Abstract
Gram-negative bacteria utilize specialized machinery to translocate drugs and protein toxins across the inner and outer membranes, consisting of a tripartite complex composed of an inner membrane secondary or primary active transporter (IMP), a periplasmic membrane fusion protein, and an outer membrane channel. We have investigated the assembly and function of the MacAB/TolC system that confers resistance to macrolides in Escherichia coli. The membrane fusion protein MacA not only stabilizes the tripartite assembly by interacting with both the inner membrane protein MacB and the outer membrane protein TolC, but also has a role in regulating the function of MacB, apparently increasing its affinity for both erythromycin and ATP. Analysis of the kinetic behavior of ATP hydrolysis indicated that MacA promotes and stabilizes the ATP-binding form of the MacB transporter. For the first time, we have established unambiguously the dimeric nature of a noncanonic ABC transporter, MacB that has an N-terminal nucleotide binding domain, by means of nondissociating mass spectrometry, analytical ultracentrifugation, and atomic force microscopy. Structural studies of ABC transporters indicate that ATP is bound between a pair of nucleotide binding domains to stabilize a conformation in which the substrate-binding site is outward-facing. Consequently, our data suggest that in the presence of ATP the same conformation of MacB is promoted and stabilized by MacA. Thus, MacA would facilitate the delivery of drugs by MacB to TolC by enhancing the binding of drugs to it and inducing a conformation of MacB that is primed and competent for binding TolC. Our structural studies are an important first step in understanding how the tripartite complex is assembled.
Gram-negative bacteria utilize transport systems composed of a tripartite assembly of proteins that span both the inner and outer membranes to pump cytotoxic compounds, such as antibiotics (1), and protein toxins (2), such as α-hemolysin, from the cell. These assemblies are composed of inner (IMP)7 and outer (OMP) membrane proteins that are connected by a periplasmic membrane fusion protein (MFP), which is anchored to the inner membrane. The systems responsible for toxin extrusion invariably utilize an ABC transporter as the IMP (2), whereas those involved in antibiotic extrusion largely utilize proton antiporters (1), but there are some that utilize ABC transporters (3). The same OMP can be utilized for both drug and toxin extrusion; for example, TolC functions with antibiotic H+ antiporters, such as AcrB that belongs to the regulation nodulation cell division family of transporters (4, 5), with ABC transporters, such as the macrolide transporter MacB (3), and with HlyB that extrudes the protein toxin α-hemolysin (2).
Although the structure of an assembled tripartite complex has not yet been determined, the structures of a few individual components have been elucidated. Most significantly, the structure of the RND transporter AcrB (6, 7) and of its cognate MFP, AcrA (8), and OMP, TolC (9), have been determined. Both AcrB and TolC are organized as homotrimers. The AcrB trimer has a periplasmic headpiece formed by the loops between helices 1 and 2 and helices 7 and 8. The headpiece has a funnel-shaped internal cavity that is connected by a pore to a large central cavity formed between the periplasmic and membrane domains. The trimeric TolC forms a cylindrical channel with a structure that is arranged into two major domains as follows: a β-barrel in the outer membrane and a periplasmic α-helical barrel. It has been proposed, and is supported by cross-linking studies (10), that the six hairpins from the upper headpiece of the AcrB trimer contact the tips of the six helical pairs of the TolC trimer (11) to form a continuous path across the periplasmic space. The structure of AcrA revealed an elongated monomer that is composed of three subdomains as follows: a β-barrel, a lipoyl domain, and a 58-Å-long α-helical hairpin (8). AcrA has been reported to interact with both TolC, via the α-helical hairpin (12), and AcrB, via the β-domains (13), to stabilize the tripartite complex (4, 5).
Several lines of evidence suggest that assembly of the tripartite complex is a dynamic process that induces conformational changes in the protein components. The structures of a number of OMPs indicate that the channel is closed at one or both sides, requiring conformational changes to allow passage of the substrate (11-14). Several studies support the hypothesis that a small number of key interactions between adjacent coiled-coils of the OMP are broken to allow the inner coils to untwist and realign with the outer coils, thereby opening the OMP entrance aperture (11, 15-17). The conformational flexibility of AcrA may enable realignment of the coiled-coil to stabilize the open state of TolC (8, 11, 15). Recently, the structures of a number of different conformations of AcrB have been determined that support a mechanistic model in which there is a functional rotation in the periplasmic domains driving transfer of drugs into the OMP (18-21). The conformational asymmetry in the IMP is also reflected in the asymmetry in the open state of the OMP TolC (11). Such conformational changes in AcrB could be transmitted to TolC via AcrA. Furthermore, studies of the RND transporter AcrD from Escherichia coli, whose typical substrates, aminoglycosides, are not expected to diffuse spontaneously across the lipid bilayer, revealed that only when AcrD was reconstituted with AcrA was it able to take up aminoglycosides, implying that AcrA is needed to “activate” AcrD (22). Collectively, these findings suggest that the MFP supports conformational changes in both the IMP and OMP.
We have sought to further investigate the assembly of the tripartite complex using the MacABTolC system responsible for the extrusion of macrolides from E. coli (3). To date this is the only tripartite pump for antibiotics that has been shown to utilize an ABC transporter, MacB, which has a novel architecture, consisting of a four-helix transmembrane domain, an N-terminal cytoplasmic nucleotide binding domain (NBD), and a large periplasmic domain formed by the loops connecting helices 1 and 2 (3, 23). The fact that the ATPase activity of the transporter can be monitored, using purified proteins, provides an additional tool for elucidating how its activity is modulated by assembly and disassembly of the tripartite complex. Indeed, recent studies, reported while this work was in progress, established that the MFP MacA modulates the steady-state ATPase activity of the IMP MacB (24). Our studies have confirmed these findings and, by undertaking pre-steady-state analysis of the kinetics, extended them by identifying how MacA modulates the ATPase mechanism of MacB. In addition, we have established that MacA also increases the capacity of MacB for binding erythromycin. This behavior provides a rationale for the retention of MacA by related systems from Gram-positive bacteria, such as Staphylococcus aureus, which lack an outer membrane and OMP (supplemental Fig. 1). Such analyses will not only have a bearing on understanding the function of tripartite drug-pumps but also of related toxin transporters.
EXPERIMENTAL PROCEDURES
Strains and Plasmids—The E. coli strains and plasmids used are described in supplemental Table 1, and the primers used for construction of plasmid vectors are described in supplemental Table 2.
Protein Overexpression and Purification—All the proteins used in this study were purified as fusion proteins with a six-histidine tag from E. coli overexpressing strains according to the protocols given in the supplemental material.
Pulldown Assays—For pulldown assays, proteins were overexpressed with a C-terminal S-tag for use with the cognate protein that was His-tagged. Membrane pellets, dissolved in detergent, or cell supernatant containing the S-tagged prey protein were mixed with 2 mg of purified His-tagged bait-protein. The mixture was loaded onto a Ni2+-charged Hitrap chelating column (GE Healthcare), so that the bait-protein could be immobilized on the column along with the prey-protein if they interact. The column was then washed with 15-20 column volumes of Tris buffer containing 75-100 mm imidazole and Triton X-100 (0.2% (v/v)) to eliminate any false-positive results because of nonspecific interactions. The bait-prey protein complex was eluted with 500 mm imidazole Tris buffer, containing Triton X-100 (0.2% (v/v)), and SDS-PAGE was used to visualize the bait- and prey-proteins. A Western blot was performed with anti-S-tag antibodies to confirm the presence of the S-tagged prey-protein.
