Abstract
The level of the MafA transcription factor is regulated by a variety of effectors of β cell function, including glucose, fatty acids, and insulin. Here, we show that phosphorylation at Ser65 of mammalian MafA influences both protein stability and transactivation potential. Replacement of Ser65 with Glu to mimic phosphorylation produced a protein that was as unstable as the wild type, whereas Asp or Ala mutation blocked degradation. Analysis of MafA chimeric and deletion constructs suggests that protein phosphorylation at Ser65 alone represents the initial degradation signal, with ubiquitinylation occurring within the C terminus (amino acids 234–359). Although only wild type MafA and S65E were polyubiquitinylated, both S65D and S65E potently stimulated transactivation compared with S65A. Phosphorylation at Ser14 also enhanced activation, although it had no impact on protein turnover. The mobility of MafA S65A was profoundly affected upon SDS-PAGE, with the S65E and S65D mutants influenced less due to their ability to serve as substrates for glycogen synthase kinase 3, which acts at neighboring N-terminal residues after Ser65 phosphorylation. Our observations not only illustrate the sensitivity of the cellular transcriptional and degradation machinery to phosphomimetic mutants at Ser65, but also demonstrate the singular importance of phosphorylation at this amino acid in regulating MafA activity.
The mammalian MafA transcription factor was originally isolated due to the significance to insulin gene expression (1, 2), with subsequent studies also demonstrating the importance of closely related MafB to hormone transcription in islet α (glucagon+) and β (insulin+) cells (3, 4). Islet β cell-specific transcription of the insulin gene appears to be mediated by interactions between MafA and other islet-enriched factors, including Pdx1 and NeuroD1 (also known as BETA2) (5). Notably, MafA is first observed during pancreatic development in the wave of insulin+ cells that eventually mature into islet β cells (6), a unique property in relation to all other islet-enriched regulators (7–10). However, MafA is not essential to β cell development, presumably due to compensation by MafB (3, 11).
MafA appears to act as a barometer of adult β cell function. For example, this factor is exclusively expressed in β cells within the context of the pancreas, and global mafA knock-out mice are diabetic due in part to compromised insulin secretion capacity (12). In addition, human embryonic stem cells differentiated to produce insulin and many islet-enriched transcription factors were neither glucose-responsive nor capable of protecting against streptozotocin-induced hyperglycemia until becoming MafA+ (13). Furthermore, MafA levels are unusually sensitive in relation to other islet regulators to metabolic effectors of islet β cell function, such as glucose (5, 14–16), fatty acids (15, 17), and insulin (18). Precisely how these effectors influence MafA expression is unclear but is likely through both transcriptional and post-transcriptional control mechanisms.
Members of the large Maf family are highly phosphorylated proteins, although how this modification influences activity has principally been examined with avian homologs. For example, alanine mutations in the ERK1/2-like4 sites at Ser14 and Ser65 of quail MafA influenced both the activity of this oncogene in transformation assays and lens α-, β-, and δ-crystalline gene transcription (19). Ser65 phosphorylation was also recently shown to be essential to GSK3 (glycogen synthase kinase 3) activity at neighboring serines and threonines in quail and mouse MafA, with these events associated with activation and protein stability (20, 21). Stimulation of quail protein activity was through recruitment of the p300/CBP-associated factor co-activator to the N-terminal activation domain (20), whereas glucose levels in islet β cells were proposed to regulate the degradation of mouse MafA (21).
Here, we examined if the protein levels and activation properties of mouse MafA could be influenced by Ser14, Ser65, and/or Thr267 phosphorylation. Phosphomimetic glutamic and aspartic acid substitution mutants were used in these studies to examine how phosphorylation potentially affected activity. The mobility of S65A was found to be faster and similar to phosphatase-treated MafA, whereas the S65E and S65D mutants behaved more like the wild type. Both the S65E and S65D mutants in MafA were also found to be substrates for GSK3. However, only the S65E mutant, and not the S65D or S65A mutants, was polyubiquitinylated and degraded in a wild type manner. In contrast, S65D, S65E, as well as S14E potentiated MafA-mediated activation. We discuss the possibility that Ser65 phosphorylation is pivotal in controlling both the degradation and activation potential of MafA.
EXPERIMENTAL PROCEDURES
DNA Constructs—The S14A, S14E, S65A, S65E, S65D, and T267A mutants were prepared in cytomegalovirus-driven Myc-MafA expression plasmid using the QuikChange™ site-directed mutagenesis kit (Stratagene, La Jolla, CA). Wild type Gal4-MafA (amino acids 1–359) and the 1–75 and 1–233 mutants were constructed by subcloning PCR-generated mouse MafA sequences into the simian virus 40 promoter/enhancer-driven Gal4 expression plasmid pSG424 (22) to create in-frame Gal4 DNA-binding domain fusion proteins. Enzyme restriction digestion and DNA sequencing analyses were utilized to determine the correctness of each construct. (Gal4)5E1bLuc (23) has been described.
Islet Isolation and Culture—Wistar rat islets were isolated as described (5) and cultured in the presence of 2.8 or 16.7 mm glucose for 24 h with or without 0.5 mm palmitate precomplexed to bovine serum albumin (palmitate:bovine serum albumin molar ratio of 5:1). Nuclear extracts were prepared as described (5) and immunoblotted using an anti-MafA antibody (1:1000; Bethyl Laboratories, Montgomery, TX).
