Abstract
Magnaporthe oryzae is a hemibiotrophic fungal pathogen that causes rice (Oryza sativa) blast. Although M. oryzae as a whole infects a wide variety of monocotyledonous hosts, no dicotyledonous plant has been reported as a host. We found that two rice pathogenic strains of M. oryzae, KJ201 and 70-15, interacted differentially with 16 ecotypes of Arabidopsis (Arabidopsis thaliana). Strain KJ201 infected all ecotypes with varying degrees of virulence, whereas strain 70-15 caused no symptoms in certain ecotypes. In highly susceptible ecotypes, small chlorotic lesions appeared on infected leaves within 3 d after inoculation and subsequently expanded across the affected leaves. The fungus produced spores in susceptible ecotypes but not in resistant ecotypes. Fungal cultures recovered from necrotic lesions caused the same symptoms in healthy plants, satisfying Koch's postulates. Histochemical analyses showed that infection by the fungus caused an accumulation of reactive oxygen species and eventual cell death. Similar to the infection process in rice, the fungus differentiated to form appressorium and directly penetrated the leaf surface in Arabidopsis. However, the pathogenic mechanism in Arabidopsis appears distinct from that in rice; three fungal genes essential for pathogenicity in rice played only limited roles in causing disease symptoms in Arabidopsis, and the fungus seems to colonize Arabidopsis as a necrotroph through the secretion of phytotoxic compounds, including 9,12-octadecadienoic acid. Expression of PR-1 and PDF1.2 was induced in response to infection by the fungus, suggesting the activation of salicylic acid- and jasmonic acid/ethylene-dependent signaling pathways. However, the roles of these signaling pathways in defense against M. oryzae remain unclear. In combination with the wealth of genetic and genomic resources available for M. oryzae, this newly established pathosystem allows comparison of the molecular and cellular mechanisms underlying pathogenesis and host defense in two well-studied model plants.
A very large number of potential plant pathogens exist in nature; however, individual plant species are susceptible to only a limited number of pathogens because of the presence of effective general defense mechanisms (Nimchuk et al., 2003; Jones and Takemoto, 2004). In some plant species, resistance against a specific pathogen is expressed in a cultivar- or accession-specific manner. This type of resistance is often mediated by the gene-for-gene interactions proposed by Flor (1971), in which direct or indirect interactions between the proteins encoded by host resistance and pathogen avirulence genes determine the outcome of infection (Chang et al., 2004; Jones and Dangl, 2006). The defense responses mediated by gene-for-gene interactions include rapid localized cell death, known as the hypersensitive response (HR; Glazebrook, 2001; Rate and Greenberg, 2001), the production of phytoalexins (Modolo et al., 2002) and other antimicrobial secondary metabolites, and the expression of pathogenesis-related (PR) proteins (Narasimhan et al., 2001). Some PR proteins exhibit antimicrobial properties. The signaling molecules implicated in these inducible defense systems include salicylic acid (SA), jasmonic acid (JA), ethylene (ET), and reactive oxygen species (ROS; Clarke et al., 2000; Kunkel and Brooks, 2002).
The molecular and cellular bases of host-pathogen interactions have been studied extensively using a small number of model systems (Roetschi et al., 2001; Bohman et al., 2004; O'Connell et al., 2004). Arabidopsis (Arabidopsis thaliana) has been used extensively as one such model because of the availability of its entire genome sequence, functional genomic resources, and an extensive collection of mutants and germplasm, in addition to the ease of genetic and molecular analyses. A number of fungal and oomycete pathogens have been reported to infect Arabidopsis naturally and/or under laboratory conditions, including obligate biotrophs (e.g. Hyaloperonospora parasitica, Albugo candida, and Erysiphe cichoracearum), hemibiotrophs (e.g. Phytophthora infestans, P. brassicae, P. cinnamomi, Colletotrichum trifolii, and C. higginsianum), and necrotrophs (e.g. Alternaria brassicicola and Botrytis cinerea; Holub et al., 1995; Rehmany et al., 2000; Roetschi et al., 2001; Robinson and Cahill, 2003; O'Connell et al., 2004; Thaler et al., 2004). Signaling pathways controlled by SA, JA, and/or ET are involved in controlling the interaction of Arabidopsis with these pathogens. The SA-dependent pathway controls the synthesis of low molecular weight antimicrobial proteins such as PR-1, PR-2, and PR-5, whereas the JA/ET-dependent pathway controls the systemic activation of a set of genes that encode antimicrobial proteins such as thionin (THI2.1), defensin (PDF1.2), PR-3, and PR-4. These signaling pathways differ in their efficacy depending on the nature of the invading pathogen. Generally, defense against biotrophic pathogens is mediated via the SA-dependent pathway, whereas the JA/ET-dependent pathway plays a key role in defense against necrotrophic pathogens (Delaney et al., 1994; Thomma et al., 1998).
Rice (Oryza sativa) blast, caused by Magnaporthe oryzae, is one of the most destructive diseases in cultivated rice, which feeds one-half of the world's population (Ford et al., 1994; Talbot and Foster, 2001). The fungus uses a hemibiotrophic infection strategy that involves initial proliferation inside living host cells before switching to a destructive necrotrophic mode. The infection of rice by M. oryzae follows a developmental process that has been observed in many foliar fungal pathogens. A germ tube produced from the conidium differentiates into a specialized infectious structure called the appressorium, which adheres tightly to the plant surface using mucilage (Howard et al., 1991). The fungus generates enormous turgor pressure inside the melanized appressorium, and a thin penetration peg pierces the host surface using this pressure to enter a leaf epidermal cell (Howard et al., 1991; De Jong et al., 1997). After penetration, the peg differentiates into bulbous and lobed infectious hyphae that grow intracellularly and intercellularly (Heath et al., 1990, 1992), resulting in blast lesions (Tucker and Talbot, 2001). The entire M. oryzae genome has been sequenced (Dean et al., 2005), and functional genomic approaches have led to the identification of hundreds of genes involved in its pathogenesis (Jeon et al., 2007).
Here, we describe the development of a new model pathosystem based on Arabidopsis as the host and M. oryzae as the pathogen. We found that two rice-pathogenic strains of M. oryzae could infect multiple ecotypes of Arabidopsis in laboratory inoculation experiments. We subsequently studied factors required for the infection of Arabidopsis by M. oryzae and how these ecotypes respond to M. oryzae at both the cellular and gene expression levels. In combination with the wealth of genetic and genomic resources available for M. oryzae, this newly established pathosystem allows comparative analyses of pathogenicity mechanisms and defense responses between Arabidopsis and rice using the same pathogen isolates.
RESULTS
Two M. oryzae Strains Interact Differentially with 16 Ecotypes of Arabidopsis
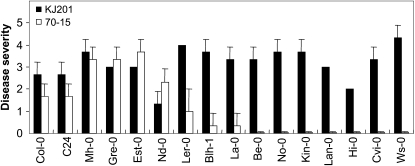
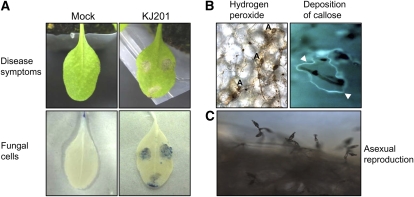
Sixteen ecotypes of Arabidopsis were inoculated with two rice pathogenic M. oryzae strains (KJ201 and 70-15; Fig. 1). All ecotypes except Niederzenz (Nd-0) exhibited a disease severity (DS) score of 2 or higher upon infection with KJ201. In contrast, most of the ecotypes infected with 70-15 (with the exception of Mühlen, Greenville, Estland, and Nd-0) had DS scores lower than 2, with seven of them exhibiting no visible symptoms (Fig. 1; Supplemental Fig. S1). These seven ecotypes, as well as Bulhary and Landsberg, which exhibited DS scores less than 1, were considered resistant to infection by 70-15. All ecotypes that exhibited DS scores higher than 1 to a strain used were considered susceptible to the strain. Disease severity among the susceptible ecotypes varied widely (Fig. 1). The responses of Columbia (Col-0) and Wassilewskija (Ws-0) to KJ201 and 70-15 illustrate the nature and progression of disease symptoms among the susceptible and resistant ecotypes (Fig. 2). Plants of Col-0, which were more susceptible to KJ201 than to 70-15, began producing small chlorotic or yellow spots within 3 days postinoculation (dpi) with KJ201 (Fig. 2A). Necrosis was also observed at 3 dpi at the center of severely chlorotic areas. These spots subsequently expanded and became highly visible by 6 dpi. The lesions covered the entire leaf surface by 9 dpi, changing the leaf color to somewhat yellow or light brown. When challenged with 70-15, the Col-0 plants developed lesions that looked similar to those caused by KJ201; however, their development was slower and it took longer for the lesions to expand. KJ201 caused chlorotic and necrotic lesions on the leaves of Ws-0 plants within 3 dpi, and by 6 dpi the highly affected leaves had withered (Fig. 2B). In contrast, Ws-0 plants exhibited resistance to 70-15, with no visible symptoms at 3 or 6 dpi. Both strains of M. oryzae sporulated on the infected leaves of the susceptible ecotypes, completing the disease cycle, but no sporulation was observed on the resistant ecotypes (Fig. 3C; Supplemental Fig. S2C). The fungus recovered from the infected leaves of susceptible plants produced the same disease symptoms in healthy plants, satisfying Koch's postulates.
