Abstract
Abscisic acid (ABA) has an important role for plant growth, development, and stress adaptation. HYPERSENSITIVE TO ABA1 (HAB1) is a protein phosphatase type 2C that plays a key role as a negative regulator of ABA signaling; however, the molecular details of HAB1 action in this process are not known. A two-hybrid screen revealed that SWI3B, an Arabidopsis thaliana homolog of the yeast SWI3 subunit of SWI/SNF chromatin-remodeling complexes, is a prevalent interacting partner of HAB1. The interaction mapped to the N-terminal half of SWI3B and required an intact protein phosphatase catalytic domain. Bimolecular fluorescence complementation and coimmunoprecipitation assays confirmed the interaction of HAB1 and SWI3B in the nucleus of plant cells. swi3b mutants showed a reduced sensitivity to ABA-mediated inhibition of seed germination and growth and reduced expression of the ABA-responsive genes RAB18 and RD29B. Chromatin immunoprecipitation experiments showed that the presence of HAB1 in the vicinity of RD29B and RAB18 promoters was abolished by ABA, which suggests a direct involvement of HAB1 in the regulation of ABA-induced transcription. Additionally, our results uncover SWI3B as a novel positive regulator of ABA signaling and suggest that HAB1 modulates ABA response through the regulation of a putative SWI/SNF chromatin-remodeling complex.
INTRODUCTION
The phytohormone abscisic acid (ABA) is a key regulator of plant growth and development as well as plant responses to decreased water availability. A fast mechanism to adjust ABA levels and respond to changing environmental cues is the hydrolysis of glucose-conjugated ABA (Lee et al., 2006). Additionally, water stress leads to the accumulation of ABA through enhanced expression of ABA biosynthetic genes, mainly 9-cis-epoxycarotenoid dioxygenase3 (Nambara and Marion-Poll, 2005; Barrero et al., 2006). ABA triggers a variety of adaptive responses, such as stomatal closure and differential gene expression, which are crucial for plant survival under stress conditions (Schroeder et al., 2001; Nambara and Marion-Poll, 2005).
Decades of research in ABA signaling have resulted in the identification of many elements of the ABA signal transduction pathway, including both negative and positive regulators (reviewed in Finkelstein et al., 2002; Himmelbach et al., 2003; Israelsson et al., 2006). Under water stress, ABA signaling leads to coordinated remodeling of gene expression, which affects more than ∼5% of the plant transcriptome (Huang et al., 2007). Downstream nuclear effects of ABA are mediated by different transcription factors (TFs) that play a positive role in ABA signaling, which comprise ABA-responsive element binding proteins (ABI5/ABF/AREB/bZIP family) (Choi et al., 2000; Finkelstein and Lynch, 2000; Uno et al., 2000; Bensmihen et al., 2002), Arabidopsis thaliana ABI3 and maize (Zea mays) VP1 TFs of the B3 domain family (McCarty et al., 1991; Giraudat et al., 1992), the ABI4 TF from the APETALA2 domain family (Finkelstein et al., 1998), and ATMYC2 and ATMYB2 TFs (Abe et al., 2003). Some TFs that function as transcriptional repressors of ABA response have also been described (Himmelbach et al., 2002; Pandey et al., 2005; Song et al., 2005). In eukaryotes, the packaging of DNA into chromatin implies that both transcriptional activators and repressors work together with large multisubunit complexes that remodel nucleosomes to regulate gene expression (Carrozza et al., 2003; Smith and Peterson, 2005). Two general classes of chromatin-modifying factors can be distinguished, those that covalently modify the N-terminal tails of histone proteins and those that utilize ATP hydrolysis to remodel or reposition nucleosomes (Carrozza et al., 2003; Smith and Peterson, 2005). The first class includes protein complexes that acetylate or deacetylate Lys residues present in the N termini of histone proteins (histone acetyltransferases) and histone deacetylases. The second class of factors is composed of ATP-dependent chromatin-remodeling complexes, which alter nucleosome structure or positioning. Among them, the yeast SWI/SNF complex was the first one to be described (Cairns et al., 1994; Peterson et al., 1994). In addition to the ATPase Swi2/Snf2, it contains a central core composed of three additional polypeptides, Swi3, Snf5, and Swp73, which are required for the assembly and activity of the complex (Cairns and Kingston, 2000; Smith and Peterson, 2005; Yang et al., 2007). Some reports of chromatin-modifying factors that affect ABA responses have been published (Song et al., 2005; Sridha and Wu, 2006); however, taking into account the deep impact of ABA on the regulation of gene expression and the many TFs involved in this process, we can envisage that many elements in this field are yet to be discovered.
Protein phosphatase type 2Cs (PP2Cs) were identified as key components of ABA signaling from pioneering work with the ABA-insensitive abi1-1 and abi2-1 mutants (Koornneef et al., 1984; Leung et al., 1994; Meyer et al., 1994; Leung et al., 1997; Rodriguez et al., 1998a). Currently, at least six Arabidopsis PP2Cs, namely ABI1, ABI2, PP2CA/AHG3, ABA-HYPERSENSITIVE GERMINATION1 (AHG1), HYPERSENSITIVE TO ABA1 (HAB1), and HAB2, are known to regulate ABA signaling. Genetic approaches indicate that these PP2Cs are negative regulators of ABA signaling (Gosti et al., 1999; Merlot et al., 2001; Tahtiharju and Palva, 2001; Gonzalez-Garcia et al., 2003; Leonhardt et al., 2004; Saez et al., 2004, 2006; Kuhn et al., 2006; Yoshida et al., 2006b; Nishimura et al., 2007). Although interacting partners for some of these PP2Cs have been described (Cherel et al., 2002; Guo et al., 2002; Himmelbach et al., 2002; Ohta et al., 2003; Miao et al., 2006; Yang et al., 2006; Yoshida et al., 2006a), the overall knowledge of their targets and their role in ABA signaling is far from complete. In this work, we have pursued a two-hybrid approach using the PP2C HAB1 as bait to identify putative interacting preys. Interestingly, a prevalent interacting partner of HAB1 was found to be the SWI3B protein, which is an Arabidopsis homolog of the SWI3 core subunit of SWI/SNF chromatin-remodeling complexes (Sarnowski et al., 2002; Zhou et al., 2003). These complexes, already characterized in yeast, Drosophila, and mammals, have not yet been biochemically characterized in plants, although genome analysis suggests that Arabidopsis contains the active components required to form such complexes (Farrona et al., 2004; Sarnowski et al., 2005). Thus, in Arabidopsis, four SWI3-like proteins (i.e., SWI3A, SWI3B, SWI3C, and SWI3D) have been identified (Sarnowski et al., 2002; Zhou et al., 2003) as well as other putative components of SWI/SNF complexes (Brzeski et al., 1999; Farrona et al., 2004; Bezhani et al., 2007). Current data on loci that encode putative components of SWI/SNF chromatin-remodeling complexes show that they operate as modifiers of transcriptional or epigenetic regulation in plant growth and development (Kwon and Wagner, 2007). Our data provide a link between a component of the ABA signaling pathway and a putative component of SWI/SNF chromatin-remodeling complexes and, therefore, suggest that these complexes are also involved in the hormonal response to abiotic stress.
RESULTS
Identification of SWI3B as a HAB1-Interacting Partner
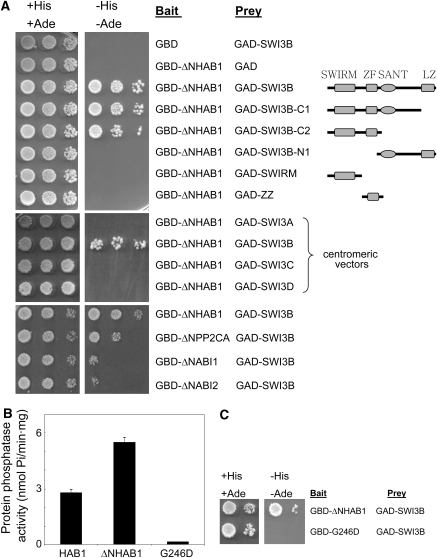
A yeast two-hybrid screen was used to identify proteins that interact with the PP2C HAB1. Preliminary experiments revealed that full-length HAB1 fused to the GAL4 DNA binding domain (GBD) resulted in the activation of HIS3 and ADE2 reporters from the AH109 yeast strain used in this study (see Supplemental Figure 1 online). N-terminal truncation of some clade A PP2Cs (Schweighofer et al., 2004) is required to reduce their potential to activate transcription (Himmelbach et al., 2002; this work). Indeed, the N-terminal 1 to 180 amino acid residues either from HAB1 (see Supplemental Figure 1 online) or from the closely related PP2C HAB2, when fused to the GBD, generated a powerful transcriptional activator. Thus, only the catalytic region (amino acid residues 179 to 511) of the PP2C HAB1 (ΔNHAB1) was used as a bait to screen an Arabidopsis expression library containing random cDNAs fused to the GAL4 activation domain (GAL) in the pACT2 vector (Nemeth et al., 1998). This N-terminal truncation of HAB1 showed approximately twofold higher phosphatase activity than full-length HAB1 (Figure 1B). From 106 colonies screened, 20 positive clones that showed autotrophic growth in medium lacking both adenine and His were selected. Sequence analysis of the recovered pACT2 clones revealed that 11 of the 20 putative interacting preys contained the full-length cDNA from SWI3B. Therefore, these results indicate that AtSWI3B is a prevalent HAB1-interacting partner in a two-hybrid screening.
Figure 1.
HAB1 and SWI3B Interact in a Yeast Two-Hybrid Assay.
Interaction was determined by growth assay on medium lacking His and adenine. Dilutions (10−1, 10−2, and 10−3) of saturated cultures were spotted onto the plates.
(A) Top, interaction assay with ΔNHAB1 as bait (fused to the GBD) and either full-length or different deletions of SWI3B as putative preys (fused to the GAD). Schemes of SWI3B domains and the different protein deletions are shown. Deletions C1 and C2 lacked C-terminal amino acid residues 346 to 469 and 221 to 469, respectively. Deletion N1 lacked N-terminal amino acid residues 1 to 220. GAD-SWIRM and GAD-ZZ comprised amino acid residues 1 to 140 and 134 to 220, respectively. Middle, interaction assay with SWI3A, SWI3B, SWI3C, and SWI3D as putative preys. Bottom, interaction assay with ΔNPP2CA, ΔNABI1, and ΔNABI2 as baits and SWI3B as prey.
(B) Protein phosphatase activity of MBP-HAB1, MBP-ΔNHAB1, and MBP-G246D ΔNhab1 fusion proteins. Values are averages ± se from three independent experiments.
(C) Interaction assay with ΔNHAB1 and G246D ΔNhab1 as baits and SWI3B as prey.