Cell Culture and Transfections—Monolayer cultures of HeLa and mouse islet βTC-3 cells were maintained as described previously (24). Gal4-MafA (0.5 μg) and (Gal4)5E1bLuc (0.5 μg) were transfected using the Lipofectamine procedure (Invitrogen) on 6-well plates. The herpes simplex virus promoter-driven Renilla luciferase expression plasmid phRL-TK (Promega) was used as a recovery marker (10 ng), with 1 μg of total DNA used for each point. The Dual-Luciferase assay (Promega) was performed 40–48 h after transfection according to the manufacturer's directions. Each experiment was repeated at least three times using at least two different plasmid preparations.
To measure the turnover rate of MafA, βTC-3 cells were cultured in 100-mm dishes and first transfected with 4 μgofthe MafA expression construct. Medium containing 0.2 mm glucose and 18 μm cycloheximide was added 48 h post-transfection (termed zero time), and cell nuclear extracts (25) were prepared at the indicated times. The MafA and Pdx1 levels were measured by Western analysis using anti-Myc (Santa Cruz Biotechnology) and anti-Pdx1 (a gift of Dr. Chris Wright, Vanderbilt University) antibodies, respectively.
Ubiquitinylation Assay—HeLa cells were transfected on 100-mm plates with 4 μg of cytomegalovirus-MafA and/or 2 μg of cytomegalovirus-human HA-ubiquitin expression vector (a gift from Dr. Hal Moses, Vanderbilt University). The cells were then washed twice after 48 h with cold phosphate-buffered saline and lysed in radioimmune precipitation assay buffer (10 mm Tris-HCl, pH 8.0, 140 mm NaCl, 0.5% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 1 mm phenylmethylsulfonyl fluoride, and 2 μg/ml aprotinin). After centrifugation, the soluble protein was incubated overnight at 4 °C with anti-MafA antibody (1:100; Bethyl Laboratories) and immunoprecipitated with protein A/G-Sepharose beads (Santa Cruz Biotechnology). The beads were washed three times with radioimmune precipitation assay buffer, and the eluted proteins were separated by SDS-PAGE and then analyzed by immunoblotting using an anti-HA antibody (Sigma).
In Vivo Labeling of MafA and 32P-Phosphoamino Acid Analysis—HeLa cells were infected with adenovirus-driven MafA (5) and then grown in phosphate-free Dulbecco's modified Eagle's medium (Invitrogen) in the presence of [32P]orthophosphate for 18 h. The washed pelleted cells were suspended for 30 min in 0.3 ml of 8 m urea (in 20 mm Tris-HCl, pH 8.0, and 100 mm NaCl), and the dissolved proteins were mixed at room temperature with 0.1 ml of nickel-agarose beads (Qiagen) for 30 min. The MafA bound beads were washed two times with 1 ml of 8 m urea, once with 1 ml of 5 mm imidazole (in 20 mm Tris-HCl, pH 8.0, and 100 mm NaCl), and once with 1 ml of buffer alone (20 mm Tris-HCl, pH 8.0, and 100 mm NaCl), and then MafA was eluted with 0.1 ml of 500 mm imidazole (in 20 mm Tris-HCl, pH 8.0, and 100 mm NaCl). 32P-Labeled MafA was localized after 10% SDS-PAGE by x-ray film exposure and cut from the gel. The eluted protein was hydrolyzed with 6 n HCl as described previously (26). 32P-Amino acids were separated by two-dimensional electrophoresis (27), and cold phosphoserine (Sigma), phosphothreonine (Sigma), and phosphotyrosine (Sigma) served as position markers.
RESULTS
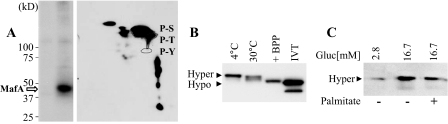
The Phosphorylation State of MafA Affects Mobility after SDS-PAGE—The DNA binding activity of mammalian MafA is inhibited by endogenous and exogenous (e.g. calf intestinal alkaline phosphatase and a rat brain-enriched phosphatase preparation (24, 28)) phosphatases. The phosphoamino acid composition of MafA was assessed in HeLa cells infected with an adenovirus-expressing mouse MafA in medium containing [32P]orthophosphate. Two-dimensional electrophoresis of acid-hydrolyzed, nickel affinity-purified 32P-labeled MafA revealed that the principal site of phosphorylation was serine, with a serine:threonine ratio of ∼12 (Fig. 1A).
FIGURE 1.
MafA mobility is affected by phosphorylation. A, adenovirus-expressed MafA was labeled with [32P]orthophosphate in HeLa cells. Left panel, 32P-labeled MafA was purified using nickel affinity chromatography and separated by SDS-PAGE. This purification strategy was utilized due to the histidine-rich region of MafA (amino acids 197–206) (1, 2). Right panel, 32P-labeled MafA was acid-hydrolyzed, and the labeled amino acids were separated by two-dimensional electrophoresis. The ratio of phosphoserine (P-S) to phosphothreonine (P-T), which are circled, was ∼12, as determined by scintillation counting. P-Y, phosphotyrosine. B, βTC-3 nuclear extract was incubated at 4 and 30 °C for 30 min or with rat brain-enriched phosphatase preparation (BPP) at 30 °C. The effect of phosphatase treatment was compared by anti-MafA Western analysis to in vitro translated (IVT) MafA. Hyper, hyperphosphorylated; Hypo, hypophosphorylated. C, MafA levels in nuclear extracts from rat islets cultured in 2.8 and 16.7 mm glucose for 24 h in the absence (–) or presence (+) of 0.5 mm palmitate and analyzed by anti-MafA Western blotting are shown. The results presented are representative of experiments performed on several independent occasions.