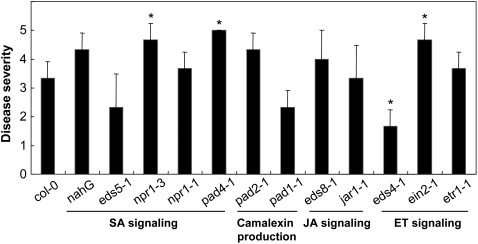
Figure 1.
Disease severity in Arabidopsis ecotypes infected with M. oryzae. Plants of 16 ecotypes were inoculated with M. oryzae strains KJ201 and 70-15. Ecotypes are as follows: Bensheim (Be-0), Bulhary (Blh-1), C24, Cape Verde Islands (Cvi-0), Columbia (Col-0), Estland (Est-0), Greenville (Gre-0), Hilversum (Hi-0), Kindalville (Kin-0), Lanark (Lan-0), Landsberg (La-0), Landsberg erecta (Ler-0), Mühlen (Mh-0), Niederzenz (Nd-0), Nossen (No-0), and Wassilewskija (Ws-0). Disease severity was measured at 6 dpi using a numerical scoring system as described in “Materials and Methods.” Black bars, KJ201; white bars, 70-15.
Figure 2.
Progression of disease symptoms caused by M. oryzae infection. The disease symptoms in two Arabidopsis ecotypes, Col-0 (A) and Ws-0 (B), infected with KJ201 and 70-15 were recorded at 0, 3, and 6 dpi.
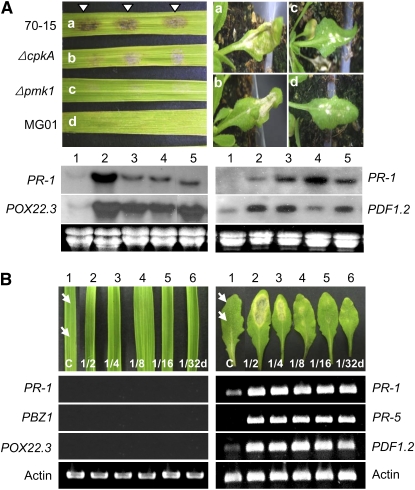
Figure 3.
Cellular responses of Arabidopsis to KJ201 infection. A, Three 5-μL drops of a fungal spore suspension (5 × 105 spores mL−1) were placed on the leaves of 28- to 30-d-old Col-0 plants. Control plants were inoculated with distilled water (Mock). Leaves at 3 dpi are shown (top row). The same leaves were stained with trypan blue and observed with a microscope (bottom row). B, H2O2 accumulation was detected in Col-0 leaves inoculated with KJ201 using DAB staining (left). The appressoria are denoted as A. Aniline blue fluorochrome staining of the leaves revealed callose accumulation (arrowheads) around the penetrated sites after 24 h (right). C, Asexual reproduction of M. oryzae on infected Col-0 leaves.
Cell Death Was Observed around the Sites of Infection
Cellular responses of Arabidopsis to infection by KJ201 were studied microscopically via histochemical staining. Inoculated leaves were cleared and stained with trypan blue to observe fungal materials (Fig. 3A). Fungal materials were found in the inoculated leaves of susceptible ecotypes at 3 dpi (Supplemental Fig. S3). No infectious hyphae were observed in resistant ecotypes when the infected leaves were stained and observed with a microscope (data not shown). The cells of susceptible ecotypes showed both increased membrane permeability (indicated by light blue staining) and collapse (dark staining and the absence of a recognizable cell shape; Koch and Slusarenko, 1990). Microscopic cell death around the sites of infection, observed via the use of Evans blue dye (which stained the dead cells blue within 30 min of treatment), was also observed at 3 dpi (Kato et al., 2007; data not shown).
Production of ROS, such as hydrogen peroxide (H2O2), and deposition of callose at the sites of fungal penetration are often associated with early host defense responses (Thordal-Christensen et al., 1997; Nishimura et al., 2003). We examined the production of H2O2 at the sites of infection by M. oryzae using diaminobenzidine (DAB; Fig. 3B). Col-0 plants infected by KJ201 accumulated a detectable level of H2O2 at 3 dpi, as indicated by reddish-brown staining. Microscopic observation revealed granulated epidermal and mesophyll cells with dark brown deposits located under or near the appressoria (Fig. 3B). Nd-0 and Ws-0 plants infected by KJ201 also produced H2O2 at infection sites. A much higher level of H2O2 accumulation was observed on Ws-0 plants (resistant to 70-15) infected by 70-15 (Supplemental Fig. S2A). A large amount of callose was deposited around the penetrated sites. This extensive callose response was observed in both the susceptible and resistant interactions (Fig. 3B; Supplemental Fig. S2B).
The Initial Developmental Process of M. oryzae in Arabidopsis Is Similar to That in Rice
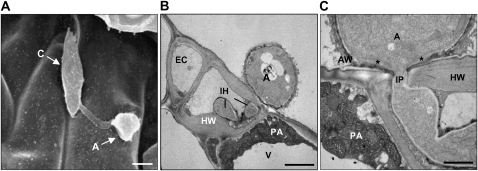
Electron microscopy was used to examine the ultrastructural features at the plant-fungus interface. Appressoria that looked similar to those formed on rice leaves were observed on the Arabidopsis leaf surface (Fig. 4A). By the time the infection peg (IP) began to emerge from the appressorium, a papillae-like structure had already formed around the infected host cells. This structure consisted of membranous fragments embedded within amorphous electron-dense materials (Fig. 4, B and C). Some of the epidermal cells invaded by the fungus appeared to be intact during the early stages of infection; however, the host cells around the fungal hyphae were thickened, and many others were collapsed. An IP emerged through a pore at the base of the appressorium and penetrated the Arabidopsis cells (Fig. 4C); however, there was no visible degradation of the host cuticle or epidermal cell wall around the IP. Host cell wall apposition material was deposited at the initial site of penetration, but this did not restrict the development of the fungus (Fig. 4C). The IP had a diameter of 0.33 μm and developed into primary infectious hyphae with a diameter of 1.5 to 3 μm within the epidermal cells (Fig. 4, B and C).
Figure 4.
Electron micrographs of Col-0 plants inoculated with M. oryzae strain KJ201. Plants inoculated with KJ201 (two 5-μL drops of a spore suspension at 5 × 105 spores mL−1 in distilled water) were observed using scanning and transmission electron microscopy at 2 dpi. A, Scanning electron micrograph showing germinated conidia (C) forming an appressorium (A) on the leaf surface. Bar = 10 μm. B, Transmission electron micrograph showing infectious hyphae (IH) penetrating an epidermal cell (EC). The host cell wall (HW), papilla-like structure (PA), and vacuole (V) are indicated. Bar = 3 μm. C, Site of initial penetration, showing part of an appressorium and IP. The outer layer of the appressorial wall (AW) is more electron dense than the inner layer; a third wall layer (marked with asterisks) forms a thickened ring around the penetration pore, continuous with the IP wall. Bar = 1 μm.
