Abstract
Despite the importance of sulfur (S) for plant nutrition, the role of the arbuscular mycorrhizal (AM) symbiosis in S uptake has received little attention. To address this issue, 35S-labeling experiments were performed on mycorrhizas of transformed carrot (Daucus carota) roots and Glomus intraradices grown monoxenically on bicompartmental petri dishes. The uptake and transfer of 35SO42− by the fungus and resulting 35S partitioning into different metabolic pools in the host roots was analyzed when altering the sulfate concentration available to roots and supplying the fungal compartment with cysteine (Cys), methionine (Met), or glutathione. Additionally, the uptake, transfer, and partitioning of 35S from the reduced S sources [35S]Cys and [35S]Met was determined. Sulfate was taken up by the fungus and transferred to mycorrhizal roots, increasing root S contents by 25% in a moderate (not growth-limiting) concentration of sulfate. High sulfate levels in the mycorrhizal root compartment halved the uptake of 35SO42− from the fungal compartment. The addition of 1 mm Met, Cys, or glutathione to the fungal compartment reduced the transfer of sulfate by 26%, 45%, and 80%, respectively, over 1 month. Similar quantities of 35S were transferred to mycorrhizal roots whether 35SO42−, [35S]Cys, or [35S]Met was supplied in the fungal compartment. Fungal transcripts for putative S assimilatory genes were identified, indicating the presence of the trans-sulfuration pathway. The suppression of fungal sulfate transfer in the presence of Cys coincided with a reduction in putative sulfate permease and not sulfate adenylyltransferase transcripts, suggesting a role for fungal transcriptional regulation in S transfer to the host. A testable model is proposed describing root S acquisition through the AM symbiosis.
Plants and arbuscular mycorrhizal (AM) fungi have been coevolving since the Devonian period (Simon et al., 1993; Brundrett, 2002). Today, the AM symbiosis involves an estimated 80% of land plant species and 92% of plant families (Wang and Qiu, 2006). This symbiosis augments nutrient uptake by roots, which deplete the rhizosphere at a rate dependent on water availability, as well as the concentration and soil mobility of the nutrient (Bhat et al., 1976; Li et al., 1991; Gahoonia et al., 1994; Wang et al., 2004). Phosphorus (P) is particularly immobile in soils (Gahoonia and Nielsen, 1991), and the AM symbiosis can increase plant growth in P-deficient soils 19-fold (Hayman and Mosse, 1971). Although P acquisition is a major benefit of the symbiosis, a growing body of research points toward a more complex role for arbuscular mycorrhizas in nutrient uptake (Liu et al., 2002; Lambers et al., 2008). Nitrogen (N) nutrition, for instance, is improved in a number of crop species (Ortiz-Ceballos et al., 2007; Perner et al., 2008), and a monoxenic culture system consisting of transformed roots in symbiosis with the mycorrhizal fungus Glomus intraradices has been used to demonstrate the capacity of this symbiosis to transfer N (Govindarajulu et al., 2005; Jin et al., 2005).
Plants take up sulfur (S) primarily as the sulfate anion (Leustek, 1996), which is often found in low concentrations in the soil. The uptake and utilization of S by plants is reviewed by Leustek et al. (2000) and Rennenberg et al. (2007). Sulfate is mobile and commonly lost through soil leaching (Eriksen and Askegaard, 2000). Because of this, typically 95% of the soil S is in organically bound forms such as sulfate esters (Tabatabai, 1986; Scherer, 2001) synthesized by soil microorganisms (Spencer and Harada, 1960; Fitzgerald, 1976). Such forms of S are not thought to be significant sources for plants (Leustek, 1996). In the past, the effects of leaching have likely been masked by the high input of S from atmospheric pollution. However, the drastic reductions in S deposition in North America and Europe over the last decade have led to an equally dramatic rise in reported cases of S deficiency in crop species (Lefohn et al., 1999; Riley et al., 2000; Baumgardner et al., 2002). Under conditions of low S availability, an AM symbiosis can increase the percentage of S in pot-grown onions (Cepa allium; Guo et al., 2007) and maize (Zea mays; Banerjee et al., 2003). The ability of mycorrhizal fungi to transfer nitrogen (N) and P from organic compounds (Joner and Jakobsen, 1994; Hawkins et al., 2000; Govindarajulu et al., 2005) also suggests the possibility that mycorrhizal plants might obtain S from organic sources.
The small number of reports on the effects of AM colonization on the uptake of S, which is present in amount equal to P in plants, have presented differing results (Gray and Gerdemann, 1973; Cooper and Tinker, 1978; Rhodes and Gerdemann, 1978a). The pioneering study by Gray and Gerdemann (1973) showing that mycorrhizal colonization in clover (Trifolium pratense) and maize increased 35S uptake compared to nonmycorrhizal plants was repeated by Rhodes and Gerdemann (1978a) in onion but was shown to be heavily influenced by P nutritional benefits. Rhodes and Gerdemann (1978b) were also the first to examine the transfer of 35SO42− through a mycorrhizal fungus exposed to an isolated source unavailable to the host plant. In a study published the same year, mycorrhizal clover (Trifolium repens) grown in sterile bicompartmental soil/agar plates failed to show a significant amount of transfer of isolated S through the fungus (Cooper and Tinker, 1978). More recent research on ectomycorrhizas is more definitive on the subject (for review, see Rennenberg, 1999). In the association between beech (Fagus sylvatica) seedlings and the ectomycorrhizal fungi Laccaria laccata or Paxillus involutus, only L. laccata was shown to increase S uptake in S-deficient conditions and only when N was supplied (Kreuzwieser and Rennenberg, 1998). Interestingly, an analysis of free-living Laccaria bicolor showed that in contrast with the results from other filamentous fungi and yeast, sulfate uptake was unaffected by the presence of S metabolites (Mansouri-Bauly et al., 2006). This may be an adaptation enabling the fungus to continue supplying sulfate to the host after its own needs are met (Mansouri-Bauly et al., 2006).
Though little is known about the mechanism of S assimilation and its regulation in mycorrhizal fungi, these pathways have been extensively characterized in other fungal species (for review, see Marzluf, 1997). Cys is the major entry point of reduced S into metabolism and is formed by the reaction of sulfide (made by the successive reduction of sulfate to sulfite and of sulfite to sulfide) with O-acetylserine catalyzed by Cys synthase. However, through the reverse trans-sulfuration pathway, both Aspergillus and Neurospora species are able to efficiently convert Met to Cys (Marzluf, 1993), making Met an efficient S source for these fungi. This pathway is crucial in Saccharomyces cerevisiae, where sulfide is assimilated not though Cys synthase, but through the sulfhydrolation of O-acetylhomoserine to form homo-Cys, from which Met is made (Thomas and Surdin-Kerjan, 1997; Ono et al., 1999). By contrast, Schizosaccharomyces pombe has been shown to be deficient in reverse trans-sulfuration pathway enzymes, making it dependent on Cys synthase for S assimilation (Brzywczy et al., 2002). This results in a reduced regulatory role for Met in S. pombe (Brzywczy et al., 2002). There is also evidence that the S assimilation enzymes are transcriptionally regulated in S. cerevisiae specifically by levels of Cys (Hansen and Johannesen, 2000). Both positive and negative global regulatory proteins have been discovered in Neurospora crassa, Aspergillus nidulans, and S. cerevisiae that share homology at the protein level (Marzluf, 1997). In N. crassa and S. cerevisiae, the expression of the S assimilatory genes is almost completely absent in the presence of Cys or Met and is greatly reduced by glutathione (GSH) addition (Ono et al., 1991, 1999; Marzluf, 1997). These same enzymes in A. nidulans, on the other hand, are not down-regulated in response to S metabolites (Clarke et al., 1997; Sienko and Paszewski, 1999; Natorff et al., 2003). The transcription of sulfate permeases is suppressed in all species studied by low concentrations of S metabolites (Bradfield et al., 1970; Ketter and Marzluf, 1988; Ketter et al., 1991; Marzluf, 1993, 1997; Cherest et al., 1997; Grynberg et al., 2001; Pilsyk et al., 2007). A putative Cys synthase transcript has been identified from the AM fungus G. intraradices (http://darwin.nmsu.edu/∼plammers/glomus/), which appears to be the only report on the identity or regulation of AM fungal genes related to S metabolism, transport, or regulation.
