Abstract
Double strand breaks (DSBs) activate the ATM kinase which promotes the accumulation of DNA damage factors in the chromatin surrounding the break. The functional significance of the resulting DNA damage foci is poorly understood. Here we show that 53BP1, a component of DNA damage foci, changes the dynamic behavior of chromatin to promote DNA repair. We used conditional deletion of the shelterin component TRF2 from mouse cells to deprotect telomeres, which, like DSBs, activate the ATM kinase, accumulate 53BP1, and are processed by non-homologous end-joining (NHEJ)1,2. Deletion of TRF2 from 53BP1-deficient cells established that NHEJ of dysfunctional telomeres is strongly dependent on the binding of 53BP1 to damaged chromosome ends. To address the mechanism by which 53BP1 promotes NHEJ, we used time-lapse microscopy to measure telomere dynamics before and after their deprotection. Imaging showed that deprotected telomeres are more mobile and sample larger territories within the nucleus. This change in chromatin dynamics was dependent on 53BP1 and ATM but did not require a functional NHEJ pathway. We propose that the binding of 53BP1 near DNA breaks changes the dynamic behavior of the local chromatin, thereby facilitating NHEJ repair reactions that involve distant sites, including joining of dysfunctional telomeres and AID-induced breaks in immunoglobulin Class-Switch Recombination (CSR).
Previous work has shown that mouse telomeres lacking TRF2 are processed by a Ku70/DNA ligase IV-dependent NHEJ reaction1,2 that requires ATM kinase signaling and is stimulated by the ATM targets γ−H2AX and MDC13,4. Here we focus on 53BP1, a third ATM target, which accumulates at DSBs and deprotected telomeres5–8. The interaction of 53BP1 with chromatin involves the binding of its Tudor domains to H4-K20diMe and an MDC1-dependent interaction with γ-H2AX9–14. While 53BP1 is not strictly required for DNA damage signaling, homology directed repair (HDR), or NHEJ in the context of V(D)J recombination, NHEJ of DSBs in CSR is severely affected by 53BP1 deficiency15,16. In the absence of 53BP1, DSBs in different switch regions fail to join successfully, resulting in a predominance of intra-switch recombination events17. It has been proposed that 53BP1 might either facilitate synapsis of DNA ends17,18 or ‘shepherd’ NHEJ factors to the break19.
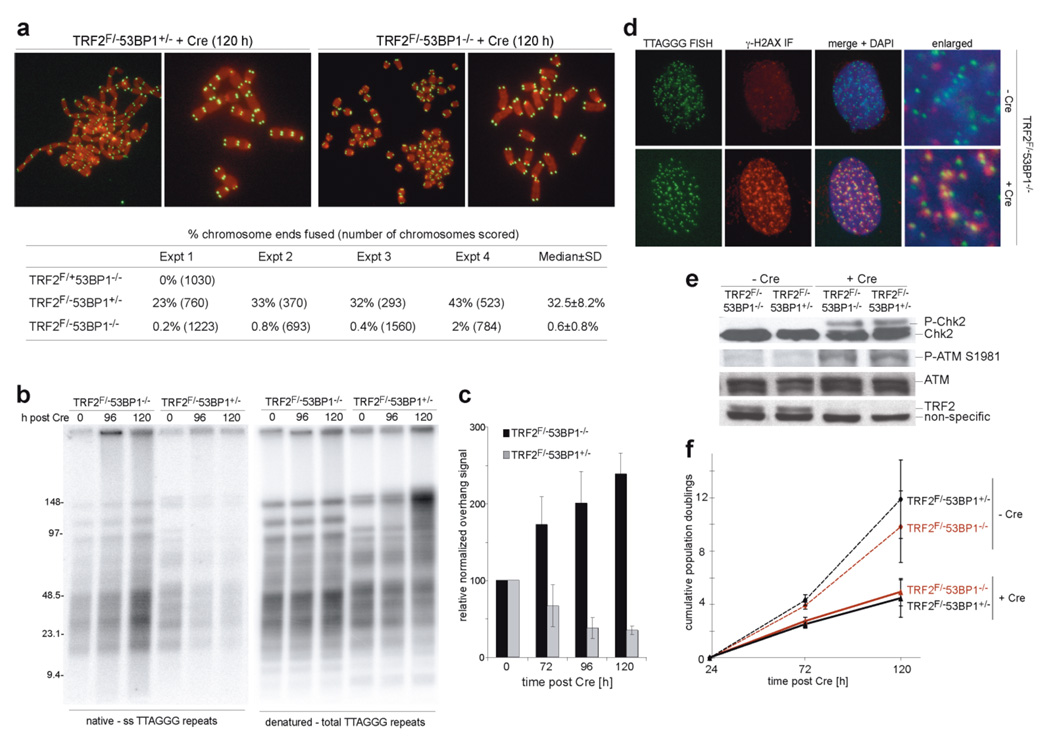
