Almost all that is known about the transition to flowering in grasses is based on studies of agronomic species. The grain produced by two tropically derived grasses, maize (Zea mays) and rice (Oryza sativa), and a temperate origin grass, wheat (Triticum aestivum), provides most of the world's food. Other grasses, such as barley (Hordeum vulgare), ryegrass species (Lolium spp.), sorghum (Sorghum bicolor), and oats (Avena sativa), are grown in lesser amounts, but also fill important food production niches. In grass species, such as sugarcane (Saccharum spp.), the vegetative portion of the plant is harvested for the Suc that accumulates in its stalks; in this crop, the inability to flower is desirable because sugar levels drop after plants make the transition to flowering as carbon assimilates are shunted to seed production. In all of these grasses, manipulation of the timing of the floral transition is a vitally important trait in maximizing yield potential. Extensive agronomic studies have been done on grass species, but studies of the small flowering dicot plant Arabidopsis (Arabidopsis thaliana) have provided an abundance of information on the genetic and molecular control of flowering. What has emerged is a complex network of genes and pathways, some parts of which are also found in the grasses. Conversely, recent discoveries show that grasses also have developed unique mechanisms to regulate flowering.
RIGHT TIME, RIGHT PLACE: FEATURES OF THE FLORAL TRANSITION
With regard to the floral transition, all higher plants share some common mechanisms that control this important switch from vegetative to reproductive growth (for review, see Baurle and Dean, 2006; Imaizumi and Kay, 2006; Turck et al., 2008). First, the shoot apical meristem (SAM), which gives rise to both vegetative and reproductive structures, is the part of the plant where the actual transition occurs. Second, the SAM must be competent to perceive inductive signals to make inflorescence and floral meristems. Third, although the SAM is the target of floral inductive signals, the signals themselves, in most cases, originate in vegetative tissues, usually the leaves. Determining the biochemical nature of this hypothetical floral inductive signal, once, and now again, called florigen, has been very difficult. However, as described below, recent studies in Arabidopsis have led to the identification of a mobile protein that fits the criteria of a long-distance florigenic signal. Finally, the floral transition can be affected by signals that feed into both environmental and endogenous (or autonomous) pathways (Fig. 1).
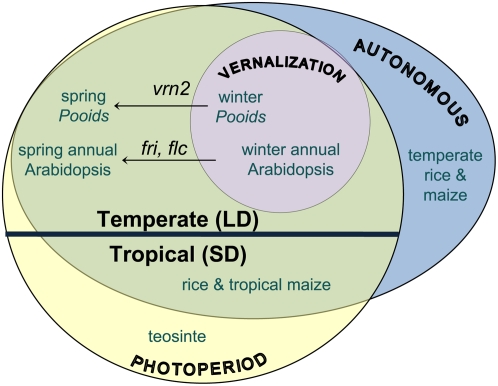
Figure 1.
Representation of overlapping floral induction pathways in agronomically important grasses and the model plant Arabidopsis. The majority of species utilize daylength cues to accelerate flowering. In general, temperate plants are induced under lengthening photoperiods, while tropical plants respond to shortening days. Often, a plant becomes competent to respond to photoperiodic signals only after undergoing a period of vernalization, as is the case with winter annual Arabidopsis and the winter pooids (wheat, barley, rye, oats). Vernalization requirement is often controlled by a small number of loci. For, example, fri or flc mutations are found in Arabidopsis with a summer annual growth habit. Similarly, spring varieties of the pooids result from loss of VRN2. Of the tropical grasses, teosinte is an obligate short-day plant, whereas rice has been shown to coordinate photoperiod and endogenous cues to initiate flowering. Similar to Arabidopsis, pooid grass flowering time is determined by the interaction of a network of signals including daylength and vernalization, as well as cues endogenous to the plant. Temperate maize is a tropical origin grass that has adapted to temperate climates, thereby becoming less sensitive to shortening days and more reliant on endogenous cues indicative of the plant's physiological status and overall readiness to flower. LD, Long days; SD, short days.
In considering the molecular chain of events that starts with the perception of signals that cause flowering and ends with the conversion of a vegetative meristem into a flower-generating reproductive meristem, it is clear that the genetic machinery that controls both ends of this chain is highly conserved in angiosperms. This is because perception of environmental signals, and floral meristem specification and flower development, are ancestral functions shared by all flowering plants. It is in the middle part of this chain—the integration of external stimuli into signals that can be interpreted as a developmental response—where plants exercise some flexibility in creating novel regulatory functions. Because the floral transition machinery is so intimately connected to the environment, a plant will use all the levers, springs, and mechanisms at its disposal or invent new ones to optimize flowering time. This is not surprising if one considers that the transition to flowering is the most critical event in the life cycle of most plants, especially monocarpic grass species that have one shot at flowering at the best time to produce seeds. Through comparison with the Arabidopsis flowering-time model, it is evident that grasses are dependent on some ancestral functions, but also have evolved their own unique mechanisms to integrate and transmit floral inductive signals.
