Abstract
Chemerin is a potent chemoattractant for cells expressing the serpentine receptor CMKLR1 (chemokine-like receptor 1), such as plasmacytoid dendritic cells and tissue macrophages. The bioactivity of chemerin is post-translationally regulated; the attractant circulates in blood in a relatively inactive form (prochemerin) and is activated by carboxyl-terminal proteolytic cleavage. We discovered that plasma carboxypeptidase N (CPN) and B (CPB or activated thrombin-activable fibrinolysis inhibitor, TAFIa) enhanced the bioactivity of 10-mer chemerin peptide NH2-YFPGQFAFSK-COOH by removing the carboxyl-terminal lysine (K). Sequential cleavages of either a prochemerin peptide (NH2-YFPGQFAFSKALPRS-COOH) or recombinant full-length prochemerin by plasmin and CPN/CPB substantially increased their chemotactic activities. Endogenous CPN present in circulating plasma enhanced the activity of plasmin-cleaved prochemerin. In addition, we discovered that platelets store chemerin protein and release it upon stimulation. Thus circulating CPN/CPB and platelets may potentially contribute to regulating the bioactivity of leukocyte chemoattractant chemerin, and further extend the molecular link between blood coagulation/fibrinolysis and CMKLR1-mediated immune responses.
Chemerin is a recently discovered chemoattractant molecule that is predicted to share structural similarity with cystatins (cysteine protease inhibitors) and cathelicidin precursors (antibacterial peptides) (1). Chemerin is present in circulating blood and several human inflammatory fluids (1). Even though chemerin is not similar to CXC and CC chemokines based on primary amino acid sequence, it functions like a chemokine in that it induces leukocyte migration and intracellular calcium mobilization. Chemerin receptor chemokine-like receptor 1 (CMKLR1,3 also named ChemR23) is a G protein-coupled receptor specifically expressed by circulating human plasmacytoid dendritic cells, natural killer cells, and tissue macrophages (1–5). In their capacity as antigen-presenting cells, plasmacytoid dendritic cells and macrophages can influence the activation of many other cell types, including monocytes, myeloid dendritic cells, B cells, T cells, and natural killer cells; thus chemerin appears to be an important chemoattractant in both innate and adaptive immune responses (2, 6, 7).
Chemerin circulates in blood in an inactive prochemerin form at low nanomolar concentrations (∼3 nm) (4). Its chemotactic activity is released following proteolytic cleavage of its carboxyl-terminal amino acids by serine proteases of the coagulation, fibrinolytic, and inflammatory cascades (4, 8). These include factor XIIa, VIIa, plasmin, neutrophil elastase, and mast cell tryptase. Of interest, staphopain B, a cysteine protease secreted by Staphylococcus aureus, also cleaves prochemerin and converts it into a potent chemoattractant (9). Interestingly, the cleavage sites in the labile carboxyl terminus (NH2-YFPGQFAFSKALPRS-COOH) are not conserved, and the cleavage products generated by chemerin-activating proteases display different potencies in bioactivity assays. Based on synthetic peptides, the 9-mer NH2-YFPGQFAFS-COOH is the most active, but it is still not as active as intact cleaved chemerin protein, indicating that the amino-terminal part of chemerin is required for maximal activity (4, 10).
Plasma carboxypeptidases CPN and CPB cleave the basic amino acids arginine or lysine from the carboxyl terminus of proteins or peptides such as bradykinin and complement proteins C3a and C5a. CPN is a constitutively active zinc metalloprotease present in plasma at a concentration of about 100 nm and is considered the major anaphylatoxins inhibitor (11), generating inactive “desArg” forms of C3a and C5a. In contrast, CPB exists in plasma as a proenzyme, proCPB, or thrombin-activable fibrinolysis inhibitor (TAFI) at a concentration of about 50 nm and is activated by thrombin in complex with thrombomodulin on the vascular endothelial surface. CPB inhibits fibrin degradation by removing carboxyl-terminal lysines from partially digested fibrin, which prevents further incorporation of fibrinolytic plasminogen and tissue plasminogen activator (12, 13). CPB is thermolabile and has a half-life of ∼15 min at 37 °C (14). We have shown that CPB also has broad substrate reactivity and is able to cleave and inactivate bradykinin, C3a, C5a, and thrombin-cleaved osteopontin (15–17). CPN and CPB may play complementary roles, with the former being constitutively active and capable of regulating systemic anaphylatoxins, and the latter activated locally at sites of vascular injury to provide site-specific anti-inflammatory control. Peptidases can also modulate the biological activity of certain chemokines (4). For example, dipeptidyl peptidase (DPP-IV/CD26), a serine protease, inactivates CXCL9, CXCL10, CXCL11, and CXCL12 by cleaving these chemokines in the amino terminus (18, 19).
Platelets store a variety of potent cytokines and chemokines within α-granules that are released upon cell activation. Platelet degranulation products, particularly the leukocyte chemoattractants, which include CXCL4 (platelet factor 4), β-thromboglobulin, CCL5 (RANTES), CCL7 (monocyte chemotactic protein 3), and CXCL12 (stromal-derived factor 1), may contribute to host defense and also play a role in pathophysiologic conditions (20, 21). For example, platelet factor 4 forms complexes with heparin in blood or some glycosaminoglycans on platelet surfaces to form the major antigen implicated in heparin-induced thrombocytopenia (22, 23). Platelets not only store CXCL12 but also express its receptor CXCR4, a coreceptor for cellular entry of human immunodeficiency virus, type 1, suggesting that platelets may be involved in host defense (24).
In this study, we found that plasma CPN or CPB can function in concert with plasmin to elicit and augment the chemotactic activity of prochemerin. Furthermore, we show that platelets could store and release partially active chemerin upon activation. Thus circulating CPN/CPB and platelets may contribute to regulating the bioactivity of leukocyte chemoattractant chemerin and further extend the molecular link between blood coagulation/fibrinolysis and CMKLR1-mediated immune responses.
