Abstract
The plant cell wall is a dynamic structure that changes in response to developmental and environmental cues through poorly understood signaling pathways. We identified two Leu-rich repeat receptor-like kinases in Arabidopsis thaliana that play a role in regulating cell wall function. Mutations in these FEI1 and FEI2 genes (named for the Chinese word for fat) disrupt anisotropic expansion and the synthesis of cell wall polymers and act additively with inhibitors or mutations disrupting cellulose biosynthesis. While FEI1 is an active protein kinase, a kinase-inactive version of FEI1 was able to fully complement the fei1 fei2 mutant. The expansion defect in fei1 fei2 roots was suppressed by inhibition of 1-aminocyclopropane-1-carboxylic acid (ACC) synthase, an enzyme that converts Ado-Met to ACC in ethylene biosynthesis, but not by disruption of the ethylene response pathway. Furthermore, the FEI proteins interact directly with ACC synthase. These results suggest that the FEI proteins define a novel signaling pathway that regulates cell wall function, likely via an ACC-mediated signal.
INTRODUCTION
The regulation of cell expansion plays a fundamental role in plant growth and development. Despite this critical role, the regulatory inputs that control this process are poorly understood. Cell expansion is regulated primarily by turgor pressure and by the properties of the plant cell wall, which is composed of a polysaccharide network of cellulose microfibrils cross-linked by hemicelluloses in a pectin matrix, along with numerous proteins (Somerville, 2006). The primary load-bearing elements of the cell wall are the cellulose microfibrils, and their orientation and cross-linking are key factors that determine both the direction and extent of cell expansion (Darley et al., 2001). In longitudinally expanding cells, the cellulose microfibrils are deposited primarily in an orientation perpendicular to the axis of expansion, thus constricting radial expansion (Green, 1980; Taiz, 1984; Baskin, 2005). Consistent with this, disruption of cellulose biosynthesis by treatment with various chemical inhibitors results in a rapid loss of growth anisotropy (Scheible et al., 2001; Desprez et al., 2002).
Cellulose microfibrils are synthesized by cellulose synthase, an enzyme that is present at the plasma membrane as a hexameric protein complex called the rosette (reviewed in Somerville, 2006). Genetic analysis and inhibitor studies indicate that cytoplasmic microtubules play an important role in guiding the orientation of the deposition of cellulose microfibrils (reviewed in Baskin, 2001), and the cellulose synthase rosette was found to move along the plasma membrane in tracks that largely coincided with the cortical microtubules (Paredez et al., 2006).
Additional components involved in regulating cell wall biosynthesis have been identified in genetic screens for mutations that alter root or hypocotyl elongation in Arabidopsis thaliana. The cobra (cob) mutant displays radial expansion in the elongation zone of the root, and this is correlated to a disorganization of microfibrils and a reduction in the level of crystalline cellulose in cells in the root elongation zone. COB encodes a putative glycosylphosphatidylinositol (GPI)-anchored extracellular protein that is localized to the longitudinal sides of root cells in a banding pattern transverse to the longitudinal axis (Schindelman et al., 2001). The sos5 mutant is a conditional mutant that displays arrested root growth and a swollen root phenotype in the presence of salt stress (Shi et al., 2003). SOS5 encodes a GPI-anchored extracellular protein with two arabinogalactan protein-like and fascilin-like domains that has been hypothesized to play a role in cell adhesion.
Several members of the receptor-like Ser/Thr protein kinase (RLK) family in Arabidopsis have been implicated in regulating cell growth in different contexts (Hématy and Höfte, 2008). The RLKs are a large, diverse family of transmembrane signaling elements in plants, only a few of which have been functionally characterized (Morillo and Tax, 2006). The Arabidopsis protein THE1, which belongs to the Cr RLK1L (for Catharanthus roseus protein kinase1–like) subfamily, has been hypothesized to sense cell wall integrity (Hématy et al., 2007). A second group of RLKs, the WAKs, are tightly bound to the cell wall and likely play an important role in regulating its function (He et al., 1996; Anderson et al., 2001). Here, we describe two Arabidopsis leucine-rich repeat (LRR) RLKs in a distinct RLK clade whose disruption results in defects in cell expansion primarily in roots. Further analysis links 1-aminocyclopropane-1-carboxylic acid (ACC) synthase (ACS) to this pathway, as well as SOS5, which together define a novel pathway regulating cell wall biosynthesis.
RESULTS
Disruption of FEI1 and FEI2 Alters Cell Expansion
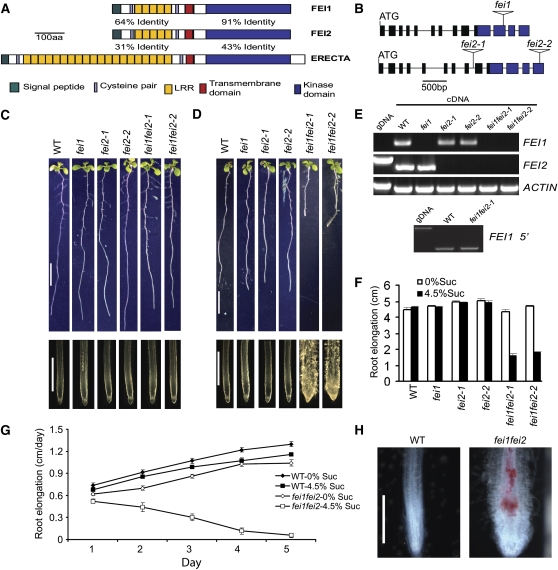
The Arabidopsis genome encodes >200 predicted LRR-RLKs, most of which have unknown functions (Morillo and Tax, 2006). We identified two highly similar LRR-RLKs (82% amino acid identity) (Figure 1A; see Supplemental Figure 1 online) that when both disrupted caused a swollen-root phenotype (Figures 1 and 2). We named these kinases FEI, after the Chinese word for fat. FEI1 (At1g31420) and FEI2 (At2g35620) are in the same RLK subfamily XIII as ERECTA (Shiu and Bleecker, 2001), which is distinct from the THE1 and WAK subfamilies. The insertions in fei1, fei2-1, and fei2-2 (Figure 1B) result in the elimination of the corresponding full-length transcript (Figure 1E). In the case of fei1, there is a truncated transcript present in the mutant plants corresponding to the region of the gene upstream of the T-DNA insertion site (Figure 1E). The single fei1 and fei2 mutants were indistinguishable from the wild type in all aspects of growth and development (Figure 1). The double fei1 fei2 mutant was nearly indistinguishable from the wild type on 1% (low) sucrose medium (Figures 1C and 1F), but in the presence of 4.5% (high) sucrose, the fei1 fei2 double mutant displayed short, radially swollen roots (Figures 1D, 1F, and 2). Root elongation was reduced in the fei1 fei2 mutant 2 d after transfer compared with wild-type seedlings (Figure 1G), and swelling was visible 3 d after transfer (see Supplemental Figure 2 online). Four days after transfer to nonpermissive conditions, the diameter of the mutant root was greater than twofold larger compared with the wild type (wild type, 163 ± 11 μm, n = 8; fei1 fei2, 316 ± 68 μm, n = 8). The F2 of a cross between fei1/fei1 and fei2/fei2 segregated seedlings displaying the mutant phenotype in a ratio consistent with two recessive loci (653 wild type, 39 swollen roots, χ2 = 0.45 for the expected 15:1 ratio). A genomic copy of FEI1 or FEI2 fused with a C-myc epitope tag (FEI1-myc or FEI2-myc) was able to fully complement the root-swelling phenotype of fei1 fei2 (Figures 3B and 3C), confirming that the phenotype was the result of disruption of these genes.
Figure 1.
fei1 fei2 Mutants Display Conditional Root Anisotropic Growth Defects.
(A) Structures of the predicted FEI and ERECTA proteins. The percentage identities between the kinase or LRR N-terminal domains of FEI1 and FEI2 or FEI2 and ERECTA are indicated. aa, amino acids.
(B) Cartoon of fei1, fei2-1, and fei2-2 alleles. Boxes represent exons (blue area represents the kinase domain), and triangles indicate the positions of T-DNA insertions.
(C) and (D) Phenotypes of indicated seedlings grown on MS medium plus 1% sucrose (C) or plus 4.5% sucrose (D) for 9 d. Bars = 1 cm in top panels and 1 mm in bottom panels.
(E) RT-PCR analysis of fei1 and fei2 mutants. Top, primers specific for the full-length open reading frame corresponding to the gene indicated at right of each gel (see Supplemental Table 1 online for the sequences of the primers used) were used to amplify the respective gene for 30 cycles from cDNA derived from the indicated line or from wild-type genomic DNA (gDNA). The ACTIN gene was amplified as a control. Bottom, primers specific for a portion of the FEI1 gene 5′ to the site of the T-DNA insertion (see Supplemental Table 1 online) were used in PCR for 30 cycles from cDNA derived from the indicated line.
(F) Quantification of root growth after transfer to permissive or nonpermissive conditions. The indicated seedlings were grown on MS medium containing 0% sucrose for 4 d and then transferred to MS medium containing either 0% or 4.5% sucrose as indicated. Root growth from the time of transfer until day 9 is indicated on the y axis. Error bars show se (n > 30).
(G) Kinetics of root elongation of wild-type and fei1 fei2 mutant seedlings. Wild-type or fei1 fei2 mutant seedlings were grown on MS medium containing 0% sucrose for 4 d and then transferred to MS medium containing either 0% or 4.5% sucrose as indicated. Root lengths were measured each day after transfer, and the amount of root growth that occurred each day after transfer was then calculated. Error bars show se (n > 15).
(H) Phloroglucinol staining for lignin (red) of seedlings grown on MS medium containing 0% sucrose for 4 d and then transferred to MS medium containing 4.5% sucrose for 5 d. Bar = 0.5 cm.
