S/S cells or pRK7 (Ctrl) or ATF4-pRK7 (ATF4) for A/A cells, respectively. Luciferase/β-galactosidase ratios of S/S cells transfected with control siRNA were normalized to 100% for every individual experiment. All graphs show mean ± S.E. of at least three experiments. Statistical analysis was performed using one-sample t test compared with 100% (A and B) or two-way ANOVA with Bonferroni posttests (D–G) to compare A/A with S/S cells or the effect of tunicamycin or tBHQ versus control A/A and S/S cells (E and F, indicated next to % increase). Relative increase of S/S and A/A in F was compared by Student's unpaired t test. ***, p < 0.0001; **, p < 0.001; n.s., not significant.
To determine whether the decreased expression of xCT in A/A MEFs is because of lower levels of transcription, we transfected A/A and S/S MEFs with xCT promoter luciferase reporter constructs of different lengths, 4700, 94, and 93 bp, respectively. Whereas the 94-bp construct contains an amino acid-response element (AARE) stretching from -94 to -86 bp, this is disrupted in the 93-bp fragment (42). When compared with S/S MEFs, A/A MEFs have severalfold lower xCT promoter activity using either the 4700- or 94-bp proximal xCT promoter constructs (Fig. 2D). This difference was abolished by reducing the proximal xCT promoter fragment to 93 bp. These results indicate that xCT is transcriptionally down-regulated in A/A MEFs via hypoactivity of the AARE consistent with a recent report showing that this AARE mediates xCT up-regulation in response to amino acid starvation via binding of ATF4 (42). Along with ATF4, xCT transcription was reported to be regulated by Nrf2 binding to antioxidant-response elements located between the 94- and 4700-bp fragments (43). However, we found that xCT promoter activity in the A/A MEFs relative to the S/S MEFs was similarly decreased regardless of the presence of an antioxidant-response element suggesting that Nrf2 was unlikely to be involved in the differential expression of xCT between the two cell lines.
We next looked at the basal and stimulated ATF4 and Nrf2 levels in nuclear extracts of both MEF lines. The S/S MEFs had readily detectable basal levels of ATF4 that were increased severalfold following treatment with the ER stress inducer, tunicamycin, whereas A/A MEFs expressed no detectable ATF4 in either the absence or presence of tunicamycin (Fig. 2E). Transfection of both MEFs with a construct containing the ATF4 5′UTR upstream of the luciferase gene demonstrated that translation of ATF4 is reduced and induction by tunicamycin abolished in A/A MEFs compared with S/S MEFs, which can be interpreted as a direct result of the absence of eIF2α phosphorylation in the A/A MEFs. The decreased relative differences in luciferase activity as compared with ATF4 protein levels in the two cell lines can be explained by the differences in half-life, which is ∼4 h for firefly luciferase (44) and 15 min for ATF4.4 In contrast, the classical Nrf2 inducer, tert-butylhydroquinone (tBHQ), increased nuclear Nrf2 equally in both cell lines, and the downstream effects of Nrf2 induction on system X-c were also similar with an ∼2-fold increase in glutamate-sensitive [35S]cystine uptake (Fig. 2F) in both cell lines.
To further substantiate the idea that ATF4 is the key factor responsible for the differential expression of system X-c in both MEF lines, we transfected S/S MEFs with the 4700-bp xCT promoter luciferase construct together with ATF4 siRNA and A/A MEFs with the 4700-bp xCT promoter luciferase construct together with a vector expressing ATF4 without the 5′UTR. In S/S MEFs, knockdown of ATF4 resulted in an ∼80% decrease in xCT promoter activity, whereas transfection of A/A MEFs with ATF4 increased xCT promoter activity to levels similar to those in S/S MEFs (Fig. 2G).
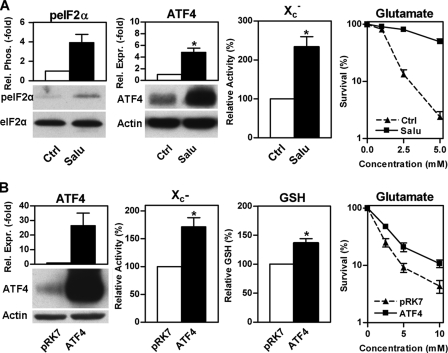
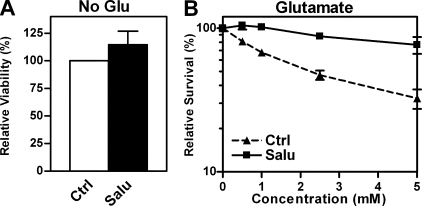
The hippocampal nerve cell line HT22 is widely used as a model for studying the response of nerve cells to oxidative stress (17). A decrease in total eIF2α increases the resistance of these cells against oxidative glutamate toxicity (oxytosis) as well as other oxidants such as hydrogen peroxide (12). Thus, we asked whether the pathway identified in MEFs is also active in HT22 cells. Induction of eIF2α phosphorylation using the specific eIF2α phosphatase inhibitor, salubrinal (45), increased ATF4 levels, system X-c activity, and resistance against oxidative glutamate toxicity in the HT22 cells (Fig. 3A). xCT protein levels remained too low to be detectable. As ATF4 is not the only protein preferentially translated under conditions of increased eIF2α phosphorylation, we asked whether the effects of salubrinal on system X-c activity and resistance against oxidative glutamate toxicity are mediated by ATF4. Transfection of HT22 cells with ATF4 also increased system X-c activity, cellular GSH, and resistance against oxidative glutamate toxicity (Fig. 3B). However, the effects of overexpression of ATF4 were not as robust as those of salubrinal on glutamate resistance suggesting that eIF2α phosphorylation may have some additional actions that are mediated independently of ATF4. We have already demonstrated that transfection with xCT alone produces similar changes (41). Thus, eIF2α phosphorylation leads to the translational up-regulation of ATF4, which transcriptionally increases xCT levels and subsequently system X-c activity, and this chain of events is sufficient for the modulation of GSH levels and an increase in the resistance of the cells to stress. Next we asked whether immature primary cortical neurons, which are also highly sensitive to oxidative glutamate toxicity (46), behave similarly as the HT22 cells in response to salubrinal. Salubrinal treatment was not only nontoxic to primary neurons (Fig. 4A) but also provided significant protection against glutamate treatment in this model (Fig. 4B).