This study was aimed at determining the forms of S taken up, metabolized, and transferred to its host roots by the fungal partner in the AM symbiosis. A second goal was to identify effectors of S transfer to host roots and to begin to identify AM fungal genes of S handling and their regulation. Using an AM symbiosis of monoxenic cultured roots, the transfer of S was shown to be nutritionally significant and to be regulated by S availability to the host and by S metabolite availability to the extraradical mycelia (ERM). The uptake and transfer of reduced S through the symbiosis was also demonstrated. Putative homologs of S uptake and metabolism genes were also identified and their transcriptional regulation examined, indicating that S uptake is transcriptionally regulated at the level of sulfate permease. These findings allow the development of a testable model of S uptake and transfer in the AM symbiosis.
RESULTS
Growth and Sulfate Uptake by Nonmycorrhizal Roots
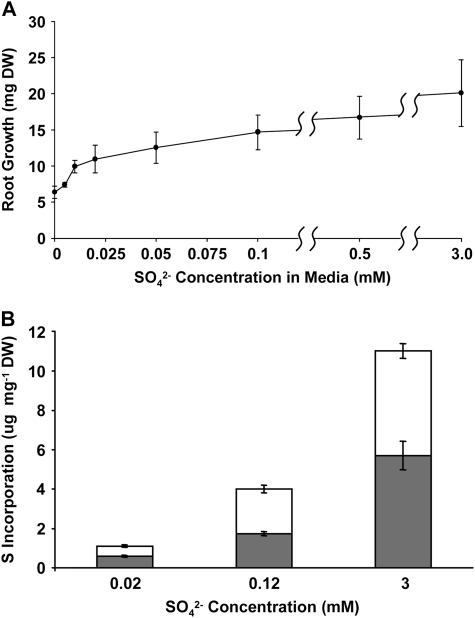
To establish conditions for studying S transfer between host roots and AM fungus, the relationship between sulfate availability, root growth, and sulfate utilization was investigated in uncolonized roots (Fig. 1). In the absence of available sulfate, there was a 3-fold decrease in root growth compared to growth under saturating sulfate levels (3 mm; Fig. 1A). Final dry weights were not significantly different when compared singly at sulfate concentrations above 0.005 mm; however, there was a consistent trend of increasing dry weight with increased S availability. Root growth was detected even in roots exposed to medium containing no S, probably due to internal S stores within the initial root segments. A much greater difference was seen in the uptake and incorporation of sulfate when roots were labeled with 35SO4−2 at high (3 mm), low (0.014 mm), or intermediate (0.114 mm) sulfate levels (Fig. 1B). There was a close to 4-fold increase in both aqueous alcohol-soluble and -insoluble S content from low to intermediate S conditions and an 11-fold increase between low and high S. A reduction in the root compartment sulfate concentration from 3 mm to 0.12 mm resulted in a reduction in uptake of 64% (Fig. 1B), with little or no reduction (27% difference in mean values, not statistically significant at the 95% confidence level) in growth (Fig. 1A). Lowering the sulfate available to mycorrhizal roots to <0.05 mm had a negative impact on the symbiosis, reducing the number of plates where the fungal ERM grew over the barrier from >80% to <20% (data not shown). There was no such reduction when 0.12 mm sulfate was utilized.
Figure 1.
The effect of sulfate concentration on the growth and S incorporation of nonmycorrhizal transformed carrot roots. A, Root growth measured as dry weight (DW) increase at different concentrations of Na2SO4 in 20 mL of modified liquid M media (St-Arnaud et al., 1996). B, The incorporation of S from solid media containing 0.02, 0.12, or 3 mm Na2SO4 labeled with 11.5 μCi of Na2[35SO4] was measured in roots after 4 weeks of labeled growth. Labeled root compounds were extracted with cold MeOH:water (70:30; white bars) before the cellular residue was solubilized in tissue solubilizer (gray bars). The fractions were analyzed by liquid scintillation counting. Error is expressed as sem.
Fungal Uptake and Transfer of Sulfate Is Influenced by Root Access to Sulfate
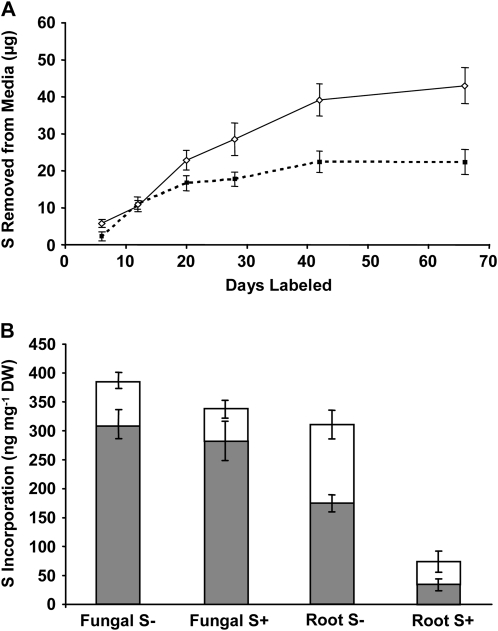
The ERM of G. intraradices takes up sulfate at significant rates (Fig. 2A) and transfers most of it to the mycorrhizal roots (Fig. 2B). The total amount of 35S in ERM tissue was <5% of the amount found in the root tissue because the root biomass is >20 times the fungal biomass (tissue mass data not shown). The availability of S in the root compartment strongly influences fungal uptake in the distal ERM compartment. More than twice as much sulfate was removed from the fungal compartment when the corresponding root compartment was exposed to 0.12 mm versus 3 mm S (Fig. 2A). When roots were grown in 0.12 mm sulfate, uptake by the fungus was linear up to day 40 (R2 = 0.98). The fungus removed 0.98 μg of S from the media per day during this time. Fungal uptake of S when 3 mm sulfate was supplied to roots was linear (0.83 μg/d of S; R2 = 0.92) only for the first 20 d before the rate of removal slowed.
Figure 2.
Increased sulfate availability in the root compartments of split plates reduces transfer of S through the distal fungal ERM. A, The uptake of S by ERM from plates labeled with 30 μCi of Na2[35SO4] in 0.12 mm Na2SO4 in the fungal compartments was measured as the removal of radioactivity from the media when the root compartments were supplied with moderate S (0.12 mm Na2SO4; solid line) or high S (3 mm Na2SO4; broken line). B, Measurements of S incorporated into the roots (Root S±) and distal fungal (Fungal S±) ERM from the above experiment after 66 d of labeling; S+ refers to high sulfate levels (3 mm) in root compartments and S− to moderate levels (0.12 mm). Radioactivity was measured by scintillation counting after a crude separation by cold MeOH:water (70:30) extraction (white bars) followed by solubilization of the remaining tissue (gray bars). The aqueous alcohol extraction dissolves sulfate, sulfite, S amino acids, and potentially also unidentified sulfated esters, while the remainder includes proteins, sulfolipids, and other unknown products. Values are presented as mean ± SEM (n ≥ 5). DW, Dry weight.
The total amount of S transferred from the fungal compartment to roots grown at high sulfate levels was about one-quarter of that in roots exposed to moderate sulfate levels (Fig. 2B). In contrast, there was no significant effect of sulfate levels in the root compartment on the incorporation of S by the distal ERM. Additionally, in roots growing at moderate S levels, the fraction containing sulfate and the S amino acids contained approximately twice as much 35S as did the same fraction in the ERM.
The amount of 35S in the media of the colonized root compartments was monitored throughout the experiment (see “Materials and Methods”) and was found to be low. The movement into the root compartment media from the fungal compartment was linear (R2 = 0.98) at 63 ng/d S when the root compartment media contained 0.12 mm sulfate. This represents 6.4% of the amount transferred to the roots. The movement into the high S root media was also linear (R2 = 0.99), with 55 ng/d S being moved across the barrier, representing 6.6% of the amount transferred to the roots.
