Abstract
The plant signaling molecule salicylic acid (SA) and/or xenobiotic chemicals like the auxin mimic 2,4-D induce transcriptional activation of defense- and stress-related genes that contain activation sequence-1 (as-1)–like cis-elements in their promoters. as-1–like sequences are recognized by basic/leucine zipper transcription factors of the TGA family. Expression of genes related to the SA-dependent defense program systemic acquired resistance requires the TGA-interacting protein NPR1. However, a number of as-1–containing promoters can be activated independently from NPR1. Here, we report the identification of Arabidopsis thaliana SCARECROW-like 14 (SCL14), a member of the GRAS family of regulatory proteins, as a TGA-interacting protein that is required for the activation of TGA-dependent but NPR1-independent SA- and 2,4-D–inducible promoters. Chromatin immunoprecipitation experiments revealed that class II TGA factors TGA2, TGA5, and/or TGA6 are needed to recruit SCL14 to promoters of selected SCL14 target genes identified by whole-genome transcript profiling experiments. The coding regions and the expression profiles of the SCL14-dependent genes imply that they might be involved in the detoxification of xenobiotics and possibly endogenous harmful metabolites. Consistently, plants ectopically expressing SCL14 showed increased tolerance to toxic doses of the chemicals isonicotinic acid and 2,4,6-triiodobenzoic acid, whereas the scl14 and the tga2 tga5 tga6 mutants were more susceptible. Hence, the TGA/SCL14 complex seems to be involved in the activation of a general broad-spectrum detoxification network upon challenge of plants with xenobiotics.
INTRODUCTION
Regulatory sequences related to the activating sequence-1 (as-1) element are found in a number of stress-responsive plant promoters including the pathogen-inducible Arabidopsis Pathogenesis Related-1 (PR-1) gene promoter (Lebel et al., 1998). Induction of this promoter by pathogens requires the plant signaling molecule salicylic acid (SA), one of the redundant class II as-1 binding proteins TGA2, TGA5, or TGA6 (Zhang et al., 2003), and the TGA-interacting ankyrin repeat protein NONEXPRESSOR OF PR genes (NPR1) (Cao et al., 1997; Ryals et al., 1997). Other as-1–containing genes, such as glutathione S-transferase F8 (GSTF8) (Chen and Singh, 1999), are expressed in an NPR1-independent manner (Uquillas et al., 2004; Blanco et al., 2005). In contrast with PR-1, GSTF8 and most of the other members of the GST gene family are not only activated by SA, but also by the auxin analog 2,4-D (Wagner et al., 2002). Likewise, synthetic promoters or promoter deletions that contain either one or multiple as-1 elements as the only regulatory upstream sequences are 2,4-D and SA inducible (Zhang and Singh, 1994; Redman et al., 2002) and do not require NPR1 (Butterbrodt et al., 2006). As relatively high doses of the two chemicals (1 mM SA and 50 μM 2,4-D) are needed for the induction, it was speculated that the response was not mediated through auxin- and/or SA-specific signal transduction pathways but rather through a more general stress response pathway (Zhang and Singh, 1994). In tobacco (Nicotiana tabacum), class II TGA factor TGA2.2 regulates NPR1- and NPR1-independent promoters, indicating that the same TGA factor can function within the two different signaling networks (Thurow et al., 2005; Butterbrodt et al., 2006). Compared with the NPR1-dependent pathway, the NPR1-independent activation of as-1–containing promoters is far less explored.
To close this gap, a yeast protein interaction screen offering Arabidopsis thaliana class II transcription factor TGA2 as bait was performed. This report describes the isolation of a TGA-interacting factor belonging to the Arabidopsis GRAS family of proteins. The acronym GRAS was coined after identification of the founding members GIBBERELLIN ACID INSENSITIVE (GAI), REPRESSOR of GA1 (RGA), and SCARECROW (SCR) (Pysh et al., 1999). GAI and RGA play important roles in gibberellic acid–dependent signal transduction processes (Silverstone et al., 1998), whereas SCR was isolated in a screen for mutations that affect root development. GAI, RGA, SCR, and SCARECLOW-like (SCL) proteins contain several conserved amino acid signatures in the so-called GRAS domain at the C terminus. By contrast, the amino acid sequence of the N-terminal domain is more variable (Di Laurenzio et al., 1996). Though direct DNA binding to a specific sequence has not been reported, GRAS proteins are classified as transcriptional regulators (Riechmann et al., 2000; Zentella et al., 2007). Our results indicate that SCL14 is recruited to target promoters through its interaction with as-1–bound TGA factors and that this recruitment is essential for the transcriptional activation of the corresponding genes.
RESULTS
Identification of TGA-Interacting Protein SCL14 by a Yeast Protein–Protein Interaction Screen
The yeast two-hybrid screen has often been applied to isolate protein interaction partners (Fields and Song, 1989). However, fusion of the bait protein with a heterologous DNA binding domain or fusion of the prey protein with an activation domain might hamper certain interactions. In the screen reported here, this potential drawback was circumvented by positioning three copies of the as-1 element (3x as-1) upstream of the HIS3 selectable marker gene (Weigel et al., 2005) and expressing TGA2 in trans. As as-1–bound TGA2 only weakly activates transcription in yeast, the resulting strain does not grow on selective medium when 15 mM 3-amino-1,2,4-triazole is added. A cDNA expression library (Minet et al., 1992) was transformed into this strain and screening for prototrophic growth was performed.
One of the five clones identified after selecting 106 yeast transformants was identical to the GRAS protein SCL14 (At1g07530, AtGRAS-2) (Bolle, 2004; Tian et al., 2004). Recovered prey plasmids were retransformed into yeast cells that contained a 3x as-1:β-galactosidase (lacZ) construct. Whereas SCL14 or TGA2 alone led to twofold elevated levels of LacZ activity, a further sevenfold activation was observed upon coexpression of both proteins (Figure 1A). This result implies that as-1–bound TGA2 recruits SCL14 to the DNA, resulting in a complex that activates transcription.
Figure 1.
Quantitative Analysis of the TGA2-SCL14 Interaction in Yeast.
(A) Yeast protein–protein–DNA interaction system. The host strain carries three copies of the as-1 element upstream of the lacZ reporter gene. SCL14 and TGA2 coding regions were expressed under the control of the MET25- and the ADH1 promoter, respectively, either alone or in combination as indicated. Interaction is measured as induction of β-galactosidase activity.
(B) Yeast two-hybrid system. β-Galactosidase activities of the yeast strain PJ69-4A, which contains the lacZ reporter gene under the control of the GAL7 promoter. Prey plasmids encode TGA2 fused to the GBD; bait plasmids encode SCL14 or NPR1 fused to the GAD.
(C) Yeast protein–protein–DNA interaction system using the same host strain as in (A). TGA2 was expressed under the control of the MET25 promoter. Fusion proteins with the GAD served as prey.
(D) Yeast two-hybrid system as in (B). Bait plasmids encode either full-length SCL14 (amino acids 1 to 769) or the SCL14 lacking the C-terminal GRAS domain (amino acids 1 to 381) fused to the GAD.
β-Galactosidase activities are indicated as Miller units. The mean value (±sd) from four independent yeast transformants is shown. The hyphens indicate the respective empty vectors.
The interaction between TGA2 and SCL14 was confirmed in the classical yeast two-hybrid system, which detects the interaction of translational fusions with the Gal4 DNA binding domain (GBD) and the Gal4 activation domain (GAD), respectively. In this assay, β-galactosidase activity yielded by the GBD-TGA2/GAD-SCL14 interaction was approximately sevenfold lower than the one obtained from the well-established interaction between GBD-TGA2 and GAD-NPR1 (Figure 1B). By contrast, the GAD-SCL14/TGA2/as-1 complex led to a higher transcriptional activation than the GAD-NPR1/TGA2/as-1 complex (Figure 1C), indicating that the presence of TGA2 influences the interaction with the two different preys in a differential manner. Complex formation between GBD-TGA2 and SCL14 was still possible, even when only the N-terminal domain (amino acids 1 to 381) of SCL14 was presented as bait (Figure 1D).
To obtain independent evidence for the formation of a TGA2/SCL14 complex, an in vitro pull-down assay with recombinant proteins was performed (Figure 2). Crude Escherichia coli extracts containing either SCL14 fused to glutathione S-transferase (GST-SCL14) or His-tagged TGA2 (His6-TGA2) were combined and loaded onto glutathione-sepharose affinity beads. After washing and subsequent elution under denaturing conditions, eluates were analyzed for the presence of His6-TGA2. Though GST-SCL14 was subject to protein degradation in E. coli and during subsequent processing steps, sufficient amounts of the protein were bound to the glutathione matrix to retain His6-TGA2. By contrast, equivalent amounts of GST protein were unable to interact with His6-TGA2.
Figure 2.
Pull-Down Assay to Demonstrate the in Vitro Interaction between TGA2 and SCL14.