EXPERIMENTAL PROCEDURES
Materials—Recombinant human chemerin21–157, polyclonal goat anti-human chemerin antibodies, and biotinylated polyclonal goat anti-human antibodies were from R & D Systems (Minneapolis, MN). Peptides 9-mer, YFPGQFAFS (chemerin149–157); 10-mer, YFPGQFAFSK (chemerin149–158); and 15-mer, YFPGQFAFSKALPRS (chemerin149–163) were synthesized by Elim Biopharmaceuticals (Hayward, CA). Human plasmin and α-thrombin were purchased from Hematologic Technologies (Essex Junction, VT). dl-2-Mercaptomethyl-3 guanidinoethylthiopropanoic acid (MGTA) was obtained from Calbiochem (La Jolla, CA). Collagen and ADP were from Chrono-log (Havertown, PA). Human plasma-derived CPB (TAFIa) and a CPB activity kit were from American Diagnostica (Stamford, CT). Thrombin receptor-activating peptide (SFLLRN peptide), d-phenylalanyl-l-prolyl-l-arginine chloromethyl ketone (PPACK), heparin-agarose, and bovine serum albumin were from Sigma. Human soluble thrombomodulin and recombinant CPN were kind gifts from Drs. John Moser and Mariko Nagashima (Berlex Biosciences, Richmond, CA). Hep3B and MEG-01 cells were from the American Type Culture Collection.
Preparation of Recombinant Prochemerin—Recombinant prochemerin was purified as previously published (8). Briefly, prochemerin with a carboxyl-terminal His6 tag was cloned into pACGP67 (BD Biosciences) and transfected into Sf-9 cells. The mature prochemerin protein has the amino acid sequence NH2-ADPELTEAQ...FAFSKALPRSPHHHHHH-COOH, where the underlined residues are not native. After viral amplification, prochemerin was expressed by adding high titer virus to shaker flasks containing Hi-5 insect cells in Ex-cell 420 medium (JRH Biosciences). After incubation for 2–3 days at 27.5 °C, the supernatant was harvested by centrifugation, filtered to 0.22 μm, and concentrated at 4 °C using a tangential flow concentrator with a 3-kDa cut-off filter (Filtron). Prochemerin was purified by nickel-Sepharose affinity chromatography (American Biosciences) and C-18 reverse phase HPLC (Vydac). The protein was lyophilized and checked for purity using electrospray mass spectrometry.
Tissue Culture—Murine pre-B lymphoma L1.2 cells were stably transfected with human CMKLR1 or empty vector pcDNA3 (Invitrogen) and maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum and 1 mg/ml geneticin (Invitrogen).
In Vitro Transwell Chemotaxis Assay—24-well plates with 5-μm pore size Transwell inserts (Costar) were used for the chemotaxis assays. 200 μl of cells (106 cells/ml) in 0.3% bovine serum albumin/Hank's solution were added to the top well, and test samples were added to the bottom well in 500 μl of solution. The cells that migrated to the lower chamber after 3 h at 37 °C were counted by flow cytometry, and the results are reported as cells/ml in the lower chamber.
Preparation of Platelet-rich Plasma, Platelet-poor Plasma, Washed Platelets, and Platelet Lysates—Blood was drawn into tubes containing 3.8% sodium citrate (9:1 v/v) and platelet-rich plasma (PRP) prepared following standard procedure (25). Platelet-poor plasma was prepared by spinning the PRP at 1200 × g for 10 min at room temperature. The platelets were washed with PIPES buffer (25 mm PIPES, 137 mm NaCl, 4 mm KCl, and 0.1% glucose) at pH 6.4 as previously described (25). Platelet lysates were obtained by lysing washed platelets with radioimmune precipitation assay lysis buffer (Upstate, NY) with protease inhibitors. The mixture was spun at 10,000 × g, and the supernatant protein concentration was determined by Bradford protein assay.
Reverse Transcription (RT)-PCR—Total RNA was prepared from human washed platelets and Hep3B and MEG-01 cells by using TRIzol reagent. Total RNA (∼1 μg) was converted to cDNAs using the oligo(dT) primer and Superscript II enzyme (Invitrogen). The specific primers used for cloning a 229-bp chemerin fragment were GAAGAAACCCGAGTGCAAAG (forward) and CTTGGAGAAGGCGAACTGTC (reverse) (the annealing temperature was 57 °C, 35 cycles).
HPLC Analysis of Chemerin Peptides Cleavage—To evaluate the efficiency of synthetic chemerin 10-mer cleavage by CPN or CPB, 50 μl of 10-mer (1 μm) was treated with either CPN or CPB (30 nm) for 30 min at 37 °C, and the reaction mixtures were loaded onto a Waters C18 (4.6 × 150 mm) column and separated with a 0–35% acetonitrile gradient in 0.1% trifluoroacetic acid (v/v) by HPLC. For 15-mer cleavage, 15-mer (1 μm) was incubated with plasmin (1 μm) at 37° for 30 min; the reaction was then terminated by PPACK (serine protease inhibitor) (10 μm). CPN or CPB (30 nm) was added and incubated for 30 min at 37 °C. 40 μl of each reaction mixture (15-mer, 15-mer plus plasmin, 15-mer plus plasmin and CPN/CPB) was analyzed by reverse phase HPLC as described above.
Kinetic Analysis of Hydrolysis of 10-mer Chemerin Peptides by CPB and CPN—Michaelis-Menten kinetics was used to determine the Km and kcat for the hydrolysis of 10-mer peptide (YFPGQFAFSK) by CPB and CPN. The concentrations of 10-mer peptides ranged from 20 to 320 μm and were digested with 50 nm CPB or 5 nm CPN for 5 min at 37 °C in assay buffer. The reactions were stopped by boiling for 5 min. Cleaved peptide was resolved by HPLC, and the nmol of peptide generated was determined from the peak area of cleaved peptide. The values for Km and kcat were determined by plotting the initial velocities of cleavage against the different substrate concentrations and then fitting to the Michaelis-Menten equation by nonlinear regression analysis as previously described (15). The experiments were performed in duplicate, and the data were pooled for analysis.
CPB Activity Assays—100 μl of CPN (10 nm) and CPB (15 nm) were added to a 96-well plate in the presence or absence of MGTA at the indicated concentrations. 50 μl of chromogenic CPB (TAFIa) substrate was used in each well as described in the Actichrome CPB kit. Activated CPB (TAFIa) ranging from 0.125 to 2 μg/ml was used to construct the standard curve. All of the tests were performed in duplicate. The plate was placed in an ELISA plate reader at 37 °C with constant mixing, and the absorbance at 420 nm was read 30 min after sample addition.