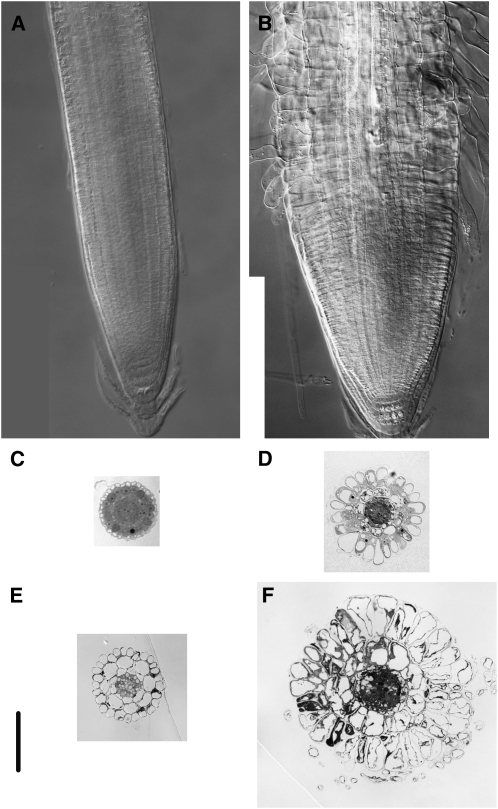
Figure 2.
Analysis of Wild-Type and fei1 fei2 Mutant Roots 4 d after Transfer from Medium Containing 0% Sucrose to Medium Containing 4.5% Sucrose.
(A) and (B) Cleared whole mount of wild-type (A) and fei1 fei2 (B) roots viewed with Nomarski optics. Note that abnormal lateral expansion in the mutant root is most apparent in the epidermis.
(C) and (D) Transverse sections through the meristems of wild-type (C) and fei1 fei2 (D) mutant roots.
(E) and (F) Transverse sections through the elongation zones of wild-type (E) and fei1 fei2 (F) mutant roots. Bar = 100 μm.
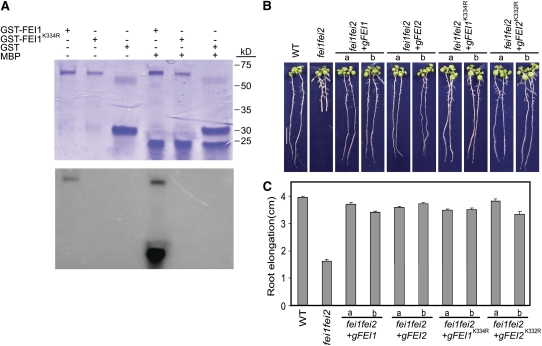
Figure 3.
Intrinsic Kinase Activity Is Not Required for FEI Function.
(A) Kinase activity of FEI1. Wild-type and FEI1K334R proteins were expressed in E. coli as GST fusion proteins, purified by glutathione-affinity chromatography, and then subjected to an in vitro kinase assay and analyzed by SDS-PAGE. Purified GST was included as a control, and myelin basic protein (MBP) was the substrate. Top, staining of the gel with Coomassie blue; bottom, autoradiograph of the gel. The positions of molecular mass markers are shown at right.
(B) Complementation of the fei1 fei2 mutant phenotype by introduction of a wild-type (gFEI1 or gFEI1) or kinase-inactive (gFEI1K334R or gFEI2K332R) version of FEI1 or FEI2. Two independent lines (a and b) are shown for each. Seedlings were grown for 4 d on MS medium containing 0% sucrose and then transferred for 4 d to MS medium containing 4.5% sucrose, and representative seedlings were photographed.
(C) Quantification of root elongation from (B). The mean (n > 15) ± se of seedling growth from days 4 to 8 is shown.
Wild-type Arabidopsis root cells undergo primarily longitudinal expansion. The increased diameter and reduced elongation observed in fei1 fei2 double mutant roots suggests that anisotropic expansion is defective in mutant root cells. Consistent with this, examination of transverse sections of root apices revealed that the fei1 fei2 epidermal cells, and to a lesser extent cells in the inner layers, displayed a high degree of radial swelling (Figure 2). The root cells of the single fei mutants appeared indistinguishable from wild-type root cells (see Supplemental Figure 3 online). The number of cells in each of the layers of the root was not appreciably altered in the fei1 fei2 mutants (i.e., there were 23 to 26 [n = 5] epidermal cells in fei1 fei2 versus 20 to 27 [n = 5] for the wild type). We conclude that the fei1 fei2 mutations cause the cells in the elongation zone to undergo a shift in expansion from longitudinal to isotropic. The fei1 fei2 mutants also displayed swollen roots on medium that contains an elevated concentration of NaCl (see Supplemental Figure 4B online). However, fei1 fei2 roots do not swell in the presence of 1 to 6% mannitol or sorbitol (see Supplemental Figure 5 online), indicating that the effect of sucrose and NaCl was not the result of a response to elevated osmolarity.
Intrinsic Kinase Activity Is Not Required for FEI Function
The sequences of the C-terminal domains of FEI1 and FEI2 have all of the features of a Ser/Thr protein kinase catalytic domain, including all 11 conserved subdomains of eukaryotic protein kinases (see Supplemental Figure 1 online) (Hanks and Quinn, 1991). To test if the FEI1 kinase has intrinsic protein kinase activity, we expressed the kinase domain of FEI1 in Escherichia coli as a glutathione S-transferase (GST) fusion protein. Purified recombinant FEI1 was active in in vitro protein kinase assays; it was able to autophosphorylate and to phosphorylate myelin basic protein (Figure 3A). Substitution of the invariant Lys residue in subdomain II in FEI1 with Arg (FEI1K334R) resulted in a complete loss of kinase activity (Figure 3A), as has been observed in other protein kinases. Interestingly, this kinase-inactive version of FEI1 (or FEI2) was able to complement a fei1 fei2 mutant (Figures 3B and 3C), although complementation was not as consistent as that observed with the wild-type FEI1 or FEI2 gene: 10 of 10 independent transformants displayed full complementation when transformed with wild-type FEI1 or FEI2, whereas 3 of 10 and 2 of 10 independent transformants were fully complemented with the respective mutant versions. This indicates that kinase activity is not essential for FEI function in vivo, although it contributes to optimal FEI function.
FEI Is Localized to the Plasma Membrane and Is Broadly Expressed
Analysis of the deduced amino acid sequence of FEI1 and FEI2 predicts a single transmembrane domain, similar to other RLKs. Consistent with this, both FEI1-myc and FEI2-myc fusion proteins were present in a microsome fraction (Figure 4I). Furthermore, a FEI2–green fluorescent protein (GFP) fusion protein, which was able to complement the fei1 fei2 mutant (see Supplemental Figure 6 online), localized to the periphery of the cell in a pattern consistent with a plasma membrane localization (Figure 4J).
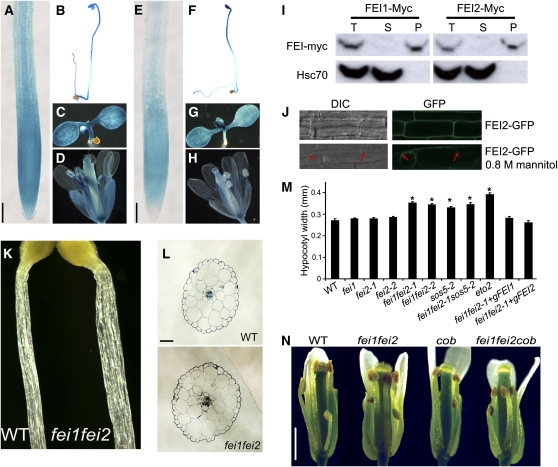
Figure 4.
FEI1 and FEI2 Expression, Localization, and Function in Hypocotyls and Flowers.
(A) to (H) Staining (blue) of transgenic lines harboring the promoter of FEI1 ([A] to [D]) or FEI2 ([E] to [H]) fused to GUS. (A), (C), (E), and (G) are from seedlings grown on MS media for 7 d. (B) and (F) show 3-d-old etiolated seedlings. (D) are (H) are flowers from plants grown in soil under long days for 3 weeks. Bars in (A) and (E) = 100 μm.
(I) Root tissue from plants expressing FEI1-myc or FEI2-myc was fractionated into soluble and microsome fractions. The total (T), soluble (S), and microsome (P) fractions were subjected to protein gel blotting and probed with an anti-C-myc (top) or anti-Hsc70 (bottom) antibody.
(J) Localization of FEI2-GFP fusion proteins. Top, Differential interference contrast (DIC) and GFP images of root cells from MS-grown seedlings; bottom, images from seedlings plasmolyzed in 0.8 M mannitol. Red arrows indicate regions of membrane that have detached from the cell wall.
(K) Images of hypocotyls from wild-type (left) and fei1 fei2 mutant (right) 3-day-old etiolated seedlings. Note that the fei1 fei2 hypocotyls are thicker.
(L) Transverse sections through hypocotyls of wild-type and fei1 fei2 mutant etiolated 3-day-old seedlings. Bar = 50 μm.
(M) Quantification of hypocotyl widths from etiolated seedlings. Asterisks indicate significant differences from the wild type (Student's t test, P < 0.05, n = 20). Error bars show se (n = 20).
(N) Stage 12 flowers from the indicated genotypes. Several petals and sepals were removed from each flower to reveal the inner parts. Note that the fei1 fei2 cob triple mutant has shorter stamen filaments. Bar = 1 mm.
Both FEI1 and FEI2 are most highly expressed in the root meristem and elongation zone, as revealed by promoter–β-glucuronidase (GUS) fusions (Figure 4). Published microarray analysis revealed that FEI1 and FEI2 are expressed at approximately equal levels in the different radial layers of the root tip, including the epidermis (Birnbaum et al., 2003). Extended staining of FEI promoter–GUS lines revealed a broader staining pattern for these two genes (Figure 4), similar to the pattern obtained from publicly available array data (Zimmermann et al., 2005).
