Abstract
Microtubules form arrays with parallel and antiparallel bundles and function in various cellular processes, including subcellular transport and cell division. The antiparallel bundles in phragmoplasts, plant-unique microtubule arrays, are mostly unexplored and potentially offer new cellular insights. Here, we report that the Physcomitrella patens kinesins KINID1a and KINID1b (for kinesin for interdigitated microtubules 1a and 1b), which are specific to land plants and orthologous to Arabidopsis thaliana PAKRP2, are novel factors indispensable for the generation of interdigitated antiparallel microtubules in the phragmoplasts of the moss P. patens. KINID1a and KINID1b are predominantly localized to the putative interdigitated parts of antiparallel microtubules. This interdigitation disappeared in double-deletion mutants of both genes, indicating that both KINID1a and 1b are indispensable for interdigitation of the antiparallel microtubule array. Furthermore, cell plates formed by these phragmoplasts did not reach the plasma membrane in ∼20% of the mutant cells examined. We observed that in the double-deletion mutant lines, chloroplasts remained between the plasma membrane and the expanding margins of the cell plate, while chloroplasts were absent from the margins of the cell plates in the wild type. This suggests that the kinesins, the antiparallel microtubule bundles with interdigitation, or both are necessary for proper progression of cell wall expansion.
INTRODUCTION
In contrast with cytokinesis in metazoan cells, cell division in land plants proceeds by generation of the cell plate, which forms the new cell wall that separates the two daughter cells (Guertin et al., 2002). A key step in formation of the cell plate is generation of the phragmoplast, which is formed at the spindle midzone in late anaphase and contains two opposing sets of microtubules together with actin filaments and Golgi-derived vesicles (Staehelin and Hepler, 1996; Jürgens, 2005). The plus ends of the phragmoplast microtubules are directed toward the cell plate, and minus ends are directed toward the divided nuclei. Vesicles containing cell plate materials are delivered toward the plus ends of microtubules and fused to form the cell plate. Thus, formation and maintenance of the antiparallel organization of phragmoplast microtubules facilitates cell plate formation.
Two types of phragmoplasts are recognized in land plants based on whether antiparallel microtubules interdigitate at the midplane of the cell plate. Endosperm cells of the monocot Haemanthus kathalinae (Bajer, 1968; Hepler and Jackson, 1968) and the eudicot Arabidopsis thaliana (Otegui et al., 2001) form the type of phragmoplast whose microtubules interdigitate. Conversely, interdigitation of antiparallel microtubules was not observed at the midplane of the cell plate of phragmoplasts in shoot and root meristems of Arabidopsis using high-pressure freezing/freeze substitution techniques (Austin et al., 2005). The authors pointed out that previously reported cells that have interdigitated microtubules in the phragmoplasts belong to a syncytial type and postulated that the interdigitation functions to stabilize the syncytial-type phragmoplasts that have fewer microtubules than those of somatic cells. However, phragmoplast organization has only been studied in a limited number of plant taxa, and it is unknown whether the phragmoplasts undergoing somatic-type cytokinesis lack overlapping microtubules in general. In addition, the proteins that function in the interdigitate-type phragmoplasts have not been studied, and the mechanism for formation of continuous interdigitation, the defining characteristic that separates the two types of phragmoplasts, is unknown.
Kinesins constitute a superfamily of microtubule-based motor proteins and are classified into more than 14 subfamilies that have their specific functions, such as transport of cargo along microtubules and regulation of microtubule dynamics (Hirokawa, 1998; Goldstein and Philp, 1999). During cytokinesis of land plants, members of the kinesin superfamily protein localize to the phragmoplast and are indispensable for its organization. For example, NPK1-activating kinesin-like protein 1 interacts with a mitogen-activated protein (MAP) kinase kinase kinase and is required for expansion of phragmoplasts in tobacco (Nicotiana tabacum) BY-2 cells (Nishihama et al., 2002). In Arabidopsis, PAKRP1/Kinesin-12A and PAKRP1L/Kinesin-12B play a critical role in the organization of phragmoplast microtubules during cytokinesis in the microspore (Lee et al., 2007).
Here, we analyzed cytokinesis of caulonemal cells in the moss Physcomitrella patens and found that antiparallel microtubules in the phragmoplast interdigitated at the equator based on observations using electron microscopy. We also found that the sister kinesins KINID1a and KINID1b are necessary for generation of the interdigitating antiparallel microtubules in the phragmoplast based on analyses of protein localization and the deletion mutant phenotype. Furthermore, we found that the mutations resulted in incomplete expansion of the aberrant phragmoplast causing a defect in cell division. These results suggest that KINID1a and KINID1b cross-link the antiparallel microtubules in the interdigitating phragmoplast and function in proper progression of cytokinesis.
RESULTS
Interdigitated Antiparallel Microtubules at the Phragmoplast Equator in Caulonemal Cells of P. patens
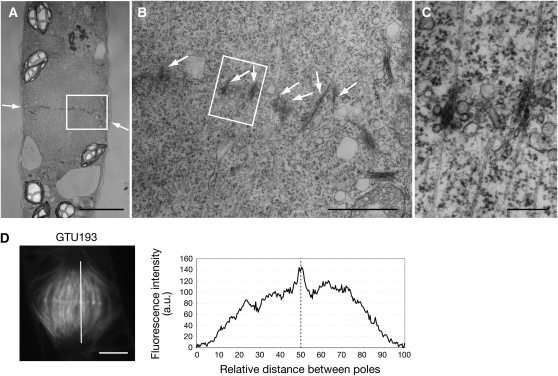
P. patens forms a filamentous structure termed the chloronemata after germination from a spore. An apical cell located at the tip of a chloronemal filament functions as a stem cell and divides continuously. Chloronemata differentiate into caulonemata, which is another filamentous structure and has thinner and more brownish chloroplasts and a more oblique cell plate than chloronemata (Cove et al., 1997). Chloronemata and caulonemata are together termed the protonemata. We observed the phragmoplasts of caulonemal apical cells using electron microscopy and found that antiparallel microtubules interdigitated at the equatorial region of the phragmoplasts (Figures 1A to 1C). The overlapped antiparallel microtubules were embedded with electron-dense materials (Figures 1B and 1C). The frequency of occurrence of interdigitated microtubules varied from cell to cell. These observations are consistent with those of endosperm cells (Hepler and Jackson, 1968).
Figure 1.
Interdigitation of Antiparallel Microtubules in the Phragmoplast of P. patens Caulonemal Cells.
(A) to (C) Electron micrographs of a cell at cytokinesis.
(A) A low magnification micrograph showing an overall view of a developing cell plate (arrows).
(B) The boxed region in (A) shows a portion of a developing cell plate. Arrows indicate microtubules overlapping with electron dense materials.
(C) The boxed region in (B) shows an enlarged image of the overlapping microtubules.
(D) A phragmoplast in cytokinesis (left) and a profile plot of fluorescence intensity (right) of the GTU193 insertion mutant line expressing sGFP-α-tubulin. Changes in fluorescence intensity along a white line connecting the two poles (left) are indicated in the right figure.
Bars = 5 μm in (A), 1 μm in (B), 200 nm in (C), and 4 μm in (D).
To visualize α-tubulin in a living P. patens cell, α-tubulin was fused to modified green fluorescent protein, sGFP (Chiu et al., 1996), and expressed under control of a constitutive promoter. A DNA fragment containing a rice (Oryza sativa) actin promoter (McElroy et al., 1990), the sGFP gene, and the P. patens α-tubulin gene TUA1 was inserted into the homeobox 7 gene (HB7) locus (Sakakibara et al., 2003) by homologous recombination (see Supplemental Figure 1A online). HB7 functions in rhizoid differentiation and therefore is dispensable for these assays. Caulonemal and chloronemal cells deficient in the HB7 gene were indistinguishable from wild-type cells (Sakakibara et al., 2003); thus, the locus was used as an insertion site. Protonemal colonies of two independent mutant lines (GTU14 and GTU193; see Supplemental Figure 1B online) could not be distinguished from wild-type colonies (see Supplemental Figure 1C online), suggesting that there was no defect in microtubule organization, which typically results in a growth deficiency (Doonan et al., 1988). The signal for sGFP-α-tubulin in caulonemal cells overlapped with that of microtubules counterstained with anti-α-tubulin antibody, indicating that sGFP-α-tubulin is incorporated into the microtubules of these mutants (see Supplemental Figures 1D and 1E online).
sGFP-α-tubulin fluorescence was more intense at the equatorial plane in the phragmoplast of a caulonemal apical cell at cytokinesis (Figure 1D). When the intensity of sGFP fluorescence was quantified along an axis connecting two poles, the sGFP signal was observed to be strongest at the equator (Figure 1D). The most intense signals were concordant with the interdigitated antiparallel microtubules observed by electron microscopy (Figure 1A). Similarly intense signal of sGFP was also detected at the phragmoplast equator in chloronemal apical cells (see Supplemental Figure 1F online). Strong signals in the equatorial region were not detected in anti-α-tubulin labeled phragmoplasts (see Supplemental Figure 1E online); the reason for these discrepant results is discussed below.