Development of Sandwich ELISA for Chemerin—Polyclonal goat anti-human chemerin antibodies were used to coat 96-well plates at a concentration of 4 μg/ml. Biotinylated polyclonal goat anti-human chemerin antibodies (0.2 μg/ml) and horseradish peroxidase-labeled streptavidin was used to detect bound chemerin protein. The lower limit of detection of chemerin in this assay was ∼0.5 ng/ml. For the determination of chemerin levels in plasma, the samples were diluted 10-fold before assay.
Mass Spectrometry—MALDI-TOF mass spectrometry was performed by the Stanford Protein and Nucleic Acid Core Facility.
Statistical Analyses—The data are expressed as the means ± S.D., and statistical evaluation was performed using Student's t test. Differences were considered to be significant when p < 0.05 (*) or 0.005 (**).
RESULTS
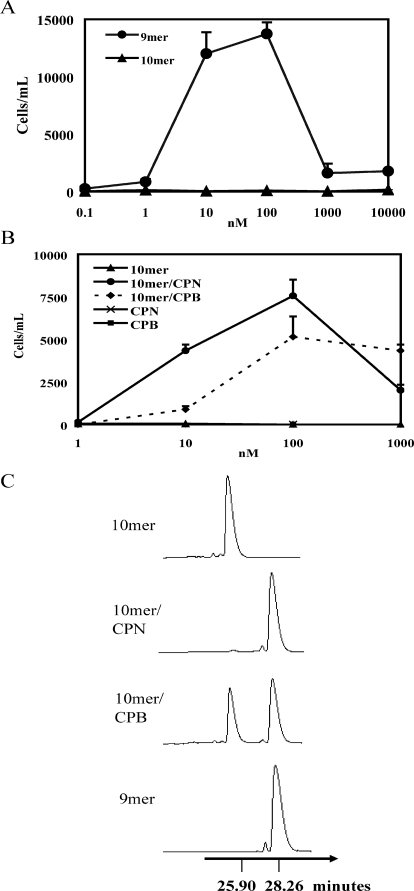
CPN and CPB Up-regulate Chemerin 10-mer Activity by Removing the Carboxyl-terminal Lysine Residue—The synthetic 9-mer chemerin peptide YFPGQFAFS (chemerin149–157) induced substantial, dose-dependent migration of human CMKLR1-L1.2 transfectants (Fig. 1A) with a peak response occurring at 10–100 nm. Empty vector transfected controls did not migrate to the 9-mer (data not shown), The 10-mer peptide YFPGQFAFSK (chemerin149–158) did not induce any significant CMKLR1-dependent chemotaxis even at 10 μm concentration (Fig. 1A). Treatment of the 10-mer peptide with CPN or CPB, however, substantially enhanced the chemotactic activity of the peptide (Fig. 1B). CPN and CPB alone (tested at 100 nm) did not induce CMKLR1/L1.2 transfectant migration (Fig. 1B). We analyzed the mixtures of 10-mer treated with either CPN or CPB by HPLC and found that the 10-mer/CPN mixture had a major peak with a retention time of 28.25 min, which is almost identical to that of the purified 9-mer (28.26 min), and different from the 10-mer (25.90 min). For the 10-mer treated with CPB, we detected two major fractions corresponding to the 10-mer (25.90 min) and 9-mer (28.28 min) (Fig. 1C). The efficiency of 10-mer cleavage by CPB at 30 nm was about 60%, likely because CPB is thermolabile under the experimental conditions used. In this work, CPN was also used at 30 nm to compare with CPB enzymatic activity. Of note, the plasma levels of proCPB and CPN are ∼50 and 100 nm, respectively (11, 14).
FIGURE 1.
CPN and CPB up-regulate chemerin 10-mer activity by removing the carboxyl-terminal lysine. A and B, in vitro transwell chemotaxis of CMKLR1/L1.2 transfectants to synthetic 9- and 10-mer chemerin peptides (A) and to CPN (30 nm) or CPB (30 nm)-treated 10-mer peptides at 37 °C for 30 min (B). The results represent one of three independent experiments and are expressed as the means ± S.D. (n = 3). C, HPLC analysis of the chemerin 10-mer cleavage products generated by CPN and CPB. 50 μl of 10-mer (1 μm) was treated with either CPN or CPB (30 nm) for 30 min at 37 °C, and the reaction mixtures were separated by HPLC.
Determination of the Kinetic Parameters for the Hydrolysis of 10-mer Peptide (Chemerin149–158) by CPB and CPN—The hydrolysis of chemerin149–158 by CPB gave estimates for Km (122.8 ± 6.4 μm), kcat (2.7 ± 0.1 s–1), and kcat/Km (2.2 × 104 m–1 s–1) (Table 1). The concentrations of chemerin149–158 ranged from 20 to 320 μm, and chemerin was digested with 50 nm CPB. The kcat/Km for chemerin cleavage was about 10-fold less efficient compared with bradykinin and C5a66–74, C3a69–77 but comparable with fibrinopeptide γ-Lys77–85. Meanwhile, the kcat/Km of 10-mer cleavage by CPN is 4.7 × 105 m–1 s–1, which is about 20-fold faster than CPB, and is about 20-fold faster than bradykinin and C5a peptide but similar to that of C3a peptide.
TABLE 1.
Hydrolysis of chemerin 10-mer peptides by CPB and CPN
Chemerin peptides ranging from 20 to 320 μm were digested with CPB or CPN as described under “Experimental Procedures.” The values for Km, kcat, and kcat/Km were compared with those obtained from CPB and CPN cleavages of peptides derived from bradykinin, C5a, C3a, and fibrinopeptides (FB) α, β, and γ (15).
