Abstract
The role of Δ4-unsaturated sphingolipid long-chain bases such as sphingosine was investigated in Arabidopsis (Arabidopsis thaliana). Identification and functional characterization of the sole Arabidopsis ortholog of the sphingolipid Δ4-desaturase was achieved by heterologous expression in Pichia pastoris. A P. pastoris mutant disrupted in the endogenous sphingolipid Δ4-desaturase gene was unable to synthesize glucosylceramides. Synthesis of glucosylceramides was restored by the expression of Arabidopsis gene At4g04930, and these sphingolipids were shown to contain Δ4-unsaturated long-chain bases, confirming that this open reading frame encodes the sphingolipid Δ4-desaturase. At4g04930 has a very restricted expression pattern, transcripts only being detected in pollen and floral tissues. Arabidopsis insertion mutants disrupted in the sphingolipid Δ4-desaturase At4g04930 were isolated and found to be phenotypically normal. Sphingolipidomic profiling of a T-DNA insertion mutant indicated the absence of Δ4-unsaturated sphingolipids in floral tissue, also resulting in the reduced accumulation of glucosylceramides. No difference in the response to drought or water loss was observed between wild-type plants and insertion mutants disrupted in the sphingolipid Δ4-desaturase At4g04930, nor was any difference observed in stomatal closure after treatment with abscisic acid. No differences in pollen viability between wild-type plants and insertion mutants were detected. Based on these observations, it seems unlikely that Δ4-unsaturated sphingolipids and their metabolites such as sphingosine-1-phosphate play a significant role in Arabidopsis growth and development. However, Δ4-unsaturated ceramides may play a previously unrecognized role in the channeling of substrates for the synthesis of glucosylceramides.
Sphingolipids are ubiquitous membrane lipids and have been shown to be essential in many different eukaryotes (Dunn et al., 2004). It is widely considered that sphingolipids play an important role in membrane structure and organization and that they are enriched in plasma membrane microdomains or so-called “lipid rafts” (Borner et al., 2005). Such properties are likely to be derived from the differences in structure (and hence biophysical properties) between sphingolipids and phosphoglycerolipids. Sphingolipids comprise a fatty acid (usually a very long chain of C22–26, but it can also be C16) N-linked to a long-chain base (LCB), usually with the addition of a polar head group to generate the mature lipid (Sperling and Heinz, 2003; Sperling et al., 2004). There is considerable diversity in both the LCB and the fatty acid components between eukaryotes, although the functional significance of this has not yet been fully explored. In addition to structural roles, sphingolipids and their metabolites also act as second messengers (Spiegel and Milstien, 2003). In animal cells, the phosphorylated sphingolipid metabolite sphingosine-1-phosphate (S-1-P) acts as a potent messenger, modulating a range of processes such as proliferation and apoptosis (Saba and Hla, 2004). A number of roles for phosphorylated long chain bases (LCB-1-P) have been observed in fungi, Drosophila, and Caenorhabditis elegans (Oskouian and Saba, 2004). In higher plants, a role for S-1-P in calcium-mediated stomatal closure has been reported for Commelina communis (Ng et al., 2001) and Arabidopsis (Arabidopsis thaliana; Coursol et al., 2003; for review, see Brownlee, 2001; Worrall et al., 2003). More recently, a similar role for phytosphingosine (4-hydroxysphinganine)-1-P has been observed in Arabidopsis (Coursol et al., 2005).
A number of the Arabidopsis genes involved in the biosynthesis of plant sphingolipids have recently been identified and functionally characterized (predominantly by complementation of Saccharomyces cerevisiae mutants and reverse genetics in Arabidopsis; reviewed by Sperling and Heinz, 2003; Dunn et al., 2004; Sperling et al., 2004). Such studies have experimentally confirmed the essential nature of sphingolipids in Arabidopsis (Chen et al., 2006; Dietrich et al., 2008; Teng et al., 2008) and also indicated the contribution of sphingolipids to many aspects of plant growth, development, and stress responses (Liang et al., 2003; Dietrich et al., 2005; Zheng et al., 2005; Ryan et al., 2007).
As part of our efforts to understand the functional consequences of sphingolipid heterogeneity, we have examined the role of sphing-4-enine (commonly known as sphingosine; d18:1Δ4t) in plants and fungi. Sphingosine is synthesized via the Δ4-trans-desaturation of sphinganine (d18:0; also known as dihydrosphingosine), although this desaturation occurs on N-acylated LCBs (i.e. using the dihydroceramide as the substrate, rather than the free dihydrosphingosine LCB). In animal cells, free sphingosine is only generated through the catabolism of sphingolipids (Spiegel and Milstien, 2003; Saba and Hla, 2004). For this reason, the dihydrosphingosine Δ4-desaturase is also known as the dihydroceramide Δ4-desaturase (Michel et al., 1997). However, in this article, we will refer to this activity as the sphingolipid Δ4-desaturase, since this broader term avoids confusion regarding the substrate of this enzyme (which remains to be defined in many organisms). The first examples of the sphingolipid Δ4-desaturase were isolated by Ternes et al. (2002), who functionally identified candidate desaturases by heterologous expression in S. cerevisiae sur2Δ mutants. Yeast cells lacking the SUR2 sphingolipid hydroxylase are unable to synthesize C4-hydroxylated LCBs such as phytosphingosine and thus prevent competition between SUR2 and the heterologous sphingolipid Δ4-desaturase for their common substrate (sphinganine; Sperling et al., 2001). This approach identified active sphingolipid Δ4-desaturases from human (MLD), mouse (mDES-1), Drosophila (DES-1), and Candida albicans, and a similar study identified the same activity from Schizosaccharomyces pombe (Garton et al., 2003). These enzymes defined a new class of desaturases (Napier et al., 2002; Hashimoto et al., 2008), with apparent higher plant orthologs present in the Arabidopsis and rice (Oryza sativa) genome sequences and other plant EST collections. However, attempts to express a tomato (Solanum lycopersicum) ortholog of the sphingolipid Δ4-desaturase in S. cerevisiae sur2Δ mutants did not result in any detectable activity (Ternes et al., 2002).
In animals, sphingosine (the direct precursor of S-1-P; Fig. 1A) is the most abundant LCB (Pyne and Pyne, 2000). However, sphingosine is a very minor component of most higher plant sphingolipids (Imai et al., 1997; Sullards et al., 2000; Dunn et al., 2004; Lynch and Dunn, 2004; Markham et al., 2006), with the predominant LCB modifications being C4-hydroxylation, Δ8-desaturation, or both, yielding phytosphingosine, sphing-8-enine, or 4-hydroxysphing-8-enine, respectively (Fig. 1A). While Δ4-unsaturated sphingolipids are abundant in some plant species (such as soybean [Glycine max]), these predominantly occur in conjunction with Δ8-unsaturation, in the form of sphinga-4,8-dienine (Markham et al., 2006). In order to better define the role of sphingosine and S-1-P in Arabidopsis, we have functionally characterized the single ortholog of the sphingolipid Δ4-desaturase. Insertional mutagenesis of the Arabidopsis sphingolipid Δ4-desaturase does not result in any phenotypic alterations to growth or development, likely indicating a very limited role for S-1-P and other Δ4-unsaturated LCBs and their phosphorylated metabolites in this organism.
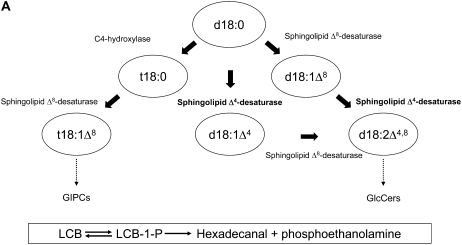
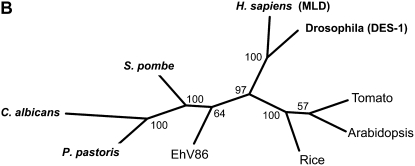
Figure 1.
