Abstract
Transcriptional reprogramming is critical for plant disease resistance responses. In potato (Solanum tuberosum), the marker gene PATHOGENESIS-RELATED-10a (PR-10a) is transcriptionally activated by pathogens, wounding, or elicitor treatment. Activation of PR-10a requires the recruitment of the activator Why1 to its promoter. In addition, PR-10a is negatively regulated by the repressor SEBF (for Silencer Element Binding Factor). Here, we show through a yeast two-hybrid screen that SEBF interacts with Pti4, which has been shown to be a transcriptional activator. SEBF recruits Pti4 via its consensus sequence–type RNA binding domain, while Pti4 is recruited to SEBF by means of its ethylene-response factor domain. In vivo plant transcription assays confirmed that SEBF interacts with Pti4 to form a repressosome, showing that Pti4 can also play a role in transcriptional repression. Chromatin immunoprecipitation revealed that both SEBF and Pti4 are recruited to the PR-10a promoter in uninduced conditions only and that the recruitment of Pti4 is dependent on the presence of SEBF, consistent with the fact that there is no Pti4 consensus binding site in PR-10a. Unexpectedly, we also demonstrated that recruitment of SEBF was dependent on the presence of Pti4, thereby explaining why SEBF, itself a repressor, requires Pti4 for its repressing function.
INTRODUCTION
To combat the invasion of potential pathogens, plants possess an immune system with which they detect elicitors and activate a battery of defense responses. As a result of defense induction, including massive transcriptional reprogramming, the spread of pathogens can be stopped and, in some cases, the plant can become resistant to subsequent invasions (Jones and Dangl, 2006). Plant defense mechanisms are linked to the upregulation of Pathogenesis-Related (PR) gene expression as well as other responses, such as the production of antimicrobial compounds and the modification of secondary cell wall composition (Stintzi et al., 1993). However, PR genes represent only a subset of the large number of genes whose expression is modified in response to pathogen attack. In the model plant Arabidopsis thaliana, for example, up to 25% of all genes are subjected to changes in regulation (Eulgem, 2005). Therefore, transcription factors fulfill a crucial role in the regulation of plant defense responses.
Several families of transcription factors involved in regulating plant defense have been characterized, including WRKY, Myb, ERF, Whirly, and TGA transcription factors (Fobert, 2006). Members of the ethylene-response factor (ERF) family bind the GCC box found in the promoter of many defense-related genes (Ohme-Takagi and Shinshi, 1995; Ohme-Takagi et al., 2000). ERF transcription factors are regulated by ethylene, jasmonic acid, salicylic acid, and some pathogen infections (Gu et al., 2000; Oñate-Sánchez and Singh, 2002; Brown et al., 2003; Lorenzo et al., 2003) as well as by abiotic stresses (Park et al., 2001; Chen et al., 2002). The tomato (Solanum lycopersicum) Pti4 is an ERF transcription factor that was first isolated by its interaction with the kinase Pto, which confers resistance to Pseudomonas syringae pv tomato expressing the avirulence gene AvrPto (Zhou et al., 1997). Pti4 controls the expression of defense-related genes, and its function is regulated at both the transcriptional and posttranscriptional levels (Gu et al., 2000; Mysore et al., 2002; Wu et al., 2002). By virtue of its ERF domain, Pti4 can bind the sequence GCCGCC (GCC box) and regulate the expression of several GCC box–containing genes (Gu et al., 2002). However, chromatin immunoprecipitation (ChIP) experiments have shown direct binding of Pti4 to some non-GCC box–containing promoters (Chakravarthy et al., 2003), leading to the hypothesis that either Pti4 is able to bind to a DNA motif other than the GCC box or it interacts with other transcription factors to regulate promoter activity (Chakravarthy et al., 2003).
The promoter of the potato (Solanum tuberosum) PATHOGENESIS-RELATED-10a (PR-10a) gene has been used as a model to understand defense-related transcriptional regulation. Several regulatory elements were characterized in this promoter, including an elicitor response element (ERE) that confers wounding- and elicitor-dependent transcriptional upregulation of PR-10a (Matton et al., 1993; Després et al., 1995). The recruitment of the transcriptional activator Why1 (formerly PBF-2) to the ERE is required for the activation of PR-10a (Desveaux et al., 2000). In unstimulated cells, Why1 is stored inactive and sequestered away from the ERE (Desveaux et al., 2000). Upon elicitation, the DNA binding activity of Why1 is released, allowing the recruitment of the protein to the ERE (Desveaux et al., 2000, 2004). PR-10a transcription is also regulated through another promoter sequence, located between positions −52 and −27, called the silencer element (SE). Binding of the transcription factor SEBF (for Silencer Element Binding Factor) to the SE represses PR-10a expression (Matton et al., 1993; Després et al., 1995; Boyle and Brisson, 2001). Like Why1, SEBF is also a single-stranded DNA binding protein. In addition, SEBF possesses a transit peptide capable of targeting the protein to the chloroplast (Boyle and Brisson, 2001). The mature protein, which is found in plastids and in the nucleus, contains two consensus sequence–type RNA binding domains (cs-RBDs) separated by a Gly-rich region. (Boyle and Brisson, 2001).
Here, we report on the interaction between the repressor SEBF and the potato homolog of the tomato transcriptional activator Pti4. We demonstrate that SEBF interacts with the SE of PR-10a through its cs-RBDII but recruits Pti4 via its cs-RBDI. We show that Pti4 is recruited to SEBF by means of its ERF domain. We also show that SEBF associates with PR-10a in unstimulated cells only and serves to draft Pti4 to the PR-10a promoter, which contains no GCC box. We provide evidence that the binding of SEBF to the promoter requires the presence of Pti4 and that the SEBF-Pti4 complex forms the core of a repressosome. The data presented here not only unravel an unprecedented and unexpected role for the activator Pti4 as an indispensable element of a repressosome but also provide concrete evidence for the previously hypothesized mechanism of recruitment of Pti4 to non-GCC box–containing genes.
RESULTS
SEBF Physically Interacts with Pti4 in Yeast and in Vitro
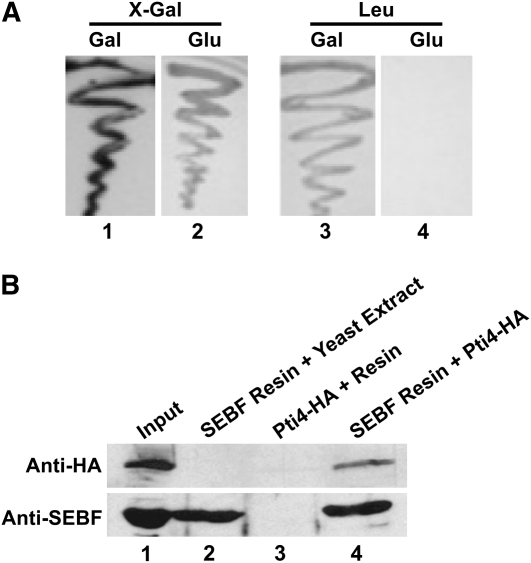
Since SEBF is one of the rare examples of a single-stranded DNA binding repressor characterized from plant systems, we sought to determine the protein composition of the SEBF-containing repressosome complex. To do so, a cDNA encoding the mature form of the potato SEBF was used as bait in a yeast two-hybrid screen against a tomato cDNA library (Zhang et al., 1999). From ∼107 transformants, 80 colonies producing blue color on X-Gal plates and capable of growth on medium lacking His, Trp, Leu, and uracil but supplemented with galactose were identified. Three of these colonies encoded tomato Pti4 (Sl Pti4). Pti4 is a transcription factor involved in plant defense signaling (Gu et al., 2000) and was chosen for further studies. The full-length coding region of potato Pti4 was amplified from a potato cDNA library (Matton and Brisson, 1989). It codes for a 26-kD protein, which is 94% identical to Sl Pti4 (see Supplemental Figure 1 online). Potato Pti4 (St Pti4, from here on referred to as Pti4) also interacted with SEBF (Figure 1A) and was used for all subsequent experiments.
Figure 1.
The Repressor SEBF Interacts with the Transcriptional Activator Pti4.