GST, GST-SCL14, or His6-TGA2 were expressed in E. coli under the control of an isopropyl-1-thio-β-d-galactopyranoside (IPTG)–inducible promoter and mixed with extracts from noninduced (non-ind.) cells to obtain input samples containing approximately equal amounts of recombinant proteins as documented by Coomassie blue staining of an SDS gel (lanes 1 to 4). Protein gel blot analysis to test for the presence of His6-TGA2 was performed with the Universal HIS detection reagent (lanes 5 to 8 and 11 and 12). Extracts containing either GST or GST-SCL14 were incubated with extracts containing His6-TGA2 and loaded onto glutathione-sepharose beads. The protein composition of the eluates was analyzed by Coomassie blue staining (lanes 9 and 10) and protein gel blot analysis (lanes 11 and 12) after separation of the proteins by SDS gel electrophoresis.
Intracellular Localization of SCL14-GFP Fusion Proteins
The subcellular localization of SCL14 was assessed by microscopy analysis of a SCL14-green fluorescent protein (GFP) fusion protein that was transiently expressed in tobacco BY-2 protoplasts. As documented in Figure 3, SCL14-GFP is localized both in the nucleus and in the cytosol. To assess whether the partial cytosolic localization of SCL14-GFP in the cell is due to incomplete nuclear import or to active export, protoplasts were treated with Leptomycin B (LMB), which blocks the function of the exportin receptor XPO1 (Kudo et al., 1999). This treatment led to the accumulation of the majority of SCL14-GFP in the nucleus, suggesting that the partial cytosolic localization of SCL14 in the absence of LMB is most likely due to XPO1-dependent nuclear export.
Figure 3.
Localization of the SCL14-GFP Fusion Protein in Protoplasts of Tobacco BY-2 Cells.
Protoplasts were transfected with plasmids encoding GFP or SCL14-GFP, incubated in the presence (2 μM LMB) or absence (mock) of the nuclear export inhibitor Leptomycin B and analyzed by fluorescence microscopy. Bright-field (right) and fluorescence images (left) are shown. Bars = 25 μm. [See online article for color version of this figure.]
Expression Analysis of Known Promoters Containing as-1–Like Elements as a Function of SCL14 Protein Levels
To analyze the in vivo effect of SCL14 on as-1–mediated gene expression, the SCL14 cDNA was cloned into a binary vector designed to express HA3-tagged proteins under the control of the Cauliflower Mosaic Virus (CaMV) 35S promoter. The construct was stably introduced into Arabidopsis plants that already contained a transgene encoding the β-glucuronidase gene (GUS) downstream of the truncated CaMV 35S promoter (as-1:GUS) (Redman et al., 2002). Transcription from this promoter fragment (+1 to −90), which contains as-1 as the only regulatory sequence, can be induced by SA and 2,4-D. Two plants (transgenic lines #5 and #6), which showed ectopic expression of HA3-SCL14 (see protein gel blot of crude extracts probed with the αSCL14 antiserum in Supplemental Figure 1A online and Figure 4A) were selected for further analysis. As shown for transgenic line #5, the overall amount of SCL14 exceeded wild-type SCL14 levels only ∼1.4-fold (see Supplemental Figure 1B online). Expression of the as-1:GUS reporter construct was enhanced under noninducing and inducing conditions, indicating that SCL14 can be a positive regulator of as-1–mediated transcription (Figures 4C and 4D). Similar results were obtained with the second HA3-SCL14 line #6 (see Supplemental Figure 2 online).
Figure 4.
Expression Analysis of as-1:GUS Plants with Different SCL14 Protein Levels.
(A) Protein gel blot analysis of as-1:GUS (WT) plants, HA3-SCL14 transgenic plant line #5 (HA3-SCL14), and scl14 mutant plants using the αSCL14 antiserum. The star marks an unspecific protein band that serves as a loading control.
(B) Position of the T-DNA insertion within the gene At1g07530 in line SALK_126931. White boxes indicate untranslated regions, and the thick black line marks the intron within the 5′ untranslated region. Right and left borders of the T-DNA are abbreviated as RB and LB, respectively.
(C) and (D) RNA gel blot analysis of GUS transcript levels in as-1:GUS plants (WT) and as-1:GUS plants expressing HA3-SCL14 (transgenic line #5) treated with either SA (C) or 2,4-D (D).
(E) RNA gel blot analysis of SCL14, GSTF8, and PR-1 transcript levels in SA-treated as-1:GUS (WT) and scl14 plants.
(F) RNA gel blot analysis of SCL14 and GSTF8 transcript levels in 2,4-D–treated as-1:GUS (WT) and scl14 plants.
Three-week-old plants were treated with 1 mM SA or 0.1 mM 2,4-D for the time spans indicated above the lanes ([C] to [F]). Ethidium bromide–stained rRNA is shown as evidence of equal loading. All plants lines, including the scl14 mutant, carry the as-1:GUS transgene; WT refers to an intact SCL14 allele. The hyphens indicate untreated plants.
Next, the Arabidopsis mutant SALK_126931, which carries a T-DNA insertion in the 5′-untranslated region of the SCL14 gene (Figure 4B), was analyzed. Homozygous mutant plants showed no detectable SCL14 mRNA and SCL14 protein levels as revealed by RNA and protein gel blot analysis, respectively (Figures 4A, 4E, and 4F). Transcript levels of the NPR1-dependent PR-1 gene and the NPR1-independent GSTF8 gene showed no difference to those found in wild-type plants after SA or 2,4-D treatment (Figures 4E and 4F), implicating that they are not subject to SCL14-dependent regulation.
Identification of Endogenous Genes Activated by SCL14
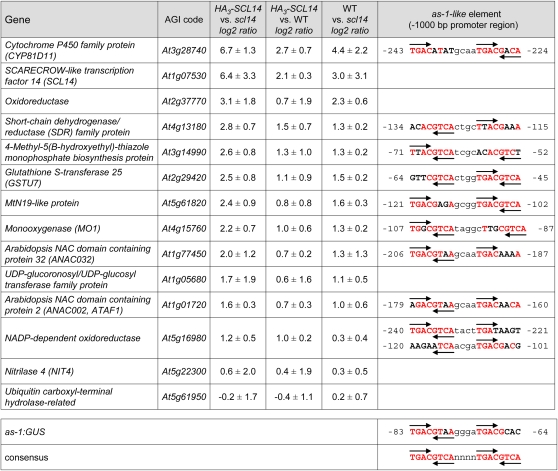
To identify endogenous candidate target genes of SCL14, the transcriptomes of wild-type, scl14 mutant, and the HA3-SCL14 transgenic plant line #5 were compared by whole-genome microarray analysis. One hundred plantlets of each genotype were grown aseptically for 25 d on membrane rafts floated on Murashige and Skoog (MS) medium. Plant material, including the roots, was harvested for RNA extraction. Hybridizations on oligonucleotide microarrays fabricated by the University of Arizona (http://www.ag.arizona.edu/microarray/) were performed in each of the three possible combinations. Fourteen genes, including SCL14, showed a more than fivefold (log2) upregulation in HA3-SCL14 transgenic plant line #5 compared with the scl14 mutant. All of them showed a more than threefold (log2) difference when comparing the expression in wild-type plants versus expression in the scl14 mutant (see Supplemental Table 1 online). Real-time RT-PCR analysis performed with RNA from aerial parts of plants grown on MS plates for 3 weeks qualitatively reproduced the microarray data for 12 of the 14 genes (Figure 5). As we hypothesized that SCL14 is recruited to the promoters of direct target genes by DNA-bound TGA factors (see above), we searched the promoter sequences for putative as-1–like elements by visual inspection. The ideal as-1–like element contains two copies of the TGAC/GTCA palindrome (Qin et al., 1994), but two imperfect palindromes with only low homology to the consensus can serve as a functional as-1–like element if the spacing between the palindromic centers is 12 bp (Krawczyk et al., 2002). Nine out of the 11 genes differentially regulated by SCL14 contain putative as-1–like elements within 250 bp upstream of the transcriptional start site (Figure 5). Interestingly, the two genes for which the original microarray data had not been confirmed (At5g22300 and At5g61950) do not contain an as-1–like element in their promoter regions.
Figure 5.
Quantitative Real-Time RT-PCR Analysis of Candidate Genes Identified by Microarray Analysis of Plants Containing Different SCL14 Protein Levels.
RNA from as-1:GUS plants (WT), as-1:GUS plants transformed with a CaMV 35S:HA3-SCL14 construct (HA3-SCL14), and scl14 mutant plants was subjected to quantitative real-time RT-PCR analysis. Plants were grown for 3 weeks on MS plates. Values denote ratios of transcription levels between the two indicated genotypes. The mean values (±sd) of three independent experiments are shown. Genes were arranged according to the expression ratios between HA3-SCL14 and scl14 plants. See Supplemental Table 1 online for expression data obtained from microarray analysis. Sequences of as-1-like elements found in the respective promoter regions are shown in the rightmost column. The numbers indicate their positions relative to the transcriptional start sites (+1). Conserved nucleotides within the two 8-bp palindromes (capital letters) are highlighted in red. Half sites of the palindromes that contain at least three of the four conserved TGAC nucleotides are marked by arrows. The sequence and the position of the as-1 element within the CaMV 35S promoter are shown.