Fungal Transfer of S to the Mycorrhizal Roots Versus Direct Uptake by Roots
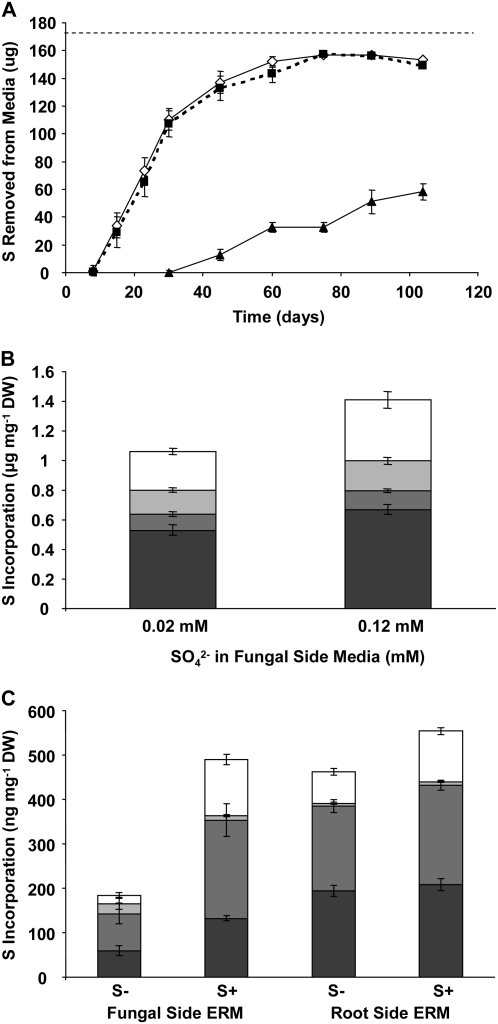
The relative contribution of S obtained from the fungal ERM to total root S was determined at intermediate S levels (0.22 mm) by comparing fungal-derived 35S in roots with 35S directly absorbed by the roots themselves. Plates were labeled with 35SO42− in the root compartment to determine the amount of S incorporated by direct root uptake. Half of these plates were also labeled in the fungal compartment with 35SO42− at 0.22 mm sulfate, while no S was added to the fungal compartments of the remaining plates. The roots took up most of the labeled sulfate initially provided within 8 weeks (Fig. 3A). This uptake was not affected by the presence of sulfate in the fungal compartments. The uptake of S from the fungal compartment media began when the fungus crossed the barrier (approximately 30 d after inoculation) and was comparable to direct root uptake rates in the next few weeks (Fig. 3A). S incorporation by the root and fungal tissue after approximately 10 weeks of growth is shown in Figure 3, B and C. When the fungal compartment contained 35SO42−, the total incorporation of S by the roots increased by approximately 25% (Fig. 3B). The largest change was in the sulfate fraction, which increased by 60%. There were no significant differences in root growth observed when sulfate was added to the fungal compartments (root weight data not shown), which is consistent with the lack of significant growth increase between moderate and high sulfate growth conditions (Fig. 1). When the fungal side was labeled, total incorporation of 35S by the fungus was similar in ERM collected from the fungal and root compartments when the fungal side was labeled (Fig. 3C), which is consistent with the absence of any known physiological differences between the ERM in the two compartments, and showing that the availability of S to the distal ERM did not affect incorporation by the ERM in the root compartment. 35S accumulated in the fungal ERM from the unlabeled fungal compartment, showing that in a low S environment (0.02 mm), S can be moved by the fungus from the root compartment to the distal ERM (Fig. 3C).
Figure 3.
Transfer of 35SO42− through the fungus versus direct root uptake. A, The uptake of S over 104 d from root and fungal compartment media. The root (proximal) compartments of split plates were labeled with 17.3 μCi of Na2[35SO4] in 0.2 mm Na2SO4 (171 μg S). One-half of the fungal (distal) compartments were labeled with 17.3 μCi of Na2[35SO4] in 0.2 mm Na2SO4 (171 μg S, white diamonds, solid line), while the others contained 0.02 mm sulfate without 35S (black squares, broken line). Distal uptake was monitored from labeled plates (black triangles, solid line). A straight broken line denotes the total initial S content of the proximal compartment media. B, Incorporation of 35S by roots when the fungal side was supplied with low unlabeled (0.02 mm) or moderate 35S-labeled (0.2 mm) sulfate was measured after 110 d of labeling by chemical fractionation into sulfate- (white bars), amino acid- (light gray bars), and protein- (medium gray bars) containing pools, and solubilized remaining tissue (dark gray bars) pools measured by liquid scintillation counting. C, Incorporation of 35S into different pools of ERM collected from the root or fungal side compartments of split plates when the fungal compartments contained 0.02 mm unlabeled sulfate (S−) or 0.2 mm of 35S-labeled (S+); as above, the root side compartments of all plates contained 0.2 mm 35S-labeled sulfate. Fractions are depicted as in A. Values represent means ± SEM (n ≥ 4). DW, Dry weight.
The Effect of S Metabolites on S Uptake and Transfer through the Mycorrhizal Symbiosis
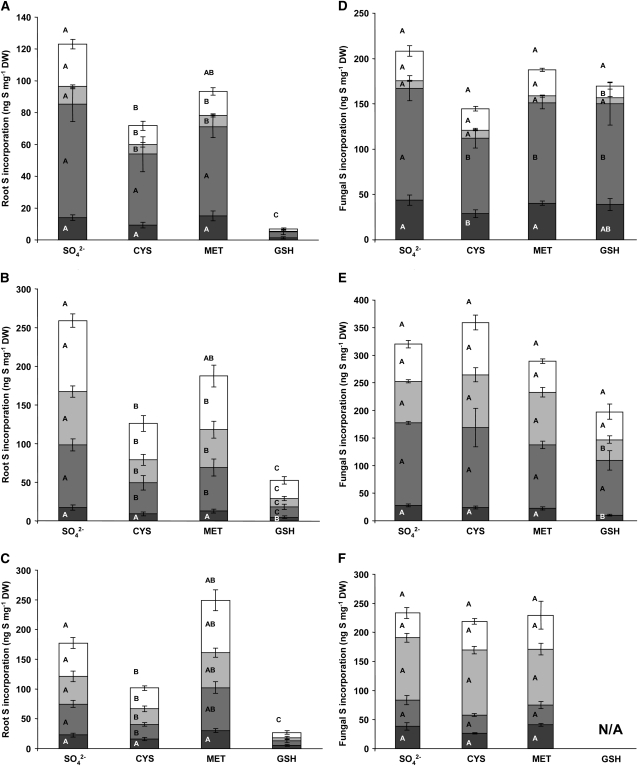
Radiolabeled sulfate was added to the fungal compartments of split plates together with Met, Cys, and GSH in amounts previously shown to suppress sulfate assimilation in other fungi (Ketter and Marzluf, 1988; Ketter et al., 1991; Kuras and Thomas, 1995; Ono et al., 1999; Grynberg et al., 2001). The distribution of 35S in root and fungal tissues was then measured after 2, 4, and 6 weeks. Substantial quantities of S were transferred to the roots over the course of the experiment despite the presence of 0.5 mm sulfate in the root compartments (Fig. 4, A–C). The presence of Cys decreased S transfer to the mycorrhizal roots by an average of 45% throughout the experiment, with the sulfate- and amino acid-containing pools of S in the roots showing larger fractional decreases. Met had no significant effect on total S transfer, although there were significant decreases in the 35S transferred to the low Mr soluble metabolite pools in mycorrhizal roots. GSH dramatically reduced 35S accumulation in the roots at all time points, with an average reduction of 80%. There were no significant effects of the presence of Cys, Met, or GSH on total S incorporation from labeled sulfate by the distal ERM and little or no effect on the partitioning of S within this tissue (Fig. 4, D–F).
Figure 4.
The effect of common S metabolite repressors Cys, Met, and GSH on the transfer and allocation of 35S in an arbuscular mycorrhiza. The incorporation of 35S into roots and fungal ERM was analyzed after labeling the distal compartments with 37.4 μCi of Na2[35SO4] in 0.12 mm Na2SO4 (91 μg S) alone (SO42−) or with 1 mm Cys, 1 mm Met, or 1 mm GSH. Root (A–C) and fungal (D–F) incorporation was analyzed after 2 (A and D), 4 (B and E), and 6 (C and F) weeks. Fungal ERM was collected from the distal compartments of the same plates as those from which root tissue was collected from the proximal compartments. Tissue was analyzed by chemical fractionation into pools containing sulfate (white bars), amino acids (light gray bars), proteins (medium gray bars), and the tissue solubilized biomass remaining after extraction (dark gray bars). Fractions were analyzed by liquid scintillation counting. Letters within the bars represent ANOVA single-factor analyses between bars of that type. Letters above the complete bar graphs are the analyses of total S uptake. Values are reported as mean ± SEM (n ≥ 5). DW, Dry weight.
GSH greatly reduced the growth of fungal ERM in the distal compartment, though not the root growth (not shown), so it is unclear whether the reduction in the transfer of S by GSH-treated ERM is due to the regulation of sulfate uptake or simply to reduced fungal growth. Neither Cys nor Met significantly affected the growth of fungal ERM or roots, with ERM biomass approximately doubling between weeks 2 and 4 (data not shown). Thus, the effect of Cys on S transfer to mycorrhizal roots does not appear to be due to growth inhibition.