Growth Curve Analyses—E. coli cells, of strain KAM3(DE3), harboring plasmids were grown at 37 °C, with shaking at 200 rpm, until the cell density gave an A600 of 0.5, when 0.05-0.1 mm isopropyl 1-thio-β-d-galactopyranoside was added to the cells. The cells were grown for a further 3 h, when the cells were diluted with 2× YT media containing 100 μg/ml erythromycin, and the growth curve was recorded over the next 12 h. For experiments to measure the loss in growth because of erythromycin, cells were grown for 3 h in the absence and presence of 50 μg/ml erythromycin, and the growth loss is the ratio of the A600 values.
Analytical Ultracentrifugation—Sedimentation equilibrium measurements were performed using a Beckman Optima XL-A analytical ultracentrifuge equipped with both absorbance and interference optics. 100 μl of MacB in buffer 1 (20 mm Tris, pH 8.0, 150 mm NaCl, 1% w/v glycerol, and 0.006% (w/v) βDDM) supplemented with 10, 25, or 50% D2O was placed in the sample compartment of a Epon double-sector centerpiece, and 110 μl of buffer 1 was placed in the reference compartment. The final protein concentrations used in the runs were between 0.5 and 1.0 mg/ml. The D2O was used to match the density of the solvent to the density of the detergent as described previously (25) The samples were centrifuged at 283 K (10 °C) and 10,000, 15,000, and 25,000 rpm using an An60-Ti rotor. Scans were acquired using the absorbance optical system 15 h after the start of the experiment and in 1-h intervals until equilibrium was attained. Sedimentation velocity measurements were performed using the same hardware at 55,000 rpm at 10 °C in buffer 1. The details of the data analysis are given as supplemental material.
Mass Spectrometry—Analyses were performed in a nanoflow ES mass spectrometer Q-ToF2 (Micromass). The following experimental parameters were used to record mass spectra of 2 mg/ml MacB in the Q-ToF2 instrument: needle voltage of 1.5 kV and MCP 2350 V.
Atomic Force Microscopy—MacB was diluted to a final concentration of 1 μg/ml, and 45 μl of the sample was allowed to adsorb to freshly cleaved mica. Imaging in air was performed with a Multimode atomic force microscope (Digital Instruments, Santa Barbara, CA) in tapping mode. The silicon cantilevers containing a diamond-like extratip had a drive frequency of ≈300 kHz and a specified spring constant of 40 newtons/m (MikroMasch, Portland, OR), and the applied imaging force was kept as low as possible (target amplitude ≈1.6-1.8 V and amplitude set-point ≈1.3-1.5 V). The molecular volumes of the protein particles were determined from particle dimensions based on AFM images (see supplemental material).
ATPase Assays—An EnzChek phosphate assay kit (Invitrogen) was used to determine the ATPase activity of MacB hydrolyzing MgATP to release phosphate, when the reactants were mixed in a stopped-flow device (see supplemental material). Generally, 2.3 μm protein was mixed with varying concentrations of ATP, up to 4 mm, in the presence of 6 mm MgCl2, to ensure that all the ATP was complexed with Mg2+. In control experiments, no ATPase activity was apparent in the absence of Mg2+. Generally, for MacB alone, the hydrolysis of ATP was characterized by a Pi burst, which was of near equivalence to the MacB concentration, consistent with the ATPase activity being attributable to MacB, rather than any contaminant proteins.
Quantification of Erythromycin Binding to Affinity-purified Mac Proteins—The equilibrium binding of [N-methyl-14C]erythromycin to purified Mac proteins was determined by rapid filtration and quantification of the radioactivity remaining on 0.2-μm filters as outlined in the supplemental material.
RESULTS
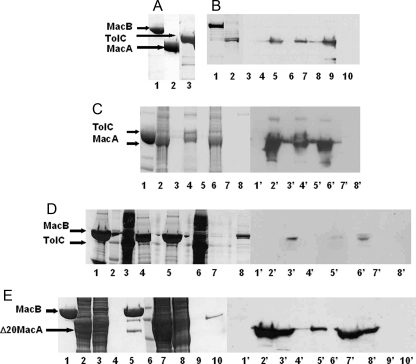
MacA Interacts with Both MacB and TolC via Its Periplasmic Domain—Interactions between E. coli MacA, MacB, and TolC were tested using detergent-solubilized proteins for pulldown assays (Fig. 1). MacA interacted with MacB (Fig. 1B), which we confirmed by cross-linking the proteins (supplemental Fig. 2A), and TolC (Fig. 1C). Although we detected an interaction between TolC and MacB (Fig. 1D), the intensity of the band suggested a weak interaction. N-terminal truncated MacA (Δ20-MacA) interacted with MacB (Fig. 1E), indicating that it is the periplasmic domains of these proteins that interact. The fact that in each case the cognate pump protein could be pulled out of a complex mixture of detergent-solubilized proteins from membranes or cells indicated that the interactions are specific.
FIGURE 1.
MacA interacts with both MacB and TolC. A, overexpression and purification of MacA, MacB, and TolC. An SDS-polyacrylamide gel of purified MacB (lane 1), MacA (lane 2), and TolC (lane 3) is shown. The purified His-tagged MacB and TolC proteins were used as bait, immobilized on a Ni2+-agarose column, over which a slurry of either detergent-solubilized membranes (e.g. from strains overexpressing S-tagged MacA or TolC) or soluble proteins (e.g. from strains overexpressing S-tagged Δ20MacA) was passed to test whether the cognate proteins from the tripartite pump could be pulled out of this complex mixture of proteins. B, pulldown of MacA by MacB. 1st and 2nd lanes, SDS-polyacrylamide gel of immobilized His-tagged MacB (1st lane) and the detergent-solubilized membranes from cells overexpressing the S-tagged MacA (2nd lane). 3rd to 11th lanes, Western blot using antibodies to the S-tag (1:5000 dilution) on MacA. The pulldown assay was performed with His-tagged MacB immobilized on a Ni2+-agarose column, over which a slurry of detergent-solubilized membranes from cells overexpressing S-tagged MacA was passed (7th to 9th lanes). The flow-through (9th lane), 100 mm imidazole wash (8th lane), and the 500 mm imidazole elution (7th lane) were tested for the presence of MacA, which was now also detected in the elution fraction, indicating that it was bound to MacB. A negative control experiment was performed in the absence of immobilized MacB in which MacA was passed through a Ni2+-agarose column (3rd to 5th lanes), and the flow-through (5th lane), 100 mm imidazole wash (4th lane), and the 500 mm imidazole elution (3rd lane) were tested for the presence of MacA, which was only found in the flow-through (5th lane), establishing that S-tagged MacA does not bind to the column. These results indicate that MacB can pull MacA from a complex mixture of membrane proteins. Purified His-tagged MacB did not cross-react with the antibodies to the S-tag (10th lane). C, pulldown of MacA by TolC. An SDS-polyacrylamide gel (lanes 1-8) for the pulldown of S-tagged MacA by His-tagged TolC and the corresponding Western blot (lanes 