Recruitment of SCL14 and TGA2 to Target Promoters
To demonstrate recruitment of TGA factors and SCL14 to promoters with as-1–like sequences (Figure 5), immunoprecipitations of three selected promoter fragments (corresponding to Arabidopsis genes CYP81D11, MtN19-like, and GSTU7) were performed on chromatin prepared from wild-type, scl14, and tga2 tga5 tga6 plants. For presentation of the data, the recovery of promoter fragments immunoprecipitated from wild-type chromatin was set to 100%, and the percentage of promoter fragments obtained from mutant chromatins was displayed accordingly. To adjust for the amount and quality of the input chromatin from different genotypes, all values were normalized to the amount of amplified ACTIN8 fragments (original CT values of the experiments are listed in Supplemental Table 2 online). As shown in Figure 6A, the promoter fragments of the genes CYP81D11, MtN19-like, GSTU7, and PR-1 are precipitated less efficiently from tga2 tga5 tga6 chromatin than from wild-type and scl14 chromatin when using the αTGA2,5 antiserum. By contrast, two reference fragments that do not contain an as-1-like motif were immunoprecipitated independently of the genotype of the plant. These results demonstrate that TGA factors bind specifically to promoters of the genes CYP81D11, MtN19-like, GSTU7, and PR-1 even in the uninduced state and independently from the presence of SCL14. When deploying the αSCL14 antiserum, chromatin of the tga2 tga5 tga6 and scl14 mutant plants both yielded significantly less CYP81D11, MtN19-like, and GSTU7 promoter fragments than chromatin from wild-type plants (Figure 6A). In combination with our protein gel blot analysis (Figure 6B), which confirmed that the SCL14 protein is present in the tga2 tga5 tga6 mutant, this result supports our hypothesis that TGA factors are required to recruit SCL14 to the target promoters analyzed here. In addition, this analysis documents that SCL14 is not recruited to the TGA factors bound to the PR-1 promoter, which is consistent with its SCL14-independent expression (Figure 4E).
Figure 6.
In Vivo SCL14 and TGA Factor Binding to the Promoters of CYP81D11, MtN19-like, and GSTU7 as Revealed by Chromatin Immunoprecipitation Analysis.
(A) Leaves from as-1:GUS (WT) plants and scl14 and tga2 tga5 tga6 (tga2,5,6) mutants were incubated in 1% formaldehyde before chromatin preparation. Chromatin samples were subjected to immunoprecipitation using 5 μL of cleared αTGA2,5 antiserum (top panel) or crude αSCL14 antiserum (bottom panel). The DNA was recovered after reversal of the cross-links and analyzed for the enrichment of promoter sequences by quantitative real-time PCR. Quantitation of relative amounts of PCR product with respect to wild-type chromatin (WT; set to 100%) with ACTIN8 sequences as a reference is shown. Values indicate means (±sd) of three independent chromatin immunoprecipitation experiments done with three independent chromatin preparations from three batches of independently grown plants. The data set obtained for the wild type and scl14 chromatin treated with the αSCL14 antiserum was derived from five independent chromatin preparations and five independent immunoprecipitation experiments. Unpaired Student's t tests were performed on the data to show significant differences compared with the reference gene GES (At1g61120), respectively (* P < 0.1, ** P < 0.05, and *** P < 0.01).
(B) Protein gel blot analysis of crude whole cell or chromatin probed either with αTGA2,5 antiserum (left panel) or crude αSCL14 antiserum (right panel), respectively. The genotype of the analyzed plants is indicated below the lanes. The star marks a nonspecific band.
Expression Pattern of Target Genes of SCL14
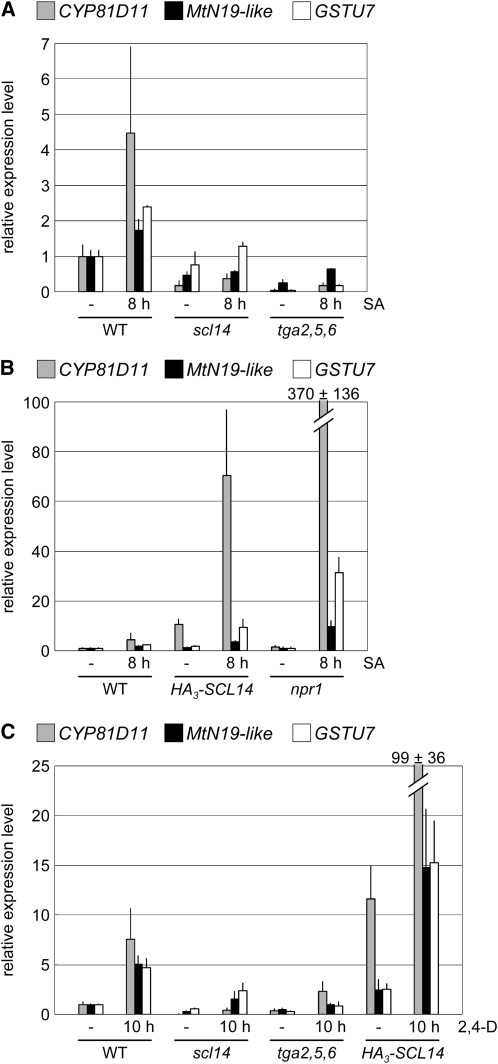
As shown in Figure 4, enhanced levels of SCL14 stimulate basal and SA- and 2,4-D–induced levels of transcription from the as-1:GUS transgene. Consistently, expression of the three selected target genes CYP81D11, MtN19-like, and GSTU7 was also inducible by SA and 2,4-D (Figure 7). Expression levels were lower in the scl14 and tga2 tga5 tga6 mutant plants and higher in the HA3-SCL14 transgenic plant line #5, demonstrating that CYP81D11, MtN19-like, and GSTU7 are regulated at least in part in an SCL14- and TGA-dependent manner. In the npr1 mutant, induction of all three genes by SA was more efficient than in wild-type plants (Figure 7B), a phenomenon that has been observed before for GSTU7 (Blanco et al., 2005). The effect was most pronounced for CYP81D11, which also has increased responsiveness to SA compared with the two other genes.
Figure 7.
Expression Analysis of Endogenous SCL14 Target Genes after SA and 2,4-D Treatment.
Quantitative real-time RT-PCR analysis of relative CYP81D11 MtN19-like and GSTU7 transcript levels in as-1:GUS (WT) plants, HA3-SCL14 transgenic plant line #5, and scl14, tga2 tga5 tga6 (tga2,5,6), and npr1-1 mutant plants. Three-week-old plants were treated with 1 mM SA ([A] and [B]) or 0.1 mM 2,4-D (C) for the indicated time spans (hyphens indicate untreated plants). Transcript values in untreated as-1:GUS plants were set to 1. The mean values (±sd) of three independent induction experiments are shown. Except for the tga2 tga5 tga6 and npr1-1 mutants, analyzed plant lines carry the as-1-GUS transgene; WT refers to an intact SCL14 allele.
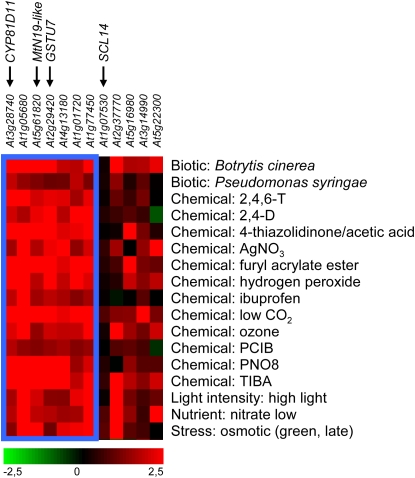
Genevestigator V3 clustering analysis (Zimmermann et al., 2004) of the genes that showed the highest difference in the expression levels between transgenic HA3-SCL14 plants and scl14 mutants (Figure 5) revealed that seven of these genes, including CYP81D11, MtN19-like, and GSTU7, belong to a cluster of genes that is upregulated after treatment with a variety of different chemicals or infection with Botrytis cinerea or Pseudomonas syringae (Figure 8). TIBA (2,4,6-triiodobenzoic acid), a compound known to inhibit auxin transport, was one of the strongest inducers of the genes identified in the microarray experiment. Similar to the results obtained with SA and 2,4-D, induction of CYP81D11, MtN19-like, and GSTU7 by TIBA depends on SCL14 and class II TGA factors (see Supplemental Figure 3 online). Expression of SCL14 (untagged) under the control of the CaMV 35S promoter rescues the scl14 mutant phenotype, as demonstrated by the restoration of CYP81D11 induction by TIBA (Figure 9). Protein gel blot analysis in the different transgenic lines confirmed accumulation of SCL14 protein at levels similar to the wild type (Figure 9).
Figure 8.
Genevestigator V3 Clustering Analysis of Genes Listed in Figure 5.
Genes (columns) were clustered based upon their expression in response to different treatments (rows) using the Biclustering method (BiMax algorithm). The color scale represents log2 ratio of fold change. The blue box marks the cluster of genes, all of which are upregulated under the indicated subset of conditions. No probe sets are available for At4g15760 and At5g61950 on the 22k Affymetrix array. 2,4,6-T, 2,4,6-trihydroxybenzamide; PCIB, p-chlorophenoxyisobutyric acid; PNO8, N-octyl-3-nitro-2,4,6-trihydroxybenzamide.
Figure 9.
Complementation of the scl14 Mutant Phenotype.