Identification of Proteins Involved in Cytokinesis
We searched gene-trap and enhancer-trap lines (Hiwatashi et al., 2001) for genes associated with cytokinesis. Trap lines showing high levels of β-glucuronidase (GUS) activity in protonemal apical cells and gametophore apices (i.e., cells that are actively dividing) were selected from 5637 gene trap and 3726 enhancer trap lines, resulting in the isolation of a line named Apical1. GUS activity was detected in both an apical cell and adjacent daughter cells in protonemata and gametophores (see Supplemental Figure 2 online).
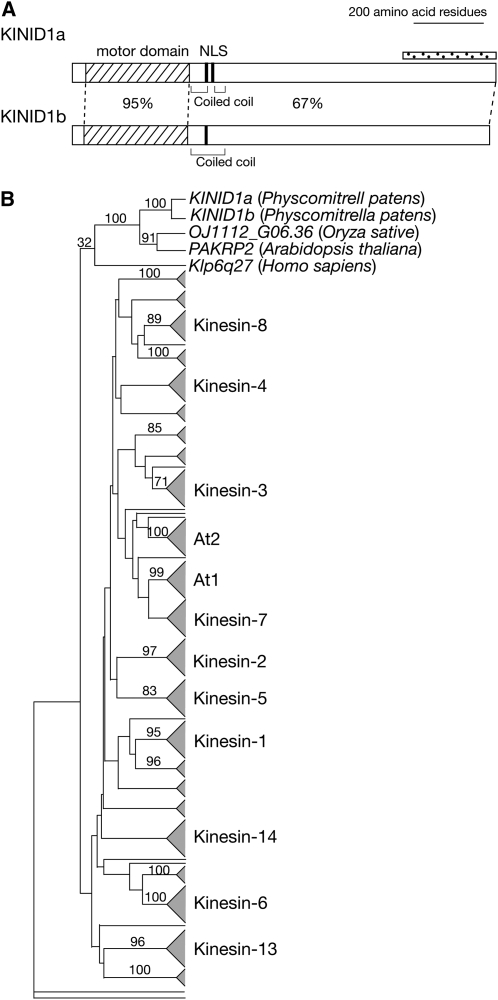
We isolated a cDNA expressed as a fusion transcript with uidA in the Apical1 line by the 5′ rapid amplification of cDNA ends (RACE) method using uidA gene-specific primers. The 3′ region was subsequently obtained using the 3′ RACE method. The resultant cDNA contained an open reading frame of 3624 bp, and the calculated molecular mass of the encoded protein was 132,619 D. The corresponding gene was named kinesin for interdigitated microtubules 1a (KINID1a; see below). Because additional bands to those corresponding to KINID1a in DNA gel blot analyses were found using wild-type genomic DNA against the KINID1a-specific probe, we performed degenerate RT-PCR using a cDNA derived from the KINID1a-disrupted line (Δkinid1a-168; see below) as a template and degenerate primers based on its amino acid sequence. We isolated a cDNA fragment whose nucleotide sequence was similar to that of KINID1a, and the encoded gene was designated KINID1b (see below). KINID1b has an open reading frame of 3591 bp, and the calculated molecular mass of the predicted protein is 131,549 D (Figure 2A). No other closely related genes were found in the P. patens genomic sequences (Rensing et al., 2008). We searched for domains in the KINID1a and b sequences using the SMART program (http://smart.embl-heidelberg.de/; Letunic et al., 2006) and the P-sort program (http://psort.nibb.ac.jp/; Nakai and Horton, 1999) and found kinesin motor domains, coiled-coil domains, and nuclear localization signals (Figure 2A).
Figure 2.
Structural and Phylogenetic Relationships of KINID1a and KINID1b.
(A) Schematics of the KINID1a and KINID1b coding regions. Kinesin motor domains (hatched boxes), nuclear localization signal sequences (NLS), and coiled-coil domains are indicated. Percentage of identical amino acid residues in kinesin motor domains and the adjacent C-terminal region are indicated. A dotted bar denotes the region used for production of the anti-KINID1a antibody.
(B) The summarized tree for the 227 kinesin superfamily genes including KINID1a and KINID1b produced using the neighbor-joining method (Saitou and Nei, 1987). The tree is rooted using Saccharomyces cerevicie Smy1 as the outgroup gene (see Methods). The original phylogenetic tree is available in Supplemental Figure 3 online. Subfamily assignments (Lawrence et al., 2004) are designated on the right. The numbers on the branches represent the local bootstrap probability. Local bootstrap values were arbitrarily selected from the original tree. Species names are designated in brackets. Branch lengths are arbitrary.
A phylogenetic analysis of the two genes with other kinesin superfamily genes showed that KINID1a and b formed a clade with the Arabidopsis kinesin gene PAKRP2 (Lee et al., 2001) and the O. sativa putative phragmoplast-associated kinesin gene OJ1112_G06.36 with 100% local bootstrap support (Figure 2B; see Supplemental Figure 3 online). This clade was not included in any previously reported subfamilies (Dagenbach and Endow, 2004; Lawrence et al., 2004). Thus, we named the two genes KINID1a and KINID1b, respectively.
KINID1a and KINID1b Are Localized to the Phragmoplast Equator
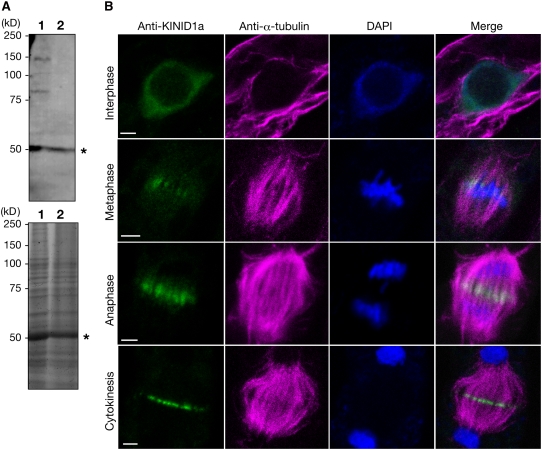
We used an anti-KINID1a antibody to immunostain caulonemal apical cells to investigate the localization of KINID1a during the cell cycle. Affinity-purified anti-KINID1a antibody against amino acid residues 903 to 1208 of KINID1a recognized a band of ∼140 kD, which is in agreement with the expected molecular mass of KINID1a (Figure 3A). Two additional bands of ∼50 and 80 kD were also detected. The 80-kD band was not detected in the KINID1a-disrupted line (Δkinid1a-168; see below), while the 50-kD band was detected in both the wild type and the KINIDa1-disrupted line. The 50-kD band is the same size as the large subunit of ribulose-1,5-bisphospate carboxylase/oxygenase, which was detected with the preimmune sera, and the 80-kD band is likely a degradation product of KINID1a.
Figure 3.
Immunodetection of KINID1a Protein.
(A) An immunoblot of wild-type whole protein extracts (lane 1) and a KINID1a deletion mutant Δkinid1a-168 extract (lane 2) was reacted against affinity-purified anti-KINID1a antibody (top panel). A Coomassie blue–stained gel is shown in the bottom panel. Asterisks mark the position of the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase.
(B) Localization of KINID1a with anti-KINID1a, microtubules with anti-α-tubulin, and DNA with 4',6-diamidino-2-phenylindole (DAPI) in caulonemal apical cells at interphase, metaphase, anaphase, and cytokinesis. The false color composite images show KINID1a in green, α-tubulin in magenta, and DNA in blue. Bars = 2 μm.
We detected immunostaining signals in the nucleus of a caulonemal apical cell during interphase (Figure 3B). Signals were also observed in the cytoplasm close to the actively growing apical tip during interphase (see Supplemental Figure 4 online). In metaphase, signals were detected in microtubules at the equatorial zone of a spindle (Figure 3B). During anaphase, signals became concentrated at the midzone of the spindle apparatus. Signals were strongest at the phragmoplast equator during cytokinesis. Signals from the chloronemal apical cells were not distinguishable from those in caulonemal apical cells (see Supplemental Figure 5 online). Signals were not detected in the KINID1a single-deletion mutant line and the KINID1a and KINID1b double-deletion mutant line (see below and Supplemental Figure 6 online).