1′-8′) probed with antibodies (1:5000 dilution) to the S-tag on MacA. Purified His-tagged TolC was immobilized on a Ni2+-agarose column (lanes 1 and 1′); a slurry of detergent-solubilized membranes from cells overexpressing S-tagged MacA was passed through the column and the proteins in the flow-through (lane 2 and 2′), released by washing the column with 100 mm imidazole (lanes 3 and 3′) and eluted with 500 mm imidazole (lanes 4 and 4′), were detected. As a negative control, S-tagged MacA was passed through the column (lanes 6 and 6′), in the absence of immobilized TolC, and the column was washed with 100 mm (lanes 7 and 7′) and 500 mm (lanes 8 and 8′). Comparing lane 4′ and 8′ demonstrates that MacA is only bound to the column in the presence of TolC, indicative of its interaction with TolC. D, pulldown of TolC by MacB. An SDS-polyacrylamide gel (lanes 1-4) for the pulldown of S-tagged TolC by His-tagged MacB and the corresponding Western blot (lanes 1′-4′) probed with antibodies (1:5000 dilution) to the S-tag on TolC. Purified His-tagged MacB was immobilized on a Ni2+-agarose column (lanes 1 and 1′), and a slurry of detergent-solubilized membranes from cells overexpressing S-tagged TolC was passed through the column (lane 3 and 3′), which was then washed with 75 mm imidazole (lanes 4 and 4′), and bound proteins were eluted with 500 mm imidazole (lanes 5 and 5′). A weak band, which was not present in the absence of immobilized MacB, was apparent in lane 5′, indicative of a weak interaction between MacB and TolC. A control experiment was performed in the absence of immobilized MacB in which TolC was passed through a Ni2+-agarose column and the flow-through (lane 6), the 100 mm imidazole wash (lane 7), and the 500 mm imidazole elution (lane 8) were tested for the presence of TolC, which was only found in the flow-through (lane 6′), establishing that S-tagged TolC does not bind to the column. A protein Mr marker was run in lane 2. E, pulldown of N-terminal truncated MacA by MacB. An SDS-polyacrylamide gel shows the His-tagged MacB (lane 1) that was immobilized on a Ni2+-agarose column (lane 1), a slurry of cytoplasmic proteins released by disruption of cells overexpressing S-tagged Δ20-MacA (lane 2), which was passed through the column, over immobilized MacB, and the proteins in the flow-through detected (lane 3); the proteins were released by washing the column with 100 mm imidazole (lane 4); and the proteins were eluted with 500 mm imidazole (lane 5). A Western blot was performed on each of the corresponding protein fractions (indicated with 1′-5′) using antibodies to the S-tag (1:5000 dilution) to detect S-tagged MacA. The elution of MacB yields an extra, low Mr, band on the SDS-polyacrylamide gel that corresponds to that expected for MacA (lane 5) and was identified as such by Western blotting (lane 5′). A control experiment was performed in the absence of immobilized MacB in which Δ20MacA was passed through a Ni2+-agarose column and the flow-through (lane 7), the first and second washes with 100 mm imidazole (lanes 8 and 9, respectively), and the 500 mm imidazole elution (lane 10) were tested for the presence of MacA, which was only found in the flow-through and first wash (lane 7′ and 8′, respectively), establishing that S-tagged MacA does not bind to the column. These results indicate that MacA does not require the N-terminal α-helix, which anchors it to the inner membrane, to interact with MacB. A protein Mr marker was run in lane 6.
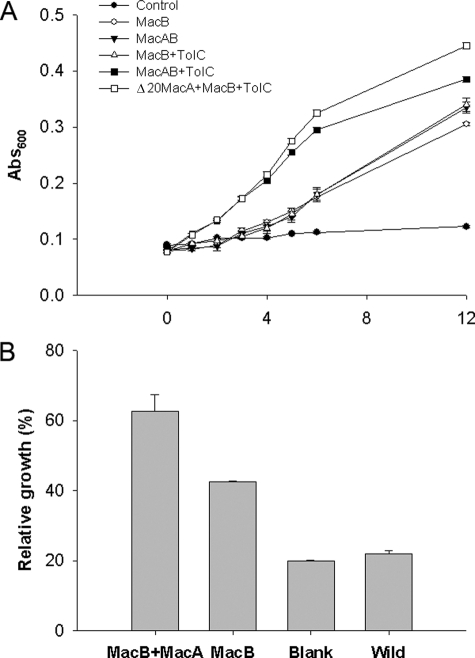
E. coli MacA and MacB Form a Functional Complex with TolC—The simultaneous expression of macA, macB, and tolC in the E. coli ΔacrAB strain KAM3 (26) conferred resistance to erythromycin, indicative of the formation of a functional complex (Fig. 2A). Cells expressing macB with tolC conferred modest resistance to erythromycin in comparison with cells expressing macB, tolC, and macA, indicating that MacA is required to couple MacB to TolC (Fig. 2A). We sought to test whether the N terminus of MacA, which incorporates an α-helix that could interact with MacB, is required for the functional assembly of the complex. A construct in which the gIII-signal sequence was fused to truncated MacA, targeting it to the periplasm, was capable of conferring resistance to erythromycin (Fig. 2A), indicating that the N-terminal domain is not essential for the assembly of the functional complex. This is consistent with a report that a truncated lipid-deficient AcrA derivative was functional as judged by resistance of the cells to erythromycin (27). MacB alone conferred elevated resistance to erythromycin, probably because of its ability to pump the antibiotic into the periplasm, but we consistently found that expressing macB with either tolC or macA conferred greater resistance. Consequently, we sought to test if MacA could enhance this ability. To overcome the difficulty in comparing the growth of cells overexpressing multiple proteins that tend to grow at different rates, we monitored the growth of cells in the presence and absence of erythromycin and determined the growth loss (for cells growing in the presence of erythromycin in comparison with cells growing in the absence of erythromycin) (Fig. 2B). This revealed a significant loss in growth of the cells expressing MacB compared with those expressing MacAB, indicating that the simultaneous expression of MacA and MacB increases the resistance of the cells to erythromycin (Fig. 2B), suggesting that MacA enhances the ability of MacB to confer antibiotic resistance. Similarly, a previous study reported that MacAB, but not MacB alone, conferred resistance to macrolides (3).
FIGURE 2.
MacAB-TolC form a tripartite complex that confers resistance to erythromycin. A, growth curves for E. coli cells, of strain KAM3 (DE3), harboring the plasmids pETDuet (•), pETDuet-MacB (○), pETDuet-MacB/MacA (▾), pETDuet-MacB/TolC (▵), pETDuet-MacB/MacA/TolC (▪) and pETDuet-MacB/gIII-SS-Δ20MacA/TolC (□) grown in the presence of 100 μg/ml erythromycin. B, bar chart showing the extent of inhibition of the growth of E. coli cells in response to 50 μg/ml erythromycin, of strain KAM3(DE3), harboring the plasmids pETDuet (blank), pETDuet-MacB (MacB), pETDuet-MacB/MacA (MacAB) or no plasmid (Wild). For each strain the A600 was determined after growth for 3 h in the absence and presence of erythromycin, and the growth inhibition was determined as the ratio of these measurements. Cells expressing both MacA and MacB suffered less from erythromycin growth inhibition than those expressing only MacB, suggesting that MacA confers elevated resistance to erythromycin on the MacB strain.