PHOTOPERIOD, COINCIDENCE, AND THE CONSTANS/FLOWERING LOCUS T REGULATORY MODULE
First defined through genetic analysis of photoperiod mutants in Arabidopsis, the CONSTANS (CO)/FLOWERING LOCUS T (FT) regulatory system (for review, see Turck et al., 2008) appears to operate in several grasses examined so far, as described below. In Arabidopsis, the CO gene integrates inductive photoperiod information via the circadian clock and activates the FT gene. Regulation of CO stability, and therefore activity, provides a particularly elegant demonstration of the external coincidence model (for review, see Imaizumi and Kay, 2006). In brief, the output from the clock, via the GIGANTEA (GI) protein, activates expression of CO, a B-box-type zinc finger transcription factor-encoding gene expressed in leaves. Levels of CO transcript oscillate in a circadian pattern, and when they coincide with a light period, indicating long days of summer, the translated CO protein is stable and directly activates its prime target, the FT gene. FT protein then acts as a transcriptional cofactor that interacts with another flowering-time transcription factor encoded by the FLOWERING LOCUS D (FD) gene to activate expression of APETALA1 (AP1) and promote floral meristem identity at the shoot apex (Abe et al., 2005; Wigge et al., 2005). Expression of CO in dark periods of a long night, however, results in degradation of CO protein and no FT activation.
Genes with similarity to GI, CO, and FT have been identified in many agronomically important grass species (Yano et al., 2000; Kojima et al., 2002; Griffiths et al., 2003; Hayama et al., 2003; Nemoto et al., 2003; Faure et al., 2007; Danilevskaya et al., 2008a). In rice, quantitative trait loci (QTL) associated with different flowering times, or Heading dates (Hd), were found to correspond to specific genes orthologous to versions of Arabidopsis flowering-time regulatory genes. Most notably, Hd1 was found to encode an ortholog of the CO gene (Yano et al., 2000), and Hd3a corresponds to a gene with the same function as FT (Kojima et al., 2002). So far, the functional significance of these genes in flowering time has been shown in rice (Yano et al., 2000) and, to some extent, in wheat (Bonnin et al., 2008). The putative CO ortholog in perennial ryegrass (Lolium perenne), a long-day species, has also been shown to cause early flowering when overexpressed in Arabidopsis (Martin et al., 2004). Miller et al. (2008) reported a putative maize CO ortholog (conz1) that maps to a location that is syntenic with the rice Hd1 and whose expression appears to vary in a circadian pattern. Similarly, a large maize FT-related gene family called ZCN (for Zea CENTRORADIALIS), after the first member of this protein discovered in Antirrhinum (Bradley et al., 1996), was described with some of the members exhibiting leaf-specific expression patterns that suggest a florigenic function (Danilevskaya et al., 2008a). Moreover, one member of this family, ZCN8, physically interacts with a shoot meristem-localized FD-like protein. The gene encoding this bZIP protein, delayed flowering1, corresponds to one of only three genes identified so far that, when mutated, cause a late-flowering phenotype in maize (Muszynski et al., 2006). Overexpression of a wheat version of FT, TaFT, in transgenic wheat caused earlier flowering in this temperate grass (Yan et al., 2006). More recently, Li and Dubcovsky (2008) showed that TaFT protein interacts with two bZIP putative orthologs of FD, called TaFDL2 and TaFDL6, and that this interaction mediates activation of the wheat VERNALIZATION1 (VRN1), a MADS-box gene with high similarity to AP1 (Yan et al., 2003).
THE MEDIUM AS THE MESSAGE: DO GRASS FT ORTHOLOGS ACT AS FLORIGENIC SIGNALS?
An interesting feature of the Arabidopsis CO/FT system is that FD protein functions at the shoot meristem, yet CO activates FT only in mature leaf tissue (An et al., 2004). Thus, the implications of the recent finding that FT protein is translated in leaf vasculature cells, enters the phloem stream, and migrates to the shoot apex to interact with FD, suggests that FT protein has properties of the long-sought florigen (Corbesier et al., 2007). Biochemical studies have shown that phloem sap is chock full of all sorts of macromolecules, including proteins, but the identification of FT as the signal provides a nice finishing touch to the chain of events that start with photoperiod induction.