Sphingolipid biosynthesis and the role of the sphingolipid Δ4-desaturase. A, A schematic representation of sphingolipid LCB modification in higher plants. Note that the actual substrates in terms of free LCBs versus N-acylated LCBs are currently undefined, as is the order of desaturation reactions to generate d18:2Δ4,8. Standard sphingolipid LCB nomenclature is used (e.g. d18:1Δ8 indicates that this is a dihydroxy-C18 LCB with a double bond at the Δ8 position; t18:0 indicates that this is a trihydroxy-C18 LCB with no double bonds). The sphingolipid class in which different LCBs are enriched is indicated (broken arrows), although it should be noted that other LCBs occur in these classes. Also shown (boxed) is the enzymatic conversion of LCBs to their phosphorylated form LCB-1-P (via LCB kinases), the dephosphorylation of LCB-1-Ps back to LCBs (via LCB-1-P phosphatases), and also the breakdown of LCB-1-Ps via the LCB-1-P lyase to generate phosphoethanolamine and hexadecanal. Free LCBs can be generated either by de novo synthesis or ceramidase-mediated release from sphingolipids (the latter is the predominant route in animals). B, Phylogenetic tree of sphingolipid Δ4-desaturase orthologs from different organisms. Functionally characterized examples are from P. pastoris (AY700778), C. albicans (XM_716863), S. pombe (NM_001022326), H. sapiens (AF002668; MLD), and Drosophila (AF466379; DES-1). Higher plant orthologs from Arabidopsis (NM_116731) and tomato (AF466378) have previously proved intractable to heterologous characterization. An unusual virally encoded ortholog is also included for comparison from Emiliania huxleyi virus 86 (EhV86; AJ890364).
RESULTS AND DISCUSSION
Functional Identification of a Sphingolipid Δ4-Desaturase Candidate Gene of Arabidopsis by Heterologous Expression in Pichia pastoris
Orthologs of the sphingolipid Δ4-desaturases identified by Ternes et al. (2002) are easily identifiable in eukaryotic genome sequences, forming a distinct clade from other microsomal lipid desaturases (Hashimoto et al., 2008). Arabidopsis contains a single ortholog of the sphingolipid Δ4-desaturase, located in proximity to the centromere of chromosome IV. This gene (At4g04930; T1J1.1) is annotated as showing similarity to the Drosophila DES-1 gene characterized by Ternes et al. (2002) and is made up of two exons split by a single intron. At4g04930 represented the only plausible detectable candidate for the sphingolipid Δ4-desaturase in the Arabidopsis genome (Fig. 1B). Therefore, functional characterization of the open reading frame (ORF) encoded by At4g04930 was attempted by heterologous expression in S. cerevisiae sur2Δ mutants, similar to that described previously (Ternes et al., 2002; Garton et al., 2003). However, despite numerous attempts, no activity was detected in association with the expression of the Arabidopsis ORF, in agreement with previous attempts to characterize plant orthologs of this enzyme (Ternes et al., 2002; our unpublished observations). This lack of activity in S. cerevisiae sur2Δ mutants most likely related to the sphingolipid substrate required by the plant desaturase, rather than to inactivity of the recombinant enzyme per se. Given that S. cerevisiae lacks some common classes of sphingolipids (such as glucosylceramides [GlcCers]) and also does not normally synthesize sphingosine, this is perhaps unsurprising, although it is important to note that this host was able to support sphingosine synthesis by other nonplant sphingolipid Δ4-desaturases (Ternes et al., 2002; Garton et al., 2003). These observations also serve to highlight our partial understanding of sphingolipid biosynthesis even in model systems.
In view of the failure of S. cerevisiae to serve as a suitable expression host for the functional characterization of higher plant sphingolipid Δ4-desaturases, an alternative system was developed. In common with many fungi, the methylotrophic yeast P. pastoris synthesizes GlcCers that contain Δ4-unsaturated LCBs, predominantly in the form of 9-methyl-sphinga-4,8-dienine. The three P. pastoris enzymes (sphingolipid C9-methyltransferase and sphingolipid Δ4- and Δ8-desaturases) responsible for the synthesis of this LCB were recently identified and characterized by heterologous expression in S. cerevisiae (Ternes et al., 2006). In this study, we deleted the intrinsic P. pastoris sphingolipid Δ4-desaturase gene to establish a mutant strain suitable as an expression host to characterize heterologously expressed sphingolipid Δ4-desaturase candidate genes. The sphingolipid Δ4-desaturase knockout strain did not contain Δ4-unsaturated LCB (data not shown) and, surprisingly, was completely devoid of GlcCers (Fig. 2A). Taking advantage of this potential screen, the Arabidopsis At4g04930 ORF was expressed in the sphingolipid Δ4-desaturase-disrupted strains of P. pastoris. As can be seen in Figure 2A, heterologous expression of the Arabidopsis ORF restores the levels of GlcCers to that observed in wild-type P. pastoris, consistent with Arabidopsis ORF At4g04930 encoding a functional sphingolipid Δ4-desaturase.
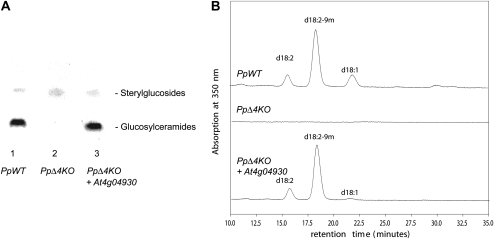
Figure 2.
Functional characterization of Arabidopsis gene At4g04930 as a sphingolipid Δ4-desaturase. A, Expression of Arabidopsis At4g04930 ORF in a P. pastoris mutant lacking sphingolipid Δ4-desaturase activity restores the synthesis of GlcCer. The P. pastoris mutant disrupted in the endogenous sphingolipid Δ4-desaturase AY700778 was transformed with either empty expression vector or vector containing the Arabidopsis ORF. Sphingolipids were extracted, separated by thin-layer chromatography, and stained by spraying with α-naphthol/sulfuric acid and subsequent heating to 160°C. The presence of GlcCer is clearly visible in P. pastoris mutants complemented with the Arabidopsis ORF (PpΔ4KO + At4g04930) but absent in mutants transformed with the empty vector (PpΔ4KO). For comparison, wild-type P. pastoris cells transformed with the empty vector are also accumulating GlcCer (PpWT). B, GlcCer from P. pastoris cells was isolated and subjected to sphingolipid LCB analysis via deacylation and derivatization with 1-fluoro-2,4-dinitrobenzene. LCBs were fractionated by HPLC and detected by A350. GlcCers from wild-type P. pastoris transformed with the empty vector (top trace) and from the P. pastoris sphingolipid Δ4-desaturase mutant transformed with Arabidopsis ORF At4g04930 (bottom trace) both contain Δ4-unsaturated LCBs (predominantly in the form of the C9-methyl-sphinga-4,8-dienine). The P. pastoris mutant transformed with the empty vector does not contain any GlcCers (middle trace).
Additional confirmation of this activity was provided by the analysis of LCBs isolated from the GlcCers of P. pastoris knockout mutant cells expressing the Arabidopsis ORF. As can be seen in Figure 2B, the LCB profiles of GlcCers isolated from wild-type P. pastoris and the P. pastoris knockout mutant disrupted in the sphingolipid Δ4-desaturase but expressing the plant ORF are similar, specifically with respect to the synthesis of the predominant 9-methyl-branched diene. As noted above, the P. pastoris knockout mutant completely lacks GlcCers (Fig. 2). On the basis of these data, we conclude that the Arabidopsis gene At4g04930 encodes the sphingolipid Δ4-desaturase responsible for the synthesis of Δ4-unsaturated LCBs such as sphingosine and sphinga-4,8-dienine. Our data also indicate the utility of P. pastoris as a new system for the identification of LCB-modifying enzymes that are not tractable using the S. cerevisiae system. The requirement for Δ4-unsaturated LCBs to initiate the synthesis of GlcCers in P. pastoris also implies that this modification is the first committed step for the biosynthesis of this class of sphingolipids in this organism, although it is equally clear that the absence of GlcCers is not essential for the normal growth of this yeast.