(A) The yeast two-hybrid interaction between the SEBF bait and the Pti4 prey was revealed through the expression of the lacZ reporter gene (β-galactosidase activity), detected as a blue color (shown here in dark gray/black) on plates containing galactose (Gal) and supplemented with X-Gal (lane 1) or by prototrophic growth (activation of the LEU2 reporter gene) in medium lacking Leu but containing galactose (lane 3). The activation of the reporter genes was dependent on the expression of the prey construct, since its suppression in medium containing glucose (Glu) does not lead to the activation of the lacZ reporter gene (lane 2) or to prototrophic growth (lane 4).
(B) The pull-down assays were performed by incubating SEBF produced in E. coli and coupled to a solid support with Pti4 expressed as an HA fusion protein in yeast (lane 4). As negative controls, pull-downs were performed by omitting SEBF (lane 3) or Pti4 (lane 2). Lane 1 contains 100% of the amount of SEBF coupled to the solid support (bottom panel) or 20% of the amount of Pti4 used in the pull-down experiments (top panel). Proteins were analyzed by immunoblotting using an anti-HA antibody for Pti4 detection (top panel) or an anti-SEBF antibody (bottom panel).
To confirm the yeast two-hybrid results and the direct physical interaction between SEBF and Pti4, we performed pull-down assays. SEBF fused to a C-terminal 6-His tag was produced in Escherichia coli and coupled to a nickel column before incubation with the C-terminal hemagglutinin (HA)-tagged Pti4 produced in yeast. Analysis of the bound fraction by immunoblot reacted with anti-HA antibodies, as presented in Figure 1B, revealed the presence of a signal (lane 4), demonstrating the existence of an in vitro interaction between Pti4 and SEBF. However, when Pti4-HA was incubated with the resin alone (lane 3) or when only a yeast extract was incubated with His-tagged SEBF bound to the column (lane 2), no signal was detected on the immunoblot, testifying to the specificity of the interaction.
SEBF Recruits Pti4 through cs-RBDI and Interacts with DNA through cs-RBDII
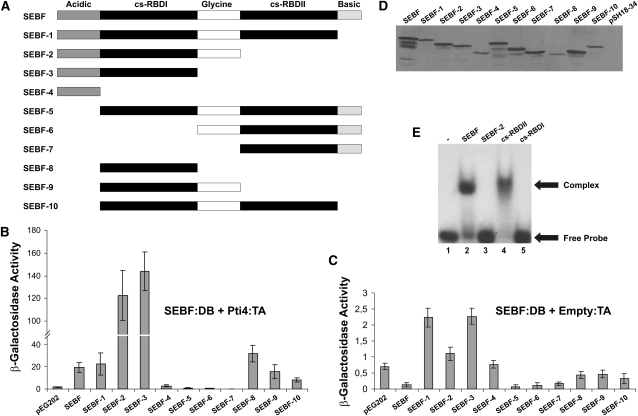
To gain some insights into the protein–protein interaction interface existing between SEBF and Pti4, we performed yeast two-hybrid assays with a series of SEBF deletions fused to the LexA DNA binding (DB) domain, as depicted in Figure 2A, and full-length Pti4 fused to the B42 transcription activation (TA) domain. Coexpression of Pti4 with SEBF-1, 2, and 3, which removed, respectively, the basic domain, cs-RBDII, and the Gly-rich domain, did not abolish β-galactosidase activity, indicating that these domains of SEBF were not required for interaction with Pti4 (Figure 2B). Of note, however, deletion of cs-RBDII in SEBF-2 and SEBF-3 stimulated the interaction with Pti4, compared with that observed with full-length SEBF, suggesting that this domain could be a negative regulator of the interaction. Further deletion, removing cs-RBDI (SEBF-4), reduced reporter gene activity to that observed with the empty vector (pEG202) expressing the DB only. This indicates that cs-RBDI is required for interaction with Pti4. Consistent with this observation, constructs SEBF-6 and SEBF-7, which lack this domain, did not interact with Pti4.
Figure 2.
SEBF Interacts with DNA through cs-RBDII and Recruits Pti4 via cs-RBDI.
(A) Schematics of SEBF and its deletion mutants analyzed in (B) to (E). Acidic and Basic indicate domains with these properties, while Glycine represents a Gly-rich region of SEBF. cs-RBDI and cs-RBDII refer to cs-RBDs I and II.
(B) Bar graph of β-galactosidase activity as the output of yeast two-hybrid assays testing the interaction of SEBF and the mutants depicted in (A) fused to the Lex-A DB domain with the full-length Pti4 fused to the B42 TA domain.
(C) Bar graph illustrating the background level of activity observed with each of the SEBF constructs depicted in (A) and coexpressed with the empty B42 TA vector.
For (B) and (C), results obtained by expressing the Lex-A DB domain alone (pEG202) along with Pti4:TA domain are shown as reference baseline values. Note the difference in scales between these panels. Values consist of n = 6 samples and represent averages ± 1 sd. Every bar represents an assay on three different colonies repeated on two independent transformation events.
(D) Immunoblot using an anti-SEBF antibody was performed to confirm the expression of SEBF and its mutant derivatives in cell lines used in (B).
(E) EMSA analyses were performed with the full-length SEBF (lane 2), SEBF-2 (lane 3), the cs-RBDI domain of SEBF (SEBF-8 in [A]; lane 4), and the cs-RBDII domain of SEBF (lane 5). All studies were done with 10 ng of purified recombinant protein and the 32P-labeled single-stranded SE oligonucleotide. Lane 1 contained only the labeled SE oligonucleotide.
In the context of full-length SEBF, deletion of the acidic domain (SEBF-5) abolished interaction with Pti4, indicating that this domain might contact Pti4 directly. Nevertheless, the acidic domain alone (SEBF-4) was not sufficient to confer interaction with Pti4. When comparison is made between SEBF-8 and SEBF-3 or between SEBF-9 and SEBF-2, the data seem to indicate that the acidic domain does play a role in the interaction with Pti4. However, in the context of a C-terminal-deleted SEBF (SEBF-8, -9, and -10), the acidic domain did not appear to play an interacting role compared with SEBF. This ambiguity suggests that the acidic domain of SEBF may interface directly with Pti4 or that it might rather serve to better expose cs-RBDI, which on its own (SEBF-8) is sufficient for the interaction with Pti4. In Figure 2C, the SEBF constructs were expressed with the empty TA vector to monitor the intrinsic level of reporter activation conferred by these proteins. The levels of reporter gene activity were not significant compared with those observed when Pti4 was coexpressed (Figure 2B), validating the conclusions that SEBF and Pti4 interact with each other. The immunoblot presented in Figure 2D demonstrates that the lack of or low reporter gene activity observed when coexpressing SEBF-4, -5, -6, or -7 with Pti4 was not the result of the absence of expression of these proteins in yeast but truly reflects a lack of interaction.
Since SEBF possesses two consensus RBDs (Boyle and Brisson, 2001), we sought to determine which one or whether both of them are required for the single-stranded DNA binding activity. Electrophoretic mobility shift assays (EMSAs), represented in Figure 2E and performed with full-length mature SEBF, demonstrated a shift indicating binding to the SE-DNA (lane 2; see Methods for description of the probe). However, deletion of cs-RBDII (SEBF-2) abolished DNA binding (lane 3). Expression of cs-RBDI and cs-RBDII followed by EMSA demonstrated that cs-RBDII (lane 4) was sufficient and was the only domain required for DNA binding activity.