Quantitative real-time RT-PCR analysis (top panel) of relative CYP81D11 transcript levels in as-1:GUS (WT) and scl14 plants and three independent transgenic lines expressing SCL14 under the control of the CaMV 35S promoter in the scl14 mutant background (scl14/35S:SCL14). Three-week-old plants grown on MS plates were treated with 0.1 mM TIBA and further incubated for 3 or 10 h as indicated below the bars (hyphens indicate untreated plants). Mean values (±sd) of three independent biological replicates are indicated. The transcript value in untreated as-1:GUS plants was set to 1. Crude protein extracts of the plants used for quantitative real-time RT-PCR analysis were subjected to protein gel blot analysis with the αSCL14 antiserum (bottom panel). The star marks an unspecific band that serves as a loading control. All plants carry the as-1:GUS transgene; WT refers to an intact SCL14 allele.
Sensitivity of Arabidopsis Plants with Altered SCL14 Levels to Xenobiotic Stress
TGA factors have been discussed previously as mediating gene expression upon xenobiotic stress (Zhang and Singh, 1994; Pascuzzi et al., 1998; Klinedinst et al., 2000; Mueller et al., 2008). Consistently, growth of tga2 tga5 tga6 and scl14 mutant plants on MS plates containing isonicotinic acid (INA; a functional analog of SA) or TIBA was reduced when compared with wild-type plants (Figure 10A). Seedlings of HA3-SCL14 transgenic plant line #5 were more tolerant, which correlates with increased expression of the selected SCL14/TGA-dependent target genes when those plants were treated with INA or TIBA (Figure 10B).
Figure 10.
Susceptibility of Plants with Decreased or Increased SCL14 Protein Levels to Toxic Chemicals.
(A) Growth phenotypes of as-1:GUS (WT) plants, HA3-SCL14 transgenic plant line #5, and scl14 and tga2 tga5 tga6 (tga2,5,6) mutant plants on MS plates containing 0.1 mM INA or 0.1 mM TIBA. Photographs were taken 2 weeks after germination. Bars = 5 mm.
(B) Quantitative real-time RT-PCR analysis of relative CYP81D11, MtN19-like, and GSTU7 transcript levels in as-1:GUS (WT) plants and HA3-SCL14 transgenic plant line #5. Plants were grown for 3 weeks on MS plates containing 25 μM INA or 12,5 μM TIBA. Transcript values in INA- or TIBA-treated wild-type plants were set to 1. Mean values (±sd) of three independent biological replicates are indicated. Except for the tga2 tga5 tga6 mutants, analyzed plant lines carry the as-1-GUS transgene; WT refers to an intact SCL14 allele.
DISCUSSION
The TGA family of transcription factors and the corresponding cis-element as-1 constitute one of the first experimental systems to be established for studying transcriptional control mechanisms in plants (Katagiri et al., 1989). The as-1 element was originally identified as the only regulatory cis-element within the truncated CaMV 35S promoter (Lam et al., 1989). This 90-bp promoter fragment confers transcriptional activation in response to 1 mM SA and 50 μM 2,4-D in tobacco (Liu and Lam, 1994; Qin et al., 1994) and Arabidopsis (Redman et al., 2002). The activating pathway is independent from NPR1 (Butterbrodt et al., 2006), a regulatory protein that interacts with TGA factors to confer expression of genes involved in systemic acquired resistance (Fan and Dong, 2002). Here, we present evidence that as-1–bound TGA factors recruit the GRAS protein SCL14 to a number of endogenous promoters that are inducible by SA, 2,4-D, and other chemicals. Activation of the corresponding genes contributes to the protection of plants against certain types of xenobiotic stress.
Domain Structure of the SCL14 Protein
Based on sequence analysis, SCL14 belongs to the plant-specific GRAS protein family that comprises 33 members in Arabidopsis (Bolle, 2004; Tian et al., 2004). GRAS proteins have been classified as transcriptional regulators and have been shown to be involved in gibberellic acid (GA) and phytochrome signaling, root and axillary shoot development, and maintenance of the shoot apical meristem. GRAS proteins typically consist of a conserved C-terminal GRAS domain that is characterized by two leucine-rich regions (LHRI and LHRII) and three distinct amino acid signatures: VHIID, PFYRE, and SAW (Pysh et al., 1999). The VHIID domain of a GRAS protein from Brassica napus interacts with a histone deacetylase, supporting the notion that GRAS proteins function in regulating gene expression at the level of transcription (Gao et al., 2004). The GRAS domain of the regulator of GA signaling GAI contributes to the interaction with the F-box protein SLEEPY1 (Dill et al., 2004), which is of major importance for the control of protein abundance as a function of GA.
Based on the occurrence of conserved amino acid sequences in the more variable N terminus of GRAS proteins, the GRAS gene family has been divided into eight subfamilies (Tian et al., 2004). SCL14 belongs to the LlSCL/SCL9 branch. Its best-characterized member is LlSCL (Lilium longiflorum SCARECROW-like) from lily, which is predominantly expressed in anthers during the premeiotic phase (Morohashi et al., 2003). By contrast, SCL14 is weakly expressed in all tissues and shows 10-fold higher levels of gene expression in dry seeds (https://www.genevestigator.ethz.ch/). When fused to the GAL4 DNA binding domain, the N terminus of the LlSCL protein confers transcriptional activation in yeast and plant cells. The activation domain was mapped to a stretch of 19 residues containing seven acidic amino acids (motif I). This motif and the motif II sequence DEDED are conserved in SCL14 at positions amino acids 83 to 101 and 309 to 313, respectively, and might be responsible for the transcriptional activation conferred by the TGA2/SCL14 complex in yeast.
Direct DNA binding has not been demonstrated for any of the GRAS proteins, but a recent study reported association of DELLA proteins with selected GA-responsive promoters (Zentella et al., 2007). SCL14 can be detected at its target promoters only in the presence of class II TGA factors (Figure 6). Presumably, TGA factors serve as sequence-specific anchor proteins to recruit SCL14 to the respective promoter regions. It remains to be shown whether a similar mechanism is operational for DELLA and other GRAS proteins.
Functional Role of SCL14 Target Genes
SCL14 target genes were identified by comparing the transcriptomes of plants expressing higher or lower levels of SCL14 (HA3-SCL14 transgenic plant line #5 versus scl14 mutant plants). Though the list might not be comprehensive, this analysis yielded suitable promoters for confirming in planta that TGA factors recruit the coactivator SCL14 to its target genes. Additionally, information on the biological context of SCL14 function were obtained. Consistent with the idea that a subset of TGA-regulated genes is related to defense responses against xenobiotic stress (Zhang and Singh, 1994; Pascuzzi et al., 1998; Klinedinst et al., 2000; Mueller et al., 2008), genes with putative functions in the inactivation of toxic compounds were found. In plants and animals, the detoxification process starts with the introduction of functional groups by enzymes like P450 monooxygenases (phase I), which are subsequently conjugated to glucose or glutathione by enzymes such as UDP:glucosyl transferases and GSTs (phase II). Such modifications result in less toxic and/or more water-soluble conjugates that are subsequently deposited either in the vacuole or the apoplast (phase III) (Sandermann, 1992). Some of the SCL14 target genes are potentially involved in phase I (cytochrome P450 family protein CYP81D11 and monooxygenase MO1) or phase II (UDP-glucosyl transferase, oxidoreductase, NADPH-dependent oxidoreductase, and GSTU7) (Figure 5). The decreased growth of the scl14 and tga2 tga5 tga6 seedlings on medium containing harmful concentrations of TIBA or INA (Figure 10) is consistent with the idea that these compounds are less efficiently detoxified because of insufficient induction of the corresponding genes. It was recently shown that the tga2 tga5 tga6 mutant is also deficient in the induction of CYP81D11 by 12-oxo-phytodienoic acid or phytoprostanes (Mueller et al., 2008). These compounds are cyclopentenone oxylipins that are formed in the plant cell via the enzymatic jasmonate pathway and a nonenzymatic, free radical–catalyzed pathway, respectively. It remains to be shown whether SCL14 is also needed for the induction of a detoxification pathway initiated by cyclopentenone oxylipins. Five of the genes listed as SCL14 dependent (CYP81D11, UDP-glucosyltransferase, NADPH-dependent oxidoreductase, GSTU7, and AtNAC002) were identified in microarray experiments designed to monitor global changes in the Arabidopsis transcriptome after treatment with the allelochemical benzooxazolinone (Baerson et al., 2005), and the CYP81D11 gene was induced by the synthetic chemical TNT (Ekman et al., 2003). Thus, the chemical warfare between neighboring plants or other organisms that produce toxic compounds might have led to the evolution of mechanisms that allow the detoxification of a wide spectrum of harmful chemicals, irrespective of whether they occur in nature or whether they are synthetic.
Genevestigator analysis indicated that the SCL14 target genes identified here are strongly induced upon infection with B. cinerea (Figure 8). It remains to be elucidated whether induction is elicited by fungal toxins, reactive oxygen species, or harmful metabolites like cyclopentenone oxylipins generated during the interaction. However, scl14 mutants were not more susceptible to Botrytis infections than wild-type plants (see Supplemental Figure 4 online), indicating that other plant defense responses determine the final outcome of this interaction.