To investigate localization of KINID1a and KINID1b in living cells, we integrated the coding regions of sGFP and Citrine, a modified yellow fluorescent protein (Griesbeck et al., 2001), respectively, in frame before a wild-type gene stop codon by homologous recombination (see Supplemental Figure 7A online). We obtained three lines (KINID1asGFP-79, 151, and 360) with a single insertion of the sGFP sequence in the KINID1a locus and two lines (KINID1bCitrine-70 and 76) with a single insertion of the Citrine sequence in the KINID1b locus (see Supplemental Figure 7B online).
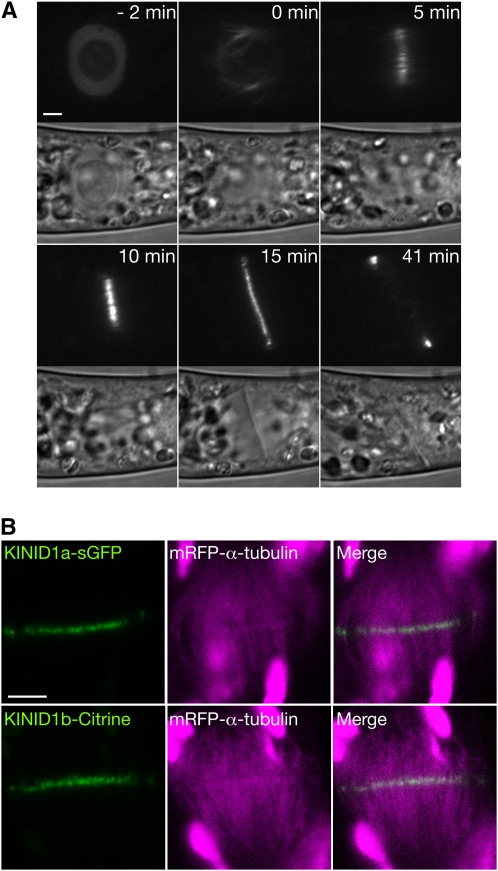
All targeted lines for both genes showed indistinguishable patterns of fluorescence. Signals accumulated in the nucleus and at the tip of protonemal apical cells during interphase (see Supplemental Figures 8A and 8B online). We took time-lapse images of KINID1a-sGFP and KINID1b-Citrine fusion proteins in caulonemal apical cells to examine the dynamics of localization during M-phase (Figure 4; see Supplemental Figure 8C online). Following nuclear envelope breakdown, the KINID1a-sGFP signals quickly accumulated in the equatorial region of the spindle. Signals remained located to the equatorial region during mitosis and cytokinesis (Figure 4A; see Supplemental Movie 1 online). The KINID1a-sGFP signals were still present during early cytokinesis (10 min in Figure 4A) and moved toward the cell periphery as cytokinesis proceeded (15 min in Figure 4A). The dynamics of localization of the KINID1b-Citrine fusion protein were indistinguishable from those of the PKINID1a-sGFP fusion protein (see Supplemental Figure 8C online).
Figure 4.
Localization of the KINID1a-sGFP Fusion Protein.
(A) Time-lapse observations of the localization dynamics of the KINID1a-sGFP fusion protein. Images were recorded every 1 min. Frames from a time-lapse movie of a caulonemal apical cell in M-phase were selected. An image showing nuclear envelope breakdown is set at 0 min. The progression of time before or after the breakdown is shown in the top right corner of the images. The top and bottom panels show KINID1a-sGFP fluorescence and bright-field images, respectively. Bar = 4 μm.
(B) Localization of KINID1a-sGFP, KINID1b-Citrine, and mRFP-α-tubulin fusion proteins at the phragmoplast equator. Autofluorescence derived from chloroplasts was detected in addition to the mRFP fluorescence at the same wavelength. Bar = 2 μm.
To examine whether KINID1a-sGFP or KINID1b-Citrine fusion proteins accumulated in the overlapping microtubules of the phragmoplast, we constitutively expressed the monomeric red fluorescent protein (mRFP) (Campbell et al., 2002)-α-tubulin fusion protein in the KINID1asGFP-360 and KINID1bCitrine-76 lines and obtained the KINID1aRTU-142 and KINID1bRTU-124 lines. Caulonemal cells of lines coexpressing both the KINID1a-sGFP or KINID1b-Citrine protein and the mRFP-α-tubulin protein were indistinguishable from those of the wild type. As observed in lines expressing the sGFP-α-tubulin fusion protein (Figure 1), the fluorescence derived from mRFP-α-tubulin was of higher intensity at the phragmoplast equator where the plus ends of the antiparallel microtubules interdigitate (Figure 4B). Fluorescent signals of KINID1a-sGFP and KINID1b-Citrine protein overlapped with the high-intensity mRFP signals (Figure 4B).
To assess whether the fusion proteins are functional in protonemata, we deleted KINID1a and KINID1b in the KINID1bCitrine-76 and KINID1asGFP-360 lines, respectively, using homologous recombination (see Supplemental Figure 9 online). Colony shape of each KINID1a and KINID1b-single deletion mutant was indistinguishable from that of the wild type (see Supplemental Figure 10 online), indicating that the fusion proteins are functional in protonemata.
The Plus Ends of Antiparallel Microtubules Do Not Interdigitate in the Phragmoplast of KINID1a and KINID1b Double-Deletion Mutants
We generated deletion mutant lines of KINID1a and KINID1b using homologous recombination to analyze the functions of those proteins (see Supplemental Figure 11A online). We obtained two KINID1a deletion mutant lines (Δkinid1a-168 and 628) and two KINID1b deletion mutant lines (Δkinid1b-1 and 10). We then deleted the KINID1b gene from the Δkinid1a-168 line producing two KINID1a and Pp KINID1b double-deletion mutant lines (Δkinid1aΔkinid1b-49 and 59). Deletion of each gene was confirmed by DNA gel blot analyses showing the expected shifts in genomic DNA bands (see Supplemental Figure 11B online). Protonemal colonies of the single-deletion mutants of each gene were indistinguishable from those of the wild type, while the size of protonemal colonies of double-deletion mutants was much reduced compared with the wild type (see Supplemental Figure 12A online).
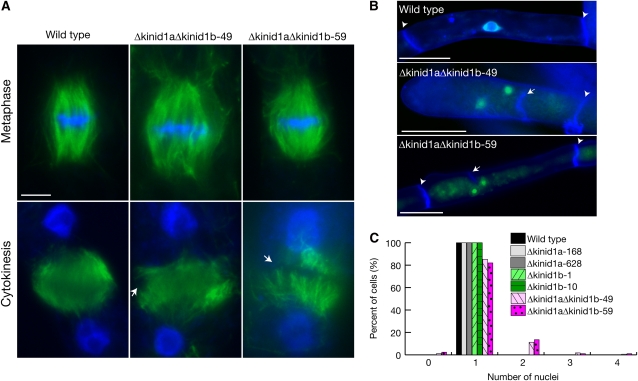
Caulonemal cells of the deletion mutant lines were immunostained with anti-α-tubulin antibody. In the four single-deletion mutant lines, the shapes of the spindles (n = 25 for each line) and phragmoplasts (n = 35 for each line) were identical to those of the wild type (n = 50; see Supplemental Figure 13 online). Spindles of the double-deletion mutants (n = 26 and 27 in the Δkinid1aΔkinid1b-49 and 59 lines, respectively) were also indistinguishable from those of the wild type (n = 50; Figure 5A). In contrast with the wild type (n = 50), the two opposing sets of microtubules in the double-deletion mutant lines were spatially separated (n = 36 in 37 phragmoplasts of the Δkinid1aΔkinid1b-49 line and 45 in 47 phragmoplasts of the Δkinid1aΔkinid1b-59 line; Figure 5A).
Figure 5.
Cytokinetic Defects in KINID1a and KINID1b Double-Deletion Mutants.
(A) A spindle (top panels) and a phragmoplast (bottom panels) in the wild type and the Δkinid1aΔkinid1b double-deletion mutant Δkinid1aΔkinid1b-49 and 59 lines. Microtubules and DNA were stained with anti-α-tubulin antibody (green) and with DAPI (blue), respectively. Arrowheads indicate the phragmoplast equator. Bar = 5 μm.
(B) Caulonemal cells of the wild type and the Δkinid1aΔkinid1b double-deletion mutants. Caulonemal cells incubated for 7 d were stained with calcofluor (blue) and SYBR green (green). Images after double staining were merged. Arrowheads and an arrow indicate normal and abnormal septa, respectively. Bars = 50 μm.
(C) Frequency of multinucleated cells. The number of nuclei per caulonemal nonapical cell was counted after double staining with calcofluor and SYBR green for the wild type (n = 170), Δkinid1a-168 (n = 185), Δkinid1a-628 (n = 170), Δkinid1b-1 (n = 180), Δkinid1b-10 (n = 211), Δkinid1aΔkinid1b-49 (n = 145), and Δkinid1aΔkinid1b-59 (n = 150).