One of the original criteria of florigen is that it is a universal signal that is common to all flowering plants. Do grass FT orthologs act as mobile flowering signals? Preliminary evidence suggests that, similar to FT, the protein encoded by Hd3a is synthesized in leaves and migrates through the phloem to the shoot apex (Tamaki et al., 2007), although direct evidence that mobile Hd3a causes flowering has yet to be shown. More recently, the report that RFT1, another FT-like homolog in rice closely related to Hd3a, has a similar or supportive role in causing flowering suggests that the story may be more complicated, and that there may be a multitude of florigenic proteins (Komiya et al., 2008). Of course, there is no a priori reason why FT and its orthologs are long-distance proteins that act the same way in all species. However, the finding that Hd3a, and FT orthologs in dicots such as tomato (Solanum lycopersicum), melon (Cucumis melo), and trees, also encode mobile long-distance signals gives credence to the original florigen criterion of universality (Bohlenius et al., 2006; Lifschitz et al., 2006; Lin et al., 2007). So far, only the role of rice Hd3a protein as a putative mobile, flower-inducing signal has been shown for any grass species. Future research will determine whether the photoperiod − circadian clock − CO-FT (leaf) → FT (apex) − FD − AP1 regulatory circuit, where → represents long-distance movement, is conserved among diverse species, including the grasses.
GRASSES SEE THE LIGHT AND TELL TIME TOO
One of the first investigations of the effects of daylength on grass flowering was done by Emerson (1924), who was trying to cross the wild progenitor of maize, teosinte (Zea mays subsp. parviglumis), into northern latitude maize. Working with various accessions of teosinte, and inspired by the recent discovery of photoperiodism by Garner and Allard (1920), Emerson found that teosinte had an absolute requirement for short-day conditions to flower. As little as 2 weeks of 10-h days could induce these plants to flower months earlier than uninduced plants, which would otherwise flower in October in response to shortening days. Teosinte plants kept under long-day conditions do not flower (J. Colasanti, unpublished data).
Migration of crop grasses into different latitudes required that they adopt other signals to induce flowering so that they could adapt to different growing seasons. Because the underlying principle of flowering time is the synchronization of the plant's internal rhythms with environmental conditions, alterations in daylength associated with seasonal changes are among the most accurate cues to determine the right time to flower. In grasses, it is clear that the core photoperiod response pathway is largely conserved with Arabidopsis, albeit less well understood (Laurie et al., 2004; Turck et al., 2008). It is interesting to note that, although major components of photoperiod sensitivity are conserved among distantly related species, optimal flowering conditions are distinctly different among the grasses. While tropical grasses, such as rice and tropical maize, respond to short days to initiate flowering, temperate cereals like wheat and barley are sensitive to lengthening days, and maize adapted to high latitude growth is largely daylength insensitive. Examples have emerged that may shed light on how these differing demands are addressed within the framework of the conserved CO/FT pathway. In rice, for example, reduced Hd1 activity results in early flowering in long days (Yano et al., 2000), an observation that led to the discovery that conserved OsGI/Hd1 genes cooperate to inhibit flowering under noninductive conditions (Hayama et al., 2003). This dual function of Hd1 as both repressor and activator of Hd3a is likely dependent on levels of active photoreceptor phytochrome B (phyB), Pfr, which accumulates during long days (Izawa et al., 2002). An observation that further supports this model of Pfr/Hd1 repressor action is the extremely early-flowering phenotype of photoperiodic sensitivity5 (se5) mutants in long days. These rice mutants also lack typical phytochrome responses, substantiating the role of SE5 in phytochrome biosynthesis (Izawa et al., 2000).
Further evidence for grass-specific modulation of CO/FT activity comes from studies with spring wheat, in which the wheat CO ortholog, TaHd1, was able to complement a rice line with a nonfunctional Hd1 allele, and resulted in early flowering under long days, which are inductive for wheat but not rice (Nemoto et al., 2003). So it seems that, although CO protein is conserved between these grasses, alterations to the mode of CO action confer grass-specific differences in flowering-time control.
Two novel regulatory genes that appear to be absent in Arabidopsis have been shown to act in the daylength control of flowering time in rice, supporting the likelihood that unique flowering-time mechanisms have evolved in grasses. First, Early heading date1 (Ehd1), which encodes a B-type response regulator, is able to activate Hd3a expression independently of Hd1 in short days (Doi et al., 2004). The expression of Ehd1 was shown recently to be kept in check by a novel CCT (for CO, CO-like, TOC1) domain protein encoded by the Ghd7 gene, which suppresses Ehd1 expression in long-day conditions (Xue et al., 2008).
The new-found variants of orthologous gene functions and the discovery of novel flowering-time genes support the notion of species-specific adaptation due to rapid migration of grasses outside their native range. Temperate accessions of maize are considered largely unresponsive to photoperiod in terms of flowering time, yet maize responds to variation in daylength. In addition to the distinct rhythms of conz1 described above, photoreceptor mutants have a minor effect on maize flowering (i.e. phyB mutants as well as elongated mesocotyl1 mutants that are deficient in functional phytochromes, flower early under long-day growth conditions; Sawers et al., 2002; Sheehan et al., 2007), a situation similar to phytochrome mutants in rice. These observations point to the possibility that, while photoperiodic input likely contributes to flowering-time variation in modern maize, endogenous signaling pathways have overridden daylength cues to optimize flowering time (Fig. 1).