FIGURE 3.
eIF2α phosphorylation increases system X-c activity and protects against oxidative glutamate toxicity via ATF4. A, 3.3 × 105 HT22 cells were plated in 60-mm dishes treated with 30 μm salubrinal (Salu) or vehicle (Ctrl) for 24 h. Similar amounts of whole cell lysates were analyzed by Western blotting for eIF2α phosphorylation (peIF2α) and ATF4 expression (ATF4). Antibodies recognizing total eIF2α (eIF2α) or actin served as loading controls. System X-c activity (X -c) was measured using Na+-independent, HCA-sensitive [3H]glutamate uptake in HT22 cells plated at a density of 2.5 × 104 cells/well in 24-well plates and treated with 30 μm salubrinal (Salu) or vehicle (Ctrl) for 24 h. For oxidative glutamate toxicity (right panel), glutamate at the indicated concentrations was added to 2.5 × 103 HT22 cells grown for 24 h with (Salu) or without (Ctrl) 30 μm salubrinal. Survival was measured by the MTT assay. B, 4.4 × 105 HT22 cells grown for 24 h were transfected with either empty vector (pRK7) or vector expressing ATF4 (ATF4). After another 24 h, cells were replated into 100-mm dishes (8.8 × 105 cells/dish) for Western blot analysis of ATF4 expression (ATF4, left panel), into 24-well plates (3 × 104/well) for system X-c activity (X -c) measured as glutamate-sensitive [35S]cystine uptake, into 60-mm dishes (1.65 × 105/dish) for GSH measurement (GSH) or into 96-well plates (2.5 × 103/well) for oxidative glutamate toxicity assays (Glutamate). Assays and treatment with glutamate were performed after 24 h. Survival was measured by the MTT assay after 24 h of glutamate treatment. All graphs represent the data from three independent experiments. For protein expression or phosphorylation, relative eIF2α phosphorylation or ATF4 expression of control cells was normalized to 1. System X-c activity and GSH were normalized to 100% in control cells. Statistical analysis of these experiments was performed by one-sample t test compared with 1 and 100, respectively; *, p < 0.05. Relative glutamate sensitivity was analyzed by two-way ANOVA. Cells treated with salubrinal or transfected with ATF4 were significantly more resistant to glutamate (column factor, p < 0.0001).
FIGURE 4.
Salubrinal is a nontoxic mediator of protection against oxidative glutamate toxicity in immature cortical neurons. 1 × 105 primary cortical neurons in 96-well plates were grown for 24 h with (Salu) or without (Ctrl) 30 μm salubrinal and then treated with glutamate at the indicated concentrations or without glutamate for another 24 h followed by an MTT assay. A, viability in the absence of glutamate and salubrinal was normalized to 100%. B, viability without glutamate with or without salubrinal was normalized to 100%. Graphs represent means of four independent experiments each done in triplicate. Statistical analysis was performed by one-sample t test compared with 100 (A) or two-way ANOVA (B). Viability of cultures treated with salubrinal alone was not different from control cells, whereas cells treated with salubrinal were significantly more resistant to glutamate (column factor, p < 0.0001).