Induction of Pathogenesis/Defense-Related Genes in Response to M. oryzae Infection
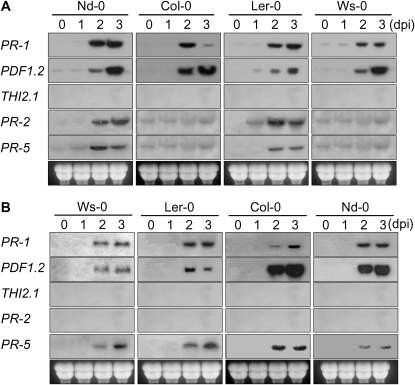
To test whether the expression patterns of pathogenesis/defense-related genes in Arabidopsis are similar to those during the interaction of rice with M. oryzae, we monitored the expression of the PR-1, PR-2, and PR-5 (Uknes et al., 1992), PDF1.2 (Penninckx et al., 1996), and THI2.1 (Vignutelli et al., 1998) genes. We selected four representative ecotypes with varying degrees of resistance to each strain (Nd-0 > Col-0 > Landsberg erecta [Ler-0] > Ws-0, KJ201 infection; Ws-0 > Ler-0 > Col-0 > Nd-0, 70-15 infection). Expression of PR-1 was noticeably increased at approximately 2 dpi in most ecotypes, regardless of their degree of resistance to the strain used. In Col-0, upon infection with KJ201, PR-1 expression was induced at 2 dpi but decreased at 3 dpi, whereas in the other ecotypes, its expression at 3 dpi was similar to or higher than that at 2 dpi. Upon infection with 70-15, in all four ecotypes, its expression was noticeable at 2 dpi and further increased at 3 dpi. PDF1.2 expression was induced in all KJ201-infected ecotypes at 2 dpi, similar to that of PR-1. However, unlike PR-1, its expression was further increased at 3 dpi in all ecotypes. In response to infection by 70-15, higher levels of PDF1.2 expression were observed in more susceptible ecotypes (Col-0 and Nd-0). Both PDF1.2 and THI2.1 are established marker genes for the JA/ET signaling pathway. However, in contrast to PDF1.2, THI1.2 transcripts were not detected in any of the ecotypes with both strains. Expression of PR-2 and PR-5 was induced only in Nd-0 and Ler-0 when infected with KJ201, but PR-2 expression was not induced in any ecotype upon infection with 70-15 (Fig. 5).
Figure 5.
Expression of PR-1, PR-2, PR-5, PDF1.2, and THI2.1 in response to M. oryzae infection. Plants of four ecotypes were inoculated with KJ201 (A) and 70-15 (B) by spraying the plants with a spore suspension (5 × 105 spores mL−1) and harvested at 0, 1, 2, and 3 dpi for northern hybridization. These plants are ordered from left to right according to their increasing disease severity upon inoculation with the two fungal isolates.
To investigate the roles of the SA-, JA-, and ET-dependent signaling pathways in defense against M. oryzae, we inoculated several Arabidopsis mutants defective in these signaling pathways with KJ201 (Fig. 6). The eds5-1, nahG, and npr1-1 mutants, which are defective in SA production, appeared to be comparable to wild-type Col-0 plants in symptom expression, but the responses of npr1-3 and pad4-1, which had been also implicated in SA signaling (Cao et al., 1997; Jirage et al., 1999), recorded higher degrees of susceptibility than Col-0 plants. In contrast to pad4-1, two other phytoalexin-deficient mutants (Glazebrook and Ausubel, 1994), pad2-1 and pad1-1, were comparable to Col-0 plants for KJ201 infection. The mutants eds8-1 and jar1-1, which are insensitive to JA (Staswick et al., 2002), were also comparable to wild-type Col-0 plants. The mutants ein2-1 and etr1-1, which exhibit defective ET signaling (Ton et al., 2002), were more susceptible and comparable to wild-type Col-0 plants, respectively. The eds4-1 mutant (Ton et al., 2002) appeared more resistant than wild-type Col-0. All data were analyzed by t test at P = 0.01.
Figure 6.
Infection of Arabidopsis mutants defective in defense-related signaling pathways with M. oryzae strain KJ201. Disease severity was recorded at 6 dpi. Observations are based on a representative of three independent experiments using 10 plants per experiment. Asterisks denote statistically significant differences between the wild type and mutant by t test (P < 0.01).
Infection by M. oryzae Increases Endogenous SA Levels
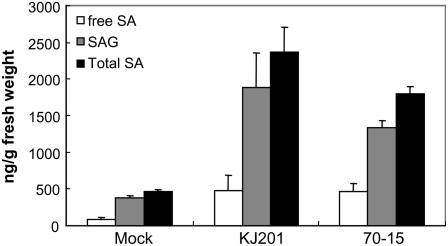
SA is a master regulatory molecule necessary for the activation of certain defense responses (Thomma et al., 2001). We examined whether the amount of endogenous SA had been altered in response to infection by M. oryzae. Intracellular SA is found predominantly as free SA and its sugar conjugate SA β-glucoside (SAG). We measured the concentrations of SA and SAG in Col-0 plants infected with KJ201 and 70-15 (Fig. 7). In the KJ201-infected plants, the amount of free SA was six times higher than that observed in the mock-treated plants; the increase in the 70-15-infected plants (five times) was comparable. The level of SAG was comparably increased upon infection with either strain.
Figure 7.
Accumulation of SA in response to fungal infection. Leaves of Col-0 plants inoculated with strains KJ201 and 70-15 were harvested at 2 dpi. Each bar represents the mean of three replicate samples. The error bar represents the sd. Free SA and SAG were assayed for each sample.
Pathogenicity Factors Essential for the Infection of Rice by M. oryzae Play Limited Roles in the Infection of Arabidopsis
To determine whether M. oryzae uses the same infection mechanism in both rice and Arabidopsis, we evaluated the pathogenicity of three mutants that are nonpathogenic in rice using Nd-0 plants. The CPKA gene, which encodes the catalytic subunit of protein kinase A, is involved in appressorium maturation (Mitchell and Dean, 1995; Xu and Hamer, 1996). Although ΔcpkA mutants form melanized appressoria, their appressoria are smaller than those of the wild type and are unable to support penetration (Xu et al., 1997). Mutants defective in PMK1, a mitogen-activated protein kinase gene, germinate and form a hook cell on hydrophobic surfaces but fail to swell and form a nascent appressorium (Xu and Hamer, 1996). The mutant MG01 is severely impaired in appressorium differentiation (Chun and Lee, 1999). Consequently, these three mutants (ΔcpkA, Δpmk1, and MG01) are unable to infect and colonize rice. However, all of these mutants were able to infect Arabidopsis leaves, producing necrotic lesions on Nd-0 leaves at 6 dpi (Fig. 8A; Supplemental Fig. S6). However, the size of the lesions and the disease severity caused by the mutants were not as severe as those caused by 70-15. The inoculation of rice (cv Nakdongbyeo) and Arabidopsis (ecotype Nd-0) with 70-15 or the mutants (ΔcpkA, Δpmk1, and MG01) induced the expression of PR-1 and POX22.3 in rice and PR-1 and PDF1.2 in Arabidopsis at 2 dpi (Fig. 8A). In rice, the expression of both genes was highest in the plants inoculated with wild type 70-15, whereas in Arabidopsis, the pattern of gene expression was more complex and did not appear to be correlated with the observed disease severity.
Figure 8.
Comparison of responses in rice and Arabidopsis to three M. oryzae mutants defective in infecting rice and M. oryzae fungal culture filtrates. A, Disease symptoms caused by 70-15 (a), ΔcpkA (b), Δpmk1 (c), and MG01 (d) at 48 h. Rice plants (cv Nakdongbyeo) were inoculated with three drops of a spore suspension (105 conidia mL−1), whereas Arabidopsis plants (ecotype Nd-0) were sprayed with the suspension. Levels of two genes in the infected leaves of each host (PR-1 and POX22.3 for rice and PR-1 and PDF1.2 for Arabidopsis) collected at 2 dpi were measured via northern hybridization: mock (lane 1), 70-15 (lane 2), ΔcpkA (lane 3), Δpmk1 (lane 4), and MG01 (lane 5). B, Comparison of the responses to a CF of KJ201 between rice and Arabidopsis. Two drops (10 μL each) of a concentrated CF of KJ201 and several dilutions (1/2, 1/4, 1/8, 1/16, and 1/32) were dropped onto rice (cv Nakdongbyeo) and Col-0 leaves (indicated by arrows). As a control, leaves were treated with a mixture of distilled water and acetone (50:50, mock; lane 1). The leaves were then monitored for 3 d. At 3 dpi, RNA was extracted from individual leaves to check the level of expression of three genes by reverse transcription-PCR: PR-1, PBZ1, and POX22.3 for rice and PR-1, PR-5, and PDF1.2 for Arabidopsis.