FEI1 and FEI2 Function in Hypocotyls and Flowers
FEI1 and FEI2 both are expressed in the hypocotyls of etiolated seedlings (Figures 4B and 4F), which, like roots, are composed of cells that undergo primarily longitudinal expansion. Thus, we examined if the fei1 fei2 double mutant had defects in hypocotyl growth. The hypocotyls of etiolated seedlings of fei1 fei2 were significantly fatter than those of the wild type or single fei mutants (Figures 4K to 4M). However, contrary to the root phenotype of fei1 fei2, this was not accompanied by a decrease in the overall length of the hypocotyl (see Supplemental Figure 7 online), and this occurred in either low or high sucrose. This swollen-hypocotyl phenotype was complemented by transgenes containing genomic copies of either FEI1 or FEI2. The modest increase in the diameter of the fei1 fei2 mutant hypocotyls (Figure 4M) was substantially less than the increased width observed in the mutant roots. Examination of transverse sections of wild-type and mutant etiolated seedlings revealed that the increased diameter of the fei1 fei2 hypocotyls was associated with an increase in cell size, not cell number (Figure 4L). We did not observe a significant change from the wild type in the level or spatial distribution of lignin in the fei1 fei2 etiolated hypocotyls, as revealed by phloroglucinol staining (see Supplemental Figure 8 online). There was no obvious swelling in any other tissues of the fei1 fei2 mutant. However, the fei1 fei2 cob triple mutant (see below), but neither fei1 fei2 nor cob, had shortened stamen filaments, and this triple mutant was partially infertile (Figure 4N), indicating a role for the FEI kinases in these tissues. Consistent with this, analysis of the promoter-GUS fusions reveals expression of both FEI1 and FEI2 in stamen filaments (Figures 4D and 4H).
The fei1 fei2 Mutant Is Defective in Cellulose Biosynthesis
The altered pattern of cell expansion in the fei1 fei2 mutants suggests a defect in cell wall function. As cortical microtubules have been implicated in regulating anisotropic growth, we examined their arrangement in epidermal cells of wild-type and fei1 fei2 roots using an anti-α-tubulin antibody. In both wild-type and fei1 fei2 double mutant root cells, the microtubules in the elongation zone were aligned primarily transversely to the axis of growth 3 d after transfer to nonpermissive conditions (see Supplemental Figure 9 online). This indicates that growth anisotropy in the fei1 fei2 mutants is not the result of disruption of the pattern of microtubules.
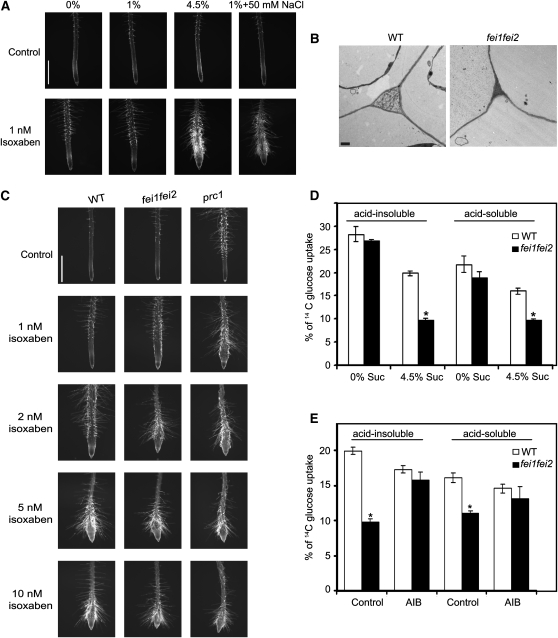
To begin to assess if the properties of the cell wall are altered in the mutant, we examined the effect of isoxaben, an inhibitor of cellulose synthase, on fei1 fei2. Growth in the presence of high sucrose rendered wild-type roots hypersensitive to isoxaben (Figure 5A), which indicates that elevated sucrose sensitizes roots to perturbations in cellulose synthesis. In the presence of low sucrose, both the prc1 mutant, which disrupts a catalytic subunit of cellulose synthase (CESA6) (Fagard et al., 2000), and fei1 fei2 displayed increased sensitivity to isoxaben (Figure 5C). This suggests that fei1 fei2 perturbs the biosynthesis or function of cellulose. Consistent with this, the roots of fei1 fei2 seedlings grown in nonpermissive conditions produced ectopic lignin (Figure 1H), which is generally correlated with a decreased level of crystalline cellulose (Humphrey et al., 2007). We further analyzed cellulose synthesis by measuring the incorporation of [14C]glucose into crystalline and noncrystalline cellulosic cell wall fractions of excised root tips. In permissive conditions, fei1 fei2 roots were similar to wild-type roots. However, in nonpermissive conditions, fei1 fei2 mutant roots displayed a striking defect in cellulose biosynthesis, as measured by the incorporation of labeled glucose into acid-insoluble material (crystalline cellulose; Peng et al., 2000) and acid-soluble material (noncrystalline cellulose and other wall polymers; Heim et al., 1998) (Figure 5D).
Figure 5.
The fei Mutants Affect Cell Wall Function.
(A) High sucrose or NaCl enhance the effect of isoxaben. Wild-type seedlings were grown on MS medium containing 0% sucrose for 4 d and then transferred to MS medium plus the indicated supplement in the presence of 0 (control) or 1 nM isoxaben as indicated. At 24 h after transfer, root tips were imaged. Bar = 1 mm.
(B) Transmission electron microscopy of cell wall junctions from wild-type and fei1 fei2 mutant epidermal root cells from seedlings grown on 4.5% sucrose. Bar = 1 μm.
(C) Response of the indicated seedlings to isoxaben. Seedlings of the indicated genotypes were grown on MS medium containing 0% sucrose for 4 d and then transferred to medium containing 1% sucrose and the indicated level of isoxaben. At 24 h after transfer, root tips were imaged. Bar = 1 mm.
(D) Incorporation of [14C]Glc into acid-soluble or acid-insoluble fractions from excised root tips from wild-type and fei mutant seedlings grown in 0% sucrose for 4 d and then transferred to 0% or 4.5% sucrose as indicated for 3 d. Asterisks indicate statistical differences between fei1 fei2 and the respective wild-type sample (Student's t test, P < 0.05). Values are means ± se from three biological replicates, and the experiments were repeated at least three times with similar results.
(E) Incorporation of [14C]Glc into acid-soluble or acid-insoluble fractions from excised roots from wild-type and fei mutant seedlings that were grown in 0% sucrose for 4 d and then transferred to 4.5% sucrose in the absence (control) or presence of AIB (1 mM) as indicated for 3 d. Asterisks indicate statistical differences between fei1 fei2 and the respective wild-type sample (Student's t test, P < 0.05). Values are means ± se from three biological replicates, and the experiments were repeated at least three times with similar results.
When viewed with a transmission electron microscope, the walls from the swollen root cells of the fei1 fei2 mutant were not appreciably altered in thickness compared with wild-type cell walls. However, the size of the intercellular spaces in the outer cell layer layers of the fei1 fei2 mutant roots was reduced in nonpermissive conditions (Figure 5B), similar to that in the sos5 and rsw1-20 mutants (Beeckman et al., 2002; Shi et al., 2003).
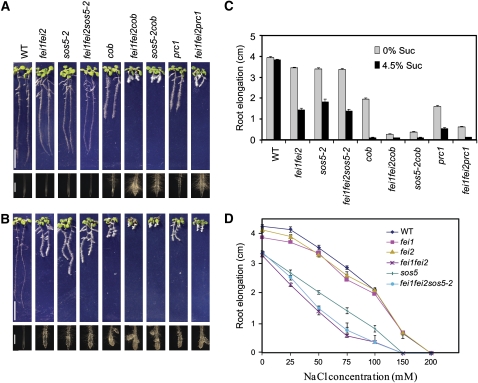
The COB gene encodes a GPI-anchored plant-specific protein of unknown function. Null cob mutants are extremely deficient in cellulose, are strongly dwarfed, and are sterile (Roudier et al., 2005). However, weak cob alleles, including the cob-1 allele used in this study, result in fertile plants that displayed a sucrose-dependent swollen-root phenotype (Figure 6). prc1-1, which is a likely null allele of CESA6, also displayed a sucrose-dependent swollen-root phenotype (Figure 6). We examined the genetic interactions of fei1 fei2 with cob and prc1. The fei1 fei2 cob and fei1 fei2 prc1 triple mutants displayed an enhanced root phenotype compared with the parental lines; the triple mutant roots were significantly shorter and more swollen in nonpermissive conditions (Figures 6B and 6C). Moreover, the fei1 fei2 cob and fei1 fei2 prc1 triple mutants displayed swollen roots even in permissive conditions, in which the single or double mutants did not display significant swelling (Figures 6A and 6C). These synergistic interactions suggest that FEI1 and FEI2 act in a pathway independent from COB or PRC1 to regulate cell wall function.
Figure 6.
Genetic Interaction of fei1 fei2 with Other Mutants Affecting Cell Elongation.
(A) and (B) Phenotypes of wild-type and various mutant seedlings grown in medium containing 0% sucrose for 4 d and then transferred to MS medium containing no (A) or 4.5% (B) sucrose for 4 d. Bars = 1 cm (top panels) and 1 mm (bottom panels; close-ups of root tips).
(C) Quantification of root elongation of various mutants grown and transferred as in (A) and (B). Values represent means of growth at 4 d after transfer to the indicated conditions. Error bars show se (n > 15).