Expression of the Sphingolipid Δ4-Desaturase Gene in Arabidopsis Is Restricted to Floral Tissues
Having identified the sole Arabidopsis ortholog of the sphingolipid Δ4-desaturase, we wished to determine the role of this enzyme in planta. Specifically, we wished to determine if ablating this enzyme activity resulted in reduced levels of Δ4-unsaturated LCBs such as sphingosine and what the functional consequences of this might be at the phenotypic level. Surprisingly, the tissue-specific expression pattern of At4g04930 is extremely restricted, as judged by analyses of microarray data sets, with high levels reported for pollen, low transcript levels being detected in floral tissue, and transcript being virtually absent in all other tissues (Supplemental Table S1). Northern-blot analysis of total RNA from Arabidopsis leaves failed to display a signal when probed with At4g04930, even after prolonged exposure (data not shown). However, such analysis cannot exclude the possibility that At4g04930 has a limited cell-specific expression pattern that is restricted to a few cell types, although it is interesting that this gene is apparently not expressed in guard cells or modulated by abscisic acid (ABA), according to transcriptomic analyses carried out by the Schroeder laboratory and available through the Arabidopsis eFP browser (Winter et al., 2007).
Identification of Insertional Mutants of Arabidopsis Disrupted in the Sphingolipid Δ4-Desaturase Gene
To better define the role of the sphingolipid Δ4-desaturase At4g04930 in sphingolipid metabolism in Arabidopsis, a reverse genetic approach was taken. Insertional mutants of At4g04930 were identified by searching the SALK (Alonso et al., 2003) and RIKEN (Kuromori et al., 2006) databases, and two suitable alleles were identified and further characterized (Supplemental Fig. S1). The first allele (RIKEN 15-1202-1) was generated as a result of the insertion of a modified Ds transposon (Kuromori et al., 2006) after the 10th residue of exon 1. The second allele (SALK_107761.42.15.x) resulted from a T-DNA insertion after 25 residues of the second exon. In both cases, the insertions were predicted to generate ORFs that lacked the essential His box motifs required for desaturase function (Hashimoto et al., 2008). Segregating T3 seeds for these two insertion mutants were obtained from BioResource Center-RIKEN and the Nottingham Arabidopsis Stock Centre, respectively, and used to isolate homozygous lines for these disruptions (via genomic PCR using primers designed against At4g04930 and the insertion cassette). Since the RIKEN Ds transposon mutants were generated in the Nössen (No-0) ecotype, we also identified and retained wild-type null lines that lacked insertions in the target gene to serve as control material for subsequent analysis. As a further confirmation of the mutant status of these two insertion alleles, quantitative (Q)-PCR was carried out on RNA isolated from developing flowers (Supplemental Fig. S1). As shown, transcripts for At4g04930 are detected in wild-type material (Columbia [Col-0] and No-0) but not in the insertion mutants, in agreement with the genomic PCR analysis. Therefore, we conclude that these two different insertion mutants have both resulted in the disruption of the synthesis and/or accumulation of transcripts derived from gene At4g04930, which encodes the sole sphingolipid Δ4-desaturase in Arabidopsis.
Sphingolipidomic Analyses of Arabidopsis Insertion Mutants
Recent advances in mass spectrometry-based approaches to measuring sphingolipids have provided powerful tools to yield quantitative and qualitative information on Arabidopsis sphingolipid composition. Using the HPLC-electrospray ionization-tandem mass spectrometry (MS/MS) protocol developed by Markham and Jaworski (2007), sphingolipidomic profiles were obtained from leaf and floral tissue of wild-type and insertion mutant alleles of the sphingolipid Δ4-desaturase At4g04930. Profiling of rosette leaf material was carried out, but comparison of the mutants with their respective wild-type backgrounds (SALK_107761.42.15.x versus Col-0; RIKEN 15-1202-1 versus No-0) failed to reveal any significant alterations to different sphingolipid classes (ceramide, hydroxyl-ceramide, GlcCer, glycosyl inositol phosphoryl ceramides [GIPCs]; Supplemental Data Set S1). Intriguingly, the levels of two phosphorylated LCBs, t18:0-1-P and t18:1Δ8-1-P, were reduced in the leaf samples of SALK_107761.42.15.x compared with Col-0 (Supplemental Data Set S2), although the reason for these changes is not obvious. No Δ4-unsaturated sphingolipids or sphingolipid metabolites were detected in these leaf samples, indicating the absence of sphingosine and sphinga-4,8-dienine in all samples, confirming previous sphingolipid analyses (Sullards et al., 2000; Markham et al., 2006) and in agreement with our observations that At4g04930 transcripts are not detected in leaf tissue.
In view of the microarray expression pattern and our own Q-PCR data (Supplemental Fig. S1), sphingolipidomic profiling was also carried out on floral material (unopened flower buds) that expresses the sphingolipid Δ4-desaturase at moderate levels. In these samples, the effect of insertional inactivation of At4g04930 was observed, specifically in the SALK insertion allele (Fig. 3, B and C). Comparison between this line and wild-type Col-0 (Fig. 3A) indicated the complete loss of Δ4-unsaturated LCBs in GlcCers (in the form of sphinga-4,8-dienine N-linked to a hydroxyl-C16 fatty acid), confirming our assignment of this gene as the sphingolipid Δ4-desaturase. Note that although the regioisomer LCB d18:1Δ8 is present in Arabidopsis floral and leaf tissues (Fig. 3D), sphingosine is not detected (Supplemental Fig. S2; Supplemental Table S2). Thus, d18:1 LCBs (Fig. 3) represent the Δ8-unsaturated form rather than the Δ4-unsaturated isomer.
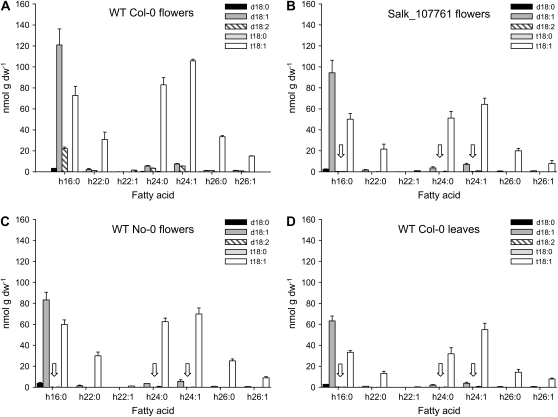
Figure 3.
Molecular species composition of GlcCer of wild-type (WT) and sphingolipid Δ4-desaturase mutant plants. Sphingolipids were extracted from wild-type Col-0 flowers (A), SALK_107761 flowers (B), wild-type No-0 flowers (C), and wild-type Col-0 leaves (D), and the amount of sphingolipid in the extract was determined by LC-MS/MS as described previously (Markham and Jaworski, 2007). Only GlcCers were found to contain appreciable amounts of d18:2 LCBs (A) with no d18:2 detectable in the other sphingolipid classes (data not shown; Markham et al., 2006). GlcCers containing d18:2 were undetectable (white arrows) in the SALK mutant line (B) and also absent from wild-type Col-0 leaf tissue (D). No d18:2 GlcCers were detected in floral tissues in wild-type No-0 (C) from which the RIKEN transposon mutant was isolated (Supplemental Table S2). Bars show averages (n = 5), and error bars show sd. dw, Dry weight.