The Interaction of HAB1 and SWI3B Requires a Functional PP2C Catalytic Domain and Maps to the N-Terminal Half of SWI3B
Protein domain analysis using the PFAM database of global domain hidden Markov models and different pattern and profile searches in Expasy (http://www.expasy.org) served to identify SWIRM (48 to 136), SANT (224 to 272), and Leu zipper domains (399 to 452) in the SWI3B amino acid sequence, in agreement with previous findings from Sarnowski et al. (2005). Additionally, we could identify a ZZ zinc finger domain (Cys-x2-Cys motifs plus a conserved YDL motif) between amino acid residues 169 and 208. A similar ZZ zinc finger domain was identified in Arabidopsis SWI3C by Hurtado et al. (2006). In order to determine specific regions of SWI3B involved in the interaction with ΔNHAB1, different deletions of the SWI3B coding sequence in the prey vector pACT2 were generated. Previously, we confirmed that a combination of the empty pGBT9 plasmid and pACT2-SWI3B did not activate transcription of the HIS3 and ADE2 reporter genes (Figure 1A); moreover, none of the deletion constructs activated transcription in the absence of bait protein interactors. In combination with the bait construct pGBT9-ΔNHAB1, the deletion constructs C1 and C2 activated transcription of the reporter genes to the same levels as full-length SWI3B (Figure 1A). This result mapped the HAB1-interacting domain to the first 220 amino acid residues of SWI3B. In agreement with this result, the prey construct N1 did not activate the reporter genes in the growth assay. Further attempts to delimit the minimal region of SWI3B that interacted with ΔNHAB1 failed, as additional deletions affecting the N-terminal half of SWI3B (SWIRM and ZZ prey constructs) eliminated the interaction with ΔNHAB1.
In order to clarify the specificity of the interaction, we examined whether other SWI3-like proteins from Arabidopsis showed interaction with ΔNHAB1. In contrast with SWI3B, none of the SWI3A, SWI3C, or SWI3D proteins interacted with ΔNHAB1 (Figure 1A, middle). This result highlights the remarkable functional diversification previously described for the four SWI3-like proteins from Arabidopsis (Sarnowski et al., 2005; Hurtado et al., 2006). HAB1 belongs to a group of PP2Cs (clade A; Schweighofer et al., 2004) in which six of the identified genes are associated with ABA signaling. Gene expression data and genetic analysis indicate that HAB1, PP2CA, ABI1, and ABI2 play a predominant role in ABA signaling in both seeds and vegetative tissue (Saez et al., 2004, 2006; Kuhn et al., 2006; http://www.genevestigator.ethz.ch). Therefore, we generated N-terminal truncations of PP2CA, ABI1, and ABI2 fused to GBD and their interaction with SWI3B was examined (Figure 1A, bottom). ΔNPP2CA, ΔNABI1, and ΔNABI2 were able to interact with SWI3B, although it was apparent in the growth assay that the interaction was weaker than that observed for ΔNHAB1. All fusion proteins were expressed at similar levels, as verified by protein gel blot analysis using antibodies against the GAD and GBD. Finally, in order to examine the role of the catalytic PP2C domain in the interaction with SWI3B, a point-mutated version of HAB1 that replaced Gly-246 for Asp (G246D hab1) was introduced in the two-hybrid test. The Gly-246 is localized in a conserved motif from eukaryotic PP2Cs, and its replacement by Asp interferes with Mg2+ binding and strongly impairs PP2C activity (Leung et al., 1994; Meyer et al., 1994). Indeed, both G246D hab1 (Robert et al., 2006) and G246D ΔNhab1 (Figure 1B) show <3% in vitro activity than the wild type. Interestingly, G246D ΔNhab1 did not interact with SWI3B in the two-hybrid assay, indicating that a functional catalytic domain of HAB1 is required for its interaction with SWI3B (Figure 1C).
Subcellular Localization of HAB1 and SWI3B
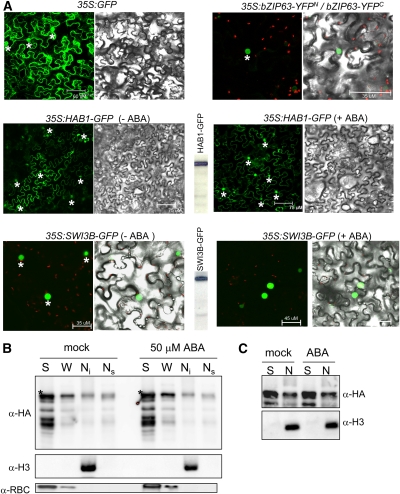
To determine the subcellular localization of HAB1 and SWI3B proteins in plant cells, we performed in vivo targeting experiments in tobacco (Nicotiana benthamiana). To this end, 35S:HAB1-GFP and 35S:SWI3B-GFP constructs were generated and delivered into leaf cells of tobacco by Agrobacterium tumefaciens infiltration (Voinnet et al., 2003). Coexpression of bZIP63-YFPN and bZIP63-YFPC served as a positive control for localization of a nuclear protein (Walter et al., 2004). In the case of SWI3B, strong green fluorescent protein (GFP) fluorescence was observed in the nucleus of tobacco cells, whereas HAB1 localized in both the nucleus and the cytosol (Figure 2A). Similar results were obtained when the coding region of HAB1 was fused to the C-terminal end of GFP (see Supplemental Figure 2 online). Finally, ABA treatment did not modify the subcellular localization of either HAB1-GFP or SWI3B-GFP under our experimental conditions (Figure 2A; see Supplemental Figure 2 online).
Figure 2.
HAB1 Localizes at Both Cytosol and Nucleus.
(A) Subcellular localization of HAB1 and SWI3B in Agrobacterium-infiltrated tobacco leaves. Epifluorescence and bright-field images of epidermal leaf cells infiltrated with a mixture of Agrobacterium suspensions harboring the indicated constructs and the silencing suppressor p19. BiFC-induced bZIP63 dimerization served to identify the nuclei of tobacco epidermal cells (asterisks). The expression of the proteins is demonstrated by immunodetection with anti-GFP for HAB1-GFP and SWI3B-GFP (center, between panels). Treatment with 50 μM ABA for 1 h (+ABA) did not change the subcellular localization of both HAB1-GFP and SWI3B-GFP.
(B) Biochemical fractionation of HAB1-dHA (full-length protein marked with asterisks). Plant material was obtained from the hab1-1∷ProHAB1-HAB1-dHA transgenic line after mock treatment or treatment with 50 μM ABA for 1 h. The soluble cytosolic fraction (S), nuclei/organelles wash fraction (W), nuclear insoluble fraction (Ni), and nuclear soluble fraction (Ns) were analyzed using anti-HA, anti-histone 3 (H3), and anti–ribulose-1,5-bis-phosphate carboxylase/oxygenase (RBC) antibodies.
(C) Relative amount of HAB1-dHA in the soluble cytosolic (S) and nuclear (N) fractions after mock treatment or treatment with 50 μM ABA for 1 h.
In addition to using GFP fusions, we examined the subcellular localization of HAB1 by standard biochemical techniques. To this end, we generated transgenic lines (in a hab1-1 background) that expressed a double hemagglutinin (HA) epitope–tagged version of HAB1 (HAB1-dHA) under the control of the HAB1 native promoter. HAB1-dHA efficiently complemented the ABA-hypersensitive phenotype of hab1-1 in germination assays (Saez et al., 2004). HAB1-dHA could be detected in both the cytosolic and nuclear fractions (Figure 2B). Washing of the nuclei/organelles with a buffer containing 0.5% Triton X-100 released a significant amount of HAB1-dHA (W fraction), which we assume to be of nuclear origin according to the localization of HAB1-GFP and GFP-HAB1 fusions. Additionally, a fraction of HAB1-dHA was associated with the nuclear insoluble fraction, which contains the major histones and is mostly composed of chromatin (Poveda et al., 2004; Cho et al., 2006). Finally, HAB1-dHA was also detected in a nuclear soluble fraction that was obtained by rupture of the nuclei and extraction with a buffer containing 0.4 M NaCl. An estimation of the cytosolic:nuclear HAB1 ratio was made based on protein blot analysis with the anti-HA antibody. Figure 2C shows an approximately threefold difference in HAB1 abundance between the cytosolic and combined nuclear fractions. Taking into account the additional fourfold enrichment during the nuclei isolation process before protein gel loading, the HAB1 cytosolic:nuclear ratio was ∼12:1. Treatment with 50 μM ABA for 1 h did not substantially modify this ratio (Figure 2C).
In Planta Interaction between HAB1 and SWI3B
Bimolecular fluorescence complementation (BiFC) assays were used to detect the interaction between HAB1 and SWI3B in plant cells. To this end, HAB1 was translationally fused to the C-terminal 84–amino acid portion of yellow fluorescent protein (YFPC) in the pSPYCE vector, which generated a HAB1-epitope HA-YFPC fusion protein (Figure 3B). For the other partner, the N-terminal half of SWI3B was translationally fused to the N-terminal 155–amino acid portion of yellow fluorescent protein (YFPN) in the pSPYNE vector, which generated a SWI3B-epitope myc-YFPN fusion protein (Figure 3B). The corresponding constructs were codelivered into leaf cells of tobacco by Agrobacterium infiltration and, as a result, fluorescence was observed in the nucleus of tobacco cells (Figure 3A, left). No fluorescence signal was observed when pSPYCE-HAB1 vector was codelivered with pSPYNE or when pSPYNE-SWI3B was codelivered with pSPYCE. Moreover, in agreement with the previous finding in the two-hybrid assay, introduction of the G246D mutation in the sequence of HAB1 abolished the interaction with SWI3B in the BiFC assay (Figure 3A, right).
Figure 3.
BiFC Visualization and Coimmunoprecipitation Experiments Show Interaction between HAB1 and SWI3B in the Nucleus of Tobacco Leaves.
(A) Introduction of the G246D substitution into HAB1 abolishes the interaction with SWI3B. Epifluorescence and bright-field images of epidermal leaf cells infiltrated with a mixture of Agrobacterium suspensions harboring constructs HAB1-HA-YFPC/SWI3BN-myc-YFPN (panel 1) or G246D-HA-YFPC/SWI3BN-myc-YFPN (panel 2) and the silencing suppressor p19. The bar corresponds to 75 μm. Green color corresponds to YFP, whereas red color is generated by chlorophyll fluorescence.
(B) Coimmunoprecipitation assay demonstrates the interaction between HAB1 and SWI3B in planta. Protein extracts obtained from tobacco leaves infiltrated with Agrobacterium suspensions harboring constructs HAB1-HA-YFPC/SWI3BN-myc-YFPN (lanes 1) or G246D-HA-YFPC/SWI3BN-myc-YFPN (lanes 2) were analyzed using anti-HA or anti-c-myc antibodies. Input levels of epitope-tagged proteins in crude protein extracts (20 μg of total protein) were analyzed by immunoblotting. Immunoprecipitated epitope HA–tagged proteins were probed with anti-c-myc antibodies to detect coimmunoprecipitation of SWI3BN-myc-YFPN with HAB1-HA-YFPC.
(C) BiFC assays show the interaction of ABI1, ABI2, and PP2CA with SWI3B in the nucleus of tobacco leaves. Cells were infiltrated with a mixture of Agrobacterium suspensions harboring constructs SWI3BN-HA-YFPC/YFPN-ABI1 (panel 3), SWI3BN-HA-YFPC/YFPN-ABI2 (panel 4), or SWI3BN-HA-YFPC/YFPN-PP2CA (panel 5) and the silencing suppressor p19.