Pti4 Is Recruited to SEBF through Its ERF Domain
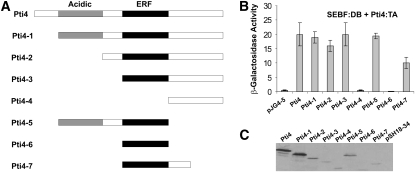
To further characterize the Pti4–SEBF interaction interface, we performed additional yeast two-hybrid assays, but this time using full-length mature SEBF fused to the LexA DB domain and a series of Pti4 deletions fused to the B42 TA domain (Figure 3A). Coexpression of SEBF with Pti4 deletions that progressively removed the N terminus up to the ERF domain (Pti4-1, -2, and -3) did not substantially alter β-galactosidase activity, indicating that these domains of Pti4 were not required for interaction with SEBF (Figure 3B). However, a further deletion, removing the ERF (Pti4-4), abolished interaction with SEBF, revealing the importance of this domain for the Pti4-SEBF complex formation. Deleting the C terminus of construct Pti4-1 up to the ERF (Pti4-5) did not alter the interaction with SEBF, further substantiating the fact that regions outside the ERF are not required for interfacing with SEBF. Attempts to demonstrate that the ERF alone (Pti4-6) was sufficient for interaction with SEBF failed. However, the immunoblot in Figure 3C (lane 7) indicates that Pti4-6 did not express well, suggesting that its low abundance may be the explanation for the apparent lack of interaction with SEBF. We thus went on to generate construct Pti4-7, which contains additional amino acids C terminal of the ERF (Figure 3A). This construct was found to be expressed in yeast (Figure 3C, lane 8) and was capable of interaction with SEBF (Figure 3B, Pti4-7). Taken together, the data point toward the ERF as the domain interfacing with SEBF. However, we cannot exclude the possibility that regions adjacent to the ERF participate in the recruitment by SEBF.
Figure 3.
The ERF Domain of Pti4 Interfaces with SEBF.
(A) Schematics of Pti4 and its deletion mutants analyzed in (B) and (C). Acidic indicates a domain with this property, while ERF stands for the ethylene-response factor domain, which contains the DNA binding region of Pti4.
(B) Bar graph of β-galactosidase activity as the output of a yeast two-hybrid assay testing the interaction of Pti4 and the mutants depicted in (A) fused to the B42 TA domain containing an HA tag along with the full-length SEBF fused to the Lex-A DB domain. Results obtained by expressing the B42 TA domain alone (pJG4-5) along with the SEBF:DB domain are shown as reference baseline values. Values consist of n = 6 samples and represent averages ± 1 sd. Every bar represents an assay on three different colonies repeated on two independent transformation events.
(C) Immunoblotting using an anti-HA antibody was performed to confirm the expression of Pti4 and its mutant derivatives in cell lines used in (B).
Recruitment of Pti4 to PR-10a Is SEBF-Dependent
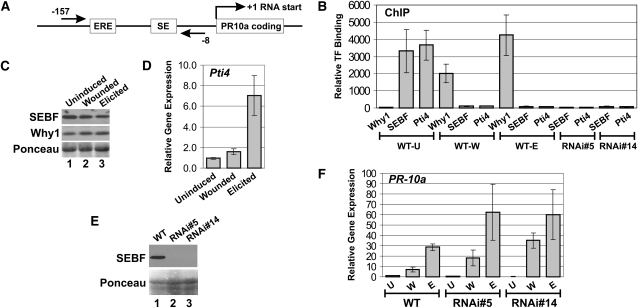
Induction of the PR-10a gene is positively controlled by Why1, which has been shown by ChIP to be recruited to the gene after wounding or elicitor treatment (Desveaux et al., 2004). The PR-10a gene has also been shown to be regulated by repressor SEBF. However, direct binding of the factor to the gene has never been addressed (Boyle and Brisson, 2001). Figure 4A is a diagram of the PR-10a gene that shows the positions of the PCR primer pair used for all of the ChIP experiments. As reported previously (Desveaux et al., 2004), Figure 4B shows that an enrichment of Why1 at the PR-10a promoter was observed following immunoprecipitations performed with the anti-Why1 antibody on wounded (WT-W) and elicited (WT-E) tissues but was absent from the ChIP performed with uninduced (WT-U) samples. Conversely, ChIP performed with the anti-SEBF antibody led to an enrichment in uninduced tissues but not in wounded or elicited tissues. This indicates that SEBF is only recruited to PR-10a in uninduced conditions, consistent with its role as a transcriptional repressor.
Figure 4.
Pti4 Binds to the PR-10a Promoter through Its Interaction with SEBF.
(A) Diagram of the PR-10a promoter showing the positions of the ERE, the SE, and the oligonucleotides used for the ChIP experiments. The straight arrows and numbers refer to the locations of the oligonucleotides with respect to the RNA start site.
(B) ChIP experiments analyzed by qPCR. ChIP was performed with an anti-Why1 antibody (Why1), an anti-SEBF antibody (SEBF), or an anti-Pti4 antibody (Pti4). The antibody used is labeled directly beneath the bar; the plant genotype and conditions are indicated below the antibody labels. WT-U, WT-W, and WT-E indicate ChIPs performed in wild-type plants that were left uninduced, or were wounded, or were treated with the elicitor, respectively. ChIP was also conducted with uninduced tissues from two independent SEBF RNAi lines (RNAi#5 and RNAi#14). The amounts of the different transcription factors (TF) binding to PR-10a were relative to their recruitment to the Actin gene PoAc97 (see Methods). Data for each bar are from three biological replicates, and errors bars are equal to 1 sd.
(C) The two top panels are immunoblot analysis of SEBF and Why1 proteins extracted from uninduced, wounded, or elicited wild-type plants. An anti-SEBF or anti-St Why1 antibody was used. The bottom panel is a Ponceau S staining of the membrane, shown in the top panels, as a loading control.
(D) Bar diagram illustrating the abundance of Pti4 transcript in uninduced, wounded, or elicited wild-type plants. Values represent means ± sd from three biological replicates.
(E) The top panel is an immunoblot analysis of SEBF proteins extracted from wild-type plants, SEBF RNAi line 5 (RNAi#5), and SEBF RNAi line 14 (RNAi#14). An anti-SEBF antibody was used. The bottom panel is a Ponceau S staining of the membrane, shown in the top panel, as a loading control.
(F) Bar graph illustrating the abundance of PR-10a transcript in uninduced (U), wounded (W), or elicited (E) plants from wild-type plants or from two SEBF RNAi lines (RNAi#5 and RNAi#14). Values represent means ± sd from three biological replicates.
We previously demonstrated, in Arabidopsis, that genes negatively regulated by Pti4 and to which Pti4 was shown to be recruited did not possess a GCC box, the cognate Pti4 DNA binding element (Chakravarthy et al., 2003). Two models were proposed to explain these observations: first, Pti4 might be recruited directly to these negatively regulated genes by binding to a novel DNA sequence; second, Pti4 might be indirectly recruited via another DNA binding protein. Since SEBF (there are nine SEBF-like genes in Arabidopsis) is a repressor binding to both PR-10a and Pti4, we saw the PR-10a gene as an opportunity to test these two models. ChIP performed with anti-Pti4 antibodies showed an enrichment of Pti4 at the PR-10a promoter in uninduced tissues but not in wounded or elicited samples (Figure 4B). This demonstrates that Pti4 can be recruited to PR-10a despite the absence of a GCC box and also that its recruitment profile is similar to that of SEBF. The immunoblot in Figure 4C and the quantitative PCR (qPCR) results in Figure 4D indicate that both SEBF and Pti4 are present in uninduced, wounded, and elicited tissues and, therefore, that their absence at the PR-10a promoter is not due to their absence from the tissue. These data thus suggest that Pti4 might be recruited to PR-10a via SEBF. To test this hypothesis, we generated two knockdown lines of SEBF through RNA interference (RNAi) technology. The immunoblot in Figure 4E indicates that levels of the SEBF protein are undetectable in these lines (lanes 2 and 3). ChIP experiments performed with the anti-SEBF antibody on uninduced tissue from the RNAi lines (RNAi#5 and RNAi#14) did not reveal any enrichment of SEBF at the PR-10a promoter (Figure 4B). This result indicates that SEBF is not recruited to PR-10a in these lines, consistent with the knockdown expression. ChIP experiments performed on the same tissue, but with the anti-Pti4 antibody, also indicated an absence of Pti4 at the PR-10a promoter (Figure 4B). These data support the model in which Pti4 is recruited to PR-10a via SEBF. To determine the effect of SEBF knockdown on PR-10a expression, we performed qPCR on PR-10a transcripts in uninduced (U) tissue and after wounding (W) and elicitor (E) treatment (Figure 4F). Silencing of SEBF did not lead to activation of PR-10a in uninduced tissues, but both wounding and elicitor treatment led to increased PR-10a transcript accumulation in the SEBF RNAi lines compared with wild-type plants.