The phosphorylation status of MafA influenced mobility after SDS-PAGE. Thus, MafA migrated noticeably faster after either rat brain-enriched phosphatase preparation treatment or upon incubation of βTC-3 cell nuclear extract in the absence of protein phosphatase inhibitors at 30 °C (Fig. 1B), the permissible temperature of the endogenous MafA phosphatase(s) (24). The slower and faster mobility forms were termed hyperphosphorylated and hypophosphorylated MafA, respectively. The migration of phosphatase-treated MafA was similar to the reticulocyte lysate-translated protein, presumably reflecting suboptimal conditions for phosphorylation in this in vitro system.
MafA levels are very sensitive to a variety of metabolic effectors of β cell function, including glucose and fatty acids (14–17). To determine the influence of these effectors on MafA mobility, rat islets were next cultured under basal (2.8 mm) or stimulatory (16.7 mm) glucose concentrations or in the presence of palmitate, which was previously shown to inhibit MafA expression (15, 17). The 46-kDa hyperphosphorylated form was observed principally under all three culture conditions (Fig. 1C), suggesting that the phosphorylation event(s) that impacts MafA mobility is not rate-limiting in β cells.
Phosphorylation at Ser65 Alone Influences MafA Mobility—The biological significance of MafA phosphorylation has been examined most extensively with the avian protein (19, 20). For example, phosphorylation at Ser14 and Ser65 appears to potentiate quail MafA activation and transformation (19). These conserved amino acids can be phosphorylated by ERK2 in vitro (19), but this kinase does not appear to be involved in (at least) Ser65 phosphorylation in vivo (21). The kinase regulating Ser65 phosphorylation is unknown; however, its actions were recently shown to be essential for recruitment of GSK3 to act on neighboring Ser61, Thr57, Thr53, and Ser49 residues (20, 21), a process necessary for p300/CBP-associated factor co-activator binding (20).
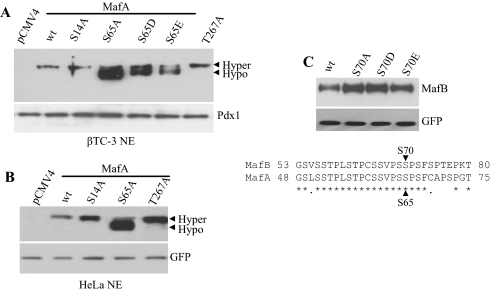
Here, we have examined how phosphorylation at Ser14, Ser65, and Thr267 could impact mammalian MafA mobility and activity. Thr267 is a conserved amino acid within the DNA-binding domain of MafA and a potential protein kinase A/protein kinase C site (29). Alanine substitution mutants in Myc-tagged MafA at Ser14, Ser65, or Thr267 were expressed in βTC-3 (Fig. 2A) and/or HeLa (Fig. 2B) cells. The migration of the S14A and T267A mutants was indistinguishable from the wild type, whereas the S65A mutant was clearly faster. An intermediate migrating form of MafA was produced with glutamic or aspartic acid mutants at Ser65 (Fig. 2A), suggesting that phosphorylation at this site directly impacts mobility. However, mutation of this conserved serine (Ser70) had no effect on the mobility of MafB (Fig. 2C), the other principal large Maf expressed in the islet (11, 30).
FIGURE 2.
MafA S65A has a faster mobility. A, βTC-3 cells were transfected with WT Myc-MafA and S14A, S65A, S65D, S65E, and/or T267A for 48 h. MafA forms in nuclear extract (NE) were analyzed via Western blotting using anti-Myc antibody. Pdx1 served as a loading control. Hyper, hyperphosphorylated; Hypo, hypophosphorylated. B, HeLa cells were co-transfected with WT Myc-MafA and S14A, S65A, or T267A with pCIG, a nuclear GFP expression construct, for 48 h. MafA forms in nuclear extract were analyzed via Western blotting using anti-MafA antibody. GFP was used as a transfection control. C, the WT MafB and S70A, S70D, S70E expression plasmids were co-transfected into HeLa cells with pCIG. MafB forms in nuclear extract were analyzed via Western blotting using anti-MafB antibody. GFP was used as a transfection control. The amino acid identity (denoted by asterisks) between MafA and MafB within the Ser65 region is shown. The Western blots shown reflect commonly obtained results.