(D) Protein gel blot analysis demonstrates the expression of SWI3BN-HA-YFPC and the corresponding YFPN-PP2Cs (asterisks). Protein extracts obtained from tobacco leaves infiltrated with Agrobacterium suspensions harboring the silencing suppressor p19 and constructs SWI3BN-HA-YFPC/YFPN-ABI1 (lane 3), SWI3BN-HA-YFPC/YFPN-ABI2 (lane 4), SWI3BN-HA-YFPC/YFPN-PP2CA (lane 5), or p19 alone (lane p19) were analyzed using anti-HA or anti-GFPN antibodies.
In addition to the observed BiFC fluorescent signal, we confirmed the interaction by coimmunoprecipitation of HAB1 and SWI3B in tobacco protein extracts prepared from the BiFC assay described above (Figure 3B). HAB1 and SWI3B can be coimmunoprecipitated, as we could detect SWI3B in the immunocomplex precipitated with an antibody to epitope HA, which pulls down the HAB1-HA-YFPC fusion protein (Figure 3B). By contrast, introduction of the G246D mutation in the sequence of HAB1 prevented the coimmunoprecipitation of SWI3B (Figure 3B). Thus, results from two in planta assays support the interaction between HAB1 and SWI3B.
Finally, BiFC assays showed that PP2CA, ABI1, and ABI2 were able to interact with SWI3B in the nucleus of tobacco cells (Figure 3C). Expression of fusion proteins was verified by protein gel blot analysis using antibodies against the epitope HA and peptide comprising amino acids 3 to 17 of GFP (anti-GFPN) (Figure 3D). ABA treatment (50 μM for 1 h) did not change the interaction of the PP2Cs and SWI3B. However, complex formation in BiFC is essentially irreversible, which prevents the imaging of changes in the protein association state (Fricker et al., 2006).
swi3b Mutants Show a Reduced Sensitivity to ABA and Reduced Expression of RD29B and RAB18
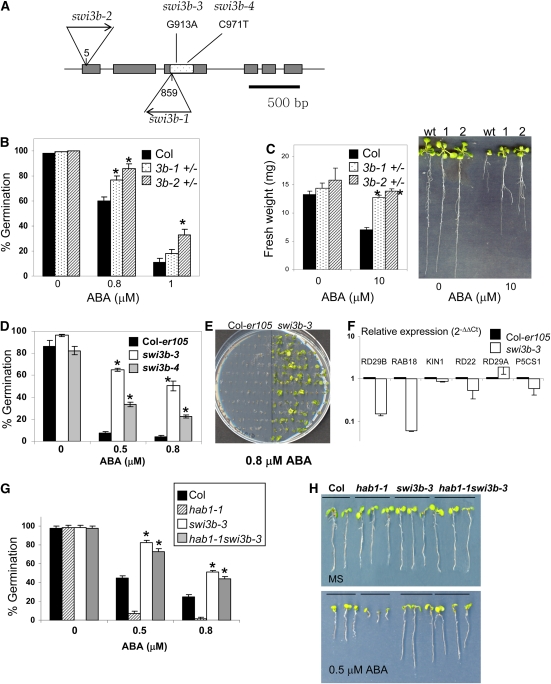
The swi3b-1 and swi3b-2 knockout mutants (Figure 4A) were previously reported to be embryo-lethal (Sarnowski et al., 2005); therefore, we decided to examine heterozygous mutants for phenotypic effects. Phenotypic effects caused by gene haploinsufficiency (monoallelic expression and heterozygosis) have been described in mutants affected in diverse components of the chromatin-remodeling machinery (Bultman et al., 2000; Roberts et al., 2000; Alarcon et al., 2004; David et al., 2006). For instance, heterozygous mice that have a single copy of either BRG1 (the mammalian orthologous gene of the yeast Swi2/Snf2 ATPase) or SNF5 (a core component of the SWI/SNF complex) are predisposed to different tumors, indicating that a full dosage of both BRG1 and SNF5 is required for proper control of gene expression and tumor suppression (Bultman et al., 2000; Roberts et al., 2000). Therefore, we decided to analyze ABA responsiveness in the progeny of Arabidopsis swi3b +/− seedlings, which represents an ∼2:1 mixture of heterozygous and wild-type seeds (Sarnowski et al., 2005). Thus, the progeny from swi3b-1 and swi3b-2 heterozygous plants were analyzed to score ABA-mediated inhibition of germination and growth. These assays revealed a reduced sensitivity to ABA of swi3b +/− seeds and seedlings compared with the wild type (Figures 4B and 4C). This phenotype was particularly apparent in growth assays, as after 10 d in 10 μM ABA both swi3b-1 and swi3b-2 +/− seedlings showed ∼80 to 90% higher weight than wild-type seedlings (Figure 4C). By contrast, water-loss assays did not show significant differences between wild-type and swi3b-1 and swi3b-2 +/− plants (see Supplemental Figure 3A online). Nevertheless, since ∼50% reduction in the expression of SWI3B (see Supplemental Figure 3B online ) led to reduced sensitivity to ABA in germination and growth assays, these results suggest that SWI3B is a positive regulator of ABA signaling.
Figure 4.
swi3b Mutants Show Reduced Sensitivity to ABA-Mediated Inhibition of Germination and Growth.
(A) T-DNA insertions in the swi3b-1 and swi3b-2 alleles and localization of ethyl methanesulfonate–induced mutations in swi3b-3 and swi3b-4 alleles. The numbering begins at the ATG translation start codon. The gray boxes represent exons. The SANT domain is spotted within the third exon.
(B) ABA effects on germination in the progeny of swi3b-1 and swi3b-2 heterozygous plants. The percentage of seeds that germinated and developed green cotyledons in the presence of the indicated concentrations of ABA is shown. Values are averages ± sd for three independent experiments (n = 200 seeds per experiment). * P < 0.01 (Student's t test) when comparing data from each genotype and the wild type in the same assay conditions.
(C) Reduced sensitivity of swi3b-1 and swi3b-2 heterozygous (+/−) seedlings to ABA-mediated growth inhibition. Fresh weight was measured in 12-d-old seedlings grown in MS medium either lacking or containing 10 μM ABA. Values are averages ± sd for three independent experiments (n = 20 seedlings per experiment). Representative seedlings of Col (wild type [wt]), swi3b-1 +/− (1), and swi3b-2 +/− (2) were removed from medium lacking or containing 10 μM ABA and rearranged on agar plates (at right).
(D) Reduced sensitivity to ABA-mediated inhibition of seed germination in swi3b-3 and swi3b-4 mutants compared with Col-er105. The percentage of seeds that showed radicle emergence at 96 h after seed stratification is shown. Values are averages ± sd for three independent experiments (n = 200 seeds per experiment). Asterisks are as described for (B).
(E) Reduced sensitivity to ABA-mediated inhibition of early growth in the swi3b-3 mutant compared with Col-er105 in medium supplemented with 0.8 μM ABA. The photograph was taken at 18 d after sowing.
(F) Reduced expression of ABA-inducible genes in swi3b-3 compared with Col-er105. Values are expression levels reached in the mutant with respect to Col-er105 (value 1) as determined by RT-qPCR analyses. Expression of gene markers was analyzed in 7-d-old seedlings grown in medium supplemented with 0.3 μM ABA. Values are averages ± sd for three independent experiments (n = 30 to 40 seedlings per experiment).
(G) The swi3b-3 phenotype is epistatic to hab1-1. The percentage of seeds that showed radicle emergence at 96 h after seed stratification is shown. Values are averages ± sd for three independent experiments (n = 200 seeds per experiment). * P < 0.01 (Student's t test) when comparing data from swi3b-3 and the wild type, or hab1-1swi3b-3 and hab1-1, in the same assay conditions.
(H) Reduced sensitivity to ABA-mediated inhibition of early growth in swi3b-3 and the hab1-1swi3b-3 double mutant. Photographs were taken at 7 d (MS) and 11 d (0.5 μM ABA) after sowing. As in (C), plants were removed from growth medium and rearranged on plates for photography.
[See online article for color version of this figure.]
Further evidence of the role of SWI3B in ABA signaling was obtained through the analysis of point mutations in swi3b alleles that were recovered by the Arabidopsis TILLING (for targeting-induced local lesions in genomes) program (http://tilling.fhcrc.org:9366/home.html) in a Columbia (Col)–er105 background. Thus, two new swi3b alleles were identified, swi3b-3 and swi3b-4, which resulted in the substitution of Asp-245 by Asn and Ser-264 by Phe, respectively. Both Asp-245→Asn and Ser-264→Phe mutations are localized in the SANT domain of SWI3B and, according to SIFT (for sorting intolerant from tolerant) software analysis, are predicted to affect protein function (SIFT score < 0.05) (Ng and Henikoff, 2001). Analysis of ABA-mediated inhibition of germination in swi3b-3 and swi3b-4 revealed that both mutants showed a reduced sensitivity to ABA in this assay compared with the Col-er105 background (where TILLING mutants were originated) (Figure 4D). In particular, the swi3b-3 mutant also showed a reduced sensitivity to ABA-mediated inhibition of early growth (Figure 4E). These results, together with those of swi3b-1 and swi3b-2 +/− seedlings, show that SWI3B is a positive regulator of ABA signaling that mediates the ABA response in seeds and vegetative tissue.
Additionally, we wondered whether SWI3B might play a role in the regulation of gene expression in response to ABA. SWI3B is a putative core component of SWI/SNF complexes, and chromatin remodelers have a well-established role in transcriptional regulation. Therefore, real-time quantitative polymerase chain reaction (RT-qPCR) was used to analyze the expression of the ABA-responsive RD29B, RAB18, KIN1, RD22, RD29A, and P5CS1 genes in the wild type and the swi3b-3 mutant (Figure 4F). In general terms, these gene markers show low expression in the absence of ABA or stress treatment, which is upregulated in response to the inductive signal. Upon ABA treatment, expression of RD29B and RAB18 in swi3b-3 was 15 and 6%, respectively, of that found in the wild type, whereas expression of the other gene markers did not differ more than twofold in both genotypes. Thus, SWI3B appears to regulate a subset of ABA-inducible genes, whereas its function seems to be partially dispensable or redundant for the expression of other ABA-responsive genes. Finally, to further characterize the genetic relationship between the ABA-hypersensitive locus hab1-1 and the ABA-insensitive locus swi3b-3, we generated a hab1-1swi3b-3 double mutant. Analysis of ABA-mediated inhibition of germination (Figure 4G) and early seedling growth (Figure 4H) revealed that hab1-1swi3b-3 showed an ABA-insensitive phenotype, in contrast with hab1-1, which indicates that SWI3B is epistatic to HAB1; therefore, HAB1 functions upstream of SWI3B in the ABA signaling pathway. In addition to reduced sensitivity to ABA in the assays described above, the swi3b-3 allele showed both impaired vegetative and reproductive growth (see Supplemental Figure 4 online), which likely reflects the key role of SWI3B in plant growth and development as a core component of diverse SWI/SNF complexes (Zhou et al., 2003; Sarnowski et al., 2005; Bezhani et al., 2007). In agreement with this role, combination of the swi3b-3 and swi3b-2 alleles was embryo-lethal (see Supplemental Figure 4 online).