| Substrate | Enzyme | Km | kcat | kcat/Km |
|---|---|---|---|---|
| μm | s-1 | m-1 s-1 | ||
| Chemerin | CPB | 122.8 ± 6.4 | 2.7 ± 0.1 | 2.2 × 104 |
| Bradykinin | CPB | 70.6 ± 4.8 | 19.7 ± 4.8 | 2.8 × 105 |
| C5a66-74 | CPB | 219.0 ± 16.2 | 29.5 ± 0.7 | 1.3 × 105 |
| C3a69-77 | CPB | 35.9 ± 6.6 | 8.4 ± 0.6 | 2.3 × 105 |
| FBα-Arg96-104 | CPB | 361.4 ± 69.2 | 1.5 ± 0.1 | 4.2 × 103 |
| FBβ-Lys125-133 | CPB | 14.3 ± 0.7 | 13.6 ± 0.2 | 9.5 × 105 |
| FBγ-Lys54-62 | CPB | 34.0 ± 4.1 | 2.6 ± 0.1 | 7.6 × 104 |
| FBγ-Lys77-85 | CPB | 238.9 ± 24.2 | 5.9 ± 0.3 | 2.5 × 104 |
| Chemerin | CPN | 170.6 ± 27.2 | 80.35 ± 5.0 | 4.7 × 105 |
| Bradykinin | CPN | 302.7 ± 29.1 | 9.1 ± 0.2 | 3.0 × 104 |
| C5a66-74 | CPN | 602.2 ± 74.3 | 9.3 ± 0.4 | 1.5 × 104 |
| C3a69-77 | CPN | 77.1 ± 11.2 | 57.9 ± 2.1 | 7.5 × 105 |
| FBα-Arg96-104 | CPN | 448.9 ± 43.8 | 2.9 ± 0.1 | 6.5 × 103 |
| FBβ-Lys125-133 | CPN | 53.2 ± 4.9 | 109.1 ± 3.6 | 2.1 × 106 |
| FBγ-Lys54-62 | CPN | 657.6 ± 20.5 | 3.5 ± 0.1 | 5.3 × 103 |
| FBγ-Lys77-85 | CPN | 3727.0 ± 408.6 | 11.8 ± 0.8 | 3.2 × 103 |
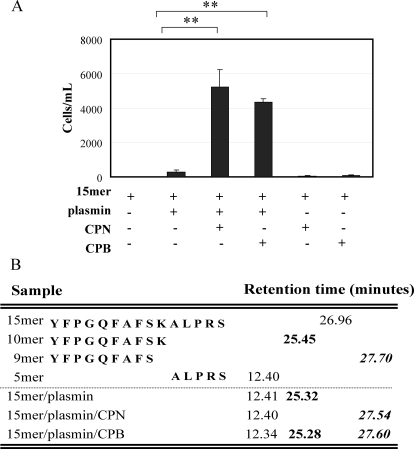
Sequential Proteolysis of Prochemerin 15-mer Peptide by Plasmin and Carboxypeptidases Synergistically Enhances Bioactivity—The synthetic chemerin 15-mer peptide, YFPGQFAFSKALPRS (chemerin149–163) is chemotactically inert (Fig. 2A). Treatment of the 15-mer with CPN or CPB alone had no effect on chemerin bioactivity. However, sequential treatment of the 15-mer with plasmin and CPN or CPB dramatically enhanced its chemotactic activity. The proteolytic products were evaluated by HPLC to determine processing sites. Plasmin first cleaved the 15-mer (retention time, 29.96 min) to 10-mer (retention time: 25.3 min; the peak with the retention time at 12.4 min is the carboxyl-terminal plasmin-generated 5-mer). CPN or CPB then converted the 10-mer to the 9-mer (retention time, ∼27.6 min) (Fig. 2B).
FIGURE 2.
Sequential proteolysis of prochemerin 15-mer by plasmin and carboxypeptidases generates bioactive chemerin 9-mer. A, in vitro transwell chemotaxis of CMKLR1/L1.2 cells to prochemerin 15-mer in the presence or absence of plasmin, CPB or CPN. 15-mer peptide (1 μm) was incubated with plasmin (1 μm) at 37 °C for 30 min; the reaction was then terminated by PPACK (10 μm). CPN or CPB (30 nm) was added and incubated for 30 min at 37 °C. The final concentration of 15-mer used for the assay was 100 nm. The results represent one of three independent experiments and are expressed as the means ± S.D. (n = 3). **, p < 0.005. B, HPLC analysis of prochemerin 15-mer cleavage by plasmin, plasmin/CPN, or plasmin/CPB.
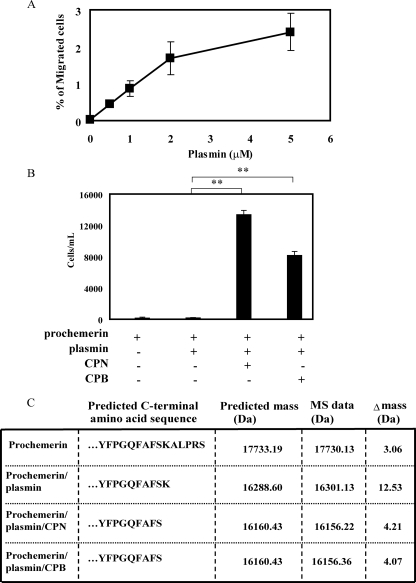
Sequential Proteolysis of Prochemerin by Plasmin and Carboxypeptidases Synergistically Enhances Bioactivity—We next asked whether sequential treatment of chemotactically inert concentrations of prochemerin by plasmin and CPN or CPB could activate the attractant and generate the NH2-YFPGQFAFS-COOH form. Plasmin alone cleaved prochemerin and increased its chemotactic bioactivity (Fig. 3A). Sequential treatment of prochemerin with plasmin and CPN or CPB, however, dramatically enhanced its chemotactic activity (Fig. 3B). The proteolytic products were evaluated by mass spectrometry to determine the processing sites (Fig. 3C). Plasmin first cleaved prochemerin (17730.12 Da) to the NH2-YFPGQFAFSK-COOH form (16301.13 Da), and then CPN or CPB removed the terminal lysine to generate a desLys form (16156.22/16156.36 Da) with enhanced activity. Prochemerin treated with CPN and CPB alone did not induce CMKLR1-mediated chemotaxis (data not shown).
FIGURE 3.
Sequential proteolysis of prochemerin protein by plasmin and carboxypeptidases generates a potent chemerin isoform. A, dose-response curve of prochemerin activation by plasmin assayed by CMKLR1/L1.2 cells chemotaxis. B, in vitro transwell chemotaxis of CMKLR1/L1.2 cells to full-length recombinant prochemerin protein, prochemerin/plasmin, prochemerin/plasmin/CPN, or CPB. The final concentration of chemerin used for the assay was 0.5 nm. The results represent one of three independent experiments and are expressed as the means ± S.D. (n = 3). **, p < 0.005. C, MALDI-TOF mass spectrometry analysis of prochemerin cleavage by plasmin, plasmin/CPN, or plasmin/CPB. The concentrations of plasmin, CPN, and CPB used in B and C were 1 μm, 30 nm, and 30 nm, respectively.