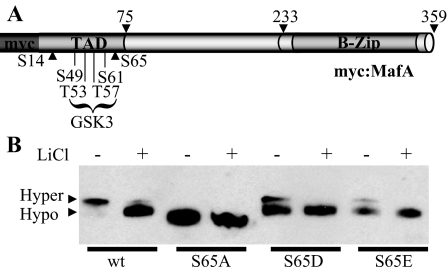
GSK3 Phosphorylates the S65E and S65D Mutants of MafA—Phosphorylation at Ser65 in MafA is necessary for the sequential actions of GSK3 on Ser49, Thr51, Thr57, and Ser61 (20, 21). To examine if S65D and S65E served as substrates for further phosphorylation, we incubated HeLa (Fig. 3) and βTC-3 (data not shown) cells expressing these mutants with LiCl, a GSK3 inhibitor (31). Notably, only the faster mobility, hypophosphorylated MafA band was detected with the wild type, S65D, and S65E forms in the presence of LiCl, although this inhibitor had no effect on S65A migration. The compression of the S65D and S65E bands after LiCl treatment indicates that both mimic Ser65 phosphorylation. However, these mutants were relatively poor GSK3 substrates as judged by the lower conversion to the slower mobility, hyperphosphorylated band (15 ± 10% conversion of S65D or S65E to the hyperphosphorylated form) (Fig. 3). Significantly, only the S65E mutant was degraded in the same manner as wild type MafA (see below).
FIGURE 3.
GSK3 phosphorylates the MafA S65D and S65E mutants, but not S65A. A, the GSK3 phosphorylation sites in the N-terminal transactivation domain (TAD) and basic leucine zipper (B-Zip) region of MafA are shown (20, 21). B, HeLa cells were transfected with plasmids encoding WT MafA and the Ser65 mutant and then treated for 8 h with 50 mm LiCl, as indicated. Nuclear extract was analyzed by Western blotting using anti-MafA antibodies, and a representative blot of this often repeated experiment is presented. Hyper, hyperphosphorylated; Hypo, hypophosphorylated.
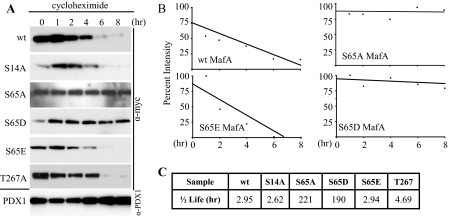
Phosphorylation at Ser65 Regulates MafA Protein Turnover—The S65A mutant affected not only MafA mobility but also apparently the steady-state level of the protein (Fig. 2, A and B). The change in mutant MafA levels was not a result of nuclear compartmentalization because S65A had the same nuclear enrichment pattern as wild type MafA (supplemental Fig. 1). To directly test whether Ser65 phosphorylation impacted MafA stability, S65A, S65D, S65E, and wild type MafA were expressed in βTC-3 cells in the presence and absence of a protein synthesis inhibitor, cycloheximide. Nuclear extracts were collected at various time points for Western blot analysis and indeed showed that turnover of S65A was profoundly reduced relative to the wild type (Fig. 4). In contrast, there was little or no effect on endogenous Pdx1 levels.
FIGURE 4.
S65E, and not S65D, has a protein turnover rate similar to WT MafA. A, the WT Myc-MafA and mutant constructs were expressed in βTC-3 cells for 48 h and then transferred into medium containing 0.2 mm glucose and 18 μm cycloheximide. Nuclear extract was prepared at the indicated times, and transfected MafA and endogenous Pdx1 protein levels were determined using anti-Myc and anti-Pdx1 antibodies, respectively. B, band intensity was measured and plotted as a percentage of the initial band intensity. No significant difference in turnover rate was found between S65A and S65D. C, the estimated half-life of WT MafA and mutants S14A, S65A, S65D, S65E, and T267A is shown (n = 3).
Strikingly, MafA S65E behaved similarly to the wild type in these protein turnover assays, whereas the S65D mutant was very stable, much like S65A (Fig. 4). Furthermore, the protein turnover rate of S14A and T267A was like that of wild type MafA, suggesting that modification of these amino acids does not impact protein stability. These results not only demonstrate that Ser65 plays a pivotal role in regulating MafA levels in β cells, but also illustrate that the protein degradation machinery can distinguish between the “phosphomimetic” S65D and S65E mutants, with recognition of only S65E.
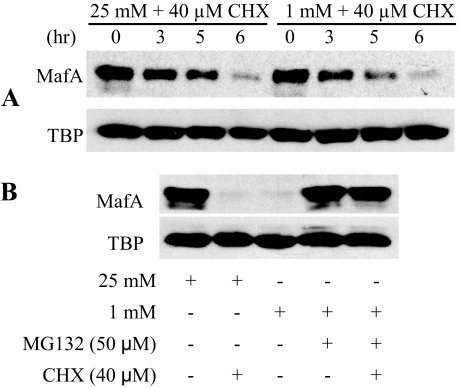
Only Wild Type MafA and the S65E Mutant Are Polyubiquitinylated—MafA levels are influenced by glucose, the most important physiological effector of islet β cell activity. Thus, increasing cellular glucose concentrations acutely stimulate (e.g. Fig. 1C) (5, 14, 16) and chronically high conditions inhibit (32) MafA mRNA and protein expression. The impact of glucose on MafA stability was examined in MIN6 β cells cultured under low-glucose (1 mm) and high-glucose (25 mm) conditions in the presence of cycloheximide; TATA-binding protein served as the internal control. In contrast to recent results suggesting that transfected MafA was less stable under low-glucose conditions (21), we found that the rate of endogenous MafA turnover was insensitive to the glucose concentration in the medium (Fig. 5A).
FIGURE 5.