Dissection of flowering-time pathways in the temperate grasses wheat and barley have identified a daylength response based on what seems to be a Triticeae lineage-specific group of pseudo response regulator Photoperiod (Ppd) genes. In barley, a two-gene system is in place. Ppd-H1 is a CCT domain-encoding gene under circadian control, which is the major determinant of barley photoperiod response and promotes flowering in inductive long days (Turner et al., 2005). Ppd-H2 expressed in winter varieties is inhibitory to flowering in noninductive photoperiods (Turner et al., 2005). In contrast to barley, dominant wheat Ppd-1 alleles are constitutively expressed, thus reducing photoperiod sensitivity and causing early flowering in short days (Worland et al., 1998). Interestingly, reduced expression of barley CO-like genes, HvCO1 and HvCO2 in the ppd-H1 mutant, combined with significantly lower levels of HvFT and a late-flowering phenotype in long days, provides a possible link between photoperiod perception by the Ppd genes and the downstream CO/FT floral induction module (Turner et al., 2005).
VERNALIZATION IN TEMPERATE GRASSES AND ARABIDOPSIS
Like many of the temperate grasses, some Arabidopsis ecotypes flower earlier in response to prolonged cold (i.e. vernalization). This is an adaptive trait that prevents seeds sown in late summer or early fall from flowering until the next spring, thereby delaying flowering until the spring rather than just before a possibly harsh winter. Analyses of mutants that interfere with this vernalization response have defined a separate pathway in the flowering model (for review, see Sung and Amasino, 2005). Vernalization does not create an inductive signal; rather, it results in the removal of a block to flowering that must be overcome so that inductive signals can cause flowering in plants that are suitably competent to undergo the floral transition. The Arabidopsis FLOWERING LOCUS C (FLC) gene has been identified as a central flowering repressor whose activity is reduced by vernalization. FLC encodes a MADS-box transcription factor that represses flowering by directly interfering with FT expression in leaves and FD expression at the SAM (Searle et al., 2006). Most Arabidopsis ecotypes that flower early without vernalization have nonfunctional versions of FLC, or its positive regulator, FRIGIDA (FRI; Fig. 1).
Vernalization in temperate cereals with a winter growth habit, such as winter wheat and barley, similarly removes a block to flowering so that the plant can perceive inductive signals, such as long-day photoperiods. However, neither FLC nor FRI orthologs have been found in grasses so the underlying molecular machinery controlling vernalization in winter cereals is different from that of Arabidopsis. Three genes that act together to maintain a winter growth habit in cold-tolerant pooid grasses have been identified: VRN1, VRN2, and VRN3 (for review, see Trevaskis et al., 2007a). VRN1 encodes an AP1-related MADS-box protein that falls into the FRUITFULL1 (FUL1) gene lineage (Preston and Kellogg, 2006). VRN1 expression is low in the absence of vernalization, but transcript levels increase in direct proportion to time of exposure to cold temperatures (Yan et al., 2003; Trevaskis et al., 2003). Modulation of VRN1 levels thus provides a quantitative measure of the length of time a plant is exposed to cold, with longer exposure resulting in earlier flowering. The finding that VRN1 expression is detected first in leaves and later at the shoot apex suggests that it has a bifunctional role; first, it accelerates flowering by mediating a systemic response to vernalization and, second, VRN1 specifies inflorescence meristem identity at the shoot apex (Trevaskis et al., 2007b; Preston and Kellogg, 2008). In maize, which does not respond to vernalization, a potential ortholog of VRN1, ZMM4, was identified in a differential expression screen comparing vegetative and reproductive shoot apices and shown to cause earlier flowering when ectopically expressed in transgenic maize (Danilevskaya et al., 2008b). Therefore, the floral meristem promotion function of this MADS-box transcription factor, along with FUL2 (Preston and Kellogg, 2008), appears to be conserved in tropical and temperate grasses.
The vernalization function of VRN1 acts through the repression of VRN2 (Hemming et al., 2008). In this respect, there is a superficial similarity between grass and Arabidopsis vernalization in that FLC and VRN2 both encode floral repressors whose activities must be overcome to allow inductive signals to act on a competent apex. However, apart from encoding different types of transcription factor proteins (FLC = MADS-box; VRN2 = CCT zinc finger), other differences exist. For example, the main role of FLC is repression of FT; during vernalization, prolonged cold exposure reduces FLC levels and allows inductive signals to activate FT. In temperate cereals, VRN2 acts as a repressor of flowering under long-day conditions, which suggests that VRN2 is not strictly a vernalization gene, but that its job is to repress flowering under long days of summer so that plants do not initiate flowering prior to winter (Trevaskis et al., 2007a). More evidence of mechanistic similarities between vernalization and photoperiod components comes from the report of rice Ghd1, which encodes a CCT-like zinc finger that is closely related to VRN2 (Xue et al., 2008). Rice does not respond to vernalization, but, similar to pooid VRN2, Ghd1 represses flowering in noninductive long days. So, in this case, similar mechanisms are used to integrate different environmental stimuli.