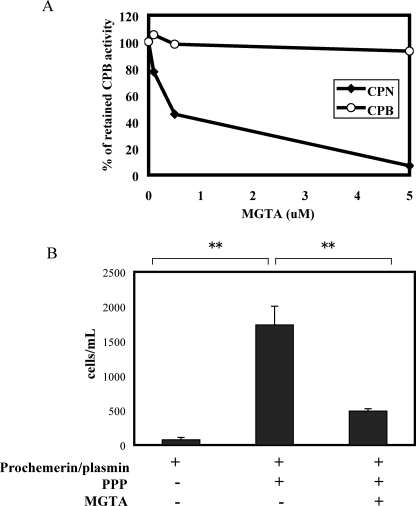
Endogenous Plasma CPN Is Critical for the Increased Activity of Plasmin-cleaved Prochemerin—Prochemerin was incubated with plasmin and then treated with PPACK to inhibit serine protease activity. Plasmin-treated prochemerin was added to normal platelet-poor plasma (PPP) as well as PPP treated with MGTA, a specific inhibitor for CPN but not CPB (Fig. 4A). Plasmin-cleaved prochemerin did not induce CMKLR1-mediated chemotaxis (Fig. 4B). Incubation with PPP, however, dramatically increased its bioactivity, which was inhibited by MGTA, indicating that endogenous plasma CPN is critical for activating low concentrations of plasmin-cleaved chemerin (Fig. 4B).
FIGURE 4.
Endogenous plasma CPN increases the bioactivity of plasmin-cleaved prochemerin. A, MGTA specifically inhibits CPN but not CPB. 1 μg/ml of either CPN or CPB was used. B, in vitro transwell chemotaxis of CMKLR1/L1.2 cells to plasmin-treated full-length recombinant prochemerin protein, prochemerin/plasmin/PPP, or prochemerin/plasmin/PPP treated with the CPN inhibitor MGTA (5 μm). Prochemerin was treated with plasmin (1 μm) at 37 °C for 30 min, and the reaction was stopped by PPACK (10 μm). PPP or PPP/MGTA was added to plasmin cleaved prochemerin. The final concentration of treated prochemerin was 0.2 nm. The results represent one of three independent experiments and are expressed as the means ± S.D. (n = 3). **, p < 0.005.
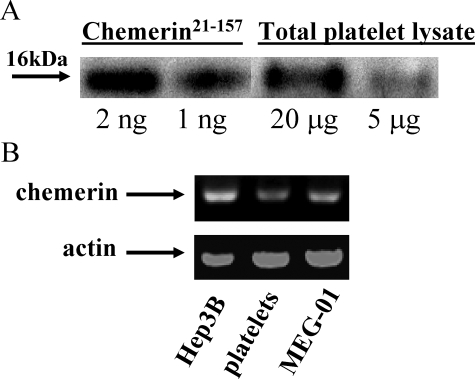
Identification of Chemerin in Platelet—Platelets store various coagulation proteins as well as inflammatory factors. To determine whether platelets are also involved in chemerin expression, Western blot analysis and RT-PCR were performed. Chemerin was detected by Western blot in total platelet lysates, with a molecular mass of ∼16 kDa, similar to recombinant chemerin21–157 (Fig. 5A). Although platelets are anuclear, long lived mRNAs are present in the cytosol, including messages for certain chemokines (26). We detected chemerin mRNA in platelets by RT-PCR. An expected 229-bp PCR product was amplified from cDNAs of platelets, as well as from Hep3B (hepatic carcinoma cell line) and MEG-01 (megakaryotic cell line) cells (Fig. 5B). Identity of the amplified chemerin PCR product was confirmed by direct sequencing.
FIGURE 5.
Platelets contain chemerin mRNA and protein. A, Western blot analysis of chemerin in platelet lysates. B, RT-PCR analysis of chemerin message in various cell lines and platelets.
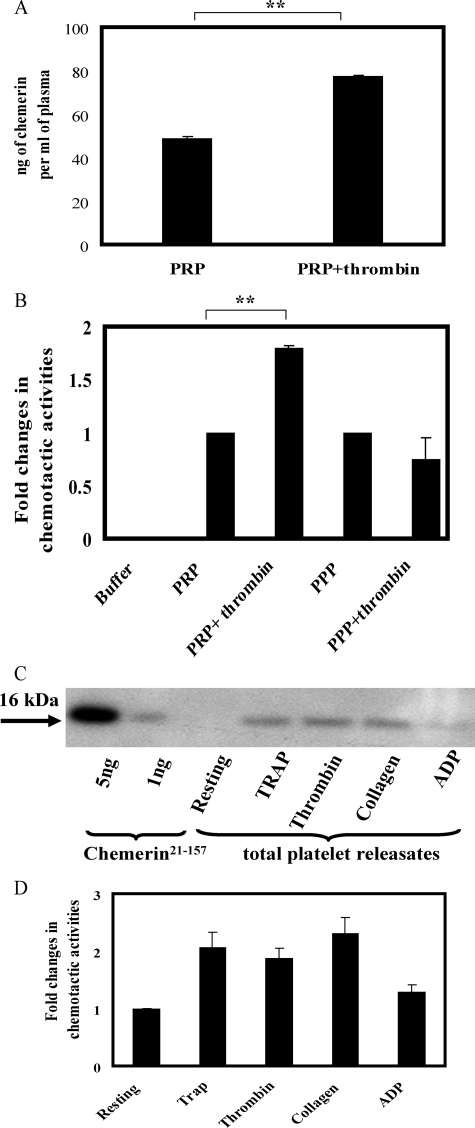
Platelets Release Chemerin upon Activation—As quantified by ELISA, the concentration of chemerin in PRP was 48 ± 1.1 ng/ml. The addition of thrombin (5 units/ml) to PRP increased the chemerin level to 78 ± 0.7 ng/ml (p < 0.005) (Fig. 6A). The addition of thrombin to PPP, on the other hand, had no effect on chemerin activities (Fig. 6B). Thrombin itself did not induce the chemotaxis of CMKLR1-transfected cells, and thrombin does not cleave and activate prochemein (8). Thus we conclude that the increased chemerin bioactivity in thrombin-treated PRP was not due to proteolysis of circulating plasma prochemerin by thrombin but rather was dependent on the release of chemerin from platelets following thrombin activation. Furthermore, platelet-activating agonists such as thrombin receptor-activating peptide, collagen, and, to a lesser extent, ADP induced the release of chemerin from washed platelets (Fig. 6C), which corresponded with an increase in chemerin bioactivity as determined by CMKLR1 transfectant migration (Fig. 6D).