Comparison of the Response of Rice and Arabidopsis to a Fungal Culture Filtrate
The ability to produce melanized functional appressoria was not an absolute requirement for the infection of Arabidopsis (Fig. 8A). This suggests that M. oryzae may use a mechanism of infection distinct from that required to infect rice. Pathogen-derived metabolites often induce lesion formation in plant leaves (Jackson and Taylor, 1996; Stone et al., 2000). A culture filtrate (CF) of M. oryzae KJ201 caused lesion formation in Arabidopsis leaves within 48 h of treatment, and the severity of the lesions was correlated with the filtrate concentration (Fig. 8B), but extracts from the culture medium itself (potato dextrose broth) did not cause phytotoxicity (Supplemental Fig. S4). The CF treatment and fungal infection produced similar patterns of symptom development in 13 ecotypes (Supplemental Figs. S7 and S8). However, the severity of symptoms caused by CF treatment did not always correlate with the degree of disease severity in certain ecotypes: although ecotypes Bulhary and Bensheim were quite susceptible to infection, the lesion formation by CF treatment in these ecotypes was limited. Col-0 plants produced visible necrotic lesions at 1 dpi. These lesions expanded over time in some experiments, but in most cases they did not expand beyond the inoculated areas. In contrast, CF placed on the leaves of rice cv Nakdongbyeo failed to produce any visible symptoms at 1 dpi; however, faint HR-like lesions were visible around the sites of inoculation at 3 dpi. Consistent with the difference in symptom expression between rice and Arabidopsis, application of CF induced the expression of three PR genes in Arabidopsis within 48 h of treatment but failed to do so in rice at the same concentration. Expression of the marker genes was induced even in plants without visible symptoms in Arabidopsis but not in rice (Fig. 8B).
Purification of Phytotoxic Compounds from M. oryzae
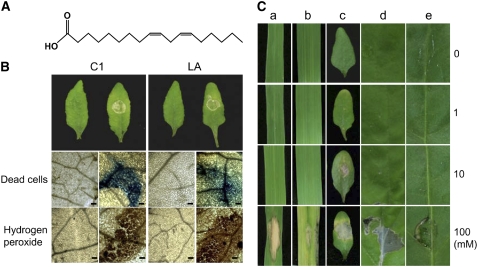
To identify the nature of the phytotoxic compounds produced by M. oryzae, a large-scale extraction and purification experiment was conducted. Five kilograms of rice grains colonized by KJ201 was extracted in ethylacetate (EtOAc). This extract exhibited phytotoxicity, but the EtOAc extract of uncolonized rice grains did not cause phytotoxicity (data not shown). The extract from the colonized rice grains (30 g dry weight) was fractionated through a silica gel column. The effectiveness of each step in increasing the biological activity of the extract was assessed through a toxicity assay on Arabidopsis leaves (Supplemental Fig. S5). We purified three compounds that exhibited high phytotoxic activity. Compound 1 (C1) was a pale yellow oil, C2 was a white solid, and C3 was colorless. Col-0 leaves treated with each of these compounds exhibited necrotic lesions within 24 h.
To determine the chemical structures of the compounds, we used a combination of NMR spectroscopy and mass spectroscopy (MS). Based on 1H-NMR, 13C-NMR, and electrospray ionization-MS data, C1 was identified as the fatty acid 9,12-octadecadienoic acid (Fig. 9A); however, we were unable to identify the other two compounds due to insufficient quantities for subsequent chemical analyses. Commercially prepared 9,12-octadecadienoic acid (L1012; Sigma) caused similar cellular changes (Fig. 9B). Leaves treated with both preparations were examined for cell death using Evans blue staining and H2O2 production using DAB staining. Small, scattered spots corresponding to dead cells were observed at the treated sites. DAB staining for H2O2 was absent in the mock-inoculated leaves but was apparent in the 9,12-octadecadienoic acid-treated leaves (Fig. 9B). The phytotoxicity of various concentrations of 9,12-octadecadienoic acid to assorted plant species was also determined (Fig. 9C). The minimal effective dose for producing necrotic lesions was lowest in Arabidopsis, in which 1 mm 9,12-octadecadienoic acid caused lesion formation at 3 dpi. At this concentration, no other plant species expressed visible symptoms. The lesions in Arabidopsis expanded on the leaves treated with 10 mm 9,12-octadecadienoic acid, whereas the other plant species still did not show visible symptoms.
Figure 9.
Phytotoxicity of 9,12-octadecadienoic acid in multiple plant species. A, Phytotoxic compound 1 (C1) was identified as 9,12-octadecadienoic acid. B, Cellular responses of ecotype Col-0 leaves treated with C1 (left) and commercially prepared 9,12-octadecadienoic acid (LA; right). A single 10-μL drop of 10 mm C1 or LA was placed on the leaves of 28- to 30-d-old plants. The control plants received a mixture of distilled water and acetone (70:30, mock). Leaves at 2 dpi are shown. The leaves were stained with Evans blue (for dead cells) and DAB (for H2O2 accumulation). C, Phytotoxic activity of three concentrations (1, 10, and 100 mm) of 9,12-octadecadienoic acid on leaves of rice (a), barley (b), Arabidopsis (c), tobacco (Nicotiana tabacum; d), and pepper (Capsicum annuum; e).
DISCUSSION
Establishment of an Arabidopsis-M. oryzae Pathosystem
To date, M. oryzae, which is well known as the causal agent of rice blast disease, has primarily been considered a pathogen of monocotyledonous plant species. In this study, we clearly demonstrate that two rice pathogenic strains of M. oryzae were able to cause disease in Arabidopsis by satisfying Koch's postulates. Although a number of pathosystems have been developed using Arabidopsis as the host, the Arabidopsis-M. oryzae pathosystem is unique in that a monocot fungal pathogen successfully colonizes and completes its disease cycle. In the case of Arabidopsis and Blumeria graminis f. sp. hordei, the barley (Hordeum vulgare) powdery mildew fungus never penetrated more than one epidermal cell, and no asexual reproduction was observed (Zimmerli et al., 2004). The M. oryzae-Arabidopsis pathosystem established here will allow comparative analyses of plant responses in monocots and dicots using the same pathogen isolates. In addition, because the two strains of M. oryzae (KJ201 and 70-15) interacted differentially with 16 ecotypes of Arabidopsis, our system will serve as a model for studying and comparing the molecular and cellular bases of compatibility within each of the two hosts. Incompatibility between rice-M. oryzae is controlled by gene-for-gene interactions (Jia et al., 2000).
Using various combinations of host and pathogen materials, we investigated the molecular and cellular bases of pathogenicity and defense. In rice-M. oryzae interactions, host penetration is mediated by a specialized cell called the appressorium. Once inside the rice cell, the fungus forms bulbous infectious hyphae that fill the infected cell and then rapidly spread to adjacent cells (Kankanala et al., 2007). The mode of penetration and initial ramification by M. oryzae in Arabidopsis appeared similar to those found in rice. The formation of wall appositions and papillae-like structures is a common response of plant cells to fungal infection (Aist and Bushnell, 1991). Papillae-like structures were produced at the site of penetration during compatible interactions in Arabidopsis (e.g. Col-0 plants with KJ201) and were localized inside the cell wall (Fig. 4, B and C). Enhanced papillae formation is correlated with the resistance of barley cultivars to B. graminis f. sp. hordei and was proposed to be the primary mechanism of resistance (Huckelhoven et al., 1999). Similar patterns have been observed in interactions between Arabidopsis and adapted and nonadapted Colletotrichum species. The development of papillae during infection by adapted C. higginsianum was reduced compared with infection by nonadapted C. lagenarium (Shimada et al., 2006). In a compatible interaction between Arabidopsis and C. distructivum isolated from Brassica campestris, similar changes were observed (O'Connell et al., 2004). These observations suggest that papillae formation may play an important role in the general defense responses of Arabidopsis against fungal infection. However, the role of papillae-like structures in defense against M. oryzae infection remains to be determined, because these structures were observed in susceptible combinations.
The rapid accumulation of ROS and hypersensitive cell death are correlated with disease resistance in many pathosystems (Rate and Greenberg, 2001). Accumulation of ROS can trigger localized hypersensitive cell death, which limits pathogen proliferation (Fellbrich et al., 2002). In rice, ROS production, initiated by the OsRac1-GTPase complex, is also required for a general role in disease resistance against M. oryzae. The appearance of localized DAB staining beneath appressoria was an early response to attack, occurring at around 6 h of attempted penetration of the cell wall by the fungal penetration peg (Ono et al., 2001; Wong et al., 2007). M. oryzae infection also induced the accumulation of ROS and phenolic compounds in Arabidopsis. Necrotic flecks reminiscent of microscopic cell death developed, and ROS accumulated at the sites of inoculation in the resistant ecotypes (e.g. Ws-0; Supplemental Fig. S2). The production of ROS was also detected in susceptible ecotypes (e.g. Col-0), but the extent of their production was less than that in resistant ecotypes (Supplemental Fig. S2), suggesting that the accumulation of ROS is correlated with resistance. However, because treatment of 9,12-octadecadienoic acid also caused the accumulation of ROS (Fig. 9), further studies are needed to determine whether ROS plays key roles in resistance or not.