(D) Quantification of total root elongation of the indicated lines at 4 d after transfer from MS medium containing 1% sucrose to the same medium with various levels of NaCl added. Error bars show se (n > 15).
sos5 was isolated as a mutant that displayed a swollen root tip in the presence of moderately high salt (Shi et al., 2003). The SOS5 gene encodes a putative cell surface adhesion protein with AGP-like and fasciclin-like domains. As the phenotype of sos5 is similar to that of the fei1 fei2 double mutant, we tested the effect of high sucrose on sos5 seedlings. Similar to fei1 fei2, growth of sos5-2 (a novel T-DNA insertion allele that is a null transcript; see Supplemental Figure 4 online) in the presence of high sucrose also resulted in a swollen-root phenotype (Figure 6B). By contrast, we did not observe a swollen-root phenotype in other sos mutants (sos1, sos2, sos3, and sos4) in response to elevated sucrose (see Supplemental Figure 10 online). Furthermore, etiolated sos5-2 seedlings displayed swollen hypocotyls similar to fei1 fei2 (Figure 4M). The roots of the sos5-2 fei1 fei2 triple mutant were indistinguishable from those of the fei1 fei2 double mutant in their response to increasing levels of NaCl (Figure 6). Likewise, the hypocotyl width of the sos5-2 fei1 fei2 triple mutant etiolated seedlings was comparable to that of the fei1 fei2 double mutant (Figure 4M). The nonadditive nature of sos5-2 and fei1 fei2 suggests that these gene products act in a linear pathway to regulate cell elongation.
ACS Plays a Role in FEI1/FEI2-Mediated Cell Expansion
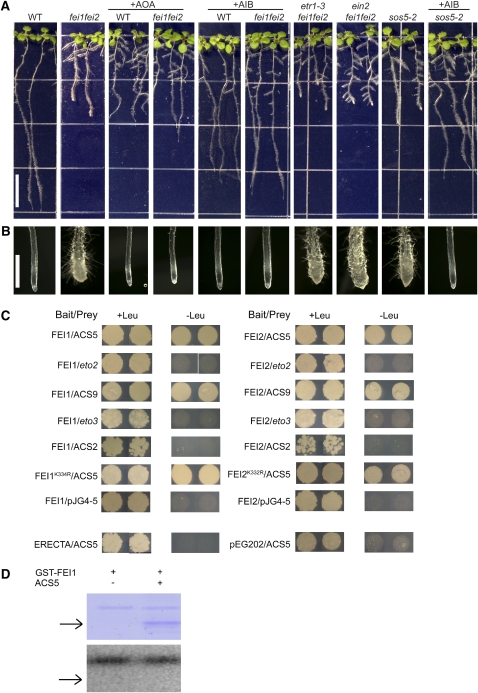
Ethylene plays an important role in regulating expansion in many plant cells, and inhibition of ethylene biosynthesis or perception can partially revert the swollen phenotypes of certain root morphology mutants, such as sabre (Aeschbacher et al., 1995) and cev1 (Ellis et al., 2002). We determined the effect of blocking ethylene biosynthesis on the fei1 fei2 swollen-root phenotype. α-Aminoisobutyric acid (AIB), which is a structural analog of ACC that blocks ACC oxidase activity by acting as a competitive inhibitor, reverted fei1 fei2 mutant roots grown in the presence of high sucrose or elevated NaCl to a nearly wild-type morphology (Figures 7A and 7B, Table 1; see Supplemental Figure 4B online). AIB also reverted the defect in cellulose synthesis in fei1 fei2 (Figure 5E). However, AIB did not revert the hypocotyl phenotype of fei1 fei2. Aminooxyacetic acid (AOA), which inhibits enzymes that require pyroxidal phosphate, including ACS, reverted the fei1 fei2 swollen-root phenotype (Figures 7A and 7B, Table 1). As AOA and AIB block ethylene biosynthesis by distinct mechanisms, it is unlikely that this phenotypic reversion of fei1 fei2 is due to off-target effects. Furthermore, this is not a general effect of AIB, as it did not revert the root-swelling phenotype of the cob mutant (see Supplemental Figure 11 online), even at higher concentrations (data not shown). Surprisingly, neither 1-methylcyclopropene (1-MCP) nor silver ion (silver thiosulfate), both of which block ethylene perception, had any appreciable effect on the root phenotype of fei1 fei2 mutants (Table 1). Likewise, neither etr1, which disrupts an ethylene receptor, nor ein2, a strong ethylene-insensitive mutant that acts downstream of ETR1, suppressed the fei1 fei2 root phenotype (Figure 7A, Table 1).
Figure 7.
Role of ACC/Ethylene on the fei Phenotype.
(A) Phenotypes of seedlings grown on MS medium containing 0% sucrose for 4 d and then transferred to MS medium containing 4.5% sucrose plus nothing, AOA (0.375 mM), or AIB (1 mM), as indicated. Bar = 1 cm. Note that the distribution of lateral roots in the fei1 fei2 mutants in the presence of high sucrose is variable; the architecture of the fei1 fei2 ein2 triple mutant is not substantially different from that of the fei1 fei2 parent.
(B) Close-ups of root tips from (A). Bar = 1 mm.
(C) Yeast two-hybrid interactions among the FEIs and ACSs. Bait and prey vectors containing the soluble kinase domains of the wild type or mutant FEI1 and FEI2 were cloned into a yeast two-hybrid bait vector and cotransformed into yeast with the indicated wild-type and eto mutant ACS preys. Positive interactions result in Leu prototrophy (growth on –Leu). The soluble kinase domains of ERECTA empty bait (pEG202) and prey (pJG4-5) vectors were used as controls.
(D) FEI1 does not phosphorylate ACS5 in vitro. Top, Coomassie blue–stained gel of purified GST-FEI and/or ACS5 protein; bottom, autoradiograph following an in vitro kinase assay. The arrows indicate the position of ACS5.
Table 1.
Root Elongation in the Absence or Presence of Ethylene Inhibitors
| Genotype | Control | +AIB | +Ag+ | +MCP |
|---|---|---|---|---|
| Wild type | 4.63 ± 0.07 | 3.67 ± 0.05 | 4.42 ± 0.05 | 4.03 ± 0.12 |
| fei1 fei2 | 1.62 ± 0.09 | 3.76 ± 0.05 | 0.99 ± 0.07 | 1.12 ± 0.08 |
| sos5-2 | 2.26 ± 0.12 | 3.68 ± 0.04 | 1.16 ± 0.09 | 1.35 ± 0.11 |
| fei1 fei2 sos5-2 | 1.50 ± 0.10 | 3.40 ± 0.04 | 0.77 ± 0.07 | 1.06 ± 0.11 |
| eto2 | 1.90 ± 0.05 | 2.78 ± 0.05 | 4.03 ± 0.06 | 3.49 ± 0.13 |
| cob | 0.12 ± 0.01 | 0.13 ± 0.01 | 0.12 ± 0.01 | nd |
| etr1-3 fei1 fei2 | 1.59 ± 0.09 | nd | nd | nd |
| ein2 fei1 fei2 | 0.98 ± 0.06 | nd | nd | nd |
| etr1-3 | 4.82 ± 0.05 | nd | nd | nd |
| ein2 | 4.84 ± 0.12 | nd | nd | nd |
Values represent means ± se of at least 15 root elongation measurements (in centimeters) between days 4 and 9. nd, not determined.
Consistent with the other similarities to the fei1 fei2 mutant, root swelling in sos5-2 seedlings grown in the presence of either high sucrose or elevated NaCl was reversed by AIB and AOA but not by blocking the response to ethylene (Figures 7A and 7B, Table 1; see Supplemental Figure 4 online). This suggests either that swelling in the absence of FEI depends on a hitherto undiscovered pathway for ethylene perception or that ACC itself acts as a signaling molecule.
We tested if the FEIs interacted with ACS using a yeast two-hybrid assay. The kinase domains of both FEI1 and FEI2 interacted with both ACS5 and ACS9, two type 2 ACS proteins (Chae and Kieber, 2005). By contrast, neither FEI1 nor FEI2 interacted with ACS2 (Figure 7C), which belongs to a distinct subclade of ACS proteins (type 1) that have divergent C-terminal domains (Chae and Kieber, 2005). Likewise, the eto2 and eto3 mutations, which alter the C-terminal domains of ACS5 and ACS9, respectively, and which block the rapid degradation of these proteins in vivo, disrupted the interaction with FEI1 and FEI2 in the yeast two-hybrid interaction (Figure 7C). Disruption of the kinase activity did not affect the interaction with ACS, as both FEI1K334R and FEI2K332R interacted with ACS5. By contrast, the ERECTA kinase domain did not interact with ACS5 in a yeast two-hybrid assay, indicating that there is specificity in the interaction with ACS5. We failed to detect an interaction between FEI1 and FEI2 with either themselves or with each other in a yeast two-hybrid assay.
We next tested the ability of FEI1 to phosphorylate purified ACS5. We were not able to detect phosphorylation of ACS5 in vitro by purified, catalytically active FEI1 (Figure 7D). The purified ACS5 used in this analysis was enzymatically active and could be phosphorylated in our conditions by a partially purified soybean (Glycine max) Ca2+-dependent protein kinase (data not shown), which had been shown previously to phosphorylate ACS (Tatsuki and Mori, 2001; Sebastià et al., 2004). Thus, the lack of phosphorylation of ACS5 by FEI1 in this analysis is not likely the result of misfolding of ACS5.
Measurements of ethylene production revealed that root tissues from wild-type and fei1 fei2 mutant seedlings grown on low or high sucrose in the light made comparable amounts of ethylene (8.9 ± 0.8 pL·cm−1 root segment−1·d−1 for the wild type versus 11.9 ± 0.2 pL·cm−1 root segment·d−1 for fei1 fei2). Likewise, ethylene production in dark-grown fei1 fei2 seedlings was similar to that in the wild type (5.6 ± 0.3 pL·seedling−1·d−1 for wild-type seedlings versus 5.8 ± 0.7 pL·seedling−1·d−1 for fei1 fei2). Thus, the FEIs do not appear to affect the overall level or catalytic activity of ACS.
