Abstract
Background
Long-term survival of heart transplants is hampered by chronic rejection (CR). Studies indicate the involvement of host macrophages in the development of CR; however, the precise role of these cells in CR is unclear. Thus, it is important to develop noninvasive techniques to serially monitor the movement and distribution of recipient macrophages in chronic cardiac allograft rejection in vivo.
Methods and Results
We have employed a rat heterotopic working-heart CR model for a magnetic resonance imaging experiment. Twenty-one allograft (PVG.1U→PVG.R8) and 9 isograft (PVG.R8→PVG.R8) transplantations were performed. Recipient macrophages are labeled via intravenous injection of micron-sized paramagnetic iron oxide particles (0.9 μm in diameter) at a dose of 4.5 mg Fe per rat 1 day before transplantation. Serial in vivo magnetic resonance images were acquired for up to 16 weeks. The migration of labeled recipient cells in our CR model, in which cardiac CR is evident at 3 weeks and most extensive by 16 weeks after transplantation, can be assessed with the use of in vivo magnetic resonance imaging for >100 days after a single micron-sized paramagnetic iron oxide injection. The location and distribution of labeled recipient cells were confirmed with magnetic resonance microscopy and histology.
Conclusions
This approach may improve our understanding of the immune cells involved in CR and the management of heart transplantation. Moreover, this study demonstrates the feasibility of noninvasively observing individual targeted cells over long time periods by serial in vivo magnetic resonance imaging.
Keywords: magnetic resonance imaging, rejection, transplantation
For decades, numerous researchers have investigated the chronic rejection (CR) that occurs after organ transplantation to better understand the cause and to develop methods to prevent or manage rejection; however, CR remains the major obstacle to long-term allograft survival after heart transplantation.1-3 Although the basic pathology of allograft hearts undergoing CR is arterial intimal thickening, comprising smooth muscle cell proliferation, fibrosis, and mononuclear cell infiltration, the precise role and contribution of immune cell infiltration in CR development are unclear. Previous studies have shown that CR is a process of ongoing recipient-donor alloimmune interaction and that cell-mediated immune responses significantly contribute to its pathology.4-6 Immunosuppressive therapies used after transplantation, although effective in controlling acute rejection, are not only ineffective in preventing CR but may also contribute to its pathology.7 Many reports have implicated host macrophages in the development of CR. The evidence for the involvement of macrophages in CR includes intense presence of mononuclear cells, particularly macrophages, in arteriosclerotic lesions8 and upregulation of cytokines associated with macrophage activation in a rat transplantation model of cardiac CR.9-11 In addition, the absence of macrophages has been found to inhibit CR,8,12 whereas early macrophage infiltration is correlated with a poor prognosis after transplantation.13 Thus, it is important to monitor the recipient immune cell response, in particular macrophages, in cardiac allografts experiencing CR, but this can be challenging in vivo.
Magnetic resonance imaging (MRI) is an established technique for noninvasive detection of cells in vivo with the use of contrast agents incorporated within target cells. Cells can be labeled with MRI contrast agents either ex vivo or in vivo. Although in vivo labeling is only effective for cells that can readily phagocytose or endocytose the contrast agents, such as macrophages, this labeling method is convenient and potentially can be readily applied to a clinical setting.14 One class of cellular contrast agents for MRI is superparamagnetic iron oxide (SPIO) particles; cells containing SPIO can generate localized hypointense spots on T2*-weighted MR images.15,16 Most cellular MRI studies rely on the superior relaxivity of SPIO or ultrasmall superparamagnetic iron oxide (USPIO) nanoparticles for image contrast.17-19 However, to observe significant signal changes, many particles need to be within an imaging voxel. This problem can be resolved with ex vivo cell labeling because a higher labeling efficiency can be achieved but is challenging for in vivo cell labeling. Recently, it has been reported that micrometer-sized para-magnetic iron oxide (MPIO) particles can be utilized for labeling a number of cell types and can have significant advantages for cell-tracking studies because a single MPIO-labeled cell can be observed in vivo by MRI.20-23
Shirwan and coworkers24 have developed a rat model for chronic cardiac allograft rejection by transplanting PVG.1U (RT1.AuBuDuCu) hearts into PVG.R8 (RT1.AaBuDuCu) rats. Cardiac CR is evident on postoperative day (POD) 20 and most extensive by POD 100 in this model. A distinct advantage of using this model is the antigenic simplicity and the development of CR without the use of immunomodulation, thus facilitating studies aimed at the elucidation of the immunological mechanisms of CR, including the effects of immunosuppressive drugs on CR.
The purpose of this study was to visualize the infiltration of individual recipient macrophages labeled with MPIO with the use of in vivo MRI during the development of cardiac CR in the PVG.1U→PVG.R8 rat model.
Methods
Animals
Breeding pairs of PVG.1U (RT1.AuBuDuCu) and PVG.R8 (RT1.AaBuDuCu) rats were obtained from the University of Louisville and successfully bred at Carnegie Mellon University. Inbred male rats weighing 260 to 280 g aged 8 to 10 weeks were used for this study. Brown Norway rats weighing 260 to 280 g were used to measure the blood half-life of the MPIO particles. All animals were housed in the animal facility at the Pittsburgh Nuclear Magnetic Resonance Center for Biomedical Research and received humane care in compliance with the Guide for the Care and Use of Laboratory Animals (National Institutes of Health publication No. 96−03, revised 1996). The animal protocol was approved by the Carnegie Mellon University Institutional Animal Care and Use Committee.