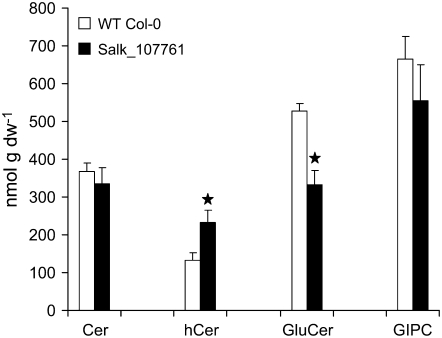
Interestingly, the loss of Δ4-unsaturated LCB in GlcCer (which represent less than 5% of the LCBs present in this class of sphingolipid) resulted in significant reduction in the total levels of GlcCer between the wild type (528 nmol g−1 dry weight) and SALK_107761 (334 nmol g−1 dry weight; Fig. 4). This was mirrored by an increase in the hydroxyl-ceramide levels in these same tissues (wild-type, 132 nmol g−1 dry weight) compared with SALK_107761 (232 nmol g−1 dry weight; Fig. 4). Given the unexpected nature of this perturbation to sphingolipid biosynthesis, we also determined by LCB analysis the levels of neutral sphingolipids (predominantly GlcCers) in a chloroform-soluble extract of floral tissues. This confirmed the decrease in these sphingolipids (322.1 nmol g−1 GlcCers in the wild type, 248.2 nmol g−1 GlcCers in SALK_107761, corresponding to 77% of the wild-type level), although this method appears to underestimate the levels of these lipids. The decrease in GlcCers observed in SALK_107761 is partially analogous to the situation in the P. pastoris mutant lacking sphingolipid Δ4-desaturase activity and consequently devoid of GlcCer. Our data, therefore, indicate a previously unsuspected role for Δ4-unsaturated LCBs in the channeling of substrate for the synthesis of GlcCers in both plants and fungi. It is perhaps relevant that in some plant species that do accumulate Δ4-unsaturated sphingolipids, such as tomato, there are also markedly high levels of GlcCers (Markham et al., 2006). Interestingly, a recent study has shown that disruption of microsomal fatty acid elongation in the Arabidopsis pas2-1 mutant results in greatly reduced levels of GlcCers but not GIPCs (Bach et al., 2008).
Figure 4.
Sphingolipid content of wild-type (WT) and sphingolipid Δ4 desaturase mutant lines. Sphingolipids were extracted from floral tissue (three samples for wild-type Col-0 and six samples for SALK_107761), and the amount of sphingolipid in the extract was determined by LC-MS/MS as described previously (Markham and Jaworski, 2007). Total amounts of each class of sphingolipid were calculated compared with added internal standards. Significant differences (P < 0.05) between the samples were determined by Student's t test and are indicated by stars. dw, Dry weight.
No other significant differences were observed in ceramides or GIPCs isolated from this same floral material. Very low levels of the free LCB sphinga-4,8-dienine were detected in the Col-0 wild type, but these were absent in the SALK insertion mutant (indicating that these LCBs are most likely derived from GlcCers; Supplemental Data Set S2). Thus, these data confirm that Δ4-unsaturated LCBs (either in sphingolipids or as free LCBs) are absent in Arabidopsis Col-0 lines in which the sphingolipid Δ4-desaturase At4g04930 has been insertionally inactivated. Moreover, this also indicates that this gene represents the sole sphingolipid Δ4-desaturating activity in Arabidopsis, since no compensatory synthesis was detected in the knockout mutant.
In the case of the RIKEN Ds insertion allele 15-1202-1, similar profiling of sphingolipids from developing flower buds indicated the complete absence of Δ4-unsaturated LCBs in both the wild type (No-0) and mutant, even in GlcCers (Supplemental Data Set S1). Thus, it appears that there is natural variation in the accumulation of different forms of sphingolipids between ecotypes, and as such, the absence of Δ4-unsaturated LCBs such as sphingosine and sphinga-4,8-dienine in No-0 would argue that such LCBs are not essential for any particular biological process (in agreement with observations of the SALK insertion line). Interestingly, both Col-0 and No-0 ecotypes showed similar (low) levels of Δ4-desaturase At4g04930 transcript accumulation (Supplemental Fig. S1). In the case of the No-0 ecotype, it is possible (based on the higher levels of phytosphingosine present) that a much more active C4-hydroxylase out-competes the sphingolipid Δ4-desaturase for the same substrates (see Fig. 1A for representation; Sperling et al., 2001). It is also noteworthy that No-0 not only lacks Δ4-unsaturated sphingolipids but also contains reduced levels of GlcCers in floral tissue, compared with Col-0. In addition to using the MS-based method described above, we also used highly optimized HPLC analysis of total LCBs (described in Tonon et al., 2005) isolated from the same floral material used for the sphingolipidomic analysis. As shown in Supplemental Figure S3, the LCB profiles of total floral sphingolipids in Col-0 and the SALK insertion mutant are very similar apart from the absence of sphinga-4,8-diene in the mutant, confirming the results presented above. Equally, it is again clear that the No-0 ecotype does not accumulate this particular LCB, irrespective of whether or not the sphingolipid Δ4-desaturase gene is insertionally inactivated (Supplemental Fig. S3). The observation that sphingolipid LCB composition is subject to natural variation may also facilitate quantitative genetic approaches to defining regulatory (homeostatic) processes.
Collectively, these data confirm the absence of Δ4-unsaturated sphingolipids from the leaves of wild-type Arabidopsis plants and that a Col-0 background insertion mutant line lacks sphinga-4,8-dienine in sphingolipids of floral tissue. It is also important to highlight that our comprehensive analysis of over 160 different sphingolipids and their metabolites failed to identify significant amounts of sphingosine in either leaf or floral tissue (Supplemental Fig. S2; Supplemental Table S2). Such analyses do have limits to their ability to detect very low abundance compounds; however, our observations are in agreement with the restricted expression pattern of the sphingolipid Δ4-desaturase gene and also with previous sphingolipid analyses in several different studies (Sullards et al., 2000; Markham et al., 2006, Markham and Jaworski, 2007).
Phenotypic Characterization of Arabidopsis Insertion Mutants
Having demonstrated that mutants disrupted in the sphingolipid Δ4-desaturase gene lacked Δ4-unsaturated sphingolipids, we wished to determine if this absence had any functional consequences. Gross phenotypic appearances of SALK_107761.42.15.x and RIKEN 15-1202-1 showed no obvious differences compared with their respective wild types (Supplemental Fig. S4); equally, a more detailed examination of a range of growth and developmental parameters showed no differences (data not shown). In the case of the RIKEN insertion mutant, these observations are in agreement with a large-scale “phenome” study that included this allele but reported no alterations to growth and development (Kuromori et al., 2006). In view of the previous reports of a role for S-1-P in Ca2+-mediated guard cell closure in Arabidopsis (Coursol et al., 2003) and Commelina (Ng et al., 2001) and that drought stress resulted in an increase of S-1-P in this latter species, we sought to establish if the insertion mutants had altered responses to drought. As discussed above, although no Δ4-unsaturated sphingolipids were detected in analysis of total rosette leaves, it was hypothetically possible that sphingosine and S-1-P might be present in specific cell types via the discrete expression of sphingolipid Δ4-desaturase At4g04930, leading to underrepresentation in microarray and Q-PCR studies of whole tissues. Thus, the availability of bona fide knockout alleles of this gene allowed this scenario to be evaluated.