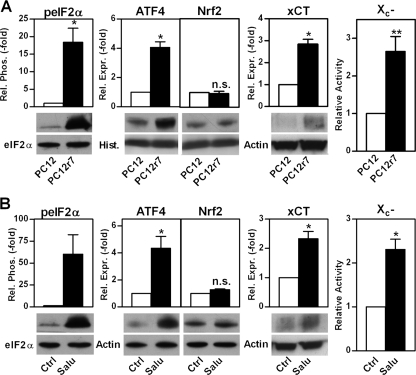
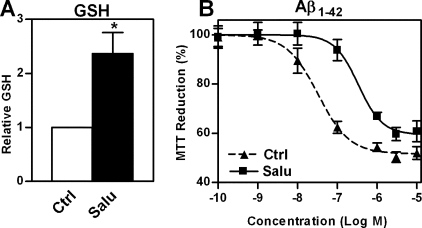
Because among the stressors to which the A/A MEFs showed increased sensitivity was the Aβ peptide, we next asked if nerve cells selected for Aβ resistance display increased levels of eIF2α phosphorylation. Indeed, clonal Aβ-resistant PC12 cells, PC12r7, which have been described previously (27–30), showed increased eIF2α phosphorylation even when grown in the absence of Aβ, and this was associated with increased levels of ATF4, but not Nrf2, xCT protein, and system X-c activity (Fig. 5A). An increased capacity for GSH recycling mediated by hypoxia-inducible factor 1β has already been described in PC12r7 (29), indicating that multiple mechanisms for increasing GSH metabolism are activated in these cells. Thus, we asked whether the increase in ATF4 and xCT via eIF2α phosphorylation is sufficient to increase GSH and reduce the response to Aβ. To specifically modulate the above described pathway in wild-type PC12 cells, we again employed salubrinal. As expected, treatment of wild-type PC12 cells for 48 h with 30 μm salubrinal significantly increased eIF2α phosphorylation, ATF4, xCT and system X-c function, whereas Nrf2 levels remained unchanged (Fig. 5B). Salubrinal treatment also increased GSH levels more than 2-fold (Fig. 6A).
FIGURE 5.
Up-regulation of system X-c activity, ATF4 protein, and eIF2α phosphorylation but not Nrf2 protein in Aβ-resistant and salubrinal-treated PC12 cells. A and B, for protein, 1.2 × 106 wild-type PC12 cells (PC12) and 6 × 105 clonal Aβ-resistant PC12 cells (PC12r7) were plated in 100-mm dishes and grown for 48 h. 30 μm salubrinal (Salu) or vehicle (Ctrl) was added at the time of plating in B. Western blots of whole cell extracts were analyzed for eIF2α phosphorylation using a phospho-specific anti-eIF2α antibody and a total eIF2α antibody as a loading control. For ATF4, Nrf2, and xCT expression, cells were fractionated. Similar amounts of nuclear extracts for ATF4 and Nrf2 or membrane protein for xCT were analyzed by Western blotting using specific antisera. Actin served as loading control with the exception of the nuclear extracts that were normalized to histone (Hist.). System X-c activity (X -c) was measured as glutamate-sensitive [35S]cystine uptake in cells plated at a density of 3 × 105 cells/well in 24-well dishes and grown for 48 h. Graphs represent quantified blots of three (A) and four (B) independent protein preparations and four (A) and three (B) independent uptake assays normalized to control conditions and shown as mean ± S.E. For statistical analysis, values for PC12r7 or salubrinal-treated PC12 cells were compared with 1 by one-sample t test, **, p < 0.01; *, p < 0.05; n.s., not significant.
FIGURE 6.
Salubrinal treatment of PC12 cells increases cellular GSH and protects against Aβ toxicity. A, 4.5 × 105 PC12 cells were plated in 60-mm dishes and grown for 48 h with 30 μm salubrinal (Salu) or vehicle (Ctrl). Cellular GSH was measured enzymatically and normalized to protein. The graph represents the mean of four experiments with the GSH levels of controls normalized to 1. Statistical analysis of values with salubrinal was performed by one-sample t test compared with 1. *, p < 0.05. B, 2 × 105 PC12 cells per well were plated into 96-well plates with either 30 μm salubrinal (Salu) or vehicle (Ctrl). After 24 h, Aβ-(1–42) was added at the indicated concentrations. MTT reduction was measured after 48 h of Aβ exposure. Each experiment was performed in triplicate, and the mean MTT value of cells without Aβ was normalized to 100%. Each data point represents the mean ± S.E. of three independent experiments. Statistical analysis was performed by nonlinear regression and F test. EC50 values of Aβ for MTT reduction are 3.3 × 10-8 and 3.3 × 10-7 m for control and salubrinal treated PC12 cells, respectively, and significantly different, p < 0.001.
The earliest and most robust sign of Aβ toxicity in cells is the loss of their ability to reduce MTT to MTT formazan (47, 48). However, MTT reduction is unaffected by Aβ in Aβ-resistant PC12 cells (27). Similarly, salubrinal treatment significantly shifted the EC50 of Aβ on MTT reduction to about 10-fold higher concentrations in wild-type PC12 cells (Fig. 6B).
In summary, eIF2α phosphorylation increases ATF4 and subsequently xCT expression in four different cellular models in two species. Thus, we asked whether the AAREs that have been described for the mouse xCT promoter (42) are evolutionarily conserved among species. Comparison of the 5′-flanking regions of the mouse, rat, human, and bovine xCT genes confirmed a 100% identity of the stretch containing the two AAREs from -94 to -68 bp (supplemental Fig. S1).