Flow Cytometry
Genetic identity of rat breeding pairs was confirmed by typing 100 μL of peripheral blood leukocytes collected from each rat tail vein with the use of antibodies specific for RT1.A in flow cytometry. Four rats from each strain were tested. Rat anti-rat RT1.Aa,b,l-FITC monoclonal antibody (mAb) was used to identify PVG.R8 rat. Two blood samples from each rat were tested by staining with and without anti-rat RT1.Aa,b,l-FITC mAb. Rat IgG 2b FITC was used as isotype control. All antibodies were purchased from Becton Dickinson (San Jose, Calif). Briefly, 50 μL of heparinized blood was incubated with 20 μL of RT1.Aa,b,l-FITC mAb solution (1:20 dilution with PBS) for 30 minutes in the dark at room temperature. Samples were then mixed with 2 mL of lysing reagent (8.26 g/L of NH4Cl, 1.0 g/L of KHCO3, and 0.037 g/L of Na4EDTA in H2O) and incubated for 5 minutes at room temperature. After washing (twice), cells were resuspended in 0.5-mL FACS medium. Flow cytometry was performed at the Transplantation Laboratory of the University of Pittsburgh Medical Center with the use of a FACScalibur (Becton Dickinson), and data were processed with the use of CellQuest (Becton Dickinson) software in a blinded manner.
Biventricular Working-Heart Transplantation Model
We employed a working-heart CR model by transplanting an en bloc heart and lung from a PVG.1U rat into the abdomen of a PVG.R8 rat as an allograft (n = 21) and PVG.R8 to PVG.R8 as an isograft (n = 9). The transplantation model and surgical procedures for our heterotopic biventricular working-heart model have been described elsewhere.25 In brief, donors were anesthetized by inhalation of isoflurane (Abbott Laboratories, North Chicago, Ill). The chest wall was opened after injecting 500 U/kg body wt of heparin sodium (Baxter, Deerfield, Ill) into the inferior vena cava (IVC). The left lung was ligated and excised. The azygos vein and the left and right superior vena cava were ligated and divided. Then the descending thoracic aorta was transected, and 10 mL of cold lactated Ringer's solution (Abbott Laboratories) was infused into the IVC followed by ligation and division of the IVC. The ascending aorta was dissected and transected at the portion between the left common carotid artery and the left subclavian artery, followed by ligation and division of the right brachiocephalic artery and the left common carotid artery. The grafts were then placed into cold lactated Ringer's solution until transplantation.
The recipients were intubated and ventilated at 1.0 mL/100 g body wt and 60 strokes per minute with a 70% O2, 30% N2O, and 2.5% isoflurane gas mixture. Both the abdominal aorta and the IVC were dissected and clamped. The graft aorta and superior vena cava were anastomosed to the recipient aorta and IVC, respectively, in an end-to-side fashion with a continuous 8−0 polypropylene suture (Ethicon Inc, Somerville, NJ). Rhythmic heartbeats commenced spontaneously after unclamping. The heart rates and ejection fractions of the transplanted hearts were similar to those of the native hearts and also exhibited good wall motion with strains at physiological ranges (determined by tagged MRI). The abdominal wall was sutured with 6−0 silk (Ethico Inc). Graft survival was monitored daily after transplantation for 1 week and then weekly by palpating the transplanted heart.
In Vivo Cell Labeling
The MPIO particles (0.9-μm classic magnetic microspheres, catalog No. MC05F) were purchased from Bangs Laboratories (Fishers, Ind) and prepared by washing and resuspension in PBS at 3 mg Fe per milliliter. Immune cells of the allograft recipients (n = 15) and isograft recipients (n = 6) were labeled by direct intravenous injection of MPIO (4.5 mg Fe per rat) 1 day before transplantation. Three PVG.R8 rats that did not undergo surgery were injected with MPIO 7 days before first in vivo MRI. Six allografts and 3 isografts without injection of MPIO particles also served as control groups.
MPIO Blood Clearance Half-Life
Brown Norway rats (n = 4) were anesthetized and ventilated as described above. The rats were given an injection of 6 mg Fe per kilogram body weight MPIO via a femoral arterial catheter, and at least six 150-μL blood samples were taken over a period of up to 30 minutes in addition to a baseline sample. After each sample, blood volume was replaced with isotonic saline. Blood samples were placed in heparin-coated tubes and mixed with 150 μL of 4% agarose. The transverse relaxation rate (1/T2) of each gel sample was measured at 7 T with a Bruker (Billerica, Mass) Biospec scanner, and the resulting data were fitted to a single exponential to determine the blood clearance half-life.
In Vivo MRI
To monitor the labeled macrophages, serial in vivo MRI of the rat hearts was started 1 week after MPIO injection, then at weekly or biweekly intervals for up to 16 weeks. For MRI, rats were intubated and mechanically ventilated as described above. The core body temperature was maintained at 36.5 ± 1°C with the use of a warm air heating system (SA Instruments, Stony Brook, NY). ECG leads were placed in the abdominal region to detect the transplanted heart rate.
MRI was performed on a Bruker AVANCE AV 4.7-T/40-cm system equipped with a 12-cm, 40-G/cm shielded gradient set. A 5.5-cm home-built surface coil was used for excitation and detection. Multislice ECG- and respiratory-gated T*2-weighted gradient-echo images were acquired with the following parameters: repetition time = 1 respiration cycle (≈ 1 s); echo time = 8 ms; field of view = 3 to 4 cm; slice thickness = 1 or 1.5 mm; in-plane resolution = 156 μm.
MR Microscopy
After in vivo MRI studies, hearts were harvested, fixed in 4% paraformaldehyde overnight, then stored in PBS. The hearts were imaged by using a Bruker AVANCE 11.7-T/89-mm system with a Micro2.5 gradient insert. High-resolution 3-dimensional images were acquired with the following parameters: repetition time = 500 ms; echo time = 8 ms; isotropic resolution = 40 μm.