The discovery that VRN3 corresponds to an ortholog of Arabidopsis FT reinforces the intimate link between the vernalization response and photoperiod induction. (Note, pooid versions of VRN3 are now known as TaFT in wheat and HvFT1 in barley.) Further, experiments conducted with doubled haploid barley have led to speculation that HvFT1 acts as a possible point of integration between the requirement for low temperature and inductive long days to cause flowering. In the absence of VRN2, flowering time becomes dependent strictly on daylength cues mediated through Ppd-H1 (Fig. 1). Thus, flowering is early if Ppd-H1 is present, whereas plants lacking both VRN2 and Ppd-H1 flower late (Hemming et al., 2008). Therefore, it seems that, in the long days of summer, VRN2 counteracts Ppd-H1 to prevent flowering prior to vernalization, and that once vernalized, a plant is competent to respond to long days through the action of Ppd-H1, which, ultimately, acts to up-regulate HvFT1.
THE SIGNAL WITHIN: ENDOGENOUS CUES THAT CAUSE FLOWERING
Arabidopsis mutants that affect flowering under both inductive and noninductive conditions are placed in the autonomous pathway. Autonomous flowering is inherently more difficult to understand compared to other pathways because the signals are linked to developmental processes rather than environmental stimuli that can be switched on and off. For example, most of what we know about flowering in temperate grasses was revealed from examining the underlying causes of vernalization, and in rice most of the genes identified have a role in photoperiod-induced flowering. This may explain why relatively little is known about flowering time in maize compared to other grasses because most studies are done with nearly day-neutral maize that relies almost exclusively on autonomous signals to control flowering. Most plants have a functioning autonomous flowering pathway because flowering usually occurs even in the absence of inductive environmental signals. Nevertheless, a few reports of autonomous flowering genes are emerging from the grasses. The difficulty in identifying autonomous pathway genes may explain why, in the long history of maize genetics, only a handful of mutants with a dramatic effect on flowering time have been identified (for review, see Colasanti and Muszynski, 2008). Although QTL analysis has identified over 300 loci associated with flowering-time differences, most of these effects are minor (Chardon et al., 2004). The indeterminate1 (id1) mutation has the most severe effect on flowering time of any maize gene, yet id1 does not seem to lie within a QTL with a large effect on flowering time. More incisive genome analyses are under way to determine whether a nearby medium effect QTL is in fact associated with id1 function (E. Buckler, personal communication). Maize id1 encodes a novel zinc finger transcriptional regulator that appears to act in the autonomous pathway (Colasanti et al., 1998; Kozaki et al., 2004). The invariability of id1 transcript and ID1 protein levels in response to diurnal light changes further suggests a role in autonomous control (Wong and Colasanti, 2007). The finding that id1 acts only in developing leaves suggests that id1 regulates either the production or transmission of a leaf-derived, florigenic signal. At present, there appears to be no connection between id1 function and putative FT-like orthologs in maize that may encode florigenic proteins.
The absence of a clear id1 ortholog in Arabidopsis suggests that id1 represents yet another regulatory gene that does not have a counterpart in all higher plants (Colasanti et al., 2006). However, recent reports reveal that id1 function may be prevalent in grasses. Ten years after the isolation of maize id1, three papers have appeared almost simultaneously describing a rice id1 ortholog. These papers confirm that a rice equivalent of id1, called RID1 (Wu et al., 2008), Ehd2 (Matsubara et al., 2008), or OsId1 (Park et al., 2008), exists in rice and functions as a key regulator of the flowering transition. (For the sake of clarity, we will call it OsID1.) Moreover, similar to maize, the highest levels of OsID1 are detected in developing leaves and its expression is unperturbed by diurnal day/night cycles. All three papers report that OsID1 acts upstream of FT ortholog Hd3a, as well as the unique Ehd1 gene. OsID1 may act independently of the CO ortholog Hd1; however, an interesting observation by Wu et al. (2008) is that loss of OsID1 function results in plants that never flower, even under inductive SD conditions. This has prompted the authors to designate OsID1 as a master regulator of flowering that stands astride both photoperiod and autonomous pathways. Nevertheless, the discovery of an id1 ortholog in rice suggests that species even closer to maize, such as sorghum and sugarcane, may have id1 equivalents as well, and this may shed some light on autonomous flowering in grasses.
In other grass species, as recently summarized by Cockram et al. (2007), other cereal loci, termed earliness per se (eps), have been shown to affect flowering time independently of environmental signals. Although many eps QTL have been mapped in wheat and barley, these sources of flowering-time variation remain poorly characterized to date, whereas the debate over whether they are truly unaffected by environmental cues remains unresolved. However, the existence of eps loci underlines the fact that many plants utilize endogenous cues to coordinate flowering time with their developmental or physiological status (Fig. 1).