The SEBF-Pti4 Complex Forms a Repressosome
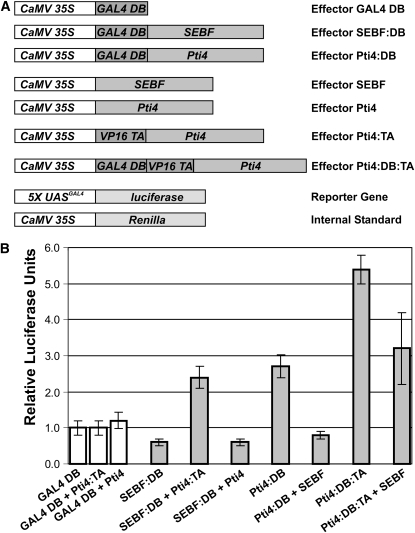
The results in Figure 4 demonstrate that Pti4 is recruited to PR-10a through interaction with SEBF. However, since Pti4 has been shown to be a transcriptional activator (Gu et al., 2002), we asked ourselves what the molecular consequences of the recruitment of Pti4 to SEBF might be. To address this question, the transcriptional properties of SEBF, Pti4, and the SEBF-Pti4 complex were examined using an in vivo plant transcription assay (Figure 5B). The constructs used in this experiment are detailed in Figure 5A. The baseline level of transcription was determined by transfecting leaves with the Gal4 DB domain (not fused to any other protein or protein domain) along with a reporter construct consisting of a firefly luciferase gene under the control of five copies of the Gal4 upstream activating sequences fused to a minimal promoter. Transfection with SEBF:DB resulted in reporter gene activation below the baseline level, consistent with the fact that SEBF is a repressor (Boyle and Brisson, 2001). Coexpression of SEBF:DB and Pti4:TA led to the activation of the reporter gene beyond baseline, confirming that the two proteins interact with one another in this plant system. We next addressed how Pti4 would modulate the transcriptional properties of SEBF. SEBF:DB was thus coexpressed with Pti4 (not fused to any foreign TA or DB domains), which resulted in activation of the reporter gene below baseline. Values were in fact not significantly different from those observed with SEBF:DB, indicating that the SEBF-Pti4 complex, like SEBF, acts as a repressor. To strengthen the argument, we tested the reciprocal constructs. First, however, Pti4:DB was transfected alone and reporter gene activation was monitored. Values were beyond baseline, confirming that Pti4 is a transcriptional activator. Conversely, expression of Pti4:DB along with SEBF (not fused to other protein domains) abolished the capacity of Pti4 to act as a transcriptional activator, as deduced by reporter gene activity falling below baseline. As an additional control, Pti4:DB was further activated by a direct fusion to the VP16 TA (Pti4:DB:TA), which led to higher values compared with Pti4:DB. Reporter gene expression mediated by this construct could also be mitigated by the addition of SEBF (Pti4:DB:TA + SEBF). However, the synergistic effect of the endogenous Pti4 transactivation domain and that of VP16 could not be fully countered by SEBF. Nevertheless, the results in Figure 5 demonstrate that when complexed with SEBF, Pti4 is no longer a transcriptional activator; that is to say, the SEBF-Pti4 complex acts as a repressosome.
Figure 5.
The SEBF-Pti4 Complex Represses Transcription in Vivo.
(A) Representations of the different constructs used in the plant two-hybrid and in vivo transcription assays. Promoters are shown in white boxes. CaMV 35S indicates the double cauliflower mosaic virus 35S:alfalfa mosaic virus promoter. 5X UASGAL4 indicates a promoter composed of a multimerized (five elements) Gal4 upstream activating sequence fused to a minimal TATA box and the Ω translational enhancer from the Tobacco mosaic virus. Coding sequences are shown in dark and light gray boxes. GAL4 DB indicates the GAL4 DNA binding domain. VP16 TA indicates the constitutive transactivation domain of viral protein 16. All constructs possess the polyadenylation signal from the nopaline synthase gene (data not shown). The 35S:Renilla construct is an internal reference to normalize transfection efficiency.
(B) Bar graph illustrating the interaction of Pti4 with SEBF as well as the transcriptional activation potential of Pti4, SEBF, and the SEBF-Pti4 complex. Each effector or pair of effectors was cotransfected with the reporter gene and the internal standard. The effector construct containing the GAL4 DB domain only was transfected into untreated leaves along with the reporter and internal standard constructs and was given an arbitrary value of 1 relative luciferase unit ± 1 sd after normalization with Renilla activity. All values are relative to the activity of Gal4 DB domain obtained in untreated leaves. Values consist of n = 25 samples and represent averages ± 1 sd. Every bar represents five bombardments repeated five times (n = 25). White bars represent Gal4 DB controls, while gray bars refer to data obtained with the proteins under investigation.
Recruitment of SEBF to PR-10a Is Pti4 Dependent
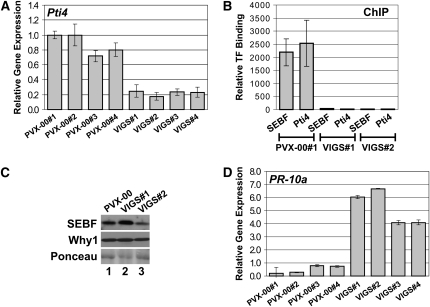
Although the results in Figure 5 indicate that the SEBF-Pti4 complex is a repressosome, one question remains unanswered: what is the role of Pti4 in the SEBF-Pti4 complex, since SEBF is itself a repressor? To tackle this question, we wanted to test whether the recruitment of SEBF to PR-10a would be in any way affected by the absence of Pti4. We thus generated two knockdown Pti4 plants. For these experiments, we used virus-induced gene silencing (VIGS) technology, since we failed to recover Pti4 knockdown lines generated by RNAi. Because Pti4 protein levels in wild-type plants are below detection levels when monitored by immunoblot analysis, we assessed the extent of knockdown by qPCR. Figure 6A indicates that levels of the Pti4 mRNA are substantially reduced in Pti4 VIGS lines (VIGS#1 to VIGS#4) compared with the empty vector VIGS controls (PVX-00#1 to PVX-00#4). ChIP experiments performed with the anti-Pti4 antibody on uninduced tissue from the Pti4 VIGS lines 1 and 2 did not reveal any enrichment (Figure 6B), confirming the absence of Pti4 protein recruitment to PR-10a and consistent with the knockdown expression. Interestingly, ChIP experiments performed on the same tissue, but with the anti-SEBF antibody, also indicated an absence of enrichment, which demonstrates that SEBF requires Pti4 for its recruitment to PR-10a. The presence of SEBF in these plants was confirmed by immunoblot analysis (Figure 6C) and indicates that the absence of SEBF at the PR-10a promoter in Pti4 knockdown lines is not due to the absence of SEBF in these tissues. As expected, ChIP from empty vector VIGS plants (PVX-00#1) revealed the presence of both SEBF and Pti4 at the PR-10a promoter after immunoprecipitation with both anti-SEBF and anti-Pti4 antibodies, respectively (Figure 6B). The effect of Pti4 knockdown on PR-10a expression was also analyzed by qPCR. In contrast with SEBF knockdown lines, activation of PR-10a was already observed in the absence of any treatment (Figure 6D).
Figure 6.
Pti4 Is Required for Binding of SEBF to the PR-10a Promoter.
(A) Pti4 transcript is less abundant in Pti4 VIGS lines. The bar diagram illustrates the abundance of Pti4 transcript in PVX-Pti4 VIGS lines (VIGS#1 to VIGS#4) relative to PVX-00 lines (PVX-00#1 to PVX-004). Data for each of the four biological replicates (each bar) are averages of three technical replicates, and errors are equal to 1 sd.