FIGURE 6.
Platelets release chemerin upon activation. A, ELISA quantification of total chemerin present in resting and thrombin (5 units/ml) activated PRP. B, in vitro transwell chemotaxis of CMKLR1/L1.2 cells to PRP, and PRP/thrombin (5 units/ml), PPP, and PPP/thrombin (5 units/ml). Thrombin at 5 units/ml was added to PRP or PPP at 37 °C for 5 min. C and D, identification of chemerin in washed platelet lysates by Western blot analysis (C) or in vitro transwell chemotaxis of CMKLR1/L1.2 cells (D). The platelets were treated with the indicated platelet-activating agonists at 37 °C for 3 min: thrombin receptor-activating peptide (20 μm), thrombin (5 units/ml), collagen (10 μg/ml), and ADP (10 μm). 200 μl of platelet releasates were tested in chemotaxis assays. The results represent one of three independent experiments and are expressed as the means ± S.D. (n = 3). **, p < 0.005.
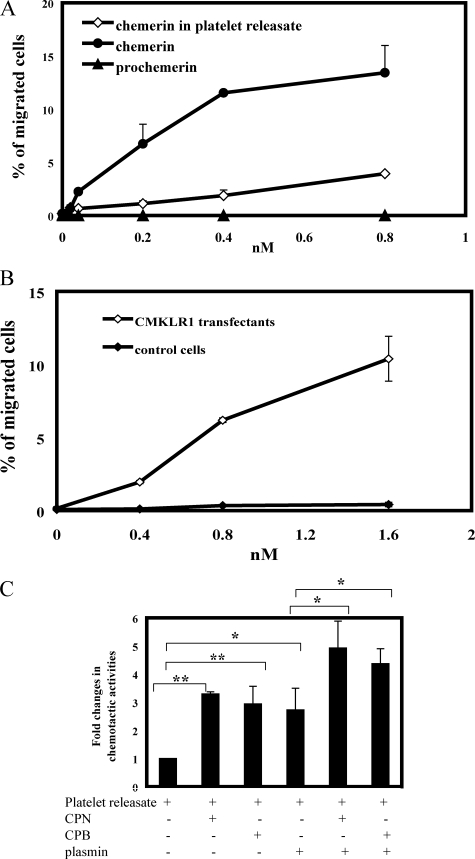
We next investigated whether the chemerin released from activated platelets is a prochemerin form or an active isoform. The same amount of prochemerin and active chemerin21–157 were used as controls for platelet-derived chemerin (as quantified by ELISA) in transwell assays using CMKLR1/L1.2 transfectants. Chemerin activity released from activated platelets is higher than prochemerin but substantially less active than chemerin21–157 (Fig. 7A). The specific response of platelet-derived chemerin to CMKLR1 transfectants was confirmed in the chemotaxis assay using nontransfected cells as controls (Fig. 7B). This suggested that platelets might release chemerin in a partially active form, or platelet-derived chemerin is a mixture of different forms of chemerin, which probably could undergo further proteolysis to fully express its biological activities. With the addition of plasmin/CPB or plasmin/CPN to platelet releasates, no enhanced chemerin activities were observed, which is likely due to the presence of various plasmin inhibitors, including α2-antiplasmin, which is known to be released from activated platelets (27) (data not shown). When platelet-derived chemerins were partially purified by heparin-agarose chromatography as previously described (8), a significantly increased chemerin activity was detected after adding plasmin, plasmin/CPB, and plasmin/CPN (Fig. 7C). Interestingly, chemerin activity was also enhanced even in the presence of CPN or CPB alone (Fig. 7C), indicating that at least a portion of the chemerins released from activated platelets has been already cleaved to a form that is accessible to CPN or CPB cleavage.
FIGURE 7.
Bioactivity of platelet-derived chemerin. A, comparison of chemotactic activity of chemerin released from activated platelets, prochemerin, and the active form chemerin21–157. B, the specific chemotactic response of platelet-derived chemerin to CMKLR1/L1.2 transfectants. C, proteolytic regulation of platelet-derived chemerin bioactivity. Platelet-derived chemerin was partially purified by heparin affinity chromatography (8). Fractions containing chemerin were eluted with 0.6 m of NaCl. The concentration of chemerin was quantitated by ELISA. The conditions of platelet-derived chemerin treated with CPN, CPB, plasmin, plasmin/CPN, or plasmin/CPB were identical to that in Fig. 3. The results represent one of three independent experiments and are expressed as the means ± S.D. (n = 3). *, p < 0.05; **, p < 0.005.
DISCUSSION
In this study, we report that plasma-derived CPN and CPB substantially up-regulated the bioactivity of plasmin-cleaved prochemerin via the removal of the carboxyl-terminal lysine residue, adding a novel mechanism to prochemerin processing and activation by proteases (Table 2). We demonstrated this in three different in vitro scenarios: 1) CPN and CPB removed the carboxyl-terminal lysine from a 10-mer chemerin peptide and converted it to a bioactive 9-mer; 2) plasmin cleaved prochemerin 15-mer to 10-mer, which was subsequently converted to the bioactive 9-mer by CPN or CPB; and 3) sequential proteolysis of recombinant prochemerin protein by plasmin followed by CPN or CPB generated a chemerin isoform with potent chemoattractant activity (carboxyl-terminal sequence NH2-YFPGQFAFS-COOH). These data show that CPN and CPB generate a highly active “desLys” form of plasmin-cleaved chemerin. Interestingly, we also found that platelets can regulate chemerin bioactivity by storing and releasing it upon stimulation. We have therefore identified additional circulating factors (Table 2) that contribute to the regulation of chemerin bioactivity and further link the processes of blood coagulation/fibrinolysis with mediators that regulate leukocyte migration.