MacB Forms Dimers—MacB has an atypical structure for an ABC transporter as it is predicted to have an N-terminal cytoplasmic NBD, which is connected to a four-helix transmembrane domain, with a large periplasmic domain formed by the loops connecting helices 1 and 2 (3, 23). If MacB resembles other ABC transporters that use a pair of NBDs to bind ATP, then it should function as a dimer. However, many transporters, including ABC transporters, have 12 membrane-spanning helices; MacB could adopt a similar topology by forming trimers. Furthermore, AcrB (4, 5) and TolC (9), which assemble into a tripartite complex with AcrA, clearly form trimers. If the trimeric arrangement of the periplasmic domains in AcrB forms a necessary scaffold for binding of AcrA, so that it can effectively interact with TolC, then by analogy the periplasmic domain of MacB might also be forced into forming trimers when interacting with MacA and TolC.
Therefore, we sought to determine the oligomeric state of MacB. Size-exclusion chromatography indicated that it forms higher order oligomers consistent with a dimer (data not shown), but such measurements are not only dependent upon the molecular weight but also the shape of the protein. Furthermore, there is a need to determine the number of detergent molecules complexed by the protein. Consequently, to determine whether the detergent-solubilized MacB was dimeric, we added a cross-linker to trap the oligomers; when we ran the cross-linked protein on an SDS-polyacrylamide gel, the most predominant band ran between the 120- and 160-kDa markers, indicative of a dimer, which has a calculated molecular mass of 145.8 kDa (supplemental Fig. 2B).
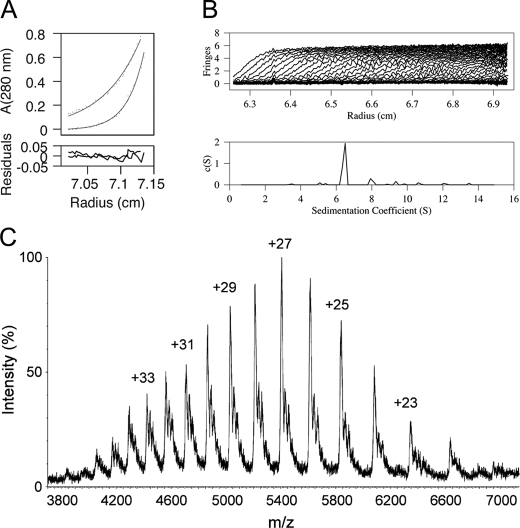
To further confirm the basic oligomeric unit as a dimer, we used two other techniques, analytical ultracentrifugation (AUC) and electrospray mass spectrometry (ES-MS). For the AUC experiments, we reduced the βDDM concentration to just below the critical micelle concentration to avoid the formation of micelles. To determine the detergent contribution in the buoyant mass of the protein-detergent complex, we used a series of different density buffers prepared using a range of D2O concentrations. The apparent molecular mass for MacB was determined from sedimentation equilibrium measurements to be 162.6 kDa (Fig. 3A) and from sedimentation velocity measurements to have a sedimentation coefficient of 6.8 S, corresponding to a molecular mass of 160.0 kDa (Fig. 3B). This molecular mass is greater than expected for monomeric and less than expected for trimeric MacB, complexed with bound detergent, but it is highly consistent with a MacB dimer to which about 16 βDDM molecules are bound. Although the amount of detergent bound to the MacB dimer appears to be lower than reported for RND (28) and MF (29) transporters, this reflects the fact that in our experimental set-up the detergent contribution was actively suppressed using a solvent density matching technique (25). Our independent measurement of bound detergent using a calorimetric assay (30) indicated that, when the detergent concentration was close to the critical micelle concentration, the amount of bound detergent was similar to that of other membrane proteins (e.g. MacB solubilized in 0.05% w/v βDDM bound 1.2 g of βDDM/g of MacB, which is equivalent to a βDDM:MacB molar ratio of 164:1). Electrospray-mass spectrometry (ES-MS) was used in nontandem configuration to determine accurately the molecular mass of the protein, under conditions that would dissociate the βDDM, revealing a peak with a molecular mass of 145,961.25 ± 20.57 Da that is consistent with a MacB dimer (Fig. 3C).
FIGURE 3.
Biophysical evidence for MacB dimer formation. A, AUC sedimentation equilibrium profiles of MacB. A representative sedimentation equilibrium profile from one of the runs (two different velocities of the same sample) is shown. Experimental data (dots) and fitted model for a 162.6-kDa particle (solid line) is shown for each. The bottom panel represents the residuals after fitting. B, AUC sedimentation velocity profiles of MacB are consistent with the formation of a stable dimer. The upper panel shows sedimentation profile curves at different time points, and the lower panel presents a c(s) size distribution analysis with solutions of the Lamm equation. The sedimentation coefficient is 6.8 S corresponding to an apparent molecular mass of 160 kDa, consistent with a dimer with about 16 detergent molecules bound. C, mass spectrum of MacB. The charge states corresponding to the peaks are graphed. The mass spectrum indicated a molecular mass for MacB of 145.96 kDa, which is consistent with a dimer.
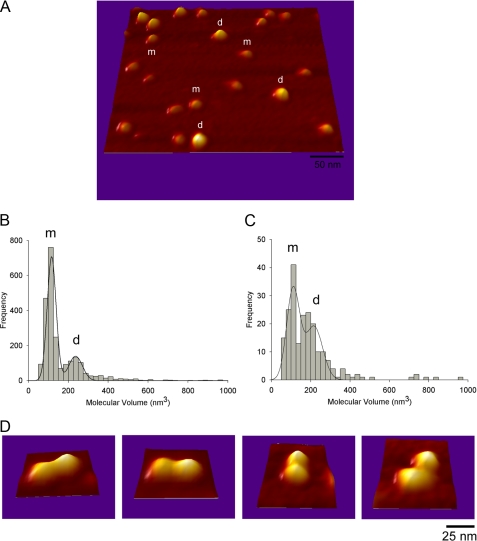
We also sought to visualize single particles of MacB by AFM (Fig. 4, A and D). Two populations were revealed with average molecular volumes of 118 and 238 nm3 (Fig. 4B). These would accommodate proteins of about 60-70 and 130-140 kDa, respectively. Furthermore, the larger particles could clearly be seen at higher resolution to consist of two protein domains that are highly suggestive of a dimer. We found that in the presence of AMP-PNP, a nonhydrolysable analogue of ATP, the ratio of dimers to monomers on the AFM grids increased (Fig. 4C), which would be consistent with the nucleotide stabilizing the interaction between monomers. Our findings are novel because detergent molecules tend to impair the resolution of AFM studies of detergent-solubilized membrane proteins; this implies that for MacB, we may be able to get information on the topology and stoichiometry of its assemblies with MacA and TolC. Indeed, under coincubation of MacA and MacB, we could clearly distinguish a significant distribution of particles with molecular volumes larger than those corresponding to MacB dimers, which is consistent with multiprotein complexes formed between both proteins (data not shown). Such promising data paves the way toward further characterization of membrane multiprotein complexes and could prove a powerful instrument for determining the stoichiometry of the tripartite assembly.
FIGURE 4.