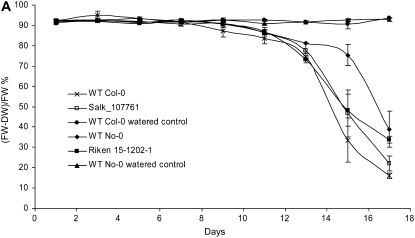
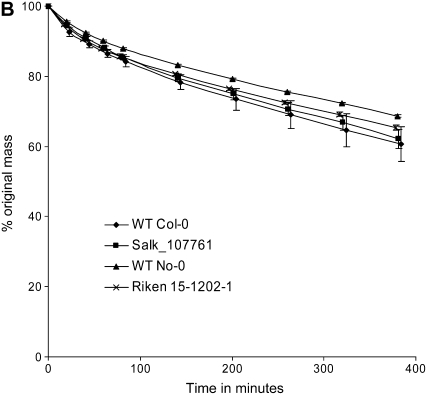
Two different protocols were used to evaluate the performance of mutant plants compared with wild-type plants under drought-induced conditions. First, in an experiment similar to that used by Ng et al. (2001), in which a 2-fold increase in S-1-P was reported in unwatered Commelina plants after 11 d, Arabidopsis plants were withheld water for 17 d, with leaf samples being harvested every 2 d. Fresh and dry weights of well-watered controls and drought-treated plants were obtained for wild-type controls and mutant lines. As can be seen in Figure 5A, there is no statistically significant difference in response to drought between the controls and mutants in terms of the plant's ability to regulate water loss. A second experimental approach was adopted to determine if maintenance of water status through guard cell closure was altered in these mutants, using well-established methods previously used to identify Arabidopsis mutants altered in their response to ABA and drought resistance (Xiong et al., 2001; Chini et al., 2004). This method determines a plant's ability to restrict water loss through (ABA-mediated) guard cell closure and has been used to characterize mutants such as aba2, aba3, abi1, and sad1 (Leon-Kloosterziel et al., 1996; Xiong et al., 2001; Chini et al., 2004). Plants were grown to just prior to bolt initiation stage under well-watered conditions, and then all aerial parts of the plants were severed from the roots with a scalpel. The fresh weights of these intact rootless plants were then determined over 7 h, indicating the rate of mass loss (water) via the transpiration stream. As can be seen in Figure 5B, no significant differences between wild-type controls and insertion mutants were observed.
Figure 5.
Phenotypic characterization of mutants for altered drought response. A, The wild type (WT) and the respective insertion mutant were grown to first bolt stage and then water was withheld. As a control, wild-type material was also fully watered. Leaves were harvested from drought-treated and control plants every 2 d for 17 d, and the (fresh weight − dry weight)/fresh weight [(FW-DW)/FW] percentage was calculated. As can be seen, there is no significant difference between the wild-type (Col-0 and No-0) and the mutant (SALK-107761 and RIKEN 15-1202-1) lines in terms of their decrease in fresh weight in response to drought. Each point represents an average (n = 3), and error bars show se. B, The wild type and the respective insertion mutants were assessed for their ability to regulate water loss through their transpiration stream via excision of roots and measurement of total mass as an indicator of water loss. Measurements were taken over a 7-h period, and data are expressed as percentages of the original starting mass (in grams) of the rootless plant. Plant material used was as for A. As can be seen, neither insertion mutant had any significant difference in mass loss. Each point represents an average (n = 3), and error bars show se.
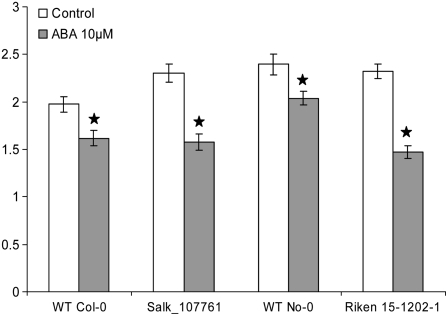
In addition to these indirect measurements of water relations in Arabidopsis, we also measured the stomatal aperture of mutants and wild-type plants in the presence or absence of ABA (10 μm; using a slightly modified method based on that described in Vahisalu et al., 2008). As can be seen in Figure 6, stomata of the mutant lines responded well to ABA compared with wild-type stomata. All four lines (Col-0, SALK_107761, No-0, and RIKEN 15-1202-1) showed a highly significant (P = 0.01) stomatal closure in response to ABA treatment, indicating that there was no difference between wild-type and mutant plants in the response to this hormone.
Figure 6.
Stomatal aperture measurements of wild-type (WT) and mutant lines after ABA treatment. Stomatal apertures were measured from wild-type and mutant leaves treated with or without ABA for 2 h. Data are from three independent experiments, and measurements were carried out double blind. Each bar represents an average (n = 3), and error bars show se. Significant differences (P < 0.01; stars) between the treated and untreated samples were determined by Student's t test.
Collectively, these data indicate that Δ4-unsaturated sphingolipids and/or their metabolites do not play a primary role in modulating water relations or response to drought stress in Arabidopsis. This is perhaps surprising given the study of Ng et al. (2001), which indicted a role for S-1-P in Commelina guard cell closure (based on exogenous application of 4–6 μm S-1-P) and also reported an increase in the endogenous levels of this phosphorylated LCB in drought-treated leaves. Our analysis of sphingolipid LCBs from Commelina leaf tissue indicates that sphingosine is a very minor component (Supplemental Fig. S2), although this does not preclude the presence of S-1-P in Commelina. Based on our current studies in Arabidopsis using insertion mutants lacking any capacity to synthesize sphingosine, it is clear that this particular LCB(-1-P) either does not play a dominant role in these processes or is functionally redundant. In that respect, it is interesting that Coursol et al. (2005) reported a different LCB-1-P, phytosphingosine-1-P (t18:0-1-P), as being involved in Arabidopsis guard cell closure. Very recently, Chen et al. (2008) demonstrated a crucial role for these C4-hydroxylated LCBs in Arabidopsis growth and development.
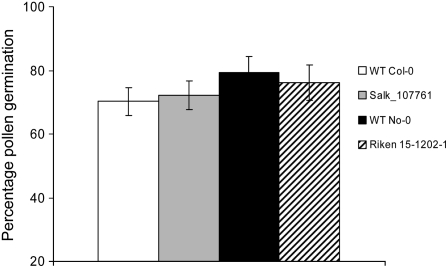
Having not observed a direct role for sphingosine and other Δ4-unsaturated sphingolipids in drought stress, control of transpiration rate, or stomatal closure, we considered any other potential roles for these LCBs. As noted above, the sphingolipid Δ4-desaturase At4g04930 is only expressed at significant levels in flowers, predominantly in pollen. Interestingly, mutations in the Drosophila DES-1 ortholog result in flies with defective spermatogenesis (Endo et al., 1996). Therefore, we investigated the formation and viability of pollen in our Arabidopsis mutants, although the initial isolation and characterization of these insertion alleles indicated that both mutant alleles (SALK and RIKEN) were fertile, implying no gross defect in pollen development or viability. Pollen from control and mutant lines were visualized by double staining with fluorescein diacetate and propidium iodide, allowing the discrimination between live and dead pollen (McConn and Browse, 1996). However, no difference was observed in the percentage of dead pollen cells between control and mutant lines (data not shown). Second, pollen from wild-type control and mutant lines was germinated in vivo in the presence of 2% Suc, and the percentage germination rate was determined according to the methods of Footitt et al. (2007). As can be seen in Figure 7, no significant differences in rates between wild-type controls and mutants were observed, indicating no role for sphingosine or Δ4-unsaturated sphingolipids in pollen viability or germination.
Figure 7.
Pollen viability of mutants. Pollen from wild-type (WT) and insertion mutant flowers was assayed for germination, with percentage germination assessed by microscopic examination. Plant material used was as for Figure 5. No significant difference between the wild type and mutants was observed. Each bar represents an average (n = 3), and error bars show se.
CONCLUSION
The aim of this study was to determine the function of the Arabidopsis gene At4g04930, assumed to encode a sphingolipid Δ4-desaturase, and also to investigate the role of Δ4-unsaturated LCBs in Arabidopsis growth and development. We have provided, to our knowledge, the first functional characterization of a higher plant sphingolipid Δ4-desaturase via complementation of a P. pastoris mutant lacking this activity. Arabidopsis insertion mutants disrupted in At4g04930 lacked any detectable transcripts for this gene and also did not contain any Δ4-unsaturated sphingolipids such as sphingosine or S-1-P. Our data, however, indicated a role for Δ4-unsaturated LCBs in the channeling of ceramides for the synthesis of GlcCers in certain tissues, although these insertion mutants displayed normal growth and development and did not indicate any perturbation to drought tolerance, transpiration rate, or pollen viability. Based on all of these data, and bearing in mind that At4g04930 transcripts are undetectable in most Arabidopsis tissues, we consider it unlikely that sphingosine and S-1-P play a significant role in the life cycle of Arabidopsis. We believe that our approach, in which the in vivo capacity to synthesize sphingosine (and hence S-1-P) is abolished by insertional gene inactivation and biochemically confirmed by a high-resolution HPLC-electrospray ionization-MS/MS analysis of Arabidopsis sphingolipids, provides definitive evidence that Δ4-unsaturated sphingolipids are nonessential in this plant species. In addition, in view of the absence of sphingosine and S-1-P in virtually all Arabidopsis tissues, we would suggest that activities recently described as “sphingosine kinase” (Worrall et al., 2008) should be renamed “LCB kinase.”