Histological Examination
Histological examinations were performed by the University of Pittsburgh Medical Center Transplantation Pathology Laboratory. Briefly, paraffin-embedded 5-μm sections underwent hematoxylin and eosin (H&E) staining for evaluation of tissue pathological changes, Perl's Prussian blue staining for detection of the presence of iron, and immunohistochemical staining with mouse anti-rat ED1+ monoclonal antibody (AbD Serotec, MorphoSys UK Ltd, Oxford, UK) for observation of macrophages. In addition, double immunofluorescent staining was performed on 6 allografts from rats that had not been injected with MPIO to identify recipient macrophages within the heart graft. Five-micrometer sections of snap-frozen allograft tissue were incubated with mouse anti-rat ED1+ mAb for 30 minutes in the dark at room temperature followed by incubation with biotinylated anti-mouse antibody and avidin D Texas red (Vector Laboratories, Peterborough, UK). Sections were then incubated with anti-rat RT1.Aa,b,l-FITC mAb in the dark at 4°C overnight for determination of recipient PVG.R8 rat cells.
Fluorescence Microscopy Assessment
Fluorescence images of infiltrating recipient cells were obtained from 6 allografts with ×200 magnification with the use of an Olympus Provis AX 70 fluorescence microscope equipped with an automatic exposure system (Olympus, Tokyo, Japan). Sections were examined with dual-channel fluorescence, ie, in FITC channel (480 nm excitation, 518 nm emission) for recipient PVG.R8 cell detection and Texas red channel (595 nm, 615 nm) for ED1-positive cell detection. Dual-channel fluorescence images were examined with the same field of view. Both ED1-positive and RT1.Aa,b,l-FITC—positive cell numbers were counted in a blinded manner as the number of cells per field of view at a magnification of ×200 on 5 randomly chosen fields per section. Double-positive (ED1+/RT1.Aa,b,l-FITC+) cells are considered recipient macrophages.
Statistical Analysis
The numbers of anti-rat ED1-positive cells (macrophages) and anti-rat RT1.Aa,b,l-FITC—positive cells (recipient cells) are expressed as the average of 5 counts ±SD. Comparisons between the recipient cells and total macrophages at 3 weeks and 16 weeks after transplantation were performed with an unpaired 2-tailed Student t test. A probability value <0.05 was considered statistically significant.
The authors had full access to and take full responsibility for the integrity of the data. All authors have read and agree to the manuscript as written.
Results
Typing PVG.R8 and PVG.1U Rats by Flow Cytometry
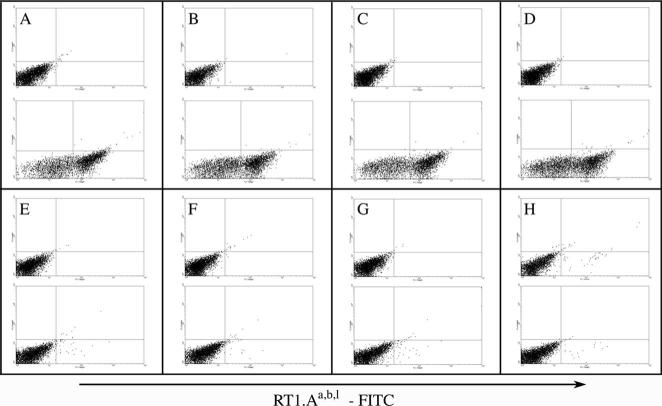
Blood samples from PVG.R8 (RT1.AaBuDuCu) and PVG.1U (RT1.AuBuDuCu) rats were analyzed by flow cytometry with the use of anti-rat RT1.Aa,b,l-FITC mAb for the purpose of distinguishing the 2 colonies of rats. Figure 1A to 1D represents blood samples from 4 PVG.R8 rats, and Figure 1E to 1H represents samples from 4 PVG.1U rats. First and third row samples were stained with an isotype control, and second and bottom row samples were stained with an anti-rat RT1.Aa,b,l-FITC antibody. Flow cytometry data indicate that only the blood samples obtained from PVG.R8 rats stained with anti-rat RT1.Aa,b,l-FITC mAb show positive staining.
Figure 1.
Typing PVG.R8 and PVG.1U rats by flow cytometry. A through D show results obtained from blood samples of 4 PVG.R8 rats, and E through H show the results obtained from 4 PVG.1U rats. For each rat, 2 blood samples were tested. The top plots in each panel show the results of samples stained with rat IgG 2b FITC as isotype control, and the lower plots show samples stained with anti-rat RT1.Aa,b,l-FITC antibody. Only PVG.R8 rats stained with anti-rat RT1.Aa,b,l-FITC antibody scored positive.
MPIO Blood Clearance Half-Life
Measured blood sample relaxation rates from each rat were fitted assuming first-order kinetics. The average blood clearance half-life (t1/2) was determined to be 1.2±0.6 minutes.
MRI Detection of Recipient Cells Labeled With MPIO
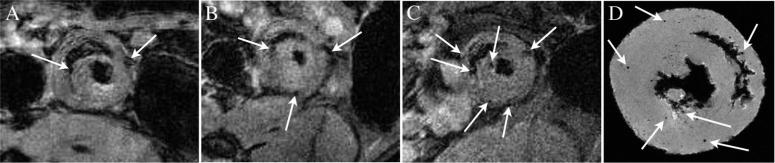
Recipient cells were labeled in vivo with 0.9-μm MPIO 1 day before transplantation. In vivo MRI of rat heart grafts was started 1 week after intravenous injection of MPIO and then subsequently imaged weekly for a period of up to 16 weeks. Representative T2*-weighted serial in vivo images are shown in Figure 2 for an allograft on POD 7 (Figure 2A), POD 14 (Figure 2B), and POD 20 (Figure 2C). One week after transplantation, the heart allografts display a few very distinct, dark spots of hypointensity (Figure 2A). This indicates that MPIO-labeled host cells have infiltrated to the graft after transplantation. At subsequent time points, more discrete regions of hypointensity appear in the same graft without an additional injection of contrast agent (Figure 2B and 2C), indicating an increase in the number of labeled cells migrating to the rejecting heart graft. In contrast, only a few discrete areas of hypointensity were seen in the isograft hearts early after transplantation, and no increase was seen at later time points (in vivo results not shown).
Figure 2.