DISCUSSION
FEIs Are Required for Anisotropic Growth in the Root
We show that the FEI1 and FEI2 LRR-RLKs are necessary for anisotropic cell expansion in Arabidopsis root cells and also play a role in cell expansion in stamen filaments and the hypocotyls of etiolated seedlings. Biochemical studies and genetic analyses with other cellulose-deficient mutants reveal that these FEI kinases modulate cell wall function, including positively regulating the biosynthesis of cellulose, a wall component necessary for anisotropic expansion. Two other divergent RLKs have been implicated in cell wall function: the WAK and THE1 kinases. The WAK kinases are involved in cell expansion in various Arabidopsis tissues, and their extracellular domains are tightly linked to the cell wall (Anderson et al., 2001; Wagner and Kohorn, 2001). Interestingly, wak2 mutants display reduced cell expansion that is sensitive to the level of sugar and salt in the medium (Kohorn et al., 2006). However, in contrast with fei1 fei2, high sugar levels suppress the cell expansion defect in wak2, and it is the extent, not the orientation, of cell expansion that is altered in wak2. THE1 has been hypothesized to be involved in monitoring the cell wall integrity, as the1 mutations suppress the short-hypocotyl phenotype, but not the cellulose-deficient phenotype, of prc1 (Hématy et al., 2007). This is distinct from the FEIs, as the fei1 fei2 double mutant significantly impairs cellulose biosynthesis. The the1 mutation also suppresses some, but not all, other mutants affecting cell expansion. Alteration of THE1 function does not have an effect in the wild-type background, suggesting perhaps genetic redundancy or that it plays a role only in conditions in which cell wall integrity is compromised. While it would be interesting to determine the interaction between the1 and fei1 fei2, the lack of suppression of the root-elongation phenotype of prc1 by the1 may render this genetic interaction noninformative.
Similar to THE1, the FEIs may also sense cell wall signals and in turn provide feedback to the cellulose biosynthesis machinery. One potential ligand for the FEIs is the extracellular protein SOS5. SOS5 encodes a putative cell surface adhesion protein that is required for normal cell expansion (Shi et al., 2003). Several lines of evidence suggest that SOS5 functions in a linear pathway with the FEIs: (1) sos5 mutants have a very similar root-elongation phenotype to fei1 fei2, including the dependence on sucrose and salt; (2) the root-swelling phenotypes of both fei1 fei2 and sos5-2 are suppressed by AIB and AOA but not by blocking the known ethylene response pathway; (3) both fei1 fei2 and sos5-2 display a thickened-hypocotyl phenotype; (4) the fei1 fei2 and sos5-2 mutations show a nonadditive genetic interaction; and (5) the patterns of expression of the FEIs and SOS5 are largely overlapping (Figure 4) (Shi et al., 2003). Thus, SOS5 acts on the same pathway as the FEIs to mediate the function of the cell wall. As SOS5 encodes an extracellular protein, it is possible that it acts as, or is involved in the production or presentation of, a FEI ligand.
In addition to fei1 fei2 and sos5-2, the cob and prc1 mutants also display root swelling that is dependent on the concentration of sucrose in the medium (Figure 6). It has been proposed that this conditional phenotype reflects defects that are apparent only at high rates of cell elongation, such as in the presence of sucrose (Benfey et al., 1993). However, our data do not support this hypothesis, as increasing sucrose above 1% actually led to a slight decrease in the rate of root elongation, at least in our growth conditions, but the root-swelling phenotype of both fei1 fei2 and sos5-2 continued to intensify. Furthermore, low levels of NaCl, which reduce the rate of root elongation, also caused swelling in the sos5-2 and fei1 fei2 mutants. The effect of sucrose/salt on fei1 fei2 mutants is not the result of increased osmotic potential of the medium, as high levels of sorbitol or mannitol do not induce the phenotype. Our results indicate that wild-type plants are more susceptible to perturbation of cellulose biosynthesis in the presence of high sucrose or salt. How these conditions affect the function of the cell wall remains to be determined.
Kinase Activity Is Dispensable for FEI Function
Consistent with their sequences, the FEIs have intrinsic kinase activity; however, kinase activity is not essential for FEI function, at least for the phenotypes that we observed. There are many examples of so-called pseudokinases (reviewed in Kroiher et al., 2001; Boudeau et al., 2006), which display clear homology to kinases but lack conservation of one or more of the catalytic residues in the kinase core. Pseudokinases are especially prevalent in plant genomes, and it has been estimated that ∼20% of Arabidopsis RLKs are kinase-deficient (Castells and Casacuberta, 2007). For example, STRUBBELIG (SUB), a member of the LRR-RLKs (class V) that is involved in the development of multiple organs, includes two alterations in residues that are highly conserved in functional kinases, and genetic and biochemical analyses indicate that the SUB kinase domain is catalytically inactive (Chevalier et al., 2005). The Arabidopsis homolog of CR4 RLK encodes an active kinase, but disruption of the kinase catalytic domain by site-directed mutagenesis does not disrupt its function in vivo (Gifford et al., 2005), similar to what we observed for FEI1 and FEI2. One model for how the FEIs and other kinase-deficient RLKs signal is that they heterodimerize with, and are then transphosphorylated by, a kinase-active member of the same protein family. An alternative possibility it that FEI signaling does not involve phosphorylation but, rather, the proteins act as scaffolds to localize other components in a protein complex or to a particular place in the cell. An example of this is the human Kinase Suppressor of Ras (KSR) protein, which is similar in sequence to protein kinases but which acts as a scaffold protein that coordinates the assembly of a multiprotein mitogen-activated protein kinase complex at the membrane (Claperon and Therrien, 2007). In any case, the kinase activity of the FEIs, while not essential, is clearly required for optimal function, as only a subset of the fei1 fei2 double mutant transformants harboring the catalytically inactive version of the FEIs were fully complemented. As kinase activity is not required for function, it is possible that the fei1 allele used in this study is not a functional null, as there is a truncated FEI1 transcript present. The similarity in the strength of the phenotype of fei1 fei2 to sos5-2, a null allele in a gene acting on the same pathway as the FEIs, argues somewhat against this.
Role of ACS5 in the SOS5/FEI Pathway
What role do ACS5 and other type 2 ACS enzymes play in regulating cell wall function in the root? ACS5 has been shown to be an enzymatically active ACS (Yamagami et al., 2003), the product of which is ACC, the immediate precursor for ethylene. Ethylene has been shown to play a role in regulating anisotropic growth. In hypocotyls, ethylene inhibits elongation primarily by altering the orientation of cell elongation, which is correlated with a change in the orientation of the microtubules (Steen and Chadwick, 1981; Lang et al., 1982; Roberts et al., 1985; Takahashi et al., 2003). In the root, ethylene strongly inhibits root elongation, but radial expansion is only modestly increased and microtubules appear to be unaffected (Baskin and Williamson, 1992). Thus, in the root, ethylene appears to primarily inhibit the overall amount of cell expansion, not its orientation. One potential mechanism for this is the elevation of ROS levels in the elongation zone of Arabidopsis roots in response to ACC, which leads to the cross-linking of Hyp-rich glycoproteins and callose deposition in the cell wall, both of which may contribute to reduced cell expansion (De Cnodder et al., 2005).
There are several mutants that affect growth anisotropy in the root that are linked to ethylene, including sabre, cev1, and lue1 (Aeschbacher et al., 1995; Ellis et al., 2002; Bouquin et al., 2003). cev1, a mutation in the cellulose synthase CesA3 gene, produces elevated levels of ethylene, and its phenotype is partially suppressed by mutations that disrupt ethylene signaling (Ellis et al., 2002). Similar to fei1 fei2, the swollen-root phenotype of the sabre mutant can be partially rescued by blocking ethylene action through the use of ethylene biosynthesis inhibitors, and the sabre mutant does not display an increase in ethylene biosynthesis (Aeschbacher et al., 1995). However, in contrast with fei1 fei2, sabre also can be rescued by inhibition of ethylene perception or by etr1. The authors propose that ethylene and SABRE counteract each other to regulate the degree of radial expansion of root cells. However, neither ethylene-overproducing mutants nor constitutive ethylene-signaling mutants have such a dramatic swollen-root phenotype, which would be predicted from such a model.
The interaction of type 2 ACSs with the FEIs and the reversion of the fei1 fei2 mutant by inhibitors of ethylene biosynthesis strongly suggest a link between ACS function and altered cell wall function in fei mutant roots. However, several lines of evidence indicate that this is not the result of altered ethylene levels: (1) mutants that increase or decrease ethylene biosynthesis do not show a root-swelling phenotype (Vogel et al., 1998); (2) the fei phenotype cannot be reversed by blocking ethylene perception; and (3) in nonpermissive conditions, ethylene production is not substantially altered in fei1 fei2 mutant roots. Thus, we conclude that the FEIs do not alter ACS activity or levels and that the FEIs do not act via ethylene. How, then, does ACS function in the FEI pathway, and how do the FEIs affect ACS function?
One possibility is that the ACS protein may perform a function distinct from the production of ACC. There are multiple examples of such so-called moonlighting proteins (Moore, 2004). However, if this were the case, it would not explain the reversion of fei1 fei2 by AIB, which is a structural analog of ACC that should not directly affect ACS function. A second model is that perhaps fei1 fei2 alter ethylene biosynthesis in a small number of critical cells, which may not be detectable in our analysis, and this elevated ethylene may be perceived by a second, independent ethylene response pathway that functions in this developmental context. This model is possible, but two lines of evidence argue somewhat against it. First, it would not explain the lack of root swelling in various ethylene biosynthesis mutants; second, it is probable that, similar to ETR1 and its paralogs, any additional ethylene receptor would be blocked by silver ion (Burg and Burg, 1967), and thus silver should, but does not, revert the fei1 fei2 phenotype. A final model that is consistent with the data is that ACC itself, rather than ethylene, acts as a signaling molecule to regulate cell expansion in the FEI/SOS5 pathway. In such a scenario, AIB, which is a structural analog of ACC, would act as a competitive inhibitor to block binding to a hypothetical ACC receptor. Disruption of ethylene binding would not affect this response, and there would be no alteration in ethylene levels in the mutant. The data are most consistent with this model, in which ACC acts as a signal, but additional studies are required to confirm this.