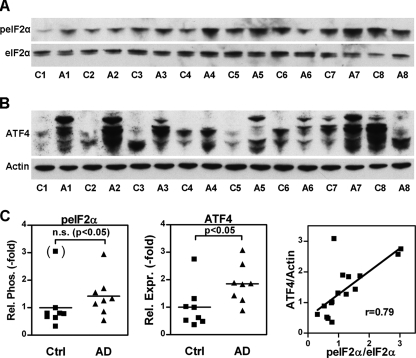
Increased eIF2α phosphorylation has been reported in the brains of transgenic mouse models of AD (24) and in human AD brains (23–25). These findings were generally interpreted as indicative of eIF2α phosphorylation-dependent programmed cell death. Alternatively, these changes might be similar to the changes seen in PC12 cells selected for Aβ resistance and thus represent an adaptive survival response. If so, we would expect that ATF4 is up-regulated in human AD brain in concert with eIF2α phosphorylation. To test this idea, we examined human frontal cortex for changes in the relative phosphorylation of eIF2α and expression of ATF4 related to AD. Western blotting of protein extracts from eight individuals with AD and age-matched controls were in line with previous reports of increased eIF2α phosphorylation (23–25) (Fig. 7, A–C). In addition, we found a significant 1.9-fold increase in cortical ATF4 levels in AD brains (Fig. 7, B and C). In the whole group of samples, the individual levels of ATF4 protein correlated linearly with relative eIF2α phosphorylation (Fig. 7C). These results strongly suggest that the phospho-eIF2α/ATF4 module is functional in human brain and is activated in AD. It was not possible to detect xCT in these samples as we did not have enough material to do membrane preparations that are essential for successful detection of xCT.
FIGURE 7.
The eIF2α phosphorylation and ATF4 translation signaling pathway in AD brains. 2% SDS extracts of midfrontal cortex of eight brains of individuals with histopathologically confirmed Alzheimer dementia (A1–A8) and eight age-matched control brains (C1–C8) were analyzed by Western blotting with antibodies against phosphorylated eIF2α (peIF2α) and an antibody to total eIF2α (A) and antibodies against ATF4 and actin (B). A and B show a representative blot of three independently performed Western blots. Each membrane was re-probed with antibodies against actin. C, for quantitative analysis of peIF2α and ATF4 expression, mean band density of controls was normalized to 1. For eIF2α and actin, mean band density of all samples was normalized to 1. For relative eIF2α phosphorylation, mean normalized peIF2α and eIF2α band density for each sample was normalized by mean normalized actin band density of the same sample. Each individual data point in C represents the relative phosphorylation as the ratio of the mean actin-normalized peIF2α band density and the mean actin-normalized eIF2α band density (left panel) or the relative ATF4 expression as the ratio of mean normalized ATF4 band density and mean normalized actin band density of three independent Western blots. Normality was tested by D'Agostino and Pearson omnibus normality test. Mean relative eIF2α phosphorylation (peIF2α) in AD brains (AD) was slightly (1.4-fold) higher than mean eIF2α phosphorylation in control brains (Ctrl). The difference was not significantly different (p = 0.083, Mann-Whitney test) unless C8, which fulfilled the criteria of an extreme outlier (3.04, Q3 + 3xIQR, 1/62), was excluded (p < 0.05, Mann-Whitney test). Mean relative ATF4 expression was higher in AD brains as compared with controls (1.9-fold, p < 0.05, Mann-Whitney test). There was a highly significant correlation of relative eIF2α phosphorylation and ATF4 expression in the whole group of samples (Spearman r 0.79, p < 0.001).
DISCUSSION
The response of cells to different forms of stress can elicit two diametrically opposed consequences, adaptation or death. Thus, when examining post-mortem tissue, especially from animals or humans suffering from a chronic disease, it is often very difficult to judge whether a specific change is associated with the adaptation of the surviving cells or their on-going death. Even in cell culture models, it can be difficult to determine whether a specific biochemical change is associated with the promotion of cell survival or cell death, particularly if there is a graded effect leading from adaptation to death.
Among the pathways about which a good deal of controversy exists is that involving the translation initiation factor eIF2α. Several studies have suggested that increased levels of eIF2α phosphorylation observed in nerve cells in response to acute toxic insults such as 6-hydroxydopamine (49) and Aβ peptide (50) as well as chronic increases detected in AD (23–25) reflect a contribution of eIF2α phosphorylation to nerve cell death. In contrast, we demonstrate that long term eIF2α phosphorylation mediates increased stress resistance in five different cell-based paradigms. First, MEFs, which are defective for eIF2α phosphorylation, are more sensitive to various acute toxic insults. Second, salubrinal-stimulated eIF2α phosphorylation induces resistance against oxidative glutamate toxicity in hippocampal HT22 cells and third in immature primary cortical neurons. Fourth, clonal PC12 cells selected for resistance against Aβ peptide, a causative factor in the development of AD, have increased basal eIF2α phosphorylation. Fifth, salubrinal-stimulated eIF2α phosphorylation diminishes the toxic effect of the Aβ peptide in wild-type PC12 cells.
We reported previously that mimicking increased eIF2α phosphorylation increases GSH levels in HT22 cells (12). In this study, we extend that observation by demonstrating the corresponding changes in fibroblasts defective for eIF2α phosphorylation, which have lower GSH, and PC12 cells treated with salubrinal, which show a greater than 2-fold increase in GSH. Thus, we conclude that the positive correlation between stress resistance and eIF2α phosphorylation is a common phenomenon and is linked to strengthened cellular antioxidant capacity by increased GSH levels.