Repetitive T2*-weighted in vivo MRI with in-plane resolution of 156 μm for an allograft on POD 7 (A), POD 14 (B), and POD 20 (C) after MPIO labeling. Few but very distinct contrast spots can be seen and were confirmed by MR microscopy (D), indicated by arrows, which represent MPIO-labeled immune cells, mainly macrophages.
High-resolution MR microscopy on fixed tissue is more sensitive in detecting MPIO-labeled cells that have infiltrated the heart. Figure 2D shows a MR microscopy image of the allograft harvested after in vivo MRI (shown in Figure 2C) on POD 20. Punctate hypointensity can be seen clearly, and each spot of hypointensity is discrete and circular, likely resulting from an individual labeled macrophage. This finding is consistent with previous phantom studies of MPIO-labeled cells and studies in which an acute rejection model is used.22,23 Figure 3A shows a representative MR microscopy of an allograft harvested at a later time point (POD 109). More MPIO-labeled macrophages have accumulated in the graft as CR progresses. A representative MR microscopy image of an isograft heart harvested on POD 119 is shown in Figure 3B. A very few spots of hypointensity are seen but significantly less than observed in allograft hearts. Two representative native heart MR microscopy images are shown in Figure 3C and 3D. The heart in Figure 3C was harvested 1 week after MPIO injection. Very sparse hypointense spots are seen. The heart shown in Figure 3D did not receive a contrast agent injection, and no hypointense spots are seen.
Figure 3.
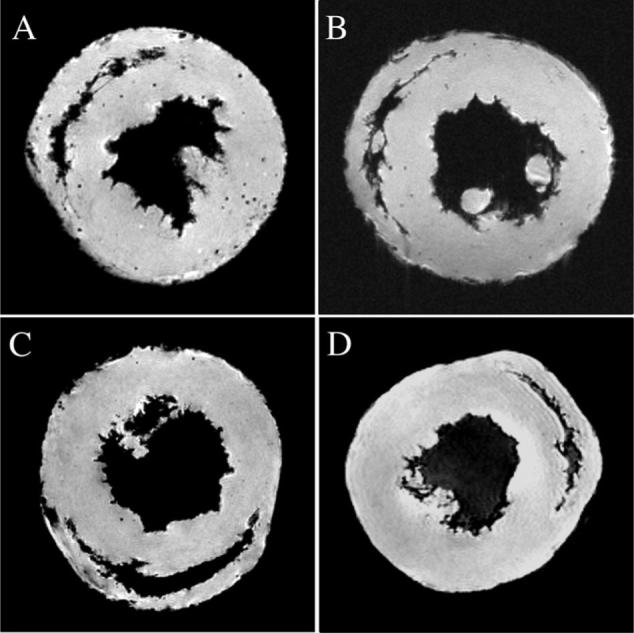
Representative MR microscopy images of transplant and native hearts. Punctate regions of hypointensity can be seen clearly in the allograft heart harvested on POD 109 (A) after in vivo labeling with MPIO. A representative isograft heart harvested on POD 119 is shown in B. Two native hearts are also shown (C and D), 1 of which was harvested 7 days after MPIO injection (C). The heart in D was not injected with MPIO.
H&E Staining for Examination of Graft Pathological Changes
H&E examination shows that allografts exhibited evidence of CR marked by mild to moderate perivascular and myocardial focal infiltration, intimal thickening, and arteriosclerotic lesions. Representative light microscopy images of H&E-stained tissues from heart grafts harvested after in vivo MRI are shown in Figure 4. Arterial intimal thickening and perivascular inflammation are present in the allograft as early as POD 22 (Figure 4A, arrows) and are more extensive by week 16 after transplantation (Figure 4B, arrows). These pathological changes are not seen in isografts (Figure 4C and 4D) harvested at the same time periods as the allografts.
Figure 4.
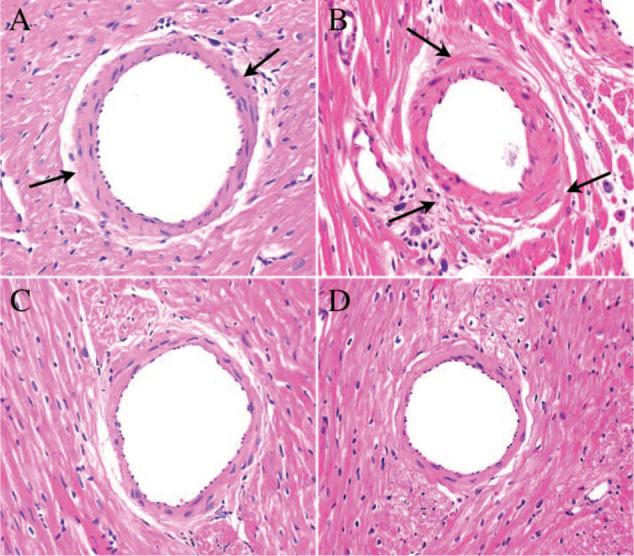
Representative light microscopy images of H&E staining on tissue from heart grafts harvested after in vivo MRI. Allograft shows arterial intimal thickening and perivascular inflammation as early as POD 22 (A, arrows), which is more extensive by week 16 after transplantation (B, arrows). These pathological changes, indicative of CR, are not seen in isografts (C and D) harvested at the same time periods as allografts. Magnification ×400.