HORMONES AND FLOWERING: IMPORTANT FOR SOME SPECIES, NOT OTHERS
Arabidopsis mutants with reduced GA synthesis are late flowering and therefore a separate GA pathway has been included in the floral regulatory model. In ryegrass, GA appears to play a major role in the floral transition, and it has been suggested that it acts as a leaf-derived, long-distance signaling molecule (King et al., 2006). Whether GA has a similar important role in other grasses has not been reported, although there could be minor effects on flowering due to reduced GA levels.
In some crop plants, hormones, or chemicals that mimic their activity, are used to alter flowering time. One example is the commercial use of ethephon (2-chloroethylphosphonic acid), which is converted to ethylene, to prevent flowering in sugarcane and increase sugar yields (Moore and Osgood, 1989). However, it is not clear whether ethylene acts by inhibiting the shoot apex from initiating further growth or by allowing vegetative growth to resume at the expense of reproductive organ formation.
PERSPECTIVES: WHAT'S NEXT?
Great progress has been made in deciphering the molecular mechanisms that regulate flowering in both Arabidopsis and agronomically significant grass species, but fundamental aspects of this important developmental transition remain unanswered. In particular, the underlying physiological changes that cause or are associated with the transition to flowering have yet to be extensively characterized. For example, vernalization pathways have been deciphered at the molecular level, but how cold-induced biochemical changes are perceived and transmitted to the regulatory network through physiological response is still unknown. Similarly, day-neutral plants, such as temperate maize and rice, flower when a developmental or physiological threshold is reached, yet the nature of these endogenous physicochemical changes is unknown. The next obvious step is to link the regulatory networks, which are controlled largely by pivotal transcription factors, with the downstream metabolic alterations that mediate the activity of these regulators.
An emerging precedent from studies of flowering, especially from research into Arabidopsis vernalization, is that epigenetic mechanisms are at work to establish a cellular memory that maintains a florally competent SAM once the stimulus (cold) is no longer present (Dennis and Peacock, 2007). In this model, the memory of winter is imprinted in SAM cells such that repression of floral inhibitors is maintained once spring returns. Future research may show that epigenetic mechanisms are more widespread, perhaps operating in the grass SAMs to maintain competency to flower.
Can knowledge gleaned from studies of monocot cereals inform us about how flowering is controlled in other grasses? Given that many diverse and unique grass-specific mechanisms are turning up, a complete understanding of flowering may require consideration on a case-by-case basis. One intriguing phenomenon concerns certain bamboos that flower synchronously decades after planting, even when offshoots derived from the original plant are separated by many degrees of latitude (Isagi et al., 2004). In this case, an autonomous signal of unknown origin must indicate when flowering will occur. Clearly a deeper understanding of flowering time mechanisms is required to answer these questions.
Acknowledgments
We thank Yukiko Mizukami (Purdue University) and two anonymous reviewers for comments on the manuscript.
This work was supported by a National Science and Engineering Research Council of Canada (NSERC) Discovery grant and the Ontario Research Fund. V.C. is a recipient of an NSERC graduate student fellowship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Joseph Colasanti (jcolasan@uoguelph.ca).
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309 1052–1056 [DOI] [PubMed] [Google Scholar]
- An HL, Roussot C, Suarez-Lopez P, Corbesler L, Vincent C, Pineiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, et al (2004) CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131 3615–3626 [DOI] [PubMed] [Google Scholar]
- Baurle I, Dean C (2006) The timing of developmental transitions in plants. Cell 125 655–664 [DOI] [PubMed] [Google Scholar]
- Bohlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, Strauss SH, Nilsson O (2006) CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312 1040–1043 [DOI] [PubMed] [Google Scholar]
- Bonnin I, Rousset M, Madur D, Sourdille P, Dupuits L, Brunel D, Goldringer I (2008) FT genome A and D polymorphisms are associated with the variation of earliness components in hexaploid wheat. Theor Appl Genet 116 383–394 [DOI] [PubMed] [Google Scholar]
- Bradley D, Carpenter R, Copsey L, Vincent C, Rothstein S, Coen E (1996) Control of inflorescence architecture in Antirrhinum. Nature 379 791–797 [DOI] [PubMed] [Google Scholar]
- Chardon F, Virlon B, Moreau L, Falque M, Joets J, Decousset L, Murigneux A, Charcosset A (2004) Genetic architecture of flowering time in maize as inferred from quantitative trait loci meta-analysis and synteny conservation with the rice genome. Genetics 168 2169–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockram J, Jones H, Leigh FJ, O'Sullivan D, Powell W, Laurie DA, Greenland AJ (2007) Control of flowering time in temperate cereals: genes, domestication, and sustainable productivity. J Exp Bot 58 1231–1244 [DOI] [PubMed] [Google Scholar]