(B) Reduced Pti4 prevents SEBF binding to the PR-10a promoter. ChIP results were analyzed by qPCR. The amounts of the different transcription factors binding to PR-10a were relative to their recruitment to the Actin gene PoAc97 (see Methods). ChIPs from two independent PVX-Pti4 VIGS lines (VIGS#1 and VIGS#2) and from a VIGS line containing the empty pGR106 vector (PVX-00) were performed with uninduced tissues. ChIP was conducted with an anti-SEBF (SEBF) or an anti-Pti4 (Pti4) antibody. Data for each bar are from two biological replicates, and errors are equal to 1 sd.
(C) SEBF is still present in the Pti4 VIGS lines. The two top panels are immunoblot analysis of SEBF and Why1 proteins extracted from the PVX-00 line and from the PVX-Pti4 VIGS lines 1 and 2 (VIGS#1 and VIGS#2). An anti-SEBF or anti-St Why1 antibody was used. The bottom panel is a Ponceau staining of the membrane, shown in the top panels, as a loading control.
(D) PR-10a transcripts are more abundant in the Pti4 VIGS lines. The bar graph illustrates the abundance of PR-10a transcript present in the PVX-00 lines (PVX-00#1 to PVX-004) and in the PVX-Pti4 VIGS lines (VIGS#1 to VIGS#4). Data for each of the four biological replicates (each bar) are averages of three technical replicates, and errors are equal to 1 sd.
DISCUSSION
This study was motivated by our interest in elucidating the protein components of the SEBF repressosome. Our data have demonstrated that SEBF is recruited to PR-10a under noninducing conditions, but after the gene is activated by wounding or elicitor treatment, the environment at the promoter is not permissive for an interaction with the repressor. Furthermore, our results established that, despite the absence of a cognate GCC box, Pti4 was recruited to PR-10a with a pattern similar to that of SEBF, and more specifically, that its interaction occurred only under uninduced conditions. These outcomes, along with the fact that Pti4 was recovered from a yeast two-hybrid screen using SEBF as bait, suggested that Pti4 is drafted to PR-10a through recruitment by SEBF. This was confirmed by experiments demonstrating the absence of Pti4 recruitment to PR-10a in SEBF knockdown plants. In vivo plant transcription assays demonstrated that the SEBF-Pti4 complex, like SEBF, repressed transcription, indicating that the complex behaves as a repressosome. Finally, Pti4 knockdown lines highlight the raison d'être for the presence of this protein in the repressosome, which is to allow SEBF to reach its DNA target. Together, our data argue that in uninduced conditions SEBF and Pti4 are components of a repressosome that assembles at the PR-10a promoter.
SEBF Recruits Pti4 to PR-10a and Not Vice Versa
Since ChIP experiments performed on Pti4 knockdown plants demonstrated that Pti4 is also required for the recruitment of SEBF to PR-10a, one could argue that Pti4 binds directly to DNA through a previously uncharacterized DNA sequence and that SEBF is drafted to the promoter via Pti4. Although this is a plausible scenario, previous data would argue against this model. Characterization of PR-10a through promoter deletion analysis revealed the presence of the SE, a negative regulatory element between positions −52 and −27 (Després et al., 1995). This identification was followed up by the biochemical purification of the factor binding to this negative element, which was coined SEBF (Boyle and Brisson, 2001). In vitro mutational analysis of the SEBF binding element by EMSA, using recombinant SEBF, allowed us to uncover mutations that disrupt SEBF DNA binding activity but also others that enhance it. Transcriptional analysis, in potato protoplasts, of modified PR-10a promoter variants bearing these mutations highlighted an inverse correlation between transcriptional activity and the recruitment capacity of SEBF to the promoter. DNA mutations that reduced the recruitment of the repressor led to higher reporter gene activity, while those that favored drafting of SEBF reduced it, compared with a wild-type promoter element or a mutated variant that did not affect SEBF binding (Boyle and Brisson, 2001). Although Pti4 might be present in potato protoplasts and potentially might assist the binding of SEBF to DNA, EMSA analyses using recombinant SEBF alone indicate that SEBF can directly bind the SE without Pti4. This clearly demonstrates that repression at the PR-10a gene is dependent on direct recruitment of SEBF to its cognate element and indicates that SEBF is indeed the DNA binding component of the SEBF-Pti4 repressosome. Although we cannot rule out the possibility that Pti4 could also be associated with a distal GCC box and interact with SEBF via DNA looping, the fact that it interfaces SEBF via the ERF domain, which coincides with its DB domain, would suggest that Pti4 may not be able to bind to DNA concomitantly with an interaction with SEBF.
Potential Mechanisms of Pti4-Assisted Recruitment of SEBF to PR-10a
The ChIP experiments in Figure 6 performed on Pti4 knockdown plants clearly identify Pti4 as mandatory to the in vivo recruitment of SEBF to PR-10a. In vitro, on the other hand, SEBF can readily interact with its cognate DNA element (Figure 2E) (Boyle and Brisson, 2001). However, in vitro, SEBF only recognizes one strand of the SE, the coding strand, but cannot bind the noncoding strand or the double-stranded DNA (Boyle and Brisson, 2001). Thus, a handful of scenarios may provide a rationale for the role of Pti4 in allowing SEBF recruitment to PR-10a, and these depend on the architecture of the SE in uninduced conditions.
First, the SE could be in a single-stranded DNA conformation. Under this condition, one would assume, however, that SEBF could bind directly to its cognate single-stranded DNA and a requirement for Pti4 would not appear obvious. Nevertheless, in this scenario, Pti4 could prevent the dismissal of SEBF from PR-10a by precluding, for example, the occurrence of posttranslational modifications or other SEBF–protein interactions that could result in decreased DNA binding affinity. The opposite setting, in which Pti4 would favor the recruitment of additional cofactors stimulating the posttranslational modification of SEBF and allowing its stronger interaction with DNA, is as probable.
Second, the SE could be in a double-stranded DNA conformation in vivo, under uninduced conditions, and prior to binding of SEBF or the SEBF-Pti4 complex. Although this seems unlikely given that SEBF cannot bind double-stranded DNA in vitro, this behavior parallels the activity of some heterogeneous nuclear ribonucleoproteins (hnRNPs). hnRNPs are among the best characterized single-stranded DNA binding proteins involved in transcriptional regulation. They can activate or repress gene expression (Hsieh et al., 1998; Ostrowski et al., 2003). Like SEBF, hnRNPs are found in many subcellular compartments (Swanson and Dreyfuss 1988; Ostrowski et al., 1991, 2002) and contain regions similar to the cs-RBD domains (Boyle and Brisson, 2001). In an exemplary case, hnRNP C1/C2 purified from an in vivo source with its associated cofactors could bind the double-stranded DNA version of its cognate sequence, while the recombinant version, devoid of cofactors, could not (Mahajan et al., 2005). The same rationale could be applied to SEBF, where Pti4 may stimulate a latent helicase activity in SEBF or help recruit such an activity to the repressosome, as is the case in the hnRNP K-Purα repressosome (Da Silva et al., 2002). Although it is unclear at present which mechanism governs the Pti4-assisted recruitment of SEBF to PR-10a, these proposed scenarios constitute future areas of potential exploration.
A Working Model of PR-10a Gene Regulation
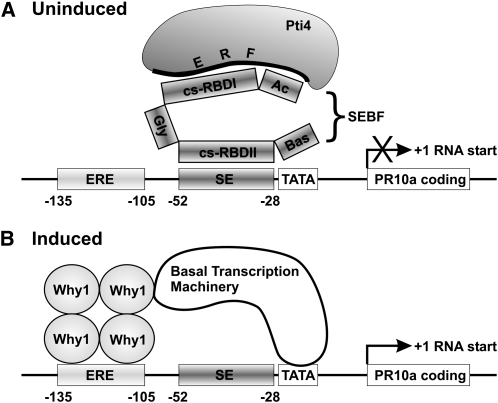
Figure 7 summarizes our results and provides a model for the regulation of PR-10a through the interplay of a repressosome and an activator. In the uninduced state, the SEBF-Pti4 repressosome complex occupies the SE, while Why1 is stored inactive and sequestered away from the ERE (Desveaux et al., 2000). Upon induction, the repressosome is dismissed from the promoter and the DNA binding activity of Why1 is induced, allowing the ERE to recruit the activator Why1 (Desveaux et al., 2000, 2004).