MRI Contrast Results From MPIO-Labeled Macrophages
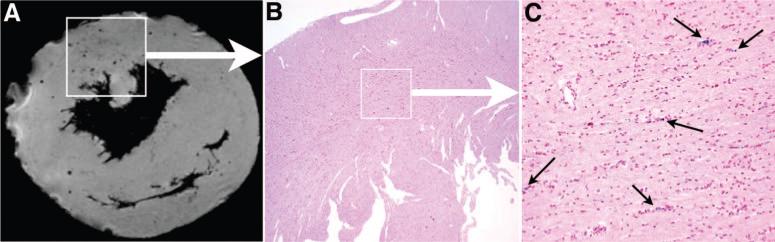
After MR microscopy assessment, grafts were sequentially sliced and analyzed with Perl's Prussian blue staining. The staining detects iron from the MPIO-labeled cells within the graft. Figure 5 shows the comparison of a MR microscopy image and staining of the corresponding section with iron stain in an allograft on POD 94. The MR microscopy image shows discrete, circular areas of hypointensity (Figure 5A). These areas of hypointensity are due to the presence of MPIO-labeled cells, which was confirmed by staining the corresponding section with Perl's Prussian blue for iron (Figure 5B). Figure 5C is the expansion of the boxed region in Figure 5B. Good agreement is present between the localization of the iron-positive cells seen with iron staining and the regions of hypointensity detected by MR microscopy.
Figure 5.
Correlation of MR microscopy and iron staining of MPIO in a POD 94 allograft. Image from MR microscopy shows the discrete and circular spots of hypointensity (A). These black spots of hypointensity effects are due to the presence of MPIO particles, which were confirmed by the matching section, that correspond to the same area as the boxed region in MR microscopy image A, stained with Perl's Prussian blue for iron (B, magnification ×40). C shows the expansion of the boxed region in B (magnification ×200).
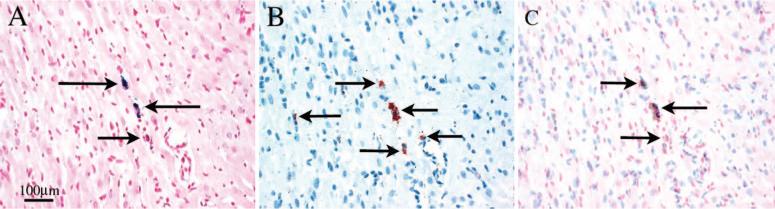
Figure 6 shows optical micrographs of a representative allograft (harvested on POD 106). Two neighboring 5-μm sections are shown: Figure 6A, stained with Perl's Prussian blue for iron, and Figure 6B, stained with anti-rat ED1+ mAb for monocytes/macrophages. Figure 6C is an overlay, illustrating that the iron-containing cells correlate well with the ED1+-stained macrophages. These results confirm that the hypointense spots observed by MRI are MPIO-labeled macrophages.
Figure 6.
The correlation between iron- and ED1-positive cell staining in a POD 106 allograft. Optical micrograph (magnification ×200) of 2 neighboring 5-μm sections stained with Perl's Prussian blue for iron (A, blue, arrows) and anti-rat ED1-positive mAb for monocytes/macrophages (B, brown, arrows). The distribution of the iron-positive cells shown correlates well with ED1-positive cells in C by overlay image of A and B.
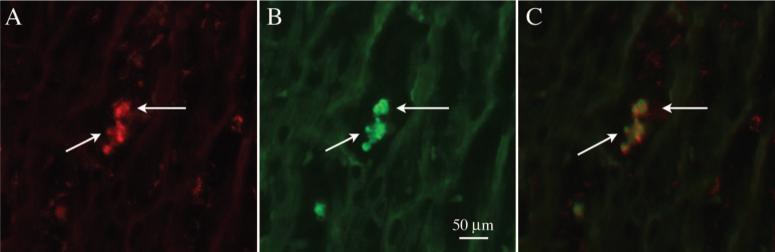
Recipient Macrophages Are Present at an Early Stage and Persistent in Chronic Cardiac Allograft Rejection
We have shown that some of the ED1-positive cells infiltrating the CR rejection site are recipient cells. The presence of host macrophages in the early stages (POD 20) of CR has been confirmed by immunofluorescence staining. Frozen tissue sections were stained with anti-rat ED1+ mAb for macrophages (Figure 7A), and the same section was then stained with anti-rat RT1.Aa,b,l mAb for rat PVG.R8 (Figure 7B). The image in Figure 7A and 7B was taken over the same field of view. Figure 7C is an overlay image of Figure 7A and 7B. Figure 8 summarizes the total number of ED1-positive cells and recipient macrophages present in the allograft at 3 weeks and 16 weeks after transplantation, respectively. The amount of ED1-positive cells and recipient macrophages found in the allograft significantly (P = 0.003 and P = 0.0026, respectively) increases during the CR process, and the cell infiltration is heterogeneous, indicated by the large SD.
Figure 7.
Fluorescent microscopic images (magnification ×400) of double immunofluorescent staining on frozen tissue from an allograft on POD 20. A, A section staining with anti-rat ED1 mAb (for macrophages, arrows, red). B, The same section as A, staining with anti-rat RT1.Aa,b,l mAb (for rat PVG.R8, arrows, green). The images shown in A and B were taken over the same field of view; C is an overlay image of A and B. These data indicate that recipient immune cells, mainly macrophages, are present at the early stages of chronic CR.
Figure 8.
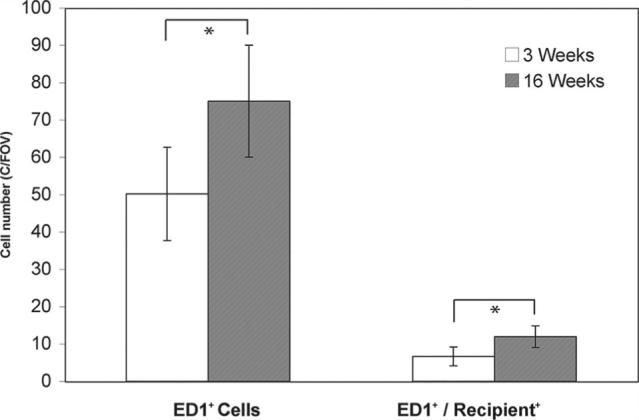
Quantification of total macrophage and recipient macrophage cells infiltrated into grafted hearts at 3 weeks and 16 weeks after transplantation. Cell numbers were counted on 5 randomly chosen fields per section of the double immunofluorescent staining images. Cells that exhibit both ED1 and RT1.Aa,b,l (recipient marker) fluorescence were considered recipient macrophages. The counting was performed in a blinded manner at a magnification of ×200. Data are graphically presented as mean±SD. *P<0.05 was considered statistically significant. The amount of ED1-positive cells and recipient macrophages present in the allograft significantly (P = 0.003 and P = 0.0026, respectively) increases as CR progresses between 3 and 16 weeks.