We generated SV40-LT immortalized TRF2F/−53BP1−/− and TRF2F/−53BP1+/− MEFs1,20 (Suppl. Fig. 1a, b) and assayed the frequency of telomere fusions in metaphase spreads collected 120 hours after Cre-mediated deletion of TRF2 (Fig. 1a). Whereas 53BP1-proficient cells showed the expected level of telomere fusions (33% of telomeres fused after 4 PD), the rate of NHEJ in 53BP1−/− cells was at least 50-fold lower, nearly as low as in DNA ligase IV deficient cells1,2. The effect of 53BP1 on telomere fusions was also obvious when assayed by in-gel hybridization assay (Fig. 1b). 53BP1-deficient cells failed to accumulate high molecular weight fusion products after deletion of TRF2 and showed no loss of the telomeric 3’ overhang, a second index for telomere fusion (Fig. 1b, c). In fact, the overhang signal increased, similar to what is observed when TRF2 is deleted from Ku70- or DNA Ligase IV-deficient cells1,2. In contrast to NHEJ, DNA damage signaling was not affected by the absence of 53BP1 (Fig. 1d, e). 53BP1 deficiency affected neither the phosphorylation of ATM and its target Chk2 nor the presence of γ-H2AX, MDC1, and NBS1 at dysfunctional telomeres (Fig. 1d, e, Suppl. Fig. 2a, b). The NHEJ defect was also not due to a change in cell cycle progression. After deletion of TRF2, the cells underwent the same number of cell divisions regardless of their 53BP1 status (Fig. 1f) and their S-phase index was not affected by 53BP1 deficiency (Suppl. Fig. 1c).
Figure 1. Requirement for 53BP1 in NHEJ of dysfunctional telomeres, but not DNA damage signaling.
a, (Top) Metaphase chromosomes after deletion of TRF2 from the indicated cells. Telomeric FISH, green; DAPI, red. (Bottom) Summary of the effect of 53BP1 on telomere fusions at 120 hours after TRF2 deletion.
b, In-gel assay for the 3’ overhang (left) and total telomeric DNA (right) after deletion of TRF2 from 53BP1-proficient and -deficient cells.
c, Quantification of overhang signals in (b) (mean±SD; n=3).
d, IF for presence of γ-H2AX at telomeres after deletion of TRF2 from 53BP1-proficent and -deficient cells.
e, Immunoblots for phosphorylated ATM and Chk2 after TRF2 deletion.
f, Proliferation of MEFs of the indicated genotype and treatment (mean±SD; n=3).
We next tested whether the Tudor domain-mediated recognition of H4-K20diMe contributed to the promotion of NHEJ. TRF2F/−53BP1−/− cells were complemented with wild type human 53BP1 or a Tudor domain mutant (53BP1-D1521A) with impaired binding to H4-K20diMe14 (Fig. 2a). Although the two forms of 53BP1 were expressed at levels comparable to endogenous 53BP1 (Fig. 2b and data not shown), the recruitment of 53BP1-D1521A to deprotected telomeres was significantly diminished (Suppl. Fig. 3). In contrast to wild type 53BP1, which reconstituted the NHEJ of dysfunctional telomeres, cells expressing the Tudor mutant of 53BP1 showed a considerable delay in the fusion of telomeres lacking TRF2 (Fig. 2c, d).