- Colasanti J, Muszynski MG (2008) The maize floral transition. In SC Hake, JL Bennetzen, eds, Handbook of Maize: Its Biology, Vol 1. Springer Science, New York, pp 41–55
- Colasanti J, Tremblay R, Wong AYM, Coneva V, Kozaki A, Mable BK (2006) The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants. BMC Genomics 7 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti J, Yuan Z, Sundaresan V (1998) The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 93 593–603 [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang SH, Fornara F, Fan QZ, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316 1030–1033 [DOI] [PubMed] [Google Scholar]
- Danilevskaya ON, Meng X, Hou ZL, Ananiev EV, Simmons CR (2008. a) A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol 146 250–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya ON, Meng X, Selinger DA, Deschamps S, Hermon P, Vansant G, Gupta R, Ananiev EV, Muszynski MG (2008. b) Involvement of the MADS-box gene ZMM4 in floral induction and inflorescence development in maize. Plant Physiol 147 2054–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis ES, Peacock WJ (2007) Epigenetic regulation of flowering. Curr Opin Plant Biol 10 520–527 [DOI] [PubMed] [Google Scholar]
- Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A (2004) Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1l. Genes Dev 18 926–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson RA (1924) Control of flowering in teosinte. J Hered 15 41–48 [Google Scholar]
- Faure S, Higgins J, Turner A, Laurie DA (2007) The FLOWERING LOCUS T-like gene family in barley (Hordeum vulgare). Genetics 176 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner WW, Allard HA (1920) Effect of the relative effect of day and night and other factors of the environment on growth and reproduction in plants. J Agric Res 18 553–606 [Google Scholar]
- Griffiths S, Dunford RP, Coupland G, Laurie DA (2003) The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol 131 1855–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K (2003) Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422 719–722 [DOI] [PubMed] [Google Scholar]
- Hemming MN, Peacock WJ, Dennis ES, Trevaskis B (2008) Low-temperature and daylength cues are integrated to regulate FLOWERING LOCUS T in barley. Plant Physiol 147 355–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Kay SA (2006) Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci 11 550–558 [DOI] [PubMed] [Google Scholar]
- Isagi Y, Shimada K, Kushima H, Tanaka N, Nagao A, Ishikawa T, Onodera H, Watanabe S (2004) Clonal structure and flowering traits of a bamboo [Phyllostachys pubescens (Mazel) Ohwi] stand grown from a simultaneous flowering as revealed by AFLP analysis. Mol Ecol 13 2017–2021 [DOI] [PubMed] [Google Scholar]
- Izawa T, Oikawa T, Sugiyama N, Tanisaka T, Yano M, Shimamoto K (2002) Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev 16 2006–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T, Oikawa T, Tokutomi S, Okuno K, Shimamoto K (2000) Phytochromes confer the photoperiodic control of flowering in rice (a short-day plant). Plant J 22 391–399 [DOI] [PubMed] [Google Scholar]
- King RW, Moritz T, Evans LT, Martin J, Andersen CH, Blundell C, Kardailsky I, Chandler PM (2006) Regulation of flowering in the long-day grass Lolium temulentum by gibberellins and the FLOWERING LOCUS T gene. Plant Physiol 141 498–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M (2002) Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol 43 1096–1105 [DOI] [PubMed] [Google Scholar]
- Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K (2008) Hd3a and RFT1 are essential for flowering in rice. Development 135 767–774 [DOI] [PubMed] [Google Scholar]
- Kozaki A, Hake S, Colasanti J (2004) The maize ID1 flowering time regulator is a zinc finger protein with novel DNA binding properties. Nucleic Acids Res 32 1710–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie DA, Griffiths S, Dunford RP, Christodoulou V, Taylor SA, Cockram J, Beales J, Turner A (2004) Comparative genetic approaches to the identification of flowering time genes in temperate cereals. Field Crops Res 90 87–99 [Google Scholar]
- Li CX, Dubcovsky J (2008) Wheat FT protein regulates VRN1 transcription through interactions with FDL2. Plant J 55 543–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschitz E, Eviatar T, Rozman A, Shalit A, Goldshmidt A, Amsellem Z, Alvarez JP, Eshed Y (2006) The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc Natl Acad Sci USA 103 6398–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MK, Belanger H, Lee YJ, Varkonyi-Gasic E, Taoka KI, Miura E, Xoconostle-Cazares B, Gendler K, Jorgensene RA, Phinney B, et al (2007) FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. Plant Cell 19 1488–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J, Storgaard M, Andersen CH, Nielsen KK (2004) Photoperiodic regulation of flowering in perennial ryegrass involving a CONSTANS-like homolog. Plant Mol Biol 56 159–169 [DOI] [PubMed] [Google Scholar]