MG132 protects MafA from proteasome-dependent degradation. A, MIN6 cells were cultured at 25 mm glucose for 18 h and then in either 1 or 25 mm glucose in the presence of 40 μm cycloheximide (CHX) for the indicated times. MafA and TATA-binding protein (TBP) levels were measured in the nuclear extracts by Western blot analysis. B, MIN6 cells were treated with MG132 for 18 h. MafA and TATA-binding protein levels in A and B were measured in the nuclear extracts by Western blot analysis.
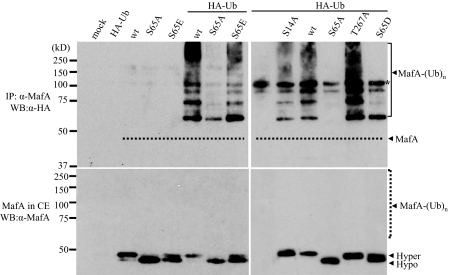
MafA degradation in cycloheximide-treated MIN6 cells was blocked by MG132 (Fig. 5B), an inhibitor of the ubiquitin-mediated proteasome pathway (33). To examine the specificity of this process in greater detail, a plasmid encoding HA-tagged ubiquitin was co-transfected with wild type MafA and S14A, S65A, S65D, S65E, and T267A mutant expression plasmids. As expected from the protein turnover experiments (Fig. 4), S65E and wild type MafA, and not the S65A and S65D mutants, were polyubiquitinylated (Fig. 6, top panels). The S14A and T267A mutants were also polyubiquitinylated, as predicted. Collectively, the data strongly indicate that phosphorylation at Ser65 regulates MafA stability. Notably, the higher molecular weight and polyubiquitinylated forms of MafA represent a very minor portion of the MafA pool in the cells, which is much less than nonubiquitinylated MafA (Fig. 6, bottom panels).
FIGURE 6.
Only S65E and WT MafA are polyubiquitinylated. HeLa cells were infected with the indicated Myc-MafA constructs in the presence or absence of a HA-ubiquitin (HA-Ub)-expressing plasmid. Top panels, to assess ubiquitinylation, immunoprecipitated (IP) MafA was subjected to anti-HA Western blotting (WB). The dashed line illustrates the location of nonubiquitinylated, hyperphosphorylated MafA, as shown directly in a MafA Western blot of whole cell extract (CE)(lower left panel). Notably, S65A and S65D are not polyubiquitinylated, whereas WT MafA and mutants S14A, S65E, and T267A are polyubiquitinylated. The asterisk denotes a nonspecific 100-kDa band. Bottom panels, MafA mutants in whole cell extract were detected by anti-MafA. The dashed line indicates the position of polyubiquitinylated MafA. Hyper, hyperphosphorylated; Hypo, hypophosphorylated.
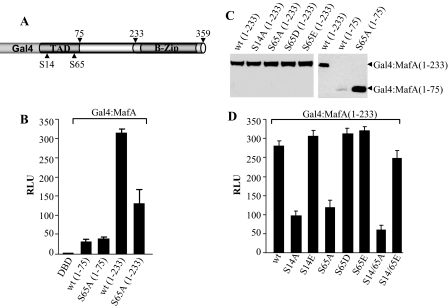
Phosphorylation at Ser65 Stimulates Transactivation—Ser65 is located within the N-terminal activation domain of large Maf proteins (19). To examine if the phosphorylation at Ser14 and Ser65 affected MafA-mediated transactivation, N-terminal sequences from 1–75 and 1–233 were fused in-frame to the DNA-binding domain of the Saccharomyces cerevisiae Gal4 transcription factor (Fig. 7A). Each of the Gal4 expression plasmids, together with a reporter plasmid containing five Gal4 DNA-binding sites upstream of the E1B TATA sequences, was transfected into HeLa cells. Strikingly, wild type Gal4-MafA-(1–75) was much less active than wild type Gal4-MafA-(1–233) (Fig. 7B). In addition, much lower levels of Gal4-MafA-(1–75) were found compared with Gal4-MafA-(1–233) as a result of Ser65-mediated degradation (compare wild type and S65A mutant Gal4-MafA-(1–75) in Fig. 7C).
FIGURE 7.
Phosphorylation at Ser14 and Ser65 potentiates MafA activation. A, the structure of the Gal4-MafA fusion proteins is shown. B and D, the (Gal4)5-driven reporter vector was co-transfected with WT and mutant Gal4-MafA-(1–75) and Gal4-MafA-(1–233), respectively, in HeLa cells. The phRL-TK-normalized results ± S.D. are shown relative to the Gal4 DNA-binding domain (DBD) vector. C, the Gal4-MafA-(1–233) or Gal4-MafA-(1–359) vectors were co-transfected with pCIG in HeLa cells. Nuclear extracts were subjected to a Gal4 DNA-binding domain Western blot. Each lane contains the same relative amount of GFP. Notably, the Ser65 mutants do not affect Gal4-MafA-(1–233) levels. The error bars depict the S.D. between experiments (n > 3). RLU, relative light units; TAD, transactivation domain; B-Zip, basic leucine zipper.