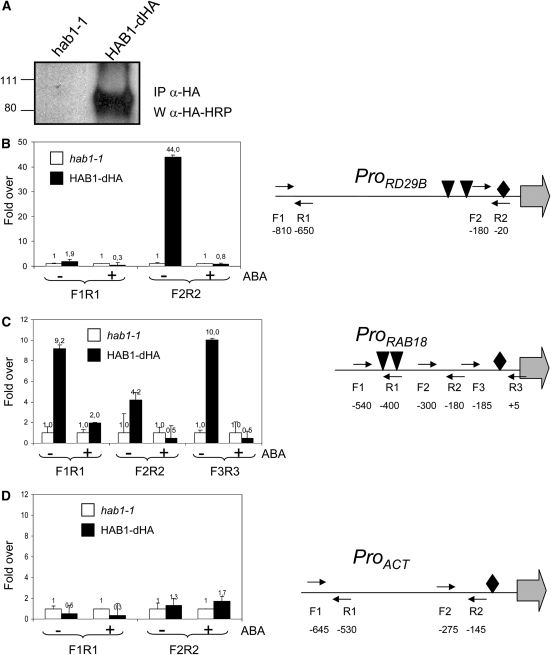
The Presence of HAB1 in the Vicinity of the ABA-Responsive RD29B and RAB18 Promoters Is Abolished by ABA
The interaction of HAB1 and SWI3B as well as the phenotype of swi3b mutants suggest that HAB1 modulates ABA response through the regulation of a putative SWI/SNF chromatin-remodeling complex. In order to analyze the presence of HAB1 in plant chromatin and the putative influence of ABA on it, we performed chromatin immunoprecipitation (ChIP) experiments. To this end, we used the hab1-1 transgenic line complemented by HAB1-dHA described above and demonstrated that HAB1-dHA could be immunoprecipitated using a monoclonal antibody to HA peptide (Figure 5A). ChIP experiments were performed on formaldehyde–cross-linked chromatin extracted from either hab1-1 or hab1-1∷ProHAB1-HAB1-dHA plants. Genomic DNA fragments that coimmunoprecipitated with HAB1-dHA were analyzed by RT-qPCR (Figures 5B to 5D). To this end, we used different primer pairs that covered the promoters of the ABA-responsive genes RD29B and RAB18 as well as a control gene, β-ACT8, which is not responsive to ABA (Saez et al., 2006). Aliquots of the total chromatin input were previously used to provide a quantitative measurement of the DNA input present in each sample, and DNA amounts present in ChIP precipitates were measured using cycle threshold values from RT-qPCR amplification curves (see Methods). Figures 5B and 5C show that the amounts of RD29B and RAB18 promoter DNAs immunoprecipitated from the HAB1-dHA transgenic plants were over 40- and 10-fold higher, respectively, than that precipitated from hab1-1 control plants, whereas the amount of β-ACT8 promoter DNA immunoprecipitated was very similar in both cases (Figure 5D). Interestingly, HAB1 was enriched in those regions of the RD29B and RAB18 promoters that were close to ABA-responsive elements and TATA boxes, and after treatment with 50 μM ABA for 1 h the presence of HAB1 in these regions was abolished (Figures 5B and 5C). 35S-HAB1 lines showed reduced expression of ABA-inducible genes compared with the wild type (Saez et al., 2004); conversely, the hab1-1 loss-of-function mutant showed twofold higher expression of ABA-inducible genes than the wild type (Saez et al., 2006). These data, together with ChIP results, suggest that HAB1 might repress ABA-induced transcription through direct chromatin interaction and that ABA treatment seems to release such inhibition.
Figure 5.
The Presence of HAB1 in the Vicinity of the ABA-Responsive RD29B and RAB18 Promoters Is Abolished by ABA.
(A) Immunoprecipitated samples (IP anti-HA) were subjected to immunoblot analysis (W anti-HA-HRP).
(B) to (D) ChIP assays of the RD29B, RAB18, and ACT8 promoters in hab1-1 or hab1-1∷ProHAB1-HAB1-dHA plants. Genomic DNA fragments that coimmunoprecipitated with HAB1-dHA were analyzed by RT-qPCR using primers of the RD29B (B), RAB18 (C), and ACT8 (D) promoters. Results are presented as ratios of the amount of DNA immunoprecipitated from HAB1-dHA samples to that from the hab1-1 control (value set to equal 1). The positions of the ABA-responsive elements (triangles) and TATA boxes (diamonds) in the sequences of the different promoters are indicated, as well as the primers used for RT-qPCR analysis. The numbering refers to the ATG translation start codon, and the beginnings of the open reading frames are indicated by arrows.
DISCUSSION
Both gain-of-function and loss-of-function phenotypes of the PP2C HAB1 are consistent with a role as a negative regulator of ABA signaling (Leonhardt et al., 2004; Saez et al., 2004). Thus, whereas constitutive expression of HAB1 (35S:HAB1) led to reduced ABA sensitivity in both seeds and vegetative tissues, the recessive hab1-1 mutant showed ABA-hypersensitive inhibition of seed germination and growth, enhanced ABA-mediated stomatal closure, and enhanced expression of ABA-responsive genes (Leonhardt et al., 2004; Saez et al., 2004, 2006). The ABA-hypersensitive phenotype of hab1-1 was strongly reinforced when combined with a loss-of-function allele of ABI1 (Saez et al., 2006). A critical aspect to improve our knowledge on HAB1 function and its role in ABA signaling is the identification of its interacting partners.
Physical Interaction of HAB1 and SWI3B
A two-hybrid assay revealed a strong interaction between the HAB1 catalytic domain and SWI3B (Figure 1A). Serial deletions of SWI3B mapped the interacting domain to the N-terminal half of the protein. Thus, both the SWIRM and ZZ zinc finger domains appeared to be required for the interaction, as deletion of either of them abolished the interaction (Figure 1A). The SWIRM (for SWI3-RSC-MOIRA) domain is a small α-helical domain of ∼85 amino acid residues found in chromosomal proteins and is predicted to mediate protein–protein interactions in the assembly of chromatin/protein complexes (Aravind and Iyer, 2002; Da et al., 2006). The ZZ zinc finger domain is also likely involved in mediating specific protein–protein interactions with transcriptional adaptors and activators (Ponting et al., 1996). Interestingly, the C-terminal half of SWI3B, which contains both the SANT (for SWI3-ADA2-NCoR-TFIIIB) and Leu zipper domains, was dispensable for the interaction with HAB1 (Figure 1A). However, the mutations found in the swi3b-3 and swi3b-4 alleles provide evidence for the importance of the SANT domain for SWI3B function. The equivalent C-terminal half of SWI3C constituted the region that interacted with the ATPase BRAHMA (Hurtado et al., 2006), which also interacted weakly with SWI3B; therefore, it seems likely that such a region plays a similar role in SWI3B. The SANT domain included in this region is structurally related to the Myb DNA binding domain; however, there is no evidence that the SANT domain directly contacts DNA. Instead, SANT domains may directly bind the N-terminal histone tails (Mohrmann and Verrijzer, 2005). Finally, it is suggested that the Leu zipper functions as a homodimerization and heterodimerization domain (Mohrmann and Verrijzer, 2005).
The HAB1 mutant allele G246D ΔNhab1, which had <3% in vitro PP2C activity than the wild type, did not interact with SWI3B. The G246D substitution affects the catalytic center of the PP2C, and according to the crystal structure of human PP2C (Das et al., 1996) such a mutation is expected to disturb the metal-coordinating residues Asp-243 and Gly-244 with concomitant reduction in catalytic activity. An alternative possibility has been postulated by Robert et al. (2006), who suggested that hab1Gly246Asp might show enhanced affinity for its substrate and therefore enhanced dephosphorylating capacity. However, using casein as a substrate, the in vitro PP2C activity of hab1Gly246Asp was severely reduced compared with that of the wild type, as was the case for G246D ΔNhab1. Additionally, the equivalent Gly-180→Asp abi1-1 and Gly-168→Asp abi2-1 mutant proteins did not show enhanced affinity (just the opposite) for their interacting partners, ATHB6/OST1 and SOS2/Prefibrillin, respectively (Himmelbach et al., 2002; Ohta et al., 2003; Yang et al., 2006; Yoshida et al., 2006a). In all of these cases, including the interaction of HAB1 and SWI3B, it appears that a functional catalytic PP2C is required for binding of the different targets.
The interaction of HAB1 and SWI3B was confirmed in planta through BiFC and coimmunoprecipitation assays (Figure 3). HAB1 is localized in both nucleus and cytosol; however, the BiFC assay clearly identified SWI3B as a nuclear target of HAB1. Interestingly, most of the targets previously identified for clade A PP2Cs were not nuclear proteins (Cherel et al., 2002; Guo et al., 2002; Ohta et al., 2003; Miao et al., 2006; Yang et al., 2006; Yoshida et al., 2006a). However, in the case of ABI1, it is supposed that the interaction with the TF ATHB6 must be nuclear (Himmelbach et al., 2002). Additionally, recent results reveal a nuclear localization signal at the very end of the C-terminal domain of ABI1 that is required for regulating ABA-dependent gene expression (Moes et al., 2008). Inspection of the C-terminal amino acid sequences of HAB1, ABI2, and PP2CA also reveals a similar short region enriched in basic amino acids (see Supplemental Figure 5 online). Additionally, the sequences of HAB1 and ABI2 display a second region that contains two positively charged clusters separated by a short linker region (see Supplemental Figure 5 online). The nuclear interaction of PP2CA, ABI1, and ABI2 with SWI3B found in BiFC assays might be physiologically relevant to regulating ABA signaling. However additional experiments (e.g., ChIP analysis) will be required to confirm the presence of these PP2Cs in plant chromatin and specifically in ABA-regulated promoters. Finally, it is noteworthy that previously described SWI3B-interacting partners connect SWI3B with different components of putative SWI/SNF complexes and, intriguingly, with the RNA and ABA binding protein FCA (Razem et al., 2006). In addition to FCA, six different SWI3B-interacting proteins have been described, namely SWI3A, SWI3C, SWI3D, BRM, SYD, and BSH, which are putative components of SWI/SNF complexes (Sarnowski et al., 2002, 2005; Bezhani et al., 2007). Analysis of the ABA response in mutants affected in these genes will be required for the identification of additional components of SWI/SNF complexes involved in ABA signaling.