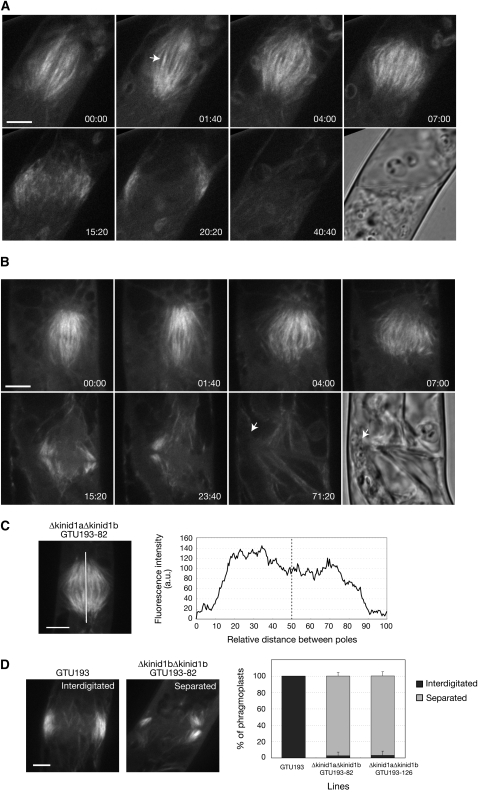
To observe microtubule dynamics during cytokinesis, we deleted both KINID1a and KINID1b genes in the GTU193 line in which the sGFP-α-tubulin fusion protein was expressed (see Supplemental Figure 14 online). We obtained two double-deletion mutant lines, Δkinid1aΔkinid1bGTU193-82 and 126. The size of the protonemal colonies of these double-deletion mutants was much reduced compared with the GTU193 line (see Supplemental Figure 15A online). Progression of mitosis and cytokinesis was observed in living apical caulonemal cells of the Δkinid1aΔkinid1bGTU193-82 and 126 lines and the GTU193 line as a control (Figure 6A; see Supplemental Movies 2 and 3 online). In the GTU193 line, after the chromosomes separated and moved toward the poles, we observed stronger sGFP fluorescence at the midzone of the anaphase spindle than in other region, corresponding to interdigitation of microtubules emanating from the poles (Figure 6A at 1 min 40 s). Upon completion of anaphase, the phragmoplast appeared between the reforming nuclei, and high-intensity sGFP fluorescence was detected at the phragmoplast equator (Figure 6A at 4 min). The phragmoplast subsequently expanded toward the cell periphery and microtubules were lost from the interior (Figure 6A after 7 min). Conversely, highly intense sGFP fluorescence at the phragmoplast equator was not observed in the Δkinid1aΔkinid1bGTU193-126 line (Figure 6B at 1 min 40 s and 4 min). As the phragmoplast expanded toward the cell periphery, the plus ends of opposite antiparallel microtubules moved away from the phragmoplast equator in the double-deletion mutant lines (Figure 6B after 7 min). Progression of mitosis and cytokinesis in the Δkinid1aΔkinid1bGTU193-82 line was similar to that in the Δkinid1aΔkinid1bGTU193-126 line. Fluorescence intensity profiles derived from a line drawn along the pole-to-pole axis of the phragmoplast were measured in the Δkinid1aΔkinid1bGTU193-82 line (Figure 6C). Whereas the GTU193 line show the highest intensity around the phragmoplast equator (Figure 1C), no peak of intensity was detected around the phragmoplast equator in the deletion mutant line (Figure 6C). The frequency of occurrence of the abnormal phragmoplasts, in which the two sets of antiparallel microtubules are separated from each other at the phragmoplast equator, was quantified. Approximately 97% of these double mutant apical caulonemal cells had abnormal phragmoplasts with the plus ends of the antiparallel microtubules separated. None of the control cells exhibited such an abnormality (Figure 6D). These results indicate that the plus ends of the antiparallel microtubules did not interdigitate at the phragmoplast equator in the double disruptants.
Figure 6.
Dynamics of Microtubule Localization during Mitosis and Cytokinesis in KINID1a and KINID1b Double-Deletion Mutants.
(A) and (B) Time-lapse observations of the dynamics of microtubule localization during mitosis and cytokinesis. A caulonema apical cell of the GTU193 line (A) and a cell of the double-deletion mutant line Δkinid1aΔkinid1bGTU193-126 (B) is shown. The images were recorded every 20 s. Selected frames are taken from a time-lapse movie (see Supplemental Movies 1 and 2 online). An image taken just prior to chromosomal separation was set at 0 min. The arrow in (A) shows the interdigitation of microtubules. Arrows in (B) show a chloroplast.
(C) A phragmoplast and a profile plot of fluorescence intensity derived from the sGFP-α-tubulin fusion product expressed in the GTU193 line and the Δkinid1aΔkinid1bGTU193-82 line. Fluorescence intensity values were measured along a line drawn on the long axis between poles (bar in the image).
(D) Proportion of phragmoplasts showing normal and abnormal shapes. The interdigitated or separated sets of phragmoplast microtubules are referred to as “Interdigitated” or “Separated,” respectively. All values are the average of three independent experiments (n > 10). Error bars indicate sd of the mean. Bars = 4 μm.
Double Disruption of KINID1a and KINID1b Causes Cytokinetic Defects
In addition to the abnormal phragmoplast formation in the double-deletion mutants, we found defects of cell plate formation and protonemal growth in the double mutants, but not in single-deletion mutants. Multinucleate caulonemal cells with incomplete septa were detected in the Δkinid1aΔkinid1b-49 and 59 lines when doubly stained with calcofluor and SYBR green (Figure 5B). Approximately 18% of caulonemal cells in the Δkinid1aΔkinid1b-49 and 59 line were multinucleate, while multinucleate cells were not observed in any of the single disruptants or in the wild-type cells (Figure 5C).
To address the underlying mechanism of the abnormal cytokinesis, we examined phragmoplast expansion in double-deletion mutants expressing sGFP-α-tubulin. We found that three of 18 phragmoplasts in the Δkinid1aΔkinid1bGTU193-82 line and three of 13 phragmoplasts in the Δkinid1aΔkinid1bGTU193-126 line showed abortion of phragmoplast expansion, while the GTU193 line showed complete expansion of the phragmoplast (n = 9). In all aborted cells, the expanding edge of the phragmoplast reached a chloroplast located adjacent to the cytokinetic apparatus and then the phragmoplast disappeared (Figure 6B; see Supplemental Movie 3 online).
DISCUSSION
Phragmoplast Microtubules Interdigitate at the Cell Plate Midplane during Cytokinesis of Protonemata
Electron microscopy revealed that the plus ends of antiparallel microtubules interdigitated at the phragmoplast equator in somatic cells of P. patens (Figures 1A and 1B). These results are in agreement with the finding of higher sGFP-α-tubulin fluorescence signals at the equator than in other regions (Figures 1D, 4B, and 6A). However, anti-α-tubulin antibody signals were significantly weaker at the equator (Figure 3B; see Supplemental Figure 1E online). This is likely due to the fact that the antibody may not be able to adequately access the interdigitated region of the phragmoplasts. Julian et al. (1993) observed a similar phenomenon in metazoan midbodies where incorporation of anti-α-tubulin antibody into interdigitated microtubules was reduced and the fluorescence intensity of anti-α-tubulin labeling did not accurately reflect the density of microtubules. Therefore, electron microscopy studies are preferable to antibody labeling, at least for the detection of interdigitation. Previously published electron micrographs of Funaria hygrometrica caulonemal cells (Schmiedel et al., 1981) and Sphagnum palustre leaflet cells (Schnepf, 1973) show interdigitated microtubules in the phragmoplast, although the authors did not comment on them. This suggests that interdigitated phragmoplasts prevail in somatic cells of mosses as well as in the endosperm cells of angiosperms. We also noted closely associated microtubules extending from both sides of the cell plate in an electron micrograph of Phaseolus vulgaris root meristem cells (Hepler and Newcomb, 1967). Presumably, interdigitated phragmoplasts may even be found in the somatic cells of angiosperms.