Discussion
MRI is an excellent imaging modality for both clinical applications as well as basic biomedical research. Recently, it has been shown that in vivo MRI can monitor cell migration and observe cellular biological processes when appropriate contrast agents are used.17-23 We have used a novel rat model and cellular MRI of labeled macrophages to study CR, which continues to be the major cause of graft loss and contributes to the morbidity and mortality in organ transplant patients. The ability to noninvasively track cells in vivo can lead to a better understanding of the complex immune mechanisms involved in chronic cardiac allograft rejection.
Iron oxide— based contrast agents, such as USPIO, SPIO, and MPIO, have been used to label and track cells in vivo by MRI. The in vivo cell labeling efficiency, by direct intravenous injection, is low because of particle dilution and accessibility. In the case of smaller particles, such as USPIO and SPIO, cells must ingest thousands of particles to create a local magnetic field gradient that is detectable by T2*-weighted MRI, especially at the level of a single cell. Each MPIO, however, can contain picogram quantities of iron, and therefore 1 or only a few MPIO particles loaded into a cell are sufficient for MRI detection.20-23 Electron micrograph studies have shown that macrophages concentrate MPIO within membrane-bound vesicles.22,23 The macrophage then becomes the basic unit of contrast, and because the superparamagnetic iron center can propagate a magnetic field gradient as much as 50 times its radius, individual cells can be easily detected with the imaging parameters used here.22,23 As well, it has been reported that single iron oxide—labeled cells can be detected at 1.5 T, and thus clinical translation of cellular tracking studies with appropriate conditions should be possible.26
In this study, cells were tracked in a rat model of CR by in vivo MRI at 4.7 T for >100 days after a single intravenous injection of MPIO to the recipient 24 hours before transplantation. We have found that the blood half-life of the 0.9-μm MPIO is short (<2 minutes) in rats, suggesting that the MPIO particles are immediately taken up by the reticuloendothelial system, including macrophages. Thus, no free particles should be circulating at the time of transplantation or at later time points. Ex vivo cellular labeling studies have also shown that macrophages phagocytose or endocytose the MPIO particles more readily than other cells types, such as B cells and T cells, and thus our in vivo labeling strategy is likely selective for macrophages.22,23 Although we cannot rule out nonmacrophage labeling, the histological correlation of iron staining and ED1-positive cells as well as our previous double fluorescence studies using an acute rejection model in rats22 supports our findings of recipient macrophage trafficking and accumulation in the graft experiencing CR. Consistent with previous results that the 0.9-μm MPIO particles do not affect cell function and proliferation, they do not appear to have any long-term toxic effects on the organism.20-23 The present study clearly demonstrates the effectiveness of in vivo macrophage labeling with MPIO for long-term longitudinal cell tracking studies by MRI.
Immunosuppressive therapy used in organ transplantation curtails acute rejection quite effectively but does not prevent CR in either humans or animals. Mononuclear cell infiltration at later stages of CR is primarily presented by macrophages.9-13 This suggests that macrophages are not only involved with acute rejection but can also participate in the promotion of CR. In our rat model of CR, MPIO-labeled macrophages are observed accumulating in the grafted heart as early as 7 days after transplantation. Serial in vivo MRI shows more accumulation as CR develops. We see some labeled-macrophage accumulation in the isograft controls early on; however, the number of labeled cells found in the isografts does not increase over time and is sparse at later time points. The early accumulation in the isograft may be in response to the transient ischemia/reperfusion injury. A few hypointensity contrast spots are seen in high-resolution MR microscopy images of the native hearts after MPIO injection. This may reflect MPIO taken up by resident-tissue macrophages.27 This mechanism is not likely responsible for MPIO accumulation in our transplant animals because the MPIO injection is 24 hours before surgery and the blood clearance of the MPIO is rapid.
Allograft hearts had fewer cells that were positive for both RT1.Aa and ED1 markers than ED1-positive only cells, suggesting that some of the ED1-positive cells may be of donor origin. Alternatively, anti-rat RT1.Aa,b,l-FITC mAb used to identify recipient cells may be less sensitive for immunohistochemical staining. Further investigation of the distribution of these cell populations will help us to understand the role they play in allograft CR. Studies of this nature could identify new targets for immunotherapy in organ transplantation.
The hallmark of CR in heart allograft is arteriosclerosis characterized by diffuse, concentric intimal thickening, resulting in narrowing and ultimate luminal occlusion of the arteries of the graft.3-5 Studies have indicated that vascular narrowing is primarily caused by the proliferation of smooth muscle cells, and recipient monocyte/macrophages seen in the vessel wall at the initial stage of chronic rejection may contribute to the luminal occlusion process.8-11 This study shows that recipient macrophages migrate to the transplanted heart at the early stages of CR and accumulate over time. This finding is in agreement with previous reports.10-13 These recipient macrophages may be involved in an initial inflammatory response that promotes the development of CR by releasing platelet-derived growth factor to prompt smooth muscle cell proliferation, producing tumor necrosis factor-α and interleukin-1 to promote inflammation and presenting transforming growth factor-β and matrix metalloprotease to induce fibrosis.8-11 Early macrophage infiltration correlates with a poor prognosis, and persistence of macrophage accumulation after resolution of acute rejection episodes is predictive of CR after transplantation.12,13 Thus, monitoring macrophage activities with the use of in vivo MRI provides a novel methodology for studying the mechanisms of CR progression. This technique may potentially lead to a clinical tool for diagnosing CR, managing treatment, and predicting outcomes after organ transplantation.