Figure 7.
Model for the Transcriptional Regulation of PR-10a by the Transcription Factors Why1, SEBF, and Pti4.
(A) In uninduced tissues, the repressosome SEBF-Pti4 is recruited to the SE through the cs-RBDII of SEBF, while the cs-RBDI and potentially the acidic domain (Ac) of SEBF are required to interface with the ERF domain of Pti4. Recruitment of the repressosome to PR-10a prevents transcription through a yet to be identified mechanism. Bas and Gly indicate a basic domain and a Gly-rich region, respectively. Their functions are unknown. TATA refers to the TATA box.
(B) In induced tissues, the SEBF-Pti4 repressosome is dismissed from the PR-10a promoter, while the ERE is populated by the transcriptional activator Why1, which is a mandatory step in the transcriptional activation of PR-10a. According to the accepted gene activation paradigm, Why1 would contact, directly or indirectly, the basal transcription machinery.
In both panels, the numbers indicate the nucleotide positions in the promoter with respect to the RNA start site.
We have previously shown that a reporter gene regulated by the first 135 bp of the PR-10a promoter and lacking the se requires wounding or elicitor treatment for its expression (Després et al., 1995). This is consistent with the observation that the expression of PR-10a in the SEBF RNAi lines (Figure 4F), where the repressosome is not recruited to the promoter, still requires induction by wounding or elicitor treatment. These observations are also in agreement with current models of gene regulation, where derepression (removal of repressing marks and/or proteins) and activation are viewed as distinct phenomena. In other words, the absence of a repressor at a promoter does not equate to gene activation. Activation requires the recruitment of activators (Roeder, 2005). Since the SEBF-Pti4 repressosome is not recruited to PR-10a after wounding or elicitor treatment, the fact that transcript accumulation is higher under these conditions in the RNAi lines, compared with wild-type plants, cannot be well explained by our simple model. However, gene regulation involves more than just transcription factor recruitment, and an important aspect of controlling genes resides in remodeling chromatin architecture (Roeder, 2005). We thus propose that a complete absence of SEBF from the promoter, whether through a knockdown of SEBF, as in our RNAi lines, or by deleting the SE element from a reporter gene (Després et al., 1995), might lead to a more open and permissive chromatin architecture, allowing easier access of the activator Why1 to the PR-10a promoter after wounding or elicitor treatment.
The observation that PR-10a was activated in the uninduced Pti4 VIGS plants is surprising, as one would have expected results similar to those obtained with the SEBF knockdown lines, since in both cases the repressosome is not recruited to PR-10a. A possible explanation is that the Pti4 VIGS plants are rendered more responsive than the control plants due to priming by the virus used to propagate the silencing construct. This priming might involve the activation of Why1, which could be recruited by PR-10a, resulting in activation of the gene. In PVX-00 plants, however, the presence of the SEBF-Pti4 repressosome at the PR-10a promoter may be sufficient to occlude the active Why1 from gaining access to the ERE. Another explanation could be that knocking down Pti4 affects not only the repressosome but also activation pathways, such as those controlling the activation of Why1. These are interesting questions that deserve further investigation.
The data presented here provide a significant advancement of our mechanistic understanding of gene regulation at the defense-associated gene PR-10a and illustrate how precarious and misleading our attempts at categorizing transcription factors as activators or repressors can be. The data presented here elevate Pti4 to the status of plant paradigm of transcription factor duality, capable of both activating and repressing transcription in a context-dependent fashion.
METHODS
Plant Material and Chemicals
Potato (Solanum tuberosum cv Kennebec) plants were grown on soil in a growth chamber at 60% humidity and under a long-day photoperiod consisting of a 16-h-light regimen with a photosynthetic photon flux density of 150 μmol photons·m−2·s−1 at 23°C followed by an 8-h-dark period at 18°C. All chemicals were from Sigma-Aldrich, unless stated otherwise.
Yeast Two-Hybrid Screening and Interaction Domain Mapping
The yeast two-hybrid transformation and screening as well as quantitative β-galactosidase activity assays were done according to the protocol of the DupLEX-A yeast two-hybrid system (OriGene Technologies). The tomato (Solanum lycopersicum) cDNA library used as prey was constructed in the laboratory of Barbara Baker and is described elsewhere (Zhang et al., 1999). The cDNA sequence encoding the mature potato SEBF protein was cloned in the BamHI/XhoI sites of the bait vector pEG202 to produce a C-terminal protein fusion with the Lex-A DB domain. Potato Pti4 was PCR-amplified from a potato cDNA library (Matton and Brisson, 1989) using the oligonucleotides 5′-AAAGCCATATGGATCAACAGTTACCACCGA-3′ and 5′-TTCGGCTCGAGAATGACCAATAGTTGATGGA-3′, which were based on the tomato Pti4 cDNA sequence. Potato Pti4 was subcloned in the NdeI and XhoI sites of plasmid pET-21a (Novagen).
For mapping the St SEBF–St Pti4 interacting domains, fusions between the Lex-A DB domain of pEG202 and either the full-length SEBF or its truncated versions were created by PCR amplification (see Supplemental Table 1 online for the sequences of the oligonucleotides used). Fusions of HA-tagged St Pti4, or deleted variants of St Pti4, with the transactivation domain B42 of pJG4-5 were also generated by PCR (see Supplemental Table 1 online for the sequences of the oligonucleotides used).
Immunoblot
The expression of fusion proteins produced in yeast was confirmed by immunoblot analysis using an anti-LexA antibody (Invitrogen) for SEBF fusions to the LexA DB and an anti-HA antibody (Santa Cruz Biotechnology) for the Pti4 constructs. Analysis of SEBF from wild-type and SEBF RNAi potato plants was performed using an anti-SEBF antibody (Boyle and Brisson, 2001).
The expression of SEBF and Whirly proteins in leaves of potato virus X (PVX)-infected plants and in treated and untreated potato tubers was determined as described (Constabel and Brisson, 1992) but using the anti-SEBF antibody at a 1:5000 dilution and the anti-St Why1 antibody at a 1:4000 dilution (Desveaux et al., 2000).
Pull-Down Assays
Mature St SEBF cDNA was cloned in pET21 (Novagen) and expressed as a C-terminal His tag fusion. St SEBF-His protein was immobilized and purified on a nickel-nitrilotriacetic acid agarose column (Qiagen) following the manufacturer's instructions. Pti4 was cloned into the yeast expression vector pJG4-5 and expressed as a B42-HA tag fusion. Yeast protein extracts expressing Pti4-B42-HA or B42-HA alone, which served as a negative control, were loaded onto an empty nickel-nitrilotriacetic acid agarose column (another negative control) or to one bound by SEBF-His. The columns were washed three times in 50 mM NaH2PO4, 300 mM NaCl, and 20 mM imidazole, pH 8.0, before eluting in the same buffer containing 500 mM imidazole. Proteins were then separated on a 12% SDS-polyacrylamide gel and transferred to a membrane for immunoblot analysis. Anti-HA (Santa Cruz Biotechnology) or anti-SEBF (Boyle and Brisson, 2001) antibodies were used to detect Pti4-HA or SEBF-His fusion proteins, respectively.
EMSA
EMSAs were performed with recombinant full-length and truncated SEBF proteins, as described previously by Boyle and Brisson (2001), using as a probe the oligonucleotide SE (5′-TCTAGACTGTCACTTGTTTTT-3′).
Stable Transformation of Potato with a Hairpin-Driven RNAi Construct
A 700-bp fragment from the cDNA of potato SEBF was amplified by PCR using the primers SEBF-BamHI (5′-TTTGTTCGGATCCTAACGCTTTC-3′) and SEBF-KpnI (5′-GTTGGGTACCATCTTCAGAATTG-3′) to generate the sense construct and primers SEBF-ScaI (5′-GGCTAAGTACTTCAGAATTGACGTC-3′) and SEBF-SacI (5′-GTTTTGAGCTCAAAGTAACCCTTTC-3′) for the antisense construct. The sense and antisense PCR products were subcloned in the pDarth vector (O'Brien et al., 2002) using restriction sites BamHI/KpnI and ScaI/SacI, respectively. Transformation in Agrobacterium tumefaciens and in potato plants was as described (Després et al., 1995).