Regulation of Gene Expression by the TGA/SCL14 Complex
In mammalian systems, transcriptional responses of genes involved in phase I, II, and III detoxification pathways can be mediated by at least three different pathways. The first two involve receptor–xenobiotic ligand interactions: (1) The aryl hydrocarbon nuclear receptor (AhR) is translocated to the nucleus after binding of the ligand and subsequently dimerizes with the coactivator Arnt to stimulate transcription (Denison and Nagy, 2003). (2) The pregnane X receptor and the androstane receptor can bind promiscuously to structurally diverse xenobiotic ligands and activate gene expression after forming heterodimers with the 9-cis-retinoic acid receptor (Kliewer et al., 2002). The third mammalian xenobiotic-sensing system involves the bZIP transcription factor Nrf2, which is retained in the cytosol by the protein Keap1. In the presence of a wide range of structurally diverse sulfhydryl-reactive electrophilic compounds, two critical Cys residues in the Keap1 protein are oxidized. This releases Nrf2, which enters the nucleus and subsequently activates target promoters, possibly in association with small bZIP transcription factors of the Maf family (Nguyen et al., 2003).
In plants, the mechanisms leading to the activation of transcriptional responses to xenobiotic stress are yet unknown. Up to now, only the TGA factors have been implicated in this response (Zhang and Singh, 1994; Pascuzzi et al., 1998; Klinedinst et al., 2000; Mueller et al., 2008). One common characteristic of the treatments that induce SCL14 target genes (Figure 8) is that they might create some sort of oxidative stress. For instance, reactive oxygene species (ROS) are generated after infection of plants with B. cinerea (Muckenschnabel et al., 2002) or P. syringae (Torres et al., 2005). Hydrogen peroxide and ozone are ROS themselves, and growth under low CO2 concentrations or high light, or treatment with the photosystem II inhibitor PNO8, enhances the ROS-forming potential of the chloroplasts (Apel and Hirt, 2004). Chemicals related to auxin action, such as 2,4-D, p-chlorophenoxyisobutyric acid, 2,4,6-trichlorophenoxyacetic acid, and TIBA, also induce SCL14 target genes. A common feature of these chemicals is that they consist of a halogen-substituted aromatic ring system that is likely to react with sulfhydryl groups. Therefore, they might activate the antixenobiotic genes by changing the redox state of the cell rather than by changing the auxin response. Consistently, the auxin transport inhibitor naphthylphthalamic acid, which does not contain strong electron-drawing substituents, does not induce expression of SCL14 target genes, supporting the hypothesis that the electrophilic character rather than the effects on auxin action are critical for eliciting the antixenobiotic stress response. Thus, redox-dependent processes either directly or indirectly influencing the activity of bZIP transcription factors may constitute a common principle between mammals and plants. Interestingly, class I factor TGA1 has been shown to be redox regulated after treatment of plants with SA (Fobert and Despres, 2005).
Protein gel blot analysis (Figure 6B) revealed that TGA factors and SCL14 are pre-existent in the cell, and chromatin immunoprecipitation analysis indicated that the TGA/SCL14/as-1 complex is preformed at the promoter (Figure 6A). However, this complex activates transcription less efficiently under noninducing conditions compared with inducing conditions. Thus, the question of how the xenobiotic stress is perceived and translated to the activation of gene expression remains to be elucidated. Concerning the mechanism of action of NPR1, a TGA-interacting protein essential for SA-specific signaling, experimental evidence for two models has been reported. In the first scenario, oxidized NPR1 resides in the cytosol as an oligomer and is translocated into the nucleus after SA-triggered reduction of critical Cys residues (Mou et al., 2003). In the nucleus, NPR1 interacts with TGA factors, leading to their recruitment to the promoter (Johnson et al., 2003), where the negative regulator SNI1 is inactivated (Li et al., 1999). In the second scenario, TGA2 and NPR1 are recruited to PR-1 independently of each other and of SA treatment (Rochon et al., 2006). After stimulation with SA, TGA2 is incorporated into a transactivating complex with NPR1, which carries a novel type of transactivation domain. At least for TGA2, these results are consistent with our chromatin immunoprecipitation analysis, which showed a considerable amount of TGA recruitment to the PR-1 promoter (Figure 6A). In both models, NPR1 activation requires an SA-induced conformational change. Consequently, overexpression of NPR1 alone does not lead to enhanced PR-1 expression in the absence of SA (Li et al., 1999), but hyperinduction of PR-1 and increased induced resistance is observed in these plants. By contrast, even a marginal increase in SCL14 (∼1.4-fold) leads to enhanced expression of target genes in the uninduced state (Figures 4C, 4D, and 7). It may be speculated that a limiting regulatory factor is outcompeted leading to low levels of uncontrolled SCL14, which confers higher background levels in the uninduced state and increased expression after chemical treatment. Still, it remains to be shown whether SCL14 is a direct target of the regulation.
In conclusion, we have identified SCL14 as regulating the induction of genes involved in the detoxification of harmful chemicals. This protein serves as a transcriptional coactivator of TGA transcription factors.
METHODS
Yeast Screen, Assays, Strains, and Plasmids
The yeast strain yTSH1 carrying the HIS3 reporter gene driven by a minimal promoter with three copies of the as-1 element is described by Weigel et al. (2005). An Arabidopsis thaliana (Col-0) cDNA library in pFL61 (Minet et al., 1992) was screened with TGA2 expressed under the control of the MET25 promoter in the modified pBridge plasmid pGBD-/TGA2 (Weigel et al., 2005). Transformation of yeast cells was performed according to Gietz et al. (1995), and transformants were selected on SD medium lacking tryptophane, uracil, and histidine, supplemented with 15 mM 3-amino-1,2,4-triazole and 1 mM methionine.
Quantitative β-galactosidase assays reflecting the transcriptional activity of the TGA2/SCL14 complex at the as-1 element were performed in yeast strain yTSZ1, which was created like yTSH1 but contains the reporter gene lacZ instead of HIS3. To construct this strain, yeast integration and reporter vector pLacZi (Clontech) was used. The 3x as-1 oligonucleotide with EcoRI and XbaI-compatible ends described by Weigel et al. (2005) was subcloned into plasmid pUC18 and ligated as an EcoRI/SalI fragment into pLacZi. Integration into the yeast strain YM4271 was performed as instructed by the manufacturer (Clontech). Because the resulting strain yTSZ1 does not allow selection for URA3-encoding plasmids, the SCL14 cDNA was inserted downstream of the MET25 promoter in a modified pBridge vector (Weigel et al., 2001), resulting in pGBD-/SCL14. TGA2 was expressed under the control of the ADH1 promoter in pGAD424 (Clontech) lacking the coding region for the GAL4 activation domain (Figure 1A). Alternatively, TGA2 was cloned into the modified pBridge vector (Figure 1C). β-Galactosidase assays to quantify the SCL14/TGA2 interaction in the classical two-hybrid system were performed in yeast strain PJ69-4a (James et al., 1996). TGA2 and SCL14 coding regions were amplified with primer pairs P1/P2 and P3/P4 (see Supplemental Table 3 online) and ligated into the BamHI site of pGBT9 (Clontech) or the SalI site of pGAD424 (Clontech) to obtain in-frame fusion proteins with GBD or GAD, respectively. The C-terminal deletion derivative of GAD-SCL14 was obtained by cutting the plasmid pGAD424-SCL14 with SpeI and subsequent religation. pGAD-NPR1 was described previously (Weigel et al., 2001). Measurement of β-galactosidase activity was performed as described by Niggeweg et al. (2000).
In Vitro Analysis of Protein–Protein Interactions
TGA2 and SCL14 were expressed in Escherichia coli BL21 as C-terminal fusions to the His6 tag or to GST using the plasmids pAC28 or pEGST (Kholod and Mustelin, 2001), respectively. Bacterial cells were grown in 200 mL Luria Broth medium at 37°C to a density of ∼1.0 A600. The expression of the His6-TGA2 protein was induced for 3 h with 0.5 mM IPTG at 37°C. For the expression of GST-SCL14 and nonfused GST (empty pEGST vector), an overnight induction with 0.1 mM IPTG at 18°C was done. The pellets of the cells were resuspended (10 mL per A600 3.5) in PBS (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4, pH 7.3) with 0.24 mg mL−1 4-(2-aminoethyl) benzenesulfonyl fluoride and 0.1 mg mL−1 lysozyme. After incubation for 1 h at 30°C and subsequent addition of 0.05% Triton X-100, cell suspensions were sonicated and spun down (10 min, 4°C, 23,000g). The supernatants were supplemented with 10% glycerol and stored at −80°C for further use.
For the pull-down assay, 100 μL of glutathione sepharose 4B (Amersham Biosciences) was added to 1 mL of the E. coli extracts containing GST-SCL14 or GST, respectively. After incubation for 1 h at room temperature, 1 mL of the His6-TGA2–containing extract was added to each of the samples followed by a second incubation under the same conditions as before. The glutathione sepharose was washed three times with PBS supplemented with 0.05% Triton X-100, and bound proteins were subsequently eluted from the material by the addition of 100 μL RPB buffer (100 mM Tris-HCl, pH 6.8, 200 mM DTT, 4% SDS, 20% glycerol, and 0.2% bromophenol blue) followed by a heat treatment for 10 min at 100°C.