AFM analyses, AFM imaging of MacB. A, three-dimensional picture of a low magnification image of MacB acquired in air in Tapping Mode with a diamond-like extra tip of resonant frequency ∼300 kHz and spring constant of 40 newtons/m. m and d show particles that belong to the first and second peak in B, respectively. B, frequency distribution of molecular volumes of MacB. The curve indicates a fitted Gaussian function. The m and d peaks correspond to volumes of 118 ± 1 nm3 (n = 1642) and 238 ± 5 nm3 (n = 665), consistent with the monomer and dimer, respectively. C, frequency distribution of molecular volumes of MacB that had been incubated with the nonhydrolysable ATP analogue AMP-PNP. The peaks correspond to volumes of 112 ± 3 nm3 (n = 94) and 218 ± 14 nm3 (n = 116). These data indicate an increase in the dimer:monomer ratio in the presence of AMP-PNP. D, high resolution images of structures where two small particles (m+m) are attached to one another, clearly indicative of dimer formation.
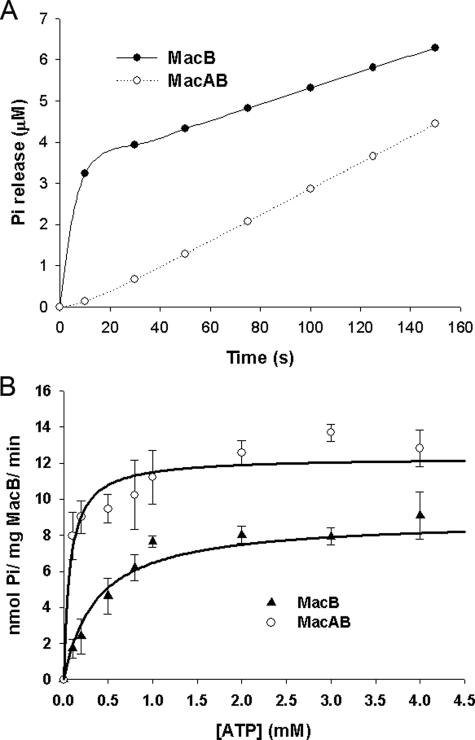
MacA Regulates the ATPase Activity of MacB—MacB retained ATPase activity when detergent-solubilized, but this was detergent-dependent, being active in Triton X-100 but not in βDDM (data not shown). The time course for hydrolysis of MgATP by MacB was determined in a stopped-flow spectrophotometer, using the dye 2-amino-6-mercapto-7-methylpurine riboside to monitor the production of inorganic phosphate (Pi). The time course was characterized by a burst in Pi production, during the first 20 s, followed by a slower steady-state rate (Fig. 5A). This kinetic behavior is consistent with the ATP being rapidly hydrolyzed, to produce Pi and ADP, but further turnovers are rate-limited either by a subsequent conformational change or the slow release of products. Because previous studies have established that the ATPase activity of MacB is inhibited by vanadate (24), which stabilizes bound ADP, this indicates that Pi is released before ADP, suggesting that ADP release is rate-limiting. The rate constant for the hydrolysis step was determined by fitting the burst phase to an exponential function, yielding a kcat value of 0.24 s-1, whereas the amplitude of the burst phase was 2.2 μm for 1 mm ATP (Fig. 5A). When the MacA and MacB concentrations were increased to 3.5 μm, the amplitude of the burst phase increased to 3.3 μm (data not shown), indicating that the burst is approximately equivalent to the MacB concentration and that both NBDs within the MacB dimer are functional. We did not notice any deviation from a single exponential that would indicate that these NBDs turn over ATP differentially. The steady-state phase was characterized by a rate of Pi production that increased in a hyperbolic manner with the ATP concentration (Fig. 5B); fitting the steady-state rate data to a hyperbolic function yielded a maximal specific activity of 8.9 nmol of ATP/min/mg MacB and a Km of 374 μm. A progressive reduction in the amplitude of the burst phase for ATP concentrations below the steady-state Km value hindered analyses because the pre-steady-state phase tended to merge with the steady-state phase; consequently, we only used the steady-state rates determined at ATP concentrations of 0.1 mm and above. For comparison, the lipid A transporter MsbA, an half-ABC transporter, was characterized by a Vmax of 37 nmol of ATP/min/mg and a Km of 878 μm (31).
FIGURE 5.
MacA regulates the ATPase activity of MacB. A, time course for the change in Pi concentration, corresponding to the absorbance change of the 2-amino-6-mercapto-7-methylpurine riboside in A, where 2.3 μm MacB was mixed with 1 mm ATP in the absence (upper trace) and presence (lower trace) of an equivalent concentration of MacA. In the absence of MacA, MacB produced a phosphate (Pi) burst, with a rate and amplitude of 0.235 (±0.001) s-1 and 2.20 (±0.01) μm, respectively. MacB did not produce a Pi burst in the presence of MacA. B, steady-state rate of Pi production by MacB as a function of the ATP concentration in the absence (lower curve) and presence (upper curve) of an equivalent concentration of MacA. The data are characterized by Vmax and Km values of 8.9 (±0.7) nmol of ATP/mg MacB/min and 374 (±126) μm, respectively, for MacB alone; and of 12.3 (±0.5) nmol of ATP/mg MacB/min and 72 (±22) μm, respectively, for MacB in the presence of an equivalent concentration of MacA.
When MacA was added to MacB, at an equivalent or higher concentration, with both proteins in detergent, no phosphate burst was observed (Fig. 5A). Considering that the detergent was present for both the ATPase assays with MacB and MacAB suggests that the burst phase is mechanistically important and cannot be attributed to the detergent modifying the behavior of MacB. Interestingly, although there was no Pi burst by MacB in the presence of MacA, there was a lag in Pi production; this could signify that a conformational change that precedes the hydrolysis step becomes rate-limiting for the first turnover. The steady-state rate of ATP hydrolysis by MacB, in the presence of MacA, was enhanced (Fig. 5B). At first glance, this behavior might appear consistent with MacA increasing the rate of ADP dissociation from MacB, so that this step was no longer rate-limiting. However, although MacA increased the specific activity of MacB from 8.9 to 12.3 nmol of ATP/mg MacB/min, which would correspond to an increase in kcat from 0.011 to 0.015 s-1, the steady-state rate did not exceed the rate for hydrolysis in the absence of MacA (e.g. 0.24 s-1) (Fig. 5B). Accordingly, the increase in the steady-state rate, which we attribute to product release, or a conformational change preceding this, would not be rapid enough to prevent the phosphate burst. It seems more likely that MacA also retards the rate of ATP hydrolysis, so that this process becomes slower than the rate of product release. Indeed, we found that the affinity of MacB for ATP was increased by more than 5-fold, from 374 to 72 μm, in the presence of MacA (Fig. 5B). If the effect of MacA was simply to enhance product release to a rate faster than that for nucleotide hydrolysis, then we would have expected a decrease in affinity for ATP. On the other hand, if MacA simply retarded nucleotide hydrolysis to a rate slower than product release, then the maximal steady-state rate would have decreased. Our data are consistent with MacA increasing the rate of product release, while decreasing the rate of hydrolysis to a similar rate to product release.
In contrast to a previous study, which indicated that the ATPase activity of reconstituted MacB was not activated by N-terminal truncated MacA (24), we found that when MacB was mixed with Δ20-MacA no Pi burst was apparent (data not shown). In the same study, and consistent with our findings, N-terminal truncated MacA was shown to interact with MacB; however, in contrast to our findings, when MacB was expressed with N-terminal truncated MacA in erythromycin-susceptible cells, there was no increase in erythromycin resistance (24). The reason for the difference with our own findings is unclear; however, in the previous study the signal sequence of OmpA was used to target MacA, truncated at position 32, to the periplasm (24), raising the possibility that the shorter MacA sequence and/or the OmpA signal sequence interfered with the ability of MacA to affect the ATPase activity of MacB.