TABLE 2.
Prochemerin cleavages by various proteases
Shown is a summary of prochemerin cleavages by various proteases. ND, not determined.
| Protease | C-terminal sequence | Amino acid order |
|---|---|---|
| Unprocessed | ...YFPGQFAFSKALPRS | 21-163 |
| Plasmin/CPB | ...YFPGQFAFS | 21-157 |
| Plasmin/CPN | ...YFPGQFAFS | 21-157 |
| Plasmin | ...YFPGQFAFSK | 21-158 |
| Elastase | ...YFPGQFAFS | 21-157 |
| ...YFPGQFA | 21-155 | |
| ...YFPG | 21-152 | |
| Tryptase | ...YFPGQFAFSK | 21-158 |
| ...YFPGQFA | 21-155 | |
| Cathepsin G | ...YFPGQFAF | 21-156 |
| Staphopain B | ...YFPGQFAFS | 21-157 |
| FVIIa | ND | |
| FXIIa | ND |
There is a growing appreciation for the role of extracellular matrix metalloproteinases and serine proteases such as DPP-IV/CD26 and cathepsin G in regulating the activity of chemokines at the post-translational level (1). These enzymes are particularly adept at modifying chemokines to dampen immune responses. For example, CCL7 is a physiological substrate of matrix metalloproteinase 2; matrix metalloproteinase 2-cleaved CCL7 acts as a general chemokine antagonist by binding to but not activating the CC-chemokine receptors-1, -2, and -3, thereby blocking leukocyte recruitment and dampening inflammation (28). Carboxypeptidases CPN and CPB are well known for their ability to inactivate a number of pro-inflammatory mediators including C5a, C3a, bradykinin, and thrombin-cleaved osteopontin (15–17). In the case of plasmin-cleaved prochemerin, CPN/CPB enhances, rather than diminishes, the chemotactic activity of the attractant under the current experimental conditions.
Cells that are chemerin-responsive include plasmacytoid dendritic cells and macrophages, leukocytes capable of functioning as “immune interpreters”; in the absence of “danger signals,” chemerin-recruited plasmacytoid dendritic cell and macrophages may play an immune suppressive role, dampening inflammation through interleukin-10 and transforming growth factor β secretion and regulating T cell responses. Thus CPN/CPB may serve to dampen inflammatory responses by inactivating anaphylatoxins and by recruiting immune suppressive CMKLR1-positive leukocytes to sites of sterile tissue injury.
The regulation of chemerin bioactivity at a site of tissue injury in vivo appears to involve a complex interplay among many enzymatic components. In our study, plasmin at 1 μm efficiently cleaves prochemerin in vitro. Although this is a substantial concentration of plasmin, human platelets have well defined plasminogen binding sites, and these binding sites are further increased by ∼5-fold upon platelet activation (29, 30). In addition, it has been reported that thrombin stimulation specifically induces plasminogen activation that is mediated by endogenous urokinase-type plasminogen activator (31). Thus given the substantial plasma concentration of plasminogen (∼2.2 μm), it is entirely plausible that prochemerin, upon release from activated platelets, will be efficiently cleaved to chemerin by plasmin generated locally at the site of vascular inflammation in vivo. Plasmin-cleaved prochemerin can be further activated by CPB and CPN. CPB is activated locally by endothelial cell bound thrombin/thrombomodulin and enhances the chemotactic activity of plasmin-cleaved prochemerin via removal of the carboxyl-terminal lysine. At the same time, CPB, in its role as a fibrinolysis inhibitor, blocks plasmin generation, which would diminish the initial activation of circulating prochemerin. Thus CPB may limit the extent of prochemerin activation but enhance the activity of the plasmin-cleaved chemerin that has already been formed. Our study provides further evidence that CPB functions not only as an inhibitor of fibrinolysis, but also as a mediator of inflammatory responses. CPB may be important for local and specific action at sites of tissue damage, whereas constitutively active CPN may regulate its targets systemically.
Platelets are critical in maintaining normal hemostasis (32, 33). Activated platelets release granular proteins that include platelet agonists, adhesive proteins, and chemoattractants at the site of vascular injury, which may play a role in tissue inflammation and remodeling (34–36). In our study, we found that platelets store chemerin. Upon activation, platelets release a mixture of chemerin isoforms with different levels of bioactivity. Some of them would undergo further proteolysis to fully express its biological activity. Our future work will investigate the role of chemerin in platelet biology and tissue remodeling.
Author's Choice—Final version full access.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 HL57530 (to L. L.) and AI59625, AI057229, and DK56339 (to E. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: CMKLR1, chemokine-like receptor 1; CPN, carboxypeptidase N; CPB, carboxypeptidase B; TAFI, thrombin-activable fibrinolysis inhibitor; MGTA, dl-2-mercaptomethyl-3 guanidinoethylthiopropanoic acid; PPACK, d-phenylalanyl-l-prolyl-l-arginine chloromethyl ketone; HPLC, high performance liquid chromatography; PRP, platelet-rich plasma; PIPES, piperazine-N,N′-bis(2-ethanesulfonic acid); RT, reverse transcription; ELISA, enzyme-linked immunosorbent assay; MALDI-TOF, matrix-assisted laser desorption ionization time-of-flight; PPP, platelet-poor plasma.