Interestingly, the Ser65 mutants did not affect Gal4-MafA-(1–233) levels (Fig. 7C), which enabled a straightforward comparison of S65A, S65D, and S65E activation ability. Both S65D and S65E stimulated Gal4-MafA-(1–233) activity, whereas S65A and S14A were relatively inactive (Fig. 7D). The increased activity of S14E Gal4-MafA-(1–233) suggests that phosphorylation throughout the activation domain is vital (Fig. 7D), as also supported by the ability of MafA to recruit the p300/CBP-associated factor co-activator after GSK3-mediated phosphorylation of Ser61, Thr57, Thr53, and Ser49 (20). Ser14, Ser65, and Thr267 were also found to influence insulin gene-driven reporter activity, with S14A (0.55 ± 0.09%) and T267A (0.52 ± 0.11%) specifically compromising expression relative to the wild type.
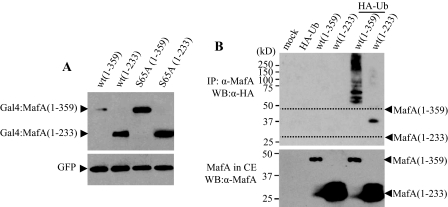
The MafA C-terminal Region (Amino Acids 234–359) Is a Ubiquitin-targeted Region—The ubiquitin-proteasome degradation pathway requires recognition of a degradation signal (e.g. Ser65 phosphorylation) by E3 ubiquitin ligase and polyubiquitinylation of lysine(s) (34). Both the relatively low lysine density in the N-terminal region of MafA and the comparable stability of the Gal4-MafA-(1–233) mutants suggested that a lysine within the Gal4 DNA-binding region may have been utilized in Gal4-MafA-(1–75) degradation (only 4 of 16 lysines in MafA are located between amino acids 1 and 233).
The level of the wild type and S65A versions of Gal4-MafA-(1–233) were compared with those of Gal4-MafA-(1–359) to examine the importance of the C-terminal lysine-rich region of MafA to protein stability. As expected of a lysine(s) necessary to ubiquitin-mediated degradation, Gal4-MafA-(1–359) protein levels were not only lower than Gal4-MafA-(1–233) protein levels but also increased in response to the S65A mutation (Fig. 8A). To further examine the significance of the C-terminal region in MafA degradation, the ubiquitinylation state of the MafA-(1–233) deletion mutant was compared with that of the wild type. This C-terminal truncation mutant was not polyubiquitinylated effectively, and much higher levels were in the cytoplasm compared with the wild type (Fig. 8B and supplemental Fig. 1).
FIGURE 8.
The C terminus (amino acids 234–359) of MafA is required for degradation. A, HeLa cells were co-transfected with WT or S65A mutant plasmids encoding Gal4-MafA-(1–233) or Gal4-MafA-(1–359) and pCIG. Nuclear extracts were analyzed by Western blotting using the anti-MafA and anti-GFP antibodies. B, HeLa cells were infected with Myc-MafA-(1–359) or Myc-MafA-(1–233) constructs in the presence or absence of a HA-ubiquitin (HA-Ub)-expressing plasmid, and immunoprecipitated (IP) MafA was subjected to anti-HA Western blotting (WB). CE, cell extract.
DISCUSSION
MafA is a key activator of adult islet β cell function, specifically through actions on genes associated with cell identity, including the insulin gene (1, 2, 12, 15). This transcription factor has been proposed to be a master regulator due to not only the importance of its target genes but also its unusual sensitivity to metabolic effectors (35). Here, we have examined the potential role of phosphorylation in regulating MafA activity and demonstrated that alanine mutations at Ser14 and Ser65 reduced activation. Notably, we found that MafA stability was specifically controlled by modification at Ser65 alone, as a glutamic acid substitution mutant was eliminated through the proteasome degradation pathway, whereas an alanine or aspartic acid mutant was not. Phosphorylation at Ser65 was also recently found to be the nucleating site for GSK3 actions at Ser49, Thr53, Thr57, and Ser61 in quail and mouse MafA (20, 21).
Two recognition signals are necessary for protein degradation in the ubiquitin-mediated proteasome pathway. The initiating event involves binding by E3 enzymes (ubiquitin ligase) to a specific degradation signal sequence (referred to as the degron), which in MafA represents Ser65 phosphorylation. In fact, the degron is commonly found within the activation domain region of transcription factors (34). Ubiquitin is covalently bound to the ε-amino group of a lysine residue(s), with polyubiquitinylation targeting substrates to the 26 S proteasome. Our results demonstrate that polyubiquitin chains are added to the lysine-rich C-terminal region of MafA, as the full-length Gal4 fusion protein was unstable and sensitive to Ser65 phosphorylation, but the truncated amino acid 1–233 chimera was stable and insensitive (Fig. 8A). Interestingly, MafA-(1–233) was principally found in the cytoplasm (supplemental Fig. 1), indicating that the C-terminal region also contributes to nuclear localization.