Possible Molecular Functions of KINID1a and KINID1b Related to the Formation of Interdigitated Phragmoplast Arrays
Interdigitation of the two antiparallel microtubule arrays of the phragmoplast was lost in the KINID1a and KINID1b double-deletion mutant lines during cytokinesis (Figures 5A and 6A to 6C). The arrays were subsequently separated at the phragmoplast equator in late cytokinesis in the mutants (Figures 6B and 6D). Together with KINID1a and KINID1b localization at the putative interdigitated regions (Figures 3B and 4B), which is similar to the localization of ATK5, which bundles antiparallel interpolar microtubules in spindles (Ambrose et al., 2005; Ambrose and Cyr, 2007), these kinesins likely function in the phragmoplast both to bundle antiparallel microtubules forming the interdigitation zone at the equator and to properly organize antiparallel microtubule arrays. As the phragmoplasts in angiosperm somatic cells organize correctly without interdigitation, it remains to be investigated whether the loss of interdigitation in the mutants is directly related to the disorganized antiparallel arrays.
The plus-end–directed homotetrameric kinesin-5 family of proteins contributes to the bipolar organization of the mitotic spindle by cross-linking and sliding the antiparallel interpolar microtubules (Kapitein et al., 2005). In yeast cells, inactivation of Cin8 and Kip1 leads to separation of two halves of the spindle to form monopolar spindles (Saunders and Hoyt, 1992), and loss of KRP125c function causes spindle collapse in Arabidopsis root cells (Bannigan et al., 2007). The kinesin-5 protein is composed of a plus-end motor, a neck domain, and a C-terminal tail containing coiled-coil domains, forming a tetramer that is structurally bipolar with two motor domains at each end (Kashina et al., 1996). The neck domain is well conserved in plus-end–directed motors (Vale and Fletterick, 1997). Although the amino acid sequences of the motor domains of KINID1a and KINID1b are not closely related to the kinesin-5 proteins (Figure 2B), KINID1a and KINID1b have a partial consensus sequence (K/RxIxNxxxV/IN) followed by a hydrophobic repeat pattern, which is conserved in the neck domain of plus-end–directed motors. Given that the putative function of the neck domain is to dictate plus-end–directed motor activity (Vale and Fletterick, 1997; Wade and Kozielski, 2000), KINID1a and KINID1b may possess plus-end–directed motor activity. Plus ends of microtubules are pointed toward the cell plate in the phragmoplast (Euteneuer and McIntosh, 1980). Bipolar plus-end–directed motors are assumed to cross-link antiparallel microtubules at the cell plate and to push the interdigitated microtubules toward the minus ends (Asada et al., 1991). The coiled-coil domains of KINID1a and KINID1b may participate in oligomerization to form a tetramer with bipolar motors that cross-link antiparallel microtubules in a fashion similar to that of the kinesin-5 proteins, although KINID1a and KINID1b do not share the amino acid sequence in the coiled-coil domain with the kinesin-5 protein.
KINID1a and KINID1b may also function to connect microtubules for transport of a protein with cross-linking activity, such as the MAP65 proteins, to the phragmoplast equator where the transported proteins then connect antiparallel microtubules. MAP65 proteins, members of the MAP65/Ase1 family, directly cross-link microtubules to form bundles in tobacco BY2 cells (Sasabe and Machida, 2006). Loss-of-function mutations of members of this protein family cause aberrant formation of antiparallel microtubules in the midzone, ultimately resulting in central spindle failure (Pellman et al., 1995; Jiang et al., 1998; Schuyler et al., 2003; Verbrugghe and White, 2004; Verni et al., 2004; Loiodice et al., 2005; Yamashita et al., 2005) as well as aberrant organization of phragmoplast microtubules (Müller et al., 2004; Sasabe and Machida, 2006). KINID1a and KINID1b themselves probably do not directly connect microtubules because of the lack of any known microtubule binding sites in their C-terminal domain. Functional analyses of proteins that interact with KINID1a and KINID1b will provide insights into the molecular functions of these kinesin family members.
Possible Mechanism of Microtubule Organization in the Phragmoplast
The shape of the mitotic spindle is formed by a balance of forces between two types of kinesin proteins. The kinesin-5 and -14 proteins function in sliding antiparallel microtubules to push the poles apart and to pull the poles together, respectively (Sharp et al., 2000). The double loss-of-function mutants PAKRP1 and PAKRP1L (kinesin-12 proteins) lose the microtubule-free zone between the opposite sets of antiparallel microtubules in the phragmoplast (Lee et al., 2007). On the other hand, the distance between two phragmoplast microtubule arrays of Arabidopsis root cells is increased in the loss-of-function mutant MAP65-3/PLE (Müller et al., 2004). In this study, we showed that KINID1a and KINID1b may function similarly to kinesin-5 in interdigitated phragmoplasts, based on the observation that the antiparallel arrays were farther apart in double-deletion mutants than in the wild type (Figure 5A). Together, the shapes of both the mitotic spindle and the phragmoplast are regulated by a balance of forces, although the proteins involved in these processes differ between the mitotic spindle and the phragmoplast.
Functional Divergence between KINID1a/KINID1b and Arabidopsis PAKRP2
Phylogenetic analysis showed that KINID1a and KINID1b form a clade with Arabidopsis PAKRP2 (Lee et al., 2001) and its rice ortholog (Figure 2B). Localization of PAKRP2 is more dispersed and scattered around the phragmoplast equator, and the distribution is affected by the secretion inhibitor brefeldin A (Lee et al., 2001). The putative function of PAKRP2 is to transport Golgi-derived vesicles to the phragmoplast, which differs from the function of KINID1a and KINID1b, which have different localization dynamics. Defects in the transport of Golgi-derived vesicles changes the amount of callose deposition onto the cell wall (Yasuhara and Shibaoka, 2000), although the effect of PAKRP2 on deposition has not been reported. We compared callose deposition on the septa between the wild type and the double-deletion mutants by staining with aniline blue (see Supplemental Figure 12E online), but no differences were detected. A C-terminal region adjacent to a motor motif, which is regarded to function for Golgi-derived vesicles transport, in PAKRP2 is 34 and 32% diverged from those of KINID1a and KINID1b, respectively. In addition, the aberrant phragmoplast of the double deletion mutants was restored by expression of KINID1a but not by PAKRP2 (see Supplemental Figure 16 online). In summary, we could not find any similarities in function for KINID1a and KINID1b to PAKRP2.
Roles of KINID1a and KINID1b in Cytokinesis
The interdigitation of endosperm cell microtubules is thought to function in mechanically stabilizing syncytial-type phragmoplasts, in which fewer microtubules are used than in somatic phragmoplasts (Austin et al., 2005). On the other hand, the number of microtubules in interdigitated moss phragmoplasts is not conspicuously different from that in the noninterdigitated phragmoplasts of angiosperms. Approximately 20% of the aberrant phragmoplasts without interdigitation in the KINID1a and KINID1b double-deletion mutants did not expand completely, resulting in the formation of multinucleated cells, although the phragmoplasts in other cells expanded toward the cell periphery and properly mediated cell plate formation to completion (Figure 5C). We noticed that a chloroplast is always located between the edge of the incomplete cell plate and the plasma membrane (Figure 6B; see Supplemental Figure 15B and Supplemental Movie 3 online). In contrast with angiosperms, chloroplasts remain near the division apparatus during cytokinesis in mosses (Jensen, 1981), making it necessary to remove them from the division plane. Therefore, KINID1a and KINID1b likely function to remove chloroplasts and complete proper cell wall formation directly or indirectly. These kinesins may bind to chloroplasts and remove them from the division plane in the direction of the phragmoplast pole. On the other hand, the loss of interdigitation may make phragmoplasts unstable through unknown mechanisms, and the expanding phragmoplast is aborted when the expanding edge approaches a chloroplast.
KINID1a and KINID1b Function in Other Cellular Processes and during Different Developmental Stages
Moss protonemal apical cells exhibit tip growth to produce filamentous cells (Cove et al., 1997). The double-deletion mutants showed retarded growth of protonemal colonies, and the length of protonemal filaments in the double mutants is shorter than in the wild type (see Supplemental Figures 12A and 12B online). The length of a protonemal filament is proportional to the rate of tip growth in a protonemal apical cell. The shorter length of protonemal filaments suggests that KINID1a and KINID1b are involved in regulating tip growth in protonemal apical cells. Microtubules converge to form foci at the tip of apical protonemal cells, and organized microtubules are essential for proper tip growth in P. patens (Doonan et al., 1988). KINID1a and KINID1b were localized to the tip of protonemal apical cells (see Supplemental Figures 4 and 8A online), suggesting that these proteins regulate the focused microtubules.
KINID1a and KINID1b likely function in other tissues, including chloronemata, gametophores, and sporophytes. The intense sGFP-α-tubulin signals at the equator in chloronemal apical cells suggest that chloronemal apical cells also have interdigitation of phragmoplast microtubules (see Supplemental Figure 1F online). KINID1a and KINID1b accumulated in spindles and phragmoplasts of chloronemal cells in a manner similar to that observed in caulonemal cells (see Supplemental Figure 5 online). Multinucleated chloronemal cells were detected in the double-deletion mutants. We also detected expression of a reporter gene in gametophores of the gene-trap line Apical1, which contains a GUS fusion to the KINID1a coding sequences (see Supplemental Figure 2A online) and abnormality of both gametophore and sporophyte shape in the double-deletion mutants (see Supplemental Figures 12C and 12D online). Thus, KINID1a and KINID1b are likely involved in phragmoplast organization in chloronemal, gametophore, and sporophyte cells as well as in caulonemal cells.