Fungal Uptake and Transfer to Host Roots of Reduced S
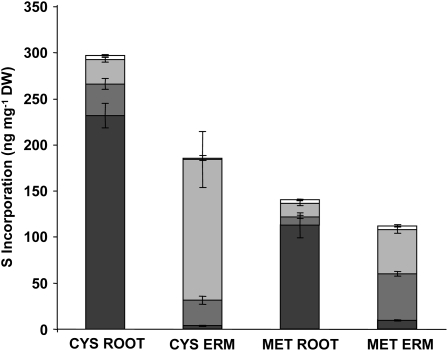
Fungal ERM imports and transfers substantial amounts of reduced S supplied as Cys or Met (Fig. 5). The amount of S transferred when 35S-labeled Cys is the sole source of S in the fungal compartment is comparable to the S transfer when plates are labeled with sulfate (Figs. 2B, 3B, and 4, A–C). Roughly one-half of this amount was transferred when Met was provided (Fig. 5). The incorporation of S by the fungal mycelium per milligram dry weight when labeled Cys is supplied to the fungal compartment is about 40% less than the level found in the roots. Utilizing Met as an S source led to a 46% increase in the amino acid fraction in the ERM (Fig. 5). There were striking differences between the relative amount of S in the solubilized fractions in both roots and fungal mycelia when reduced S was supplied compared to sulfate. Labeling with Cys and Met resulted in a much higher percentage of labeling in the solubilized root tissue pool (78% and 88%, respectively) than when sulfate was supplied as the primary S source (7%–21%; Fig. 4, A–D). The opposite relationship was found in fungal mycelia, where applying Cys or Met as primary sources of S led to solubilized tissue fractions that were at most 8% of the total S incorporated (Fig. 5). When labeled sulfate was the primary S source, 5% to 22% of the total S incorporation is found in the tissue solubilized fraction (Fig. 4, D–F).
Figure 5.
Transfer of reduced S through a mycorrhizal symbiosis. Mycorrhizal roots were grown in 0.5 mm Na2SO4 and the fungal partner allowed to grow into distal compartments containing either 1 mm Cys labeled with 25.7 μCi [35S]Cys or 1 mm Met labeled with 54.2 μCi of [35S]Met as the sole S source. Roots and fungal distal ERM were collected after 4 weeks of labeling and the radioactive counts split into different biochemical fractions. Fractions included SO42− (white bars), amino acids (light gray bars), proteins (medium gray bars), and the remaining solubilized cellular material (dark gray bars) and were obtained through scintillation counting. Values are reported as mean ± SEM (n = 9). DW, Dry weight.
Gene Expression Measurements
The expression of fungal transcripts encoding putative genes involved in sulfate assimilation was analyzed in relation to the application of Cys to the distal ERM. Partial sequences of transcripts representing putative S assimilatory genes were identified by high throughput 454 sequencing of cDNA from fungal ERM grown in split plates in M medium (3 mm sulfate). These included a 234-bp sequence with 67% identity at the amino acid level to SUL1 from S. cerevisiae and a 247-bp sequence with 67% and 70% identities at the amino acid level to S adenylyltransferase from A. nidulans and A. terreus, respectively. Sequences with homology to enzymes defining the reverse trans-sulfuration pathway were also identified. These included a 403-bp sequence with 79% and 71% identities at the amino acid level to cystathionine β-synthase from A. fumigatus and CYS4 from S. cerevisiae. Additionally, a 220-bp sequence was identified with 58% and 69% amino acid identities to CYS3 from S. cerevisiae and cystathionine γ-lyase from A. fumigatus. The presence of transcripts for these putative S genes is consistent with sulfate uptake and assimilation via the usual assimilation pathway and also strongly suggests the presence of the reverse trans-sulfuration pathway in AM fungi. The expression of the sulfate permease and reverse trans-sulfuration enzyme candidates was analyzed in ERM after exposure to 0.12 mm sulfate with or without the presence of 1 mm Cys. The conditions were chosen to enable the comparison between gene expression changes and the uptake and transfer of radiolabeled sulfate with Cys addition (Fig. 4, A–C). There was a reduction in the expression of the candidate high affinity sulfate permease sequence by a factor of 3.15 ± 1.15. The expression of sequences with homology to sulfate adenylyltransferase and cystathionine γ-lyase was not significantly different between treatments. The expression with Cys addition of the putative cystathionine γ-lyase sequence studied was significantly different. However, the difference was small (<2-fold).
DISCUSSION
The uptake of 35S by nonmycorrhizal roots was dependent on the external sulfate concentration (Fig. 1B), as has been reported for tomato (Solanum lycopersicum) seedlings and carrot (Daucus carota) storage root sections (Cram, 1983; Lopez et al., 2002). In carrot, Cram et al. (1983) showed a proportional increase up to an external concentration of 50 mm, leading the authors to conclude that feedback inhibition of sulfate uptake is absent. Vegetative root growth was less affected by alterations in available sulfate concentrations than was uptake or reduction (Fig. 1A). Subsequent measurements of mycorrhizal uptake activity were conducted using sulfate concentrations that increased potential differences in root uptake and contents and minimized the limitation of growth by S.
Bicompartmental petri plate cultures are a well-established model mycorrhizal system (St. Arnaud et al., 1996) used extensively in studies of mycorrhizal metabolism and nutrient transfer (e.g. Govindarajulu et al., 2005). This system made possible the use of defined media and the isolation of the majority of ERM from the rhizosphere, allowing S movement through the ERM to be quantified. With moderate S levels available to the roots (0.12 mm), uptake and transfer by the fungus increased the amount of S in the roots by 25% (Fig. 3B). The movement of 35S into roots was also observed in all the experiments regardless of the S source utilized in quantities much higher than those found in fungal ERM. Fungal uptake and transfer of S was influenced by the concentration of sulfate available to the host roots. Increasing the sulfate concentration in the root compartment from 0.12 mm to 3 mm resulted in a halving of fungal uptake (Fig. 2A), while approximately one-fifth of the amount of transferred S was found in roots grown in 3 mm sulfate compared to the amount found in roots grown in 0.12 mm sulfate (Fig. 2B). These reductions may be due to host signaling and/or to changes in the concentration gradient of S at the root-fungal interface.
After supplying 35S-sulfate to the fungal compartment under a range of conditions, labeled sulfate was invariably found in roots in amounts that dwarfed the total quantities in the fungal mycelium. For example, after 6 weeks of labeling the fungal compartment with 35SO42−, total sulfate in the fungus was 5.1% of the amount in the roots (Fig. 4, C and F). Roots in that experiment were grown in 0.5 mm sulfate, and the distal ERM was supplied with 0.12 mm, and the same was true (5.3%) when the roots were grown in 0.22 mm sulfate and the fungus was also labeled with 0.22 mm sulfate (Fig. 3, B and C). Based on this evidence, one may conclude that the sulfate anion is transferred by G. intraradices to host roots. However, as shown in Figure 5, the fungus is clearly also capable of the uptake and transfer of reduced forms of S at rates comparable to sulfate. Thus, the ERM can supply the host plant with organic forms of S from the soil. Because an estimated 95% of the S in soils is in organic forms, the ability of mycorrhizal plants to access S from this source is potentially important for plant nutrition.
The first steps toward understanding the regulation of sulfate uptake by a mycorrhizal fungus in the symbiotic state were taken by analyzing the effect of common S metabolite repressors Cys, Met, and GSH on the uptake and transfer of radiolabeled sulfate through the fungus. The addition of 1 mm met was shown to suppress the expression of S assimilation pathway genes in yeast (Kuras and Thomas, 1995) and the complete suppression of uptake has been demonstrated in the presence of 10 times lower levels (Breton and Surdin-Kerjan, 1977). This finding was later refined by mutant analyses and the suppression shown to be due to the synthesis of Cys through the reverse trans-sulfuration pathway (Ono et al., 1999). The sulfate permeases sB in A. nidulans and Cys-14 in N. crassa are also strongly suppressed by Met (Ketter and Marzluf, 1988; Ketter et al., 1991; Grynberg et al., 2001). In contrast to these organisms, the addition of 1 mm Met to the fungal compartments of mycorrhizal split plates had no significant effect on total sulfate uptake and transfer (Fig. 4).
By contrast, the application of 1 mm Cys to the ERM resulted in S transfer from fungus to mycorrhizal roots being approximately halved (Fig. 4, A–C). The amount by which sulfate transfer is reduced in the presence of Cys (Fig. 4) is similar to the level of S that is taken up as Cys and transferred to the roots (Fig. 5). Labeled GSH has been shown to be imported by ectomycorrhizal oak (Quercus rober) trees at rates comparable to sulfate (Seegmuller and Rennenberg, 2002). However, the addition of 1 mm GSH to endomycorrhizal distal ERM resulted in a suppression of the growth of this tissue. While the transfer of sulfate continued and the assimilation by the ERM was largely unaffected on a per-milligram basis, S transfer was greatly diminished. Because GSH also suppressed the expression of the putative sulfate permease (data not shown), it is unclear how much of the effect of GSH was due to regulation of transport and how much to repression of ERM growth. Thus, the regulation of S uptake and metabolism in G. intraradices is different from that in other filamentous fungi studied to date.