Both SA- and JA/ET-Dependent Signaling Pathways Are Activated during Arabidopsis-M. oryzae Interactions
In Arabidopsis, the expression pattern of certain marker genes has been used as an indicator of the activation of defensive signaling pathways mediated by plant hormones such as SA, ET, and JA, and the speed and magnitude of their expression are often correlated with resistance. Typically, genes under the control of the SA-dependent signaling pathway, such as PR-1, PR-2, and PR-5, are induced in response to biotrophic pathogens (McDowell et al., 2005; Flors et al., 2008). In contrast, the JA/ET-dependent signaling pathway, for which PDF1.2 is a marker gene, is typically activated in response to necrotrophic pathogens (Glazebrook, 2001).
M. oryzae is a hemibiotroph because the fungus initially grows intracellularly without causing host cell death but promotes necrosis later. Both signaling pathways, therefore, could be involved in regulating resistance responses against M. oryzae. The expression of PR-1 and PDF1.2 was induced in both the resistant and susceptible ecotypes of Arabidopsis in response to M. oryzae infection (Fig. 5). Notably, their expression patterns were similar regardless of the degree of resistance with some ecotype-specific patterns, suggesting that the activation of these genes is not causally related to resistance but is instead part of the general defense response to cellular damage. Similar patterns of expression were also observed for PR-1 and PDF1.2 in Arabidopsis in response to infection by Leptosphaeria maculans (Bohman et al., 2004). However, the rapid induction of PR genes during 24 h has been observed in incompatible rice-M. oryzae interactions, whereas in compatible interactions, induction of PR expression was significantly slower and weaker. In rice, a strong induction of PR gene expression during an incompatible interaction precedes the first visible signs of HR, and late induction during a compatible interaction coincides with the formation of water-soaked lesions (Ahn et al., 2005).
When Col-0 plants were inoculated with KJ201 and 70-15, the level of SA was increased more than five times compared with the mock-treated plants. Infection by Botrytis cinerea, a typical necrotrophic fungal pathogen, also increased the SA level about three times and induced PR-1 gene expression in Arabidopsis (Veronese et al., 2006). To further assess the contribution of the SA and JA/ET signaling pathways to resistance against M. oryzae, several Arabidopsis mutants defective in these pathways were infected with M. oryzae (Fig. 6). Most of the mutants were more susceptible than wild-type Col-0, with the exception of eds4-1. It appears that a complex network of signaling events is involved in controlling resistance against hemibiotrophic pathogens, which undergo both biotrophic and necrotrophic phases during proliferation in planta. Discrepancies between the expression patterns of marker genes and defense responses of signaling mutants were observed in Arabidopsis-C. higginsianum and Arabidopsis-L. maculans interactions (Bohman et al., 2004; Narusaka et al., 2004).
M. oryzae Uses Multiple Factors to Colonize Arabidopsis, Some of Which Appear Specific to Arabidopsis
Pathogens have evolved various strategies to overcome the barriers that they encounter during the infection of potential hosts (Mendgen et al., 1996). On rice leaves, M. oryzae goes through a series of well-defined developmental steps during infection, including the formation of a specialized infectious structure (appressorium). Our data show that M. oryzae undergoes a developmental sequence in Arabidopsis similar to that in rice. Conidia of strain 70-15 germinated and formed melanized appressoria on the surface of Col-0 leaves, and penetration pegs entered the epidermal cells directly. Interestingly, nonpathogenic mutants of 70-15 defective in appressorium formation/function exhibited reduced virulence compared with 70-15 but still managed to cause disease symptoms (Fig. 8; Supplemental Fig. S6), suggesting that appressoria may enhance disease severity but are not essential for the infection of Arabidopsis.
In response to treatment with a crude CF of M. oryzae, lesion formation and induction of PR gene expression were observed in Arabidopsis but not in rice, suggesting that M. oryzae employs virulence factors (i.e. phytotoxins) unique to infecting Arabidopsis. Recent studies have reported that Fusarium phytotoxins have elicitor-like activity in Arabidopsis, causing the induction of defensive genes, the accumulation of SA and ROS, and lesion formation (Nishiuchi et al., 2006). The involvement of such toxins may explain why M. oryzae mutants defective in appressorium formation/function managed to cause disease symptoms in Arabidopsis. However, the production of phytotoxins does not appear to be the sole factor determining virulence in Arabidopsis, and other virulence factors are probably involved in successfully colonizing Arabidopsis. The severity of symptoms caused by CF treatment did not always correlate with the degree of disease severity in certain ecotypes (Supplemental Figs. S7 and S8). This observation suggests that several mechanisms operate during pathogenesis in Arabidopsis.
The CF of M. oryzae strain KJ201 was sufficient to induce cell death in Arabidopsis within 48 h of application, but rice cells were unaffected at the same concentration, suggesting the existence of previously unknown compounds highly toxic to Arabidopsis cells. One compound responsible for cell death in Arabidopsis was identified as 9,12-octadecadienoic acid; notably, it caused cell death even when it was applied topically without wounding or infiltration. The cell death elicited by 9,12-octadecadienoic acid led to lesions similar to those caused by M. oryzae. These data suggest that 9,12-octadecadienoic acid may play a role in pathogenesis in Arabidopsis. ROS accumulation, which was observed when Arabidopsis plants were infected with M. oryzae, was also induced by 9,12-octadecadienoic acid treatment. Commercially prepared 9,12-octadecadienoic acid also induced lesions in Arabidopsis. Finally, the phytotoxic activity of 9,12-octadecadienoic acid was significantly higher in Arabidopsis than in other plant species (Fig. 9). These results suggest that during Arabidopsis-M. oryzae interactions, the fungus may mostly behave as a necrotroph, obtaining nutrients from dead cells through the secretion of this and other compounds, which is distinct from the mechanisms of infection observed in rice.
Certain lipids regulate a wide range of important cellular processes in plants, including the regulation of ROS production (Sang et al., 2001) and the activation of defense gene expression (Farmer and Ryan, 1992). A previous report indicated that necrotrophic pathogens use oxidative bursts to invade and destroy plant tissues (Deighton et al., 1999). The 9,12-octadecadienoic acid may be a precursor of biologically active oxylipins in plants with JA as a terminal signal (Blechert et al., 1995). The production of excess 9,12-octadecadienoic acid by M. oryzae may disrupt cellular homeostasis, leading to cell death.
In summary, we established a novel pathosystem based on M. oryzae and Arabidopsis and found that rice pathogenic M. oryzae strains KJ201 and 70-15 were able to infect the dicot Arabidopsis. The pathogen appears to require both conserved and host-specific virulence factors during the infection of rice and Arabidopsis. We also produced evidence suggesting that fungal metabolites are important determinants of the pathogenicity of M. oryzae in Arabidopsis. This novel pathosystem provides a valuable new model for studying the function and evolution of fungal pathogenicity factors as well as defense mechanisms.
MATERIALS AND METHODS
Plant and Fungal Materials
The 16 ecotypes of Arabidopsis (Arabidopsis thaliana) used in this study were obtained from the Arabidopsis Biological Resource Center at Ohio State University: Bensheim, Bulhary, C24, Cape Verde Islands, Col-0, Estland, Greenville, Hilversum, Kindalville, Lanark, Landsberg, Ler-0, Mühlen, Nd-0, Nossen, and Ws-0. In addition to these ecotypes, we used the following mutants derived from Col-0: eds4-1, eds5-1, eds8-1, ein2-1, npr1-1, npr1-3, pad1-1, pad2-1, and pad4-1. The plants were grown in a mixture of commercial potting soil and perlite (3:1) or on Murashige and Skoog agar medium (Murashige and Skoog, 1962) in a growth chamber with a 16-h-day/8-h-night photoperiod at 22°C and 80% relative humidity. Rice plants (Oryza sativa ‘Nakdongbyeo’) were grown in test tubes in a growth chamber with a 16-h-day/8-h-night photoperiod at 25°C.
M. oryzae strains KJ201 and 70-15 (Leung et al., 1988; Chao and Ellingboe, 1991) were grown on oatmeal agar medium (OMA; 50 g oatmeal L−1) at 25°C under constant fluorescent light to promote conidiation. We used three M. oryzae mutants defective in pathogenicity: ΔcpkA (Mitchell and Dean, 1995), Δpmk1 (Xu and Hamer, 1996), and MG01 (Chun and Lee, 1999).