In summary, our results show the following: (1) the infiltration of MPIO-labeled recipient immune cells, mainly macrophages, can be detected by in vivo MRI for >100 days; (2) it is primarily the macrophages that contained the MPIO particles, as confirmed by histology and fluorescence microscopy; (3) the biventricular working-heart CR model is a good rat model to mimic the clinical situation for the purpose of studying the migration of recipient cells as well as the process of ongoing recipient-donor alloimmune interaction in the cardiac allograft undergoing CR; and (4) recipient macrophage cells are present at the early stages of CR and remain in the graft for an extended period of time.
In conclusion, our approach of tracking immune cells noninvasively by in vivo MRI has great potential to further our understanding of the cellular mechanisms involved in CR. Moreover, this study demonstrates the feasibility of noninvasively observing individual targeted cells over long periods of time with the use of in vivo MRI.
CLINICAL PERSPECTIVE
Numerous studies have shown that immune cells including macrophages play crucial roles in the development of organ rejection. The ability to monitor the migration and localization of specific cell types in vivo, noninvasively, and in real time will greatly improve our understanding of the complex roles that different cells play in cardiac allograft rejection. This study presents the feasibility of imaging individual recipient macrophages in vivo by magnetic resonance imaging over long periods of time in a rodent heterotopic working-heart transplantation model with the use of a sensitive contrast agent, micrometer-sized paramagnetic iron oxide particles. In this study, recipient cells, mainly macrophages, have been labeled in vivo by direct intravenous administration of micrometer-sized paramagnetic iron oxide particles before heart transplantation. This cell-labeling procedure is convenient for clinical application. Thus, this approach provides a novel methodology for studying the mechanisms of cardiac allograft rejection in both animals and humans. Moreover, the current gold standard for diagnosing and staging rejection after organ transplantation is biopsy, which is not only invasive but also prone to sampling errors because rejection sites are highly heterogeneous. The activated macrophages have been found to be the primary cells in the cellular infiltrate of rejecting grafts. Thus, this imaging modality using magnetic resonance imaging to monitor cell migration in real time, with whole-heart visualization of cellular infiltration, could potentially lead to a powerful clinical tool, providing information for noninvasive evaluation of graft rejection, managing treatment, and predicting outcomes after heart transplantation.
Acknowledgments
We thank Lisa McGaw for assistance with animals and Dr Haosen Zhang for helpful comments.
Sources of Funding
This study was supported by grants from the National Institutes of Health (P41EB-001977, R01EB-00318, R01HL-081349 to Dr Ho; RO1AI-47864 to Dr Shirwan) and a grant from the Pennsylvania Department of Health's Commonwealth Universal Research Enhancement Program.
Footnotes
Disclosures
None.
References
- 1.Hornick P, Rose M. Chronic rejection in the heart. Methods Mol Biol. 2006;333:131–144. doi: 10.1385/1-59745-049-9:131. [DOI] [PubMed] [Google Scholar]
- 2.Hertz MI, Boucek MM, Deng MC, Edwards LB, Keck BM, Kirklin JK, Naftel DC, Rowe AW, Taylor DO, Trulock EP. Scientific Registry of the International Society for Heart and Lung Transplantation: introduction to the 2005 annual reports. J Heart Lung Transplant. 2005;24:939–944. doi: 10.1016/j.healun.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 3.Demetris AJ, Murase N, Ye Q, Galvao FHF, Richert C, Saad R, Pham S, Duquesnoy R, Zeevi A, Fung JJ, Starzl TE. Obliterative arteriopathy and Quilty lesions: leukocyte trafficking, microchimerism and the immunolymphatic theory of chronic rejection. Am J Pathol. 1997;150:563–578. [PMC free article] [PubMed] [Google Scholar]
- 4.Shirwan H. Chronic allograft rejection: do the Th2 cells preferentially induced by indirect alloantigen recognition play a dominant role? Transplantation. 1999;68:715–726. doi: 10.1097/00007890-199909270-00001. [DOI] [PubMed] [Google Scholar]
- 5.Adams DH, Russell ME, Hancock WW, Sayegh MH, Wyner LR, Karnovsky MJ. Chronic rejection in experimental cardiac transplantation: studies in the Lewis-F344 model. Immunol Rev. 1993;134:5–19. doi: 10.1111/j.1600-065x.1993.tb00637.x. [DOI] [PubMed] [Google Scholar]
- 6.Ichikawa N, Demetris AJ, Starzl TE, Ye Q, Okuda T, Chun HJ, Liu K, Kim YM, Murase N. Donor and recipient leukocytes in organ allografts of recipients with variable donor-specific tolerance: with particular reference to chronic rejection. Liver Transpl. 2000;6:686–702. doi: 10.1053/jlts.2000.19029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shirwan H, Wu GD, Barwari L, Liu A, Cramer DV. Induction of allograft nonresponsiveness after intrathymic inoculation with donor class I allopeptides, II: evidence for persistent chronic rejection despite high levels of donor microchimerism. Transplantation. 1997;64:1671–1676. doi: 10.1097/00007890-199712270-00007. [DOI] [PubMed] [Google Scholar]