Figure 2. Optimal NHEJ of dysfunctional telomeres requires interaction of 53BP1 with H4-K20diMe.
a, Schematic of the domain structure of 53BP1.
b, Immunoblot for 53BP1 expression in TRF2F/−53BP1−/− cells complemented with empty vector, wild type (WT), or the Tudor mutant 53BP1-D1521A.
c, Telomere fusions in metaphase spreads of TRF2F/−53BP1−/− MEFs complemented as indicated and treated with Cre as specified.
d, Quantification of telomere fusions in (c) (mean±SD; n=3).
The finding that the Tudor domain mutation in 53BP1 did not abrogate NHEJ suggested that a second, H4-K20diMe-independent, interaction of 53BP1 with chromatin contributes to NHEJ. As MDC1 is required for the stable association of 53BP1 with DSBs9–14, we generated TRF2F/FMDC1−/− MEFs18 and determined their ability to execute the telomere fusion reaction. As expected, the binding of 53BP1 near deprotected telomeres was impaired by MDC1 deficiency, despite normal levels of H2AX phosphorylation (Suppl. Fig. 4a, b). MDC1 deficiency resulted in a delay in the NHEJ of telomeres but did not abrogate this process (Suppl. Fig. 5a, b), confirming our previous findings with MDC1 shRNAs3. Furthermore, inhibition of MDC1 with shRNAs affected the residual NHEJ of telomeres in the context of the 53BP1 Tudor domain mutant (Suppl. Fig. 5c–e).
These data establish an important role for chromatin-bound 53BP1 in the NHEJ of deprotected telomeres. Previous data showed that 53BP1 contributes to NHEJ in CSR15,16 but not in other settings such as V(D)J recombination. We argued that the crucial difference between the 53BP1-dependent and –independent NHEJ reactions might be the distance between the DNA ends involved. Whereas the two ends generated by RAG1/2 or chromosome-internal DNA damage are close together, the DNA ends generated by AID in CSR are often far apart as are dysfunctional telomeres, which are processed by NHEJ in G121 when chromosome ends are dispersed throughout the nucleus.
We turned to time-lapse microscopy to address whether 53BP1 altered the synapsis and/or dynamics of deprotected telomeres. To image telomeres, we used an EGFP-tagged version of the shelterin component TRF1, which remains associated with telomeres when TRF2 is removed (Fig. 3a–c, Suppl. Fig. 6). Overexpression of this and other forms of TRF1 does not affect the protective function of telomeres22 (data not shown). To mark sites of DNA damage signaling, we fused mCherry to aa 1220–1711 of 53BP1 (mCherry-BP1-2, see Fig. 2a and Fig. 3a, b), creating a fusion protein that lacks most of the functional domains of 53BP1. Upon deletion of TRF2, mCherry-BP1-2 re-localized to telomeric sites containing EGFP-TRF1 (Fig. 3c). The mCherry-BP1-2 allele did not affect the frequency of telomere fusions in 53BP1-proficient cells nor did it restore the NHEJ defect in 53BP1-deficient cells (Suppl. Fig. 7a), establishing that it is a neutral marker for DNA damage in this context. Furthermore, key imaging experiments repeated in cells lacking mCherry-BP1-2 gave the same outcome (Suppl. Fig. 7b,c and see below).
Figure 3. 53BP1 promotes mobility of dysfunctional telomeres.
a, Schematic of the live-cell imaging experiments.
b, Immunoblotting for EGFP-TRF1, mCherry-BP1-2, and TRF2.
c, Co-localization of EGFP-TRF1 and mCherry-BP1-2 fluorescence signals.
d, Representative traces of telomeres in indicated MEFs expressing EGFP-TRF1 and mCherry-BP1-2. Snapshots taken at indicated timepoints.
e, 10 representative telomeres from indicated MEFs, treated as specified, were tracked as in (d); cumulative distance at each time-point was plotted against time.
f, Frequency distribution of the cumulative distance traveled in one representative experiment in MEFs of the indicated genotype and treatment (median±SD.; n=55; p calculated using a two-tailed Mann-Whitney test).
g, Frequency distribution of the maximum displacement from the starting point registered by individual telomeres during the imaging session described in (f). (median±SD and p-values calculated as in (f)).
h, i, Analysis as in (f) in MEFs of the indicated genotypes and treatments, expressing EGFP-TRF1 marker only (n=50).