This conclusion is supported by a number of published reports. For example, a recent study demonstrated that treatment of rats with salubrinal significantly reduced the nerve cell death seen following treatment with kainic acid (51). Long term systemic administration of salubrinal also minimized hypoglossal and facial motor neuron damage in a murine model of sleep apnea (52). In addition, mutations in the AD-associated protein presenilin-1 can decrease eIF2α phosphorylation in nerve cells in culture (53), and expression of mutant PS-1 in mutant Alzheimer precursor protein-expressing mice is known to lead to a significantly accelerated pathology (54). Finally, eIF2α phosphorylation decreases in many tissues, including the brain, during aging, and this decrease is associated with an increase in the levels of pro-apoptotic proteins (8).
eIF2α phosphorylation is known to positively regulate the levels of the transcription factor ATF4 (55). The molecular basis for this consists of regulated translation initiation via small open reading frames within the 5′UTR of ATF4 and is highly conserved across species from Aplysia to humans (1). Other proteins induced by eIF2α phosphorylation via similar or closely related mechanisms include the lysine/arginine transporter Cat-1 (56) and nuclear factor ATF5 (57).
ATF4 was shown recently to induce the expression of the specific subunit, xCT, of the glutamate/cystine antiporter system X-c in response to cystine starvation via interaction with an amino acid-response element within the proximal xCT gene promoter region (42). Our data strongly suggest that decreased ATF4 levels in fibroblasts defective for eIF2α phosphorylation mediate decreased xCT expression and system X-c activity via the AARE. Furthermore, our data indicate that basal xCT expression is determined by ATF4, whereas Nrf2, a well known inducer of xCT (43, 58), only plays the role of an additional multiplier of basal expression levels as relative increases in system X-c activity via Nrf2 induction were similar in A/A and S/S MEFs. Furthermore, we show that ATF4 is sufficient to mediate the changes in xCT expression, GSH content, and stress resistance as ATF4 transfection of HT22 cells mimics the effects of increased eIF2α phosphorylation. We already reported that transfection with xCT alone also induces increases in GSH and resistance to oxidative glutamate toxicity in HT22 cells (41). Thus, our data strongly support the idea that a signaling module consisting of eIF2α phosphorylation, ATF4, and xCT regulates cysteine availability and subsequently cellular GSH synthesis and antioxidant capacity in non-neuronal and neuronal cells in vitro. Furthermore, we show that the molecular basis for the ATF4-dependent xCT induction is completely conserved across four mammalian species from mice to humans, supporting the idea that not only the phospho-eIF2α/ATF4 module (1) but also the ATF4/xCT module is evolutionarily conserved.
Although a recent study using ATF4 knock-out mice suggested that ATF4 promoted cell death (59), these results may rather be more indicative of the consequences of an adaptive response to chronic ATF4 depletion. Mice deficient in ATF4 are >50% smaller than their wild-type counterparts and suffer from a variety of developmental defects (60–62).
Aβ, the peptide involved in the pathophysiology of AD, induces oxidative stress in vitro (27). Correspondingly, markers of oxidative stress are also enhanced in AD brains (for review see Refs. 63, 64). We show that the phospho-eIF2α/ATF4/xCT module is activated in resistance against Aβ and regulates toxic cellular responses to Aβ in fibroblasts and PC12 cells. This observation is consistent with previous studies (40) that have shown that Aβ treatment of fibroblasts can elicit a variety of changes that are associated with AD. Increased eIF2α phosphorylation in AD brains and murine AD models has been reported by others (23–25). Because acute treatment with Aβ or transfection with mutant Alzheimer precursor protein induces eIF2α phosphorylation as well as cell death (24, 65), AD-associated eIF2α phosphorylation has been interpreted as an expression of cellular damage and cell death. However, after chronic neurodegeneration over many years, an AD brain consists mainly of cells that have escaped neurodegeneration. This observation coupled with our findings that indicate that the phospho-eIF2α/ATF4 module is activated in AD brains strongly contradicts the idea that increased eIF2α phosphorylation is associated with cell death. Indeed, in light of our in vitro data, we believe that these changes should rather be interpreted as an adaptive neuroprotective response.
Several lines of evidence suggest that the early clinical hall-mark of AD, failure of memory, might, at least in part, result from dysfunction rather than loss of neurons (for review see Ref. 66). Most interestingly, recent reports demonstrate that eIF2α phosphorylation negatively regulates memory formation as well as long term potentiation, the proposed physiological correlate of memory, in mice (11). This mechanism seems to be mediated via AFT4 rather than by decreased protein synthesis. Thus, it seems possible that defective memory formation in AD might be accelerated by the protective response of the brain against Aβ-induced oxidative stress.
As AD is relentlessly progressive and finally fatal, this protective response ultimately fails. Of note, increased system X-c activity can be a double-edged sword as it inevitably mediates the release of glutamate (16), which can be toxic to neurons. Normally, extracellular glutamate is rapidly taken up by astrocytic glutamate transporters in the brain (67). Recent findings in ischemic brain tissue as well as in astrocytomas invading brain tissue suggest that up-regulated system X-c activity with concomitant down-regulation of glutamate transporters might increase neuronal loss (68, 69). Down-regulation of astrocytic glutamate transporters has been demonstrated repeatedly in AD (70, 71). Thus, as a detrimental side effect, activation of the phospho-eIF2α/ATF4/xCT module might even accelerate the selective neuronal loss observed in AD.