However, it should also be emphasized that we believe that it is very likely that other LCBs and LCB-1-Ps play important roles in plant-specific processes. Such data are starting to emerge regarding the role of phytosphingosine (Coursol et al., 2005; Shi et al., 2007; Chen et al., 2008) and also Δ8-unsaturated LCBs (Ryan et al., 2007). The significance of sphingolipids and their phosphorylated metabolites is still an evolving story in many eukaryotic systems, and it is hoped that studies such as this can contribute to the development of more precise models for their form and function in different organisms. In addition, our observation of the unexpected role of Δ4-unsaturated N-acylated LCBs in GlcCer synthesis highlights our still only partial understanding of sphingolipid metabolism in plants and yeast.
MATERIALS AND METHODS
Construction of a Phylogenetic Tree
The amino acid sequences of sphingolipid Δ4-desaturase genes from Arabidopsis (Arabidopsis thaliana) were aligned using the EMBOSS multiple alignment program EMMA. The phylogenetic tree was created using the PHYLIP software package. To test the reliability of the final tree, 100 bootstrap samples were made of the alignment and distance matrices created for each bootstrap sample using PROTDIST. The distance matrices were converted into trees using the neighbor-joining method, and a consensus tree was created with numbers indicating how many times each branch was observed in the bootstrap sets.
Generation of a Sphingolipid Δ4-Desaturase Deletion Mutant of Pichia pastoris
Generation of the knockout cassette for the deletion of the Δ4-desaturase was performed by restriction enzyme-based cloning. The ORF including 500 bp of flanking upstream and downstream sequences was amplified from genomic DNA with PfuTurbo DNA polymerase (Stratagene). The primers used were Delta4-F-EcoRI and Delta4-R-HindIII (Supplemental Table S3). The primers included adapter sequences (underlined in Supplemental Table S3) containing the indicated restriction sites. The PCR products were purified by gel electrophoresis and ligated into the multiple cloning site of the Litmus38i vector (New England Biolabs) using the restriction sites EcoRI and HindIII. A part of the ORF was subsequently deleted by digestion with XbaI and XmnI. The zeocin resistance cassette from the GAPZ-B vector (Invitrogen) was amplified using the primers Zeo-F-XbaI and Zeo-R-XmnI and ligated into the interrupted ORF using the indicated restriction sites (underlined in Supplemental Table S3).
Transformation of P. pastoris Cells by Electroporation
The vector containing the knockout cassette was linearized and used for the transformation of electrocompetent GS115 (his4) cells (Invitrogen). Transformants were selected on yeast potato dextrose media plates containing 100 mg L−1 zeocin. Correct integration of the knockout cassette was tested by PCR with the primer pairs Delta4-Test-F/Zeo-int-R and Zeo-int-F/Delta4-Test-R (data not shown).
Cloning of the Arabidopsis Δ4-Desaturase
The complete coding sequence of the putative Δ4-desaturase from Arabidopsis was amplified with Pwo polymerase (Peqlab) using a standard PCR program and the specific primers d4At-F and d4At-R on cDNA (generated from RNA isolated from floral tissue). PCR products were ligated in the P. pastoris expression vector pPIC3.5 (Invitrogen) in the EcoRI and NotI restriction sites of the multiple cloning site. The cloned complete coding sequence was checked by sequencing.
Transformation of P. pastoris
Chemical competent cells from P. pastoris strain GS115-Δ4-desaturase-KO were transformed with the ScaI and NdeI linearized construct described above. The strains GS115 (= wild-type) and GS115-Δ4-desaturase-KO were also transformed with linearized empty vector pPIC3.5. Single colonies were isolated and used for lipid analysis.
Liquid Culture
A preculture was grown in liquid minimal glycerol medium (1.34% yeast nitrogen base without amino acids, 1% glycerol) for 24 h at 30°C with shaking (180 rpm). A total of 200 mL of minimal glycerol medium was inoculated 1:100 from the preculture and incubated again for 24 h at 30°C. Gene expression was induced by the addition of methanol to a final concentration of 0.5% and further incubation for 24 h. Cells were harvested by centrifugation.
Isolation of GlcCer
Approximately 5 g fresh weight of P. pastoris cells was suspended in 5 mL of water and boiled in a water bath for 15 min. The cells were sedimented by centrifugation, and the lipids were extracted by shaking in 10 mL of chloroform:methanol (1:1) overnight at 8°C, followed by 9 mL of chloroform:methanol (2:1) for at least 4 h at 8°C. The lipid extract was washed by phase partitioning with chloroform:methanol:0.45% (w/v) NaCl (8:4:3), and the solvents subsequently evaporated. The lipids were redissolved in chloroform, and GlcCer was purified by preparative thin-layer chromatography on silica gel 60 plates (Merck) developed in chloroform:methanol (85:15, v/v).
Analysis of the Sphingolipid LCB Composition
The LCB composition of P. pastoris glucosylceramides was analyzed as described by Ternes et al. (2006). Similar methods were used for the analyses of Arabidopsis and Commelina communis sphingolipid LCBs, according to the methods of Borner et al. (2005) and Tonon et al. (2005). Briefly, samples (yeast pellet or plant tissues) were subjected to strong alkaline hydrolysis in 10% barium hydroxide in dioxane. The resulting free LCBs were converted to dinitrophenyl derivatives and analyzed by reverse-phase HPLC with detection by absorbance at 350 nm. Identification was via comigration with known standards (Avanti Polar Lipids) and atmospheric pressure chemical ionization-MS (Tonon et al., 2005).
Sphingolipidomic Analysis
Sphingolipid analysis was carried out exactly as described by Markham and Jaworski (2007). Analyses were carried out on pooled material (leaves and flowers) from a number of individual plants grown in the same growth cabinet. A minimum of three technical replicates were run from each pooled sample.
Growth of Arabidopsis and Identification of Arabidopsis Δ4-Desaturase Insertion Mutants
Arabidopsis plants were grown and maintained as described previously (Footitt et al., 2007) in soil, with plants grown to maturity in controlled-environment rooms (16 h of light at 23°C and 70% relative humidity/8 h of dark at 18°C and 80% relative humidity). During the light phase, the incident photosynthetically active radiation was 150 to 175 μmol m−2 s−1 at the soil level. The position of plant trays was rotated to minimize light effects. Arabidopsis seed stocks were obtained from the Nottingham Arabidopsis Stock Centre (Salk_107761.42.15.x, Col-0 ecotype) and the RIKEN BioResource Centre (insertion mutant RIKEN 15-1202-1, No-0 ecotype). PCR was used to identify and isolate homozygous mutants from segregating populations using a gene-specific primer in conjunction with a primer to the insertion module. Genomic DNA was extracted from leaf material, and PCR was performed at 94°C for 15 min, followed by 35 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1.5 min, and then 72°C for 7 min using HotStart Taq Pol master mix (Qiagen). The following primers were used for the identification of RIKEN 15-1202-1 homozygous individuals: 930F (5′-TCTCTCTCGTTTGACTTTCC-3′), 930R (5′-TGCTAAGAAGAGATTGTGGTT-3′), and Ds3-2a (5′-CCGGATCGTATCGGTTTTCG-3′). The following primer combination was used for the identification of Salk_107761.42.15.x homozygous individuals: SalkLP (5′-AGGTTGTCGAAGAAACAAACG-3′), SalkRP (5′-TGATGAACTCCCAGTAGCCAG-3′), and LBb1 (5′-GCGTGGACCGCTTGCTGCAACT-3′).