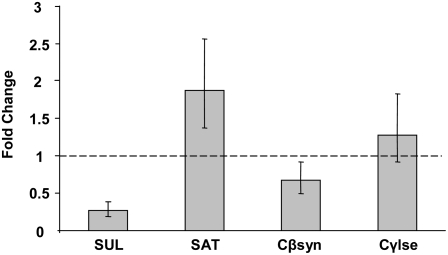
The changes in expression of genes putatively involved in sulfate transport and assimilation indicates a role for transcriptional regulation of sulfate permease in the reduction in sulfate transport in the presence of Cys. The reduction in S transfer of 45% with the application of 1 mm Cys to distal ERM (Fig. 4C) correlated with a 3.8- ± 1.2-fold reduction in the expression of mRNA encoding a putative sulfate permease. The lack of a significant change in the expression of a cDNA sequence with high homology to S-adenosyl transferases suggests that the reduction in S transfer is a function of reduced uptake rather than sulfate reduction. However, the analysis of more S assimilation pathway genes and compartment-specific sulfate permease isozymes is needed before concluding that the transcriptional regulation of this sulfate permease gene explains the reduction in S transfer with the addition of Cys. Sequences encoding putative genes with homology to cystathionine β-synthase and cystathionine γ-lyase were also identified. Their expression is indicative of a functional reverse trans-sulfuration pathway in G. intraradices, akin to A. nidulans and S. saccharomyces, but unlike S. pombe (Paszewski and Grabski, 1975; Ono et al., 1992a, 1992b; Brzywczy et al., 2002). The putative cystathionine β-synthase sequence was slightly down-regulated (1.49- ± 0.41-fold) in the presence of Cys, while the sequence with homology to cystathionine γ-lyase showed no significant change.
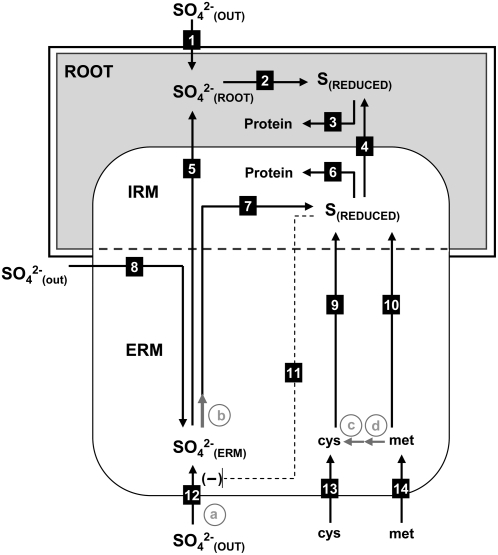
Figure 6 presents a working model consistent with the findings presented. We have demonstrated uptake, assimilation, and transfer of sulfate (Figs. 3B and 4, A–C) and of reduced S (Fig. 5) by the fungal partner to the host roots in physiologically significant quantities, along with bidirectional transport of sulfate within the fungal ERM (Fig. 3C). The inverse relationship between the rhizospheric sulfate concentration and the amount of sulfate uptake from the fungal compartment (Fig. 2, A and B) suggests that an increase in root intracellular or apoplastic sulfate can suppress both the transfer of sulfate from the intraradical hyphae to the host and the uptake by the extraradical hyphae. This relationship is depicted in the model as the negative regulation of sulfate transfer across the root-hyphal interface by root sulfate levels, which subsequently results in an increase in the fungal intracellular sulfate concentration and suppression of uptake in the distal ERM. The relationship between the transfer of a reduced form of S when Cys or Met are imported by the fungus and the uptake of sulfate in the presence of Cys and Met suggest a simultaneous transfer of sulfate and reduced S and/or a regulatory response. Relative real-time PCR measurements of expression for a putative high affinity sulfate permease gene revealed an approximately 73% reduction in transcript levels when the ERM was exposed to 1 mm Cys, indicating a regulatory effect on sulfate uptake (Figs. 6 and 7). The working model proposed is a testable interpretation of the findings presented that invites future experiments to verify gene identities, localize expression within the host roots, and analyze regulation in mycorrhizas of whole plants.
Figure 6.
Fungal gene expression changes in response to the addition of 1 mm Cys to ERM. Expression relative to control plates of sequences with homology to high affinity sulfate permease (SUL), S-adenosyl transferase (SAT), cystathionine β-synthase (Cβsyn), and cystathionine γ-lyase (Cγlse) was analyzed by quantitative real-time PCR using a ribosomal protein sequence as a control as previously reported (Govindarajulu et al., 2005; see “Materials and Methods”). RNA was extracted from ERM tissue collected after 24 hr incubation with 0.12 mm SO42− and with or without 1 mm Cys added to the distal compartments of five split plates per sample. Values are reported as mean ± SEM (n = 3 biological replicates each consisting of tissue from five to seven plates).
Figure 7.
Model depicting S transfer through an endomycorrhizal symbiosis. Roots and fungal mycelium both import SO42− from external sources (1 and 12), and fungal uptake can supply isolated mycelium (8). The transfer of SO42− through the mycorrhizal symbiosis is inversely related to root uptake (1 and 5). Cys and Met are imported by the fungus (13 and 14), resulting in a reduction of fungal uptake of SO42− (11) and the transfer of a reduced form of S to the root (4). The reduction of SO42− (7 and 2) and uptake of reduced S (13 and 14) lead to incorporation in the protein pools in both roots (3) and fungus (6). Putative steps in the assimilation pathway based on sequence data are depicted as gray arrows. Steps involving putative S assimilation genes are labeled in gray letters as follows: a, high affinity sulfate permease; b, sulfate adenylyltransferase; c, γ-cystathionine lyase; d, β-cystathionine synthetase. IRM, Intraradical mycelium.
MATERIALS AND METHODS
Chemicals and Reagents
Gel-Gro gellan (MP Biomedical) was used for the solidification of growth media. Radioactively labeled sulfate was obtained as Na235SO4 from MP Biomedicals. Labeled Cys and Met were separated from a crude mixture of 35S-labeled compounds (TRAN35S-LABEL; MP Biomedicals) containing ≤70% [35S]Met and 15% [35S]Cys. The compounds were separated using an amino acid analyzer (Hitachi L8800). Retention times for the amino acids were determined using post-column ninhydrin bonding and UV absorption. The radioactive compounds were then collected in a ninhydrin-free environment by manual collection of the appropriate fractions.
Growth Media
All experimental procedures used M medium (Fortin et al., 2002) with modifications to reduce the sulfate content. This was autoclaved and, for culture plates, solidified with 3.5 gL−1 Gel Gro gellan (MP Biomedicals). The 3 mm MgSO4 normally present in M medium was replaced with 0.1 mm MgCl2. An additional 2 mm of calcium was added as CaCl2 to replace the divalent cation concentration needed for the solidification of Gel-Gro. Sulfate was supplied as Na2SO4 at different concentrations as indicated. Normal microelement concentrations for M media containing 0.02 mm sulfate were used for each experiment. These were included in all experiments with the exception of the measurement of nonmycorrhizal root growth (Fig. 1A.). The medium in the root, but not fungal, compartments contained 10 gL−1 (29.2 mm) Suc. Na2SO4 was added to the fungal compartments at the time of labeling as 0.2 mL of concentrated solution.
Growth of Roots and Mycorrhizas
Uncolonized Ri T-DNA-transformed roots of carrot (Daucus carota; DC2; Diop et al., 1992) were grown at 25°C with 0.02, 0.12, 0.5, or 3 mm Na2SO4 in modified liquid M media. Roots were inoculated at plate inception as previously described (Pfeffer et al., 1999). The roots and fungus were allowed to proliferate on both sides of bicompartmented petri plates at 25°C until the fungal ERM was well developed (approximately 6 weeks). The colonized roots and media in each compartment were transferred to empty compartments of new plates in which the other compartment contained new medium. At the time of transfer, the media in which the mycorrhizal roots were growing was supplemented with one-quarter of the original nutrient contents, including Suc and P, but excluding CaCl2, added as approximately 0.5 to 1 mL of a sterile solution. Root growth over the barrier after transplantation was prevented by pruning, and the fungal ERM typically grew over the barrier within 1 week of the transfer, colonizing the empty compartment. Fungal compartments were labeled when ERM had growth into at least one-half of the media or within 2 weeks of crossing the barrier, whichever was first.