Role of HAB1, SWI3B, and a Putative SWI/SNF Complex in ABA Signaling
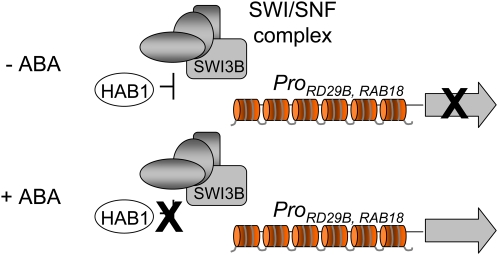
No SWI/SNF complex has been biochemically purified in plants, although comparative genome analysis indicates that plants encode a remarkably high number of potential components of such a complex (Sarnowski et al., 2005). In yeast, Drosophila, and mammals, it is well known that an important subset of highly inducible genes requires a SWI/SNF complex as a transcriptional activator (Mohrmann and Verrijzer, 2005). It has been reported previously that hab1-1 mutants show twofold higher expression of ABA-responsive genes than wild-type plants (Saez et al., 2006), whereas 35S:HAB1 plants show reduced expression of ABA-inducible genes (Saez et al., 2004); therefore, HAB1 negatively regulates the expression of these genes. HAB1 is localized in both nucleus and cytosol and, therefore, could influence ABA signaling at different steps. ChIP experiments reveal the presence of HAB1 in the vicinity of the ABA-responsive RAB18 and RD29B promoters, and ABA treatment eliminates HAB1 from these regions (Figure 5). These results, taken together with the negative effect of HAB1 on the expression of ABA-inducible genes, strongly suggest a direct regulatory effect of HAB1 on ABA-mediated transcriptional regulation. Thus, the presence of HAB1 in the vicinity of ABA-responsive promoters correlates with the inhibition of their transcription under basal conditions, whereas ABA-mediated removal of HAB1 from these regions appears to be required for full induction of them (Figure 6). In this context, both the HAB1–SWI3B interaction and the impaired upregulation by ABA of RAB18 and RD29B in swi3b-3 suggest that HAB1 might regulate a putative SWI/SNF complex targeted to some ABA-responsive promoters (Figure 6). The phenotypes described in this work for swi3b-1 and swi3b-2 +/− seedlings as well as swi3b-3 and swi3b-4 mutants are consistent with SWI3B acting as a positive regulator of ABA signaling. Taking into account the opposed roles of HAB1 and SWI3B in this signaling pathway, it is reasonable to postulate that HAB1 negatively regulates SWI3B function, modulating its role as a positive regulator of ABA signaling. Alternatively, SWI3B might anchor HAB1 to a putative SWI/SNF complex, where the phosphatase activity of HAB1 might dephosphorylate a component required for proper function of the chromatin remodeler. Taking into account that the presence of HAB1 in the vicinity of the ABA-responsive RD29B and RAB18 promoters is abolished by ABA (Figure 5), we speculate that ABA must inhibit HAB1 function, which releases its inhibitory effect on a yet unknown SWI/SNF complex involved in the transcriptional activation of ABA-responsive genes (Figure 6). Finally, it will be an exciting challenge for the future understanding of how the dynamic structure of the chromatin is modulated in response to ABA to regulate gene expression as well as to characterize the cell signaling events that lead to chromatin remodeling.
Figure 6.
A Model for the Involvement of HAB1, SWI3B, and a Putative Plant SWI/SNF Complex in the Regulation of Plant Transcriptional Response to ABA on the Chromatin Template.
HAB1 is a negative regulator of ABA signaling that interacts with SWI3B, which is a positive regulator of ABA signaling. SWI3B must play a key role as a core subunit of an unidentified SWI/SNF complex, which is predicted to regulate nucleosomal structure in response to ABA. In the absence of ABA, HAB1 is localized in the vicinity of the RAB18 and RD29B promoters and negatively regulates the expression of these genes. ABA inhibits HAB1 and releases its inhibitory effect on a putative SWI/SNF complex. [See online article for color version of this figure.]
METHODS
Plant Material
Arabidopsis thaliana (ecotype Col) and tobacco (Nicotiana benthamiana) plants were routinely grown under greenhouse conditions in pots containing a 1:3 perlite:soil mixture. For in vitro culture, Arabidopsis seeds were surface-sterilized by treatment with 70% ethanol for 20 min, followed by commercial bleach (2.5%) containing 0.05% Triton X-100 for 10 min, and finally, four washes with sterile distilled water. Stratification of the seeds was conducted during 3 d at 4°C. Afterward, seeds were sown on Murashige and Skoog (MS) plates containing solid medium composed of MS basal salts and 1% sucrose, solidified with 1% agar, and pH was adjusted to 5.7 with KOH before autoclaving. Plates were sealed and incubated in a controlled-environment growth chamber at 22°C under a 16-h-light/8-h-dark photoperiod at 80 to 100 μE·m−2·s−1.
The swi3b-1 (Koncz_2208) and swi3b-2 (GABI_302G08) alleles are T-DNA mutants in the Col background. They were kindly provided by G. Rios and have been described previously (Sarnowski et al., 2005). TILLING mutants were obtained through the Arabidopsis TILLING project, which performed a high-throughput reverse genetic screening to identify ethyl methanesulfonate–induced mutations in the Col-er105 background (Till et al., 2003). As a result, two alleles were identified, swi3b-3 and swi3b-4, which showed changes with SIFT score < 0.05 and, therefore, were predicted to be deleterious to the gene product (Ng and Henikoff, 2001). These mutants were backcrossed once with Col-er105, and F2 homozygous mutants were genotyped by PCR amplification and DNA sequencing using the primers F1261 and R1560. In the case of swi3b-3, a second backcross was done with Col, and F2 swi3b-3 mutants lacking the er105 mutation were selected. In order to generate the hab1-1swi3b-3 double mutant, we transferred pollen of swi3b-3 (Col background) to the stigmas of emasculated flowers of hab1-1 (Col background). The resulting F2 individuals were genotyped by PCR for the presence of the double mutant.
Yeast Two-Hybrid Screening
The HAB1 coding sequence was excised from a pSK-HAB1 construct (Rodriguez et al., 1998b) using EcoRI-SalI double digestion and subcloned into EcoRI-SalI doubly digested pGBT9 to generate an in-frame fusion with the GBD. To generate the HAB1 deletion, the HAB1 sequence encoding the catalytic PP2C region (amino acid residues 179 to 511; ΔNHAB1) was amplified by PCR and blunt-end-cloned into the EcoRV site from pBluescript SK+ (Stratagene). The ΔNHAB1 coding sequence was excised with EcoRI-SalI and subcloned into pGBT9. The pGBT9-ΔNHAB1 bait was transformed into the yeast strain AH109 (BD Biosciences). An oligo(dT)-primed cDNA library prepared in plasmid pACT2 using mRNA from an Arabidopsis cell suspension was kindly provided by K. Salchert (Nemeth et al., 1998). Yeast host AH109 carrying the pGBT9-ΔNHAB1 bait was transformed with 100 μg of DNA from the pACT2 cDNA library, then the cells were plated on SCD medium lacking Leu and Trp. Approximately 106 clones were obtained, and upon plating in SCD medium lacking Leu, Trp, His, and adenine, 20 clones containing putative interacting preys were selected. Yeast DNA was recovered and electroporated into Escherichia coli strain MC1065. pACT2 clones containing putative interacting preys were sequenced and retransformed into yeast strain AH109 carrying either the empty vector pGBT9 or pGBT9-ΔNHAB1 bait in order to verify true positives.
The PP2CA cDNA was obtained from the ABRC (clone M76G17STM). The PP2CA sequence encoding the catalytic PP2C region (amino acid residues 90 to 399) was amplified using the primers FDNPP2CA and RPP2CA. The PCR product was cloned into the pCR8/GW/TOPO entry vector (Invitrogen), and the ΔNPP2CA-coding sequence was excised with EcoRI-SalI and subcloned into pGBT9. The ABI1 and ABI2 cDNAs were kindly provided by Erwin Grill and have been described previously (Meyer et al., 1994; Rodriguez et al., 1998a). The ABI1 sequence encoding the catalytic PP2C region (amino acid residues 122 to 433) was excised using EcoRI-PstI double digestion and subcloned into pGBKT7 to generate pGBKT7-ΔNABI1. The ABI2 sequence encoding the catalytic PP2C region (amino acid residues 96 to 423; ΔNHAB2) was excised using ScaI-SalI double digestion and subcloned into pGBT9 to generate pGBT9-ΔNABI2.
Construction of Plasmids
pACT2-SWI3B-C1 was generated from the pACT2-SWI3B full-length cDNA recovered from the two-hybrid screening through XhoI digestion and subsequent religation. pACT2-SWI3B-C2, pACT2-SWI3B-N1, pACT2-SWIRM, and pACT2-ZZ were generated through PCR-mediated amplification using the following primer pairs, respectively: FATG and R660, F661 and R1410, FATG and R420, and F400 and R660. Constructs that express fusion proteins between the GAD and SWI3A, SWI3B, SWI3C, or SWI3D in the centromeric vector pPC86 were kindly provided by J.C. Reyes (CABIMER), and they have been described by Hurtado et al. (2006). Protein fusion between the GBD and ΔNHAB1 were generated in the multicopy vector pGBT9 for the yeast two-hybrid screening or the centromeric vector pDBLeu for targeted interaction assays with SWI3-like proteins. The G246D mutation was introduced into the pGBT9-ΔNHAB1 construct through replacement of a BglII-EcoRV fragment of HAB1 with a PCR-mutagenized version (see below).
Constructs to investigate the subcellular localization of HAB1 and SWI3B were generated in Gateway-compatible vectors. To this end, the coding sequences of HAB1 and SWI3B were PCR-amplified using the following primer pairs, respectively: FBamHI and Rno-stop, and FATG and R1407no-stop. The PCR products were cloned into the pCR8/GW/TOPO entry vector (Invitrogen) and recombined by LR reaction into the pMDC83 destination vector (Curtis and Grossniklaus, 2003).
Constructs to investigate in planta interaction using BiFC assays were made in the pSPYNE-35S and pSPYCE-35S vectors (Walter et al., 2004) as well as the Gateway vector pYFPN43 (kindly provided by A. Ferrando, Universidad de Valencia). The coding sequences of HAB1 and G246D hab1 were excised from pCR8/GW/TOPO constructs using double digestion with BamHI-StuI and subcloned into pSPYCE doubly digested BamHI-SmaI. The N-terminal half of SWI3B was excised from a pSK-SWI3B construct using double digestion with BamHI-DraI and subcloned into pSPYNE and pSPYCE doubly digested BamHI-SmaI. Constructs in which the basic Leu zipper TF bZIP63 is cloned in pSPYNE-35S and pSPYCE-35S were kindly provided by J. Kudla (University of Münster). The coding sequences of ABI1, ABI2, and PP2CA were PCR-amplified and cloned into pCR8/GW/TOPO and recombined by LR reaction into the pYFPN43 destination vector.
Expression and Purification of MBP-HAB1, MBP-ΔNHAB1, and MBP-G246D ΔNhab1
The coding region of the HAB1 cDNA (Rodriguez et al., 1998b) was PCR-amplified using the primers FSphI and RSphISacI. The PCR product was cloned subsequently into the EcoRV site of pBluescript SK (Stratagene), generating pSK-HAB1. Next, an EcoRI-SalI DNA fragment was excised from pSK-HAB1 and subcloned into the pMal-c2 vector (New England Biolabs). In order to obtain an N-terminal deletion of HAB1 (ΔNHAB1), a HindIII DNA fragment encompassing the amino acid residues 116 to 511 was excised from pSK-HAB1 and subcloned into the pMal-c2 vector. HAB1 cDNA was mutagenized by PCR in order to engineer a G246D substitution (Ho et al., 1989). To this end, the following oligonucleotides were used as primers: FPCR1 (5′-TATGATGGTCATGACGGCCATAAGGTT-3′), in which the codon for Gly-246 (GGA) was changed to Asp (GAC), RATT380, FATTATG, and RPCR2. Once the pMalc2-based constructs were verified by sequencing, expression of recombinant MBP-HAB1, MBP-ΔNHAB1, and MBP-G246D ΔNhab1 was induced with 1 mM isopropylthio-β-galactoside in E. coli DH5α cells. The fusion proteins were purified by amylose affinity chromatography according to the manufacturer's instructions (New England Biolabs).