Confirmation of the absence of Δ4-desaturase transcript was obtained by real-time PCR. RNA was extracted from three independent biological replicates comprising 100 mg of inflorescence tissue (Supplemental Fig. S1) using an RNeasy plant RNA isolation kit with on-column DNase treatment (Qiagen) followed by RNA cleanup using the RNeasy plant RNA isolation kit. One microgram of total RNA was treated with the Turbo DNA-free kit (Ambion) and used as a template to synthesize cDNA using the SuperScript III Platinum Two-Step qRT-PCR Kit with SYBR Green (Invitrogen). PCR was performed on the ABI 7500 Real Time PCR System (Applied Biosystems) using Platinum SYBR Green qPCR SuperMix-UDG reagents (Invitrogen) according to the manufacturer's specification, with the cDNA equivalent of 17.5 ng of RNA in a 25-μL reaction volume. Reactions were performed in duplicate, and the absence of genomic DNA and primer dimers was confirmed by analysis of RT-minus and water control samples and by examination of dissociation curves. The primers for the Arabidopsis Δ4-desaturase, T1J1F2.2 (5′-TGGAGATCTTTCGCGTATCTAATC-3′) and T1J1R2.2 (5′-CATACCGCCTCCAACAAATG-3′), were designed using Primer Express version 2.0 (Applied Biosystems). Primers for the reference gene At4g34270 have been described (Czechowski et al., 2005). Calculation of normalized expression values was done after the method of Livak and Schmittgen (2001).
Drought Stress Experiments
Leaves were detached and weighed (one from a representative plant on each day; many plants were used in rotation so as not to defoliate them). Then the leaf was dried at 80°C and reweighed. Rosette plants were detached from the soil surface, placed under 30% relative humidity at 19°C, and weighed at designated time points. Percentage of fresh weight was calculated based on the initial weight of the plants as described by Chini et al. (2004).
Pollen Germination in Vitro
Pollen germination assays were carried out as described by Footitt et al. (2007). For each line, pollen from two flowers was cultured in suspended drops in control medium [16% (w/v) polyethylene glycol-3550, 2% (w/v) Suc, 1 mm CaCl2, 1 mm Ca(NO3)2, 1 mm MgSO4, and 0.015% (w/v) boric acid, pH 6.5]. Pollen was incubated in a humid chamber for 16 h in hanging drops on microscope slides. Germination was scored by microscopic examination. Tubes of germinated pollen grains were visualized with a Carl Zeiss Axiovert 135 inverted microscope.
Stomatal Bioassays
Stomatal assays were performed by floating leaves for 2.5 h under continuous illumination (60–100 μE m−2 s−1) in MES/KCl buffer (5 mm KCl, 10 mm MES, and 50 μm CaCl2, pH 6.15). Following the opening of stomata, leaves were treated with 10 μm ABA for another 2 h. The leaves were subsequently homogenized individually in a Waring blender for 30 s, and the epidermal fragments were collected on a 100-μm nylon mesh (SpectraMesh; Merck). Stomatal apertures from epidermal fragments were then measured using a calibrated light microscope attached to an imaging system (QWin software; Leica). Experiments were repeated three times and by counting at least 20 stomata per leaf per treatment; this analysis was performed double blind, meaning that neither the experimenter measuring the apertures nor the researcher who provided the material was aware of the identity (i.e. genotype) of the four samples undergoing the ABA treatment.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Validation of insertion mutants by Q-PCR.
Supplemental Figure S2. LCB profile of plant sphingolipids showing resolution of Δ8 and Δ4 regioisomers and the absence of sphingosine in Arabidopsis.
Supplemental Figure S3. LCB profiles of the wild type and insertion mutants.
Supplemental Figure S4. Phenotypic appearance of wild-type and mutant Arabidopsis.
Supplemental Table S1. Microarray-derived tissue-specific expression profile for At4g04930.
Supplemental Table S2. LCB analysis of wild-type and mutant Arabidopsis.
Supplemental Table S3. List of PCR primers used in construction of the P. pastoris expression system.
Supplemental Data Set 1. Sphingolipidomic analysis of sphingolipids from the wild type and insertion mutants (leaves and floral tissue).
Supplemental Data Set 2. Sphingolipidomic analysis of free LCBs and LCP-1-Ps from the wild type and insertion mutants (leaves and floral tissue).
Supplementary Material
Acknowledgments
This paper is dedicated to the memory of our colleague Petra Sperling. J.N. thanks Teresa Dunn for helpful discussions. We thank Allison van de Meene and Mitch Le Bloa of the Rothamsted Research Centre for Bioimaging for advice with microscopy. We thank the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants and RIKEN Genomic Sciences Center for providing the Ds insertion mutant. We also thank Enid MacRobbie for the gift of the Commelina seeds.
This work was supported by the Deutsche Forschungsgemeinschaft (grant no. SP 967/1–1 to P.S. and S.Z.) and the Biotechnology and Biological Sciences Research Council (to Rothamsted Research).
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) are: Dirk Warnecke (warnecke@botanik.uni-hamburg.de) and Johnathan A. Napier (johnathan.napier@bbsrc.ac.uk).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Bach L, Michaelson LV, Haslam R, Bellec Y, Gissot L, Marion J, Da Costa M, Boutin JP, Miquel M, Tellier F, et al (2008) The very-long-chain hydroxy fatty acyl-CoA dehydratase PASTICCINO2 is essential and limiting for plant development. Proc Natl Acad Sci USA 105 14727–14731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GH, Sherrier DJ, Weimar T, Michaelson LV, Hawkins ND, Macaskill A, Napier JA, Beale MH, Lilley KS, Dupree P (2005) Analysis of detergent-resistant membranes in Arabidopsis: evidence for plasma membrane lipid rafts. Plant Physiol 137 104–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlee C (2001) Intracellular signalling: sphingosine-1-phosphate branches out. Curr Biol 11 R535–R538 [DOI] [PubMed] [Google Scholar]
- Chen M, Han G, Dietrich CR, Dunn TM, Cahoon EB (2006) The essential nature of sphingolipids in plants as revealed by the functional identification and characterization of the Arabidopsis LCB1 subunit of serine palmitoyltransferase. Plant Cell 18 3576–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Markham JE, Dietrich CR, Jaworski JG, Cahoon EB (2008) Sphingolipid long-chain base hydroxylation is important for growth and regulation of sphingolipid content and composition in Arabidopsis. Plant Cell 20 1862–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Grant JJ, Seki M, Shinozaki K, Loake GJ (2004) Drought tolerance established by enhanced expression of the CC-NBS-LRR gene, ADR1, requires salicylic acid, EDS1 and ABI1. Plant J 38 810–822 [DOI] [PubMed] [Google Scholar]
- Coursol S, Fan LM, Le Stunff H, Spiegel S, Gilroy S, Assmann SM (2003) Sphingolipid signalling in Arabidopsis guard cells involves heterotrimeric G proteins. Nature 423 651–654 [DOI] [PubMed] [Google Scholar]
- Coursol S, Le Stunff H, Lynch DV, Gilroy S, Assmann SM, Spiegel S (2005) Arabidopsis sphingosine kinase and the effects of phytosphingosine-1-phosphate on stomatal aperture. Plant Physiol 137 724–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich CR, Han G, Chen M, Berg RH, Dunn TM, Cahoon EB (2008) Loss-of-function mutations and inducible RNAi suppression of Arabidopsis LCB2 genes reveal the critical role of sphingolipids in gametophytic and sporophytic cell viability. Plant J 54 284–298 [DOI] [PubMed] [Google Scholar]