Protein analysis was done by separating 20 μL of the eluates side by side with the input E. coli extracts on two 10% SDS polyacrylamide gels. One of the gels was stained with Coomassie Brilliant Blue (0.01% Coomassie Brilliant Blue F 250 in 10% glacial acetic acid), and the second gel was used to transfer the proteins onto a polyvinylidene difluoride membrane (Millipore) as described by Kyhse-Andersen (1984). For the detection of the His-tagged TGA2, the Universal HIS Western Blot Kit (Clontech) was used according to the manufacturer's instructions. Finally, the membrane was exposed to autoradiography film (Cronex 10T; Agfa).
Transient Expression of SCL14-GFP in BY-2 Protoplasts
To express SCL14 as a fusion protein with GFP, a modified vector carrying the 35SC4PPDK-sGFP(S65T) chimeric gene (Chiu et al., 1996; Thurow et al., 2005) was used. For the localization of unfused GFP, plasmid pAVA319 (Siemering et al., 1996) was used. Protoplasts were prepared from tobacco (Nicotiana tabacum) BY-2 cells and transformed in the presence of polyethylene glycol according to Haasen et al. (1999). For the inhibition of nuclear export, protoplasts were incubated for 16 h after DNA transfer with 2 μM LMB (LC Laboratories; 2 mM stock solution in ethanol) for 4 h. Controls contained 0.1% ethanol. The transformed cells were subjected to microscopy analysis using a BX-60 fluorescence microscope (Olympus).
Plant Material, Growth Conditions, and Chemical Treatments
Arabidopsis plants (Col-0 background) containing an as-1:GUS reporter construct (Redman et al., 2002) were provided by J. Arias (University of Maryland, Baltimore, MD). This line served as wild-type control in all analysis. The Arabidopsis tga2 tga5 tga6 mutant was a kind gift of Y. Zhang (University of British Columbia, Vancouver, Canada). Seeds of the T-DNA insertion line SALK_126931 (scl14) and npr1-1 were obtained from the Nottingham Arabidopsis Stock Centre.
Plants were grown under controlled environmental conditions (21/19°C, 100 μmol photons m−2 s−1, 16-h-light/8-h-dark cycle, 60% relative humidity). For chemical treatment (Figures 4 and 7C), 3-week-old plants grown on soil were carefully uprooted. The roots were washed twice in beakers containing tap water until the soil was completely removed. Fifteen plants were subsequently transferred to 30 mL of 50 mM sodium phosphate buffer containing 1 mM SA or 0.1 mM 2,4-D in 0.1% ethanol and allowed to float for the indicated time points. Alternatively, plants grown in soil were sprayed with 1 mM SA (Figures 7A and 7B). For treatment with INA and TIBA, plants were grown on MS plates supplemented with 0.1 mM or 25 μM INA or with 0.1 mM or 12,5 μM TIBA in 0.1% dimethyl sulfoxide (Figure 10). Alternatively, plants were sprayed with 0.1 mM TIBA (Figure 9).
Binary Vectors and Plant Transformation
To generate a binary vector for the expression of an HA3-tagged SCL14 under the control of the CaMV 35S promoter, Gateway technology (Invitrogen) was used. The coding region of the SCL14 was amplified with primers P5 and P6 (see Supplemental Table 3 online), cloned into the pDONR207 vector (Invitrogen), and subsequently recombined into the binary destination vector pAlligator2 (Bensmihen et al., 2004) that contains the Gateway cassette downstream of an HA3-tag encoding sequence.
For generation of transgenic plants, binary plasmids were electroporated (GenePulser II; Bio-Rad) into Agrobacterium tumefaciens strain GV3101 (pMP90). The resulting agrobacteria were used to transform as-1:GUS plants with the HA3-SCL14–encoding T-DNA by the floral dipping method (Clough and Bent, 1998). Transgenic lines with ectopic expression of the transgene were identified by protein gel blot analysis with the αSCL14 antiserum.
Antibodies and Protein Gel Blot Analysis of Plant Extracts
For the generation of an antiserum directed against SCL14, the above-mentioned GST-SCL14 fusion protein, expressed in E. coli BL21, was used as antigen. Because of the poor solubility of the recombinant protein, the protein was purified from the pellet obtained after sonication and centrifugation of bacterial cells. The pellets were washed two times in 20 mM Tris-HCl, pH 7.5, resuspended in 1% SDS, and incubated at 65°C for 5 min. Purification of the GST-SCL14 fusion was done by preparative gel electrophoresis and electroelution using the Elutrap apparatus (Schleicher and Schüll). Approximately 250 to 300 μg of the fusion protein per injection was used for rabbit immunization according to the standard 60-d immunization procedure protocol by Eurogentec. Antisera were collected 8 weeks after the first immunization.
Preparation of denaturing plant extracts (Schiermeyer et al., 2003), protein gel blot analysis (Thurow et al., 2005), and the αTGA2,5 antiserum (Ndamukong et al., 2007) were described before, although in this work the resolving gel contained 8% rather than 10% polyacrylamide. Both the αSCL14 and the αTGA2,5 antisera were diluted 1:1000.
RNA Blot Analysis and Quantitative Real-Time RT-PCR Analysis
For RNA gel blot analysis, total RNA was extracted from 150 to 300 mg of plant material using the TRIZOL method (Chomczynski and Mackey, 1995). RNA gel blots were prepared and hybridized as described by Heinekamp et al. (2002). Probes were amplified from either cDNA or plasmid templates using appropriate primers for SCL14/At1g07530, GUS, PR-1/At2g14610, and GST6/At2g47730 (see Supplemental Table 4 online). The purified PCR products were subsequently labeled with 32P.
For quantification of transcript levels with real-time RT-PCR, RNA was extracted from plant leaf material as described above. cDNA synthesis was performed with 1 μg of total RNA, 20 pmol of oligo(dT) (18 dT), and 200 pmol of random nonamer oligonucleotides. Water was added to a final reaction volume of 12.5 μL. The mixture was heated to 70°C for 10 min, 20 nmol deoxynucleotide triphosphate, 4 μL 5× reaction buffer (Fermentas), and 30 units of ribonuclease inhibitor (Eppendorf) were added (final volume 20 μL), and the mixture was incubated at 37°C for 10 min. One hundred units of RevertAid H Minus M-MuLV reverse transcriptase (Fermentas) was added (final volume 20 μL), and the mixture was incubated at 42°C for 70 min and then heated to 70°C for 10 min. Amplification and quantification of cDNA using QuantiTect primers from Qiagen for the genes listed in Figure 5 and At1g13320 (protein phosphatase type 2) as a reference (Czechowski et al., 2005) (see Supplemental Table 5 online for primer sequences) was done in the iCycler System (Bio-Rad). The amplification mix consisted of 1× NH4-reaction buffer (Bioline), 2 mm MgCl2, 100 μM deoxynucleotide triphosphate, 0.4 μM primers, 0.25 units BIOTaq DNA polymerase (Bioline), 10 nm Fluorescein (Bio-Rad), 100,000 times diluted SYBR Green I solution (Cambrex), 1 μL of a 1:10 dilution of cDNA as template, and doubly distilled water filled to a total volume of 25 μL. PCR consisted of a 3-min denaturation step at 95°C followed by 40 cycles of 20 s at 95°C, 20 s at 55°C, and 40 s at 72°C. Calculations were done according to the 2−ΔΔCT method (Livak and Schmittgen, 2001).
Chromatin Immunoprecipitations
Chromatin immunoprecipitations and subsequent real-time PCR analysis were done as specified by Ndamukong et al. (2007) but with a cleared αTGA2,5 antiserum depleted for αGST antibodies. (The antiserum was generated against a mixture of GST-TGA2 and GST-TGA5 antigen). Eight milligrams of recombinant GST protein was coupled to 1 mL prewashed glutathione-sepharose 4B beads (Amersham Biosciences). After addition of 3 mL PBS to the loaded beads (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4, pH 7.3), 500 μL of the slurry were filled into microcolumns (Micro Bio-Spin columns; Bio-Rad). Columns were washed twice with 500 μm PBS before loading with 500 μL of the antiserum. After incubation of the sealed column for 1 h at 4°C, the flow-through was collected by gentle centrifugation (1 min, 2500 rpm). The process was repeated twice with fresh GST-loaded beads, and 5 μL of the final flow-through was added to the chromatin samples. After this enrichment, more consistent immunoprecipitation results with respect to the reference gene were obtained when comparing different mutant chromatins. The clearing was not necessary for the αSCL14 antiserum. Primers for the quantitative real-time PCR are given in Supplemental Table 6 online. Calculation was done according to the 2−ΔΔCT method (Livak and Schmittgen, 2001), taking ACTIN8 reference sequences for normalization.