ChIP
Two grams of tissue per experiment was treated and processed for ChIP analysis as described previously (Chakravarthy et al., 2003; Desveaux et al., 2004). The antibodies used for the ChIPs were anti-St Why1 (Desveaux et al., 2000), anti-SEBF (Boyle and Brisson, 2001), and anti-Pti4 (Chakravarthy et al., 2003). The sequences of the oligonucleotides used to amplify the PR-10a promoter were 5′-AAGAAGGCACATTTCAAGAAC-3′ and 5′-ACCTATAAATACCATCGAACA-3′.
Biological replicates of ChIP were performed from each genotype/treatment sample, and three qPCR experiments were done with each sample. The qPCRs were performed with 40 cycles of a two-temperature protocol in a total volume of 20 μL using the Bio-Rad iQ SYBR Green Supermix kit according to the manufacturer's instructions. To amplify the PR-10a promoter with the oligonucleotides described above, an annealing temperature of 55°C was used. The sequences of the oligonucleotides used to amplify the Actin promoter were 5′-ACTATTATTCAATTTATCTGCGGCC-3′ and 5′-AAAAATGGCAGGCCAACTCT -3′, and an annealing temperature of 64°C was used. For each immunoprecipitation (IP), binding of a transcription factor (SEBF, Why1, or Pti4) to the PR-10a promoter relative to its binding at the Actin promoter was determined with the following formula: amount PR-10a promoter (IP)/amount Actin promoter (IP). The amount of target DNA was defined as 2−Ct, where Ct is the threshold cycle.
In Vivo Plant Transcription Assays
The construction of the reporter gene and the internal standard vectors as well as the methodology for the in vivo plant transcription assays were described previously (Després et al., 2003). N-terminal protein fusions of Pti4 and SEBF with the Gal4 DB and VP16 TA and the unfused versions were created by PCR amplification and cloned into pBI524 as described previously (Després et al., 2003).
VIGS in Potato
The protocol for VIGS in potato (cv Kennebec) was based on that previously described by Faivre-Rampant and collaborators (2004) with some modifications. The PVX vector pGR106 (Lu et al., 2003) was obtained from David Baulcombe (Sainsbury Laboratory). To construct the PVX-Pti4, a 335-bp PCR fragment was amplified from potato cDNA using the oligonucleotides StPti4NotI-F (5′-AGCGGCCGCGAAACACCGAAGGGAAGACA-3′) and StPti4AscI-R (5′-AGGCGCGCCCTCCACTCCTCCGTCACATT-3′) and subcloned into the NotI/AscI restriction sites of pGR106. Each of the PVX constructs (PVX-Pti4 and the empty vector PVX-00) was cotransformed with the helper plasmid pSoup (Hellens et al., 2000) into Agrobacterium strain LB4404 by electroporation.
Potato plants were propagated in vitro as described (Faivre-Rampant et al., 2004), but with some modifications. Five stem pieces per Magenta box were cultivated in 100 mL of MS medium (Murashige and Skoog, 1962) supplemented with 0.4 mg/mL thiamine, 0.5 mg/mL pyridoxine, 0.5 mg/mL nicotinic acid, 100 mg/mL myo-inositol, 2 mg/mL Gly, 30 g/L sucrose, 1 mg/L indolebutyric acid, 0.2 mg/L kinetin, 12 mM AgNO3, and 96 mM Na2S2O3.
Ten-day- to 2-week-old in vitro plants were agroinoculated with the PVX vectors. The different Agrobacterium strains were grown for 2 d at 28°C with shaking and then incubated in an induction buffer (10 mM MES, 10 mM MgCl2, and 200 μM acetosyringone) for at least 3 h at room temperature. After induction, the bacterial suspension was pelleted and used to inoculate the surface of leaves, which were previously wounded with a razor blade to facilitate bacterial penetration. Two weeks later, the plants were transferred to soil in controlled-environment chambers with a 16-h photoperiod. Five to 8 weeks later, the plants were analyzed by real-time PCR for St Pti4 levels as indicated below. Plants demonstrating no or highly reduced Pti4 transcript levels were chosen for ChIP experiments and PR-10a expression analysis. Four biological replicates were used for qPCR experiments.
RNA Extractions and Real-Time qPCR
RNA from four biological replicates of potato leaves was extracted using the Tri reagent method (Sigma-Aldrich) following the manufacturer's instructions specific for high-polysaccharide-containing samples. First-strand cDNA was synthesized from 2 μg of total RNA using SuperScript II reverse transcriptase (Invitrogen) and a poly(T) oligonucleotide, following the manufacturer's instructions. For each biological replicate, a pool of five leaves was used.
RNA from three biological replicates of potato tubers was extracted as described previously (Boyle and Brisson, 2001). For each biological replicate, a pool of three tuber discs was used. qPCR was performed using the SYBR Green method. SYBR Green PCRs were performed using 2 μL of cDNA samples (50 ng), 5 μL of the Fast SYBR Green Master Mix (Applied Biosystem), and 10 pmol of each primer in a total volume of 10 μL. Melting curves were determined using the dissociation curve software SDS 2.2.2 to ensure that only a single product was amplified. For quantification of Pti4 transcript levels in VIGS plants, a forward primer (St-Pti4qPCR-F1, 5′-TCACCGCCGGCGAAGTAAA-3′) located outside of the sequence targeted for silencing and a reverse primer (StPti4qPCR-R1, 5′-CGTTAGACAGCGGCCGTGG-3′) located inside the sequence targeted for silencing were used. PR10-a quantification was performed using the primers PR10a-F (5′-TGACAATCTTATTCCTAAGTTGATGC-3′) and PR10a-R (5′-AGGTCATCTTCTTGATGCTTCC-3′). As an endogenous control, the primers Ubiq-F (5′-CTCCGTGGTGGTATGCAGAT-3′) and Ubiq-R (5′-CACGTTGTCAATGGTGTCG-3′) were designed for quantification of the ubiquitin gene (accession number BQ045862). The ABI PRISM 7900HT sequence detection system (Applied Biosystems) was used to detect the amplification level and was programmed with an initial step of 10 min at 95°C followed by 45 cycles alternating between 15 s at 95°C and 1 min at 60°C. All reactions were run in technical triplicate for each biological replicate, and the average values were used for quantification. The relative quantification of target genes was determined using the ΔΔCT method. Briefly, the Ct (threshold cycle) values of target genes were normalized to an endogenous control gene (ubiquitin) (ΔCT = Cttarget – Ctubiquitin) and compared with a calibrator (ΔΔCT = ΔCtsample − ΔCtcalibrator). Relative expression (RQ) was calculated using the sequence detecion system SDS 2.2.2 software (Applied Biosystems) and the formula RQ = 2−ΔΔCT.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AF389431 (St SEBF), U89255 (Sl Pti4), EU851735 (St Pti4), and X55751 (actin gene PoAc97).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Protein Sequence Alignment between Tomato and Potato Pti4.
Supplemental Figure 2. Data from the ChIP Experiments of Figure 4B.
Supplemental Figure 3. Data from the ChIP Experiments of Figure 6B.
Supplemental Table 1. Sequences of the Primers Used to Construct the Protein Fusions Analyzed in Yeast Two-Hybrid Assays.
Supplementary Material
Acknowledgments
We thank Barbara Baker for the tomato cDNA library and David Baulcombe and the Sainsbury Laboratory for the pGR106 vector. We also thank Louise Cournoyer for generating the SEBF-RNAi potato transgenic plants, Myriam Beauchemin for help with the immunoblots, and Jee Yan Chu for technical and editorial assistance. P.B., V.R., and A.D. were supported by scholarships from the Natural Science and Engineering Research Council (NSERC) of Canada. Research in the K.B., C.D., and N.B. laboratories was supported by the NSERC Discovery Grant Program.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Normand Brisson (normand.brisson@umontreal.ca).
Online version contains Web-only data.