To analyze the effect of 53BP1 on the movement of dysfunctional telomeres, time-lapse microscopy was performed at an early stage after deletion of TRF2 (72–84 hr post-Cre) when most TRF1-marked sites still represent free chromosome ends3. Initial analysis of EGFP and mCherry signals indicated that dysfunctional telomeres became more mobile but only when 53BP1 was present (Suppl. Video 1 and 2). We observed occasional apparent fusion events in 53BP1-proficient cells (Suppl. Video 3) but no potential fusions were observed in 53BP1-deficient cells even for telomeres that were closely apposed (Suppl. Video 4).
To obtain a quantitative measure of telomere mobility, individual EGFP-TRF1/mCherry-BP1-2-labeled dysfunctional telomeres were traced in deconvolved and projected images (Fig. 3a, d; Suppl. Fig. 8a, b). In parallel, EGFP-TRF1-marked functional telomeres were analyzed in cells not treated with Cre. Functional telomeres in 53BP1-proficient and -deficient cells traveled in a constrained random walk over a total path of approximately 3.7–3.9±0.7 µm at a median speed of 180–190 nm/min−1, which is comparable to movement of human telomeres (Fig. 3e, f; Suppl. Figs. 7b, 9a)23. In contrast, dysfunctional telomeres were significantly more mobile, traveling at a speed of 270–360 nm/min over a cumulative distance ranging from 5.4 to 7.2 µm (Fig. 3e, f; Suppl. Figs. 7b, 9a). The mobility increase associated with telomere dysfunction was significantly attenuated in cells lacking 53BP1 (Fig. 3e), resulting in a median cumulative distance traveled of 4.4±0.2 µm. The presence of the mCherry-BP1-2 DNA damage marker did not affect the outcome; the same results were obtained after deletion of TRF2 from 53BP1+/− or 53BP1−/− cells that only contained the EGFP-TRF1 marker (Suppl. Fig 7b, c). Importantly, in both settings the dysfunctional telomeres in 53BP1-proficient cells sampled larger territories than functional telomeres (maximal displacement 1.2±0.3 µm versus 0.51±0.29 µm; Fig. 3g, Suppl. Figs. 7c, 9b). Assuming that telomeres sample spheres within the nucleus, a 2-fold change in the radius of the 2D territory will increase the volume sampled by a dysfunctional telomere by a factor 8. Furthermore, a substantial fraction (> 10%) of the dysfunctional telomeres roamed well beyond 2 µm whereas none of the 115 functional telomeres analyzed moved beyond that distance (Fig. 3g, Suppl. Fig. 7c). When 53BP1 was not present, the median maximal displacement of the dysfunctional telomeres was only 0.8±0.2 µm, indicating movement within a significantly smaller territory and only one out of 115 dysfunctional telomeres sampled an area beyond 2 µm (Fig. 3g, Suppl. Fig. 7c). These data establish that telomeres become more mobile and sample larger territories when they are deprived of their normal protection and that this change in their dynamic behavior is promoted by 53BP1. Assuming that the rate of NHEJ correlates with the probability of an encounter between two telomeres, the ability of 53BP1 to expand the 3 dimensional space visited by dysfunctional telomeres is a likely explanation for its effect on telomere fusion. We confirmed that the absence of the NHEJ reaction itself could not account for the slower movement as telomeres in DNA ligase IV-deficient cells still showed a considerable increase in their mobility upon TRF2 deletion (Fig. 3h, Suppl. Fig. 9c, d).
The ability of 53BP1 to promote the mobility of dysfunctional telomeres could explain why ATM kinase, its upstream regulator, is required for telomere fusions4. Indeed, similar to 53BP1-deficient cells, dysfunctional telomeres failed to gain their maximal mobility in ATM-deficient cells (Fig. 3i, Suppl. Fig. 9c, d).
Next, we tested whether 53BP1 led to global chromatin mobility in response to DNA damage. We tracked functional telomeres in wild type or Lig4−/− cells immediately after treatment with γ-irradiation or after allowing the cells to recover. In all cases, we found that the mobility of the telomeres was unaffected by the induction of damage elsewhere in the genome (Suppl. Fig. 10) arguing that increased mobility is a local event, taking place at the site of damage where 53BP1 accumulates.