- Matsubara K, Yamanouchi U, Wang Z-X, Minobe Y, Izawa T, Yano M (2008) Ehd2, a rice ortholog of the maize ID1 gene, promotes flowering by up-regulating Ehd1. Plant Physiol 148: 1425–1435 [DOI] [PMC free article] [PubMed]
- Miller TA, Muslin EH, Dorweiler JE (2008) A maize CONSTANS-like gene, conz1, exhibits distinct diurnal expression patterns in varied photoperiods. Planta 227 1377–1388 [DOI] [PubMed] [Google Scholar]
- Moore PH, Osgood RV (1989) Prevention of flowering and increasing sugar yield of sugarcane by application of ethephon (2-chloroethylphosphonic acid). J Plant Growth Regul 8 205–210 [Google Scholar]
- Muszynski MG, Dam T, Li B, Shirbroun DM, Hou ZL, Bruggemann E, Archibald R, Ananiev EV, Danilevskaya ON (2006) delayed flowering1 encodes a basic leucine zipper protein that mediates floral inductive signals at the shoot apex in maize. Plant Physiol 142 1523–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto Y, Kisaka M, Fuse T, Yano M, Ogihara Y (2003) Characterization and functional analysis of three wheat genes with homology to the CONSTANS flowering time gene in transgenic rice. Plant J 36 82–93 [DOI] [PubMed] [Google Scholar]
- Park SJ, Kim SL, Lee S, Je BI, Piao HL, Park SH, Kim CM, Ryu CH, Park SH, Xuan XH, et al (2008) Rice Indeterminate 1 (OsId1) is necessary for the expression of Ehd1 (Early heading date 1) regardless of photoperiod. Plant J 10.1111/j.1365-313X.2008.03667.x [DOI] [PubMed]
- Preston JC, Kellogg EA (2006) Reconstructing the evolutionary history of paralogous APETALA1/FRUITFULL-like genes in grasses (Poaceae). Genetics 174 421–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston JC, Kellogg EA (2008) Discrete developmental roles for temperate cereal grass VERNALIZATION1/FRUITFULL-like genes in flowering competency and the transition to flowering. Plant Physiol 146 265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawers RJH, Linley PJ, Farmer PR, Hanley NP, Costich DE, Terry MJ, Brutnell TP (2002) elongated mesocotyl1, a phytochrome-deficient mutant of maize. Plant Physiol 130 155–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I, He YH, Turck F, Vincent C, Fornara F, Krober S, Amasino RA, Coupland G (2006) The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev 20 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan MJ, Kennedy LM, Costich DE, Brutnell TP (2007) Subfunctionalization of PhyB1 and PhyB2 in the control of seedling and mature plant traits in maize. Plant J 49 338–353 [DOI] [PubMed] [Google Scholar]
- Sung S, Amasino RM (2005) Remembering winter: toward a molecular understanding of vernalization. Annu Rev Plant Biol 56 491–508 [DOI] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K (2007) Hd3a protein is a mobile flowering signal in rice. Science 316 1033–1036 [DOI] [PubMed] [Google Scholar]
- Trevaskis B, Bagnall DJ, Ellis MH, Peacock WJ, Dennis ES (2003) MADS box genes control vernalization-induced flowering in cereals. Proc Natl Acad Sci USA 100 13099–13104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B, Hemming MN, Dennis ES, Peacock WJ (2007. a) The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci 12 352–357 [DOI] [PubMed] [Google Scholar]
- Trevaskis B, Tadege M, Hemming MN, Peacock WJ, Dennis ES, Sheldon C (2007. b) Short vegetative phase-like MADS-box genes inhibit floral meristem identity in barley. Plant Physiol 143 225–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59 573–594 [DOI] [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA (2005) The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310 1031–1034 [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309 1056–1059 [DOI] [PubMed] [Google Scholar]
- Wong AYM, Colasanti J (2007) Maize floral regulator protein INDETERMINATE1 is localized to developing leaves and is not altered by light or the sink/source transition. J Exp Bot 58 403–414 [DOI] [PubMed] [Google Scholar]
- Worland AJ, Borner A, Korzun V, Li WM, Petrovic S, Sayers EJ (1998) The influence of photoperiod genes on the adaptability of European winter wheats (reprinted from Wheat: Prospects for global improvement, 1998). Euphytica 100 385–394 [Google Scholar]
- Wu C, You C, Li C, Long T, Chen G, Byrne ME, Zhang QF (2008) RID1, encoding a Cys2/His2-type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice. Proc Natl Acad Sci USA 105 12915–12920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue WY, Xing YZ, Weng XY, Zhao Y, Tang WJ, Wang L, Zhou HJ, Yu SB, Xu CG, Li XH, et al (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40 761–767 [DOI] [PubMed] [Google Scholar]
- Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J (2006) The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA 103 19581–19586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J (2003) Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci USA 100 6263–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, et al (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12 2473–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]