In accord with previous studies (24), we could not detect an effect of erythromycin on the kinetics of MacB ATPase either in the absence or presence of MacA (data not shown). Recent studies have established that the ATPase activity of Pdr5 is uncoupled from substrate binding; this basal ATPase activity might be required to constantly cycle the transporter between conformations, so as to maintain the accessibility of the cytosolic substrate-binding site (32). In the case of MacB, its ATPase activity might instead be stimulated by TolC, to reset its conformation following drug transfer to TolC.
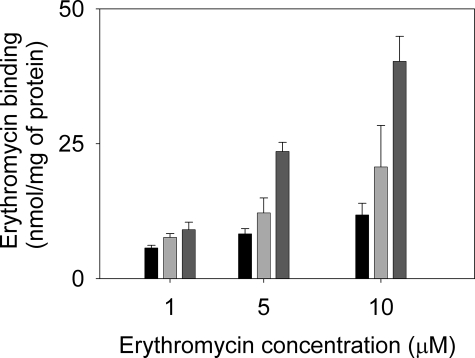
MacA Increases the Capacity of MacB to Bind Erythromycin—Although we could not detect any effect of erythromycin on the ATPase activity of MacB, we could detect the binding of [14C]erythromycin to detergent-solubilized MacB (Fig. 6). Importantly, we found that the MacAB complex bound more erythromycin than MacB alone (which binds considerably more than MacA alone) and that the amount bound increased in a concentration-dependent manner, as would be expected if MacA increased the affinity of MacB for erythromycin. However, because of the insolubility of the antibiotic in aqueous solutions, it was not possible to test a full range of erythromycin concentrations that might saturate MacAB. Consequently, we cannot exclude the possibility of the formation of additional sites within the MacAB complex that are not apparent in either MacB or MacA alone. Our data indicate that MacA not only modulates the ATPase activity of MacB but also enhances its capacity to bind erythromycin (be this due to an increase in affinity of MacB for drugs or the formation of additional drug-binding sites within the MacAB complex).
FIGURE 6.
MacA increases the capacity of MacB to bind erythromycin. Purified proteins (50 μg of MacA or MacB, or 25 μg of MacA plus 25 μg of MacB) were incubated in the presence of [N-methyl-14C]erythromycin at concentrations as indicated (1, 5, or 10 μm), after which drug binding was measured by rapid filtration. The bars represent the erythromycin bound by MacA (left, black), MacB (middle, light gray), and MacAB (right, dark gray). The data indicate that MacA enhances the binding of erythromycin to MacB.
DISCUSSION
Gram-negative bacteria possess tripartite pumps that facilitate the extrusion of protein toxins and cytotoxic compounds, such as antibiotics, from the cell. In these tripartite assemblies the IMP is coupled to an OMP by a periplasmic MFP, forming a pump that can translocate molecules across both the inner and outer membranes. Intriguingly, Gram-positive bacteria that lack an outer membrane also possess MFPs, suggesting that they play a role in addition to stabilizing the interaction of the IMP with the OMP. Indeed, we have noted in some of these MFPs a large deletion corresponding to the coiled-coil hairpin that would interact with the OMP (supplemental Fig. 1). To investigate this possibility, we sought to determine the role of MacA, the MFP that couples the IMP MacB, an ATP-driven transporter, with the OMP TolC in E. coli to extrude macrolide antibiotics (3). We established that MacA interacts with both MacB and TolC (Fig. 1) to form a functional tripartite complex (Fig. 2). However, we also found that MacA enhanced the resistance to erythromycin conferred by MacB alone (Fig. 2B), suggesting that it modulated the transport activity of MacB, which is consistent with our data demonstrating that MacA regulates the drug binding and ATPase activity of MacB (Figs. 5 and 6). The role of the MFP BesA in the activity of the BesABC pump in Borrelia burgdorferi, the causative agent of Lyme disease, has recently been highlighted (33). It is quite remarkable that in this system BesA also lacks the α-helical hairpin. As such this periplasmic protein is unable to unlock the periplasmic entry site of the OMP, a function that is attributed to the hairpin (11). To compensate, the OMP has evolved to be constitutively leaky. The reason for the retention of BesA in the Borrelia system is most likely associated with its function in the activation of the inner membrane component, in that case an RND transporter. It is conceivable then, that a similar role is also present in the ABC transporters and their associated MFPs, and it could explain the preservation of the hairpin-lacking MFPs in Gram-positive bacteria, such as S. aureus (supplemental Fig. 1).
Although E. coli MacB has an NBD, which incorporates Walker A and B motifs and an ABC signature sequence that are characteristic features of members of the ABC superfamily, it is not a classic ABC transporter. It has a large periplasmic domain, reminiscent of that found in RND transporters, which form trimers in which these domains form substantive sites of contact between the protomers. Consequently, we sought to determine the oligomeric state of MacB and in doing so have developed a novel ES-MS approach to unambiguously establish that MacB forms dimers (Fig. 3C). ES-MS is now widely accepted as a powerful method to determine accurately the stoichiometry of intact protein complexes (34). However membrane proteins, solubilized by detergent or adsorbed in micelles, have remained difficult to analyze under similar MS conditions. The development of strategies to tackle this field is challenging, primarily because the large quantities of detergent suppress the protein signal, whereas the poor solubility of membrane proteins in aqueous buffers often causes the electrospray needle to block. To date only a few MS studies have reported the observation of membrane proteins or their complexes by MS (35-39). Here we report on the use of a miniaturized form of ES with reduced flow rates (nano-ES), and a high collision energy that facilitates the desolvation process and induces dissociation of detergent-proteins clusters, to determine the oligomeric state of an integral homomeric membrane protein. In our protocol we used lower quantities of βDDM, than have been reported previously, without apparent detrimental effects on the stability of the protein. Under our experimental conditions, we have successfully maintained the noncovalent subunit interactions, such that the oligomeric state of the protein complex could be determined without ambiguity. As would be expected for an ABC transporter in which the NBDs interact, our biophysical studies of MacB indicate that it forms dimers (Figs. 3 and 4 and supplemental Fig. 3). This is an interesting observation because it raises the following question. How does dimeric MacB interact with trimeric TolC and the number of MacA molecules needed to stabilize the tripartite complex? Extension of our AFM studies will be vital for determining the stoichiometry of the interactions to provide an understanding of the assembly of the tripartite complex, which may be difficult to address by other methods, such as crystallization of the complex.