METHODS
Plant Materials, Culture Conditions, and Transformation
The wild-type strain of Physcomitrella patens subsp patens (Ashton and Cove, 1977) was used. Culture conditions and polyethylene glycol–mediated transformation were performed as previously described (Nishiyama et al., 2000).
RACE and RT-PCR
Primer sequences are listed in Supplemental Table 1 online. 5′ RACE of the gene-trap line Apical1 was performed as previously described to obtain the 5′ region of the corresponding gene (Hiwatashi et al., 2001). Total RNA was used to synthesize cDNA using SuperScriptII reverse transcriptase (Invitrogen) with the uidA-specific primer GUS-R2. The 3′ region was obtained using the 3′ RACE method with cDNA synthesized from the adapter primer. PCR was performed using the cDNA as a template with the universal amplification primer (UAP) and the 229-2p1 primer, which hybridizes at the 5′ end of the API1 5′ DNA fragment. PCR cycling conditions consisted of 3 min at 94°C, 30 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 3 min. Nested PCR was performed in the same PCR cycling conditions with the UAP and the 229-2p2 primer. The amplified PCR product was purified using an agarose gel QIAquick gel extraction kit (Qiagen) and cloned into pGEM-T (Promega). This plasmid was named p229-2-4.
RT-PCR using the degenerate primers API1MF1, API1MF2, API1MR1, and API1MR2 based on conserved domains between KINID1a and PAKRP2 was performed to isolate the KINID1b cDNA. First-strand cDNA synthesis was performed using Superscript II (Invitrogen) with total RNA extracted from the Δkinid1a-168 protonemal cells and an adaptor primer containing a polyT stretch (Invitrogen). PCR was performed using Blend Taq DNA polymerase (TOYOBO) using the API1MF1 and the API1MR1 primers. PCR cycling conditions consisted of 2 min at 94°C, 35 cycles of 94°C for 30 s, 44°C for 30 s, and 72°C for 30 s. Additional PCR was performed with the API1MF2 and the API1MR2 primers. PCR conditions were as above with the exception that the annealing temperature and the number of cycles was 50°C and 40 s, respectively. The PCR product was purified from an agarose gel using a QIAquick gel extraction kit (Qiagen) and cloned into pGEM-T (Promega).
A full-length KINID1b cDNA was obtained by PCR using KOD Plus (TOYOBO) with the api1LF5 and the api1LR9 primers. PCR cycling conditions consisted of 2 min at 94°C, 35 cycles of 94°C for 15 s, 58°C for 30 s, and 68°C for 4 min. The PCR product was isolated from an agarose gel using a QIAquick gel extraction kit (Qiagen) and cloned into pCR4Blunt-TOPO (Invitrogen).
Phylogenetic Analysis
We used a data set of kinesin homologs (Dagenbach and Endow, 2004) and 104 Oryza sativa genes with significant similarity (E-value < 1e-4), which were obtained using the amino acid sequence of the KINID1a kinesin-motor domain as a query for a BLASTP search (Altschul et al., 1997) against the nonredundant protein data set from the National Center for Biotechnology Information. Deduced amino acid sequences were aligned with those of the kinesin motor domains of KINID1a and KINID1b using MAFFT ver. 5.861 (Katoh et al., 2002) and then revised manually. After exclusion of short or redundant sequences, 113 amino acid residues were used to calculate evolution distances for 227 genes using the JTT model (Jones et al., 1992) to construct a neighbor-joining tree (Saitou and Nei, 1987). The obtained tree was rooted using Saccharomyces cerevisiae Smy1 as an outgroup, based on a previously reported kinesin phylogenetic tree (Dagenbach and Endow, 2004). Statistical support for internal branches by bootstrap analyses was calculated using 1000 replications with the PROTDIST, NEIGHBOR, SEQBOOT, and CONSENSE programs in the PHYLIP version 3.65 software package (distributed by the author, J. Felsenstein, 2005, University of Washington, Seattle).
Antibody Preparation and Immunoblotting
A DNA fragment encoding KINID1a amino acid residues 903 to 1208 was PCR amplified with the api1F3010SalI-1 and the api1R3940STOPEco52I primers using the p229-2-4 plasmid as a template and then cloned into pET28 (Merck). Recombinant KINID1a protein was expressed in BL21 (Merck), purified on Chelating Sepharose Fast Flow (GE Healthcare) according to the supplier's instructions and used as antigen to raise polyclonal antibody in rabbits. Anti-KINID1a antibody was purified using a HiTrap NHS-activated HP column (GE Healthcare) to which recombinant KINID1a protein was adsorbed. Antibody was eluted with 100 mM glycine, pH 2.7, and subsequently neutralized with 1 M Tris-HCl, pH 9.0, and dialyzed against PBS.
Proteins were extracted from protonemal cells regenerated for 3 d after protoplast isolation according to Yokota and Shimmen (1994). Extracted protein was separated by SDS-PAGE (Laemmli, 1970) and transferred to Hybond-P membranes (GE Healthcare). After blocking with 3% (w/v) skim milk in TTBS (TBS containing 0.1% [w/v] Tween-20) at 4°C overnight, the membrane was incubated with anti-KINID1a diluted 1000-fold with TTBS for 1 h at 37°C. The membrane was then washed with TTBS, followed by incubation with peroxidase-conjugated anti-rabbit IgG (GE Healthcare) diluted 50,000-fold with TTBS for 1 h at room temperature. After washing with TTBS, signals from the secondary antibody were detected with ECL Plus Western Blotting Detection reagents (GE Healthcare).
Light and Fluorescence Microscopy
For immunostaining, dissected protonemata were homogenized with a polytron (KINEMATICA) and cultured on BCDATG medium for 7 d under white light. The procedure for immunostaining as described by Sato et al. (2001) was slightly modified. Briefly, protonemata were fixed in PME (100 mM PIPES, 5 mM EGTA, and 2 mM MgSO4, pH 6.8) with 8% (w/v) paraformaldehyde, 1% (v/v) DMSO, and 0.01% (v/v) Nonidet P-40 for 1 h. After three washings in PME supplemented with 0.01% (v/v) Nonidet P-40 (PMEN0.01), protonemata were attached to polyethylene-imine-coated cover glasses. The attached protonemata were treated with an enzyme solution containing 2% (w/v) driserase, 0.4 M mannitol, and 1× proteinase inhibitor (Complete Mini EDTA-free; Roche Diagnostics) for 10 min. After washing with PMEN0.01 three times, protonemata were soaked in cold methanol for 10 min at –20°C. After three rinses with PMEN0.01, protonemata were subjected to further permeabilization with 0.05% (v/v) Triton X-100 and 0.05% (w/v) BSA for 10 min. After rinsing with PBS, the protonemata were incubated with anti-α-tubulin monoclonal antibody (Ab-1; Oncogene Research Products) diluted 100-fold in PBS overnight at 4°C. Following three rinses in PBS, protonemata were treated for 3 h at 37°C with Alexa Fluor 488 rabbit anti-mouse IgG (Invitrogen). An additional two rinses in PBS were followed by staining with 0.2 mg/L DAPI for 10 min. After washing with PBS, the protonemata were mounted with a Slow Fade Light Antifade Kit (Invitrogen).
For calcofluor and SYBR Green staining, protonemata were fixed in 3.7% (v/v) formaldehyde and 0.1% (v/v) Nonidet P-40 in 50 mM PIPES, pH 6.8, for 1 h. After three washes with PBS, cells were stained with CFSB solution (10 μg/mL calcofluor and 1× SYBR Green I in 50 mM PIPES, pH 6.8) for 1 h and then mounted in CFSB solution.
For time-lapse observations, protonemal cells were cultured on BCDATG agar medium in a glass-bottom dish for 7 d. Time-lapse observations for the KINID1a-sGFP, KINID1b-Citrine, and GFP-α-tubulin fusion proteins were performed using a UPLSAPO 60XW lens mounted on an Olympus IX70 microscope equipped with a spinning disk confocal unit (CSU21; Yokogawa) and a 488-nm excitation laser. Red fluorescence from chlorophyll was subtracted by an additional barrier filter. Images were captured with Cool SNAP HQ (Roper Scientific) running Meta Morph version 4.6 software (Molecular Devices). Measurements of fluorescence intensity were performed using ImageJ 1.36b (http://rsb.info.nih.gov/ij/).