PP2C Activity Assays
Phosphatase activity was measured using 33P-labeled casein as a substrate. Dephosphorylated casein (P-4765; Sigma-Aldrich) was 33P-labeled with bovine heart cAMP-dependent protein kinase (P-5511; Sigma-Aldrich) in a 500-μL reaction volume containing 50 mM Tris-HCl, pH 7.6, 10 mM MgCl2, 1 mM DTT, 60 μM cAMP, 50 μM unlabeled ATP, and 0.1 μCi/μL [γ-33P]ATP. The radiolabeled casein was precipitated with 20% trichloroacetic acid, and after two washings with 10% trichloroacetic acid, the casein was dissolved in 200 mM Tris-HCl, pH 7.6. Phosphatase assays were performed in a 50-μL reaction volume containing 20 mM Tris-HCl, pH 7.6, 10 mM MgCl2, 1 mM DTT, and ∼10,000 cpm of 33P-labeled casein. After incubation for 30 min at 30°C, the reaction was stopped with 100 μL of 20% trichloroacetic acid, samples were centrifuged, and the release of 33Pi in the supernatant was determined by scintillation counting.
Transient Protein Expression in Tobacco
Experiments were performed basically as described by Voinnet et al. (2003). The different binary vectors described above were introduced into Agrobacterium tumefaciens C58C1 (pGV2260) (Deblaere et al., 1985) by electroporation, and transformed cells were selected on Luria-Bertani plates supplemented with kanamycin (50 μg/mL). Then, they were grown in liquid Luria-Bertani medium to late exponential phase and cells were harvested by centrifugation and resuspended in 10 mM MES-KOH, pH 5.6, containing 10 mM MgCl2 and 150 μM acetosyringone to an OD600 of 1. These cells were mixed with an equal volume of Agrobacterium C58C1 (pCH32 35S:p19) expressing the silencing suppressor p19 of Tomato bushy stunt virus (Voinnet et al., 2003) so that the final density of Agrobacterium solution was ∼1. Bacteria were incubated for 3 h at room temperature and then injected into young fully expanded leaves of 4-week-old tobacco plants. Leaves were examined after 3 to 4 d with a Leica TCS-SL confocal microscope and a laser scanning confocal imaging system. Samples for immunoblot and immunoprecipitation assays were harvested, frozen in liquid nitrogen, and stored at −80°C.
Germination and Growth Assays
To measure ABA sensitivity, seeds (∼200 seeds per experiment) were plated on solid medium composed of MS basal salts, 1% sucrose, and increasing concentrations of ABA. In order to score seed germination, the percentage of seeds that had germinated and developed fully green expanded cotyledons was determined. ABA-resistant growth from swi3b +/− heterozygous seedlings (∼20 seedlings per experiment) was scored by weighing whole plants after 12 d of the transfer of 5-d-old seedlings grown on 0.5 μM ABA onto MS plates supplemented with 10 μM ABA. Heterozygous individuals from the swi3b-1 or swi3b-2 progeny were identified by their hygromycin or sulfadiazine resistance, respectively.
Protein Extraction, Protein Blot Analysis, and Immunoprecipitation
Protein extracts for immunodetection experiments were prepared from either tobacco leaves infiltrated with Agrobacterium or transgenic lines from Arabidopsis. Plant material (∼200 mg) for protein gel blot analysis was directly extracted in 2× Laemmli buffer (125 mM Tris-HCl, pH 6.8, 4% SDS, 20% glycerol, 2% mercaptoethanol, and 0.001% bromphenol blue), and proteins were run on a 10% SDS-PAGE gel and analyzed by immunoblotting. Plant material (∼1 g) for immunoprecipitation experiments was extracted in 3 volumes of PBS supplemented with 1 mM EDTA, 0.05% Triton X-100, and protease inhibitor cocktail (Roche). Protein concentration in each lysate was adjusted to the same value, and equal volumes of lysates (1 mL) were incubated with 1 μg/mL anti-HA high-affinity rat monoclonal antibody (clone 3F10; Roche) for 4 h at 4°C. After incubation, 20 μL of protein G–agarose beads (Roche) was added to precipitate the antigen/antibody complex. The protein G–agarose beads were collected after 1 h of incubation at 4°C by centrifugation and washed three times with extraction buffer. The antigen/antibody complex was eluted by boiling in Laemmli buffer and run on a 10% SDS-PAGE gel. Proteins immunoprecipitated with anti-HA antibodies were transferred onto Immobilon-P membranes (Millipore) and probed with either anti-HA-peroxidase or anti-c-myc-peroxidase conjugate (Roche), and detection was performed using the ECL advance protein gel blotting detection kit (GE Healthcare). The imaging of the chemiluminiscent signal was achieved using a highly efficient cooled CCD camera system (LAS-3000 luminiscent image analyzer from Fuji Photo Film). The signal intensities of the digitalized images were quantified using Image-Gauge version 4.0 software (Fuji Photo Film) according to the manufacturer's conditions. Immunodetection of GFP fusion proteins was performed with an anti-GFP monoclonal antibody (clone JL-8; Clontech) as primary antibody and ECL anti-mouse peroxidase (GE Healthcare) as secondary antibody. A rabbit antibody against peptide comprising amino acids 3 to 17 of GFP (anti-GFPN) was employed to detect YFPN fusion proteins (G1544; Sigma-Aldrich).
RNA Analyses
Plants were grown on MS plates supplemented with 1% sucrose either in the absence or presence of 0.3 μM ABA. After 7 d, ∼30 to 40 seedlings were collected and frozen in liquid nitrogen. Total RNA was extracted using a Qiagen RNeasy plant mini kit, and 1 μg of the RNA solution obtained was reverse-transcribed using 0.1 μg of oligo(dT)15 primer and Moloney murine leukemia virus reverse transcriptase (Roche), to finally obtain a 40-μL cDNA solution. RT-qPCR amplifications and measurements were performed using an ABI PRISM 7000 sequence detection system (Perkin-Elmer Applied Biosystems). RT-qPCR amplifications were monitored using the Eva-Green fluorescent stain (Biotium). Relative quantification of gene expression data was performed using the 2−ΔΔCT (or comparative CT) method (Livak and Schmittgen, 2001). Expression levels were normalized using the CT values obtained for the β-actin8 gene. The presence of a single PCR product was further verified by dissociation analysis in all amplifications. All quantifications were made in triplicate on RNA samples obtained from three independent experiments. The sequences of the primers used for PCR amplifications are indicated at Supplemental Table 1 online.
Generation of Epitope HA-Tagged HAB1 Transgenic Lines
The pBluescriptSK-ProHAB1:HAB1 construct was described by Saez et al. (2004). Two copies of the HA epitope sequence encoding YPYDVPDYA were cloned at the C-terminal sequence of HAB1 cDNA in the construct mentioned above. The complete expression cassette comprising the HAB1 promoter, the double HA epitope–tagged HAB1 coding sequence, and the NOS terminator was subcloned into SacI-SalI doubly digested pCAMBIA 1300 (hygromycin-resistant). The resulting construct was named pCAMBIA1300-ProHAB1:HAB1-dHA and used to transform hab1-1 (kanamycin-resistant) plants as described by Saez et al. (2004). Transgenic plants were screened in vitro on MS medium (M5524; Sigma-Aldrich) with 20 mg/L hygromycin B (H9773; Sigma-Aldrich).
Biochemical Fractionation of Epitope HA-Tagged HAB1
This protocol is based on fractionation techniques described by Bowler et al. (2004), Poveda et al. (2004), and Cho et al. (2006). Rosette leaves from 3- to 4-week-old plants were mock-treated or treated with 50 μM ABA for 1 h, harvested, and frozen in liquid nitrogen. Next, plant material was ground in lysis buffer (20 mM Tris-HCl, pH 7.4, 25% glycerol, 20 mM KCl, 2 mM EDTA, 2.5 mM MgCl2, 250 mM sucrose–containing protease inhibitor cocktail [Roche], and 1 mM phenylmethylsulfonyl fluoride [PMSF]). The lysate was filtered through four layers of Miracloth and centrifuged at 1000g for 10 min at 4°C to pellet the nuclei. The soluble cytosolic fraction was removed, and the pellet was washed in nuclei resuspension buffer, 20 mM Tris-HCl, 25% glycerol, 2.5 mM MgCl2, and 0.5% Triton X-100. After centrifugation at 1000g for 30 s at 4°C, a nuclear pellet was obtained, which was resuspended in 5 volumes of medium salt buffer (Bowler et al., 2004), 20 mM Tris-HCl, 0.4 M NaCl, 1 mM EDTA, 5% glycerol, 1 mM 2-mercaptoethanol, 0.1% Triton X-100, 0.5 mM PMSF, and protease inhibitor cocktail (Roche) and then frozen and thawed. After incubation with gentle mixing for 15 min at 4°C, the nuclear insoluble fraction, containing the major nuclear protein histones, was precipitated by centrifugation at 10,000g for 10 min, whereas the supernatant contained the nuclear soluble fraction. Detection of HAB1 was performed using anti-HA peroxidase conjugate (Roche). The purity of the different fractions was demonstrated using rabbit antibodies against histone H3 (Abcam) and ribulose-1,5-bisphosphate carboxylase.
ChIP
The ChIP protocol described here is a variation of the previously published protocols from Johnson et al. (2002) and Pascual-Ahuir et al. (2006). A transgenic line of Arabidopsis expressing a double HA epitope–tagged HAB1 in a hab1-1 background was used as starting plant material. In parallel, plant material from the hab1-1 mutant was used as a control for the experiment. Rosette leaves from 3- to 4-week-old plants were mock-treated or treated with 50 μM ABA for 1 h and then harvested and immersed in buffer A (0.4 M sucrose, 10 mM Tris, pH 8, 1 mM EDTA, 1 mM PMSF, and 1% formaldehyde) under vacuum for 10 min. Gly was added to a final concentration of 0.1 M, and incubation was continued for an additional 5 min under vacuum. Next, the plant material was washed with TBS (20 mM Tris-HCl, pH 8, and 150 mM NaCl) and frozen in liquid nitrogen. Cross-linked material (∼1 g) was ground with a mortar with pestle, after which it was resuspended in 1 mL of ice-cold lysis buffer (50 mM HEPES/KOH, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% deoxycholate, 0.1% SDS, and 1 mM PMSF) and transferred to a 2-mL screw-cap vial. Approximately 0.5 mL of zirconia/silica beads was added, and plant material was disrupted at 4°C for 10 min in the Mini Bead Beater 8 (Biospec Products; maximal speed, three rounds of 1 min). The lysate was collected into a 1.5-mL microtube and centrifuged for 1 min at 4°C. The pellet was collected and washed once in 1 mL of ice-cold lysis buffer. Next, the pellet was resuspended in 0.5 mL of cold lysis buffer and sonicated three times for 20 s (Branson Sonifier; output, 50%; needle, 5). Sonication resulted in the fragmentation of the chromatin into soluble pieces in the range of 300 to 500 bp. After centrifugation for 30 min at 4°C, the supernatant containing the soluble chromatin fragments (chromatin input) was transferred to 1.5-mL microtubes and stored at −80°C.