What is the nature of the interaction of the FEI and ACS proteins? The FEI proteins do not appear to phosphorylate ACS5, which is consistent with the lack of requirement for kinase activity for FEI function. Furthermore, ethylene levels are not altered in fei1 fei2 mutants, suggesting that there is no change in ACS levels or activity. One model consistent with the data is that the FEIs act as a scaffold to localize a fraction of ACS protein to a subdomain of the plasma membrane and/or to assemble ACS into a protein complex. This would be similar to KSR, a protein kinase that acts as a scaffold in a mitogen-activated protein kinase cascade. This localized ACS would then generate a localized signal to regulate cell wall biosynthesis.
We propose that the FEI kinases play a role in regulating cell wall architecture, possibly mediating interactions between the cell wall and intracellular signaling pathways. The FEI RLKs may act as a scaffold to localize ACS or may complex ACS with other proteins. The extracellular SOS5 protein also feeds into this pathway. Exactly how ACS functions in this pathway, and how this pathway interacts with the biosynthetic machinery of the cell wall and with other regulatory inputs into cell wall function, are important questions for the future.
METHODS
Plant Material
The Columbia (Col-0) ecotype of Arabidopsis thaliana was used in this study. The fei1 insertion (SALK_080073) (Alonso et al., 2003) was localized to position +2599 (relative to the translational start site). The fei2-1 insertion was isolated by PCR screening (using primers FEI2-S5, FEI1-A5, and T-DNA left border primer; see Supplemental Table 1 online) of a T-DNA insertion library made in a Col-0 gl1 line (http://www.dartmouth.edu/∼tjack/et.html). The fei2-1 insertion was localized to position +2012. The fei2-2 insertion (SALK_044226) (Alonso et al., 2003) was localized to position +3386. The insertion sites all were confirmed by DNA sequencing of PCR-amplified products using gene-specific and left border primers (see Supplemental Table 1 online) from the respective lines. The fei1 fei2-1 double mutant line was used in all experiments unless noted otherwise. The sos5-2 (SALK_125874) (Alonso et al., 2003) and ein2-50 (SALK_106282) (Alonso et al., 2003) alleles were obtained from the SALK T-DNA insertion collection. The cob-1 and prc1-1 alleles used in this study were obtained from the Arabidopsis Stock Center. The eto2 (Kieber et al., 1993) and etr1-3 (Chang et al., 1993) mutants have been described previously.
Growth Conditions and Measurements
For growth in soil, plants were grown at 23°C in ∼75 μE constant light. For growth in vitro, seeds were surface-sterilized and cold-treated at 4°C for 3 d in the dark and then treated with white light for 3 h. Seedlings were grown on vertical plates containing 1× Murashige and Skoog (MS) salts, 1% sucrose, and 0.6% phytagel (Sigma-Aldrich) at 22°C in ∼100 μE constant light. For measurements of root elongation, seedlings were grown for 4 d on vertical plates containing no sucrose or in some cases 1% sucrose, as noted in the figure legends, and then transferred to MS medium supplemented with the indicated additions. For the ethylene inhibitor studies, AIB (1 mM), AOA (0.375 mM), and MCP (20 mg Ethylbloc; Floralife) were added to a 6-liter container or silver thiosulfate (0.02 mM) was added to the high-sucrose MS agar.
RT-PCR
Total RNA was isolated from 7-d-old seedlings using the RNeasy kit (Qiagen). First-strand cDNA was synthesized from 1 μg of the total RNA pretreated with RNase-free DNase (Promega) using the SuperScript II kit (Invitrogen) with random hexamers, according to the manufacturer's instructions. Quantitative RT-PCR was performed with SYBR Premix Ex-Taq according to the manufacturer's instructions (Takara Bio) using gene-specific primers (see Supplemental Table 1 online).
FEI Constructs and Transgenic Plants
Genomic fragments comprising the entire coding region of FEI1 or FEI2 and 1 kb of the respective 5′ flanking DNA were amplified from BAC T8E3 or T20F21 DNA, respectively, by PCR (primers FEI1-S7/FEI1-A3 and FEI2-S7/FEI2-A4; see Supplemental Table 1 online) using Pfu DNA polymerase as described by the manufacturer (Stratagene), and the fragments were cloned into pENTR-TOPO-D (Invitrogen). The resultant entry plasmid was used in an LR reaction (as described by the manufacturer; Invitrogen) to introduce the respective genes into the binary pGWB16 (Nakagawa et al., 2007) vector for complementation. The kinase domain of FEI1 were amplified from cDNA by RT-PCR using first-strand cDNA generated from wild-type Col RNA and gene-specific primers (FEI1-C2/FEI1-A5; see Supplemental Table 1 online). Kinase-deficient versions of FEI1 or FEI2 were obtained by site-directed mutagenesis using primers containing the desired point mutation (FEI1-M2F/FEI1-M2R and FEI2-M2F/FEI2-M2R; see Supplemental Table 1 online). For expression of a GFP fusion protein, a FEI2 genomic fragment (amplified using primers FEI2-S/FEI2-A4; see Supplemental Table 1 online) was cloned into pENTR-TOPO-D (Invitrogen) and then introduced into the binary vector pGWB5 (Nakagawa et al., 2007). For promoter-GUS fusions, genomic fragments comprising 3 kb of 5′ flanking DNA of FEI1 or FEI2 were amplified from wild-type genomic DNA (using primers FEI1-PROM-F1/FEI1-PROM-R1 and FEI2-PROM-F2/FEI2-PROM-R2; see Supplemental Table 1 online), cloned into pENTR4 vector, and then introduced into the binary vector pGWB2 (Nakagawa et al., 2007). All clones were confirmed by DNA sequencing. The resulting plasmids were transformed into Agrobacterium tumefaciens strain GV3101. Transgenic plants were generated by the floral dip method (Clough and Bent, 1998) and selected on MS medium containing 50 mg/L kanamycin and 30 mg/L hygromycin. All destination binary vectors were kindly provided by Tsuyoshi Nakagawa from the Research Institute of Molecular Genetics in Matsue, Japan.
Protein Kinase Assays
The FEI1 and FEI1K334R kinase domains in pENTR-TOPO-D (see above) were introduced into the plasmid pDEST15 by Gateway cloning (Invitrogen). The respective GST fusion proteins were isolated using Glutathione Sepharose 4 Fast Flow medium according to the manufacturer's directions (Amersham Biosciences). ACS5 was purified as described (Chae et al., 2003). Myelin basic protein was purchased from Sigma-Aldrich. The in vitro kinase assays were performed in kinase reaction buffer (50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 10 mM MnCl2, 10 mM DTT, 10 μM ATP, and 5 μCi of [γ-32P]ATP [2 mCi/mL; Perkin-Elmer Life Science]). The reaction was incubated at room temperature for 1 h and then terminated by adding 10 μL of 6× SDS sample buffer. The reaction was then incubated at 97°C for 5 min and run on 12% SDS-PAGE. The gel was stained with Coomassie Brilliant Blue G 250, dried, and subjected to autoradiography.
Phloroglucinol Staining
Phloroglucinol staining was performed as described (Caño-Delgado et al., 2003). Seedlings were fixed in a solution of three parts ethanol to one part acetic acid and then cleared in a solution of chloral hydrate:glycerol:water (8:1:2). The seedlings were then stained with lignin in a 2% phloroglucinol-HCl solution.
Analysis of FEI Expression Patterns
Tissue from transgenic lines harboring the FEI1 or FEI2 promoter–GUS fusions was stained in 100 mM sodium phosphate buffer (pH 7.0) with 10 mM EDTA (pH 8.0), 0.5 mM potassium ferricyanide, 0.5 mM potassium ferrocyanide, 1 mM 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, and 0.1% Triton X-100. The tissue was stained either for 1 h or overnight at 37°C as indicated. Chlorophyll was removed with 95% ethanol. Ten independent transgenic lines were analyzed, and a representative line was photographed.
Localization of FEI2-GFP
Root apices from 7-d-old transgenic plants harboring 35S:FEI2-GFP were used for confocal analyses. A Zeiss LSM510 confocal microscope filtered with a FITC10 set (excitation at 488 nm with emission at 505 to 530 nm and 530 to 560 nm) was used for this analysis. Mannitol (0.8 M) was applied to the root tip on the slides for plasmolysis.
Membrane Fractionation of FEI1-myc Fusion Proteins
FEI1-myc and FEI2-myc homozygous transgenic lines were grown on 1% sucrose MS plates for 7 d. Membrane proteins were fractionated by grinding 200 mg of root tissue per 500 μL of buffer (20 mM Tris [pH 8.0], 0.33 M sucrose, 1 mM EDTA, and plant protease inhibitor cocktail [Roche Applied Science]), and insoluble debris was pelleted by centrifugation at 2000g for 10 min at 4°C. The supernatant from the spin was designated the total fraction. Some of the total fraction (150 μL) was further centrifuged at 20,000g for 45 min at 4°C. The supernatant from this spin was designated the soluble fraction, and the pellet was resuspended in 100 μL of buffer to form the microsome fraction. Proteins were separated by 12% SDS-PAGE and analyzed by protein gel blotting. The anti-myc antibody was obtained from Roche Applied Science. Anti-Hsc antibody used as a loading control was obtained from Stressgen, and chicken anti-mouse secondary antibody was obtained from Santa Cruz Biotechnology.