References
- 1.Zabel, B. A., Zuniga, L., Ohyama, T., Allen, S. J., Cichy, J., Handel, T. M., and Butcher, E. C. (2006) Exp. Hematol. 34 1106–1114 [DOI] [PubMed] [Google Scholar]
- 2.Zabel, B. A., Silverio, A. M., and Butcher, E. C. (2005) J. Immunol. 174 244–251 [DOI] [PubMed] [Google Scholar]
- 3.Wittamer, V., Franssen, J. D., Vulcano, M., Mirjolet, J. F., Le Poul, E., Migeotte, I., Brézillon, S., Tyldesley, R., Blanpain, C., Detheux, M., Mantovani, A., Sozzani, S., Vassart, G., Parmentier, M., and Communi, D. (2003) J. Exp. Med. 198 977–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zabel, B. A., Ohyama, T., Zuniga, L., Kim, J. Y., Johnston, B., Allen, S. J., Guido, D. G., Handel, T. M., and Butcher, E. C. (2006) Exp. Hematol. 34 1021–1032 [DOI] [PubMed] [Google Scholar]
- 5.Parolini, S., Santoro, A., Marcenaro, E., Luini, W., Massardi, L., Facchetti, F., Communi, D., Parmentier, M., Majorana, A., Sironi, M., Tabellini, G., Moretta, A., and Sozzani, S. (2007) Blood 109 3625–3632 [DOI] [PubMed] [Google Scholar]
- 6.Zhang, Z., and Wang, F. S. (2005) Cell Mol. Immunol. 2 411–417 [PubMed] [Google Scholar]
- 7.Wittamer, V., Bondue, B., Guillabert, A., Vassart, G., Parmentier, M., and Communi, D. (2005) J. Immunol. 175 487–493 [DOI] [PubMed] [Google Scholar]
- 8.Zabel, B. A., Allen, S. J., Kulig, P., Allen, J. A., Cichy, J., Handel, T. M., and Butcher, E. C. (2005) J. Biol. Chem. 280 34661–34666 [DOI] [PubMed] [Google Scholar]
- 9.Kulig, P., Zabel, B. A., Dubin, G., Allen, S. J., Ohyama, T., Potempa, J., Handel, T. M., Butcher, E. C., and Cichy, J. (2007) J. Immunol. 178 3713–3720 [DOI] [PubMed] [Google Scholar]
- 10.Wittamer, V., Grégoire, F., Robberecht, P., Vassart, G., Communi, D., and Parmentier, M. (2004) J. Biol. Chem. 279 9956–9962 [DOI] [PubMed] [Google Scholar]
- 11.Matthews, K. W., Mueller-Ortiz, S. L., and Wetsel, R. A. (2004) Mol. Immunol. 40 785–793 [DOI] [PubMed] [Google Scholar]
- 12.Redlitz, A., Tan, A. K., Eaton, D. L., and Plow, E. F. (1995) J. Clin. Investig. 96 2534–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bajzar, L., Morser, J., and Nesheim, M. (1996) J. Biol. Chem. 271 16603–16608 [DOI] [PubMed] [Google Scholar]
- 14.Ceresa, E., Van de Borne, K., Peeters, M., Lijnen, H. R., Declerck, P. J., and Gils, A. (2006) J. Biol. Chem. 281 15878–15883 [DOI] [PubMed] [Google Scholar]
- 15.Myles, T., Nishimura, T., Yun, T. H., and Leung, L. L. K. (2003) J. Biol. Chem. 278 51059–51067 [DOI] [PubMed] [Google Scholar]
- 16.Nishimura, T., Myles, T., Piliponsky, A. M., Kao, P. N., Berry, G. J., and Leung, L. L. K. (2007) Blood 109 1992–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myles, T., and Leung, L. L. (2008) J. Biol. Chem. 283 17789–17796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hildebrandt, M., and Schabath, R. (2008) Front. Biosci. 13 1774–1779 [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez-Gronow, M., Kaczowka, S., Gawdi, G., and Pizzo, S. V. (2008) Front. Biosci. 13 1610–1618 [DOI] [PubMed] [Google Scholar]
- 20.Boehlen, F., and Clemetson, K. J. (2001) Transfus. Med. 11 403–417 [DOI] [PubMed] [Google Scholar]
- 21.Brandt, E., Ludwig, A., Petersen, F., and Flad, H. D. (2000) Immunol. Rev. 177 204–216 [DOI] [PubMed] [Google Scholar]
- 22.Warkentin, T. E., Chong, B. H., and Greinacher, A. (1998) Thromb. Haemostasis 79 1–7 [PubMed] [Google Scholar]
- 23.Cines, D. B., Rauova, L., Arpelly, G., Reilly, M. P., McKenzie, S. E., Sachais, B. S., and Poncz, M. (2007) J. Clin. Apher. 22 31–36 [DOI] [PubMed] [Google Scholar]
- 24.Clemetson, K. J., Clemetson, J. M., Proudfoot, A. E., Power, C. A., Baggiolini, M., and Wells, T. N. (2000) Blood 96 4046–4054 [PubMed] [Google Scholar]
- 25.Du, X. Y., Clemetson, J. M., Navdaev, A., Magnenat, E. M., Wells, T. N., and Clemetson, K. J. (2002) J. Biol. Chem. 277 35124–35132 [DOI] [PubMed] [Google Scholar]
- 26.Power, C. A., Clemetson, J. M., Clemetson, K. J., and Wells, T. N. (1995) Cytokine 7 479–482 [DOI] [PubMed] [Google Scholar]
- 27.Plow, E. F., and Collen, D. (1981) Blood 58 1069–1074 [PubMed] [Google Scholar]
- 28.McQuibban, G. A., Gong, J. H., Tam, E. M., McCulloch, C. A., Clark-Lewis, I., and Overall, C. M. (2000) Science 289 1202–1206 [DOI] [PubMed] [Google Scholar]
- 29.Miles, L. A., and Plow, E. F. (1985) J. Biol. Chem. 260 4303–4311 [PubMed] [Google Scholar]
- 30.Miles, L. A., Ginsberg, M. H., White, J. G., and Plow, E. F. (1986) J. Clin. Investig. 77 2001–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lenich, C., Liu, J. N., and Gurewich, V. (1997) Blood 90 3579–3586 [PubMed] [Google Scholar]
- 32.Andrews, R. K., and Berndt, M. C. (2004) Thromb. Res. 114 447–453 [DOI] [PubMed] [Google Scholar]
- 33.Clemetson, K. J., and Clemetson, J. M. (2007) Curr. Pharm. Des. 13 2673–2683 [DOI] [PubMed] [Google Scholar]
- 34.Du, X. Y., Sim, D. S., Lee, W. H., and Zhang, Y. (2006) Blood Cells Mol. Dis. 36 414–421 [DOI] [PubMed] [Google Scholar]
- 35.von Hundelshausen, P., Petersen, F., and Brandt, E. (2007) Thromb. Haemostasis 97 704–713 [DOI] [PubMed] [Google Scholar]
- 36.Stellos, K., and Gawaz, M. (2007) Thromb. Haemostasis 98 922–929 [DOI] [PubMed] [Google Scholar]