References
- Boyle, B., and Brisson, N. (2001). Repression of the defense gene PR-10a by the single-stranded DNA binding protein SEBF. Plant Cell 13 2525–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, R.L., Kazan, K., McGrath, K.C., Maclean, D.J., and Manners, J.M. (2003). A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol. 132 1020–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy, S., Tuori, R.P., D'Ascenzo, M.D., Fobert, P.R., Després, C., and Martin, G.B. (2003). The tomato transcription factor Pti4 regulates defense-related gene expression via GCC box and non-GCC box cis elements. Plant Cell 15 3033–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W., et al. (2002). Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14 559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constabel, C.P., and Brisson, N. (1992). The defense-related STH-2 gene product of potato shows race-specific accumulation after inoculation with low concentrations of Phytophthora infestans zoospores. Planta 188 289–295. [DOI] [PubMed] [Google Scholar]
- Da Silva, N., Bharti, A., and Shelley, C.S. (2002). hnRNP-K and Pur(alpha) act together to repress the transcriptional activity of the CD43 gene promoter. Blood 100 3536–3544. [DOI] [PubMed] [Google Scholar]
- Després, C., Chubak, C., Rochon, A., Clark, R., Bethune, T., Desveaux, D., and Fobert, P.R. (2003). The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15 2181–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després, C., Subramaniam, R., Matton, D.P., and Brisson, N. (1995). The activation of the potato PR-10a gene requires the phosphorylation of the nuclear factor PBF-1. Plant Cell 7 589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desveaux, D., Després, C., Joyeux, A., Subramaniam, R., and Brisson, N. (2000). PBF-2 is a novel single-stranded DNA binding factor implicated in PR-10a gene activation in potato. Plant Cell 12 1477–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desveaux, D., Subramaniam, R., Després, C., Mess, J.N., Lévesque, C., Fobert, P.R., Dangl, J.L., and Brisson, N. (2004). A “Whirly” transcription factor is required for salicylic acid-dependent disease resistance in Arabidopsis. Dev. Cell 6 229–240. [DOI] [PubMed] [Google Scholar]
- Eulgem, T. (2005). Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci. 10 71–78. [DOI] [PubMed] [Google Scholar]
- Faivre-Rampant, O., Gilroy, E.M., Hrubikova, K., Hein, I., Loake, G.J., Birch, P., Taylor, M., and Lacomme, C. (2004). Potato virus X-induced gene silencing in leaves and tubers of potato. Plant Physiol. 134 1308–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fobert, P. (2006). Transcription factors regulating plant defense responses. In Model Plants and Crop Improvement. R. Koebner and R. Varshney, eds (Boca Raton, FL: CRC Press), pp. 159–205.
- Gu, Y.Q., Wildermuth, M.C., Chakravarthy, S., Loh, Y.T., Yang, C., He, X., Han, Y., and Martin, G.B. (2002). Tomato transcription factors Pti4, Pti5 and Pti6 activate defense responses when expressed in Arabidopsis. Plant Cell 14 817–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Y.Q., Yang, C., Thara, V.K., Zhou, J., and Martin, G.B. (2000). Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell 12 771–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens, R.P., Edwards, E.A., Leyland, N.R., Bean, S., and Mullineaux, P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42 819–832. [DOI] [PubMed] [Google Scholar]
- Hsieh, T.Y., Matsumoto, M., Chou, H.C., Schneider, R., Hwang, S.B., Lee, A.S., and Lai, M.M. (1998). Hepatitis C virus core protein interacts with heterogeneous nuclear ribonucleoprotein K. J. Biol. Chem. 273 17651–17659. [DOI] [PubMed] [Google Scholar]
- Jones, J.D., and Dangl, J.L. (2006). The plant immune system. Nature 444 323–329. [DOI] [PubMed] [Google Scholar]
- Lorenzo, O., Piqueras, R., Sánchez-Serrano, J.J., and Solano, R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R., Malcuit, I., Moffett, P., Ruiz, M.T., Wu, A.J., Rathjen, J.P., Bendahmane, A., Day, L., and Baulcombe, D.C. (2003). High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 22 5690–5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan, M.C., Narlikar, G.J., Boyapaty, G., Kingston, R.E., and Weissman, S.M. (2005). Heterogeneous nuclear ribonucleoprotein C1/C2, MeCP1, and SWI/SNF form a chromatin remodelling complex at the beta-globin locus control region. Proc. Natl. Acad. Sci. USA 102 15012–15017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matton, D.P., and Brisson, N. (1989). Cloning, expression, and sequence conservation of pathogenesis-related gene transcripts of potato. Mol. Plant Microbe Interact. 6 325–331. [DOI] [PubMed] [Google Scholar]
- Matton, D.P., Prescott, G., Bertrand, C., Camirand, A., and Brisson, N. (1993). Identification of cis-acting elements involved in the regulation of the pathogenesis-related gene STH-2 in potato. Plant Mol. Biol. 22 279–291. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Plant Physiol. 15 473–497. [Google Scholar]
- Mysore, K.S., Crasta, O.R., Tuori, R.P., Folkers, O., Swirsky, P.B., and Martin, G.B. (2002). Comprehensive transcript profiling of Pto- and Prf-mediated host defense responses to infection by Pseudomonas syringae pv. tomato. Plant J. 32 299–315. [DOI] [PubMed] [Google Scholar]
- O'Brien, M., Kapfer, C., Major, G., Laurin, M., Bertrand, C., Kondo, K., Kowyama, Y., and Matton, D.P. (2002). Molecular analysis of the stylar-expressed Solanum chacoense small asparagine-rich protein family related to the HT modifier of gametophytic self-incompatibility in Nicotiana. Plant J. 32 985–996. [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi, M., and Shinshi, H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi, M., Suzuki, K., and Shinshi, H. (2000). Regulation of ethylene-induced transcription of defense genes. Plant Cell Physiol. 41 1187–1192. [DOI] [PubMed] [Google Scholar]
- Oñate-Sánchez, L., and Singh, K.B. (2002). Identification of Arabidopsis ethylene-responsive element binding factors with distinct induction kinetics after pathogen infection. Plant Physiol. 128 1313–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski, J., Kawata, Y., Schullery, D.S., Denisenko, O.N., and Bomsztyk, K. (2003). Transient recruitment of the hnRNP K protein to inducibly transcribed gene loci. Nucleic Acids Res. 31 3954–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski, J., Sims, J.E., Sibley, C.H., Valentine, M.A., Dower, S.K., Meier, K.E., and Bomsztyk, K. (1991). A serine/threonine kinase activity is closely associated with a 65-kDa phosphoprotein specifically recognized by the kappa B enhancer element. J. Biol. Chem. 266 12722–12733. [PubMed] [Google Scholar]
- Ostrowski, J., Wyrwicz, L., Rychlewski, L., and Bomsztyk, K. (2002). Heterogeneous nuclear ribonucleoprotein K protein associates with multiple mitochondrial transcripts within the organelle. J. Biol. Chem. 277 6303–6310. [DOI] [PubMed] [Google Scholar]
- Park, J.M., Park, C.J., Lee, S.B., Ham, B.K., Shin, R., and Paek, K.H. (2001). Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell 13 1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder, R.G. (2005). Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 579 909–915. [DOI] [PubMed] [Google Scholar]
- Stintzi, A., Heitz, T., Prasad, V., Wiedemann-Merdinoglu, S., Kauffmann, S., Geoffroy, P., Legrand, M., and Fritig, B. (1993). Plant “pathogenesis-related” proteins and their role in defense against pathogens. Biochimie 75 687–706. [DOI] [PubMed] [Google Scholar]
- Swanson, M.S., and Dreyfuss, G. (1988). Classification and purification of proteins of heterogenous nuclear ribonucleoprotein particles by RNA-binding specificities. Mol. Cell. Biol. 8 2237–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, K., Tian, L., Hollingworth, J., Brown, D.C., and Miki, B. (2002). Functional analysis of tomato Pti4 in Arabidopsis. Plant Physiol. 128 30–37. [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Fan, W., Kinkema, M., Li, X., and Dong, X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. USA 96 6523–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J., Tang, X., and Martin, G.B. (1997). The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis-element of pathogenesis-related genes. EMBO J. 16 3207–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.