Whole-Genome Array
Microarrays spotted with the Arabidopsis Genome Oligo Set version 3.0 (26.000 genes; Qiagen) were obtained from D. Galbraith (University of Arizona, Tucson, AZ). Total RNA was extracted from wild-type, scl14, and HA3-SCL14 plants according to the TRIZOL method (Invitrogen) and purified using the RNeasy mini kit (Qiagen). Slides were rehydrated at 60°C and UV cross-linked according to the supplier's website (http://www.ag.arizona.edu/microarray/). The Amino Allyl MessageAmp II aRNA amplifcation kit (Ambion) was used for cDNA synthesis, in vitro transcription, and Cy3/Cy5-labeling of the 5-(3-aminoallyl)-UTP–containing aRNAs with the following modifications: Purification and concentration of double-stranded cDNA was done using the DNAclear kit (Ambion), and the large-scale transcription reaction was purified with the MEGAclaer kit (Ambion). Each RNA was labeled with Cy3 and Cy5, and the following hybridizations were performed using six slides: Cy3wt/Cy5scl14, Cy3wt/Cy5HA3-SCL14, Cy5wt/Cy3scl14, Cy5wt/Cy3HA3-SCL14, Cy3scl14/Cy5/HA3-SCL14, and Cy5scl14/Cy3HA3-SCL14. Hybridization and washing was done as recommended on the supplier's website (http://www.ag.arizona.edu/microarray/). The slides were scanned with a G2505B microarray scanner (Agilent Technologies). Image processing, including spot finding and quantification of signal intensity, was done using the software Automatic Imageprocessing for Microarrays (Katzer, 2004). Normalization of the local background-corrected raw intensity data was done with nonlinear lowess regression. Each normalized data set was scaled by division with its standard deviation. The preprocessing of the data was done using R (http://can.r-project.org, http://www.bioconductor.org). Differentially expressed genes were identified by an analysis of variance mixed-effects model using SAS PROC MIXED, where false discovery rate–adjusted P values were obtained by the Benjamini-Hochberg method (Bretz et al., 2005). Normalization and statistical computation was done independently for a high and a low gain data set, allowing the recovery of lost data from saturated spots.
Clustering Analysis
To identify groups of genes with similar expression profiles in a subset of conditions, the Biclustering tool (BiMax algorithm; Prelic et al., 2006) of Genevestigator V3 (https://www.genevestigator.ethz.ch; Zimmermann et al., 2004) was used. The putative SCL14-dependent genes obtained by microarray analysis (Figure 5) were compared with publicly available 22k Affymetrix microarray data sets at the Genevestigator database. The bicluster of upregulated genes for the profile stimulus was received using the following parameters: discretization, 1.0; minimum rows, 2; minimum columns, 6.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database or GenBank/EMBL databases under the following accession numbers: At1g07530 (SCL14), At5g06950 (TGA2), At1g64280 (NPR1), At2g14610 (PR-1), At2g47730 (GST6), At1g61120 (GES), At1g49240 (ACTIN8), and At1g13320 (protein phosphatase type II). Accession numbers for genes identified in the microarray experiment are listed in Figure 5.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Protein Gel Blot Analysis of HA3-SCL14 Expressing Plants.
Supplemental Figure 2. Analysis of GUS Reporter Gene Activity in as-1:GUS Plants Expressing HA3-SCL14.
Supplemental Figure 3. Expression of Endogenous SCL14 Target Genes after TIBA Treatment.
Supplemental Figure 4. Infection of Wild-Type and scl14 Mutant Plants with Botrytis cinerea.
Supplemental Table 1. Genes Differentially Expressed in Plants with Different SCL14 Protein Levels According to Microarray Analysis.
Supplemental Table 2. Original CT Values of Chromatin Immunoprecipitation Experiments.
Supplemental Table 3. List of Primers Used for Cloning.
Supplemental Table 4. List of Primers Used for Amplification of RNA Gel Blot Probes.
Supplemental Table 5. List of Primers for Quantitative Real-Time RT-PCR Analysis.
Supplemental Table 6. List of Primers for Chromatin Immunoprecipitation Experiments.
Supplementary Material
Acknowledgments
We thank A. Hermann and R. Scholz (Georg-August-Universität Göttingen) for excellent technical assistance, U. Süthoff (Georg-August-Universität Göttingen) for help with the pull-down experiment, Y. Zhang (University of British Columbia, Vancouver, Canada) for the tga2 tga5 tga6 mutant, B. Davies and B. Causier (Centre of Plants Sciences, University of Leeds, UK) for yeast vectors, and J. Arias (University of Maryland, Baltimore, MD) for transgenic Arabidopsis seeds encoding as-1:GUS. This work was supported by the graduate program of the Deutsche Forschungsgemeinschaft (GRK 521).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Christiane Gatz (cgatz@gwdg.de).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Apel, K., and Hirt, H. (2004). Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55 373–399. [DOI] [PubMed] [Google Scholar]
- Baerson, S.R., Sanchez-Moreiras, A., Pedrol-Bonjoch, N., Schulz, M., Kagan, I.A., Agarwal, A.K., Reigosa, M.J., and Duke, S.O. (2005). Detoxification and transcriptome response in Arabidopsis seedlings exposed to the allelochemical benzoxazolin-2(3H)-one. J. Biol. Chem. 280 21867–21881. [DOI] [PubMed] [Google Scholar]
- Bensmihen, S., To, A., Lambert, G., Kroj, T., Giraudat, J., and Parcy, F. (2004). Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsis seeds. FEBS Lett. 561 127–131. [DOI] [PubMed] [Google Scholar]
- Blanco, F., Garreton, V., Frey, N., Dominguez, C., Perez-Acle, T., Van der Straeten, D., Jordana, X., and Holuigue, L. (2005). Identification of NPR1-dependent and independent genes early induced by salicylic acid treatment in Arabidopsis. Plant Mol. Biol. 59 927–944. [DOI] [PubMed] [Google Scholar]
- Bolle, C. (2004). The role of GRAS proteins in plant signal transduction and development. Planta 218 683–692. [DOI] [PubMed] [Google Scholar]
- Bretz, F., Landgrebe, J., and Brunner, E. (2005). Multiplicity issues in microarray experiments. Methods Inf. Med. 44 431–437. [PubMed] [Google Scholar]
- Butterbrodt, T., Thurow, C., and Gatz, C. (2006). Chromatin immunoprecipitation analysis of the tobacco PR-1a- and the truncated CaMV 35S promoter reveals differences in salicylic acid-dependent TGA factor binding and histone acetylation. Plant Mol. Biol. 61 665–674. [DOI] [PubMed] [Google Scholar]
- Cao, H., Glazebrook, J., Clarke, J.D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88 57–63. [DOI] [PubMed] [Google Scholar]
- Chen, W., and Singh, K.B. (1999). The auxin, hydrogen peroxide and salicylic acid induced expression of the Arabidopsis GST6 promoter is mediated in part by an ocs element. Plant J. 19 667–677. [DOI] [PubMed] [Google Scholar]
- Chiu, W., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H., and Sheen, J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6 325–330. [DOI] [PubMed] [Google Scholar]
- Chomczynski, P., and Mackey, K. (1995). Short technical reports. Modification of the TRI reagent procedure for isolation of RNA from polysaccharide- and proteoglycan-rich sources. Biotechniques 19 942–945. [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabodopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Czechowski, T., Stitt, M., Altmann, T., Udvardi, M.K., and Scheible, W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison, M.S., and Nagy, S.R. (2003). Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu. Rev. Pharmacol. Toxicol. 43 309–334. [DOI] [PubMed] [Google Scholar]
- Di Laurenzio, L., Wysocka-Diller, J., Malamy, J.E., Pysh, L., Helariutta, Y., Freshour, G., Hahn, M.G., Feldmann, K.A., and Benfey, P.N. (1996). The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86 423–433. [DOI] [PubMed] [Google Scholar]
- Dill, A., Thomas, S.G., Hu, J., Steber, C.M., and Sun, T.P. (2004). The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16 1392–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekman, D.R., Lorenz, W.W., Przybyla, A.E., Wolfe, N.L., and Dean, J.F. (2003). SAGE analysis of transcriptome responses in Arabidopsis roots exposed to 2,4,6-trinitrotoluene. Plant Physiol. 133 1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, W., and Dong, X. (2002). In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. Plant Cell 14 1377–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields, S., and Song, O. (1989). A novel genetic system to detect protein-protein interactions. Nature 340 245–246. [DOI] [PubMed] [Google Scholar]
- Fobert, P.R., and Despres, C. (2005). Redox control of systemic acquired resistance. Curr. Opin. Plant Biol. 8 378–382. [DOI] [PubMed] [Google Scholar]
- Gao, M.J., Parkin, I., Lydiate, D., and Hannoufa, A. (2004). An auxin-responsive SCARECROW-like transcriptional activator interacts with histone deacetylase. Plant Mol. Biol. 55 417–431. [DOI] [PubMed] [Google Scholar]
- Gietz, R.D., Schiestl, R.H., Willems, A.R., and Woods, R.A. (1995). Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11 355–360. [DOI] [PubMed] [Google Scholar]
- Haasen, D., Kohler, C., Neuhaus, G., and Merkle, T. (1999). Nuclear export of proteins in plants: AtXPO1 is the export receptor for leucine-rich nuclear export signals in Arabidopsis thaliana. Plant J. 20 695–705. [DOI] [PubMed] [Google Scholar]
- Heinekamp, T., Kuhlmann, M., Lenk, A., Strathmann, A., and Droge-Laser, W. (2002). The tobacco bZIP transcription factor BZI-1 binds to G-box elements in the promoters of phenylpropanoid pathway genes in vitro, but it is not involved in their regulation in vivo. Mol. Genet. Genomics 267 16–26. [DOI] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E.A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, C., Boden, E., and Arias, J. (2003). Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell 15 1846–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri, F., Lam, E., and Chua, N.H. (1989). Two tobacco DNA-binding proteins with homology to the nuclear factor CREB. Nature 340 727–730. [DOI] [PubMed] [Google Scholar]
- Katzer, M. (2004). Automatic Segmentation of Microarray Images. PhD dissertation (Bielefeld, Germany: Universität Bielefeld).