Plant Inoculation and Disease Rating
Inoculation with M. oryzae was performed using 4-week-old Arabidopsis plants. After harvesting the conidia from fungal cultures on OMA using sterile water, the concentration was adjusted to 5 × 105 conidia mL−1. Ten plants were sprayed with 20 mL of the conidial suspension using an air brush. After incubating in a dew chamber for 16 h at 25°C under 100% relative humidity, the inoculated plants were transferred to a growth chamber set at 22°C and 80% relative humidity. Each inoculation experiment was repeated three times.
To quantify the disease symptoms, a numerical scoring system based on the DS was used. The DS at 6 dpi was rated on a scale of 0 to 5, with 0 indicating no necrotic or chlorotic flecks on the leaves (the controls continuously exhibited a score of 0). The numerical scale reflects the percentage of the leaf area exhibiting necrosis/chlorosis: 1, 1%–20%; 2, 21%–40%; 3, 41%–60%; 4, 61%–80%; and 5, 81%–100%.
Light Microscopy
Three drops of a conidial suspension (5 μL per drop; 5 × 105 conidia mL−1 in water) were placed on each leaf of 4-week-old plants. The inoculated plants were then kept at 25°C for 16 h in a moist chamber. Microscopic lesions and fungal hyphae were visualized by staining the infected leaves as described previously (Vogel and Somerville, 2000). The leaves were cleared in alcoholic lactophenol, consisting of 1 volume of lactophenol (phenol:glycerol:lactic acid:water, 1:1:1:1, v/v) and 2 volumes of ethanol and rinsed with lactophenol. The treated leaves were stained with 250 μg mL−1 trypan blue in lactophenol. The tubes containing the leaf samples were then placed in boiling water for 2 min, followed by cooling for 1 h. The leaves were then destained in lactophenol for 1 h. Finally, the samples were mounted in 50% glycerol and examined under bright-field illumination.
Cell death, indicated by a loss of plasma membrane integrity, was detected by staining the infected cells with Evans blue solution (0.25% [w/v] in 0.1 mm CaCl2, pH 5.6; Sigma) for 30 min. Evans blue was infiltrated into the leaves as a 0.25% aqueous solution 72 h after pathogen inoculation (Baker and Mock, 1994; Ahn et al., 2007). After at least 30 min of staining, the leaves were rinsed with water and observed with a light microscope. Leaf areas damaged during physical handling were not included in the evaluation of the proportion of dead cells.
For the detection of H2O2, endogenous peroxidase-dependent in situ histochemical staining using DAB was performed according to the method of Rusterucci et al. (2001). Leaves inoculated with an M. oryzae conidial suspension were excised after 72 h and kept in a solution of 1 mg mL−1 DAB for 8 h in the dark. After replacing the DAB solution with water, the leaves were incubated under the same conditions for an additional 8 h. The leaves were then cleared in a mixture of ethanol and acetic acid (96:4, v/v), mounted on a glass slide in 50% glycerol, and examined with a light microscope. Callose formation was detected using aniline blue (Stone et al., 2000). The stained leaves were observed with an Axioplan Universal microscope (Carl Zeiss) with a fluorescein filter set with excitation at 395 nm and emission at 495 nm.
Electron Microscopy
For scanning electron microscopy, infected leaves at 48 h after infection were fixed in 4% paraformaldehyde and then washed in phosphate buffer, followed by a series of ethanol washes (30%, 50%, 70%, 96%, and 100%). After drying the fixed leaves in a Samdri-PVT-3B critical point drying apparatus (Tousimis), they were mounted on stubs and covered with 20–25 μm of gold-palladium in a Hummer II Sputter Coater (Anatech). The coated samples were observed with an AMRAY 1000 scanning electron microscope (AMRAY). For transmission electron microscopy, 2- to 3-mm pieces of infected leaves were fixed in 0.05 m sodium phosphate buffer (pH 7.5) containing 2.5% (v/v) glutaraldehyde for 18 h at 4°C, including 5 min of vacuum infiltration, and incubated with 2% (w/v) osmium tetroxide in the same buffer for 2 h at 20°C. After embedding in Spurr's epoxy resin, ultrathin sections were cut using an ultramicrotome (MT-X; RMC) and collected on carbon-coated grids. The sections were stained with 2% uranyl acetate for 3 min and with Reynold's lead solution (Reynolds, 1963) for 3 min and then observed using a JEM-1010 electron microscope (JEOL) operating at 70 kV.
Northern Hybridization
Leaves were harvested for RNA isolation at 0, 1, 2, and 3 dpi. The samples were preserved at –70°C until RNA extraction. Total RNA was extracted using the lithium chloride precipitation method. For northern hybridization, 15 μg of total RNA per lane was separated electrophoretically on a denaturing formaldehyde-agarose gel (8% formaldehyde, 0.5× MOPS, and 1.5% agarose) and then blotted onto a Hybond-N+ membrane (Amersham Pharmacia Biotech; Sambrook et al., 1989). Uniform sample loading was confirmed by staining for rRNA with ethidium bromide. The blots were hybridized and washed as described previously (Kim et al., 2001) and exposed to x-ray film (AGFA). The probes were labeled with [α-32P]dCTP by random primer labeling (Boehringer Mannheim). The probes included the genes that encode PR-1, PR-2, PR-5, PDF1.2, and THI1.2.
Preparation of the Fungal CF
Fungal conidia collected from a 7-d-old OMA culture were inoculated into 300 mL of potato dextrose broth (Difco) in 500-mL conical flasks and cultured on an orbital shaker at 125 rpm for 7 d at 25°C in the dark. The liquid culture was then filtered through sterilized Whatman No. 2 filter paper to remove mycelia and subsequently through a 0.22-μm Millipore filter to eliminate the conidia. After vacuum drying, the culture filtrate was dissolved in 5 mL of acetone and dropped onto rice and Arabidopsis leaves. The filtrate of fresh potato dextrose broth was also applied as a negative control. The plants were then monitored for 3 d.
Purification and Identification of the Phytotoxins Produced by KJ201
To determine the chemical structure and biological activity of the phytotoxic metabolites produced by M. oryzae, a large-scale culture of strain KJ201 on grains of cv Annam rice was set up in 20 flasks, each containing 250 g of rice and 200 mL of distilled water. The flasks, plugged loosely with cotton, were allowed to sit overnight (12 h) and were then autoclaved for 1 h. Each flask was then inoculated with 5 mL of a conidial suspension in water (109 spores mL−1) and incubated at 25°C for 3 weeks in the dark, after which the cultures were harvested and dried in a forced-air hood. The dried cultures (approximately 5 kg total) were then ground into a fine flour-like powder using a laboratory mill and extracted three times with 10 L of EtOAc. The extracts were then combined, filtered through Whatman No. 4 filter paper, and dried using a rotary evaporator.
The dried extract (approximately 30 g) was resuspended in CHCl3 and separated through a silica gel column (60 × 750 mm; 3.0 cm [i.d.] × 43 cm, Kiesel gel 60, 100 g, 230–400 mesh) using CHCl3:n-Hex (1:1, v/v) as the elutant. The fractions exhibiting phytotoxic activity were pooled and applied to a reverse-phase column (30 × 300 mm) of C18 silica (Sep-Pak; Waters) and eluted with 70% methanol. The structure of a purified compound (approximately 350 mg) was identified using a combination of 600 MHz NMR spectroscopy (Bruker) and electron ionization MS (JEOL).
Quantification of SA and SAG in the Leaf Tissues
Total SA (free SA + SAG) was extracted and quantified as described previously (Enyedi et al., 1992). Leaf tissue samples (0.5 g fresh weight) were frozen in liquid nitrogen, ground to a fine powder, and extracted sequentially with 90% and 100% methanol. After vacuum drying of the pooled methanol extracts, the residue was resuspended in 5 mm sodium acetate buffer (pH 5.5) containing 80 units of β-glucosidase (Sigma) per gram of plant tissue (fresh weight). Following enzymatic hydrolysis (90 min at 37°C), the reaction was stopped by the addition of 10% TCA. The solution was then partitioned using EtOAc:cyclopentane:isopropanol (100:99:1, v/v/v). SA was quantified by measuring the fluorescence (excitation, 301 nm; emission, 412 nm) after separation through a C18 reverse-phase HPLC column (Waters). The HPLC column was maintained at 40°C and equilibrated with 0.5% glacial acetic acid:methanol (75:25, v/v) at a flow rate of 1.5 mL min−1. Three minutes after injection, a methanol gradient (25%–60%) was applied over 7 min, after which the methanol concentration was returned to 25%. All data were corrected based on the recovery rate of spiked samples.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Responses of six ecotypes to 70-15 at 6 dpi.