Multiwavelength confocal microscopy images for sGFP, Citrine, and mRFP were obtained with a Leica TCS-SP2 inverted confocal laser microscope (Leica Microsystems). Acquisition was along one focal line, using 488 nm (laser power: 16%) and 546 nm (laser power: 100%) and collecting the 500- to 535-nm emission spectrum for sGFP and Citrine and the 640- to 685-nm emission spectrum for mRFP.
Electron Microscopy
Protonemata were cultured in a thin sheet of BCDAT agar medium overlaid onto the same agar medium, separated by cellophane. After differentiation of caulonemata, the sheet was excised and put into a fixative containing 2.5% (v/v) glutaraldehyde, 0.5% (w/v) formaldehyde, 0.001% (v/v) Triton X-100, and 0.05 M sodium phosphate, pH 7.4. After incubation for 1 h at room temperature, the sheets were rinsed with 0.05 M sodium phosphate and postfixed with 1% (w/v) aqueous osmium tetroxide for 2 h at room temperature. The sheets were then dehydrated with acetone and embedded in Epon resin (TAAB Laboratories). Ultrathin sections (60 nm thick) were stained with 2% uranyl acetate in 70% (v/v) methanol followed by Raynold's lead citrate solution. The stained sections were observed using an H-7600 transmission electron microscope (Hitachi High-Technologies) at 80 kV.
Accession Numbers
Sequence data from this article can be found in GenBank/EMBL data libraries under the following accession numbers: P. patens genes KINID1a (AB434497), KINID1b (AB434498), TUA1 (AB096718); HB7 (AB028078); Arabidopsis PAKRP2 (At4g14330), PAKRP1 (At4g14150), PAKRP1L (At3g23670); O. sativa OJ1112_G06.36 (XM_468330).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Generation of the Lines Expressing the sGFP and α-Tubulin Fusion Proteins.
Supplemental Figure 2. GUS Activity in the Gene-Trap Line Apical1.
Supplemental Figure 3. Neighbor-Joining Tree of Kinesin Superfamily Genes Including KINID1a and KINID1b.
Supplemental Figure 4. Immunodetection of KINID1a Protein with Anti-KINID1a Antibody in a Caulonemal Apical Cell Indicating a Signal in Cytoplasm.
Supplemental Figure 5. Immunodetection of KINID1a Protein with Anti-KINID1a Antibody in Chloronemal Apical Cells.
Supplemental Figure 6. Immunodetection of KINID1a Protein with Anti-KINID1a Antibody in Caulonemal Apical Cells of the KINID1a and KINID1b Double-Deletion Mutant Line Expressing sGFP-α-tubulin Fusion Protein.
Supplemental Figure 7. Construction of the KINID1a-sGFP and KINID1b-Citrine Lines.
Supplemental Figure 8. Localized Fluorescent Signals of KINID1a-sGFP and KINID1b-Citrine Fusion Proteins in Interphase.
Supplemental Figure 9. Generation of KINID1a-Deletion Mutant Lines Expressing the KINID1b-Citrine Fusion Protein and KINID1b-Deletion Mutant Lines Expressing the KINID1a-sGFP Fusion Protein.
Supplemental Figure 10. Colony of the KINID1a-Deletion Mutant Line Expressing the KINID1b-Citrine Fusion Protein and KINID1b-Deletion Mutant Line Expressing the KINID1a-sGFP Fusion Protein.
Supplemental Figure 11. Construction of KINID1a and KINID1b Deletion Mutant Lines.
Supplemental Figure 12. Phenotypes of the KINID1a and KINID1b Deletion Mutants and the Wild Type.
Supplemental Figure 13. A Spindle and Phragmoplast of the KINID1a and KINID1b Single Deletion Mutants and the Wild Type.
Supplemental Figure 14. Generation of the KINID1a and KINID1b Double-Deletion Mutant Lines Expressing the sGFP-α-Tubulin Fusion Protein.
Supplemental Figure 15. Phenotypes of the KINID1a and KINID1b Double-Deletion Mutant Lines Expressing the sGFP-α-Tubulin Fusion Protein.
Supplemental Figure 16. Complementation Test with KINID1a and PAKRP2 in the KINID1a and KINID1b Double-Deletion Mutant Line.
Supplemental Table 1. Primers Used in This Work.
Supplemental Movie 1. Dynamics of Localization of the KINID1a-sGFP Fusion Protein.
Supplemental Movie 2. Dynamics of Spindle and Phragmoplast Formation in the GTU193 Line.
Supplemental Movie 3. Dynamics of Spindle and Phragmoplast Formation in the Δkinid1aΔkinid1bGTU193-126 Line.
Supplemental Data Set 1. Alignment Used to Create the Phylogenetic Tree of Kinesin Superfamily Genes Including KINID1a and KINID1b.
Supplemental Methods. Construction of Plasmids for Gene Targeting and DNA Gel Blot Analysis.
Supplemental References.
Supplementary Material
Acknowledgments
We thank T. Kagawa for providing p35S-sGFP-ad2-talin; T. Nakagawa for providing pUGW0; R. Tsien for providing Citrine and mRFP1 in pRSETB; Futamura Chemical Industries for providing the cellophane; Kyowa Hakko Kogyo for providing driselase; B. Liu for advice on antibody preparation and immunoblotting; E. Aoki, M. Goto, and M. Tanaka for moss transformation; and C. Ono and Y. Ichikawa for technical assistance. The computations were partly done on an SGI Origin 2000 in the Computer Lab of the National Institute for Basic Biology. The National Institute for Basic Biology Center for Analytical Instruments provided sequence and microscope facilities. This research was partly supported by grants from the Ministry of Education, Science, Culture, and Sports of Japan (M.H.) and the Japan Society for the Promotion of Science (M.H., T.M., and T.F.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Mitsuyasu Hasebe (mhasebe@nibb.ac.jp).
Online version contains Web-only data.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose, J.C., and Cyr, R. (2007). The kinesin ATK5 functions in early spindle assembly in Arabidopsis. Plant Cell 19 226–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose, J.C., Li, W., Marcus, A., Ma, H., and Cyr, R. (2005). A minus-end-directed kinesin with plus-end tracking protein activity is involved in spindle morphogenesis. Mol. Biol. Cell 16 1584–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada, T., Sonobe, S., and Shibaoka, H. (1991). Microtubule translocation in the cytokinetic apparatus of cultured tobacco cells. Nature 350 238–241. [Google Scholar]
- Ashton, N.W., and Cove, D.J. (1977). The isolation and preliminary characterization of auxotrophic and analogue resistant mutants in the moss Physcomitrella patens. Mol. Gen. Genet. 154 87–95. [Google Scholar]
- Austin II, J.R., Segui-Simarro, J.M., and Staehelin, L.A. (2005). Quantitative analysis of changes in spatial distribution and plus-end geometry of microtubules involved in plant-cell cytokinesis. J. Cell Sci. 118 3895–3903. [DOI] [PubMed] [Google Scholar]
- Bajer, A.S. (1968). Fine structure studies on phragmoplast and cell plate formation. Chromosoma 24 383–417. [Google Scholar]
- Bannigan, A., Scheible, W.R., Lukowitz, W., Fagerstrom, C., Wadsworth, P., Somerville, C., and Baskin, T.I. (2007). A conserved role for kinesin-5 in plant mitosis. J. Cell Sci. 120 2819–2827. [DOI] [PubMed] [Google Scholar]
- Campbell, R.E., Tour, O., Palmer, A.E., Steinbach, P.A., Baird, G.S., Zacharias, D.A., and Tsien, R.Y. (2002). A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99 7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, W., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H., and Sheen, J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6 325–330. [DOI] [PubMed] [Google Scholar]
- Cove, J.L., Knight, C.D., and Lamparter, T. (1997). Mosses as model systems. Trends Plant Sci. 2 99–105. [Google Scholar]
- Dagenbach, E.M., and Endow, S.A. (2004). A new kinesin tree. J. Cell Sci. 117 3–7. [DOI] [PubMed] [Google Scholar]
- Doonan, J.H., Cove, D.J., and Lloyd, C.W. (1988). Microtubules and microfilaments in tip growth - Evidence that microtubules impose polarity on protonemal growth in Physcomitrella patens. J. Cell Sci. 89 533–540. [Google Scholar]
- Euteneuer, U., and McIntosh, J.R. (1980). Polarity of midbody and phragmoplast microtubules. J. Cell Biol. 87 509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, L.S., and Philp, A.V. (1999). The road less traveled: emerging principles of kinesin motor utilization. Annu. Rev. Cell Dev. Biol. 15 141–183. [DOI] [PubMed] [Google Scholar]
- Griesbeck, O., Baird, G.S., Campbell, R.E., Zacharias, D.A., and Tsien, R.Y. (2001). Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J. Biol. Chem. 276 29188–29194. [DOI] [PubMed] [Google Scholar]