As 53BP1 is the first DNA damage factor to be implicated in the movement of sites of DNA damage, it will be important to determine the mechanism by which it affects chromatin mobility. Latrunculin A did not alter the gain in mobility of deprotected telomeres (Suppl. Fig. 11a), arguing against an actin-driven process. Similarly, the HDAC inhibitor trichostatin A had no effect on the mobility of the deprotected telomeres, making it unlikely that 53BP1 acts through its interaction with HDAC424 (Suppl. Fig. 11b). We imagine that 53BP1 dislodges a factor that restricts chromatin mobility or that 53BP1 endows the chromatin with dynamic features, for instance by altering higher order interactions that affect the thermal persistence length.
These data reveal that the DNA damage factor 53BP1 is crucial for the NHEJ reaction at dysfunctional telomeres and suggest a mechanism by which 53BP1 promotes telomere fusions. Telomeres, like other chromosomal sites, have a limited range of motion, showing constrained diffusion within a territory with a radius of ≤0.5 µm (this work and refs. 23 and 2523,25). When telomeres become damaged, their mobility increases significantly, which is expected to increase the chance that two dysfunctional telomeres become closely apposed, thereby allowing the NHEJ machinery to engage the two DNA ends. Given that 53BP1 is also important for CSR, we suggest that the change in chromatin dynamics also promotes the joining of AID-induced breaks. In an accompanying paper by Nussenzweig and colleagues, this paradigm is further extended to distant DSBs in V(D)J recombination. We therefore suggest that 53BP1 is a facilitator of NHEJ at all distant DNA ends, acting by increasing the mobility of the local chromatin. As joining of distant DNA ends promotes chromosome rearrangements, we consider it unlikely that the main role of 53BP1 is to promote NHEJ at DNA ends that are far apart. It appears more likely that the increased chromatin mobility we observe reflects an attribute of 53BP1-containing chromatin that is important for DNA repair at all DSBs, including those that are closely apposed and breaks that are processed by HDR. We imagine that at telomeres, this chromatin change leads to increased mobility, whereas at chromosome-internal DSBs the same change in the chromatin may facilitate their repair in other ways.
Methods Summary
MEFs from E13.5 embryos obtained from crosses between TRF2F/− and 53BP1+/− mice1,20 were isolated and immortalized at passage 2 with pBabeSV40LT (a gift from G. Hannon). Cre was introduced by retroviral infection with pMMP Hit&Run Cre26. For time-lapse microscopy, TRF2F/−53BP1+/− and TRF2F/−53BP1−/− cells, expressing EGFP-TRF1 and mCherry-BP1-2; or TRF2F/−Lig4−/−p53−/−1 and TRF2F/−ATM−/−4 cells expressing EGFP-TRF1 only, were imaged untreated or 72 h after Cre-mediated deletion of TRF2. Time-lapse images were acquired using a DeltaVision RT microscope system (Applied Precision) in 3D in both EGFP and mCherry channels using SoftWoRx software every 30 seconds over 20 minutes. Images were deconvolved, projected in two dimensions and tracking analysis was performed with ImageJ. Data were collected from 3 independent experiments (n≥50) and Mann-Whitney statistical analysis was performed using Prism software.
Full methods accompany this paper.
Supplementary Material
Acknowledgments
We are extremely grateful to Junjie Chen for his generous gift of 53BP1 and MDC1 deficient mice. We thank Devon White for expert mouse husbandry. We thank the Bio-Imaging Facilities at CSHL and at RU. John Petrini is thanked for his gift of NBS1 antibody, Agnel Sfeir for providing the TRF1 antibody and Akimitsu Konishi for the BP1-2 construct. Mladen Dimitrov (UFR de Mathematiques, Univ. Paris 7) is thanked for mathematical advice on data analysis and Santiago Jaramillo and Yu Fu for Matlab programming. Members of the de Lange lab are thanked for comments on the manuscript. This work was supported by a grant from the NIH (GM049046) and by the NIH Director’s Pioneer Award (OD000379) to TdL and grants from the NIH to DLS (EY18244 and GM42694). ND was supported by an HHMI Pre-doctoral fellowship.