Significantly, the S65A mutant blocked degradation of MafA (Fig. 4) as well as the amino acid 1–359 (Fig. 8A) and amino acid 1–75 (Fig. 7C) chimeras. We conclude from these results that the MafA degron is defined by phosphorylation of Ser65 alone, as further supported by the instability of the S65E mutant. In contrast, it was recently suggested that MafA stability was regulated by GSK3 phosphorylation of Ser49, Thr53, Thr57, and Ser61 (20, 21). The principal utilization of multiple alanine site mutants within these studies likely contributes to this discrepancy, although it is noteworthy that only the S65A mutation was shown to stabilize MafA and eliminate MG132 sensitivity (compare S65A with T57A and T53A in Fig. 4D of Ref. 21). We believe that the ∼2-fold reduction in the rate of degradation of compound Ser49, Thr53, Thr57, and Ser61 mutants reflects poor recognition by E3 ligase (compare mutant 4A with WT in Fig. 4A of Ref. 20), as the stability of S65D illustrated the exquisite sensitivity of the conjugation machinery (Fig. 4). Notably, this 4A mutant only reduced the rate of MafA degradation, whereas S65A and S65D prevented degradation entirely. Furthermore, both S65E and S65D mutants of MafA were phosphorylated by GSK3 (Fig. 3), yet only S65E was subjected to the ubiquitin-mediated proteasome pathway. In addition, MafA was recently reported to be less stable at low- compared with high-glucose concentrations in β cells (21), which we did not observe (Fig. 5). Our focus on the regulation of endogenous MafA possibly explains the discrepancy with their transfection data. It is likely that glucose-induced MafA expression is regulated at the transcriptional level, as it shares many of the factors involved in glucose-stimulated insulin gene expression (15, 36).
Phosphorylation at Thr53 and Thr57 in MafA was confirmed using phosphosite-specific antibodies (20). We have subjected nickel affinity chromatography-purified MafA to mass spectrometry analysis to directly identify sites of phosphorylation, an approach that previously illustrated quail MafA Ser272 phosphorylation (i.e. equivalent to mouse Ser342 (37)). Presently, Ser14, Thr53, Ser56, Thr132, Ser234, Ser290, Ser297, and Ser342 phosphorylation has been found by mass spectrometry of mouse MafA (data not shown). Our inability to observe Ser65 phosphorylation probably reflects properties of the proteinase-released peptide, as even the unmodified form was undetectable. Importantly, the ability of S65E and S65D to influence the mobility of MafA by SDS-PAGE (Fig. 2) and their impact on its destruction and activation properties strongly suggest that this site is phosphorylated in vivo.
Ser14 phosphorylation contributed to transactivation by MafA (Fig. 7) but did not impact protein turnover (Fig. 4). The S14E mutant was equally active compared with the wild type and S14E/S65E in Gal4-MafA-(1–233), whereas the S14A/S65A double mutant was less active than the individual alanine site mutants (Fig. 7D). Collectively, our results demonstrate that phosphorylation at Ser65 alone influences both the steady-state levels and transactivation potential of MafA in β cells. Although Ser65 phosphorylation does not appear to be regulated in vivo (Fig. 1) (21), it is likely that the action of GSK3 (and other kinases) on the activation domain will be regulated to potentiate transactivation capabilities and reduce degradation. Evidence supporting such an idea was provided by showing that GSK3-mediated recruitment of the p300/CBP-associated factor co-activator impacted MafA stability (20).
Supplementary Material
Acknowledgments
We thank Dr. William Tansey for scientific comments and encouragement.
This work was supported, in whole or in part, by National Institutes of Health Grant P01 DK42502 (to R. S.), Molecular Endocrinology Training Program Postdoctoral Training Grant 5T32 DK07563-20 (to R. B.), Postdoctoral Research Training Grant in Diabetes and Endocrinology 5T32 DK007061-34 to (N. L. V.), Grant R01 DK58096 (to V. P.), Postdoctoral Fellowship F32 DK070406 (to D. K. H.), Grant R01 DK067581 (to S. O.), and Grant DK70787 (to B. E. W.). This work was also supported by an American Heart Association Predoctoral fellowship (to N. L. V.). Partial support was also provided to the Molecular Biology Core Laboratory by the Vanderbilt University Diabetes Research and Training Center (United States Public Health Service Grant P60DK20593). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Footnotes
The abbreviations used are: ERK, extracellular signal-regulation protein kinase; HA, hemagglutinin; WT, wild type; GFP, green fluorescent protein.
References
- 1.Matsuoka, T.-A., Zhao, L., Artner, I., Jarrett, H. W., Friedman, D., Means, A., and Stein, R. (2003) Mol. Cell. Biol. 23 6049–6062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olbrot, M., Rud, J., Moss, L. G., and Sharma, A. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 6737–6742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Artner, I., Blanchi, B., Raum, J. C., Guo, M., Kaneko, T., Cordes, S., Sieweke, M., and Stein, R. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 3853–3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishimura, W., Rowan, S., Salameh, T., Maas, R. L., Bonner-Weir, S., Sell, S. M., and Sharma, A. (2008) Dev. Biol. 314 443–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao, L., Guo, M., Matsuoka, T.-A., Hagman, D. K., Parazzoli, S. D., Poitout, V., and Stein, R. (2005) J. Biol. Chem. 280 11887–11894 [DOI] [PubMed] [Google Scholar]