To immunoprecipitate HAB1-dHA cross-linked to chromatin fragments, samples were incubated with 10 μg/mL anti-HA high-affinity rat monoclonal antibody (clone 3F10; Roche) for 30 min on a roller at room temperature. In the meantime, protein G–agarose beads were washed twice with lysis buffer, 25 μL was added to each sample, and the incubation was continued for 60 min. The agarose beads were recovered by centrifugation and then washed with 1 mL of each of the following buffers: 2× lysis buffer, 2× lysis buffer and 0.5 M NaCl, 1× buffer B (10 mM Tris-HCl, pH 8, 0.25 M LiCl, 1 mM EDTA, 0.5% Nonidet P-40, and 0.5% deoxycholate), and 1× TE (10 mM Tris-HCl, pH 8, and 1 mM EDTA). The immunocomplexes were eluted from the beads by incubation for 10 min at 65°C in 250 μL of buffer containing 50 mM Tris-HCl, pH 8, 10 mM EDTA, and 1% SDS. After centrifugation, the supernatant was transferred to a microtube containing 250 μL of TE buffer and 20 μg of Pronase (Roche), and the samples were incubated for 1 h at 42°C followed by 5 h at 65°C to reverse formaldehyde-induced cross-links. In addition to the immunoprecipitated samples, aliquots (50 μL) of the total chromatin input that were not subjected to immunoprecipitation were also treated with Pronase and de-cross-linked to provide a quantitative measurement of the DNA input present in each sample. Finally, genomic DNA fragments were purified by the addition of 50 μL of 4 M LiCl and extraction with 300 μL of phenol:chloroform:isoamyl alcohol and ethanol precipitation (adding 20 μg of glycogen as carrier). DNA pellets were washed with 70% ethanol, dissolved in 100 μL of TE buffer, and stored at −20°C. RT-qPCR was used to determine the amounts of genomic DNA immunoprecipitated in the ChIP experiment. The sequences of the primers used for PCR amplifications are indicated at Supplemental Table 1 online.
Accession Numbers
The Arabidopsis Genome Initiative locus identifiers for HAB1 and SWI3B are At1g72770 and At2g33610, respectively. RD29B, RAB18, KIN1, RD22, RD29A, and P5CS1 correspond to At5g52300, At5g66400, At5g15960, At5g25610, At5g52310, and At2g39800, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The N-Terminal Region of HAB1 Has Transcriptional Activation Function in Yeast When Fused to the Gal4 DNA Binding Domain.
Supplemental Figure 2. Treatment with 50 μM ABA for Different Periods of Time (5 min to 1 h) Does Not Change the Subcellular Localization of GFP-HAB1.
Supplemental Figure 3. Detached Leaves Water Loss Assays Do Not Show Significant Differences between Wild-Type and swi3b-1 and swi3b-2 +/− Plants.
Supplemental Figure 4. Reduced Vegetative and Reproductive Growth of the swi3b-3 Mutant.
Supplemental Figure 5. Putative Nuclear Localization Signals in HAB1, PP2CA, and ABI2.
Supplemental Table 1. Primers Used for PCR Amplifications.
Supplemental Data Set 1. Text File of the Alignment Corresponding to Supplemental Figure 5.
Supplementary Material
Acknowledgments
We thank K. Salchert (Max Planck Institute) for providing the pACT2 Arabidopsis cDNA library, J.C. Reyes (CABIMER) for SWI3-like constructs, and J. Kudla (University of Münster) for BiFC vectors. We thank the Sainsbury Laboratory (John Innes Centre) for providing Agrobacterium C58C1 (pCH32 35S:p19 and pCH32 35S:GFP). We thank M. Proft and A. Pascual-Ahuir (Instituto de Biología Molecular y Celular de Plantas) for invaluable help with ChIP experiments. We thank G. Rios (Instituto Valenciano de Investigaciones Agrarias) and C. Koncz (Max Planck Institute) for providing the swi3b-1 and swi3b-2 mutants and the ABRC/Nottingham Arabidopsis Stock Centre for distributing seeds from TILLING lines. A.R. was supported by a fellowship from Portuguese Agency for Science and Technology. J.S. was supported by a Formacion Personal Investigador fellowship from Spanish Agency for Science and Education. S.R. was supported by the European Social Fund through an I3P fellowship from Consejo Superior de Investigaciones Científicas. This work was supported by Grants BIO2005-01760 and BIO2008-0221 from the Ministerio de Educación y Ciencia and Fondo Europeo de Desarrollo Regional.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Pedro L. Rodriguez (prodriguez@ibmcp.upv.es).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon, J.M., Malleret, G., Touzani, K., Vronskaya, S., Ishii, S., Kandel, E.R., and Barco, A. (2004). Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: A model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron 42 947–959. [DOI] [PubMed] [Google Scholar]
- Aravind, L. and Iyer, L. M. (2002). The SWIRM domain: A conserved module found in chromosomal proteins points to novel chromatin-modifying activities. Genome Biol. 3 RESEARCH0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero, J.M., Rodriguez, P.L., Quesada, V., Piqueras, P., Ponce, M.R., and Micol, J.L. (2006). Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3 and ABA1 in response to salt stress. Plant Cell Environ. 29 2000–2008. [DOI] [PubMed] [Google Scholar]
- Bensmihen, S., Rippa, S., Lambert, G., Jublot, D., Pautot, V., Granier, F., Giraudat, J., and Parcy, F. (2002). The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 14 1391–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezhani, S., Winter, C., Hershman, S., Wagner, J.D., Kennedy, J.F., Kwon, C.S., Pfluger, J., Su, Y., and Wagner, D. (2007). Unique, shared, and redundant roles for the Arabidopsis SWI/SNF chromatin remodeling ATPases BRAHMA and SPLAYED. Plant Cell 19 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler, C., Benvenuto, G., Laflamme, P., Molino, D., Probst, A.V., Tariq, M., and Paszkowski, J. (2004). Chromatin techniques for plant cells. Plant J. 39 776–789. [DOI] [PubMed] [Google Scholar]
- Brzeski, J., Podstolski, W., Olczak, K., and Jerzmanowski, A. (1999). Identification and analysis of the Arabidopsis thaliana BSH gene, a member of the SNF5 gene family. Nucleic Acids Res. 27 2393–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman, S., Gebuhr, T., Yee, D., La Mantia, C., Nicholson, J., Gilliam, A., Randazzo, F., Metzger, D., Chambon, P., Crabtree, G., and Magnuson, T. (2000). A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 6 1287–1295. [DOI] [PubMed] [Google Scholar]
- Cairns, B.R., Kim, Y.J., Sayre, M.H., Laurent, B.C., and Kornberg, R.D. (1994). A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc. Natl. Acad. Sci. USA 91 1950–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns, B.R., and Kingston, R.E. (2000). The SWI/SNF family of remodelling complexes. In Chromatin Structure and Gene Expression, S.C.R Elgin and J.L. Workman, eds (Oxford, UK: Oxford University Press), pp. 97–110.
- Carrozza, M.J., Utley, R.T., Workman, J.L., and Cote, J. (2003). The diverse functions of histone acetyltransferase complexes. Trends Genet. 19 321–329. [DOI] [PubMed] [Google Scholar]
- Cherel, I., Michard, E., Platet, N., Mouline, K., Alcon, C., Sentenac, H., and Thibaud, J.B. (2002). Physical and functional interaction of the Arabidopsis K(+) channel AKT2 and phosphatase AtPP2CA. Plant Cell 14 1133–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, Y.H., Yoo, S.D., and Sheen, J. (2006). Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 127 579–589. [DOI] [PubMed] [Google Scholar]
- Choi, H., Hong, J., Ha, J., Kang, J., and Kim, S.Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275 1723–1730. [DOI] [PubMed] [Google Scholar]
- Curtis, M.D., and Grossniklaus, U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da, G., Lenkart, J., Zhao, K., Shiekhattar, R., Cairns, B.R., and Marmorstein, R. (2006). Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes. Proc. Natl. Acad. Sci. USA 103 2057–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, A.K., Helps, N.R., Cohen, P.T., and Barford, D. (1996). Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 A resolution. EMBO J. 15 6798–6809. [PMC free article] [PubMed] [Google Scholar]
- David, G., Dannenberg, J.H., Simpson, N., Finnerty, P.M., Miao, L., Turner, G.M., Ding, Z., Carrasco, R., and Depinho, R.A. (2006). Haploinsufficiency of the mSds3 chromatin regulator promotes chromosomal instability and cancer only upon complete neutralization of p53. Oncogene 25 7354–7360. [DOI] [PubMed] [Google Scholar]
- Deblaere, R., Bytebier, B., De Greve, H., Deboeck, F., Schell, J., Van Montagu, M., and Leemans, J. (1985). Efficient octopine Ti plasmid-derived vectors for Agrobacterium-mediated gene transfer to plants. Nucleic Acids Res. 13 4777–4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrona, S., Hurtado, L., Bowman, J.L., and Reyes, J.C. (2004). The Arabidopsis thaliana SNF2 homolog AtBRM controls shoot development and flowering. Development 131 4965–4975. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R.R., Gampala, S.S., and Rock, C.D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14 (suppl.): S15–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R., and Lynch, T.J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R., Wang, M.L., Lynch, T.J., Rao, S., and Goodman, H.M. (1998). The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricker, M., Runions, J., and Moore, I. (2006). Quantitative fluorescence microscopy: From art to science. Annu. Rev. Plant Biol. 57 79–107. [DOI] [PubMed] [Google Scholar]
- Giraudat, J., Hauge, B.M., Valon, C., Smalle, J., Parcy, F., and Goodman, H.M. (1992). Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4 1251–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Garcia, M.P., Rodriguez, D., Nicolas, C., Rodriguez, P.L., Nicolas, G., and Lorenzo, O. (2003). Negative regulation of abscisic acid signaling by the Fagus sylvatica FsPP2C1 plays a role in seed dormancy regulation and promotion of seed germination. Plant Physiol. 133 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti, F., Beaudoin, N., Serizet, C., Webb, A.A., Vartanian, N., and Giraudat, J. (1999). ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11 1897–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Y., Xiong, L., Song, C.P., Gong, D., Halfter, U., and Zhu, J.K. (2002). A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev. Cell 3 233–244. [DOI] [PubMed] [Google Scholar]
- Himmelbach, A., Hoffmann, T., Leube, M., Hohener, B., and Grill, E. (2002). Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J. 21 3029–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach, A., Yang, Y., and Grill, E. (2003). Relay and control of abscisic acid signaling. Curr. Opin. Plant Biol. 6 470–479. [DOI] [PubMed] [Google Scholar]