- Dietrich CR, Perera MA, Yandeau-Nelson M, Meeley RB, Nikolau BJ, Schnable PS (2005) Characterization of two GL8 paralogs reveals that the 3-ketoacyl reductase component of fatty acid elongase is essential for maize (Zea mays L.) development. Plant J 42 844–861 [DOI] [PubMed] [Google Scholar]
- Dunn TM, Lynch DV, Michaelson LV, Napier JA (2004) A post-genomic approach to understanding sphingolipid metabolism in Arabidopsis thaliana. Ann Bot (Lond) 93 483–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo K, Akiyama T, Kobayashi S, Okada M (1996) Degenerative spermatocyte, a novel gene encoding a transmembrane protein required for the initiation of meiosis in Drosophila spermatogenesis. Mol Gen Genet 253 157–165 [DOI] [PubMed] [Google Scholar]
- Footitt S, Dietrich D, Fait A, Fernie AR, Holdsworth MJ, Baker A, Theodoulou FL (2007) The COMATOSE ATP-binding cassette transporter is required for full fertility in Arabidopsis. Plant Physiol 144 1467–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garton S, Michaelson LV, Beaudoin F, Beale MH, Napier JA (2003) The dihydroceramide desaturase is not essential for cell viability in Schizosaccharomyces pombe. FEBS Lett 538 192–196 [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Yoshizawa AC, Okuda S, Kuma K, Goto S, Kanehisa M (2008) The repertoire of desaturases and elongases reveals fatty acid variations in 56 eukaryotic genomes. J Lipid Res 49 183–191 [DOI] [PubMed] [Google Scholar]
- Imai H, Ohnishi M, Hotsubo K, Kojima M, Ito S (1997) Sphingoid base composition of cerebrosides from plant leaves. Biosci Biotechnol Biochem 61 351–353 [Google Scholar]
- Kuromori T, Wada T, Kamiya A, Yuguchi M, Yokouchi T, Imura Y, Takabe H, Sakurai T, Akiyama K, Hirayama T, et al (2006) A trial of phenome analysis using 4000 Ds-insertional mutants in gene-coding regions of Arabidopsis. Plant J 47 640–651 [DOI] [PubMed] [Google Scholar]
- Leon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JA, Koornneef M (1996) Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J 10 655–661 [DOI] [PubMed] [Google Scholar]
- Liang H, Yao N, Song JT, Luo S, Lu H, Greenberg JT (2003) Ceramides modulate programmed cell death in plants. Genes Dev 17 2636–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). method. Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
- Lynch DV, Dunn TM (2004) An introduction to plant sphingolipids. New Phytol 161 677–702 [DOI] [PubMed] [Google Scholar]
- Markham JE, Jaworski JG (2007) Rapid measurement of sphingolipids from Arabidopsis thaliana by reversed-phase high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Rapid Commun Mass Spectrom 21 1304–1314 [DOI] [PubMed] [Google Scholar]
- Markham JE, Li J, Cahoon EB, Jaworski JG (2006) Separation and identification of major plant sphingolipid classes from leaves. J Biol Chem 281 22684–22694 [DOI] [PubMed] [Google Scholar]
- McConn M, Browse J (1996) The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel C, van Echten-Deckert G, Rothers J, Sandhoff K, Wang E, Merril AH (1997) Characterization of ceramide synthesis: a dihydroceramide desaturase introduces the 4,5-trans-double bond of sphingosine at the level of dihydroceramide. J Biol Chem 272 22432–22437 [DOI] [PubMed] [Google Scholar]
- Napier JA, Michaelson LV, Dunn TM (2002) A new class of lipid desaturase central to sphingolipid biosynthesis and signalling. Trends Plant Sci 7 475–478 [DOI] [PubMed] [Google Scholar]
- Ng CK, Carr K, McAinsh MR, Powell B, Hetherington AM (2001) Drought-induced guard cell signal transduction involves sphingosine-1-phosphate. Nature 410 596–599 [DOI] [PubMed] [Google Scholar]
- Oskouian B, Saba JD (2004) Death and taxis: what non-mammalian models tell us about sphingosine-1-phosphate. Semin Cell Dev Biol 15 529–540 [DOI] [PubMed] [Google Scholar]
- Pyne S, Pyne NJ (2000) Sphingosine 1-phosphate signalling in mammalian cells. Biochem J 349 385–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan PR, Liu Q, Sperling P, Dong B, Franke S, Delhaize E (2007) A higher plant Δ8 sphingolipid desaturase with a preference for (Z)-isomer formation confers aluminum tolerance to yeast and plants. Plant Physiol 144 1968–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saba JD, Hla T (2004) Point-counterpoint of sphingosine 1-phosphate metabolism. Circ Res 94 724–734 [DOI] [PubMed] [Google Scholar]
- Shi L, Bielawski J, Mu J, Dong H, Teng C, Zhang J, Yang X, Tomishige N, Hanada K, Hannun YA, Zuo J (2007) Involvement of sphingoid bases in mediating reactive oxygen intermediate production and programmed cell death in Arabidopsis. Cell Res 17 1030–1040 [DOI] [PubMed] [Google Scholar]
- Sperling P, Heinz E (2003) Plant sphingolipids: structural diversity, biosynthesis, first genes and functions. Biochim Biophys Acta 1632 1–15 [DOI] [PubMed] [Google Scholar]
- Sperling P, Ternes P, Moll H, Franke S, Zahringer U, Heinz E (2001) Functional characterization of sphingolipid C4-hydroxylase genes from Arabidopsis thaliana. FEBS Lett 494 90–94 [DOI] [PubMed] [Google Scholar]
- Sperling P, Warnecke D, Heinz E (2004) Plant sphingolipids. In M Hohmann, ed, Topics in Current Genetics, Vol 6. Lipid Metabolism and Membrane Biogenesis. Springer Verlag, Berlin, pp 337–380
- Spiegel S, Milstien S (2003) Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat Rev Mol Cell Biol 4 397–407 [DOI] [PubMed] [Google Scholar]
- Sullards MC, Lynch DV, Merrill AH, Adams J (2000) Structure determination of soybean and wheat glucosylceramides by tandem mass spectrometry. J Mass Spectrom 35 347–353 [DOI] [PubMed] [Google Scholar]
- Teng C, Dong H, Shi L, Deng Y, Mu J, Zhang J, Yang X, Zuo J (2008) Serine palmitoyltransferase, a key enzyme for de novo synthesis of sphingolipids, is essential for male gametophyte development in Arabidopsis. Plant Physiol 146 1322–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ternes P, Franke S, Zahringer U, Sperling P, Heinz E (2002) Identification and characterization of a sphingolipid Δ4-desaturase family. J Biol Chem 277 25512–22518 [DOI] [PubMed] [Google Scholar]
- Ternes P, Sperling P, Albrecht S, Franke S, Cregg JM, Warnecke D, Heinz E (2006) Identification of fungal sphingolipid C9-methyltransferases by phylogenetic profiling. J Biol Chem 281 5582–5592 [DOI] [PubMed] [Google Scholar]
- Tonon T, Sayanova O, Michaelson LV, Qing R, Harvey D, Larson TR, Li Y, Napier JA, Graham IA (2005) Fatty acid desaturases from the microalga Thalassiosira pseudonana. FEBS J 272 3401–3412 [DOI] [PubMed] [Google Scholar]
- Vahisalu T, Kollist H, Wang YF, Nishimura N, Chan WY, Valerio G, Lamminmäki A, Brosché M, Moldau H, Desikan R, Schroeder JI, Kangasjärvi J (2008) SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 452 487–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provert NJ (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2 e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrall D, Liang YK, Alvarez S, Holroyd GH, Spiegel S, Panagopulos M, Gray JE, Hetherington AM (2008) Involvement of sphingosine kinase in plant cell signalling. Plant J 28 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrall D, Ng CK, Hetherington AM (2003) Sphingolipids, new players in plant signaling. Trends Plant Sci 8 317–320 [DOI] [PubMed] [Google Scholar]
- Xiong L, Gong Z, Rock CD, Subramanian S, Guo Y, Xu W, Galbraith D, Zhu JK (2001) Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev Cell 6 771–781 [DOI] [PubMed] [Google Scholar]
- Zheng H, Rowland O, Kunst L (2005) Disruptions of the Arabidopsis enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. Plant Cell 17 1467–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.