- 6.Matsuoka, T.-A., Artner, I., Henderson, E., Means, A., Sander, M., and Stein, R. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 2930–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aramata, S., Han, S.-I., and Kataoka, K. (2007) Endocr. J. 54 659–666 [DOI] [PubMed] [Google Scholar]
- 8.Babu, D. A., Deering, T. G., and Mirmira, R. G. (2007) Mol. Genet. Metab. 92 43–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Habener, J. F., Kemp, D. M., and Thomas, M. K. (2005) Endocrinology 146 1025–1034 [DOI] [PubMed] [Google Scholar]
- 10.Kim, S. K., and MacDonald, R. J. (2002) Curr. Opin Genet. Dev. 12 540–547 [DOI] [PubMed] [Google Scholar]
- 11.Artner, I., Le Lay, J., Hang, Y., Elghazi, L., Schisler, J. C., Henderson, E., Sosa-Pineda, B., and Stein, R. (2006) Diabetes 55 297–304 [DOI] [PubMed] [Google Scholar]
- 12.Zhang, C., Moriguchi, T., Kajihara, M., Esaki, R., Harada, A., Shimohata, H., Oishi, H., Hamada, M., Morito, N., Hasegawa, K., Kudo, T., Engel, J. D., Yamamoto, M., and Takahashi, S. (2005) Mol. Cell. Biol. 25 4969–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kroon, E., Martinson, L. A., Kadoya, K., Bang, A. G., Kelly, O. G., Eliazer, S., Young, H., Richardson, M., Smart, N. G., Cunningham, J., Agulnick, A. D., D'Amour, K. A., Carpenter, M. K., and Baetge, E. E. (2008) Nat. Biotechnol. 26 443–452 [DOI] [PubMed] [Google Scholar]
- 14.Kataoka, K., Han, S.-I., Shioda, S., Hirai, M., Nishizawa, M., and Handa, H. (2002) J. Biol. Chem. 277 49903–49910 [DOI] [PubMed] [Google Scholar]
- 15.Poitout, V., Hagman, D., Stein, R., Artner, I., Robertson, R. P., and Harmon, J. S. (2006) J. Nutr. 136 873–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vanderford, N. L., Andrali, S. S., andÖzcan, S. (2007) J. Biol. Chem. 282 1577–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagman, D. K., Hays, L. B., Parazzoli, S. D., and Poitout, V. (2005) J. Biol. Chem. 280 32413–32418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ueki, K., Okada, T., Hu, J., Liew, C. W., Assmann, A., Dahlgren, G. M., Peters, J. L., Shackman, J. G., Zhang, M., Artner, I., Satin, L. S., Stein, R., Holzenberger, M., Kennedy, R. T., Kahn, C. R., and Kulkarni, R. N. (2006) Nat. Genet. 38 583–588 [DOI] [PubMed] [Google Scholar]
- 19.Benkhelifa, S., Provot, S., Nabais, E., Eychene, A., Calothy, G., and Felder-Schmittbuhl, M. P. (2001) Mol. Cell. Biol. 21 4441–4452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rocques, N., Abou Zeid, N., Sii-Felice, K., Lecoin, L., Felder-Schmittbuhl, M. P., Eychene, A., and Pouponnot, C. (2007) Mol. Cell 28 584–597 [DOI] [PubMed] [Google Scholar]
- 21.Han, S.-I., Aramata, S., Yasuda, K., and Kataoka, K. (2007) Mol. Cell. Biol. 27 6593–6605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lillie, J. W., and Green, M. R. (1989) Nature 338 39–44 [DOI] [PubMed] [Google Scholar]
- 23.Wang, J.-C., Stafford, J. M., and Granner, D. K. (1998) J. Biol. Chem. 273 30847–30850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao, L., Cissell, M. A., Henderson, E., Colbran, R., and Stein, R. (2000) J. Biol. Chem. 275 10532–10537 [DOI] [PubMed] [Google Scholar]
- 25.Schreiber, E., Matthias, P., Muller, M. M., and Schaffner, W. (1989) Nucleic Acids Res. 17 6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunter, T., and Sefton, B. M. (1980) Proc. Natl. Acad. Sci. U. S. A. 77 1311–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ostrovsky, P. C., and Maloy, S. (1995) Genes Dev. 9 2034–2041 [DOI] [PubMed] [Google Scholar]
- 28.Matsuoka, T.-A., Zhao, L., and Stein, R. (2001) J. Biol. Chem. 276 22071–22076 [DOI] [PubMed] [Google Scholar]
- 29.Civil, A., van Genesen, S. T., and Lubsen, N. H. (2002) Nucleic Acids Res. 30 975–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishimura, W., Kondo, T., Salameh, T., El Khattabi, I., Dodge, R., Bonner-Weir, S., and Sharma, A. (2006) Dev. Biol. 293 526–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies, S. P., Reddy, H., Caivano, M., and Cohen, P. (2000) Biochem. J. 351 95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harmon, J. S., Stein, R., and Robertson, R. P. (2005) J. Biol. Chem. 280 11107–11113 [DOI] [PubMed] [Google Scholar]
- 33.Kim, D., Kim, S. H., and Li, G. C. (1999) Biochem. Biophys. Res. Commun. 254 264–268 [DOI] [PubMed] [Google Scholar]
- 34.Muratani, M., and Tansey, W. P. (2003) Nat. Rev. Mol. Cell Biol. 4 192–201 [DOI] [PubMed] [Google Scholar]
- 35.Wang, H., Brun, T., Kataoka, K., Sharma, A. J., and Wollheim, C. B. (2007) Diabetologia 50 348–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raum, J. C., Gerrish, K., Artner, I., Henderson, E., Guo, M., Sussel, L., Schisler, J. C., Newgard, C. B., and Stein, R. (2006) Mol. Cell. Biol. 26 5735–5743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sii-Felice, K., Pouponnot, C., Gillet, S., Lecoin, L., Girault, J. A., Eychene, A., and Felder-Schmittbuhl, M. P. (2005) FEBS Lett. 579 3547–3554 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.