Supplementary Material
Acknowledgments
We thank Dr. Hideyo Sato (Yamagata University, Tsuruoka, Japan) for the xCT promoter constructs, Dr. Sylvia Smith (Medical College of Georgia, Augusta, GA) for the xCT antibody, Drs. Randal J. Kaufman and Donalyn Scheuner (Howard Hughes Medical Institute, University of Michigan, Ann Arbor, MI) for the S/S and A/A mouse embryonic fibroblasts, and Dr. David Ron (Skirball Institute, New York University School of Medicine) for the ATF4 5′-UTR and ATF4 plasmids. We also acknowledge the assistance of Dr. Chi-un Choe and Doerthe Behrends (University Medical Center Hamburg-Eppendorf, Hamburg, Germany) for their help in the preparation of the primary cortical cultures.
This work was supported, in whole or in part, by National Institutes of Health Grant AG025337 (to P. M.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: AD, Alzheimer disease; Aβ, amyloid β; UTR, untranslated region; ANOVA, analysis of variance; MEF, mouse embryonic fibroblast; DMEM, Dulbecco's modified Eagle's medium; siRNA, short interfering RNA; BCNU, 1,3-bis(2-chloroethyl)-N-nitrosourea; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; HBSS, Hanks' balanced salt solution; HCA, homocysteic acid; TBS, Tris-buffered saline; AARE, amino acid-response element; tBHQ, tert-butylhydroquinone.
J. Lewerenz, unpublished observations.
References
- 1.Wek, R. C., Jiang, H.-Y., and Anthony, T. G. (2006) Biochem. Soc. Trans. 34 7-11 [DOI] [PubMed] [Google Scholar]
- 2.Proud, C. G. (2005) Semin. Cell Dev. Biol. 16 3-12 [DOI] [PubMed] [Google Scholar]
- 3.DeGracia, D. J., Kumar, R., Owen, C. R., Krause, G. S., and White, B. C. (2002) J. Cereb. Blood Flow Metab. 22 127-141 [DOI] [PubMed] [Google Scholar]
- 4.Novoa, I., Zeng, H., Harding, H. P., and Ron, D. (2001) J. Cell Biol. 153 1011-1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin, J. H., Li, H., Yasumura, D., Cohen, H. R., Zhang, C., Panning, B., Shokat, K. M., Lavail, M. M., and Walter, P. (2007) Science 318 944-949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scheuner, D., Van der Mierde, D., Song, B., Flamez, D., Creemers, J. W., Tsukamoto, K., Ribick, M., Schuit, F. C., and Kaufman, R. J. (2005) Nat. Med. 11 757-764 [DOI] [PubMed] [Google Scholar]
- 7.Gietzen, D. W., Ross, C. M., Hao, S., and Sharp, J. W. (2004) J. Nutr. 134 717-723 [DOI] [PubMed] [Google Scholar]
- 8.Hussain, S. G., and Ramaiah, K. V. A. (2007) Biochem. Biophys. Res. Commun. 355 365-370 [DOI] [PubMed] [Google Scholar]
- 9.Grallert, B., and Boye, E. (2007) Cell Cycle 6 2768-2772 [DOI] [PubMed] [Google Scholar]
- 10.Scheuner, D., Song, B., McEwan, E., Liu, C., Laybutt, R., Gillespie, P., Saunders, T., Bonner-Weir, S., and Kaufman, R. J. (2001) Mol. Cell 7 1165-1176 [DOI] [PubMed] [Google Scholar]
- 11.Costa-Mattioli, M., Gobert, D., Stern, E., Gamache, K., Colina, R., Cuello, C., Sossin, W., Kaufman, R. J., Pelletier, J., Rosenblum, K., Krnjevic, K., Lacaille, J. C., Nader, K., and Sonenberg, N. (2007) Cell 129 195-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan, S., Somia, N., Maher, P., and Schubert, P. (2001) J. Cell Biol. 152 997-1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu, P. D., Jousse, C., Marciniak, S. J., Zhang, Y., Novoa, I., Scheuner, D., Kaufman, R. J., Ron, D., and Harding, H. P. (2004) EMBO J. 23 169-179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maher, P. (2005) Ageing Res. Rev. 4 288-314 [DOI] [PubMed] [Google Scholar]
- 15.Sato, H., Tamba, M., Ishii, T., and Bannai, S. (1999) J. Biol. Chem. 274 11455-11458 [DOI] [PubMed] [Google Scholar]
- 16.Bannai, S. (1986) J. Biol. Chem. 261 2256-2263 [PubMed] [Google Scholar]
- 17.Tan, S., Schubert, D., and Maher, P. (2001) Curr. Top. Med. Chem. 1 497-506 [DOI] [PubMed] [Google Scholar]
- 18.Baker, D. A., McFarland, K., Lake, R. W., Shen, H., Tang, X.-C., Toda, S., and Kalivas, P. W. (2003) Nat. Neurosci. 6 743-749 [DOI] [PubMed] [Google Scholar]