Supplemental Figure S2. Cellular responses of Arabidopsis to infection by KJ201 and 70-15 at 3 dpi.
Supplemental Figure S3. Microscopic observation of fungal structures in planta using trypan blue staining.
Supplemental Figure S4. Treatment of plants with a control extract from PD broth.
Supplemental Figure S5. Schematic diagram summarizing the steps involved in purifying the phytotoxic compounds.
Supplemental Figure S6. Cellular responses of Arabidopsis to Δpmk1 infection.
Supplemental Figure S7. Disease severity and lesion size caused by M. oryzae and a crude culture filtrate, respectively.
Supplemental Figure S8. Cellular responses of Arabidopsis to a crude culture filtrate of M. oryzae.
Supplementary Material
Acknowledgments
We are grateful to Soonok Kim and Sook-Young Park for their valuable comments and suggestions on the manuscript. We also thank members of our laboratories for advice or contributions to improving this paper.
This work was supported by the Crop Functional Genomics Center's 21st Century Frontier Research Program funded by the Ministry of Science and Technology (grant no. CG1141), by the Biogreen21 Project funded by the Rural Development Administration (grant no. 20080401–034–044–008–01–00 to Y.-H.L.), and by the U.S. Department of Agriculture-National Research Initiative (grant no. 2002–02367 to S.K.). J.-Y.P. was supported by a graduate fellowship from the Ministry of Education through the Brain Korea 21 Program.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Yong-Hwan Lee (yonglee@snu.ac.kr).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Ahn IP, Kim S, Lee YH, Suh SC (2007) Vitamin B1-induced priming is dependent on hydrogen peroxide and the NPR1 gene in Arabidopsis. Plant Physiol 143 838–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn IP, Kim SO, Kang SC, Suh SC, Lee YH (2005) Rice defense mechanisms against Cochliobolus miyabeanus and Magnaporthe grisea are distinct. Phytopathology 95 1248–1255 [DOI] [PubMed] [Google Scholar]
- Aist JR, Bushnell WR (1991) Invasion of plants by powdery mildew fungi, and cellular mechanisms of resistance. In GT Cole, HC Hoch, eds, The Fungal Spore and Disease Initiation in Plants and Animals. Plenum Press, New York, pp 321–345
- Baker CJ, Mock NM (1994) An improved method for monitoring cell death in cell suspension and leaf disc assays using Evans blue. Plant Cell Tissue Organ Cult 39 7–12 [Google Scholar]
- Blechert S, Brodschelm W, Holder S, Kammerer L, Kutchan TM, Mueller MJ, Xia ZQ, Zenk MH (1995) The octadecanoic pathway: signal molecules for the regulation of secondary pathways. Proc Natl Acad Sci USA 92 4099–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohman S, Staal J, Thomma BP, Wang M, Dixelius C (2004) Characterisation of an Arabidopsis-Leptosphaeria maculans pathosystem: resistance partially requires camalexin biosynthesis and is independent of salicylic acid, ethylene and jasmonic acid signalling. Plant J 37 9–20 [DOI] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88 57–63 [DOI] [PubMed] [Google Scholar]
- Chang JH, Goel AK, Grant SR, Dangl JL (2004) Wake of the flood: ascribing functions to the wave of type III effector proteins of phytopathogenic bacteria. Curr Opin Microbiol 7 11–18 [DOI] [PubMed] [Google Scholar]
- Chao CT, Ellingboe AH (1991) Selection for mating competence in Magnaporthe grisea pathogenic to rice. Can J Bot 69 2130–2134 [Google Scholar]
- Chun SJ, Lee YH (1999) Genetic analysis of a mutation on appressorium formation in Magnaporthe grisea. FEMS Microbiol Lett 173 133–137 [Google Scholar]
- Clarke JD, Volko SM, Ledford H, Ausubel FM, Dong X (2000) Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell 12 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, Orbach MJ, Thon M, Kulkarni R, Xu JR, Pan H, et al (2005) The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434 980–986 [DOI] [PubMed] [Google Scholar]
- Deighton N, Muckenschnabel II, Goodman BA, Williamson B (1999) Lipid peroxidation and the oxidative burst associated with infection of Capsicum annuum by Botrytis cinerea. Plant J 20 485–492 [DOI] [PubMed] [Google Scholar]
- De Jong JC, McCormack BJ, Smirnoff N, Talbot NJ (1997) Glycerol generates turgor in rice blast. Nature 389 244–245 [Google Scholar]
- Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, et al (1994) A central role of salicylic acid in plant disease resistance. Science 266 1247–1250 [DOI] [PubMed] [Google Scholar]
- Enyedi AJ, Yalpani N, Silverman P, Raskin I (1992) Localization, conjugation, and function of salicylic acid in tobacco during the hypersensitive reaction to tobacco mosaic virus. Proc Natl Acad Sci USA 89 2480–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer EE, Ryan CA (1992) Octadecanoid precursors of jasmonic acid activate the synthesis of wound-inducible proteinase inhibitors. Plant Cell 4 129–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellbrich G, Romanski A, Varet A, Blume B, Brunner F, Engelhardt S, Felix G, Kemmerling B, Krzymowska M, Nurnberger T (2002) NPP1, a Phytophthora-associated trigger of plant defense in parsley and Arabidopsis. Plant J 32 375–390 [DOI] [PubMed] [Google Scholar]
- Flor HH (1971) Current status of the gene-for-gene concept. Annu Rev Phytopathol 9 275–296 [Google Scholar]
- Flors V, Ton J, van Doorn R, Jakab G, Garcia-Agustin P, Mauch-Mani B (2008) Interplay between JA, SA and ABA signalling during basal and induced resistance against Pseudomonas syringae and Alternaria brassicicola. Plant J 54 81–92 [DOI] [PubMed] [Google Scholar]
- Ford TL, Cooley JT, Christou P (1994) Current status for gene transfer into rice utilizing variety-independent delivery systems. In RS Ziegler, SA Leong, PS Teng, eds, Rice Blast Disease. CAB International, Wallingford, UK, pp 195–208
- Glazebrook J (2001) Genes controlling expression of defense responses in Arabidopsis: 2001 status. Curr Opin Plant Biol 4 301–308 [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Ausubel FM (1994) Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc Natl Acad Sci USA 91 8955–8959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath MC, Howard RJ, Valent B, Chumley FG (1992) Ultrastructural interactions of one strain of Magnaporthe grisea with goosegrass and weeping lovegrass. Can J Bot 70 779–787 [Google Scholar]
- Heath MC, Valent B, Howard RJ, Chumley FG (1990) Correlations between cytologically detected plant-fungal interactions and pathogenicity of Magnaporthe grisea toward weeping lovegrass. Phytopathology 80 1382–1386 [Google Scholar]
- Holub EB, Brose E, Tor M, Clay C, Crute IR, Beynon JL (1995) Phenotypic and genotypic variation in the interaction between Arabidopsis thaliana and Albugo candida. Mol Plant Microbe Interact 8 916–928 [DOI] [PubMed] [Google Scholar]
- Howard RJ, Ferrari MA, Roach DH, Money NP (1991) Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc Natl Acad Sci USA 88 11281–11284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckelhoven R, Fodor J, Preis C, Kogel KH (1999) Hypersensitive cell death and papilla formation in barley attacked by the powdery mildew fungus are associated with hydrogen peroxide but not with salicylic acid accumulation. Plant Physiol 119 1251–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AO, Taylor CB (1996) Plant-microbe interactions: life and death at the interface. Plant Cell 8 1651–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J, Park SY, Chi MH, Choi J, Park J, Rho HS, Kim S, Goh J, Yoo S, Choi J, et al (2007) Genome-wide functional analysis of pathogenicity genes in the rice blast fungus. Nat Genet 39 561–565 [DOI] [PubMed] [Google Scholar]
- Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B (2000) Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J 19 4004–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, Ausubel FM, Glazebrook J (1999) Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc Natl Acad Sci USA 96 13583–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DA, Takemoto D (2004) Plant innate immunity: direct and indirect recognition of general and specific pathogen-associated molecules. Curr Opin Immunol 16 48–62 [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444 323–329 [DOI] [PubMed] [Google Scholar]
- Kankanala P, Czymmek K, Valent B (2007) Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19 706–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Miura E, Matsushima R, Sakamoto W (2007) White leaf sectors in yellow variegated2 are formed by viable cells with undifferentiated plastids. Plant Physiol 144 952–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Ahn IP, Park CH, Park SG, Park SY, Jwa NS, Lee YH (2001) Molecular characterization of the cDNA encoding an acidic isoform of PR-1 protein in rice. Mol Cells 11 115–121 [PubMed] [Google Scholar]