Footnotes
Statement of contributions
ND and TdL planned and designed the experiments. ND performed all experiments. YMC and DLS provided assistance with the imaging experiments. ND and TdL wrote the paper and made the figures.
References
- 1.Celli G, de Lange T. DNA processing not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- 2.Celli GB, Lazzerini Denchi E, de Lange T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat Cell Biol. 2006;8:885–890. doi: 10.1038/ncb1444. [DOI] [PubMed] [Google Scholar]
- 3.Dimitrova N, de Lange T. MDC1 accelerates non-homologous end-joining of dysfunctional telomeres. Genes Dev. 2006;20:3238–3243. doi: 10.1101/gad.1496606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lazzerini Denchi E, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 5.DiTullio RA, Jr, et al. 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nat Cell Biol. 2002;4:998–1002. doi: 10.1038/ncb892. [DOI] [PubMed] [Google Scholar]
- 6.Wang B, Matsuoka S, Carpenter PB, Elledge SJ. 53BP1, a mediator of the DNA damage checkpoint. Science. 2002;298:1435–1438. doi: 10.1126/science.1076182. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez-Capetillo O, Liebe B, Scherthan H, Nussenzweig A. H2AX regulates meiotic telomere clustering. J Cell Biol. 2003;163:15–20. doi: 10.1083/jcb.200305124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 9.Goldberg M, et al. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 2003;421:952–956. doi: 10.1038/nature01445. [DOI] [PubMed] [Google Scholar]
- 10.Lou Z, Minter-Dykhouse K, Wu X, Chen J. MDC1 is coupled to activated CHK2 in mammalian DNA damage response pathways. Nature. 2003;421:957–961. doi: 10.1038/nature01447. [DOI] [PubMed] [Google Scholar]
- 11.Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 12.Ward IM, Minn K, Jorda KG, Chen J. Accumulation of checkpoint protein 53BP1 at DNA breaks involves its binding to phosphorylated histone H2AX. J Biol Chem. 2003;278:19579–19582. doi: 10.1074/jbc.C300117200. [DOI] [PubMed] [Google Scholar]
- 13.Bekker-Jensen S, Lukas C, Melander F, Bartek J, Lukas J. Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J Cell Biol. 2005;170:201–211. doi: 10.1083/jcb.200503043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Botuyan MV, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manis JP, et al. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat Immunol. 2004;5:481–487. doi: 10.1038/ni1067. [DOI] [PubMed] [Google Scholar]
- 16.Ward IM, et al. 53BP1 is required for class switch recombination. J Cell Biol. 2004;165:459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reina-San-Martin B, Chen J, Nussenzweig A, Nussenzweig MC. Enhanced intra-switch region recombination during immunoglobulin class switch recombination in 53BP1−/− B cells. Eur J Immunol. 2007;37:235–239. doi: 10.1002/eji.200636789. [DOI] [PubMed] [Google Scholar]
- 18.Lou Z, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 19.Xie A, et al. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol Cell. 2007;28:1045–1057. doi: 10.1016/j.molcel.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ward IM, Minn K, van Deursen J, Chen J. p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol Cell Biol. 2003;23:2556–2563. doi: 10.1128/MCB.23.7.2556-2563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Konishi A, de Lange T. Cell cycle control of telomere protection and NHEJ revealed by a ts mutation in the DNA-binding domain of TRF2. Genes Dev. 2008;22:1221–1230. doi: 10.1101/gad.1634008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 23.Molenaar C, et al. Visualizing telomere dynamics in living mammalian cells using PNA probes. Embo J. 2003;22:6631–6641. doi: 10.1093/emboj/cdg633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kao GD, et al. Histone deacetylase 4 interacts with 53BP1 to mediate the DNA damage response. J Cell Biol. 2003;160:1017–1027. doi: 10.1083/jcb.200209065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abney JR, Cutler B, Fillbach ML, Axelrod D, Scalettar BA. Chromatin dynamics in interphase nuclei and its implications for nuclear structure. J Cell Biol. 1997;137:1459–1468. doi: 10.1083/jcb.137.7.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silver DP, Livingston DM. Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol Cell. 2001;8:233–243. doi: 10.1016/s1097-2765(01)00295-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.