- 8.Kitchens WH, Chase CM, Uehara S, Cornell LD, Colvin RB, Russell PS, Madsen JC. Macrophage depletion suppresses cardiac allograft vasculopathy in mice. Am J Transplant. 2007;7:2675–2682. doi: 10.1111/j.1600-6143.2007.01997.x. [DOI] [PubMed] [Google Scholar]
- 9.Azuma H, Nadeau KC, Ishibashi M, Tilney NL. Host leukocytes and their products in chronic kidney allograft rejection in rats. Transpl Int. 1994;7(Suppl 1):S325–S327. doi: 10.1111/j.1432-2277.1994.tb01382.x. [DOI] [PubMed] [Google Scholar]
- 10.Heemann UW, Tullius SG, Tamatami T, Miyasaka M, Milford E, Tilney NL. Infiltration patterns of macrophages and lymphocytes in chronically rejecting rat kidney allografts. Transpl Int. 1994;7:349–355. doi: 10.1007/BF00336711. [DOI] [PubMed] [Google Scholar]
- 11.Russell ME, Wallace AF, Hancock WW, Sayegh MH, Adams DH, Sibinga NE, Wyner LR, Karnovsky MJ. Upregulation of cytokines associated with macrophage activation in the Lewis-to-F344 rat transplantation model of chronic cardiac rejection. Transplantation. 1995;59:572–578. [PubMed] [Google Scholar]
- 12.Azuma H, Nadeau KC, Ishibashi M, Tilney NL. Prevention of functional, structural, and molecular changes of chronic rejection of rat renal allografts by a specific macrophage inhibitor. Transplantation. 1995;60:1577–1582. doi: 10.1097/00007890-199560120-00034. [DOI] [PubMed] [Google Scholar]
- 13.Pilmore HL, Painter DM, Bishop GA, McCaughan GW, Eris JM. Early up-regulation of macrophages and myofibroblasts: a new marker for development of chronic renal allograft rejection. Transplantation. 2000;69:2658–2662. doi: 10.1097/00007890-200006270-00028. [DOI] [PubMed] [Google Scholar]
- 14.Ho C, Hitchens TK. A non-invasive approach to detecting organ rejection by MRI: monitoring the accumulation of immune cells at the transplanted organ. Curr Pharm Biotechnol. 2004;5:551–566. doi: 10.2174/1389201043376535. [DOI] [PubMed] [Google Scholar]
- 15.Weissleder R, Elizondo G, Wittenberg J, Rabito CA, Bengele HH, Josephson L. Ultrasmall superparamagnetic iron oxide: characterization of a new class of contrast agents for MR imaging. Radiology. 1990;175:489–493. doi: 10.1148/radiology.175.2.2326474. [DOI] [PubMed] [Google Scholar]
- 16.Yeh TC, Zhang W, Ildstad ST, Ho C. Intracellular labeling of T-cells with superparamagnetic contrast agents. Magn Reson Med. 1993;30:617–625. doi: 10.1002/mrm.1910300513. [DOI] [PubMed] [Google Scholar]
- 17.Ye Q, Yang DW, Williams M, Williams D, Pluempitiwiriyawej C, Moura JMF, Ho C. In-vivo detection of acute rat renal allograft rejection by MRI with USPIO particles. Kidney Int. 2002;61:1124–1135. doi: 10.1046/j.1523-1755.2002.00195.x. [DOI] [PubMed] [Google Scholar]
- 18.Bulte JW, Duncan ID, Frank JA. In-vivo magnetic resonance tracking of magnetically labeled cells after transplantation. J Cereb Blood Flow Metab. 2002;22:899–907. doi: 10.1097/00004647-200208000-00001. [DOI] [PubMed] [Google Scholar]
- 19.Oude Engberink RD, van der Pol SM, Döpp EA, de Vries HE, Blezer EL. Comparison of SPIO and USPIO for in vitro labeling of human monocytes: MR detection and cell function. Radiology. 2007;243:467–474. doi: 10.1148/radiol.2432060120. [DOI] [PubMed] [Google Scholar]
- 20.Hinds KA, Hill JM, Shapiro EM, Laukkanen MO, Silva AC, Combs CA, Varney TR, Balaban RS, Koretsky AP, Dunbar CE. Highly efficient endosomal labeling of progenitor and stem cells with large magnetic particles allows magnetic resonance imaging of single cells. Blood. 2003;102:867–872. doi: 10.1182/blood-2002-12-3669. [DOI] [PubMed] [Google Scholar]
- 21.Shapiro EM, Skrtic S, Sharer K, Hill JM, Dunbar CE, Koretsky AP. MRI detection of single particles for cellular imaging. Proc Natl Acad Sci USA. 2004;101:10901–10906. doi: 10.1073/pnas.0403918101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu YL, Ye Q, Foley LM, Hitchens TK, Sato K, Williams JB, Ho C. In situ labeling of immune cells with iron oxide particles: an approach to detect organ rejection by cellular MRI. Proc Natl Acad Sci U S A. 2006;103:1852–1857. doi: 10.1073/pnas.0507198103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams JB, Ye Q, Hitchens TK, Kaufman CL, Ho C. MRI detection of macrophages labeled using micrometer-sized iron oxide particles. J Magn Reson Imaging. 2007;25:1210–1218. doi: 10.1002/jmri.20930. [DOI] [PubMed] [Google Scholar]
- 24.Shirwan H, Mhoyan A, Yolcu ES, Que X, Ibrahim S. Chronic cardiac allograft rejection in a rat model disparate for one single class I MHC molecule is associated with indirect recognition by CD4(+) T cells. Transpl Immunol. 2003;11:179–185. doi: 10.1016/S0966-3274(03)00004-2. [DOI] [PubMed] [Google Scholar]
- 25.Wu YJ, Sato K, Ye Q, Ho C. MRI investigations of graft rejection following organ transplantation using rodent models. Methods Enzymol. 2004;386:73–105. doi: 10.1016/S0076-6879(04)86003-8. [DOI] [PubMed] [Google Scholar]
- 26.Heyn C, Ronald JA, Mackenzie LT, MacDonald IC, Chambers AF, Rutt BK, Foster PJ. In vivo magnetic resonance imaging of single cells in mouse brain with optical validation. Magn Reson Med. 2006;55:23–29. doi: 10.1002/mrm.20747. [DOI] [PubMed] [Google Scholar]
- 27.Paul LC, Grothman GT, Benediktsson H, Davidoff A, Rozing J. Macrophage subpopulations in normal and transplanted heart and kidney tissues in the rat. Transplantation. 1992;53:157–162. doi: 10.1097/00007890-199201000-00032. [DOI] [PubMed] [Google Scholar]