Cellulose Synthesis Assays
Cellulose synthesis was determined by [14C]Glc labeling as described (Fagard et al., 2000) with the following modifications. Seedlings were grown on 0% sucrose MS plates for 4 d and then transferred to MS medium containing various supplements (as indicated in the figure legends) for 3 d. Root tips (1.5 cm) were cut and washed three times with 3 mL of glucose-free MS medium. Forty root tips were then incubated in 1 mL of MS medium containing 0.1 μCi/mL [14C]Glc (NEN Research) for 1 h in the dark at 22°C in glass tubes. After treatment, the roots were washed three times with 3 mL of glucose-free MS medium. Next, the roots were extracted three times with 3 mL of boiling absolute ethanol for 20 min, and total aliquots were collected (ethanol-soluble fraction). Roots were then resuspended in 3 mL of chloroform:methanol (1:1, v/v), extracted for 20 min at 45°C, and finally resuspended in 3 mL of acetone for 15 min at room temperature with gentle shaking. The remaining material was resuspended in 500 μL of an acetic acid:nitric acid:water solution (8:1:2, v/v/v) for 1 h in a boiling-water bath. Acid-soluble material and acid-insoluble material were separated by glass microfiber filters (GF/A; 2.5 cm diameter; Whatman), after which the filters were washed with 5 mL of water. The acid wash and water wash constitute the acid-soluble fraction. The filters yield the acid-insoluble fraction. The amount of label in each fraction was determined by scintillation counting using liquid scintillation fluid (Scintiverse BD cocktail; SX 18-4; Fisher). The percentage of label incorporation was expressed as 100× the ratio of the amount of label in each fraction to the total amount of label (ethanol plus acid-soluble plus acid-insoluble fractions).
Microscopy
Arabidopsis root tips were fixed in 2% paraformaldehyde and 2.5% glutaraldehyde in phosphate buffer (0.1 M sodium phosphate, pH 7.4). After rinsing with phosphate buffer, the samples were postfixed with 1% osmium tetroxide in sodium phosphate buffer for 30 min. Samples were dehydrated through an increasing ethanol series followed by propylene oxide and infiltrated and embedded in Polybed 812 epoxy resin (Polysciences). For light microscopy, 1-μm cross sections of the root tips were cut using a glass knife and a Leica Ultracut S ultramicrotome (Leica Microsystems), mounted on glass slides, and stained with 1% toluidine blue in 1% borax. For transmission electron microscopy, selected blocks were further trimmed and ultrathin sections (70 nm) were cut using a diamond knife. Ultrathin sections were mounted on 200-mesh copper grids and stained with 4% uranyl acetate and Reynolds' lead citrate. Sections were examined using a LEO EM-910 transmission electron microscope operating at 80 kV (Carl Zeiss), and digital images were taken using an Orius SC1000 CCD camera (Gatan).
Whole root tips were visualized by first fixing in an ethanol:acid (9:1) solution overnight, followed by two washes in 90 and 70% ethanol. Roots were then cleared with a chloral hydrate:glycerol:water solution (8:1:2), and the tips were visualized using Nomarski optics using a Nikon Eclipse 80i microscope.
Analysis of Microtubules
Seedlings were grown for 5 d on 1% sucrose and then transferred onto plates containing 1% sucrose, 4.5% sucrose, or 1% sucrose plus 50 mM NaCl for 3 d. Seedlings were fixed, stained for microtubules, and imaged, all as described (Bannigan et al., 2006). Briefly, the fixative contained 4% paraformaldehyde, 1% glutaraldehyde, 50 mM PIPES, and 1 mM CaCl2. Seedlings were permeabilized by mild digestion of pectin and brief incubation in ice-cold methanol. After rehydration in PBS, roots were incubated with 1:1000 mouse monoclonal anti-tubulin antibody (Sigma-Aldrich) at 37°C overnight. The secondary antibody used was Cy3-conjugated goat anti-mouse antibody (1:200; Jackson ImmunoResearch). The imaging of whole roots was performed using a confocal microscope (510 Meta; Carl Zeiss) equipped with a 63× oil-immersion objective. Projections were assembled using Zeiss software.
Measurement of Ethylene Production
Approximately 30 seedlings were grown on 1% sucrose MS plates for 3 d and then transferred to 4.5% sucrose plates for 3 d. Root tips (1 cm) were excised and placed in 22-mL gas chromatography vials that contained 3 mL of 4.5% liquid MS medium. The vials were capped and incubated for 24 h at 23°C in the dark, and the accumulated ethylene was measured as described by Vogel et al. (1998). For etiolated tissue, seedlings (∼40 per vial) were grown in 22-mL gas chromatography vials containing 3 mL of MS medium in the dark for 4 d. The accumulated ethylene was measured by gas chromatography as described (Vogel et al., 1998).
Yeast Two-Hybrid Analysis
The open reading frames corresponding to the various tested genes were cloned into the bait plasmid (pEG202) or prey plasmid (pJG4-5) by Gateway cloning from the respective entry clones made with the primers shown in Supplemental Table 1 online. The plasmids were transformed into the yeast strain EGY48 via LiOAc transformation as described (Chen et al., 1992).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: FEI1, At1g31420; FEI2, At2g35620; and SOS5, At3g46550.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sequence Alignment of FEI1 and FEI2.
Supplemental Figure 2. Time Course of Root Swelling following Transfer from 0 to 4.5% Sucrose Media.
Supplemental Figure 3. Transverse Sections through the Elongation Zone of the Root from Various Single, Double, and Triple Mutants.
Supplemental Figure 4. Analysis of the sos5-2 Mutant.
Supplemental Figure 5. The fei1 fei2 Mutant Phenotype in Response to Sucrose Is Not the Result of Increased Osmoticum.
Supplemental Figure 6. The FEI2-GFP Fusion Is Functional.
Supplemental Figure 7. Hypocotyl Length Is Not Affected in the fei Mutants.
Supplemental Figure 8. Phloroglucinol Staining for Lignin in Wild-Type and fei1 fei2 Seedlings Grown on MS Medium for 3 d in the Dark.
Supplemental Figure 9. Organization of Microtubules Is Not Altered in the fei1 fei2 Mutant.
Supplemental Figure 10. Growth in the Presence of Elevated Sucrose Does Not Affect Other sos Mutants.
Supplemental Figure 11. Effect of Inhibition of Ethylene Biosynthesis on the cob Mutant.
Supplemental Table 1. Primers Used in This Study.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation Grant IOB-0444347 to J.J.K. We thank Victoria Madden and Elena Davis for help in transmission electron microscopy and Jason Reed, Jayson Punwani, and Cris Argueso for critically reading the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Joseph J. Kieber (jkieber@unc.edu).
Online version contains Web-only data.
References
- Aeschbacher, R., Hauser, M.-T., Feldmann, K.A., and Benfey, P.N. (1995). The SABRE gene is required for normal cell expansion in Arabidopsis. Genes Dev. 9 330–340. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Anderson, C.M., Wagner, T.A., Perret, M., He, Z.H., He, D., and Kohorn, B.D. (2001). WAKs: Cell wall-associated kinases linking the cytoplasm to the extracellular matrix. Plant Mol. Biol. 47 197–206. [PubMed] [Google Scholar]
- Bannigan, A., Wiedemeier, A.M.D., Williamson, R.E., Overall, R.L., and Baskin, T.I. (2006). Cortical microtubule arrays lose uniform alignment between cells and are oryzalin resistant in the Arabidopsis mutant, radially swollen 6. Plant Cell Physiol. 47 949–958. [DOI] [PubMed] [Google Scholar]
- Baskin, T.I. (2001). On the alignment of cellulose microfibrils by cortical microtubules: A review and a model. Protoplasma 215 150–171. [DOI] [PubMed] [Google Scholar]
- Baskin, T.I. (2005). Anisotropic expansion of the plant cell wall. Annu. Rev. Cell Dev. Biol. 21 203–222. [DOI] [PubMed] [Google Scholar]
- Baskin, T.I., and Williamson, R.E. (1992). Ethylene, microtubules and root morphology in wild-type and mutant Arabidopsis seedlings. Plant Biochemistry and Physiology Symposium 11 118–130. [Google Scholar]
- Beeckman, T., Przemeck, G.K.H., Stamatiou, G., Lau, R., Terryn, N., De Rycke, R., Inze, D., and Berleth, T. (2002). Genetic complexity of cellulose synthase A gene function in Arabidopsis embryogenesis. Plant Physiol. 130 1883–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey, P.N., Linstead, P.J., Roberts, K., Schiefelbein, J.W., Hauser, M.-T., and Aeschbacher, R. (1993). Root development in Arabidopsis: Four mutants with dramatically altered root morphogenesis. Development 119 57–70. [DOI] [PubMed] [Google Scholar]
- Birnbaum, K., Shasha, D.E., Wang, J.Y., Jung, J.W., Lambert, G.M., Galbraith, D.W., and Benfey, P.N. (2003). A gene expression map of the Arabidopsis root. Science 302 1956–1960. [DOI] [PubMed] [Google Scholar]
- Boudeau, J., Miranda-Saavedra, D., Barton, G.J., and Alessi, D.R. (2006). Emerging roles of pseudokinases. Trends Cell Biol. 16 443–452. [DOI] [PubMed] [Google Scholar]
- Bouquin, T., Mattsson, O., Naested, H., Foster, R., and Mundy, J. (2003). The Arabidopsis lue1 mutant defines a katanin p60 ortholog involved in hormonal control of microtubule orientation during cell growth. J. Cell Sci. 116 791–801. [DOI] [PubMed] [Google Scholar]
- Burg, S., and Burg, E. (1967). Molecular requirements for the biological activity of ethylene. Plant Physiol. 42 144–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caño-Delgado, A., Penfield, S., Smith, C., Catley, M., and Bevan, M. (2003). Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J. 34 351–362. [DOI] [PubMed] [Google Scholar]