Our findings and previous studies, which have shown that disruption of the Walker A and B motifs not only inhibits the ATPase activity but also blocks the capacity of MacB to confer macrolide resistance (24), suggest that MacB operates by a similar mechanism to typical ABC transporters. Our understanding of how ABC transporters couple ATP hydrolysis to transport is still rudimentary, but the determination of the crystal structures of several ATPase subunits and complete ABC transporters has suggested conservation of key steps in the molecular mechanism. The binding of ATP to both soluble ATPase subunits (40), solubilized NBDs from ABC transporters (41, 42), and to the NBDs within a complete ABC transporter (43, 44) has been shown to promote dimerization as the ATP is bound at the interface of these nucleotide-binding sites, sandwiched between the Walker A motif of one NBD and the ABC signature motif of the other NBD. Our AFM studies indicated that there is a higher proportion of MacB dimers in the presence of the nonhydrolysable nucleotide AMP-PNP (Fig. 4). In complete ABC transporters, the binding of ATP causes the transporter to adopt a conformation in which the substrate-binding site is outward-facing, because ATP bridges the two NBDs, closing off the inward-facing substrate-binding site (45). Conversely, the release of the hydrolysis products ADP and phosphate is thought to promote an inward-facing conformation, as the structural constraint imposed by binding of the nucleotide is released. Consistent with this proposal, several studies indicate that NBD dimerization cannot be induced by ADP (46), because interactions between the γ-phosphate of ATP and the signature sequence catalyze these events (47). We found that MacA modulates these processes in MacB (Fig. 5), and so our kinetic data have clear implications in terms of such a mechanism. Most significantly, MacA increases the apparent affinity of MacB for ATP, while decreasing the rate of ATP hydrolysis, so as to promote and stabilize the ATP binding conformation, which we presume to be the conformation in which the antibiotic-binding site is outward-facing. In this manner, MacA would play a direct role in driving antibiotic translocation between MacB and TolC.
During the course of our studies another investigation reported on the effect of MacA on the steady-state ATPase activity of MacB (24). In this study, MacB, solubilized in Triton X-100, appeared to have a specific activity that was an order of magnitude higher than we had found; however, a discontinuous assay was used to determine the ATPase activity, and this could well have been influenced by pooling the time points from both the burst and steady-state phases. Indeed, kcat was marginally slower than the hydrolysis rate determined from the Pi burst phase in our experiments (e.g. 0.17 versus 0.24 s-1). Possibly for similar reasons, this assay did not detect the effect of MacA on MacB solubilized in Triton X-100. However, they did find that MacB reconstituted into liposomes was characterized by a reduced kcat (e.g. kcat = 0.10 s-1) and decreased affinity of MacB for ATP (e.g. Km = 2.30 mm). Co-reconstitution of MacB with MacA had the effect of increasing both kcat (e.g. kcat = 0.78 s-1) and the affinity of MacB for ATP (e.g. Km = 0.38 mm). This behavior is similar to the effect of MacA on the steady-state kinetics of the ATPase activity of solubilized MacB in our study. To know if the reconstituted protein behaves in an identical manner to the solubilized protein would require the reconstitution of sufficient amounts of MacB to define any Pi burst, which is technically demanding. Consequently, although these earlier studies support our conclusion that MacA affects the ATPase activity of MacB, they do not give the detailed insight into the ATPase mechanism provided by our pre-steady-state analyses.
Our studies established that conformational changes can be propagated from the periplasmic domain of MacA to the cytoplasmic NBD of MacB (Figs. 1 and 2), and consequently, it is plausible that TolC, by interacting with MacA, can detect the nucleotide state of MacB. The binding of ATP to MacB would stimulate MacA to interact with TolC, inducing the latter to adopt the open state. Because ATP and MacA stabilize in the outward-facing conformation of MacB, drug transfer from MacB to TolC would be facilitated. TolC would then communicate with the NBD of MacB (again, such communication is probably conveyed via MacA) to stimulate ATP hydrolysis, which would be required to reset MacB in the inward-facing conformation. If ATP hydrolysis is controlled by TolC, so that the MacB conformation is only reset after the interaction with TolC and productive transfer of drugs, this would provide an explanation of the apparent insensitivity of the ATPase activity of MacB to drugs. Indeed, without such a feedback control mechanism of drug export, it would appear that drug stimulation of the ATPase activity of MacB would be counter-productive because the MacB conformation could be reset before drug transfer to TolC. There is an analogy in the mechanism of operation of ABC transporters that work in conjunction with a periplasmic binding protein (47). In the E. coli maltose transporter, the binding of ATP to the ATPase subunit MalK induces conformational changes, detected by tryptic digestion, in the periplasmic loops of the membrane subunits MalF and MalG (48), whilst EPR studies revealed that ATP binding, but not ADP binding, caused an increase in affinity between the transporter and the maltose-binding protein (MBP), and “forcing open” the bound MBP to release its substrate (49). The transporter is reset in its original inward-facing conformation by ATP hydrolysis, which is stimulated by the MBP (50). In our model TolC replaces the MBP and, because the open state requires disruption of the second selectivity filter and subsequent twisting of the helices of the TolC channel to keep it fully open, there is a need for the MacA hairpins to stabilize this conformation, which could not be achieved by interaction with MacB alone. Although we have interpreted our findings in terms of an alternating conformation model for MacB, in which the drug-binding sites are inwardly and outwardly exposed, such a model could easily be refined to account for drug binding to a fixed periplasmic site, in analogy to AcrB, with ATP binding and hydrolysis coupled to conformational changes that induce TolC association and dissociation. Clearly the work presented here provides an important framework for further studies that will enable the elucidation of the mechanism underlying the dynamics of assembly of the MacABTolC tripartite pump in the future.
Supplementary Material
Acknowledgments
We thank Dr. Martin Moncrieffe (University of Cambridge) for help with the AUC data collection and analysis.
This work was supported by grants from the Wellcome Trust (Durham group). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Methods, Figs. 1 and 2, Tables 1 and 2, and additional references.
Author's Choice—Final version full access.
Footnotes
The abbreviations used are: IMP, inner membrane protein; βDDM, β-dodecyl maltoside; MFP, membrane fusion protein; OMP, outer membrane protein; AUC, analytical ultracentrifugation; ES-MS, electrospray-mass spectrometry; AMP-PNP, adenosine 5′-(β,γ-imino)triphosphate; S-tag, for proteins fused to the 15-amino acid polypeptide (e.g. Lys-Glu-Thr-Ala-Ala-Ala-Lys-Phe-Glu-Arg-Gln-His-Met-Asp-Ser) derived from RNase A; NBD, nucleotide binding domain; MBP, maltose binding-protein; AFM, atomic force microscopy.