Collection of Roots and ERM
Root material was collected with forceps and rinsed for 5 min in deionized water to remove external, and reduce apoplastic, 35S. The collected roots were rinsed again with deionized water in a separate container, blotted dry, frozen in liquid N, and lyophilized. ERM was collected by blending the solidified medium at high speed in 10 mm sodium citrate buffer, pH 6.0, at an approximate gel:buffer ratio of 1:2.5 for 2 min, which dissolved the gellan (Pfeffer et al., 1999). Tissue was collected on a sieve that was rinsed with four 30-mL aliquots of cold 10 mm Na2SO4, followed by an equal quantity of cold deionized water. The final flow-through was checked for radioactivity, which was not detectable by scintillation counting in a 0.5-mL aliquot. To collect ERM from root compartments, the roots were first removed from the media under a dissecting microscope. Fungal mycelium collected from the sieve was blotted briefly and frozen in liquid N. Tissue was weighed after overnight lyophilization.
Extraction Procedure
For biochemical fractionation, lyophilized fungal mycelium was pulverized with two 3-mm stainless steel beads using a bead mill (Retsch MM301). Due to the high lipid content, 0.1 mL of cold methanol:water (70:30) was added to aid in disruption. The samples were shaken at 30 Hz for 4 min, and 2-μL samples were analyzed by dissecting microscope to ensure that hyphae and any spores had been broken. After disruption, 0.9 mL of cold methanol:water (70:30) was added, and the sample was vortexed for 5 min. Samples were then centrifuged and supernatants collected. The cold aqueous methanol extraction was repeated twice more using 1 mL each time and the supernatants pooled. Then 0.5 mL of supernatant solution was scintillation counted after adding to 5 mL of BioSafe II (MP Biomedicals) scintillation cocktail. To determine the amount of sulfate in the samples, the sulfate was precipitated from an aliquot of the aqueous alcohol extract by adding 0.1 mL of 100 mm Na2SO4 and 0.3 mL of 10 mm HCl solution to 0.5 mL of the sample followed by vortexing and the addition of 0.1 mL of 100 mm BaCl2. Samples were then incubated for 30 min at 100°C. The resulting barium sulfate precipitate was removed by centrifugation, and the sulfate-free supernatant was scintillation counted. The amount of sulfate in each sample was determined by the subtraction of these counts from those obtained from an aliquot of the original aqueous alcohol extract. Tests with standards showed that 99.8% ± 0.1% of sulfate was removed by this procedure (three trials with three samples each) and that 96.4% ± 1.3% of 35S-Cys and 95.6% ± 1.6% of 35S-Met remained in solution.
Residues after aqueous ethanol extraction were extracted using 1 mL of protein extraction buffer containing 9 m urea, 1% SDS, 25 mm Tris-HCl, pH 6.8, 1 mm EDTA, and 0.7 m 2-mercaptoethanol as described by Osherov and May (1998). After vortexing and centrifugation at 10,000 rpm for 5 min, 0.5 mL of the supernatant was added to 5 mL of scintillation cocktail and counted. The remaining cellular debris was transferred to sealed glass vials containing 0.5 mL TS-2 tissue solubilizer (MP Biomedicals) and incubated at 70°C for 1 to 2 weeks until solubilized. The solubilized solution was mixed with 5 mL of scintillation fluid and titrated to pH 7 with HCl. This solution was diluted 10-fold to diminish color quenching and analyzed by scintillation counting.
Dried roots were pulverized in 15-mL centrifuge tubes with 10 5-mm metal beads for 30 min using a paint shaker. Five milliliters of cold methanol:water (70:30) was added to the powder, and the tubes were vortexed for 5 min. While keeping particulates suspended, a 1-mL aliquot of the solution was transferred to a microcentrifuge tube and centrifuged. Subsequent steps were as for fungal samples.
Measurements of 35S in Media
Three 1-cm diameter wells were excavated in the gel at the time of labeling with a sterilized cork borer as far from one another as possible. Then 0.1 to 0.2 mL of media of the same composition as that in the compartment being analyzed was added to each well, and this volume was maintained through weekly refilling as needed. At the start of the experiment, label was added to one of the wells, and 35S in each of the three wells were measured after 1 week to ensure efficient diffusion of the radioactivity through the plate. 35S contents were measured by adding 50 μL of liquid media to 5 mL of scintillation fluid, followed by scintillation counting.
Nonmycorrhizal Root Measurements
Roots were grown on solidified M media containing 0.12 mm S as Na2SO4 for 4 weeks, and approximately 5-cm root segments were aseptically removed, weighed, and added to 20 mL of liquid M media without S (ZnSO4 7H2O and CuSO4 5H2O replaced by 1.2 mg of ZnCl and 0.5 mg of CuCl2) for measurements of S-mediated growth limitation. For each sulfate concentration (0, 0.005, 0.01, 0.02, 0.05, 0.1, 0.5, and 3 mm), roots were weighed and then placed on 20 mL of solidified media per side of three bicompartmental petri plates and allowed to grow for 3 months. The roots were collected, dried at 70°C overnight, and weighed. To measure S uptake, roots were grown at 0.12 mm sulfate as above and transferred into 20 mL of S media solidified with gellan containing 0.02, 0.12, or 3 mm sulfate labeled with 11.5 μCi of Na2[35SO4]. After 4 weeks, the roots were removed from the media, rinsed several times in deionized water to remove external radioactivity, lyophilized, and weighed. The extraction procedure for the radiolabeled tissue was simplified to include only the aqueous alcohol and solubilization steps.
Addition of S Metabolic Regulators
Cys, Met, and GSH were added to a final concentration of 1 mm (as 0.2–0.5 mL of sterile aqueous solutions) to the fungal compartments of split plates with 10-week-old roots growing with 0.52 mm Na2SO4. The uptake of 37.4 μCi of Na2[35SO4], which was added to fungal compartments simultaneously with metabolic regulators and 0.12 mm Na2SO4, was monitored by analyzing aliquots of media from a liquid-filled well as described above. The plates were collected at 2, 4, and 6 weeks. The fungal mycelium was collected by dissolving the gellan in a blender with 10 mm sodium citrate, pH 6, then sieving the blended solution. The roots were extracted from the media using forceps and rinsed in deionized water for 5 min before freezing. The fungal and root tissues were extracted as described above.
Using cultures grown as for the above experiment, 1 mm of Cys labeled with 25.7 μCi of [35S]Cys or 1 mm Met labeled with 54.2 μCi of [35S]Met was applied to the fungal sides of 15 plates as the sole S source. The plates were incubated at 25°C for 1 month prior to collecting and extracting the biochemical fractions as described above.
RNA Extraction and Putative Gene Fragment Isolation
Sequences of putative S gene fragments were identified from an EST database (Jun et al., 2002) and from additional high throughput sequencing data (P.J. Lammers, M. DeJong, and Y. Shachar-Hill, unpublished data) from cDNA made from ERM. To confirm their identity, total RNA was extracted using the RNeasy Plant Mini kit (Qiagen) from frozen Glomus intraradices germinating spore tissue, disrupted as already described using a bead mill, followed by DNA removal using RNase-free DNase (Turbo DNA-free; Ambion). Samples were split into two aliquots following quantification by absorbance for the synthesis of cDNA and a control without reverse transcriptase. cDNA was synthesized using SuperScript II reverse transcriptase (Invitrogen). Primer sets were developed from EST and high throughput sequencing data as follows: putative sulfate permease forward primer, 5′-TAGCAATAATTACAAGAATACCAG-3′, and reverse primer 5′-GGGATTCTTATCTTGGAA-3′; putative cystathionine β-synthase forward primer, 5′-CTAGCCACTCCTGTAATTGTTCCTC-3′, and reverse primer, 5-GAGAAAGCAGGAATACTTAAACCAGG-3′; putative cystathionine γ-lyase forward primer, 5′-GGTGGAATGTTAAGTTTTAGGATTAAGG-3′, and reverse primer, 5′- CATCAACATCTTCGATACCGATG-3′. PCR products were separated by and isolated from agarose gel using the QIAquick Gel Extraction kit (Qiagen) and sequenced by an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems). Resulting sequences showed single open reading frames.