- Ho, S.N., Hunt, H.D., Horton, R.M., Pullen, J.K., and Pease, L.R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77 51–59. [DOI] [PubMed] [Google Scholar]
- Huang, D., Jaradat, M.R., Wu, W., Ambrose, S.J., Ross, A.R., Abrams, S.R., and Cutler, A.J. (2007). Structural analogs of ABA reveal novel features of ABA perception and signaling in Arabidopsis. Plant J. 50 414–428. [DOI] [PubMed] [Google Scholar]
- Hurtado, L., Farrona, S., and Reyes, J.C. (2006). The putative SWI/SNF complex subunit BRAHMA activates flower homeotic genes in Arabidopsis thaliana. Plant Mol. Biol. 62 291–304. [DOI] [PubMed] [Google Scholar]
- Israelsson, M., Siegel, R.S., Young, J., Hashimoto, M., Iba, K., and Schroeder, J.I. (2006). Guard cell ABA and CO2 signaling network updates and Ca2+ sensor priming hypothesis. Curr. Opin. Plant Biol. 9 654–663. [DOI] [PubMed] [Google Scholar]
- Johnson, L., Cao, X., and Jacobsen, S. (2002). Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 12 1360–1367. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Reuling, G., and Karssen, C.M. (1984). The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol. Plant. 61 377–383. [Google Scholar]
- Kuhn, J.M., Boisson-Dernier, A., Dizon, M.B., Maktabi, M.H., and Schroeder, J.I. (2006). The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol. 140 127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, C.S., and Wagner, D. (2007). Unwinding chromatin for development and growth: A few genes at a time. Trends Genet. 23 403–412. [DOI] [PubMed] [Google Scholar]
- Lee, K.H., Piao, H.L., Kim, H.Y., Choi, S.M., Jiang, F., Hartung, W., Hwang, I., Kwak, J.M., Lee, I.J., and Hwang, I. (2006). Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126 1109–1120. [DOI] [PubMed] [Google Scholar]
- Leonhardt, N., Kwak, J.M., Robert, N., Waner, D., Leonhardt, G., and Schroeder, J.I. (2004). Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16 596–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, J., Bouvier-Durand, M., Morris, P.C., Guerrier, D., Chefdor, F., and Giraudat, J. (1994). Arabidopsis ABA response gene ABI1: Features of a calcium-modulated protein phosphatase. Science 264 1448–1452. [DOI] [PubMed] [Google Scholar]
- Leung, J., Merlot, S., and Giraudat, J. (1997). The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J., and Schmittgen, T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25 402–408. [DOI] [PubMed] [Google Scholar]
- McCarty, D.R., Hattori, T., Carson, C.B., Vasil, V., Lazar, M., and Vasil, I.K. (1991). The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 66 895–905. [DOI] [PubMed] [Google Scholar]
- Merlot, S., Gosti, F., Guerrier, D., Vavasseur, A., and Giraudat, J. (2001). The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 25 295–303. [DOI] [PubMed] [Google Scholar]
- Meyer, K., Leube, M.P., and Grill, E. (1994). A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264 1452–1455. [DOI] [PubMed] [Google Scholar]
- Miao, Y., Lv, D., Wang, P., Wang, X.C., Chen, J., Miao, C., and Song, C.P. (2006). An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell 18 2749–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moes, D., Himmelbach, A., Korte, A., Haberer, G., and Grill, E. (2008). Nuclear localization of the mutant protein phosphatase abi1 is required for insensitivity towards ABA responses in Arabidopsis. Plant J. 54 806–819. [DOI] [PubMed] [Google Scholar]
- Mohrmann, L., and Verrijzer, C.P. (2005). Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim. Biophys. Acta 1681 59–73. [DOI] [PubMed] [Google Scholar]
- Nambara, E., and Marion-Poll, A. (2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56 165–185. [DOI] [PubMed] [Google Scholar]
- Nemeth, K., et al. (1998). Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev. 12 3059–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, P.C., and Henikoff, S. (2001). Predicting deleterious amino acid substitutions. Genome Res. 11 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, N., Yoshida, T., Kitahata, N., Asami, T., Shinozaki, K., and Hirayama, T. (2007). ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J. 50 935–949. [DOI] [PubMed] [Google Scholar]
- Ohta, M., Guo, Y., Halfter, U., and Zhu, J.K. (2003). A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc. Natl. Acad. Sci. USA 100 11771–11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, G.K., Grant, J.J., Cheong, Y.H., Kim, B.G., Li, L., and Luan, S. (2005). ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiol. 139 1185–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual-Ahuir, A., Struhl, K., and Proft, M. (2006). Genome-wide location analysis of the stress-activated MAP kinase Hog1 in yeast. Methods 40 272–278. [DOI] [PubMed] [Google Scholar]
- Peterson, C.L., Dingwall, A., and Scott, M.P. (1994). Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc. Natl. Acad. Sci. USA 91 2905–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting, C.P., Blake, D.J., Davies, K.E., Kendrick-Jones, J., and Winder, S.J. (1996). ZZ and TAZ: New putative zinc fingers in dystrophin and other proteins. Trends Biochem. Sci. 21 11–13. [PubMed] [Google Scholar]
- Poveda, A., Pamblanco, M., Tafrov, S., Tordera, V., Sternglanz, R., and Sendra, R. (2004). Hif1 is a component of yeast histone acetyltransferase B, a complex mainly localized in the nucleus. J. Biol. Chem. 279 16033–16043. [DOI] [PubMed] [Google Scholar]
- Razem, F.A., El Kereamy, A., Abrams, S.R., and Hill, R.D. (2006). The RNA-binding protein FCA is an abscisic acid receptor. Nature 439 290–294. [DOI] [PubMed] [Google Scholar]
- Robert, N., Merlot, S., N'guyen, V., Boisson-Dernier, A., and Schroeder, J. I. (2006). A hypermorphic mutation in the protein phosphatase 2C HAB1 strongly affects ABA signaling in Arabidopsis. FEBS Lett. 580 4691–4696. [DOI] [PubMed] [Google Scholar]
- Roberts, C.W., Galusha, S.A., McMenamin, M.E., Fletcher, C.D., and Orkin, S.H. (2000). Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc. Natl. Acad. Sci. USA 97 13796–13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, P.L., Benning, G., and Grill, E. (1998. a). ABI2, a second protein phosphatase 2C involved in abscisic acid signal transduction in Arabidopsis. FEBS Lett. 421 185–190. [DOI] [PubMed] [Google Scholar]
- Rodriguez, P.L., Leube, M.P., and Grill, E. (1998. b). Molecular cloning in Arabidopsis thaliana of a new protein phosphatase 2C (PP2C) with homology to ABI1 and ABI2. Plant Mol. Biol. 38 879–883. [DOI] [PubMed] [Google Scholar]
- Saez, A., Apostolova, N., Gonzalez-Guzman, M., Gonzalez-Garcia, M.P., Nicolas, C., Lorenzo, O., and Rodriguez, P.L. (2004). Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J. 37 354–369. [DOI] [PubMed] [Google Scholar]
- Saez, A., Robert, N., Maktabi, M.H., Schroeder, J.I., Serrano, R., and Rodriguez, P.L. (2006). Enhancement of abscisic acid sensitivity and reduction of water consumption in Arabidopsis by combined inactivation of the protein phosphatases type 2C ABI1 and HAB1. Plant Physiol. 141 1389–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnowski, T.J., Rios, G., Jasik, J., Swiezewski, S., Kaczanowski, S., Li, Y., Kwiatkowska, A., Pawlikowska, K., Kozbial, M., Kozbial, P., Koncz, C., and Jerzmanowski, A. (2005). SWI3 subunits of putative SWI/SNF chromatin-remodeling complexes play distinct roles during Arabidopsis development. Plant Cell 17 2454–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnowski, T.J., Swiezewski, S., Pawlikowska, K., Kaczanowski, S., and Jerzmanowski, A. (2002). AtSWI3B, an Arabidopsis homolog of SWI3, a core subunit of yeast Swi/Snf chromatin remodeling complex, interacts with FCA, a regulator of flowering time. Nucleic Acids Res. 30 3412–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder, J.I., Allen, G.J., Hugouvieux, V., Kwak, J.M., and Waner, D. (2001). Guard cell signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52 627–658. [DOI] [PubMed] [Google Scholar]
- Schweighofer, A., Hirt, H., and Meskiene, I. (2004). Plant PP2C phosphatases: Emerging functions in stress signaling. Trends Plant Sci. 9 236–243. [DOI] [PubMed] [Google Scholar]
- Smith, C.L., and Peterson, C.L. (2005). ATP-dependent chromatin remodeling. Curr. Top. Dev. Biol. 65 115–148. [DOI] [PubMed] [Google Scholar]
- Song, C.P., Agarwal, M., Ohta, M., Guo, Y., Halfter, U., Wang, P., and Zhu, J.K. (2005). Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17 2384–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridha, S., and Wu, K. (2006). Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J. 46 124–133. [DOI] [PubMed] [Google Scholar]
- Tahtiharju, S., and Palva, T. (2001). Antisense inhibition of protein phosphatase 2C accelerates cold acclimation in Arabidopsis thaliana. Plant J. 26 461–470. [DOI] [PubMed] [Google Scholar]
- Till, B.J., et al. (2003). Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res. 13 524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno, Y., Furihata, T., Abe, H., Yoshida, R., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 97 11632–11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet, O., Rivas, S., Mestre, P., and Baulcombe, D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33 949–956. [DOI] [PubMed] [Google Scholar]
- Walter, M., Chaban, C., Schutze, K., Batistic, O., Weckermann, K., Nake, C., Blazevic, D., Grefen, C., Schumacher, K., Oecking, C., Harter, K., and Kudla, J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40 428–438. [DOI] [PubMed] [Google Scholar]
- Yang, X., Zaurin, R., Beato, M., and Peterson, C.L. (2007). Swi3p controls SWI/SNF assembly and ATP-dependent H2A–H2B displacement. Nat. Struct. Mol. Biol. 14 540–547. [DOI] [PubMed] [Google Scholar]
- Yang, Y., Sulpice, R., Himmelbach, A., Meinhard, M., Christmann, A., and Grill, E. (2006). Fibrillin expression is regulated by abscisic acid response regulators and is involved in abscisic acid-mediated photoprotection. Proc. Natl. Acad. Sci. USA 103 6061–6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, R., Umezawa, T., Mizoguchi, T., Takahashi, S., Takahashi, F., and Shinozaki, K. (2006. a). The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J. Biol. Chem. 281 5310–5318. [DOI] [PubMed] [Google Scholar]
- Yoshida, T., Nishimura, N., Kitahata, N., Kuromori, T., Ito, T., Asami, T., Shinozaki, K., and Hirayama, T. (2006. b). ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol. 140 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, C., Miki, B., and Wu, K. (2003). CHB2, a member of the SWI3 gene family, is a global regulator in Arabidopsis. Plant Mol. Biol. 52 1125–1134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.