- 19.Burdo, J., Dargusch, R., and Schubert, D. (2006) J. Histochem. Cytochem. 54 549-557 [DOI] [PubMed] [Google Scholar]
- 20.Shih, A. Y., Erb, H., Sun, X., Toda, S., Kalivas, P., and Murphy, T. H. (2006) J. Neurosci. 26 10514-10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.La Bella, V., Valentino, F., Piccoli, T., and Piccoli, F. (2007) Neurochem. Res. 32 1081-1090 [DOI] [PubMed] [Google Scholar]
- 22.Sato, H., Shiiya, A., Kimata, M., Maebara, K., Tamba, M., Sakakura, Y., Makino, N., Sugiyama, F., Yagami, K., Moriguchi, T., Takahashi, S., and Bannai, S. (2005) J. Biol. Chem. 280 37423-37429 [DOI] [PubMed] [Google Scholar]
- 23.Chang, R. C., Wong, A. K., Ng, H. K., and Hugon, J. (2002) Neuroreport 13 2429-2432 [DOI] [PubMed] [Google Scholar]
- 24.Kim, H. S., Choi, Y. J., Shin, K. Y., Joo, Y., Lee, Y. K., Jung, S. Y., Suh, Y. H., and Kim, J. H. (2007) J. Neurosci. Res. 85 1528-1537 [DOI] [PubMed] [Google Scholar]
- 25.Unterberger, U., Hoftberger, R., Gelpi, E., Flicker, H., Budka, H., and Voigtlander, T. (2006) J. Neuropathol. Exp. Neurol. 65 348-357 [DOI] [PubMed] [Google Scholar]
- 26.Page, G., A., Rioux, B., Ingrand, S., Lafay-Chebassier, C., Pain, S., Perault Pochat, M. C., Bouras, C., Bayer, T., and Hugon, J. (2006) Neuroscience 139 1343-1354 [DOI] [PubMed] [Google Scholar]
- 27.Behl, C., Davis, J. B., Lesley, R., and Schubert, D. (1994) Cell 77 817-822 [DOI] [PubMed] [Google Scholar]
- 28.Sagara, Y., Dargusch, R., Klier, F. G., Schubert, D., and Behl, C. (1996) J. Neurosci. 16 497-505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soucek, T., Cumming, R., Dargusch, R., Maher, P., and Schubert, D. (2003) Neuron 39 43-56 [DOI] [PubMed] [Google Scholar]
- 30.Cumming, R. C., Dargusch, R., Fischer, W. H., and Schubert, D. (2007) J. Biol. Chem. 282 30523-30534 [DOI] [PubMed] [Google Scholar]
- 31.Davis, J. B., and Maher, P. (1994) Brain Res. 652 169-173 [DOI] [PubMed] [Google Scholar]
- 32.Lewerenz, J., Letz, J., and Methner, A. (2003) J. Neurochem. 87 522-531 [DOI] [PubMed] [Google Scholar]
- 33.Schubert, D., and Piasecki, D. (2001) J. Neurosci. 21 7455-7462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maher, P., Lewerenz, J., Lozano, C., and Torres, J. L. (2008) J. Neurochem. 107 690-700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cordey, M., and Pike, C. J. (2006) J. Neurochem. 96 204-217 [DOI] [PubMed] [Google Scholar]
- 36.Griffith, O. W., and Meister, A. (1979) J. Biol. Chem. 254 7558-7560 [PubMed] [Google Scholar]
- 37.Burdo, J., Schubert, D., and Maher, P. (2008) Brain Res. 1189 12-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schafer, F. Q., and Buettner, G. R. (2001) Free Radic. Biol. Med. 30 1191-1212 [DOI] [PubMed] [Google Scholar]
- 39.Ravindranath, V., and Reed, D. J. (1990) Biochem. Biophys. Res. Commun. 169 1075-1079 [DOI] [PubMed] [Google Scholar]
- 40.Khan, T. K., and Alkon, D. L. (2008) Neurobiol. Aging, in press [DOI] [PubMed]
- 41.Lewerenz, J., Klein, M., and Methner, A. (2006) J. Neurochem. 98 916-925 [DOI] [PubMed] [Google Scholar]
- 42.Sato, H., Nomura, S., Maebara, K., Sato, K., Tamba, M., and Bannai, S. (2004) Biochem. Biophys. Res. Commun. 325 109-116 [DOI] [PubMed] [Google Scholar]
- 43.Sasaki, H., Sato, H., Kuriyama-Matsumura, K., Sato, K., Maebara, K., Wang, H., Tamba, M., Itoh, K., Yamamoto, M., and Bannai, S. (2002) J. Biol. Chem. 277 44765-44771 [DOI] [PubMed] [Google Scholar]
- 44.Leclerc, G. M., Boockfor, F. R., Faught, W. J., and Frawley, L. S. (2000) BioTechniques 29 590-598 [DOI] [PubMed] [Google Scholar]
- 45.Boyce, M., Bryant, K. F., Jousse, C., Long, K., Harding, H. P., Scheuner, D., Kaufman, R. J., Ma, D., Coen, D. M., Ron, D., and Yuan, J. (2005) Science 307 935-939 [DOI] [PubMed] [Google Scholar]
- 46.Murphy, T. H., Schnaar, R. L., and Coyle, J. T. (1990) FASEB J. 4 1624-1633 [PubMed] [Google Scholar]
- 47.Abe, K., and Kimura, H. (1996) J. Neurochem. 67 2074-2078 [PubMed] [Google Scholar]