Quantitative Real-Time PCR Measurements
Mycorrhizal split plates were grown until the fungal compartment was approximately one-half colonized. To the fungal compartment, 1 mL of a filter sterilized solution containing either sodium sulfate or sodium sulfate and Cys was applied to give a final concentration of 0.12 mm sulfate and 1 mm Cys. Plates were incubated for 24 h before tissue from five to seven plates was collected as described above and immediately frozen in liquid N. RNA was extracted and converted to cDNA as described above. The initial quantitative real-time PCR (qRT-PCR) reaction mixture containing primers at a concentration of 300 nm and 1 ng of cDNA template was made to 0.015 mL in a 96-well plate, 0.01 mL of which was transferred to a 384-well plate, and the PCR reactions were monitored using an ABI Prism 7900 HT Sequence Detection system (Applied Biosystems) with the following cycling program: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, and 60°C for 1 min. Power SYBR Green 2-Step Master mix (Applied Biosystems) was used for all real-time PCR assays. The ΔΔCT, and comparative CT, methods were utilized for the determination of relative gene expression (Livak and Schmittgen, 2001). The expression of an S4 ribosomal protein was used to normalize relative gene expression data as described by Govindarajulu et al. (2005). Other primer sequences (IDT, Syntegen) for qRT-PCR were designed using Primer Express software from Applied Biosystems as follows: putative high affinity sulfate permease forward, 5′-TTGGATCATTCTTTCATGCGTATC-3′, reverse, 5′- GACACCGGCTAATGGAGTACGT-3′; putative S-adenosyl transferase forward, 5′-CCGGAGTTGATGATCCTTACG-3′, reverse, 5′-ACTGACACACTGTTTACTAACATCAACAA-3′; putative cystathionine β-synthase forward, 5′-TGCTTCAGTTGGTGTACGAACAA-3′, reverse, 5′- AAATGAGCCAGGAAAAGGTTGA-3′; putative cystathionine γ-synthase forward, 5′-GACCAGCGTGAGTCATTTTAGAAG-3′, reverse, 5′-AGCTGAATCTTTGGGTGGTGTT-3′. Experimental samples were measured using six technical replicates for each of three biological replicates per condition, and errors were expressed as se of the mean (SEM).
The sequences studied were submitted to GenBank and given the following accession numbers: putative high affinity sulfate permease (FJ161947); putative sulfate adenylyltransferase (FJ161948); putative cystathionine γ-lyase (FJ161949); and putative cystathionine β-synthase (FJ161950).
Acknowledgments
We thank Inga Krassovskaya for help with molecular data analysis and protocols, Joseph Leykam for the HPLC separation of [35S]Cys and [35S]Met, and the Research Technology Support Facility at Michigan State University for sequencing and qRT-PCR analyses.
This work was supported by the National Science Foundation (award no. 0616016).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: James W. Allen (allenj28@msu.edu).
References
- Banerjee R, Evande R, Kabil O, Ojha S, Taoka S (2003) Reaction mechanism and regulation of cystathionine beta-synthase. Biochim Biophys Acta 1647 30–35 [DOI] [PubMed] [Google Scholar]
- Baumgardner RE, Lavery TF, Rogers CM, Isil SS (2002) Estimates of the atmospheric deposition of sulfur and nitrogen species: Clean Air Status and Trends Network, 1990-2000. Environ Sci Technol 36 2614–2629 [DOI] [PubMed] [Google Scholar]
- Bhat KKS, Nye PH, Baldwin JP (1976) Diffusion of phosphate to plant roots in soil. 4. Concentration distance profile in rhizosphere of roots with root hairs in a low-p soil. Plant Soil 44 63–72 [Google Scholar]
- Bradfield G, Somerfield P, Meyn T, Holby M, Babcock D, Bradley D, Segel IH (1970) Regulation of sulfate transport in filamentous fungi. Plant Physiol 46 720–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton A, Surdin-Kerjan Y (1977) Sulfate uptake in Saccharomyces cerevisiae: biochemical and genetic study. J Bacteriol 132 224–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brundrett MC (2002) Coevolution of roots and mycorrhizas of land plants. New Phytol 154 275–304 [DOI] [PubMed] [Google Scholar]
- Brzywczy J, Sienko M, Kucharska A, Paszewski A (2002) Sulphur amino acid synthesis in Schizosaccharomyces pombe represents a specific variant of sulphur metabolism in fungi. Yeast 19 29–35 [DOI] [PubMed] [Google Scholar]
- Cherest H, Davidian JC, Thomas D, Benes V, Ansorge W, Surdin-Kerjan Y (1997) Molecular characterization of two high affinity sulfate transporters in Saccharomyces cerevisiae. Genetics 145 627–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke DL, Newbert RW, Turner G (1997) Cloning and characterisation of the adenosyl phosphosulphate kinase gene from Aspergillus nidulans. Curr Genet 32 408–412 [DOI] [PubMed] [Google Scholar]
- Cooper KM, Tinker PB (1978) Translocation and transfer of nutrients in vesicular-arbuscular mycorrhizas. 2. Uptake and translocation of phosphorus, zinc and sulfur. New Phytol 81 43–52 [Google Scholar]
- Cram J (1983) Characteristics of sulfate transport across plasmalemma and tonoplast of carrot root-cells. Plant Physiol 72 204–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diop T, Becard G, Pichey Y (1992) Long-term invitro culture of an endomycorrhizal fungus, Gigaspora margarita, on Ri T-dna transformed roots of carrot. Symbiosis 12 249–259 [Google Scholar]
- Eriksen J, Askegaard M (2000) Sulphate leaching in an organic crop rotation on sandy soil in Denmark. Agric Ecosyst Environ 78 107–114 [Google Scholar]
- Fitzgerald JW (1976) Sulfate ester formation and hydrolysis: potentially important yet often ignored aspect of sulfur cycle of aerobic soils. Bacteriol Rev 40 698–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin JA, Becard G, Declerck S, Dalpe Y, St-Arnaud M, Coughlan AP, Piche Y (2002) Arbuscular mycorrhiza on root-organ cultures. Can J Bot 80 1–20 [Google Scholar]
- Gahoonia TS, Nielsen NE (1991) A method to study rhizosphere processes in thin soil layers of different proximity to roots. Plant Soil 135 143–146 [Google Scholar]
- Gahoonia TS, Raza S, Nielsen NE (1994) Phosphorus depletion in the rhizosphere as influenced by soil-moisture. Plant Soil 159 213–218 [Google Scholar]
- Govindarajulu M, Pfeffer PE, Jin HR, Abubaker J, Douds DD, Allen JW, Bucking H, Lammers PJ, Shachar-Hill Y (2005) Nitrogen transfer in the arbuscular mycorrhizal symbiosis. Nature 435 819–823 [DOI] [PubMed] [Google Scholar]
- Gray LE, Gerdemann JW (1973) Uptake of sulphur-35 by vesicular-arbuscular mycorrhizae. Plant Soil 39 687–689 [Google Scholar]
- Grynberg M, Piotrowska M, Pizzinini E, Turner G, Paszewski A (2001) The Aspergillus nidulans metE gene is regulated by a second system independent from sulphur metabolite repression. Biochim Biophys Acta 1519 78–84 [DOI] [PubMed] [Google Scholar]
- Guo T, Zhang JL, Christie P, Li XL (2007) Pungency of spring onion as affected by inoculation with arbuscular mycorrhizal fungi and sulfur supply. J Plant Nutr 30 1023–1034 [Google Scholar]
- Hansen J, Johannesen PF (2000) Cysteine is essential for transcriptional regulation of the sulfur assimilation genes in Saccharomyces cerevisiae. Mol Gen Genet 263 535–542 [DOI] [PubMed] [Google Scholar]
- Hawkins HJ, Johansen A, George E (2000) Uptake and transport of organic and inorganic nitrogen by arbuscular mycorrhizal fungi. Plant Soil 226 275–285 [Google Scholar]
- Hayman DS, Mosse B (1971) Plant growth responses to vesicular-arbuscular mycorrhiza. 1. Growth of endogone-inoculated plants in phosphate-deficient soils. New Phytol 70 19–27 [Google Scholar]
- Jin H, Pfeffer PE, Douds DD, Piotrowski E, Lammers PJ, Shachar-Hill Y (2005) The uptake, metabolism, transport and transfer of nitrogen in an arbuscular mycorrhizal symbiosis. New Phytol 168 687–696 [DOI] [PubMed] [Google Scholar]