References
- 1.Borges-Walmsley, M. I., McKeegan, K. S., and Walmsley, A. R. (2003) Biochem. J. 376 313-338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holland, I. B., Schmitt, L., and Young, J. (2005) Mol. Membr. Biol. 22 29-39 [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi, N., Nishino, K., and Yamaguchi, A. (2001) J. Bacteriol. 183 5639-5644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Touze, T., Eswaran, J., Bokma, E., Koronakis, E., Hughes, C., and Koronakis, V. (2004) Mol. Microbiol. 53 697-706 [DOI] [PubMed] [Google Scholar]
- 5.Tikhonova, E. B., and Zgurskaya, H. I. (2004) J. Biol. Chem. 279 32116-32124 [DOI] [PubMed] [Google Scholar]
- 6.Murakami, S., Nakashima, R., Yamashita, E., and Yamaguchi, A. (2002) Nature 419 587-593 [DOI] [PubMed] [Google Scholar]
- 7.Yu, E. W., McDermott, G., Zgurskaya, H. I., Nikaido, H., and Koshland, D. E., Jr. (2003) Science 300 976-980 [DOI] [PubMed] [Google Scholar]
- 8.Mikolosko, J., Bobyk, K., Zgurskaya, H. I., and Ghosh, P. (2006) Structure (Lond.) 14 577-587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koronakis, V., Sharff, A., Koronakis, E., Luisi, B., and Hughes, C. (2000) Nature 405 914-919 [DOI] [PubMed] [Google Scholar]
- 10.Tamura, N., Murakami, S., Oyama, Y., Ishiguro, M., and Yamaguchi, A. (2005) Biochemistry 44 11115-11121 [DOI] [PubMed] [Google Scholar]
- 11.Bavro, V. N., Pietras, Z., Furnham, N., Pérez-Cano, L., Fernández-Recio, Pei, X. Y., Misra, R., and Luisi, B. (2008) Mol. Cell 30 114-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lobedanz, S., Bokma, E., Symmons, M. F., Koronakis, E., Hughes, C., and Koronakis, V. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 4612-4617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elkins, C. A., and Nikaido, H. (2003) J. Bacteriol. 185 5349-5356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Federici, L., Du, D., Walas, F., Matsumura, H., Fernandez-Recio, J., McKeegan, K. S., Borges-Walmsley, M. I., Luisi, B. F., and Walmsley, A. R. (2005) J. Biol. Chem. 280 15307-15314 [DOI] [PubMed] [Google Scholar]
- 15.Andersen, C., Koronakis, E., Bokma, E., Eswaran, J., Humphreys, D., Hughes, C., and Koronakis, V. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 11103-11108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bokma, E., Koronakis, E., Lobedanz, S., Hughes, C., and Koronakis, V. (2006) FEBS Lett. 580 5339-5343 [DOI] [PubMed] [Google Scholar]
- 17.Vediyappan, G., Borisova, T., and Fralick, J. A. (2006) J. Bacteriol. 188 3757-3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murakami, S., Nakashima, R., Yamashita, E., Matsumoto, T., and Yamaguchi, A. (2006) Nature 443 173-179 [DOI] [PubMed] [Google Scholar]
- 19.Seeger, M. A., Schiefner, A., Eicher, T., Verrey, F., Diederichs, K., and Pos, K. M. (2006) Science 313 1295-1298 [DOI] [PubMed] [Google Scholar]
- 20.Sennhauser, G., Amstutz, P., Briand, C., Storchenegger, O., and Grutter, M. G. (2007) PLoS Biol. 5 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seeger, M. A., von Ballmoos, C., Eicher, T., Brandstätter, L., Verrey, F., Diederichs, K., and Pos, K. M. (2008) Nat. Struct. Mol. Biol. 15 199-205 [DOI] [PubMed] [Google Scholar]
- 22.Aires, J. R., and Nikaido, H. (2005) J. Bacteriol. 187 1923-1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi, N., Nishino, K., Hirata, T., and Yamaguchi, A. (2003) FEBS Lett. 546 241-246 [DOI] [PubMed] [Google Scholar]
- 24.Tikhonova, E. B., Devroy, V. K., Lau, S. Y., and Zgurskaya, H. I. (2007) Mol. Microbiol. 63 895-910 [DOI] [PubMed] [Google Scholar]
- 25.Reynolds, J. A., and Tanford, C. (1976) Proc. Natl. Acad. Sci. U. S. A. 73 4467-4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morita, Y., Kodama, K., Shiota, S., Mine, T., Kataoka, A., Mizushima, T., and Tsuchiya, T. (1998) Antimicrob. Agents Chemother. 42 1778-1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zgurskaya, H. I., and Nikaido, H. (1999) J. Mol. Biol. 285 409-420 [DOI] [PubMed] [Google Scholar]
- 28.Stroebel, D., Sendra, V., Cannella, D., Helbig, K., Nies, D. H., and Covès, J. (2007) Biochim. Biophys. Acta 1768 1567-1573 [DOI] [PubMed] [Google Scholar]
- 29.Heuberger, E. H., Veenhoff, L. M., Duurkens, R. H., Friesen, R. H., and Poolman, B. (2002) J. Mol. Biol. 317 591-600 [DOI] [PubMed] [Google Scholar]
- 30.Butler, P. J., Ubarretxena-Belandia, I., Warne, T., and Tate, C. G (2004) J. Mol. Biol. 340 797-808 [DOI] [PubMed] [Google Scholar]
- 31.Doerrler, W. T., and Raetz, C. R. (2002) J. Biol. Chem. 277 36697-36705 [DOI] [PubMed] [Google Scholar]
- 32.Ernst, R., Kueppers, P., Klein, C. M., Schwarzmueller, T., Kuchel, K., and Schmitt, L. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 5069-5074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bunikis, I., Denker, K., Ostberg, Y., Andersen, C., Benz, R., and Bergstrom, S. (2008) PLoS Pathog. 4 e1000009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharon, M., and Robinson, C. V. (2007) Annu. Rev. Biochem. 76 167-193 [DOI] [PubMed] [Google Scholar]
- 35.Hanson, C. L., Ilag, L. L., Malo, J., Hatters, D. M., Howlett, G. J., and Robinson, C. V. (2003) Biophys. J. 85 3802-3812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ilag, L. L., Ubarretxena-Belandia, I., Tate, C. G., and Robinson, C. V. (2004) J. Am. Chem. Soc. 126 14362-14363 [DOI] [PubMed] [Google Scholar]
- 37.Lengqvist, J., Svensson, R., Evergren, E., Morgenstern, R., and Griffiths, W. J. (2004) J. Biol. Chem. 279 13311-13316 [DOI] [PubMed] [Google Scholar]
- 38.Meier, T., Morgner, N., Matthies, D., Pogoryelov, D., Keis, S., Cook, G. M., Dimroth, P., and Brutschy, B. (2007) Mol. Microbiol. 65 1181-1192 [DOI] [PubMed] [Google Scholar]
- 39.Barrera, N. P., Di Bartolo, N., Booth, P. J., and Robinson, C. V. (2008) Science 321 243-246 [DOI] [PubMed] [Google Scholar]
- 40.Chen, J., Lu, G., Lin, J., Davidson, A. L., and Quiocho, F. A. (2003) Mol. Cell 12 651-661 [DOI] [PubMed] [Google Scholar]
- 41.Smith, P. C., Karpowich, N., Millen, L., Moody, J. E., Rosen, J., Thomas, P. J., and Hunt, J. F. (2002) Mol. Cell 10 139-149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaitseva, J., Oswald, C., Jumpertz, T., Jenewein, S., Wiedenmann, A., Holland, I. B., and Schmitt, L. (2006) EMBO J. 25 3432-3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dawson, R. J., and Locher, K. P. (2007) FEBS Lett. 581 935-938 [DOI] [PubMed] [Google Scholar]
- 44.Ward, A., Reyes, C. L., Yu, J., Roth, C. B., and Chang, G. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 19005-19010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dawson, R. J., Hollenstein, K., and Locher, K. P. (2007) Mol. Microbiol. 65 250-257 [DOI] [PubMed] [Google Scholar]
- 46.Lu, G., Westbrooks, J. M., Davidson, A. L., and Chen, J. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 17969-17974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oloo, E. O., Fung, E. Y., and Tieleman, D. P. (2006) J. Biol. Chem. 281 28397-28407 [DOI] [PubMed] [Google Scholar]
- 48.Daus, M. L., Landmesser, H., Schlosser, A., Muller, P., Herrmann, A., and Schneider, E. (2006) J. Biol. Chem. 281 3856-3865 [DOI] [PubMed] [Google Scholar]
- 49.Chen, J., Sharma, S., Quiocho, F. A., and Davidson, A. L. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 1525-1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oldham, M. L., Khare, D., Quiocho, F. A., Davidson, A. L., and Chen, J. (2007) Nature 450 515-521 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.