- 48.Liu, Y., and Schubert, D. (1997) J. Neurochem. 69 2285-2293 [DOI] [PubMed] [Google Scholar]
- 49.Ryu, E. J., Harding, H. P., Angelastro, J. M., Vitolo, O. V., Ron, D., and Greene, L. A. (2002) J. Neurosci. 22 10690-10698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang, R. C.-C., Suen, K.-C., Ma, C.-H., Elyaman, W., Ng, H.-K., and Hugon, J. (2002) J. Neurochem. 83 1215-1225 [DOI] [PubMed] [Google Scholar]
- 51.Sokka, A.-L., Putkonen, N., Mudo, G., Pryazhnikov, E., Reijonen, S., Khiroug, L., Belluardo, N., Lindholm, D., and Korhonen, L. (2007) J. Neurosci. 27 901-908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu, Y., Fenik, P., Zhan, G., Sanfillipo-Cohn, B., Naidoo, N., and Veasay, S. C. (2008) J. Neurosci. 28 2168-2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kudo, T., Katayama, T., Imaizumi, K., Yasuda, Y., Yatera, M., Okochi, M., Tohyama, M., and Takeda, M. (2002) Ann. N. Y. Acad. Sci. 977 349-355 [DOI] [PubMed] [Google Scholar]
- 54.Eriksen, J. L., and Janus, C. G. (2007) Behav. Genet. 37 79-100 [DOI] [PubMed] [Google Scholar]
- 55.Harding, H. P., Zhang, Y., Zeng, H., Novoa, I., Lu, P. D., Calfon, M., Sadri, N., Yun, C., Popko, B., Paules, R., Stojdl, D. F., Bell, J. C., Hettmann, T., Leiden, J. M., and Ron, D. (2003) Mol. Cell 11 619-633 [DOI] [PubMed] [Google Scholar]
- 56.Yaman, I., Fernandez, J., Liu, H., Caprara, M., Komar, A. A., Koromilas, A. E., Zhou, L., Snider, M. D., Scheuner, D., Kaufman, R. J., and Harzoglou, M. (2003) Cell 113 519-531 [DOI] [PubMed] [Google Scholar]
- 57.Zhou, D., Palam, L. R., Jiang, L., Narasimhan, J., Staschke, K. A., and Wek, R. C. (2008) J. Biol. Chem. 283 7064-7073 [DOI] [PubMed] [Google Scholar]
- 58.Shih, A. Y., Johnson, D. A., Wong, G., Kraft, A. D., Jiang, L., Erb, H., Johnson, J. A., and Murphy, T. H. (2003) J. Neurosci. 23 3394-3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lange, P. S., Chavez, J. C., Pinto, J. T., Coppola, G., Sun, C.-W., Townes, T. M., Geschwind, D. H., and Ratan, R. R. (2008) J. Exp. Med. 205 1227-1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hettmann, T., Barton, K., and Leiden, J. M. (2000) Dev. Biol. 222 110-123 [DOI] [PubMed] [Google Scholar]
- 61.Matsuoka, H. C., and Townes, T. M. (2002) Blood 99 736-745 [DOI] [PubMed] [Google Scholar]
- 62.Yang, X., Matsuda, K., Bialek, P., Jacquot, S., Masuoka, H. C., Schinke, T., Li, L., Brancorsini, S., Sassone-Corsi, P., Townes, T. M., Hanauer, A., and Karsenty, G. (2004) Cell 117 387-398 [DOI] [PubMed] [Google Scholar]
- 63.Sonnen, J. A., Breitner, J. C., Lovell, M. A., Markesbery, W. R., Quinn, J. F., and Montine, T. J. (2008) Free Radic. Biol. Med. 45 219-230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Butterfield, D. A., Reed, T., Newman, S. F., and Sultana, R. (2007) Free Radic. Biol. Med. 43 658-677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suen, K.-C., Yu, M.-S., So, K.-F., Chang, R. C.-C., and Hugon, J. (2003) J. Biol. Chem. 278 49819-49827 [DOI] [PubMed] [Google Scholar]
- 66.Venkitaramani, D. V., Chin, J., Netzer, W. J., Gouras, G. K., Lesne, S., Malinow, R., and Lombroso, P. J. (2007) J. Neurosci. 27 11832-11837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kanai, Y., and Hediger, M. A. (2004) Pfluegers Arch. 447 469-479 [DOI] [PubMed] [Google Scholar]
- 68.Fogal, B., Li, J., Lobner, D., McCullough, L. D., and Hewett, S. J. (2007) J. Neurosci. 27 10094-10105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sontheimer, H. (2003) Trends Neurosci. 26 543-549 [DOI] [PubMed] [Google Scholar]
- 70.Li, S., Mallory, M., Alford, M., Tanaka, S., and Masliah, E. (1997) J. Neuropathol. Exp. Neurol. 56 901-911 [DOI] [PubMed] [Google Scholar]
- 71.Jacob, C. P., Koutsilieri, E., Bartl, J., Neuen-Jacob, E., Arzberger, T., Zander, N., Ravid, R., Roggendorf, W., Riederer, P., and Grunblatt, E. (2007) J. Alzheimer Dis. 11 97-116 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.