- Joner EJ, Jakobsen I (1994) Contribution by 2 arbuscular mycorrhizal fungi to P-uptake by cucumber (Cucumis sativus l) from P-32 labeled organic-matter during mineralization in soil. Plant Soil 163 203–209 [Google Scholar]
- Jun J, Rehrer C, Pfeffer PE, Shachar-Hill Y, Lammers PJ (2002) Expression in an arbuscular mycorrhizal fungus of genes putatively involved in metabolism, transport, the cytoskeleton and the cell cycle. Plant Soil 244 141–148 [Google Scholar]
- Ketter JS, Jarai G, Fu YH, Marzluf GA (1991) Nucleotide-sequence, messenger-rna stability, and dna recognition elements of cys-14, the structural gene for sulfate permease-II in Neurospora crassa. Biochemistry 30 1780–1787 [DOI] [PubMed] [Google Scholar]
- Ketter JS, Marzluf GA (1988) Molecular-cloning and analysis of the regulation of cys-14+, a structural gene of the sulfur regulatory circuit of Neurospora crassa. Mol Cell Biol 8 1504–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzwieser J, Rennenberg H (1998) Sulphate uptake and xylem loading of mycorrhizal beech roots. New Phytol 140 319–329 [DOI] [PubMed] [Google Scholar]
- Kuras L, Thomas D (1995) Functional-analysis of met4, a yeast transcriptional activator responsive to S-adenosylmethionine. Mol Cell Biol 15 208–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambers H, Raven JA, Shaver GR, Smith SE (2008) Plant nutrient-acquisition strategies change with soil age. Trends Ecol Evol 23 95–103 [DOI] [PubMed] [Google Scholar]
- Lefohn AS, Husar JD, Husar RB (1999) Estimating historical anthropogenic global sulfur emission patterns for the period 1850-1990. Atmos Environ 33 3435–3444 [Google Scholar]
- Leustek T (1996) Molecular genetics of sulfate assimilation in plants. Physiol Plant 97 411–419 [Google Scholar]
- Leustek T, Martin MN, Bick JA, Bick JA, Davies JP (2000) Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu Rev Plant Physiol 51 145–165 [DOI] [PubMed] [Google Scholar]
- Li XL, George E, Marschner H (1991) Extension of the phosphorus depletion zone in VA-mycorrhizal white clover in a calcareous soil. Plant Soil 136 41–48 [Google Scholar]
- Liu A, Hamel C, Elmi A, Costa C, Ma B, Smith DL (2002) Concentrations of K, Ca and Mg in maize colonized by arbuscular mycorrhizal fungi under field conditions. Can J Plant Sci 82 271–278 [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
- Lopez J, Bell CI, Tremblay N, Dorais M, Gosselin A (2002) Uptake and translocation of sulphate in tomato seedlings in relation to sulphate supply. J Plant Nutr 25 1471–1485 [Google Scholar]
- Mansouri-Bauly H, Kruse J, Sykorova Z, Scheerer U, Kopriva S (2006) Sulfur uptake in the ectomycorrhizal fungus Laccaria bicolor S238N. Mycorrhiza 16 421–427 [DOI] [PubMed] [Google Scholar]
- Marzluf GA (1993) Regulation of sulfur and nitrogen-metabolism in filamentous fungi. Annu Rev Microbiol 47 31–55 [DOI] [PubMed] [Google Scholar]
- Marzluf GA (1997) Molecular genetics of sulfur assimilation in filamentous fungi and yeast. Annu Rev Microbiol 51 73–96 [DOI] [PubMed] [Google Scholar]
- Natorff R, Sienko M, Brzywczy J, Paszewski A (2003) The Aspergillus nidulans metR gene encodes a bZIP protein which activates transcription of sulphur metabolism genes. Mol Microbiol 49 1081–1094 [DOI] [PubMed] [Google Scholar]
- Ono B, Hazu T, Yoshida S, Kawato T, Shinoda S, Brzvwczy J, Paszewski A (1999) Cysteine biosynthesis in Saccharomyces cerevisiae: a new outlook on pathway and regulation. Yeast 15 1365–1375 [DOI] [PubMed] [Google Scholar]
- Ono B, Heike C, Yano Y, Inoue T, Naito K, Nakagami S, Yamane A (1992. a) Cloning and mapping of CYS4 gene of Saccharomyces cerevisiae. Curr Genet 21 285–289 [DOI] [PubMed] [Google Scholar]
- Ono B, Tanaka K, Naito K, Heike C, Shinoda S, Yamamoto S, Ohmori S, Oshima T, Toh-e A (1992. b) Cloning and characterization of CYS3(CYI1) gene of Saccharomyces cerevisiae. J Bacteriol 174 3339–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono BI, Naito K, Shirahige YI, Yamamoto M (1991) Regulation of cystathionine gamma-lyase in Saccharomyces cerevisiae. Yeast 7 843–848 [DOI] [PubMed] [Google Scholar]
- Ortiz-Ceballos AI, Pena-Cabriales JJ, Fragoso C, Brown GG (2007) Mycorrhizal colonization and nitrogen uptake by maize: combined effect of tropical earthworms and velvet bean mulch. Biol Fertil Soils 44 181–186 [Google Scholar]
- Osherov N, May GS (1998) Optimization of protein extraction from Aspergillus nidulans for gel electrophoresis. Fungal Genet Rep 45 41–42 [Google Scholar]
- Paszewski A, Grabski J (1975) Enzymatic lesions in methionine mutants of Aspergillus nidulans: role and regulation of an alternative pathway od cysteine and methionine synthesis. J Bacteriol 124 893–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perner H, Rohn S, Driemel G, Batt N, Schwarz D, Kroh LW, George E (2008) Effect of nitrogen species supply and mycorrhizal colonization on organosulfur and phenolic compounds in onions. J Agric Food Chem 56 3538–3545 [DOI] [PubMed] [Google Scholar]
- Pfeffer PE, Douds DD, Becard G, Shachar-Hill Y (1999) Carbon uptake and the metabolism and transport of lipids in an arbuscular mycorrhiza. Plant Physiol 120 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilsyk S, Natorff R, Sienko M, Paszewski A (2007) Sulfate transport in Aspergillus nidulans: a novel gene encoding alternative sulfate transporter. Fungal Genet Biol 44 715–725 [DOI] [PubMed] [Google Scholar]
- Rennenberg H (1999) The significance of ectomycorrhizal fungi for sulfur nutrition of trees. Plant Soil 215 115–122 [Google Scholar]
- Rennenberg H, Herschbach C, Haberer K, Kopriva S (2007) Sulfur metabolism in plants: Are trees different? Plant Biol 9 620–637 [DOI] [PubMed] [Google Scholar]
- Rhodes LH, Gerdemann JW (1978. a) Influence of phosphorus nutrition on sulfur uptake by vesicular-arbuscular mycorrhizae of onion. Soil Biol Biochem 10 361–364 [Google Scholar]
- Rhodes LH, Gerdemann JW (1978. b) Hyphal translocation and uptake of sulfur by vesicular-arbuscular mycorrhizae of onion. Soil Biol Biochem 10 355–360 [Google Scholar]
- Riley NG, Zhao FJ, McGrath SP (2000) Availability of different forms of sulphur fertilisers to wheat and oilseed rape. Plant Soil 222 139–147 [Google Scholar]
- Scherer HW (2001) Sulphur in crop production. Eur J Agron 14 81–111 [Google Scholar]
- Seegmuller S, Rennenberg H (2002) Transport of organic sulfur and nitrogen in the roots of young mycorrhizal pedunculate oak trees (Quercus robur L.). Plant Soil 242 291–297 [Google Scholar]
- Sienko M, Paszewski A (1999) The metG gene of Aspergillus nidulans encoding cystathionine beta-lyase: cloning and analysis. Curr Genet 35 638–646 [DOI] [PubMed] [Google Scholar]
- Simon L, Bousquet J, Levesque RC, Lalonde M (1993) Origin and diversification of endomycorrhizal fungi and coincidence with vascular land plants. Nature 363 67–69 [Google Scholar]
- Spencer B, Harada T (1960) Role of choline sulphate in the sulphur metabolism of fungi. Biochem J 77 305–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Arnaud M, Hamel C, Vimard B, Caron M, Fortin JA (1996) Enhanced hyphal growth and spore production of the arbuscular mycorrhizal fungus Glomus intraradices in an in vitro system in the absence of host roots. Mycol Res 100 328–332 [Google Scholar]
- Tabatabai MA (1986) Sulfur in Agriculture. American Society of Agronomy, Madison, WI, pp 207–226
- Thomas D, Surdin-Kerjan Y (1997) Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 61 503–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Qiu YL (2006) Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza 16 299–363 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Kelly JM, Kovar JL (2004) In situ dynamics of phosphorus in the rhizosphere solution of five species. J Environ Qual 33 1387–1392 [DOI] [PubMed] [Google Scholar]