- Guertin, D.A., Trautmann, S., and McCollum, D. (2002). Cytokinesis in eukaryotes. Microbiol. Mol. Biol. Rev. 66 155–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler, P.K., and Jackson, W.T. (1968). Microtubules and early stages of cell-plate formation in the endosperm of Haemanthus katherinae Baker. J. Cell Biol. 38 437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa, N. (1998). Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science 279 519–526. [DOI] [PubMed] [Google Scholar]
- Hiwatashi, Y., Nishiyama, T., Fujita, T., and Hasebe, M. (2001). Establishment of gene-trap and enhancer-trap systems in the moss Physcomitrella patens. Plant J. 28 105–116. [DOI] [PubMed] [Google Scholar]
- Jensen, L.C.W. (1981). Division, growth and branch formation in moss protonema: studies of sequential cytological changes in living cells. Protoplasma 107 301–308. [Google Scholar]
- Jiang, W., Jimenez, G., Wells, N.J., Hope, T.J., Wahl, G.M., Hunter, T., and Fukunaga, R. (1998). PRC1: A human mitotic spindle-associated CDK substrate protein required for cytokinesis. Mol. Cell 2 877–885. [DOI] [PubMed] [Google Scholar]
- Jones, D.T., Taylor, W.R., and Thornton, J.M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8 275–282. [DOI] [PubMed] [Google Scholar]
- Julian, M., Tollon, Y., Lajoie-Mazenc, I., Moisand, A., Mazarguil, H., Puget, A., and Wright, M. (1993). γ-Tubulin participates in the formation of the midbody during cytokinesis in mammalian cells. J. Cell Sci. 105 145–156. [DOI] [PubMed] [Google Scholar]
- Jürgens, G. (2005). Plant cytokinesis: Fission by fusion. Trends Cell Biol. 15 277–283. [DOI] [PubMed] [Google Scholar]
- Kapitein, L.C., Peterman, E.J., Kwok, B.H., Kim, J.H., Kapoor, T.M., and Schmidt, C.F. (2005). The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature 435 114–118. [DOI] [PubMed] [Google Scholar]
- Kashina, A.S., Baskin, R.J., Cole, D.G., Wedaman, K.P., Saxton, W.M., and Scholey, J.M. (1996). A bipolar kinesin. Nature 379 270–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K., Misawa, K., Kuma, K., and Miyata, T. (2002). MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Lawrence, C.J., et al. (2004). A standardized kinesin nomenclature. J. Cell Biol. 167 19–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.R., Giang, H.M., and Liu, B. (2001). A novel plant kinesin-related protein specifically associates with the phragmoplast organelles. Plant Cell 13 2427–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.R., Li, Y., and Liu, B. (2007). Two Arabidopsis phragmoplast-associated kinesins play a critical role in cytokinesis during male gametogenesis. Plant Cell 19 2595–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic, I., Copley, R.R., Pils, B., Pinkert, S., Schultz, J., and Bork, P. (2006). SMART 5: Domains in the context of genomes and networks. Nucleic Acids Res. 34 D257–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiodice, I., Staub, J., Setty, T.G., Nguyen, N.P., Paoletti, A., and Tran, P.T. (2005). Ase1p organizes antiparallel microtubule arrays during interphase and mitosis in fission yeast. Mol. Biol. Cell 16 1756–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy, D., Zhang, W., Cao, J., and Wu, R. (1990). Isolation of an efficient actin promoter for use in rice transformation. Plant Cell 2 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, S., Smertenko, A., Wagner, V., Heinrich, M., Hussey, P.J., and Hauser, M.T. (2004). The plant microtubule-associated protein AtMAP65-3/PLE is essential for cytokinetic phragmoplast function. Curr. Biol. 14 412–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai, K., and Horton, P. (1999). PSORT: A program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24 34–36. [DOI] [PubMed] [Google Scholar]
- Nishihama, R., Soyano, T., Ishikawa, M., Araki, S., Tanaka, H., Asada, T., Irie, K., Ito, M., Terada, M., Banno, H., Yamazaki, Y., and Machida, Y. (2002). Expansion of the cell plate in plant cytokinesis requires a kinesin-like protein/MAPKKK complex. Cell 109 87–99. [DOI] [PubMed] [Google Scholar]
- Nishiyama, T., Hiwatashi, Y., Sakakibara, I., Kato, M., and Hasebe, M. (2000). Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Res. 7 9–17. [DOI] [PubMed] [Google Scholar]
- Otegui, M.S., Mastronarde, D.N., Kang, B.H., Bednarek, S.Y., and Staehelin, L.A. (2001). Three-dimensional analysis of syncytial-type cell plates during endosperm cellularization visualized by high resolution electron tomography. Plant Cell 13 2033–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellman, D., Bagget, M., Tu, Y.H., Fink, G.R., and Tu, H. (1995). Two microtubule-associated proteins required for anaphase spindle movement in Saccharomyces cerevisiae. J. Cell Biol. 130 1373–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing, S.A., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319 64–69. [DOI] [PubMed] [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Sakakibara, K., Nishiyama, T., Sumikawa, N., Kofuji, R., Murata, T., and Hasebe, M. (2003). Involvement of auxin and a homeodomain-leucine zipper I gene in rhizoid development of the moss Physcomitrella patens. Development 130 4835–4846. [DOI] [PubMed] [Google Scholar]
- Sasabe, M., and Machida, Y. (2006). MAP65: A bridge linking a MAP kinase to microtubule turnover. Curr. Opin. Plant Biol. 9 563–570. [DOI] [PubMed] [Google Scholar]
- Sato, Y., Wada, M., and Kadota, A. (2001). Choice of tracks, microtubules and/or actin filaments for chloroplast photo-movement is differentially controlled by phytochrome and a blue light receptor. J. Cell Sci. 114 269–279. [DOI] [PubMed] [Google Scholar]
- Saunders, W.S., and Hoyt, M.A. (1992). Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell 70 451–458. [DOI] [PubMed] [Google Scholar]
- Schmiedel, G., Reiss, H.D., and Schnepf, E. (1981). Associations between membranes and microtubules during mitosis and cytokinesis in caulonema tip cells of the moss Funaria hygrometrica. Protoplasma 108 173–190. [Google Scholar]
- Schnepf, E. (1973). Mikrotubulus-Anordnug und -Umordnung, Wandbildung und Zellmorphogenese in jungen Sphagnum-Blättechen. Protoplasma 78 145–173. [Google Scholar]
- Schuyler, S.C., Liu, J.Y., and Pellman, D. (2003). The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. J. Cell Biol. 160 517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, D.J., Rogers, G.C., and Scholey, J.M. (2000). Roles of motor proteins in building microtubule-based structures: a basic principle of cellular design. Biochim. Biophys. Acta 1496 128–141. [DOI] [PubMed] [Google Scholar]
- Staehelin, L.A., and Hepler, P.K. (1996). Cytokinesis in higher plants. Cell 84 821–824. [DOI] [PubMed] [Google Scholar]
- Vale, R.D., and Fletterick, R.J. (1997). The design plan of kinesin motors. Annu. Rev. Cell Dev. Biol. 13 745–777. [DOI] [PubMed] [Google Scholar]
- Verbrugghe, K.J., and White, J.G. (2004). SPD-1 is required for the formation of the spindle midzone but is not essential for the completion of cytokinesis in C. elegans embryos. Curr. Biol. 14 1755–1760. [DOI] [PubMed] [Google Scholar]
- Verni, F., Somma, M.P., Gunsalus, K.C., Bonaccorsi, S., Belloni, G., Goldberg, M.L., and Gatti, M. (2004). Feo, the Drosophila homolog of PRC1, is required for central-spindle formation and cytokinesis. Curr. Biol. 14 1569–1575. [DOI] [PubMed] [Google Scholar]
- Wade, R.H., and Kozielski, F. (2000). Structural links to kinesin directionality and movement. Nat. Struct. Biol. 7 456–460. [DOI] [PubMed] [Google Scholar]
- Yamashita, A., Sato, M., Fujita, A., Yamamoto, M., and Toda, T. (2005). The roles of fission yeast ase1 in mitotic cell division, meiotic nuclear oscillation, and cytokinesis checkpoint signaling. Mol. Biol. Cell 16 1378–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhara, H., and Shibaoka, H. (2000). Inhibition of cell-plate formation by brefeldin A inhibited the depolymerization of microtubules in the central region of the phragmoplast. Plant Cell Physiol. 41 300–310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.