- Kholod, N., and Mustelin, T. (2001). Novel vectors for co-expression of two proteins in E. coli. Biotechniques 31 322–323, 326–328. [DOI] [PubMed] [Google Scholar]
- Kliewer, S.A., Goodwin, B., and Willson, T.M. (2002). The nuclear pregnane X receptor: A key regulator of xenobiotic metabolism. Endocr. Rev. 23 687–702. [DOI] [PubMed] [Google Scholar]
- Klinedinst, S., Pascuzzi, P., Redman, J., Desai, M., and Arias, J. (2000). A xenobiotic-stress-activated transcription factor and its cognate target genes are preferentially expressed in root tip meristems. Plant Mol. Biol. 42 679–688. [DOI] [PubMed] [Google Scholar]
- Krawczyk, S., Thurow, C., Niggeweg, R., and Gatz, C. (2002). Analysis of the spacing between the two palindromes of activation sequence-1 with respect to binding to different TGA factors and transcriptional activation potential. Nucleic Acids Res. 30 775–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo, N., Matsumori, N., Taoka, H., Fujiwara, D., Schreiner, E.P., Wolff, B., Yoshida, M., and Horinouchi, S. (1999). Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 96 9112–9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyhse-Andersen, J. (1984). Electroblotting of multiple gels: A simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J. Biochem. Biophys. Methods 10 203–209. [DOI] [PubMed] [Google Scholar]
- Lam, E., Benfey, P.N., Gilmartin, P.M., Fang, R.X., and Chua, N.H. (1989). Site-specific mutations alter in vitro factor binding and change promoter expression pattern in transgenic plants. Proc. Natl. Acad. Sci. USA 86 7890–7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel, E., Heifetz, P., Thorne, L., Uknes, S., Ryals, J., and Ward, E. (1998). Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J. 16 223–233. [DOI] [PubMed] [Google Scholar]
- Li, X., Zhang, Y., Clarke, J.D., Li, Y., and Dong, X. (1999). Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell 98 329–339. [DOI] [PubMed] [Google Scholar]
- Liu, X., and Lam, E. (1994). Two binding sites for the plant transcription factor ASF-1 can respond to auxin treatments in transgenic tobacco. J. Biol. Chem. 269 668–675. [PubMed] [Google Scholar]
- Livak, K.J., and Schmittgen, T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)).Methods 25 402–408. [DOI] [PubMed] [Google Scholar]
- Minet, M., Dufour, M.E., and Lacroute, F. (1992). Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J. 2 417–422. [DOI] [PubMed] [Google Scholar]
- Morohashi, K., Minami, M., Takase, H., Hotta, Y., and Hiratsuka, K. (2003). Isolation and characterization of a novel GRAS gene that regulates meiosis-associated gene expression. J. Biol. Chem. 278 20865–20873. [DOI] [PubMed] [Google Scholar]
- Mou, Z., Fan, W., and Dong, X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113 935–944. [DOI] [PubMed] [Google Scholar]
- Muckenschnabel, I., Goodman, B.A., Williamson, B., Lyon, G.D., and Deighton, N. (2002). Infection of leaves of Arabidopsis thaliana by Botrytis cinerea: Changes in ascorbic acid, free radicals and lipid peroxidation products. J. Exp. Bot. 53 207–214. [DOI] [PubMed] [Google Scholar]
- Mueller, S., Hilbert, B., Dueckershoff, K., Roitsch, T., Krischke, M., Mueller, M.J., and Berger, S. (2008). General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell 20 768–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndamukong, I., Al Abdallat, A., Thurow, C., Fode, B., Zander, M., Weigel, R., and Gatz, C. (2007). SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J. 50 128–139. [DOI] [PubMed] [Google Scholar]
- Nguyen, T., Sherratt, P.J., and Pickett, C.B. (2003). Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu. Rev. Pharmacol. Toxicol. 43 233–260. [DOI] [PubMed] [Google Scholar]
- Niggeweg, R., Thurow, C., Weigel, R., Pfitzner, U., and Gatz, C. (2000). Tobacco TGA factors differ with respect to interaction with NPR1, activation potential and DNA-binding properties. Plant Mol. Biol. 42 775–788. [DOI] [PubMed] [Google Scholar]
- Pascuzzi, P., Hamilton, D., Bodily, K., and Arias, J. (1998). Auxin-induced stress potentiates trans-activation by a conserved plant basic/leucine-zipper factor. J. Biol. Chem. 273 26631–26637. [DOI] [PubMed] [Google Scholar]
- Prelic, A., Bleuler, S., Zimmermann, P., Wille, A., Bühlmann, P., Gruissem, W., Hennig, L., Thiele, L., and Zitzler, E. (2006). Comparison of biclustering methods: A systematic comparison and evaluation of biclustering methods for gene expression data. Bioinformatics 22 1122–1129. [DOI] [PubMed] [Google Scholar]
- Pysh, L.D., Wysocka-Diller, J.W., Camilleri, C., Bouchez, D., and Benfey, P.N. (1999). The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 18 111–119. [DOI] [PubMed] [Google Scholar]
- Qin, X.F., Holuigue, L., Horvath, D.M., and Chua, N.H. (1994). Immediate early transcription activation by salicylic acid via the cauliflower mosaic virus as-1 element. Plant Cell 6 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redman, J., Whitcraft, J., Gulam, H., and Arias, J. (2002). Abiotic and biotic stress differentially stimulate as-1 element activity in Arabidopsis. Plant Cell Rep. 21 180–185. [Google Scholar]
- Riechmann, J.L., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290 2105–2110. [DOI] [PubMed] [Google Scholar]
- Rochon, A., Boyle, P., Wignes, T., Fobert, P.R., and Despres, C. (2006). The coactivator function of Arabidopsis NPR1 requires the core of Its BTB/POZ domain and the oxidation of C-terminal cysteines. Plant Cell 18 3670–3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals, J., Weymann, K., Lawton, K., Friedrich, L., Ellis, D., Steiner, H.Y., Johnson, J., Delaney, T.P., Jesse, T., Vos, P., and Uknes, S. (1997). The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor I kappa B. Plant Cell 9 425–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandermann, H., Jr. (1992). Plant metabolism of xenobiotics. Trends Biochem. Sci. 17 82–84. [DOI] [PubMed] [Google Scholar]
- Schiermeyer, A., Thurow, C., and Gatz, C. (2003). Tobacco bZIP factor TGA10 is a novel member of the TGA family of transcription factors. Plant Mol. Biol. 51 817–829. [DOI] [PubMed] [Google Scholar]
- Siemering, K.R., Golbik, R., Sever, R., and Haseloff, J. (1996). Mutations that suppress the thermosensitivity of green fluorescent protein. Curr. Biol. 6 1653–1663. [DOI] [PubMed] [Google Scholar]
- Silverstone, A.L., Ciampaglio, C.N., and Sun, T. (1998). The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurow, C., Schiermeyer, A., Krawczyk, S., Butterbrodt, T., Nickolov, K., and Gatz, C. (2005). Tobacco bZIP transcription factor TGA2.2 and related factor TGA2.1 have distinct roles in plant defense responses and plant development. Plant J. 44 100–113. [DOI] [PubMed] [Google Scholar]
- Tian, C., Wan, P., Sun, S., Li, J., and Chen, M. (2004). Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol. Biol. 54 519–532. [DOI] [PubMed] [Google Scholar]
- Torres, M.A., Jones, J.D.G., and Dangl, J.L. (2005). Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat. Genet. 37 1130–1134. [DOI] [PubMed] [Google Scholar]
- Uquillas, C., Letelier, I., Blanco, F., Jordana, X., and Holuigue, L. (2004). NPR1-independent activation of immediate early salicylic acid-responsive genes in Arabidopsis. Mol. Plant Microbe Interact. 17 34–42. [DOI] [PubMed] [Google Scholar]
- Wagner, U., Edwards, R., Dixon, D.P., and Mauch, F. (2002). Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Mol. Biol. 49 515–532. [DOI] [PubMed] [Google Scholar]
- Weigel, R.R., Bauscher, C., Pfitzner, A.J., and Pfitzner, U.M. (2001). NIMIN-1, NIMIN-2 and NIMIN-3, members of a novel family of proteins from Arabidopsis that interact with NPR1/NIM1, a key regulator of systemic acquired resistance in plants. Plant Mol. Biol. 46 143–160. [DOI] [PubMed] [Google Scholar]
- Weigel, R.R., Pfitzner, U.M., and Gatz, C. (2005). Interaction of NIMIN1 with NPR1 modulates PR gene expression in Arabidopsis. Plant Cell 17 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella, R., Zhang, Z.L., Park, M., Thomas, S.G., Endo, A., Murase, K., Fleet, C.M., Jikumaru, Y., Nambara, E., Kamiya, Y., and Sun, T.P. (2007). Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19 3037–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B., and Singh, K.B. (1994). ocs element promoter sequences are activated by auxin and salicylic acid in Arabidopsis. Proc. Natl. Acad. Sci. USA 91 2507–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y.L., Tessaro, M.J., Lassner, M., and Li, X. (2003). Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15 2647–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.