- Koch E, Slusarenko A (1990) Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM (2002) Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 5 325–331 [DOI] [PubMed] [Google Scholar]
- Leung H, Borromeo S, Bernardo MA, Notteghem JL (1988) Genetic analysis of virulence in the rice blast fungus Magnaporthe grisea. Phytopathology 78 1227–1233 [Google Scholar]
- McDowell JM, Williams SG, Funderburg NT, Eulgem T, Dangl JL (2005) Genetic analysis of developmentally regulated resistance to downy mildew (Hyaloperonospora parasitica) in Arabidopsis thaliana. Mol Plant Microbe Interact 18 1226–1234 [DOI] [PubMed] [Google Scholar]
- Mendgen K, Hahn M, Deising H (1996) Morphogenesis and mechanisms of penetration by plant pathogenic fungi. Annu Rev Phytopathol 34 367–386 [DOI] [PubMed] [Google Scholar]
- Mitchell TK, Dean RA (1995) The cAMP-dependent protein kinase catalytic subunit is required for appressorium formation and pathogenesis by the rice blast pathogen Magnaporthe grisea. Plant Cell 7 1869–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modolo LV, Cunha FQ, Braga MR, Salgado I (2002) Nitric oxide synthase-mediated phytoalexin accumulation in soybean cotyledons in response to the Diaporthe phaseolorum f. sp. meridionalis elicitor. Plant Physiol 130 1288–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15 473–497 [Google Scholar]
- Narasimhan ML, Damsz B, Coca MA, Ibeas JI, Yun DJ, Pardo JM, Hasegawa PM, Bressan RA (2001) A plant defense response effector induces microbial apoptosis. Mol Cell 8 921–930 [DOI] [PubMed] [Google Scholar]
- Narusaka Y, Narusaka M, Park P, Kubo Y, Hirayama T, Seki M, Shiraishi T, Ishida J, Nakashima M, Enju A, et al (2004) RCH1, a locus in Arabidopsis that confers resistance to the hemibiotrophic fungal pathogen Colletotrichum higginsianum. Mol Plant Microbe Interact 17 749–762 [DOI] [PubMed] [Google Scholar]
- Nimchuk Z, Eulgem T, Holt BF III, Dangl JL (2003) Recognition and response in the plant immune system. Annu Rev Genet 37 579–609 [DOI] [PubMed] [Google Scholar]
- Nishimura MT, Stein M, Hou BH, Vogel JP, Edwards H, Somerville SC (2003) Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301 969–972 [DOI] [PubMed] [Google Scholar]
- Nishiuchi T, Masuda D, Nakashita H, Ichimura K, Shinozaki K, Yoshida S, Kimura M, Yamaguchi I, Yamaguchi K (2006) Fusarium phytotoxin trichothecenes have an elicitor-like activity in Arabidopsis thaliana, but the activity differed significantly among their molecular species. Mol Plant Microbe Interact 19 512–520 [DOI] [PubMed] [Google Scholar]
- O'Connell R, Herbert C, Sreenivasaprasad S, Khatib M, Esquerre-Tugaye MT, Dumas B (2004) A novel Arabidopsis-Colletotrichum pathosystem for the molecular dissection of plant-fungal interactions. Mol Plant Microbe Interact 17 272–282 [DOI] [PubMed] [Google Scholar]
- Ono E, Wong HL, Kawasaki T, Hasegawa M, Kodama O, Shimamoto K (2001) Essential role of the small GTPase Rac in disease resistance of rice. Proc Natl Acad Sci USA 98 759–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx IA, Eggermont K, Terras FR, Thomma BP, De Samblanx GW, Buchala A, Metraux JP, Manners JM, Broekaert WF (1996) Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8 2309–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rate DN, Greenberg JT (2001) The Arabidopsis aberrant growth and death2 mutant shows resistance to Pseudomonas syringae and reveals a role for NPR1 in suppressing hypersensitive cell death. Plant J 27 203–211 [DOI] [PubMed] [Google Scholar]
- Rehmany AP, Lynn JR, Tor M, Holub EB, Beynon JL (2000) A comparison of Peronospora parasitica (Downy mildew) isolates from Arabidopsis thaliana and Brassica oleracea using amplified fragment length polymorphism and internal transcribed spacer 1 sequence analyses. Fungal Genet Biol 30 95–103 [DOI] [PubMed] [Google Scholar]
- Reynolds ES (1963) The use of lead citrate at high pH as an electron opaque stain in electron microscopy. J Cell Biol 17 208–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson LH, Cahill DM (2003) Ecotypic variation in the response of Arabidopsis thaliana to Phytophthora cinnamomi. Australas Plant Pathol 32 53–64 [Google Scholar]
- Roetschi A, Si-Ammour A, Belbahri L, Mauch F, Mauch-Mani B (2001) Characterization of an Arabidopsis-Phytophthora pathosystem: resistance requires a functional PAD2 gene and is independent of salicylic acid, ethylene and jasmonic acid signalling. Plant J 28 293–305 [DOI] [PubMed] [Google Scholar]
- Rusterucci C, Aviv DH, Holt BF III, Dangl JL, Parker JE (2001) The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell 13 2211–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sang Y, Cui D, Wang X (2001) Phospholipase D and phosphatidic acid-mediated generation of superoxide in Arabidopsis. Plant Physiol 126 1449–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada C, Lipka V, O'Connell R, Okuno T, Schulze-Lefert P, Takano Y (2006) Nonhost resistance in Arabidopsis-Colletotrichum interactions acts at the cell periphery and requires actin filament function. Mol Plant Microbe Interact 19 270–279 [DOI] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I, Rowe ML (2002) Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14 1405–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JM, Heard JE, Asai T, Ausubel FM (2000) Simulation of fungal-mediated cell death by fumonisin B1 and selection of fumonisin B1-resistant (fbr) Arabidopsis mutants. Plant Cell 12 1811–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot N, Foster A (2001) Genetics and genomics of the rice blast fungus Magnaporthe grisea: developing an experimental model for understanding fungal diseases of cereals. Adv Bot Res 34 263–287 [Google Scholar]
- Thaler JS, Owen B, Higgins VJ (2004) The role of the jasmonate response in plant susceptibility to diverse pathogens with a range of lifestyles. Plant Physiol 135 530–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma B, Eggermont K, Penninckx I, Mauch-Mani B, Vogelsang R, Cammue BPA, Broekaert WF (1998) Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA 95 15107–15111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BP, Penninckx IA, Broekaert WF, Cammue BP (2001) The complexity of disease signaling in Arabidopsis. Curr Opin Immunol 13 63–68 [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen H, Zhang Z, Wei Y, Collinge D (1997) Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 11 1187–1194 [Google Scholar]
- Ton J, De Vos M, Robben C, Buchala A, Metraux JP, Van Loon LC, Pieterse CM (2002) Characterization of Arabidopsis enhanced disease susceptibility mutants that are affected in systemically induced resistance. Plant J 29 11–21 [DOI] [PubMed] [Google Scholar]
- Tucker SL, Talbot NJ (2001) Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annu Rev Phytopathol 39 385–417 [DOI] [PubMed] [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J (1992) Acquired resistance in Arabidopsis. Plant Cell 4 645–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veronese P, Nakagami H, Bluhm B, Abuqamar S, Chen X, Salmeron J, Dietrich RA, Hirt H, Mengiste T (2006) The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18 257–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignutelli A, Wasternack C, Apel K, Bohlmann H (1998) Systemic and local induction of an Arabidopsis thionin gene by wounding and pathogens. Plant J 14 285–295 [DOI] [PubMed] [Google Scholar]
- Vogel J, Somerville S (2000) Isolation and characterization of powdery mildew-resistant Arabidopsis mutants. Proc Natl Acad Sci USA 97 1897–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong HL, Pinontoan R, Hayashi K, Tabata R, Yaeno T, Hasegawa K, Kojima C, Yoshioka H, Iba K, Kawasaki T, et al (2007) Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell 19 4022–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JR, Hamer JE (1996) MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev 10 2696–2706 [DOI] [PubMed] [Google Scholar]
- Xu JR, Urban M, Sweigard JA, Hamer JE (1997) The CPKA gene of Magnaporthe grisea is essential for appressoria penetration. Mol Plant Microbe Interact 10 187–194 [Google Scholar]
- Zimmerli L, Stein M, Lipka V, Schulze-Lefert P, Somerville S (2004) Host and non-host pathogens elicit different jasmonate/ethylene responses in Arabidopsis. Plant J 40 633–646 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.