- Castells, E., and Casacuberta, J.M. (2007). Signalling through kinase-defective domains: The prevalence of atypical receptor-like kinases in plants. J. Exp. Bot. 58 3503–3511. [DOI] [PubMed] [Google Scholar]
- Chae, H.S., Faure, F., and Kieber, J.J. (2003). The eto1, eto2, and eto3 mutations and cytokinin treatment increase ethylene biosynthesis in Arabidopsis by increasing the stability of ACS protein. Plant Cell 15 545–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae, H.S., and Kieber, J.J. (2005). Eto Brute? The role of ACS turnover in regulating ethylene biosynthesis. Trends Plant Sci. 10 291–296. [DOI] [PubMed] [Google Scholar]
- Chang, C., Kwok, S.F., Bleecker, A.B., and Meyerowitz, E.M. (1993). Arabidopsis ethylene-response gene ETR1: Similarity of product to two-component regulators. Science 262 539–544. [DOI] [PubMed] [Google Scholar]
- Chen, D.C., Yang, B.C., and Kuo, T.T. (1992). One-step transformation of yeast in stationary phase. Curr. Genet. 21 83–84. [DOI] [PubMed] [Google Scholar]
- Chevalier, D., Batoux, M., Fulton, L., Pfister, K., Yadav, R.K., Schellenberg, M., and Schneitz, K. (2005). STRUBBELIG defines a receptor kinase-mediated signaling pathway regulating organ development in Arabidopsis. Proc. Natl. Acad. Sci. USA 102 9074–9079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claperon, A., and Therrien, M. (2007). KSR and CNK: Two scaffolds regulating RAS-mediated RAF activation. Oncogene 26 3143–3158. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Darley, C.P., Forrester, A.M., and McQueen-Mason, S.J. (2001). The molecular basis of plant cell wall extension. Plant Mol. Biol. 47 179–195. [PubMed] [Google Scholar]
- De Cnodder, T., Vissenberg, K., Van Der Straeten, D., and Verbelen, J.P. (2005). Regulation of cell length in the Arabidopsis thaliana root by the ethylene precursor 1-aminocyclopropane-1-carboxylic acid: A matter of apoplastic reactions. New Phytol. 168 541–550. [DOI] [PubMed] [Google Scholar]
- Desprez, T., Vernhettes, S., Fagard, M., Refrégier, G., Desnos, T., Aletti, E., Py, N., Pelletier, S., and Höfte, H. (2002). Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiol. 128 482–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, C., Karafyllidis, I., Wasternack, C., and Turner, J.G. (2002). The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard, M., Desnos, T., Desprez, T., Goubet, F., Refregier, G., Mouille, G., McCann, M., Rayon, C., Vernhettes, S., and Hofte, H. (2000). PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12 2409–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford, M.L., Robertson, F.C., Soares, D.C., and Ingram, G.C. (2005). ARABIDOPSIS CRINKLY4 function, internalization, and turnover are dependent on the extracellular crinkly repeat domain. Plant Cell 17 1154–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, P.B. (1980). Organogenesis—A biophysical view. Annu. Rev. Plant Physiol. 31 51–82. [Google Scholar]
- Hanks, S.K., and Quinn, A.M. (1991). Protein kinase catalytic domain sequence database: Identification of conserved features of primary structure and classification of family members. Meth. Enzymol. 200 38–62. [DOI] [PubMed] [Google Scholar]
- He, Z.-H., Fujiki, M., and Kohorn, B.D. (1996). A cell wall-associated, receptor-like protein kinase. J. Biol. Chem. 271 19789–19793. [DOI] [PubMed] [Google Scholar]
- Heim, D.R., Larrinua, I.M., Murdoch, M.G., and Roberts, J.L. (1998). Triazofenamide is a cellulose biosynthesis inhibitor. Pestic. Biochem. Physiol. 59 163–168. [Google Scholar]
- Hématy, K., and Höfte, H. (2008). Novel receptor kinases involved in growth regulation. Curr. Opin. Plant Biol. 11 321–328. [DOI] [PubMed] [Google Scholar]
- Hématy, K., Sado, P.-E., Van Tuinen, A., Rochange, S., Desnos, T., Balzergue, S., Pelletier, S., Renou, J.-P., and Höfte, H. (2007). A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr. Biol. 17 922–931. [DOI] [PubMed] [Google Scholar]
- Humphrey, T.V., Bonetta, D.T., and Goring, D.R. (2007). Sentinels at the wall: Cell wall receptors and sensors. New Phytol. 176 7–21. [DOI] [PubMed] [Google Scholar]
- Kieber, J.J., Rothenburg, M., Roman, G., Feldmann, K.A., and Ecker, J.R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell 72 427–441. [DOI] [PubMed] [Google Scholar]
- Kohorn, B.D., Kobayashi, M., Johansen, S., Riese, J., Huang, L.F., Koch, K., Fu, S., Dotson, A., and Byers, N.R. (2006). An Arabidopsis cell wall-associated kinase required for invertase activity and cell growth. Plant J. 46 307–316. [DOI] [PubMed] [Google Scholar]
- Kroiher, M., Miller, M.A., and Steele, R.E. (2001). Deceiving appearances: signaling by “dead” and “fractured” receptor protein-tyrosine kinases. Bioessays 23 69–76. [DOI] [PubMed] [Google Scholar]
- Lang, J., Eisinger, W., and Green, P. (1982). Effects of ethylene on the orientation of microtubules and cellulose microfibrils of pea epicotyl cells with polylamellate cell walls. Protoplasma 110 5–14. [Google Scholar]
- Moore, B. (2004). Bifunctional and moonlighting enzymes: Lighting the way to regulatory control. Trends Plant Sci. 9 221–228. [DOI] [PubMed] [Google Scholar]
- Morillo, S.A., and Tax, F.E. (2006). Functional analysis of receptor-like kinases in monocots and dicots. Curr. Opin. Plant Biol. 9 460–469. [DOI] [PubMed] [Google Scholar]
- Nakagawa, T., et al. (2007). Improved Gateway binary vectors: High-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71 2095–2100. [DOI] [PubMed] [Google Scholar]
- Paredez, A.R., Somerville, C.R., and Ehrhardt, D.W. (2006). Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312 1491–1495. [DOI] [PubMed] [Google Scholar]
- Peng, L., Hocart, C.H., Redmond, J.W., and Williamson, R.E. (2000). Fractionation of carbohydrates in Arabidopsis root cell walls shows that three radial swelling loci are specifically involved in cellulose production. Planta 211 406–414. [DOI] [PubMed] [Google Scholar]
- Roberts, I.N., Lloyd, C.W., and Roberts, K. (1985). Ethylene-induced microtubule reorientations: Mediation by helical arrays. Planta 164 439–447. [DOI] [PubMed] [Google Scholar]
- Roudier, F., Fernandez, A.G., Fujita, M., Himmelspach, R., Borner, G.H.H., Schindelman, G., Song, S., Baskin, T.I., Dupree, P., Wasteneys, G.O., and Benfey, P.N. (2005). COBRA, an Arabidopsis extracellular glycosyl-phosphatidyl inositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. Plant Cell 17 1749–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible, W.R., Eshed, R., Richmond, T., Delmer, D., and Somerville, C. (2001). Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis ixr1 mutants. Proc. Natl. Acad. Sci. USA 98 10079–10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelman, G., Morikami, A., Jung, J., Baskin, T.I., Carpita, N.C., Derbyshire, P., McCann, M.C., and Benfey, P.N. (2001). COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev. 15 1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastià, C.H., Hardin, S.C., Clouse, S.D., Kieber, J.J., and Huber, S.C. (2004). Identification of a new motif for CDPK phosphorylation in vitro that suggests ACC synthase may be a CDPK substrate. Arch. Biochem. Biophys. 428 81–91. [DOI] [PubMed] [Google Scholar]
- Shi, H., Kim, Y., Guo, Y., Stevenson, B., and Zhu, J.-K. (2003). The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell 15 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu, S.-H., and Bleecker, A.B. (2001). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 98 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville, C. (2006). Cellulose synthesis in higher plants. Annu. Rev. Cell Dev. Biol. 22 53–78. [DOI] [PubMed] [Google Scholar]
- Steen, D.A., and Chadwick, A. (1981). Ethylene effects in pea stem tissue. Evidence for microtubule mediation. Plant Physiol. 67 460–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiz, L. (1984). Plant cell expansion: Regulation of cell wall mechanical properties. Annu. Rev. Plant Physiol. 35 585–657. [Google Scholar]
- Takahashi, H., Kawahara, A., and Inoue, Y. (2003). Ethylene promotes the induction by auxin of the cortical microtubule randomization required for low-pH-induced root hair initiation in lettuce (Lactuca sativa L.) seedlings. Plant Cell Physiol. 44 932–940. [DOI] [PubMed] [Google Scholar]
- Tatsuki, M., and Mori, H. (2001). Phosphorylation of tomato 1-aminocyclopropane-1-carboxylic acid synthase, LE-ACS2, at the C-terminal region. J. Biol. Chem. 276 28051–28057. [DOI] [PubMed] [Google Scholar]
- Vogel, J.P., Woeste, K.W., Theologis, A., and Kieber, J.J. (1998). Recessive and dominant mutations in the ethylene biosynthetic gene ACS5 of Arabidopsis confer cytokinin insensitivity and ethylene overproduction, respectively. Proc. Natl. Acad. Sci. USA 95 4766–4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, T.A., and Kohorn, B.D. (2001). Wall-associated kinases are expressed throughout plant development and are required for cell expansion. Plant Cell 13 303–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagami, T., Tsuchisaka, A., Yamada, K., Haddon, W.F., Harden, L.A., and Theologis, A. (2003). Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the Arabidopsis gene family. J. Biol. Chem. 278 49102–49112. [DOI] [PubMed] [Google Scholar]
- Zimmermann, P., Hennig, L., and Gruissem, W. (2005). Gene-expression analysis and network discovery using Genevestigator. Trends Plant Sci. 10 407–409. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.